Abstract
Real time in vivo methods are needed to better understand the interplay between diet and the gastrointestinal microbiota. Therefore, a rodent indirect calorimetry system was equipped with hydrogen (H2) and methane (CH4) sensors. H2 production was readily detected in C57BL/6J mice and followed a circadian rhythm. H2 production was increased within 12 hours after first exposure to a lowly-digestible starch diet (LDD) compared to a highly-digestible starch diet (HDD). Marked differences were observed in the faecal microbiota of animals fed the LDD and HDD diets. H2 was identified as a key variable explaining the variation in microbial communities, with specific taxa (including Bacteroides and Parasutterella) correlating with H2 production upon LDD-feeding. CH4 production was undetectable which was in line with absence of CH4 producers in the gut. We conclude that real-time in vivo monitoring of gases provides a non-invasive time-resolved system to explore the interplay between nutrition and gut microbes in a mouse model, and demonstrates potential for translation to other animal models and human studies.
Introduction
Carbohydrates are a major dietary constituent of humans and rodents. Not all carbohydrates are metabolically equal. Most dietary carbohydrates, including several sugars and starches high in amylopectin content, are readily digested and thus absorbed early in the gastro-intestinal tract, making them quickly available to the organism1. Other carbohydrates, such as amylose-rich starches, are only available to the organism after fermentation by the intestinal microbiota2, which results in a more gradual release to the organism. Microbial fermentation results in a variety of metabolic products, including short-chain fatty acids (SCFA), which are thought to mediate the beneficial health effects of the intestinal microbial community3. Glucose and other monosaccharides, present as such in the diet or becoming available from highly-digestible carbohydrates, are readily taken up via transporters from the small intestinal lumen into the body. This occurs primarily in the jejunum, the proximal part of the small intestine4. Carbohydrates that are less readily digestible reach the caecum and colon, where most of the intestinal microbiota reside5. Specific microbial communities utilize these substrates, in the process generating metabolites that are absorbed by the body, or are excreted as gases or in the faeces. Major digestion products are SCFA, which are known to influence host physiology, acting as energy substrates and as signalling molecules3. Other digestion products are the microbial fermentation gases hydrogen (H2), methane (CH4), and hydrogen sulphide (H2S)6.
Since the studies of Gordon et al.7, it is increasingly realized that the small and large intestinal microbiota not only plays a major role in gastrointestinal health but also in the host’s metabolic health8,9. However, how the microbial community affects metabolic health and how this can be beneficially modulated by nutrition and specific nutrients is far less well established. While a variety of cross-sectional methods can be applied to analyse changes in intestinal microbiota in rodents at specific time points, longitudinal measurements in rodent and human studies mainly rely on sampling of the faeces, long after food-microbiota interactions have already taken place throughout the gastrointestinal tract. Continuous measurements of fermentation gas emissions are already in place for ruminants like cattle and sheep10–12, as they are known to fully rely on microbiota fermentation in rumen and hindgut to digest cellulose, being distinct from monogastric organisms including rodents and humans. Furthermore, recent studies showed strong correlations between dynamics of metabolite production and microbiota composition and activity in dairy cows13,14. Measurements of H2 and CH4 as indicators of human gut microbial activity in vivo have been used before15–19, but these are in fact single-time-point gas measurements that lack the information that continuous analysis can provide.
Therefore, our study objective was to apply a simple non-invasive method to monitor the effect of diet on intestinal microbiota in real time using a human-relevant model, which we envisioned as a powerful tool to better understand the direct impact of nutrition on the microbiota and by extension of diet-microbiota interactions on human health.
C57BL/6J mice are the most widely used model in medical and nutritional health research and have shown their validity in dissecting microbe-host interactions and causality testing. However, analysis of fermentation gases in mice and other rodent models is a largely unexplored area. As is the case in humans, single-time-point measurements of H2 (refs20–23) and CH4 (refs24–26) have been reported for mice and rats. This is critical, because only continuous measurements allow to faithfully study the time-resolved kinetics of digestion and metabolism of nutrients reaching the gut microbiota.
Indirect calorimetry makes use of the measurement of oxygen (O2) and carbon dioxide (CO2), as well as food and water intake and locomotor activity, to analyse energy metabolism. We have equipped a commercially available indirect calorimetry system with sensors for H2 and CH4, allowing continuous measurements of release of these gases non-invasively in real time. We applied this extended system to explore the adaptation of gut microbiota to highly- and lowly-digestible carbohydrates. To the best of our knowledge, this is the first time that food-microbiota interactions have been studied continuously, non-invasively and in real time in a murine model.
Results
In vitro reflects in vivo diet digestibility
To confirm the difference in digestibility of the two starches incorporated into our experimental diets (Table 1), an in vitro model that mimics food digestion for the oral, gastric and small intestinal phases was used. The lowly-digestible starch diet (LDD) showed a slower and 14% less complete carbohydrate digestion than the highly-digestible starch diet (HDD; Fig. 1). In addition, we quantified food intake and faecal energy content in female and male mice habituated to the experimental diets (Table 2). Daily faecal mass was increased in all mice fed LDD, whereas faecal energy density was increased in LDD females only. LDD mice lost on average twice as much energy in faeces compared to HDD mice. With similar food and energy intake, the diet digestibility was 6% lower in LDD vs HDD fed mice (Table 2). Taken together, both in vitro and in vivo analyses showed a reduced digestibility of the LDD vs HDD.
Table 1.
Diet composition.
| Component | Diet | |
|---|---|---|
| HDD | LDD | |
| Casein | 212.2 | 212.2 |
| L-Cysteine | 3.0 | 3.0 |
| Amylose mix (AmyloGel 03003) | 0.0 | 568.6 |
| Amylopectin (C*Gel 04201) | 568.6 | 0.0 |
| Coconut oil | 21.4 | 21.4 |
| Sunflower oil | 83.1 | 83.1 |
| Flaxseed oil | 14.2 | 14.2 |
| Cholesterol | 0.03 | 0.03 |
| Cellulose | 50.0 | 50.0 |
| Mineral mix (AIN-93G-MX) | 35.0 | 35.0 |
| Vitamin mix (AIN-93-VX) | 10.0 | 10.0 |
| Choline bitartrate | 2.5 | 2.5 |
| Total (g) | 1000.0 | 1000.0 |
| Gross energy density (kJ g−1)a | 18.9 | 19.5 |
| Calculated energy density (kJ g−1)b | 17.9 | 17.9 |
| Protein (en%)b | 20 | 20 |
| Carbohydrate (en%)b | 55 | 55 |
| Fat (en%)b | 25 | 25 |
Values are g kg−1 of diet unless otherwise specified. aMeasured by bomb calorimetry, bcalculated based on Atwater’s nutritional values. HDD, highly-digestible starch diet; LDD, lowly-digestible starch diet.
Figure 1.
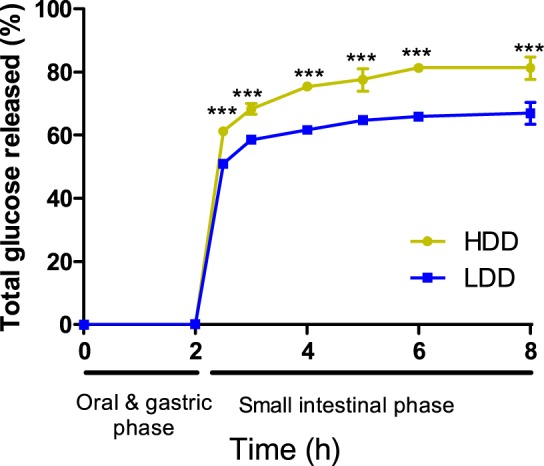
In vitro digestibility of starches in experimental diets. Triplicate samples of the lowly- and highly-digestible starch diets (LDD and HDD, respectively) were digested in vitro, and free glucose concentrations were determined at indicated time points. Statistical comparisons were made with two-way ANOVA with Bonferroni’s post hoc test; ***P ≤ 0.001. Values are plotted as mean ± s.d.
Table 2.
Dietary in vivo digestibility of the experimental diets.
| Females | Males | |||||
|---|---|---|---|---|---|---|
| HDD | LDD | P value | HDD | LDD | P value | |
| Food intake (g) | 2.53 ± 0.05 | 2.71 ± 0.24 | 0.1942 | 2.82 ± 0.21 | 2.86 ± 0.37 | 0.8489 |
| Gross energy intake (kJ) | 48.01 ± 0.92 | 53.08 ± 4.76 | 0.0816 | 53.36 ± 4.01 | 55.88 ± 7.18 | 0.5634 |
| Faeces weight (g) | 0.20 ± 0.01 | 0.41 ± 0.06 | 0.0006 | 0.24 ± 0.02 | 0.45 ± 0.05 | 0.0002 |
| Faeces gross energy (kJ g−1) | 15.48 ± 0.26 | 16.18 ± 0.24 | 0.0072 | 15.94 ± 0.27 | 16.01 ± 0.23 | 0.7227 |
| Faeces energy loss (kJ) | 3.10 ± 0.12 | 6.68 ± 1.08 | 0.0006 | 3.74 ± 0.22 | 7.26 ± 0.78 | 0.0001 |
| Digestible energy intake (kJ) | 44.91 ± 0.91 | 46.40 ± 3.70 | 0.4645 | 49.63 ± 3.80 | 48.62 ± 6.44 | 0.7967 |
| Diet digestibility (%) | 93.6 ± 0.2 | 87.5 ± 0.9 | <0.0001 | 93.0 ± 0.2 | 87.0 ± 0.6 | <0.0001 |
Energy balance calculated over the third week of exposure to the diets in females and males (n = 4 per sex and diet) and expressed per day. Statistical comparisons by Student’s t-test within sex. Data reported as mean ± s.d.
