Abstract
The thioredoxin (Trx)-coupled arsenate reductase (ArsC) is a family of enzymes that catalyzes the reduction of arsenate to arsenite in the arsenic detoxification pathway. The catalytic cycle involves a series of relayed intramolecular and intermolecular thiol-disulfide exchange reactions. Structures at different reaction stages have been determined, suggesting significant conformational fluctuations along the reaction pathway. Herein, we use two state-of-the-art NMR methods, the chemical exchange saturation transfer (CEST) and the CPMG-based relaxation dispersion (CPMG RD) experiments, to probe the conformational dynamics of B. subtilis ArsC in all reaction stages, namely the enzymatic active reduced state, the intra-molecular C10–C82 disulfide-bonded intermediate state, the inactive oxidized state, and the inter-molecular disulfide-bonded protein complex with Trx. Our results reveal highly rugged energy landscapes in the active reduced state, and suggest global collective motions in both the C10–C82 disulfide-bonded intermediate and the mixed-disulfide Trx-ArsC complex.
Introduction
Protein thiol-disulfide exchange reactions play fundamental roles in living systems, represented by the thioredoxin (Trx) and glutaredoxin (Grx) systems that maintain the cytoplasmic reducing environment, the protein DsbA that catalyzes the formation of protein disulfide bonds in bacterial periplasm, as well as the protein disulfide isomerase (PDI) proteins that facilitate correct disulfide bonding1–5. Although extensive studies have been carried out to elucidate the underlying mechanism for protein thiol-disulfide exchange reactions1–3,6–10, there still remain much controversies as well as challenges, particularly in the case of inter-protein disulfide exchanges involving transiently formed protein-protein mixed disulfide complexes3,4. The Trx-coupled arsenate reductase (ArsC) in Gram-positive bacteria like Bacillus subtilis and Staphylococcus aureus catalyzes the reduction of arsenate to arsenite as part of the arsenic detoxification pathway, and the oxidized inactive form of ArsC is reactivated by the upstream Trx system11–16. The enzymatic reaction of arsenate reduction occurs through an intramolecular cascade of thiol-disulfide exchanges involving three catalytic active cysteines in ArsC, whereas the regeneration of ArsC activity is accomplished by an inter-protein thiol-disulfide exchange with Trx. The involvement of both intra- and inter-protein thiol-disulfide exchange reactions in a single catalytic cycle makes the Trx-coupled ArsC an attractive model system for investigating protein thiol-disulfide exchange mechanisms. Moreover, the high-resolution NMR structure of B. subtilis Trx-ArsC mixed-disulfide intermediate17 is one of the few protein-protein complex structures of Trx with a downstream protein determined thus far, providing the structural basis for investigating the inter-protein disulfide exchanges.
It is becoming commonly accepted that fluctuations of the protein conformational dynamics play a critical role in driving the enzymatic catalysis reactions. For thiol-disulfide exchange reactions, in particular, protein conformational dynamics play an essential role in regulating cysteine reactivity, such as affecting the thiol pKa values3,4,18. Via combined efforts from both X-ray crystallography and solution NMR spectroscopy, the structures of Trx-coupled ArsC (either B. subtilis ArsC or S. aureus pI258 ArsC, abbreviated as Bs_ArsC and Sa_ArsC) have been elucidated in various reaction stages, including the transiently formed Trx-ArsC mixed-disulfide complex15–17,19,20. However, information concerning the changes of protein dynamics and conformational landscapes along the reaction pathway is still limited, and the underlying mechanism of how each of the five cysteines involved (three from ArsC and two from Trx) become sequentially activated at different reaction steps remains to be further elucidated. We have previously performed the backbone 15N NMR relaxation measurements of Bs_ArsC in both the reduced and oxidized states20. Herein we use a combination of the Carr-Purcell-Meiboom-Gill (CPMG)-based R2 relaxation dispersion (CPMG RD) and chemical exchange saturation transfer (CEST) NMR methods21–24 to investigate the conformational dynamics of Trx-coupled ArsC during the catalytic cycle, and to gain further understanding of protein thiol-disulfide exchanges. In brief, the CPMG RD method measures the changes of the effective transverse relaxation rates (R2eff) at different frequencies νCPMG (νCPMG = 1/(4τcp) and τcp is the delay between the refocusing pulses in the CPMG pulse train), and can be used to extract information such as the exchange rate kex, the populations of the ground and invisible states pA and pB, and the absolute value of chemical shift differences |Δω| between the two states22. It is sensitive to exchange processes on the 200–2000 s−1 regime24, but not to motions on slower timescales. The CEST method, which uses the saturation transfer effect to probe the exchanges between a ground state conformation and possible sparsely-populated excited state conformation(s), is complementary to CPMG RD in that it can report on motions on the timescale of ~20–200 s−1, and it also has the advantage of being able to directly observe the chemical shifts of the invisible states21,24. For clarity, we show a brief summary of the arsenate reduction catalytic cycle in Fig. 1. The ArsC enzyme in different reaction states will be abbreviated as “re-ArsC” for the reduced state, “int-ArsC” for the C10–C82 disulfide bonded intermediate state, “ox-ArsC” for the oxidized state, and “c-ArsC” for ArsC in the Trx-complexed state throughout the manuscript. Unless otherwise mentioned, all states of the ArsC sample in the manuscript refer to Bs_ArsC.
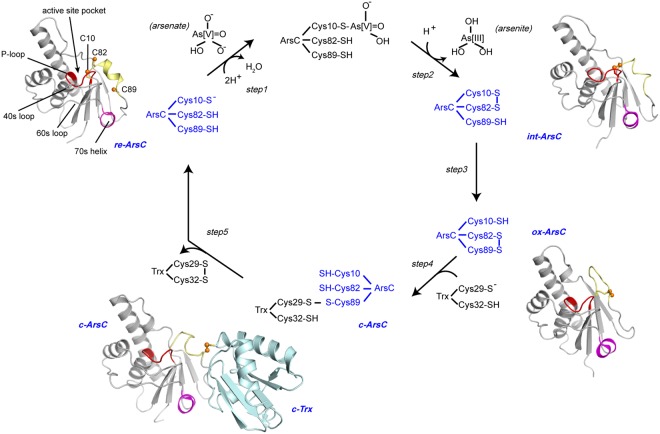
Figure 1.
The catalytic mechanism of B. subtilis ArsC. Schematic presentation of the catalytic and re-activation cycle of B. subtilis ArsC. The four essential redox states re-ArsC, int-ArsC, ox-ArsC and c-ArsC are colored in blue. The structures of re-ArsC, ox-ArsC and c-ArsC (PDB entries 1z2d, 1z2e, and 2ipa) and a structure model of the C10–C82 disulfide-bonded int-ArsC generated using AMBER54 are shown. The active site P-loop, the three active cysteine residues and other important structural regions are shown in the structure of re-ArsC.
Results
Sulfate binding of re-ArsC captured by CEST experiments
The re-ArsC is the active state of the enzyme with all three essential cysteine residues (C10, C82 and C89) in the free thiol state. It binds the substrate arsenate via the oxyanion-binding P-loop and carries out the initial nucleophilic attack on arsenate using the active C10, forming a covalent C10-HAsO3− intermediate (step 1 in Fig. 1). Since the re-ArsC structure requires oxyanion binding to become stabilized19,20,25, the available solution structure of re-ArsC, which was determined in the presence of sulfate, should be regarded as a mimic of the enzyme non-covalently bound with the substrate. To clearly distinguish between the sulfate-bound and free forms of re-ArsC, we herein denote the two as re-ArsC·sulfate and re-ArsCfree, respectively (see Fig. S1 for details of the assignments of re-ArsCfre).
The 15N CEST experiment was carried out on both re-ArsC·sulfate and re-ArsCfree. In re-ArsC·sulfate, a total of 19 non-overlapping residues were identified to undergo exchanges by the CEST experiments, including the segment F8-M19 covering the P-loop region and a number of residues in the adjacent 40 s and 60 s loops, as well as in the 80 s segment (Fig. 2A,B). The depths of the minor dips (corresponding to the excited state) in the CEST profiles are dependent on the sulfate concentrations for all residues in the P-loop and for many residues in neighboring regions (Fig. S2), suggesting that the exchange is probably related to the binding and releasing of the sulfate ion. This scenario is further supported by the high similarity of the Δδfree-sulfate (chemical shifts differences between re-ArsCfree and re-ArsC·sulfate) and Δω values (chemical shift differences between the major and minor dips in the CEST profiles) for the majority of residues (Fig. 2E).