Measuring H2 production in real time
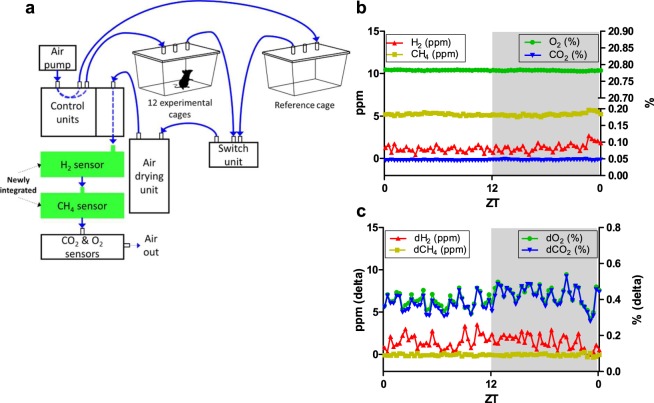
Reduced digestibility likely also affects colonic fermentation, for which H2 has been used as a marker in mice20. However, measurement of its continuous production in response to the diet has not yet been possible. We therefore adapted and extended an indirect calorimetry system to allow H2 and CH4 production to be studied in real time, by introducing the respective sensors in series with the O2 and CO2 sensors already present in the system (Fig. 2a). To determine if the small quantities of H2 originating from microbial carbohydrate fermentation in mice could be detected by our system, we measured gas concentrations in cages with and without chow-fed mice over 24 h. Stable signals for all gases were seen in the absence of mice (Fig. 2b), and the concentrations were clearly decreased for O2 and increased for CO2 in mouse-occupied cages, as expected (Fig. 2c). H2 increased (Fig. 2c), while CH4 concentrations were not altered by the presence of a chow-fed mouse in the cage. The adapted indirect calorimetry system was therefore suitable for simultaneous respirometry and H2 production measurements in real time, however under the conditions tested, CH4 production appeared to be absent based on measured ambient levels well above the lower detection limit of the CH4 sensor (Fig. 2b).
Figure 2.
Real-time measurements of hydrogen (H2) and methane (CH4) production in mice within indirect calorimetry system. (a) Illustration of the indirect calorimetry system extended for H2 and CH4 measurements. Direction of air flow in the tubing is shown in blue, new gas sensors are shown in green. For clarity, tube lengths are not to scale (all equal) and food and drink containers with sensors are not shown, nor are the infrared beam bars for activity measurements. (b) Ambient concentrations of H2 and CH4 (left y-axis, ppm) and O2 and CO2 (right y-axis, %) were recorded in an empty (reference) cage at 20 min intervals for 24 h. (c) Gas concentrations in a cage occupied by a chow-fed female adult mouse were measured and compared to the corresponding concentrations in the reference cage and expressed as delta values. White and grey areas in panels b and c represent the inactive light and active dark phase for the animal, respectively. ZT, Zeitgeber time.
H2 production indicates extent of carbohydrate digestibility
Since the contrasting digestibility of the experimental diets was expected to result in sustained differences in H2 production as a consequence of fermentation in the large intestine, we fed female and male mice, as a proof-of-concept, either the HDD or the LDD for three weeks and measured H2, CH4, O2, and CO2 levels continuously during several days (Study 1). Calculation of energy expenditure, based on 24 h O2 consumption and CO2 production, revealed no differences between dietary groups (females 1.59 ± 0.08 vs 1.63 ± 0.08 kJ h−1 in HDD and LDD respectively, P = 0.2094; males 1.80 ± 0.13 vs 1.76 ± 0.13 kJ h−1 in HDD and LDD respectively, P = 0.5470). However, 24 h mean respiratory exchange ratio (RER) was lower in LDD- vs HDD-fed male mice (0.85 ± 0.03 vs 0.88 ± 0.03 respectively, P = 0.0097), indicating higher fat oxidation and lower carbohydrate oxidation in LDD mice. Overall, these observations agree with indirect calorimetry data reported for mice fed diets containing carbohydrates similar to the carbohydrates used here27.
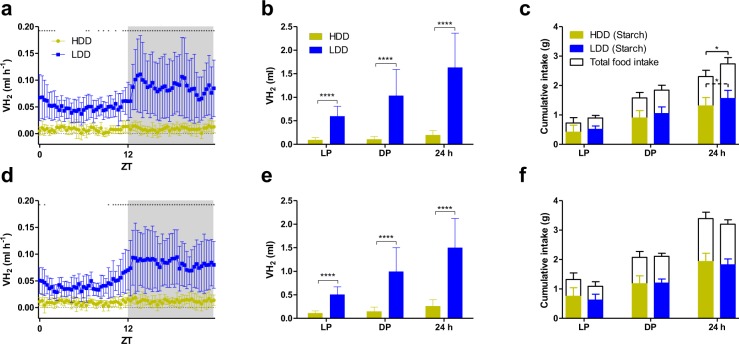
Both LDD-fed females (Fig. 3a,b) and males (Fig. 3d,e) constantly produced more H2 than HDD-fed mice. A distinct pattern of H2 production became apparent in LDD-fed mice, with H2 levels being higher in the active dark phase and lower, but still clearly present, in the inactive light phase (Fig. 3a,b,d,e). This was fully consistent with the circadian food and starch intake (Fig. 3c,f). Importantly, the difference in H2 production between HDD- and LDD-fed mice was explained by the type of starch rather than the amount of starches ingested, as cumulative starch consumption was similar between the groups (Fig. 3c,f). Together, this data provides proof-of-concept for measuring H2 production in real time as an indicator of carbohydrate digestibility.
Figure 3.
H2 production in mice reflects starch digestibility. Female (a) and male (d) mice were fed either HDD or LDD for three weeks and volume of H2 produced (VH2) was recorded for 24 h in the adapted indirect calorimetry system. Cumulative H2 production in females (b) and males (e) quantified during the 12 h light phase (LP), 12 h dark phase (DP) or the complete 24 h photoperiod. Cumulative starch and total food intake in females (c) and males (f) over the measuring period calculated from food intake records. White and grey areas represent the light and the dark phase, respectively. Time course data was analysed by repeated measures two-way ANOVA with Bonferroni’s test for multiple comparisons and time points where P < 0.05 are indicated with black asterisks (panels a and d). Other statistical comparisons made by Student’s t-test or Mann-Whitney U test; *P ≤ 0.05, ****P < 0.0001 (n = 11 LDD females, n = 12 remaining groups). Data shown as mean ± s.d. ZT, Zeitgeber time.
H2 evolution reflects adaptation to dietary carbohydrates
As we could show that H2 production can be sensitively and continuously measured, we next questioned whether it would be possible to measure adaptation to the diet in vivo in real time.
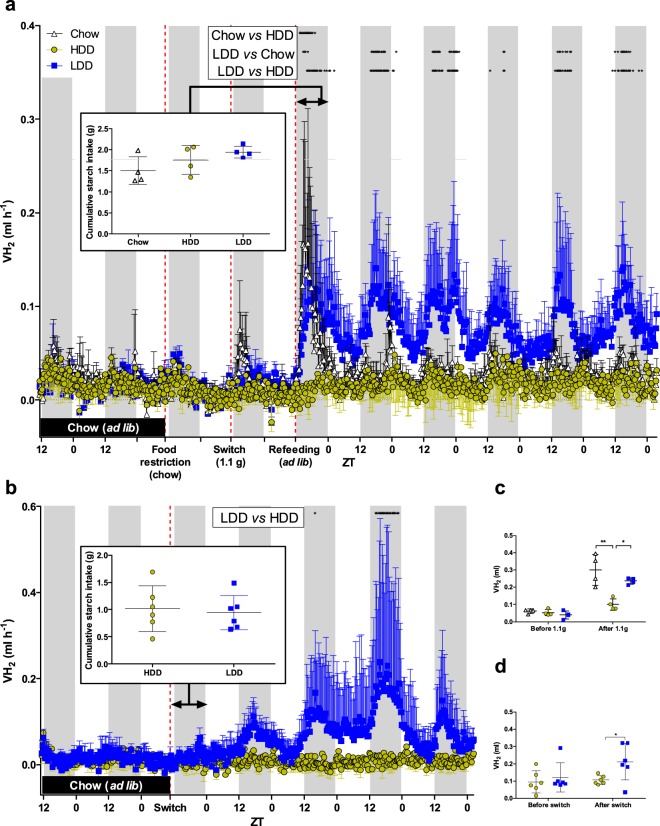
For this, we provided HDD or LDD to mice that had no previous exposure to these diets and we followed H2 production continuously. We introduced the new diets in one of two conditions; the first condition was as a single meal challenge given to fasted mice, followed by ad libitum access to the diet the next day, as a second fasting-refeeding challenge (Study 2, Fig. 4a). The second condition was by replacing the standard chow diet directly with HDD or LDD ad libitum (Study 3, Fig. 4b). H2 production was significantly increased in LDD- compared to HDD-fed mice as early as 4 h after fasted mice gained ad libitum access to the experimental diet (Fig. 4a). The direct switch from chow to HDD or LDD without fasting had similar results, with LDD-fed mice producing significantly more H2 after 53 h of access to the LDD compared to mice receiving HDD (2-way ANOVA, Fig. 4b). In both conditions, i.e. fasted or directly switched to HDD or LDD, cumulative H2 production became significantly higher already within 12 h upon access to LDD vs HDD (Fig. 4c,d), and H2 production patterns in LDD-fed mice closely followed the patterns of LDD intake (Additional File 1: Fig. S1). Interestingly, mice that continued on the chow diet after a period of food restriction exhibited a spike in H2 production (Fig. 4a), while consuming similar amounts of starches compared to the HDD and the LDD groups. H2 production in HDD-fed mice remained lower compared to mice on LDD or chow, as expected. Importantly, LDD-induced H2 production increased gradually before reaching its maximal levels (up to 0.89 ml h−1), revealing the process of adaptation to the lowly-digestible starch. LDD- vs HDD-fed mice thus showed a differential adaptation, likely in their microbiota, based on increased H2 production.
Figure 4.