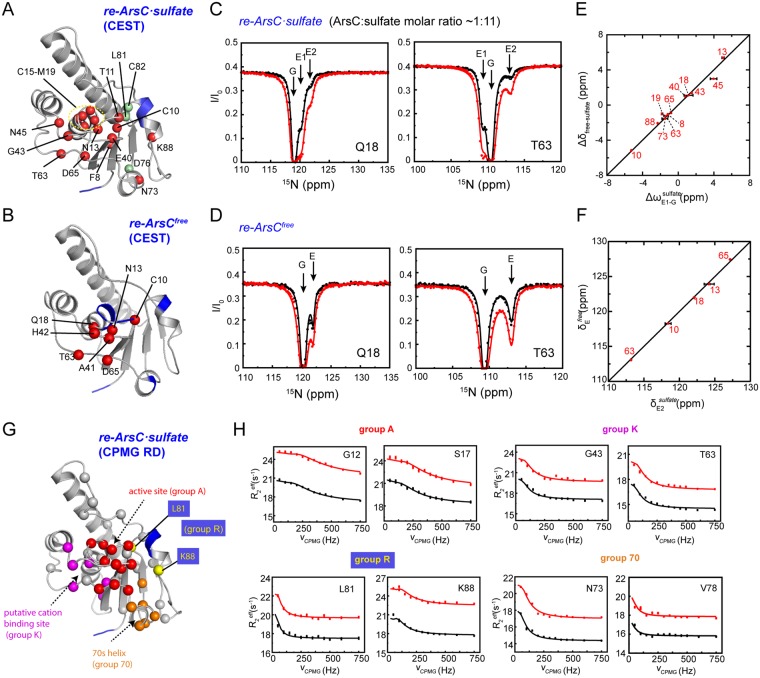
Figure 2.
15N CEST and CPMG RD results of re-ArsC. (A,B) Mapping of residues showing conformational exchanges in re-ArsC·sulfate (A) and re-ArsCfree (B) identified by CEST experiments onto the structure (shown as spheres). Residues showing sulfate concentration-dependent CEST profiles are shown as red balls in (A). Residues with missing backbone amide resonances are colored blue. (C,D) Representative 15N CEST profiles in re-ArsC·sulfate (C) and re-ArsCfree (D) obtained on a 600-MHz spectrometer using B1 fields of 8.4 Hz (black) and 13.5 Hz (red). The re-ArsC·sulfate sample was prepared with ArsC concentration of 1.8 mM and sulfate concentration of 20 mM. (E) Comparison between the Δω values extracted from CEST experiment of re-ArsC·sulfate and the chemical shift differences Δδfree − sulfate for residues showing sulfate concentration-dependent CEST profiles. Here ΔωE1-Gsulfate = δE1sulfate − δGsulfate, with δGsulfate, δE1sulfate corresponding to the chemical shifts of the major and first minor dips in re-ArsC·sulfate CEST profiles, respectively, and Δδfree-sulfate = δfree − δsulfate, with δfree, δsulfate corresponding to the chemical shifts in re-ArsCfree and re-ArsC·sulfate, respectively. Residue numbers are denoted. (F) Comparison between the chemical shift of the E2 state in re-ArsC·sulfate (δE2sulfate) and E state in re-ArsCfree (δEfree), with residue numbers denoted. (G) Mapping of residues showing conformational exchanges in the CPMG RD experiments in re-ArsC·sulfate onto the structure (shown as spheres). Residues that are included in the “A”, “K”, “R” and “70” groups are shown in red, magenta, yellow and orange, respectively. Residues with missing backbone amide resonances are colored blue. (H) Representative RD profiles of the four groups obtained on 600-MHz (black) and 800-MHz (red) spectrometers.
In addition, a number of residues in re-ArsC·sulfate (including C10, T11, A15-Q18, T63, D65, N73, C82 and K88) harbor at least two distinct minor dips in the CEST profile (Figs 2C and S3A). Analyses using different three-state exchange models26 favor a G ⇌ E1 ⇌ E2 exchange processes (G is the ground state, E1 and E2 are two different excited states corresponding to the larger and smaller minor dips) (Fig. S4). The observation that the depths of the third dip are also affected by sulfate concentrations (Fig. S2) further supports this exchange model rather than E1 ⇌ G ⇌ E2 or G ⇌ E2 ⇌ E1 model. Since the G ⇌ E1 exchange reflects the sulfate binding and releasing event, it is rational to assume that E1 corresponds to re-ArsCfree. Thus, the E1 ⇌ E2 exchange process observed in re-ArsC·sulfate is expected to also exist in re-ArsCfree. As expected, six residues (N13, Q18, A41, H42, T63 and D65) show two-state G ⇌ E exchanges in re-ArsCfree (Figs 2D and S3B), and four (N13, Q18, T63 and D65) of these show three-state G ⇌ E1 ⇌ E2 exchanges in re-ArsC·sulfate. Furthermore, residue C10 is observed to undergo three-state exchanges in re-ArsCfree and even more complex exchanges in re-ArsC·sulfate (Fig. S3A,B). The chemical shifts of the E state in re-ArsCfree and the E2 state in re-ArsC·sulfate are essentially similar (Fig. 2F). Taken together, the CEST data reveal two exchange processes around the active site, one corresponding to the sulfate binding with kex of ~120 s−1, and the other reflecting conformational exchanges between an empty substrate binding pocket with an excited state with kex of ~300 s−1, where the exchange rates were estimated from the global fitting of residues R16-Q18, G43, T63 and D65 in re-ArsC·sulfate using the G ⇌ E1 ⇌ E2 three-state exchange model (Table S1 and Fig. S3C). Moreover, the dissociation constant Kd for sulfate binding to re-ArsC is estimated to be ~1 mM based on the CEST results, which is confirmed by isothermal titration calorimetry (ITC) measurements with an estimated Kd of 0.7 mM.
Multiple conformational exchanges in re-ArsC
To further characterize the motions of re-ArsC·sulfate, we employed the CPMG RD method and identified about forty residues undergoing conformational exchanges in re-ArsC·sulfate (Figs 2G,H and S5). These residues cluster in four regions surrounding the active site pocket: the P-loop, the 80 s segment, the 40 s and 60 s loops, and the 70 s helix, which is consistent with our previous 15N relaxation measurements20.
Data analysis indicates that residues in different structural regions are involved in separate local motions (Table 1). A subgroup of 12 residues (including T11-N13, C15-S17, G38-E40, and D65-I67) located on a continuous surface around the active site pocket can be described by a two-state exchange model with global exchange parameters kex = 870 ± 58 s−1 and pE = 0.52 ± 0.01%. These residues are designated as group A for clarity (“A” for “active site”). A second group of residues (Q18, V34, S36, G43, N45 and T63) undergo a local exchange process with kex = 328 ± 36 s−1 and pE = 1.28 ± 0.09%. These residues locate around a putative potassium binding pocket that was identified in S. aureus pI258 ArsC but appears to be inactive in B. subtilis ArsC27,28, and are designated as group K (“K” for potassium). Residues in the 80 s redox functional segment also exhibit motions different from group A residues. Three residues (G83, D84, A85) are missing in the 1H-15N HSQC spectrum, indicative of conformational exchanges on the intermediate timescale, whereas both residues L81 and K88 (which we designate as group “R” for “redox”) are estimated to show exchange rates of ~40 s−1, which is much slower compared to group A and falls outside the time regime that could be reliably characterized by the RD method. Note that the relationship between kex and |Δω| determines whether the conformational exchange falls into the fast exchange (when kex > |Δω|) or intermediate exchange (when kex ≈ |Δω|) regime on the NMR timescale29. Since the |Δω| values are distinct among different residues, the above-mentioned residues in the 80 s segment may still be involved in the same collective motion (with a same global kex) while displaying different |Δω| values. The ones with |Δω| close to kex would be significantly broadened and undetectable. Furthermore, residues D68, D70, L72-N74 and V78 around the 70 s helix (designated as group 70) can be fitted using a two-state exchange model with exchange parameters kex = 269 ± 48 s−1 and pE = 1.9 ± 0.2%.
Table 1.