H2 evolution upon first exposure to starches of different digestibility. (a) Standard chow-fed mice within indirect calorimetry were food-restricted leading to fasting (dotted line), which was followed by feeding 1.1 g of chow (black), HDD (yellow), or LDD (blue; n = 4 per group) prior to the dark phase as a single meal test (2nd dotted line). As a result, they were fasted the next day, and received prior to dark phase ad libitum access to the same diet (3rd dotted line) for an additional 5.5 days. Inset: First 12 h cumulative starch-intake of ad libitum feeding with experimental diets. (b) Chow-fed mice (n = 6 per group) were switched to LDD or HDD without prior food restriction and measurements continued for another 4.5 days. Inset: First 12 h cumulative starch-intake after diet switch. (c) Cumulative H2 production over 12 h before (while food-restricted on chow) and after feeding 1.1 g of chow, HDD, or LDD (n = 4 per group). (d) Cumulative H2 production over 12 h before and after switching directly from chow to HDD or LDD (n = 6 per group). All mice received no other diet than chow during their whole lifetime prior to these experiments and the dietary switch (black bar), but colour usage reflects subgroups after first exposure to new diets. White and grey areas represent light and dark phases, respectively. Time course data was analysed by repeated measures two-way ANOVA with Bonferroni’s test for multiple comparisons (chow vs HDD, LDD vs chow, and LDD vs HDD), and time points where P < 0.05 are indicated with black stars (panels a,b). Cumulative data was statistically compared using unpaired two-tailed Student’s t-test (between HDD and LDD) and one-way ANOVA with Bonferroni’s multiple comparisons post hoc test (between chow, HDD, and LDD); *P ≤ 0.05, **P ≤ 0.01. Data is presented as mean ± s.d. For clarity, either upper or lower error bars are shown. ZT, Zeitgeber time.
Alterations in intestinal microbiota by dietary carbohydrates
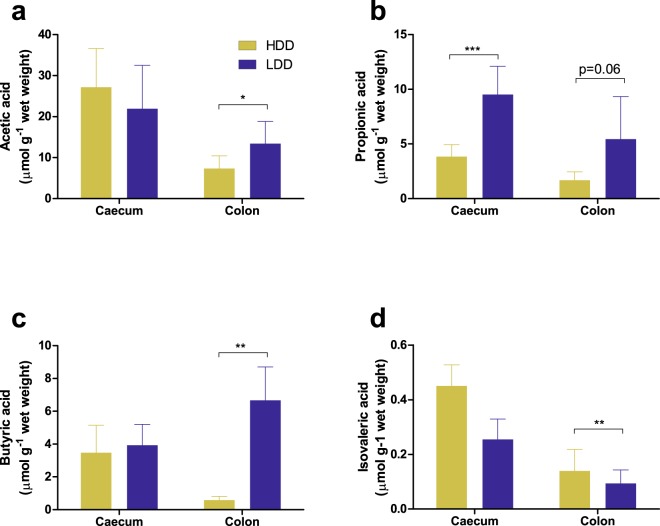
Since the production of H2 fully depends on intestinal microbial communities and their metabolism, we further investigated the changes in the microbiota induced by the LDD to validate our observations. As an additional parameter of fermentation, we first assessed SCFA levels in intestinal digesta after 3 weeks of exposure to the HDD or the LDD (Study 1). Total caecal SCFA levels were similar between LDD- and HDD-fed mice (35.6 ± 13.9 vs 34.9 ± 11.9 μmol g−1, respectively), including valeric and isobutyric levels (data not shown), whereas total SCFA in colon were higher in LDD- compared to HDD-fed mice (25.6 ± 9.6 vs 9.6 ± 4.1 μmol g−1, P = 0.0059). Acetic acid (Fig. 5a) and propionic acid (Fig. 5b) were the two most abundant SCFA, and both were significantly elevated in LDD-fed mice in colon and caecum contents, respectively. Butyric acid was the most differentially produced SCFA, enriched by 13.8-fold in LDD colon content (Fig. 5c). Finally, isovaleric acid, a product of microbial protein fermentation, was the least abundant of the measured SCFA in all groups and was significantly lower in caecum of LDD- vs HDD-fed mice (Fig. 5d).
Figure 5.
Short-chain fatty acid (SCFA) concentrations in intestinal digesta of mice fed starches of different digestibility. (a) Acetic acid, (b) propionic acid, (c) butyric acid and (d) isovaleric acid concentrations in mouse caecum (n = 6 per group) and colon (n = 5 HDD, n = 7 LDD) contents obtained after three weeks of feeding HDD (yellow bars) or LDD (blue bars). Statistical comparisons were made using unpaired two-tailed Student’s t-test; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001. Data shown as mean ± s.d.
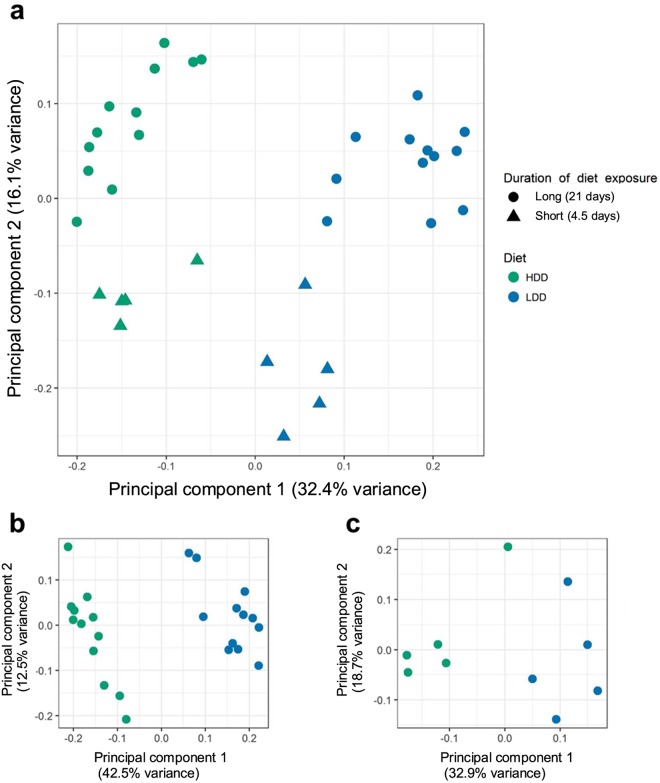
We next compared the overall changes in faecal microbiota communities induced by HDD or LDD after exposure to the diets for 3 weeks (Study 1) and 4.5 d (Study 3). Principal coordinates analysis (PCoA) using the UniFrac unweighted distance matrix revealed a clear separation between the two dietary groups (Fig. 6a). These observations were supported by Adonis analysis, using either weighted or unweighted UniFrac distances, as diet explained a large part of the variation in microbiota composition (20% and 29%, respectively, P < 0.001, Table 3). H2 volume was the second most important variable, followed by duration of intervention and age, and body weight, with minor but significant effects (Table 3). Of note, duration of intervention and age of mice are dependent variables due to study design. In order to control for the effects of duration of dietary exposure, we also analysed Studies 1 and 3 separately. After 3 weeks of intervention, diet and H2 production were the only significant variables, with H2 explaining up to 34% of the variation (Fig. 6b and Table 3). However, only diet contributed significantly to the variation after 4.5 d of intervention in adult mice (Fig. 6c and Table 3). Additionally, α-diversity appeared to decrease with duration of intervention irrespective of the dietary intervention, with no consistent effects of the diet (Additional File 2: Fig. S2). This is in line with the differences in age of these mice, namely the young mice showing lower α-diversity than the older mice.
Figure 6.
Starch digestibility primarily determines faecal microbiota composition. Principal coordinates analysis (PCoA) plot illustrating the unweighted UniFrac distances of the intestinal microbiota of mice after long- and short-term exposure to HDD and LDD (a, Studies 1 and 3 combined), and only long-term (b, Study 1) and short-term (c, Study 3) exposures. Each data point represents a sample of faecal pellets of one individual mouse (n = 12 long-term exposure per diet, n = 5 short-term exposure per diet).
Table 3.
Faecal microbiota composition of mice fed HDD or LDD and other host and environmental variables.
| Weighted UniFrac | Unweighted UniFrac | |||
|---|---|---|---|---|
| R2 | P value | R2 | P value | |
| Studies 1 and 3 combined (long- and short-term exposures) | ||||
| Diet | 0.198 | 0.001 | 0.287 | 0.001 |
| Cumulative H2 production | 0.098 | 0.023 | 0.173 | 0.001 |
| Duration of intervention* | 0.094 | 0.015 | 0.141 | 0.001 |
| Age* | 0.094 | 0.02 | 0.141 | 0.002 |
| Sex | 0.057 | 0.12 | 0.05 | 0.018 |
| Body weight | 0.08 | 0.042 | 0.1 | 0.002 |
| Food intake | 0.06 | 0.08 | 0.043 | 0.142 |
| Starch intake | 0.068 | 0.079 | 0.043 | 0.135 |
| Study 1 (long-term exposure, post-weaning, n = 12) | ||||
| Diet | 0.26 | 0.003 | 0.4 | 0.001 |
| Cumulative H2 production | 0.198 | 0.005 | 0.344 | 0.001 |
| Sex | 0.062 | 0.195 | 0.03 | 0.658 |
| Body weight | 0.046 | 0.289 | 0.02 | 0.87 |
| Food intake | 0.099 | 0.062 | 0.049 | 0.3 |
| Starch intake | 0.1 | 0.077 | 0.05 | 0.279 |
| Study 3 (short-term exposure, adult, n = 5) | ||||
| Diet | 0.24 | 0.056 | 0.293 | 0.004 |
| Cumulative H2 production | 0.14 | 0.217 | 0.108 | 0.5 |
| Body weight | 0.08 | 0.594 | 0.137 | 0.228 |
| Food intake | 0.06 | 0.748 | 0.192 | 0.044 |
| Starch intake | 0.06 | 0.761 | 0.192 | 0.05 |
Results are obtained using Adonis Permutational Multivariate Analysis of Variance. *Duration of intervention (short- and long-term) and age of animals (young vs adult) are not independent variables.