Summary of the kinetic parameters of ArsC from CPMG RD data.
| re-ArsC·sulfate | |||
| Group A | residues 11–13, 15–17, 38–40, 65–67 | ||
| kex = 870 ± 58 s−1 | pE = 0.52 ± 0.01% | kGE = 4.5 ± 0.1 s−1 | kEG = 866 ± 58 s−1 |
| Group K | residues 18, 34, 36, 43, 45, 63 | ||
| kex = 328 ± 36 s−1 | pE = 1.28 ± 0.09% | kGE = 4.2 ± 0.1 s−1 | kEG = 324 ± 36 s−1 |
| Group R | residues 81, 88 | ||
| kex = 40 ± 20 s−1 | pE = 6.6 ± 3.0% | kGE = 2.6 ± 0.7 s−1 | kEG = 37 ± 20 s−1 |
| Group 70 | residues 68, 70, 72–74, 78 | ||
| kex = 269 ± 48 s−1 | pE = 1.9 ± 0.2% | kGE = 5.1 ± 0.2 s−1 | kEG = 264 ± 48 s−1 |
| int-ArsC | |||
| Conformation-1 | residues 7-9, 20, 22, 24, 34, 35, 50, 62, 68-1, 73, 74, 78, 79-1, 85, 96, 98-1, 102, 104 | ||
| kex = 685 ± 24 s−1 | pE = 4.9 ± 0.3% | kGE = 33.6 ± 0.1 s−1 | kEG = 651 ± 24 s−1 |
| Conformation-2 | residues 7-9, 20, 22, 24, 34, 35, 50, 62, 68-2, 73, 74, 78, 79-2, 85, 96, 98-2, 99-2, 102, 104 | ||
| kex = 681 ± 23 s−1 | pE = 4.9 ± 0.3% | kGE = 33.4 ± 0.1 s−1 | kEG = 648 ± 24 s−1 |
| c-ArsC | |||
| residues 10, 12, 13, 15–18, 20, 36, 39,45, 65, 67, 80, 82, 83, 87–89, 95–97, 100–102 | |||
| kex = 443 ± 22 s−1 | pE = 2.8 ± 0.1% | kGE = 12.4 ± 0.1 s−1 | kEG = 431 ± 22 s−1 |
A number of residues located around the P-loop region are identified to exhibit conformational exchanges in re-ArsC·sulfate by both CEST and CPMG RD methods, and a subset of these display three-state exchanges in the CEST profile. Comparison of the exchange parameters including kex, pE and |Δω| derived from the two methods suggests that for group A residues forming the active site pocket, the CPMG RD and CEST experiments report on different exchange processes. For group K residues (Q18, G43 and T63), although the kex value derived from CPMG RD data is close to the kex,2 value (the apparent exchange rate between E1 and E2 in the three-state exchange model), we did not find apparent correlations between the corresponding chemical shift differences. On the other hand, the CEST profiles of many residues in the P-loop region as well as the 70 s and 80 s segments could not be well fitted globally, therefore prohibiting accurate determination of the exchange parameters, which also indicates that the motions in re-ArsC·sulfate may be more complex (Table S1 and Fig. S3). Taken together, re-ArsC·sulfate displays highly complex dynamics involving multiple conformational exchange processes for different structural regions. In particular, the active P-loop undergoes at least three different exchange processes, namely the binding of sulfate and exchanges with two different excited states on different timescales.
Structural characterization of int-ArsC
The C10–C82 disulfide-bonded int-ArsC is an important but short-lived reaction intermediate during the intra-molecular disulfide cascade, and could only be trapped by mutating the C89 residue to quench the reaction. To facilitate dynamic studies, we completed the chemical shift assignments of int-ArsC and characterized its structural properties (Fig. 3). Twenty-four out of the total of 139 residues are missing in the 1H-15N HSQC spectra of int-ArsC, and quite a few residues around the active site show broadened resonances, suggesting significant conformational exchanges on the intermediate timescale. Furthermore, a number of residues located near the 80 s segment show two sets of peaks in the HSQC spectrum, indicating dual conformations in slow exchange with each other (Figs 3A–C and S6). Judging from the intensities of the two sets of peaks, the two conformations have a population ratio of approximately 1:1 at 25 °C, whereas decreasing the temperature results in the population increase of one set of the peaks (Fig. 3C). We designate the set of peaks that is dominant at lower temperatures as conformation-1, and the other set as conformation-2.
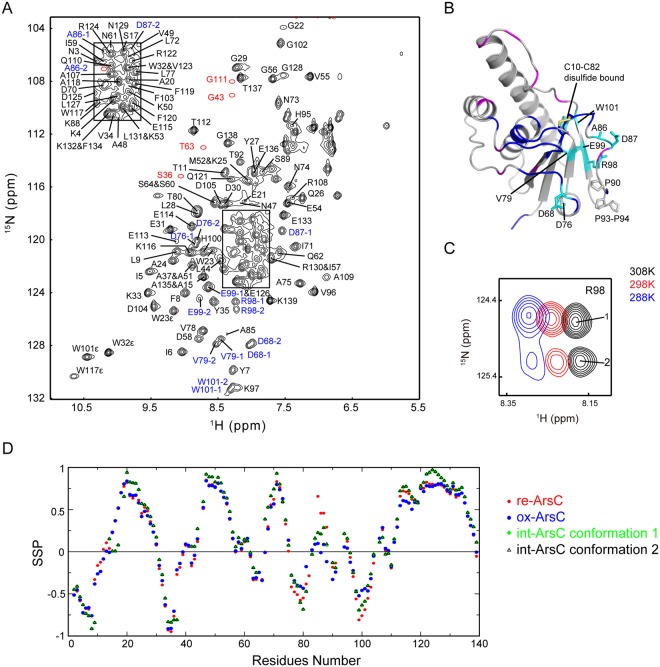
Figure 3.
Assignments and structural characterization of int-ArsC. (A) The 1H-15N HSQC spectrum of int-ArsC labeled with backbone resonance assignments. The positions of the weak resonances are indicated by red circles in the spectrum. Residues showing two sets of peaks are labeled in blue with ‘−1’ and ‘−2’ designating two conformations. (B) Mapping of residues with backbone amide resonances missing (blue) or significantly broadened (magenta), and residues with two sets of backbone amide resonances (shown as cyan sticks) onto the structure model of int-ArsC. The C10–C82 disulfide bond and the three proline residues in this region are also shown. (C) Representative local spectral region showing the two sets of signals from R98 at different temperatures. (D) Secondary structure propensity derived from the chemical shifts from 15N, 13Cα, 13Cβ, 13CO, 1Hα and 1HN atoms using the program package SSP30 for re-ArsC, ox-ArsC and int-ArsC with the two sets of conformations. SSP values close to 1 indicate high propensity of α-helices, whereas SSP values close to −1 indicate high propensity of β-strands.
Previous structural studies showed that the 80 s redox functional segment (C82–C89) undergoes a “helix-to-loop” transition coupled to its self-oxidization during the thiol-disulfide cascade19, and int-ArsC is the reaction intermediate during this transition from the reduced to oxidized state. Inspection of the 3D NOESY spectra of int-ArsC identified no characteristic NOEs of helical conformations in the 80 s segment, whereas secondary structure propensity (SSP) analysis using the assigned 13Cα/13Cβ/13CO/1Hα/1HN/15N chemical shifts30 estimated helical-forming propensities for residues A85-K88 in between the predicted values of re-ArsC·sulfate (with ~50% of helical propensity) and ox-ArsC states (with close to 0 helical propensity) (Fig. 3D). This indicates that the 80 s segment in int-ArsC adopts an “intermediate” conformation, or that the chemical shifts reflect an average over different populations with higher and lower helical contents.
Global collective motions in int-ArsC
While the CEST experiment detected no conformational exchanges in int-ArsC, the CPMG RD data identified about thirty residues exhibiting millisecond timescale dynamics. These residues spread over different regions of the protein structure, including the central β-sheet and the surrounding α-helices (Fig. 4A,B), reflecting an extremely dynamic and transient nature of this intermediate state. Data analysis revealed that the RD profiles of a total of 21 residues could be described using a global two-state exchange process. These residues are located around the active site pocket and extend to nearly all the regular secondary structural elements, including all strands in the central β-sheet, indicating a global collective motion involving different structural regions.
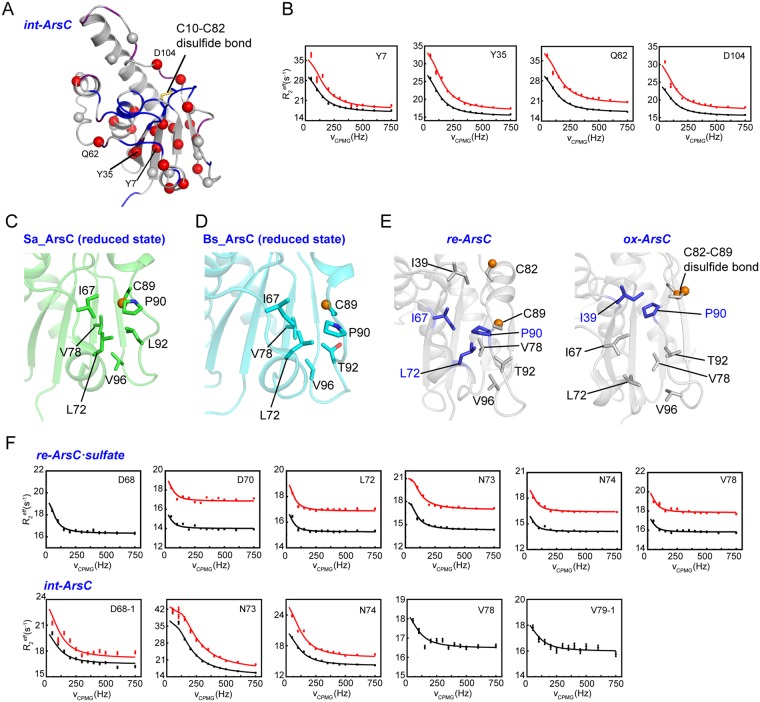
Figure 4.
CPMG RD results of int-ArsC. (A) Mapping of residues showing conformational exchanges in int-ArsC identified by CPMG RD experiments onto the structure model (shown as spheres). Residues with missing backbone amide resonances are colored blue, and residues with weak signals are colored purple. (B) Representative RD profiles of residues in int-ArsC obtained on 600-MHz (black) and 800-MHz (red) spectrometers. (C,D) The local conformation of the hydrophobic pocket that C89 occupies in the reduced state of Sa_ArsC (C, PDB code: 1ljl) and Bs_ArsC (D, PDB code: 1z2d). (E) Comparison of the local structural changes of the hydrophobic pocket in re-ArsC and ox-ArsC showing the packing of P90 either with I67, I72 in re-ArsC (left) or with I39 in ox-ArsC (right). (F) Representative RD profiles of residues in the 70 s segment in re-ArsC·sulfate (upper panel) and int-ArsC (lower panel) obtained on 600-MHz (black) and 800-MHz (red) spectrometers. The data for D68 in re-ArsC·sulfate and V78 and V79 in int-ArsC obtained at B0 field of 800 MHz were excluded from analysis due to relatively large experimental errors.
For residues showing two sets of peaks, both conformation-1 and 2 show similar relaxation dispersion phenomenon and the estimated exchange parameters are essentially similar when using the data set from either conformation. The global exchange parameters for the 21 residues are estimated to be kex = 685 ± 24 s−1, pE = 4.9 ± 0.3% when using the data from conformation-1, and kex = 681 ± 23 s−1, pE = 4.9 ± 0.3% when using the data from conformation-2. The extracted absolute shift differences |Δω| for residues with two sets of peaks are different from the 15N chemical shift differences |Δδ| between the conformations, which is expected since the RD experiment is sensitive to exchanges on fast timescales whereas the existence of double peaks reflects exchanges on slower timescales.
C10–C82 disulfide bond induces faster motions in the 70s helix
In the previous crystallographic study of Sa_ArsC in the C10–C82 disulfide-bonded intermediate state (int-ArsC), Messens and co-workers pointed out that C89 needs to come out of a hydrophobic pocket and move over a distance of ~10 Å to carry out the nucleophilic attack on C8219. In the reduced state (re-ArsC), this local hydrophobic pocket is formed by several hydrophobic residues in the 70 s segment (I67, L72 and V78 in Sa_ArsC) and the 90 s loop (P90, L92 and V96 in Sa_ArsC) (Fig. 4C). In Bs_ArsC, the corresponding residues are I67, L72, V78, P90, T92 and V96 (Fig. 4D). Though T92 in Bs_ArsC is a polar residue, it uses its methyl group to contact the hydrophobic core, while its hydroxyl group points outward. In the oxidized state (ox-ArsC), C89 moves out of the hydrophobic pocket, accompanied by changes of the relative positions of the hydrophobic residues that can be viewed as a sliding between two hydrophobic tracks, one formed by residues L72, V78, I67 and I39, and the other by P90, T92, V96 (Fig. 4E). In particular, the P90 side chain forms contacts with both I67 and L72 in re-ArsC, whereas it slides toward the active site and interacts with I39 in the ox-ArsC, undergoing the largest conformational change among these hydrophobic residues. Notably, the amide signals of both I39 and I67 are missing in the HSQC spectrum of int-ArsC, indicative of intermediate conformational exchanges on the NMR timescale, which might be related with the switching of P90 between two conformations, one with P90 docking at the I67 site (resembling re-ArsC) and the other docking at the I39 site (resembling ox-ArsC). Moreover, the CPMG RD data show that residues D68, N73, N74, V78 and V79 in the 70 s segment undergo conformational exchanges in int-ArsC, which may also be related to the local structural rearrangement that could facilitate the movement of P90 and C89.
Intriguingly, millisecond timescale motion for the 70 s helix is also observed in re-ArsC (Table 1 and Fig. 4F), but not in either ox-ArsC or the Trx-complexed c-ArsC (vide infra). In re-ArsC·sulfate, the residues in group 70 exhibit local motions with kex of ~269 s−1, and is uncoupled from the motions at the active site (kex ~ 870 s−1). Under the assumption of a two-state exchange process (where G and E stand for the ground and the excited states), the apparent exchange rate constant kex is the sum of kGE and kEG, and the rate constant kGE for the transition from the ground state to excited state can be calculated by kGE = kex * pE (pE is the relative population of the excited state). Therefore, the forward transition rate constant kGE for group 70 in re-ArsC·sulfate is 5 s−1. In int-ArsC, however, the motions of group 70 residues become coupled to the active site, with a global kex of ~680 s−1 and pE is ~4.9%. Thus, the rate constant kGE increases to 33 s−1 in int-ArsC, which is about seven-fold faster than in re-ArsC·sulfate.
Taken together, the data indicate local millisecond dynamics of the 70 s segment pre-exist in the reduced state, whereas formation of the C10–C82 disulfide bond in int-ArsC results in acceleration of the motions, as well as coupling of this local dynamics with the active site pocket. The increased rate for the transition to excited state in int-ArsC is favorable for the movement of C89 out of the hydrophobic pocket, and the coupling of motions between the 70 s site and the active site may be important for relaying the large-scale movement of C89 with its subsequent nucleophilic attack on the C10–C82 disulfide.
ox-ArsC shows limited dynamics
The ox-ArsC corresponds to the inactivated state of the enzyme after completing the reduction of one molecule of arsenate. Both CEST and CPMG RD data demonstrate that the dynamics in ox-ArsC are largely diminished. Only four residues (G12, S14, N45 and T80) show varying R2eff depending on the νCPMG field in the RD profiles, whereas the observed deviations of R2eff are quite small (Fig. S7A). Individual fitting results indicate that the four residues are not involved in a collective motion and cannot be fitted globally. The relatively large fitting errors also prohibited reliable estimation of the kinetic parameters. Apparently, the P-loop residues in ox-ArsC are significantly rigidified on the millisecond timescale, showing little R2eff variations (Fig. S7B). It is also observed that the R2eff values in this loop are systematically higher in the active re-ArsC·sulfate state than the inactive ox-ArsC state, indicative of conformational dynamics in re-ArsC but not ox-ArsC (Fig. S7C). Furthermore, the 80 s segment also gains higher rigidity, and the 70 s segment also shows no conformational exchanges in ox-ArsC.
Global collective motions in c-ArsC
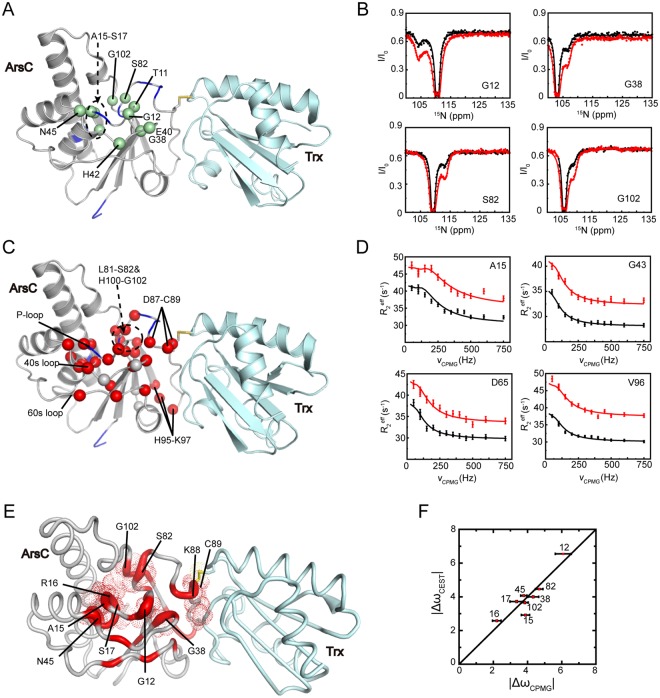
During the regeneration of ArsC by Trx, a mixed-disulfide Trx-ArsC complex is transiently formed. The CEST method identified a total of eleven residues in c-ArsC to undergo conformational exchanges (Fig. 5A,B). These residues are located near the active site pocket and can be analyzed using a two-state exchange model, with a global exchange rate of 440 s−1 and a minor state population pE of ~3.8%. Further, the CPMG RD method also identified about thirty residues to show νCPMG-dependent R2eff values, distributed in a large structural area surrounding the P-loop and the Cys82–Cys89 redox functional segment (Fig. 5C,D). The RD profile for the majority of these residues can be described by a global two-state exchange process, with the estimated exchange parameters of kex = 443 ± 22 s−1 and minor state pE = 2.80 ± 0.10% (Table 1). These values are highly similar to the values obtained from the CEST data, suggesting that the two methods may be reporting on the same exchange process. This scenario is further supported by the essential similarity between the absolute chemical shift differences |Δω| extracted from the two methods (Fig. 5F).