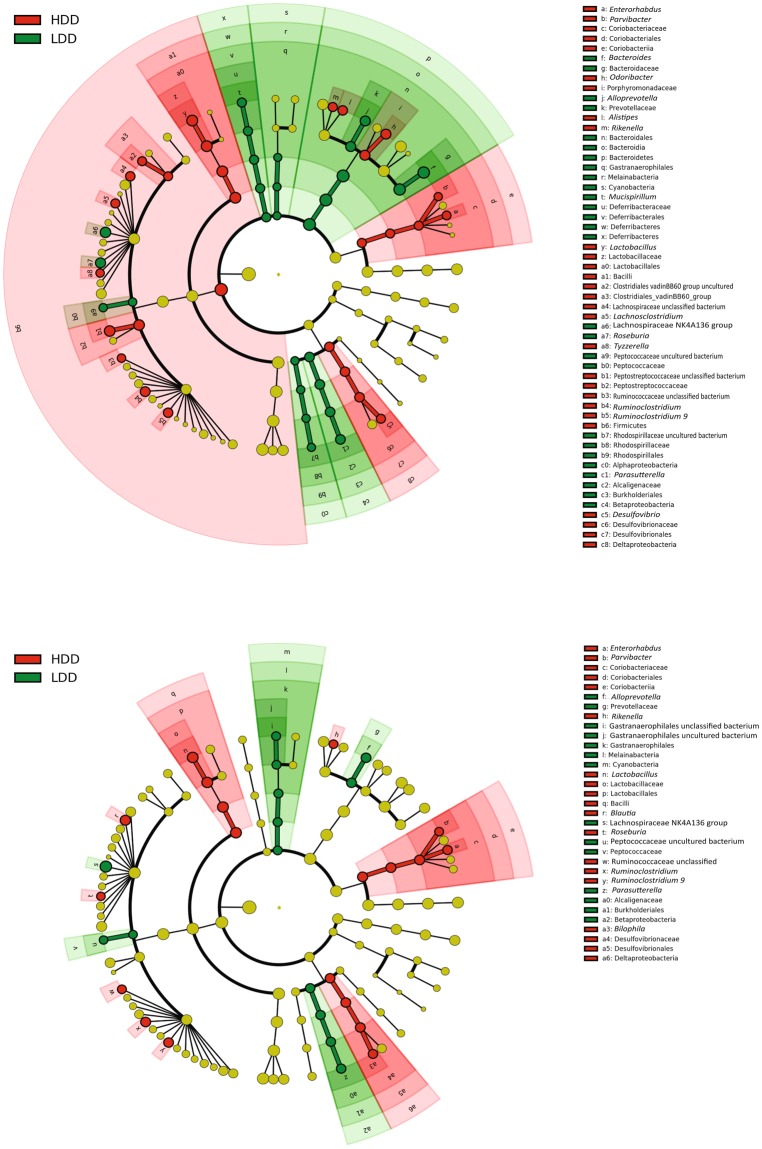
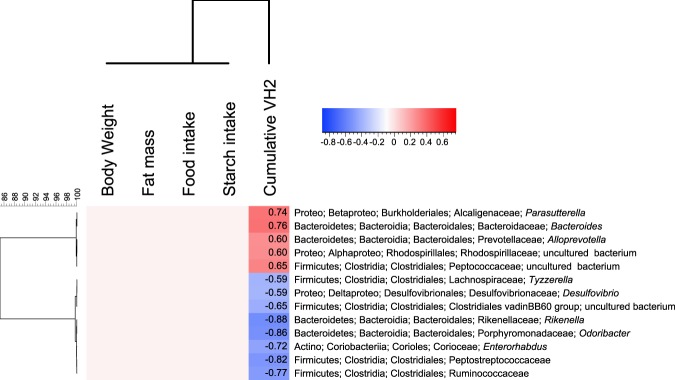
We then aimed to identify which microbial taxa were significantly associated with the observed differences in β-diversity. The microbiota of mice fed LDD vs HDD for 3 weeks was enriched in Bacteroides, Parasutterella, Roseburia, and Alloprevotella, along with two other families (Fig. 7a and Additional File 3: Fig. S3a). In comparison, Lactobacillus, Rikenella, Odoribacter, Enterorhabdus, and Desulfovibrio among others appeared enriched in HDD- vs LDD-fed mice (Fig. 7a and Additional File Fig. S3b). Similar differences were seen after 4.5 d of exposure (Fig. 7b and Additional file 3: Fig. S3c,d). While fewer taxa were affected by the short-term dietary intervention, changes in genus level were consistent for both groups (Additional file 4: Fig. S4). Moreover, H2 production was the only (environmental) variable that was significantly correlated with specific bacteria taxa after three weeks of intervention, with five genera correlating positively with H2 production and eight genera showing a negative correlation (Fig. 8 and Additional file 5: Fig. S5). Eleven of these 13 genera were also significantly influenced by diet (Fig. 7a and Additional file 3: Fig. 3a,b). Finally, Archaea (some of which are CH4 producers) could not be detected in any of the samples despite the use of primers targeting both bacterial and archaeal 16S rRNA genes equally well. This agrees with the absence of CH4 detection in these mice and under these nutritional challenges.
Figure 7.
Exposure to starches of different digestibility induces distinct microbial taxa. (a) Cladogram representing bacteria genera that were significantly enriched by LDD or HDD after 3 weeks of exposure to the diets (n = 12 per diet, Study 1). (b) Bacterial genera that were significantly increased by LDD or HDD after 4.5 d of exposure to the diets (n = 5 per diet, Study 3). Comparisons were done using the linear discriminant analysis effect size (LEfSe) method. LDA scores are shown in Additional file 3: Fig. 3e,f. Nomenclature of microbial genus level taxa is based on highest achievable taxonomic resolution at phylum, class, order, family or genus level.
Figure 8.
Specific bacterial genera correlate only with in vivo H2 production. Spearman’s rank correlation coefficients of faecal microbiota, H2 production, food and starch intake, body weight, and fat mass of mice exposed to HDD or LDD for 3 weeks after weaning (n = 12 per diet, Study 1). Non-red and non-blue cells all have a Spearman’s correlation value of 0 with FDR P value > 0.13. Nomenclature of microbial genus level taxa is based on highest achievable taxonomic resolution at phylum, class, order, family or genus level.
Discussion
The goal of this study was to measure real-time interactions between diet, gut microbes, and the host. We implemented H2 and CH4 detection in an indirect calorimetry system to track fermentation continuously in mice. H2 production revealed a time frame for microbiota adaptation to starch of low digestibility, which corresponded with shifts in microbial community composition induced by diet. Thus, measuring H2 production allowed us to non-invasively study effects of the diet on the intestinal microbiota in real-time.
The difference in starch digestibility as part of the experimental diets was confirmed both by in vitro and in vivo measurements, but did not significantly alter total intake of digestible energy (gross energy minus faecal energy losses) between dietary groups within a sex. The lower digestibility of the starch in the LDD thus suggests that partially undigested starch reached the large intestine which was subsequently partially fermented by the intestinal microbiota providing energy substrates, e.g. SCFA, to the host. Energy of undigested starch can be lost after fermentation in the form of products not utilizable by the host, such as H2. However, previous studies considered energy loss in the form of H2 and CH4 negligible, representing less than 0.2% of total energy expenditure in humans consuming non-starch polysacharides28. Studies in rats fed various types of resistant starch also indicated that energy loss through fermentation is minimal, although the actual H2 output was not measured directly29. Here, our data show that H2 is produced constantly on a lowly-digestible starch diet. Although the volume of H2 produced by the mice in our study may be little in terms of energy loss, it is plausible that carbohydrates that give a higher level of fermentation could further increase the H2 output, which might represent a significant factor to take into account over a lifetime.
H2 production was detected in mice under all conditions tested, with the amounts produced clearly being influenced by the form of carbohydrate consumed. Mice fed moderately fermentable carbohydrates have been shown to produce H2 (ref.20). Even in conditions where little fermentation is expected, such as feeding corn starch-based chow30 or pure sucrose31, H2 production has been seen in rats. In line with our data on three diets with a different carbohydrate profile, this illustrates that H2 production can directly reflect subtle changes in carbohydrate fermentation. Interestingly, H2 production was clearly associated with the food intake pattern. This is in contrast with data reported in humans, where H2 and CH4 peaked at rather unpredictable times after food intake despite the proper control of the meal schedule28. This might be due to e.g. differences in dietary meal composition, time resolution of sampling, intestinal transit time, or other differences in intestinal physiology between humans and mice. More recently, using gut capsule technology, a similar H2 pattern as in our mice was also observed in a human pilot trial based on dietary fibre differences32.
We demonstrated that real-time monitoring of H2 production can be used to investigate transient effects of diet in time and explore the process of adaptation rather than the end stage only. So far, studies mainly investigated, by measuring H2 and other fermentation parameters at selected time points, longer timeframes ranging from 1 day to several weeks25,33–35. In our study, significant differences in H2 production appeared within 12 h upon access to LDD. This timeframe was clearly influenced by fasting and whether the diet was provided ad libitum or in a restricted amount. We speculate these early increases in H2 output to reflect immediate effects of diet on microbial metabolism preceding changes in community structure. Another observation was that mice fed chow produced H2, although at low levels. Real-time monitoring newly revealed that a period of food restriction followed by refeeding led to a marked and acute increase in H2 production once chow became available again. A likely explanation is excessive eating after food deprivation, causing a larger amount of not fully digested chyme to enter the large intestine and thus increasing substrate availability to the microbiota. In addition, a 24 h fasting period alone has been shown to produce shifts in microbial diversity36 and microbiota configuration37. Such changes could in turn alter fermentation stoichiometry and microbial function in response to the diet and ultimately lead to a higher H2 output. Our analysis indicates (short-term) effects of fasting and refeeding on microbial activity, which should be careful taken into account in nutritional studies focussing on changes in microbiota composition and function.