Figure 5.
CEST and CPMG RD results of c-ArsC. (A,C) Residues showing conformational exchanges in c-ArsC identified by the CEST (A) and CPMG RD (C) experiments are mapped onto the Trx-ArsC complex structure (PDB ID: 2ipa), shown as green and red spheres, respectively. (B) Representative 15N CEST profiles of c-ArsC obtained on a 600-MHz spectrometer using B1 fields of 8.4 Hz (black) and 13.5 Hz (red). (D) Representative RD profiles of c-ArsC obtained on 600-MHz (black) and 800-MHz (red) spectrometers. (E) Mapping of the |Δω| values based on the CPMG RD results onto the c-ArsC structure. Residues undergoing collective motions are colored in red, and the thickness of the cartoon tubes reflects the relative values of |Δω|. Residues with |Δω| > 2.0 are shown as dotted spheres. (F) Comparison between the absolute Δω values extracted from CEST and CPMG RD experiments for residues in c-ArsC with residue numbers denoted.
The extensive conformational exchanges observed near the active site in c-ArsC is quite different from ox-ArsC, and more closely resembles re-ArsC. Because a stable Trx-ArsC sample needs to be prepared using a C10S/C15A/C82S triple mutant of ArsC and the C89 residue forms an intermolecular disulfide bond with Trx (see Methods), the chemical environment of the residues around the active site pocket are largely affected. Residues exhibiting millisecond dynamics are mostly located within close proximity to the C10, C15, C82 and C89 residues, and therefore a direct comparison of the |Δω| values with the chemical shift differences |Δδ| between re-ArsC and c-ArsC, or between ox-ArsC and c-ArsC, is not feasible. Instead, we mapped the |Δω| values onto the c-ArsC structure to show the residues that have the largest chemical/conformational differences between the ground and invisible states (Fig. 5E). Residues G12, A15 (mutated from C15), R16, S17 in the P-loop, G38 and N45 in the 40 s loop, S82 (mutated from C82), K88 and C89 in the 80 s segment, and G102 in the 100 s loop show the largest |Δω| values, indicating that they undergo the most significant structural rearrangement in the excited state. The G12, A15-S17, G38, S82 and G102 residues form a circle surrounding the active site pocket, whereas the K88 and C89 residues form the center of the interaction surface with Trx.
Inter-protein coupling of motions in the Trx-ArsC complex
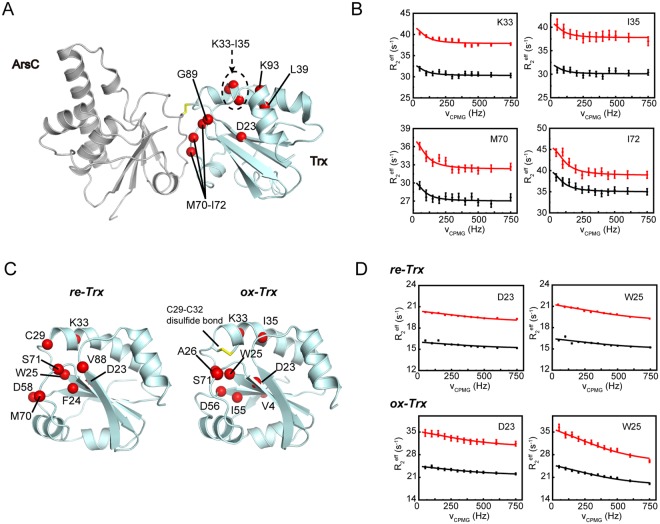
The Trx-ArsC complex is transiently formed during the catalytic cycle and a subsequent nucleophilic attack from Trx-C32 quickly resolves the mixed disulfide bond. The global collective motions in c-ArsC intrigued us to ask whether the coupling of dynamics is further extended to residues in Trx (designated as c-Trx). 15N CPMG RD experiments were collected using a Trx-ArsC complex sample with Trx uniformly 15N-labeled and ArsC unlabeled. A total of ten residues (D23, K33-I35, L39, M70-I72, G89 and K93) were found to exhibit conformational exchanges in c-Trx (Fig. 6A,B). Among these, M70-I72 and G89 locate on the interaction surface that directly contacts ArsC in the complex structure. In particular, M70 side chain plays a main role in complex formation by inserting into a hydrophobic groove on the surface of ArsC17. The other residues are clustered around the cysteine residue C32 in Trx and include the acidic D23 which was suggested to play a role in the activation of C3231–33. Moreover, the resonance of C29 is weak and that of G30 is missing, indicative of intermediate exchanges.
Figure 6.
CPMG RD results of Trx in the complex and free states. (A,C) Residues showing conformational exchanges in c-Trx (A) or re-Trx, ox-Trx (B) identified by the CPMG RD experiments are mapped onto the structures (shown as red spheres). (B) Representative RD profiles of c-Trx (B), re-Trx and ox-Trx (D) obtained on 600-MHz (black) and 800-MHz (red) spectrometers.
RD data analysis of c-Trx suggests exchange parameters similar to c-ArsC, and a combined global fitting using the data from both c-ArsC and c-Trx estimates the exchange rate constant kex = 464 ± 21 s−1 and minor state population pE = 2.70 ± 0.10%, which is essentially identical to the values extracted from the c-ArsC data alone. For comparison, we also measured the CPMG RD data of Trx in its reduced and oxidized states (designated as re-Trx and ox-Trx), both detecting a number of residues exhibiting conformational exchanges around the two catalytic active cysteine residues (Fig. 6C,D). However, the exchange rate constant kex is much higher (kex ~ 2000–3000 s−1) in both re-Trx and ox-Trx compared to c-Trx. The results indicate the formation of the mixed-disulfide complex significantly changes the motions of Trx active site residues and also results in an inter-protein coupling of dynamics.
Discussion
During the cascade of thiol-disulfide exchange in the arsenate reduction, a total of five catalytic active cysteine residues from two proteins are sequentially activated to perform a series of nucleophilic attacks. It is therefore intriguing to elucidate the structural basis for the regulation of this highly ordered chain of reactions. The CPMG RD and CEST experiments together demonstrate highly complex conformational spaces sampled by ArsC in different reaction stages. In particular, the re-ArsC·sulfate shows the most complex dynamics behavior around the active site, with P-loop residues undergoing three or more exchange processes, indicating that the local conformational energy landscape is extremely rugged. In a previous reported kinetic study of ArsC, it was shown that the presence of excess tetrahedral oxyanions such as sulfate is essential for obtaining the optimal enzymatic activity, whereas in the absence of tetrahedral oxyanions the enzyme is unstable and displays low enzymatic activity25. Therefore, rather than the apo re-ArsCfree state, a sample with the P-loop stabilized by tetrahedral oxyanions is expected to represent the active state of the enzyme. While the best stabilizing oxyanion is the substrate arsenate itself, it could not be used because it would quickly react with the enzyme to generate the oxidized form. Despite the lower binding affinity (Kd ~ 0.7 mM) to re-ArsC compared to the native substrate arsenate (Km ~ 47 μM for Bs_ArsC)28, the sulfate ion has been shown to directly bind the P-loop and act as a good stabilizing agent20,25,28. The interaction between sulfate and the P-loop residues could modulate the local dynamics and help shape the active site conformation to gain optimal geometry for the highest catalytic efficiency. In the re-ArsC·sulfate state, the P-loop harboring the catalytic essential C10 residue undergoes fast internal motions with kex of ~870 s−1, which may be important for priming the initial nucleophilic attack. On the other hand, the 80 s segment harboring the C82 and C89 residues undergoes slower motions in the re-ArsC·sulfate state, and becomes involved in a fast global collective motion with kex of ~680 s−1 only after forming the C10–C82 disulfide bond in the int-ArsC state. The faster motions in the 80 s segment and the related 70 s helix in int-ArsC may also be important in facilitating the subsequent nucleophilic attack by C89. In contrast, in the ox-ArsC state in which all three catalytic cysteine residues (C10, C82 and C89) are inactivated, the protein conformational dynamics become largely diminished. Taken together, the conformational dynamics of ArsC in different state are highly correlated with the enzymatic reaction as also suggested by previous studies17,19,20.
On the other hand, it remains less well understood how the inter-protein disulfide exchanges between Trx and ArsC are regulated by protein dynamics. In particular, among the two cysteines in Bs_Trx, C29 is active in the free reduced state for attacking downstream protein substrates while C32 is inactive. Only upon formation of the mixed-disulfide Trx-substrate complex can C32 become activated. The activation mechanism of C32 and how this cysteine residue resolves the mixed-disulfide complex has been investigated and under debate in the past decades. This process has proven difficult to study primarily due to the transient nature of the mixed-disulfide complexes. Such complexes usually can only be obtained by trapping the reaction using Trx mutants with the second cysteine (C32 in Bs_Trx) mutated and therefore measurements of the biophysical properties (such as pKa values) of this resolving cysteine cannot be made directly in the mixed-disulfide complexes. Moreover, the mixed-disulfide Trx-substrate complexes are usually dynamic and are not easy to be crystallized34. The solution NMR structure of B. subtilis Trx-ArsC complex previously determined in our lab is one of the few high-resolution structure of Trx complexed with a substrate protein17,35–38. The structure reveals an extended hydrophobic interaction surface between the two proteins, but could not provide direct answer as to how Trx-C32 becomes activated in this complex. On the other hand, besides the large scarcity of structural information, there has been a lack of information concerning protein dynamics that has been proven essential for enzyme activity.
In a computational study starting from the solution structure of Trx-ArsC complex, Roos and co-workers suggested an intriguing mechanism for C32Trx activation which is affected by the hydrogen bonding and protonation state of C82ArsC 18. In brief, the thiol group of C82ArsC in Trx-ArsC complex has a pKa value of 7.9 when it is free of hydrogen-bonding interactions, whereas its pKa drops to 6.3 and becomes more easily deprotonated when it is hydrogen bonded to the side chain of R16ArsC. Further, when C82ArsC is deprotonated, the thiol group of C32Trx has higher possibility of forming hydrogen bonds with either the amide of C29Trx or W28Trx, with C32TrxSγ-C29TrxN and C32TrxSγ-W28TrxN hydrogen-bonds formed during 60% and 23% of the time when C82ArsC is in the deprotonated state, as compared to the significantly smaller fraction of 1.6% and 0.5% when C82ArsC is neutral18. Simultaneous formation of both C32TrxSγ-C29TrxN and C32TrxSγ-W28TrxN hydrogen-bonds could help lowering the pKa of C32Trx, facilitating its deprotonation and thus activation. Our current study of the Trx-ArsC complex dynamics reveals a global collective motion involving the catalytic essential residues from both c-ArsC and c-Trx, as well as residues across the complex interface, which provides strong experimental support for the long-range coupling of conformational fluctuations that enables the propagation of structural changes from one protein to the other. In particular, among the residues involved in collective motions in c-ArsC, residues G38ArsC, I39ArsC and D87ArsC locate between the active site pocket and the protein-protein interface. Residues I67ArsC and K88ArsC form direct hydrophobic contacts c-Trx. Likewise, residues M70-I72Trx in c-Trx locate in the protein-protein interface, whereas G89Trx locates in between the interface region and the active site region surrounding C29Trx and C32Trx. In addition, residue D23Trx that has been previously proposed to activate C32Trx via a bond water molecule31–33,39 is also involved in the collective motion.
Furthermore, the kcat for the reduction of ox-ArsC by re-Trx has been determined to be approximately 1.9 s−1 under a buffer conditions of 50 mM Tris (pH 8.0), 50 mM K2SO4 and 0.1 mM EDTA34. Our current CEST and CPMG RD data of c-ArsC demonstrate a two-state exchange process with the forward rate constant kGE = 13 s−1. Assuming the excited state E resembles the conformation in which C82ArsC becomes hydrogen-bonded with R16ArsC, and using the values of 60% and 23% from the computational results as the probability of C32Trx in forming hydrogen bonds with C29Trx or W28Trx in this conformational state, we could estimate the rate constant of C32Trx simultaneously forming two hydrogen bonds with C29Trx and W28Trx to be 13 * 0.6 * 0.23 = 1.8 s−1, which is highly similar to the kcat value. This further suggests that inter-protein coupling of dynamics could indeed play a central role in regulating the activation of cysteine thiol groups during disulfide exchanges, and may represent the rate-limiting step in the case of Trx-ArsC redox reaction.
The coupling of motions among different structural regions is not only observed in the Trx-ArsC complex, but also in the C10–C82 disulfide-bonded int-ArsC state. The millisecond timescale motion of the 70 s helix, which is important for activating C89ArsC, is coupled to the active site only in this intermediate state but not in the reduced state. C89ArsC, as well as C32Trx, functions as a resolving cysteine (CR) that resolves an unstable disulfide bond (C10–C82 in int-ArsC, and C89ArsC-C29Trx in the Trx-ArsC complex) in transiently formed, energetically unfavorable reaction intermediates. These CR residues are inactive in the reduced state of the enzyme and becomes rapidly activated in the transiently-formed intermediate. The intra-protein and inter-protein coupling of motions observed in int-ArsC and the Trx-ArsC complex may reflect a common scenario of protein dynamics-regulated activation of cysteine thiol groups in thiol-disulfide exchange reactions.
As discussed earlier, the mechanism for protein thiol-disulfide transfer reaction is difficult to study and remains in debate for the past twenty years. Moreover, the transient nature of Trx-substrate mixed-disulfide intermediate makes experimental measurement of the pKa of the resolving cysteine unfeasible by current available methods. Our current study highlights both intra- and inter-protein coupling of millisecond motions, strongly suggesting the cysteine thiol activities could be regulated by long-range propagation of enzyme dynamics. The results are also highly supportive of the working model of Trx-catalyzed thiol-disulfide exchange reaction as proposed by Roos et al. by computational methods. The advance NMR methods in detecting the sparsely populated excited states and extracting the kinetic parameters for the exchange processes shed new light on understanding thiol-disulfide exchange reactions, and when further combined with more detailed computational investigations, would hopefully provide a more comprehensive understanding in this field.
Methods
Sample Preparations
Protein expression, labeling and purification of B. subtilis ArsC and Trx proteins (including mutants) were similar to previously reported17,20. The C10–C82 disulfide-bonded int-ArsC sample was prepared using the ArsC_C15AC89S mutant via incubation with 0.3% H2O2 at room temperature for 1 h followed by gel filtration chromatography. The protein concentration was kept below 1 mg/ml during the incubation to minimize intermolecular disulfide bond formation. Formation of the C10–C82 disulfide bond was verified by Ellman’s test40 using 5, 5′-dithio-bis 2-nitrobenzoic acid (DTNB) as the reagent, which reacts with free thiols to yield TNB2− and can be monitored by absorption at 412 nm. The Trx-ArsC mixed-disulfide complex was prepared using the Trx_C32S and ArsC_C10SC15AC82S mutants and following the previously reported 5,5-dithiobis(2-nitrobenzoic acid) incubation protocol17,34. The Trx-ArsC complex samples were prepared with 15N-labeled ArsC and unlabeled Trx or 15N-labeled Trx and unlabeled ArsC. The purities of the protein samples were determined to be greater than 95% as judged by SDS-PAGE. The NMR samples of re-ArsC·sulfate, ox-ArsC and Trx-ArsC complexes were prepared in a buffer containing 20 mM Tris-HCl (pH 6.85), 40 mM KCl, 20 mM urea and 20 mM Na2SO4, the samples of re-ArsCfree and int-ArsC were prepared in a buffer containing 20 mM Tris-HCl (pH 6.85), 40 mM KCl, 20 mM urea and 40 mM NaCl, and the samples of re-Trx and ox-Trx were prepared in a buffer containing 20 mM Tris-HCl (pH 6.85). Excess dithiothreitol (DTT) was added in the samples of the re-ArsC and re-Trx to ensure a reducing environment. D2O was added to 5% for field lock and 2,2-dimethyl-2-silapen-tanesulfonic acid was used as the internal chemical shift reference.
Backbone resonance assignments of re-ArsCfree and int-ArsC
The NMR experiments for re-ArsCfree and int-ArsC backbone assignments were carried out at 25 °C on Bruker Avance 500-MHz and 600-MHz spectrometers equipped with four RF channels and cryogenic triple resonance probes with pulsed field gradients. The backbone chemical shift assignments were obtained by using the conventional three-dimensional HNCA, HNCACB, HNCO, CBCA(CO)NH experiments41. All NMR spectra were processed using NMRPipe42 and analyzed using NMRView43.