As could be expected, the driver of the experimental differences, the dietary starch digestibility, was the most important factor explaining the variation in microbiota, showing colinearity with our measured in vivo H2 production. Although current knowledge of the dynamics of H2 within the gastrointestinal tract is limited, it is well documented that H2 is exclusively produced during fermentation by hydrogenogens6. Among the major hydrogenogenic bacteria are Bacteroidetes and clostridial members of Firmicutes38. In line with this, we observed that LDD, a source of carbohydrates for caecum and colon, stimulated the fermentative Bacteroidetes bacteria, more specifically the genus Bacteroides. This is consistent with the dose-dependent increase in caecal Bacteroidetes density in response to amylose39 and similar findings for amylose on a high-fat background40. Here we extend these findings and show, for the first time in vivo, a positive correlation between Bacteroides and H2 production in mice.
Interestingly, after the short-term exposure to LDD in adult mice, Bacteroidetes were not significantly increased compared to the HDD group. This might be associated with the shorter duration of the treatment and possibly with more firmly established microbial communities in adulthood. However, most genera induced by diet in adult mice after 4.5 days correspond to those induced in mice after weaning, which were exposed for 3 weeks.
Another consistent shift in microbial community composition was the promotion of Deltaproteobacteria, particularly Desulfovibrio and Bilophila, in HDD-fed mice. Deltaproteobacteria are the major representatives of colonic sulphate reducing bacteria (SRB)41 including Desulfovibrio. SRB along with methanogens and acetogenic bacteria are the only gut microbes able to use H2 as an electron donor to produce H2S and acetate. Although not a SRB itself, taurine-respiring Bilophila species can also produce H2S. Additionally, there is evidence of CH4 production in rats26 and mice, with the presence of methanogens in humanized microbiota mouse models42 and by high fat dietary feeding43. The fact that we neither detected CH4 nor Archaea suggests that H2 was preferentially used to produce H2S in mice fed readily digestible starch. H2S is a potentially toxic product of bacterial metabolism44–46 and it has been implicated in human health and disease19 and, more recently, thermogenesis47. Moreover, H2S has been reported to inhibit the production of SCFA and specifically to impair butyrate oxidation, depriving colonic cells from their main energy source45,48. In line, we report a dramatic difference in colonic butyrate in HDD-fed mice. Apart from Deltaproteobacteria, we observed increased abundances of Odoribacter, a known H2S producer49 and Rikenella, a desulphatase-secreting bacterium50, under HDD-feeding. Members of the genus Rikenella are able to cleave sulphate from mucin glycans, making them available for microbial degradation51 and potentially acting as a donor of sulphate to H2S producers. Based on these facts we speculate that the lack of fermentable carbohydrates favoured the presence of hydrogenotrophs associated with the production of H2S, which could have led to the decreased H2 output and colonic SCFA levels that was observed in mice fed highly-digestible starch.
The major taxon increased in HDD-fed mice in our study belonged to the genus Lactobacillus. In contrast, diets supplemented with resistant starch tended to enrich the Lactobacillus population in mouse caecum, but much less at high doses of resistant starch39. Incidentally, hydrogenase genes, which encode enzymes for the reversible oxidation of H2, were recently shown to be completely absent in Bacilli and bifidobacteria38. Considering the lack of a correlation between H2 production and Lactobacillus in our study, new questions emerge about the ability of Lactobacillus to thrive in H2-poor environments.
The increase in isovaleric acid, a product of branched-chain amino acid catabolism52, in HDD-fed mice, suggests a shift of microbiota towards protein fermentation. Bacteria from the genera Enterorhabdus53 and Parvibacter54, both significantly induced by HDD-feeding, have the ability to ferment amino acids. Additionally Olsenella, only present in two samples in the HDD group, is documented to grow on tyrosine and produce p-cresol55, supporting our hypothesis for a shift to protein fermentation. This might have important implications for the host, since products of protein fermentation such as phenols, ammonia, certain amines, and H2S, are considered to play important roles in the initiation or progression of bowel diseases, inflammation, DNA damage, and cancer56.
Altogether, our results emphasize H2 as a key factor within the intestinal microenvironment and the usefulness of knowing its production dynamics to understand the interplay between host, diet, and the intestinal microbiota. At the same time, we are aware that our approach to study such interactions may have conceivable limitations. It has been argued that changes in gas evolution (and other indirect markers of fermentation) cannot accurately indicate changes in fermentation57, and even “real-time”, carefully controlled measurements have failed to show quantitative changes in H2 and CH4 production proportionally linked to consumption of fermentable carbohydrates15,28. We completely agree with these authors that the measured outcomes, H2 and CH4, not only reflect the type of carbohydrate consumed, but are the end result of a very complex fermentation stoichiometry that depends on the host’s capacity to digest and absorb nutrients, the dominance and metabolic activity of microbial species, and their interactions. However, the conclusion that fermentation gases are extremely limited parameters to study carbohydrate fermentation is largely based on human data, where eating pattern, environment, genetic variation, and the gut microbe interact and ultimately determine an individual’s response to the diet58,59. When these and other factors can be better controlled, as it is the case with animal models, the analysis of carbohydrate fermentation through H2 and CH4 quantification has much to offer. The fact that in vitro models to measure H2 and CH4 evolution are still developing and proposed as a tool to unravel the mechanisms behind the association between microbiota and host health60 is encouraging.
Overall, the applications of gas analysis within an indirect calorimetry system go beyond the arena of carbohydrate quality and nutritional studies, and may be used as a diagnosis tool in clinical practice19,61,62. It opens up new avenues not only in preclinical research in rodents, but also has potential in human-diet-microbiota interaction studies if such sensor technology is incorporated into indirect calorimetry chambers or ventilated hood systems.
Conclusions
Using our customized indirect calorimetry system we were able to continuously quantify H2 production in mice as a reflection of the starch digestibility of the diet. H2 monitoring also allowed us to catch the earliest stages in the adaptation to carbohydrates of different digestibility, revealing a nuanced process with high inter-individual variation. Importantly, in vivo H2 production was significantly correlated with specific microbial taxa, including Bacteroides and Parasutterella. The implemented H2 and CH4 sensor-technology described here opens yet unmet avenues to study the effects of nutrition on microbiota in real time, not only in rodents, but potentially also in humans.
Methods
Coupling of hydrogen (H2) and methane (CH4) sensors into indirect calorimetry system
A PhenoMaster indirect calorimetry system (TSE Systems, Bad Homburg, Germany) was extended by coupling a Sensepoint XCD H2 gas analyser (Honeywell Analytics, Hegnau, Switzerland) and a CH4 gas analyser (ABB Automation GmbH, Frankfurt am Main, Germany) in a closed circuit in series in front of a Siemens High-Speed Sensor Unit containing the O2 and CO2 sensors. This order was chosen to prevent dilution of the sample with reference air, which is required by the Siemens unit. The H2 sensor has a stability of <±2% full scale deflection (fsd)/yr representing <2 ppm/yr as it was adjusted to a measuring range from 0 to 100 ppm. The CH4 sensor has a zero drift of ≤1% of span per week and a measuring range from 0 to 500 ppm. A two point calibration of both H2 and CH4 analysers was performed within 24 h before each animal experiment. The calibration procedure was carried out using three gas mixtures (Linde Gas Benelux BV, Dieren, The Netherlands): zero (20.947% O2 and N2), span H2 (98.8 ppm H2 and synthetic air), and span CH4 (0.521% CO2, 450 ppm CH4, and N2). The zero calibration mixture was flushed through the system for 10 min and ADC signals were assigned H2 and CH4 values of 0 ppm. Thereafter, each of the span gas mixtures was run for 10 min and ADC signals assigned 98.8 ppm H2 and 450 ppm CH4, accordingly. For O2 and CO2 calibration, the routine indicated in the TSE manufacturer’s manual was followed, using an additional gas mixture (0.999% CO2 and N2) for the span calibration point. Animals were measured as previously described63 with minor adjustments for the newly coupled sensors. These include the adjustment of airflow to 0.431 min−1 and the measuring time per cage set to 1.5 min. Data was recorded using an updated, customized, version of the TSE PhenoMaster software (V5.8.0) specially developed for the incorporation of H2 and CH4 measurements.
Animal experiments and sample harvest
All animal experiments were approved by the Animal Experiments Committee (DEC 2014085.h) of Wageningen, The Netherlands, and performed in accordance to EU directive 2010/63/EU. Female and male C57BL/6JRccHsd mice (Harlan Laboratories BV, Horst, The Netherlands) were housed in Makrolon II cages enriched with wood chips and wood shavings, with free access to drinking water, at 23 °C ± 1 °C and a 12:12 h light:dark cycle. Standard rodent chow (RMH-B, AB Diets, Woerden, The Netherlands) was provided exclusively and continuously since weaning, unless specified. Three different studies were conducted to investigate diet-host-microbiota interactions upon provision of diets containing starches with differences in digestibility (the experimenter was not blinded to the diets that the animals were given).
Study 1 (long-term exposure, post-weaning)
Mice were mated and the offspring reassigned to a foster dam 1 or 2 days after birth to obtain standardized litters. Males and females were stratified by body weight at post-natal day (PN) 21, housed individually and randomly assigned to be fed a highly- or a lowly-digestible starch diet (HDD and LDD, respectively; see below). The randomisation was achieved by generating a column of random numbers in a spreadsheet and sorting each diet and animal number according to the column of random numbers from smallest to largest. From PN36-42, a subgroup of mice was measured in the indirect calorimetry system with ad libitum access to the experimental diets (males: n = 12 per diet, females HDD n = 12, LDD n = 11). Fresh faecal pellets were sampled on PN39 (n = 6 per diet and sex) and stored at −80 °C for intestinal microbiota analysis. Another subgroup of female mice was culled on PN42 for collection of caecum (n = 6) and colon contents (n = 5 HDD, n = 7 LDD), and the faeces produced during the last week before sacrifice were collected for gross energy measurements (see In vivo diet digestibility). Before sacrifice, food was removed 1 h after the start of the light phase and animals decapitated 2–6 h after removal of food. Caecum and colon contents were immediately frozen in liquid nitrogen, and stored at −80 °C until analysis.