Backbone 15N CPMG RD measurements
15N-labeled ArsC samples were prepared with protein concentrations of 0.7 mM and were argon-flushed. The 15N CPMG RD experiments44 were carried out at 25 °C on Bruker Avance 600- and 800-MHz spectrometers equipped with cryogenic probes. A constant transverse relaxation time of TCPMG = 60 ms and 80 ms were used for ArsC and Trx, respectively, whereas TCPMG = 40 ms was used for the Trx-ArsC complex. Data were recorded for fourteen different νCPMG values of 50, 100 (x2), 150, 200, 250, 300, 350, 400, 450, 500, 600 and 750 Hz. Here νCPMG = 1/(4τcp), where τcp is the delay between refocusing pulses during the CPMG pulse train. All data were recorded at three different 15N carrier frequencies to circumvent off-resonance effect and to obtain reliable relaxation dispersion profiles for signals located in different regions of the spectrum. The spectra were processed using NMRPipe42 and the peak intensities were measured using NMRView43. Residues exhibiting conformational exchanges on appropriate timescales would show a dispersion profile of R2eff values dependent on νCPMG. The effective transverse relaxation rates R2eff were determined using the equation R2eff = (−1/TCPMG)ln(IνCPMG/I0), where TCPMG is the constant transverse relaxation time, I0 is the intensity measured in the reference spectrum, and IνCPMG is the intensity measured at different CPMG field strengths νCPMG45. Uncertainties in R2eff were calculated as ΔR2eff = (1/TCPMG)(ΔI/IνCPMG), where ΔI is the average standard deviation of peak intensities estimated from repeat measurements46. By using the cpmg_fit software from L. Kay and D. Korzhnev47, all dispersion data were fitted to the Richard-Carver equation48 assuming a two-state exchange model without assumption regarding the exchange regime. Fitting uncertainties were extracted using the covariance matrix method49. The dispersion curves for individual residues were generated using the GLOVE50 program by fixing the kex, pGpE and Δω parameters. For global data fitting, clustering of residues were based on comparisons of the estimated kex, pE parameters obtained from individual fittings, while taking into account of their locations in the three-dimensional protein structure. In addition, for residues showing three-state exchanges in the CEST data, we also tried using three-state exchange models to fit the CPMG data but did not improve the results.
Backbone 15N CEST experiments
15N-labeled ArsC samples were prepared with protein concentrations of 1.8 mM and were argon-flushed. 15N CEST experiments21 were carried out at 25 °C on a Bruker Avance 600-MHz spectrometer. A total of 191 2D data sets were acquired with the 15N carrier frequencies positioned from 100 ppm to 138 ppm at a spacing of 0.2 ppm (12.16 Hz) during the irradiation time of TEX = 800 ms. In all experiments, irradiation field strengths B1 of 8.4 ± 0.2 Hz and 13.5 ± 0.2 Hz were used, and a 2.7 kHz field 1H decoupling composite pulse sequence (90x-240y-90x) was applied during the TEX period. Data without using the B1 field during the TEX period was recorded as the reference experiment. B1 calibration was carried out following the previously reported methods51. All the data sets were processed using the NMRPipe program42, and peak intensities were obtained by NMRView43. The CEST profiles for the individual residues were generated by calculating the intensity ratios I/I0 versus the varied 15N carrier frequencies, where I0 is the intensity measured in the reference spectrum, and I is the intensity measured with the application of the B1 field. The uncertainties of peak intensities were estimated from repeat measurements. For residues showing only one minor dip, the data were fitted to a two-state exchange model using the python program ChemEx (https://github.com/gbouvignies/chemex) as described previously21, and the fitting uncertainties were extracted using the covariance matrix method49. Unlike c-ArsC, the re-ArsC·sulfate state displays highly complex dynamics and many residues show two or more minor dips. For these residues, the data were individually fitted using the three-state exchange models with an in-house written Matlab script from B. Yu and D. Yang26. Briefly, the CEST data were fitted using three different three-state models (model 1: G ⇌ E1 ⇌ E2; model 2: G ⇌ E2 ⇌ E1; model 3: E1 ⇌ G ⇌ E2), and the G ⇌ E1 ⇌ E2 model was found to best describe the profiles (Fig. S4). The detailed equations for fitting the data using three-state models were described in ref.26, and the fitting uncertainties were extracted by calculating the inverse of the Jacobian matrix52. Furthermore, 15N CEST experiments were collected using protein samples at 0.5 mM and 2 mM concentrations by employing a residue-selective 1D CEST pulse scheme53 to exclude the possibility that the observed CEST effects were due to protein oligomerization at high concentrations (Fig. S8).
Electronic supplementary material
Acknowledgements
All NMR experiments were performed at the Beijing NMR Center and the NMR facility of National Center for Protein Sciences at Peking University. This research was supported by Grant 2016YFA0501201 from the National Key R&D Program of China and Grant 31170681 from the National Natural Science Foundation of China to C. J.
Author Contributions
W.Z. prepared the samples, carried out the NMR experiments and performed data analysis. X.N. and J.D. contributed to the NMR experiments and assist the data analysis. Y.H. and C.J. conceived and designed the research, and wrote the manuscript.
Data Availability
The assigned chemical shifts of re-ArsCfree and int-ArsC have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu/) under the accession number of 27283 and 27329.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yunfei Hu, Email: yunfei@pku.edu.cn.
Changwen Jin, Email: changwen@pku.edu.cn.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33766-4.
References
- 1.Yi MC, Khosla C. Thiol-disulfide exchange reactions in the mammalian extracellular environment. Annu Rev Chem Biomol Eng. 2016;7:197–222. doi: 10.1146/annurev-chembioeng-080615-033553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert HF. Thiol/disulfide exchange equilibria and disulfide bond stability. Method Enzymol. 1995;251:8–28. doi: 10.1016/0076-6879(95)51107-5. [DOI] [PubMed] [Google Scholar]
- 3.Netto LE, de Oliveira MA, Tairum CA, da Silva Neto JF. Conferring specificity in redox pathways by enzymatic thiol/disulfide exchange reactions. Free Radic Res. 2016;50:206–245. doi: 10.3109/10715762.2015.1120864. [DOI] [PubMed] [Google Scholar]
- 4.Cheng ZY, Zhang JF, Ballou DP, Williams CH. Reactivity of thioredoxin as a protein thiol-disulfide oxidoreductase. Chem Rev. 2011;111:5768–5783. doi: 10.1021/cr100006x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bechtel Tyler J., Weerapana Eranthie. From structure to redox: The diverse functional roles of disulfides and implications in disease. PROTEOMICS. 2017;17(6):1600391. doi: 10.1002/pmic.201600391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mossner E, Iwai H, Glockshuber R. Influence of the pK(a) value of the buried, active-site cysteine on the redox properties of thioredoxin-like oxidoreductases. FEBS letters. 2000;477:21–26. doi: 10.1016/S0014-5793(00)01738-5. [DOI] [PubMed] [Google Scholar]
- 7.Carvalho AT, et al. Mechanism of thioredoxin-catalyzed disulfide reduction. Activation of the buried thiol and role of the variable active-site residues. J Phys Chem B. 2008;112:2511–2523. doi: 10.1021/jp7104665. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi N, Creighton TE. On the reactivity and ionization of the active site cysteine residues of Escherichia coli thioredoxin. Biochemistry. 1996;35:8342–8353. doi: 10.1021/bi960465v. [DOI] [PubMed] [Google Scholar]
- 9.Jeng MF, Dyson HJ. Direct measurement of the aspartic acid 26 pKa for reduced Escherichia coli thioredoxin by 13C NMR. Biochemistry. 1996;35:1–6. doi: 10.1021/bi952404n. [DOI] [PubMed] [Google Scholar]
- 10.Chivers PT, Laboissiere MCA, Raines RT. The CXXC motif: Imperatives for the formation of native disulfide bonds in the cell. EMBO J. 1996;15:2659–2667. doi: 10.1002/j.1460-2075.1996.tb00626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Messens J, Silver S. Arsenate reduction: thiol cascade chemistry with convergent evolution. J Mol Biol. 2006;362:1–17. doi: 10.1016/j.jmb.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Rosen BP. Biochemistry of arsenic detoxification. FEBS Lett. 2002;529:86–92. doi: 10.1016/S0014-5793(02)03186-1. [DOI] [PubMed] [Google Scholar]
- 13.Sato T, Kobayashi Y. The ars operon in the skin element of Bacillus subtilis confers resistance to arsenate and arsenite. J Bacteriol. 1998;180:1655–1661. doi: 10.1128/jb.180.7.1655-1661.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Messens J, Hayburn G, Desmyter A, Laus G, Wyns L. The essential catalytic redox couple in arsenate reductase from Staphylococcus aureus. Biochemistry. 1999;38:16857–16865. doi: 10.1021/bi9911841. [DOI] [PubMed] [Google Scholar]
- 15.Zegers I, Martins JC, Willem R, Wyns L, Messens J. Arsenate reductase from S. aureus plasmid pI258 is a phosphatase drafted for redox duty. Nat Struct Biol. 2001;8:843–847. doi: 10.1038/nsb1001-843. [DOI] [PubMed] [Google Scholar]
- 16.Bennett MS, Guan Z, Laurberg M, Su XD. Bacillus subtilis arsenate reductase is structurally and functionally similar to low molecular weight protein tyrosine phosphatases. Proc Natl Acad Sci USA. 2001;98:13577–13582. doi: 10.1073/pnas.241397198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, et al. Conformational fluctuations coupled to the thiol-disulfide transfer between thioredoxin and arsenate reductase in Bacillus subtilis. J Biol Chem. 2007;282:11078–11083. doi: 10.1074/jbc.M700970200. [DOI] [PubMed] [Google Scholar]
- 18.Roos G, et al. How thioredoxin dissociates its mixed disulfide. PLoS computational biology. 2009;5:e1000461. doi: 10.1371/journal.pcbi.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Messens J, et al. All intermediates of the arsenate reductase mechanism, including an intramolecular dynamic disulfide cascade. Proc Natl Acad Sci USA. 2002;99:8506–8511. doi: 10.1073/pnas.132142799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo X, et al. Solution structures and backbone dynamics of arsenate reductase from Bacillus subtilis: reversible conformational switch associated with arsenate reduction. J Biol Chem. 2005;280:39601–39608. doi: 10.1074/jbc.M508132200. [DOI] [PubMed] [Google Scholar]
- 21.Vallurupalli P, Bouvignies G, Kay LE. Studying “invisible” excited protein states in slow exchange with a major state conformation. J Am Chem Soc. 2012;134:8148–8161. doi: 10.1021/ja3001419. [DOI] [PubMed] [Google Scholar]
- 22.Loria JP, Rance M, Palmer AG. A relaxation-compensated Carr–Purcell–Meiboom–Gill sequence for characterizing chemical exchange by NMR spectroscopy. J Am Chem Soc. 1999;121:2331–2332. doi: 10.1021/ja983961a. [DOI] [Google Scholar]
- 23.Palmer AG, 3rd, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/S0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 24.Sekhar A, Kay LE. NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers. Proc. Natl. Acad. Sci. USA. 2013;110:12867–12874. doi: 10.1073/pnas.1305688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Messens J, et al. Kinetics and active site dynamics of Staphylococcus aureus arsenate reductase. J Biol Inorg Chem. 2002;7:146–156. doi: 10.1007/s007750100282. [DOI] [PubMed] [Google Scholar]
- 26.Yu B, Yang D. Coexistence of multiple minor states of fatty acid binding protein and their functional relevance. Sci Rep. 2016;6:34171. doi: 10.1038/srep34171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lah N, Lah J, Zegers I, Wyns L, Messens J. Specific potassium binding stabilizes pI258 arsenate reductase from Staphylococcus aureus. J Biol Chem. 2003;278:24673–24679. doi: 10.1074/jbc.M303194200. [DOI] [PubMed] [Google Scholar]
- 28.Roos G, et al. Interplay between ion binding and catalysis in the thioredoxin-coupled arsenate reductase family. J Mol Biol. 2006;360:826–838. doi: 10.1016/j.jmb.2006.05.054. [DOI] [PubMed] [Google Scholar]
- 29.Kleckner IR, Foster MP. An introduction to NMR-based approaches for measuring protein dynamics. Biochim Biophys Acta. 2011;1814:942–968. doi: 10.1016/j.bbapap.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: Implications for fibrillation. Protein Sci. 2006;15:279–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katti SK, Lemaster DM, Eklund H. Crystal structure of thioredoxin from Escherichia coli at 1.68Å resolution. J Mol Biol. 1990;212:167–184. doi: 10.1016/0022-2836(90)90313-B. [DOI] [PubMed] [Google Scholar]
- 32.Chivers PT, Raines RT. General acid/base catalysis in the active site of Escherichia coli thioredoxin. Biochemistry. 1997;36:15810–15816. doi: 10.1021/bi971504l. [DOI] [PubMed] [Google Scholar]
- 33.Menchise V, et al. Crystal structure of the wild-type and D30A mutant thioredoxin h of Chlamydomonas reinhardtii and implications for the catalytic mechanism. Biochem J. 2001;359:65–75. doi: 10.1042/bj3590065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Messens J, et al. How thioredoxin can reduce a buried disulphide bond. J Mol Biol. 2004;2339:527–537. doi: 10.1016/j.jmb.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 35.Chartron J, Shiau C, Stout CD, Carroll KS. 3′-phosphoadenosine-5′-phosphosulfate reductase in complex with thioredoxin: A structural snapshot in the catalytic cycle. Biochemistry. 2007;46:3942–3951. doi: 10.1021/bi700130e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai SD, et al. Structural snapshots along the reaction pathway of ferredoxin-thioredoxin reductase. Nature. 2007;448:92–102. doi: 10.1038/nature05937. [DOI] [PubMed] [Google Scholar]
- 37.Lian FM, et al. Structural snapshots of yeast alkyl hydroperoxide reductase Ahp1 peroxiredoxin reveal a novel two-cysteine mechanism of electron transfer to eliminate reactive oxygen species. J Biol Chem. 2012;287:17077–17087. doi: 10.1074/jbc.M112.357368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma XX, et al. Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1. J Biol Chem. 2011;286:13430–13437. doi: 10.1074/jbc.M110.205161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LeMaster DM. Structural determinants of the catalytic reactivity of the buried cysteine of Escherichia coli thioredoxin. Biochemistry. 1996;35:14876–14781. doi: 10.1021/bi961607o. [DOI] [PubMed] [Google Scholar]
- 40.llman GL. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 41.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectros. 1999;34:93–158. doi: 10.1016/S0079-6565(98)00025-9. [DOI] [Google Scholar]
- 42.Delaglio F, et al. NMRPipe - a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 43.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 44.Tollinger M, Skrynnikov NR, Mulder FA, Forman-Kay JD, Kay LE. Slow dynamics in folded and unfolded states of an SH3 domain. J Am Chem Soc. 2001;123:11341–11352. doi: 10.1021/ja011300z. [DOI] [PubMed] [Google Scholar]
- 45.Mulder FA, Skrynnikov NR, Hon B, Dahlquist FW, Kay LE. Measurement of slow (micros-ms) time scale dynamics in protein side chains by (15)N relaxation dispersion NMR spectroscopy: application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J Am Chem Soc. 2001;123:967–975. doi: 10.1021/ja003447g. [DOI] [PubMed] [Google Scholar]
- 46.Korzhnev DM, et al. Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature. 2004;430:586–590. doi: 10.1038/nature02655. [DOI] [PubMed] [Google Scholar]
- 47.Korzhnev DM, Kloiber K, Kay LE. Multiple-quantum relaxation dispersion NMR spectroscopy probing millisecond time-scale dynamics in proteins: theory and application. J Am Chem Soc. 2004;126:7320–7329. doi: 10.1021/ja049968b. [DOI] [PubMed] [Google Scholar]
- 48.Carver JP, Richards RE. General 2-site solution for chemical exchange produced dependence of T2 upon Carr-Purcell pulse separation. J Magn Reson. 1972;6:89–105. [Google Scholar]
- 49.Taylor, J. Introduction to error analysis: The study of uncertainties in physical measurements. University Science Books (1997).
- 50.Sugase K, Konuma T, Lansing JC, Wright PE. Fast and accurate fitting of relaxation dispersion data using the flexible software package GLOVE. J Biomol NMR. 2013;56:275–283. doi: 10.1007/s10858-013-9747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuwen T, Kay LE. Longitudinal relaxation optimized amide 1H-CEST experiments for studying slow chemical exchange processes in fully protonated proteins. J Biomol NMR. 2017;67:295–307. doi: 10.1007/s10858-017-0104-y. [DOI] [PubMed] [Google Scholar]
- 52.Walter, R. 1976. Principles of Mathematical Analysis. Third edition, McGraw-Hill.
- 53.Niu X, et al. Residue selective N-15 CEST and CPMG experiments for studies of millisecond timescale protein dynamics. J Magn Reson. 2018;293:47–55. doi: 10.1016/j.jmr.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 54.Case DA, et al. The AMBER biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The assigned chemical shifts of re-ArsCfree and int-ArsC have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu/) under the accession number of 27283 and 27329.