Study 2 (short-term exposure with fasting, adult)
Eight-month-old female mice were individually housed in indirect calorimetry cages. After a 2-day adaptation period, mice were allowed a restricted amount (1.1 g) of chow 1 h before the onset of the dark phase to induce a fasting state in early morning, as published64. At the end of the light phase at 18.00 h, mice were re-fed with a restricted amount (1.1 g) of chow, or first-time HDD or LDD (the refeeding diet was assigned randomly; n = 4 per dietary group). Shortly before the following dark phase mice received access to the same diet they were allocated the day before, but now ad libitum. Indirect calorimetry measurements continued for an additional 5.5 d.
Study 3 (short-term exposure without fasting, adult)
Ten-month-old female mice were individually housed in indirect calorimetry cages. After a 2-day adaptation period, mice were provided clean bedding and given ad libitum first-time access to either HDD or LDD (random assignment, n = 6) shortly before the dark phase and for the remaining experimental period. Measurements continued for an additional 4.5 d. Faecal pellets produced after the introduction of the new diets were collected from the bedding at the end of the experiment and stored at −80 °C.
Experimental diets
Both the HDD and the LDD satisfy the nutrient requirements for rodent growth and lactation (AIN-93G)65, with appropriate levels of mono- and poly-unsaturated fatty acids. The macronutrient composition was 20.1 energy percentage (en%) protein, 54.9 en% carbohydrates, and 25 en% fat (Table 1), with starch being the sole source of carbohydrates. The starch fraction (Cargill BV, Sas van Gent, The Netherlands) of the HDD was composed of 100% amylopectin (which is highly digestible), while that of the LDD was a mixture of 60% amylose (which resists complete digestion) and 40% amylopectin. The diets were pelleted by Research Diet Services BV, Wijk bij Duurstede, The Netherlands.
In vivo diet digestibility
Total faeces produced from PN 35–42 (Study 1) were recovered from the bedding of a subgroup of randomly selected animals (n = 4 per sex and diet). Food intake was recorded over the same period. Gross energy in faeces and food was determined in blinded samples using a C7000 bomb calorimeter (IKA, Staufen, Germany) and diet digestibility was calculated as published66.
In vitro carbohydrate digestibility
The in vitro digestibility of starches in the experimental diets was determined in blinded samples in triplicate, as published67. Briefly, 5 intact pellets of each diet were cryoground to homogeneous particle size and weighed separately into 3 tubes (70 mg). Each sample was digested in a 15-ml tube by adding cocktail solutions (modified from Versantvoort et al.68) and digestive enzymes at 37 °C in three sequential steps to represent the oral (5 min), gastric (2 h), and duodenal (6 h) phases. Two blanks containing only enzymes and solutions were included. Samples were taken at several time points during the gastric and duodenal phases and centrifuged. Clean supernatants were recovered and free glucose content was determined by the glucose oxidase peroxidase method69. Starch digestion was expressed as the percentage of total glucose released based on the amount of starches in the diets.
Quantification of SCFA in intestinal digesta by gas chromatography (GC)
Short-chain fatty acids in caecum- and colon-contents were determined as previously reported70, with some modifications. Samples (about 25 mg) were weighed, thawed, homogenized in 100 μl of ultrapure water, and centrifuged for 3 min at 21,382 g. To 50 μl of supernatant, 100 μl of 2-ethylbutyric acid solution (0.45 mg ml−1) were added as internal standard. An external standard curve was prepared containing 50 μl of a mixture of acetic, propionic, butyric, valeric, isobutyric, and isovaleric acid at concentrations ranging from 0.002 mg ml−1 to 0.8 mg ml−1, to which 100 μl of internal standard were added. Blanks containing only water or water and internal standard were included for quality control. HCl and oxalic acid were added to all samples, blanks, and standards in order to protonate the SCFA. Gas chromatography was performed on a FOCUS GC apparatus coupled to a flame ionization detector (Interscience, Breda, The Netherlands). Samples were injected (1 μl) into an CP-FFAP CB column (25 m × 0.53 mm × 1.00 μm; Agilent Technologies, Santa Clara, CA, USA). Helium served as carrier gas at a pressure of 40 kPa. The initial oven temperature was 100 °C with 0.5 min hold, ramped to 180 °C at 8 °C min−1 with 1 min hold, and finally ramped to 200 °C at 20 °C min−1 with 5 min hold. Peak identities and areas were analysed with Xcalibur software (version 2.2; Thermo Scientific, Waltham, MA, USA). Concentrations of SCFA were normalised to the internal standard and expressed relative to original sample weight.
Microbiota analysis
Microbial DNA was isolated from faecal pellets using the Maxwell® 16 Instrument (Promega, Leiden, The Netherlands). Faecal pellets were added to a bead-beating tube with 350 μl Stool Transport and Recovery (STAR) buffer, 0.25 g of sterilized zirconia beads (0.1 mm), and three glass beads (2.5 mm). Faecal pellets were homogenized by bead-beating three times (60 s × 5.5 ms) and incubation for 15 min at 95 °C at 100 rpm. Samples were then centrifuged for 5 min at 4 °C and 14,000 g and supernatants transferred to sterile tubes. Pellets were re-processed using 200 μl STAR buffer and both supernatants were pooled. DNA purification was performed with a customized kit (AS1220; Promega) using 250 μl of the final supernatant pool. DNA was eluted in 50 μl of DNAse- RNAse-free water and its concentration measured using a DS-11 FX+ Spectrophotometer/Fluorometer (DeNovix Inc., Wilmington, USA). The V4 region of 16S ribosomal RNA (rRNA) gene was amplified in duplicate PCR reactions for each sample in a total reaction volume of 50 μl. The master mix contained 1 μl of a unique barcoded primer, 515F-n and 806R-n (10 μM each per reaction), 1 μl dNTPs mixture, 0.5 μl Phusion Green Hot Start II High-Fidelity DNA Polymerase (2 U/μl; Thermo Scientific, Landsmeer, The Netherlands), 10 μl 5× Phusion Green HF Buffer, and 36.5 μl DNAse- RNAse-free water. The amplification program included 30 s of initial denaturation step at 98°C, followed by 25 cycles of denaturation at 98 oC for 10 s, annealing at 50 °C for 10 s, elongation at 72 °C for 10 s, and a final extension step at 72 °C for 7 min. The PCR product was visualised in 1% agarose gel (~290 bp) and purified with CleanPCR kit (CleanNA, Alphen aan den Rijn, The Netherlands). The concentration of the purified PCR product was measured with Qubit dsDNA BR Assay Kit (Invitrogen, California, USA) and 200 ng of microbial DNA from each sample were pooled for the creation of the final amplicon library which was sequenced (150 bp, paired-end) on the Illumina HiSeq. 2000 platform (GATC Biotech, Constance, Germany).
Microbiota data processing and analysis
Data filtering and taxonomy assignment were performed using the NG-Tax pipeline71. Briefly, an OTU table was created for each sample with the most abundant sequences. Low abundance OTUs were discarded, using a minimum relative abundance threshold of 0.1%. Two distinct in-house assembled mock communities were included in the library and were compared with their theoretical composition for quality control (Additional file 6: Fig. 6). Calculations for α- and β-diversity analyses were performed using the publicly available Microbiome R package (version 1.2.1)72. Adonis permutational multivariate analyses of variance (PERMANOVA) using either the weighted or unweighted Unifrac distances were performed with the Vegan package (version 2.5-2) and were used to determine the amount of variation explained by the grouping variables. Linear Discriminant Analysis (LDA) Effect Size (LEfSe) was applied to determine the differences between the microbial communities of HDD- and LDD-fed mice using a publicly available pipeline (http://huttenhower.sph.harvard.edu/galaxy/)73; the threshold for the logarithmic LDA score was set to 2.0. P values for Kruskal-Wallis and Wilcoxon tests for the LEfSe analysis were set to 0.05. For non-parametric Student’s t-tests, reads were transformed to their relative abundances and tests were carried out with 999 permutations using QIIME (version 1; http://qiime.org/index.html)74. Statistical significance was determined using the Benjamini-Hochberg false discovery rate (FDR) adjustment.
Data analysis
Statistical analysis was performed using GraphPad Prism 5.04 (GraphPad, San Diego, CA, USA), unless stated otherwise. All data was tested for normality using the D’Agostino and Pearson omnibus test and its distribution was normalized by log transformation when applicable. Comparisons between two groups were made using unpaired two-tailed Student’s t-tests (for data with normal distribution) or two-tailed Mann-Whitney U tests (VH2 during light phase between HDD and LDD). Comparisons between more than two groups were made by one-way analysis of variance (ANOVA) with post hoc Bonferroni’s test for multiple comparisons. Time course data (H2 evolution) was analysed by repeated measures two-way ANOVA with Bonferroni’s post hoc test. When sample sizes being compared were similar and relatively large (n > 5), similarity of variances was not taken into account. All data is reported as mean ± s.d. Statistical significance was set at 5%, with levels indicated as *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P < 0.0001. Sample size was not determined statistically as the effect size was unknown, but it was based on our previous results on the use of indirect calorimetry to assess metabolic flexibility64,75.
Ethics approval and consent to participate
All animal experiments were approved by the Animal Experiments Committee (DEC 2014085.h) and performed in accordance to EU directive 2010/63/EU.
Electronic supplementary material
Acknowledgements
We acknowledge the help of the personnel at the CARUS animal facility. We also thank Inge van der Stelt and students Laura Dewitte and Lini Sholihah for their help in collecting data. This project was funded by the Dutch Technology Foundation STW (13509).
Author Contributions
J.K. and E.M.v.S. conceived, designed, and supervised the project. J.M.S.F.-C., H.J.M.S. and L.M.S.B. conducted animal experiments. H.J.M.S. and E.M.v.S. integrated the H2 and CH4 sensors into the indirect calorimetry system. L.M.S.B., A.O. and H.S. provided input for experimental design and interpretation of results. N.B. and V.G.-C. analysed and interpreted in vitro digestion data. P.K. and H.S. carried out all microbiome analyses and interpreted the data. J.M.S.F.-C., P.K., E.M.v.S., J.K., and H.S. interpreted data and prepared the manuscript. All authors critically revised and approved the manuscript.
Data Availability
The 16S rRNA gene sequencing dataset supporting the conclusions of this article is available in the European Nucleotide Archive (ENA) database with accession code PRJEB23475 at http://www.ebi.ac.uk/ena/data/view/prjeb23475. The authors declare that all other data supporting the findings of this study are available within the paper and its additional files 1–6, or from the corresponding authors upon reasonable request.
Competing Interests
N.B. and V.G.-C. are employed by Cargill. A.O. is employed by Danone Nutricia Research.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33619-0.
References
- 1.Englyst KN, Liu S, Englyst HN. Nutritional characterization and measurement of dietary carbohydrates. Eur. J. Clin. Nutr. 2007;61(Suppl 1):S19–39. doi: 10.1038/sj.ejcn.1602937. [DOI] [PubMed] [Google Scholar]
- 2.Elia M, Cummings JH. Physiological aspects of energy metabolism and gastrointestinal effects of carbohydrates. Eur. J. Clin. Nutr. 2007;61(Suppl 1):S40–74. doi: 10.1038/sj.ejcn.1602938. [DOI] [PubMed] [Google Scholar]
- 3.den Besten G, et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid. Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silk DB, Webb JP, Lane AE, Clark ML, Dawson AM. Functional differentiation of human jejunum and ileum: a comparison of the handling of glucose, peptides, and amino acids. Gut. 1974;15:444–449. doi: 10.1136/gut.15.6.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012;9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 6.Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooper LV, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 8.Cox LM, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Nood E, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Fernandez G, Denman SE, Cheung J, McSweeney CS. Phloroglucinol Degradation in the Rumen Promotes the Capture of Excess Hydrogen Generated from Methanogenesis Inhibition. Front. Microbiol. 2017;8:1871. doi: 10.3389/fmicb.2017.01871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olijhoek DW, et al. Effect of dietary nitrate level on enteric methane production, hydrogen emission, rumen fermentation, and nutrient digestibility in dairy cows. J. Dairy Sci. 2016;99:6191–6205. doi: 10.3168/jds.2015-10691. [DOI] [PubMed] [Google Scholar]
- 12.Rooke JA, et al. Hydrogen and methane emissions from beef cattle and their rumen microbial community vary with diet, time after feeding and genotype. Br. J. Nutr. 2014;112:398–407. doi: 10.1017/S0007114514000932. [DOI] [PubMed] [Google Scholar]
- 13.Sollinger, A. et al. Holistic Assessment of Rumen Microbiome Dynamics through Quantitative Metatranscriptomics Reveals Multifunctional Redundancy during Key Steps of Anaerobic Feed Degradation. mSystems3, 10.1128/mSystems.00038-18 (2018). [DOI] [PMC free article] [PubMed]
- 14.van Lingen HJ, et al. Diurnal Dynamics of Gaseous and Dissolved Metabolites and Microbiota Composition in the Bovine Rumen. Front. Microbiol. 2017;8:425. doi: 10.3389/fmicb.2017.00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christl SU, Murgatroyd PR, Gibson GR, Cummings JH. Production, metabolism, and excretion of hydrogen in the large intestine. Gastroenterology. 1992;102:1269–1277. doi: 10.1016/0016-5085(92)90765-Q. [DOI] [PubMed] [Google Scholar]
- 16.Symonds EL, Kritas S, Omari TI, Butler RN. A combined 13CO2/H2 breath test can be used to assess starch digestion and fermentation in humans. J. Nutr. 2004;134:1193–1196. doi: 10.1093/jn/134.5.1193. [DOI] [PubMed] [Google Scholar]
- 17.Ong DK, et al. Manipulation of dietary short chain carbohydrates alters the pattern of gas production and genesis of symptoms in irritable bowel syndrome. J. Gastroenterol. Hepatol. 2010;25:1366–1373. doi: 10.1111/j.1440-1746.2010.06370.x. [DOI] [PubMed] [Google Scholar]
- 18.Valeur J, Puaschitz NG, Midtvedt T, Berstad A. Oatmeal porridge: impact on microflora-associated characteristics in healthy subjects. Br. J. Nutr. 2016;115:62–67. doi: 10.1017/S0007114515004213. [DOI] [PubMed] [Google Scholar]
- 19.Carbonero F, Benefiel AC, Gaskins HR. Contributions of the microbial hydrogen economy to colonic homeostasis. Nat. Rev. Gastroenterol. Hepatol. 2012;9:504–518. doi: 10.1038/nrgastro.2012.85. [DOI] [PubMed] [Google Scholar]
- 20.Isken F, Klaus S, Osterhoff M, Pfeiffer AF, Weickert MO. Effects of long-term soluble vs. insoluble dietary fiber intake on high-fat diet-induced obesity in C57BL/6J mice. J. Nutr. Biochem. 2010;21:278–284. doi: 10.1016/j.jnutbio.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Hartmann L, Taras D, Kamlage B, Blaut M. A new technique to determine hydrogen excreted by gnotobiotic rats. Lab. Anim. 2000;34:162–170. doi: 10.1258/002367700780457617. [DOI] [PubMed] [Google Scholar]
- 22.Ostrander CR, Stevenson DK, Neu J, Kerner JA, Moses SW. A sensitive analytical apparatus for measuring hydrogen production rates. I. Application to studies in small animals. Evidence of the effects of an α-glucosidehydrolase inhibitor in the rat. Anal. Biochem. 1982;119:378–386. doi: 10.1016/0003-2697(82)90601-7. [DOI] [PubMed] [Google Scholar]
- 23.Dufourlescoat C, Lecoz Y, Andrieux C, Szylit O. Effects of nature, size and level of incorporation of dietary fibers on colonic functions in germ-free rats and in heteroxenic rats inoculated with a human flora. Food. Hydrocoll. 1995;9:9–15. doi: 10.1016/S0268-005x(09)80189-6. [DOI] [Google Scholar]
- 24.Rodkey FL, Collison HA, O’Neal JD. Carbon monoxide and methane production in rats, guinea pigs, and germ-free rats. J. Appl. Physiol. 1972;33:256–260. doi: 10.1152/jappl.1972.33.2.256. [DOI] [PubMed] [Google Scholar]
- 25.Walter DJ, Eastwood MA, Brydon WG, Elton RA. An experimental design to study colonic fibre fermentation in the rat: the duration of feeding. Br. J. Nutr. 1986;55:465–479. doi: 10.1079/bjn19860054. [DOI] [PubMed] [Google Scholar]
- 26.Tuboly E, et al. Determination of endogenous methane formation by photoacoustic spectroscopy. J. Breath Res. 2013;7:046004. doi: 10.1088/1752-7155/7/4/046004. [DOI] [PubMed] [Google Scholar]
- 27.Scribner KB, Pawlak DB, Aubin CM, Majzoub JA, Ludwig DS. Long-term effects of dietary glycemic index on adiposity, energy metabolism, and physical activity in mice. Am. J. Physiol. Endocrinol. Metab. 2008;295:E1126–1131. doi: 10.1152/ajpendo.90487.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poppitt SD, et al. Circadian patterns of total 24-h hydrogen and methane excretion in humans ingesting nonstarch polysaccharide (NSP) diets and the implications for indirect calorimetric and D2 18O methodologies. Eur. J. Clin. Nutr. 1996;50:524–534. [PubMed] [Google Scholar]
- 29.Tulley RT, et al. Comparative methodologies for measuring metabolizable energy of various types of resistant high amylose corn starch. J. Agric. Food Chem. 2009;57:8474–8479. doi: 10.1021/jf900971c. [DOI] [PubMed] [Google Scholar]
- 30.Nishimura N, Tanabe H, Yamamoto T. Isomaltodextrin, a highly branched alpha-glucan, increases rat colonic H2 production as well as indigestible dextrin. Biosci. Biotechnol. Biochem. 2016;80:554–563. doi: 10.1080/09168451.2015.1104237. [DOI] [PubMed] [Google Scholar]
- 31.Tonouchi H, et al. Studies on absorption and metabolism of palatinose (isomaltulose) in rats. Br. J. Nutr. 2011;105:10–14. doi: 10.1017/S0007114510003193. [DOI] [PubMed] [Google Scholar]
- 32.Kalantar-Zadeh K, et al. A human pilot trial of ingestible electronic capsules capable of sensing different gases in the gut. Nat. Electron. 2018;1:79–87. doi: 10.1038/s41928-017-0004-x. [DOI] [Google Scholar]
- 33.Levrat MA, Remesy C, Demigne C. Very acidic fermentations in the rat cecum during adaptation to a diet rich in amylase-resistant starch (crude potato starch) J. Nutr. Biochem. 1991;2:31–36. doi: 10.1016/0955-2863(91)90046-8. [DOI] [Google Scholar]
- 34.Key FB, Mathers JC. Digestive adaptations of rats given white bread and cooked haricot beans (Phaseolus vulgaris): large-bowel fermentation and digestion of complex carbohydrates. Br. J. Nutr. 1995;74:393–406. doi: 10.1079/bjn19950143. [DOI] [PubMed] [Google Scholar]
- 35.Walker AW, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohl KD, Amaya J, Passement CA, Dearing MD, McCue MD. Unique and shared responses of the gut microbiota to prolonged fasting: a comparative study across five classes of vertebrate hosts. FEMS Microbiol. Ecol. 2014;90:883–894. doi: 10.1111/1574-6941.12442. [DOI] [PubMed] [Google Scholar]
- 37.Crawford PA, et al. Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc. Natl. Acad. Sci. USA. 2009;106:11276–11281. doi: 10.1073/pnas.0902366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolf PG, Biswas A, Morales SE, Greening C, Gaskins HR. H2 metabolism is widespread and diverse among human colonic microbes. Gut Microbes. 2016;7:235–245. doi: 10.1080/19490976.2016.1182288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tachon S, Zhou J, Keenan M, Martin R, Marco ML. The intestinal microbiota in aged mice is modulated by dietary resistant starch and correlated with improvements in host responses. FEMS Microbiol. Ecol. 2013;83:299–309. doi: 10.1111/j.1574-6941.2012.01475.x. [DOI] [PubMed] [Google Scholar]
- 40.Barouei Javad, Bendiks Zach, Martinic Alice, Mishchuk Darya, Heeney Dustin, Hsieh Yu-Hsin, Kieffer Dorothy, Zaragoza Jose, Martin Roy, Slupsky Carolyn, Marco Maria L. Microbiota, metabolome, and immune alterations in obese mice fed a high-fat diet containing type 2 resistant starch. Molecular Nutrition & Food Research. 2017;61(11):1700184. doi: 10.1002/mnfr.201700184. [DOI] [PubMed] [Google Scholar]
- 41.Muyzer G, Stams AJ. The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 2008;6:441–454. doi: 10.1038/nrmicro1892. [DOI] [PubMed] [Google Scholar]
- 42.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 43.Laverdure R, Mezouari A, Carson MA, Basiliko N, Gagnon J. A role for methanogens and methane in the regulation of GLP-1. Endocrinol. Diabetes Metab. 2018;1:e00006. doi: 10.1002/edm2.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Attene-Ramos MS, Wagner ED, Plewa MJ, Gaskins HR. Evidence that hydrogen sulfide is a genotoxic agent. Mol. Cancer Res. 2006;4:9–14. doi: 10.1158/1541-7786.MCR-05-0126. [DOI] [PubMed] [Google Scholar]
- 45.Roediger WE, Duncan A, Kapaniris O, Millard S. Sulphide impairment of substrate oxidation in rat colonocytes: a biochemical basis for ulcerative colitis? Clin. Sci. 1993;85:623–627. doi: 10.1042/cs0850623. [DOI] [PubMed] [Google Scholar]
- 46.Attene-Ramos MS, Wagner ED, Gaskins HR, Plewa MJ. Hydrogen sulfide induces direct radical-associated DNA damage. Mol. Cancer. Res. 2007;5:455–459. doi: 10.1158/1541-7786.MCR-06-0439. [DOI] [PubMed] [Google Scholar]
- 47.Soriano RN, et al. Endogenous peripheral hydrogen sulfide is propyretic: its permissive role in brown adipose tissue thermogenesis in rats. Exp. Physiol. 2018;103:397–407. doi: 10.1113/EP086775. [DOI] [PubMed] [Google Scholar]
- 48.Segata N, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13:R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goker M, et al. Complete genome sequence of Odoribacter splanchnicus type strain (1651/6) Stand. Genomic. Sci. 2011;4:200–209. doi: 10.4056/sigs.1714269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bomar L, Maltz M, Colston S, Graf J. Directed culturing of microorganisms using metatranscriptomics. mBio. 2011;2:e00012–00011. doi: 10.1128/mBio.00012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai HH, Hart CA, Rhodes JM. Production of mucin degrading sulphatase and glycosidases by Bacteroides thetaiotaomicron. Lett. Appl. Microbiol. 1991;13:97–101. doi: 10.1111/j.1472-765X.1991.tb00580.x. [DOI] [Google Scholar]
- 52.Dai ZL, Wu GY, Zhu WY. Amino acid metabolism in intestinal bacteria: links between gut ecology and host health. Front. Biosci. (LandmarkEd.) 2011;16:1768–1786. doi: 10.2741/3820. [DOI] [PubMed] [Google Scholar]
- 53.Clavel T, et al. Isolation of bacteria from the ileal mucosa of TNFdeltaARE mice and description of Enterorhabdus mucosicola gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 2009;59:1805–1812. doi: 10.1099/ijs.0.003087-0. [DOI] [PubMed] [Google Scholar]
- 54.Clavel T, Charrier C, Wenning M, Haller D. Parvibacter caecicola gen. nov., sp. nov., a bacterium of the family Coriobacteriaceae isolated from the caecum of a mouse. Int. J. Syst. Evol. Microbiol. 2013;63:2642–2648. doi: 10.1099/ijs.0.045344-0. [DOI] [PubMed] [Google Scholar]
- 55.Li X, Jensen RL, Hojberg O, Canibe N, Jensen BB. Olsenella scatoligenes sp. nov., a 3-methylindole- (skatole) and 4-methylphenol- (p-cresol) producing bacterium isolated from pig faeces. Int. J. Syst. Evol. Microbiol. 2015;65:1227–1233. doi: 10.1099/ijs.0.000083. [DOI] [PubMed] [Google Scholar]
- 56.Windey K, De Preter V, Verbeke K. Relevance of protein fermentation to gut health. Mol. Nutr. Food. Res. 2012;56:184–196. doi: 10.1002/mnfr.201100542. [DOI] [PubMed] [Google Scholar]
- 57.Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 2001;81:1031–1064. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- 58.Zoetendal EG, Akkermans ADL, Akkermans-van Vliet WM, de Visser JAGM, de Vos WM. The host genotype affects the bacterial community in the human gastronintestinal tract. Microb. Ecol. Health Dis. 2009;13:129–134. doi: 10.1080/089106001750462669. [DOI] [Google Scholar]
- 59.Zeevi D, et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 60.Carter EA, Barr RG. Preliminary studies demonstrating acetoclastic methanogenesis in a rat colonic ring model. J. Nutr. Metab. 2013;2013:540967. doi: 10.1155/2013/540967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pimentel M. Breath testing for small intestinal bacterial overgrowth: should we bother? Am. J. Gastroenterol. 2016;111:307–308. doi: 10.1038/ajg.2016.30. [DOI] [PubMed] [Google Scholar]
- 62.Pimentel M, Mathur R, Chang C. Gas and the microbiome. Curr. Gastroenterol. Rep. 2013;15:356. doi: 10.1007/s11894-013-0356-y. [DOI] [PubMed] [Google Scholar]
- 63.Hoevenaars FP, et al. Effects of dietary history on energy metabolism and physiological parameters in C57BL/6J mice. Exp. Physiol. 2013;98:1053–1062. doi: 10.1113/expphysiol.2012.069518. [DOI] [PubMed] [Google Scholar]
- 64.Duivenvoorde LP, et al. Oxygen restriction as challenge test reveals early high-fat-diet-induced changes in glucose and lipid metabolism. Pflugers Arch. 2015;467:1179–1193. doi: 10.1007/s00424-014-1553-8. [DOI] [PubMed] [Google Scholar]
- 65.Reeves PG, Nielsen FH, Fahey GC., Jr. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 1993;123:1939–1951. doi: 10.1093/jn/123.11.1939. [DOI] [PubMed] [Google Scholar]
- 66.Isken F, et al. Impairment of fat oxidation under high- vs. low-glycemic index diet occurs before the development of an obese phenotype. Am. J. Physiol. Endocrinol. Metab. 2010;298:E287–295. doi: 10.1152/ajpendo.00515.2009. [DOI] [PubMed] [Google Scholar]
- 67.Garcia-Campayo Vicenta, Han Sonia, Vercauteren Ronny, Franck Anne. Digestion of Food Ingredients and Food Using an <i>In Vitro</i> Model Integrating Intestinal Mucosal Enzymes. Food and Nutrition Sciences. 2018;09(06):711–734. doi: 10.4236/fns.2018.96055. [DOI] [Google Scholar]
- 68.Versantvoort CH, Oomen AG, Van de Kamp E, Rompelberg CJ, Sips AJ. Applicability of an in vitro digestion model in assessing the bioaccessibility of mycotoxins from food. Food Chem. Toxicol. 2005;43:31–40. doi: 10.1016/j.fct.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 69.Morin LG, Prox J. Single glucose oxidase-peroxidase reagent for two-minute determination of serum glucose. Clin. Chem. 1973;19:959–962. [PubMed] [Google Scholar]
- 70.Ladirat SE, et al. Impact of galacto-oligosaccharides on the gut microbiota composition and metabolic activity upon antibiotic treatment during in vitro fermentation. FEMS Microbiol. Ecol. 2014;87:41–51. doi: 10.1111/1574-6941.12187. [DOI] [PubMed] [Google Scholar]
- 71.Ramiro-Garcia J, et al. NG-Tax, a highly accurate and validated pipeline for analysis of 16S rRNA amplicons from complex biomes. F1000Research. 2016;5:1791. doi: 10.12688/f1000research.9227.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lahti, L. & Shetty, S. Tools for microbiome analysis in R. Version 0.99.90. http://microbiome.github.io/microbiome/. (2017).
- 73.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Duivenvoorde LP, van Schothorst EM, Swarts HJ, Keijer J. Assessment of metabolic flexibility of old and adult mice using three noninvasive, indirect calorimetry-based treatments. J. Gerontol. A Biol. Sci. Med. Sci. 2015;70:282–293. doi: 10.1093/gerona/glu027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA gene sequencing dataset supporting the conclusions of this article is available in the European Nucleotide Archive (ENA) database with accession code PRJEB23475 at http://www.ebi.ac.uk/ena/data/view/prjeb23475. The authors declare that all other data supporting the findings of this study are available within the paper and its additional files 1–6, or from the corresponding authors upon reasonable request.