Abstract
Microbial communities primarily consist of multiple species that affect one another’s fitness both directly and indirectly. This study showed that the cocultivation of Paenibacillus amylolyticus and Xanthomonas retroflexus exhibited facultative mutualistic interactions in a static environment, during the course of which a new adapted phenotypic variant of X. retroflexus appeared. Although the emergence of this variant was not directly linked to the presence of P. amylolyticus, its establishment in the coculture enhanced the productivity of both species due to mutations that stimulated biofilm formation. The mutations were detected in genes encoding a diguanylate cyclase predicted to synthesise cyclic-di-GMP. Examinations of the biofilm formed in cocultures of P. amylolyticus and the new variant of X. retroflexus revealed a distinct spatial organisation: P. amylolyticus only resided in biofilms in association with X. retroflexus and occupied the outer layers. The X. retroflexus variant therefore facilitated increased P. amylolyticus growth as it produced more biofilm biomass. The increase in X. retroflexus biomass was thus not at the expense of P. amylolyticus, demonstrating that interspecies interactions can shape diversification in a mutualistic coculture and reinforce these interactions, ultimately resulting in enhanced communal performance.
Introduction
Several studies have examined how bacterial species adapt and shown how bacterial genotype and phenotype change in monospecies cultures [1–5]. Nevertheless, there is limited empirically-based information on how the inclusion of additional species affects this adaptive process. The acquisition of such knowledge should be a priority, as it is widely acknowledged that almost all bacteria reside alongside other species in their natural environment. It has been demonstrated that when multiple species co-exist in biofilms, interspecies interactions result in emergent activities and functions, thus making the study of whole communities highly relevant [6–9]. It is thus to be expected that the inclusion of more species will affect adaptation of the individual species [10]. By using a small spatial scale and the rapid generation time of bacteria, coexisting species can be examined for several generations within a relatively short timeframe. This provides the possibility of examining intraspecies and interspecies adaptation to interactions in a community. Interspecies bacterial interactions are often categorised into two broad categories as either cooperation or competition. Several studies have in detail described these interactions, and their functional implications [11–14]. Especially competition between microorganisms has drawn extensive attention and has been suggested to dominate over cooperation in microbial communities [15].
In monospecies populations, intraspecific variation (phenotypic and/or genotypic) may result in competition between the distinct variants [16, 17]. Models have shown how the production of extracellular polymeric substances can be a strong competitive advantage in biofilm [18]. Such intraspecific variation is not yet well understood in multispecies communities, although it has been suggested to influence interspecies interactions across a wide range of biological systems [19–22]. Living in multispecies communities has been found to affect the productivity and evolution of the individual species [23]. The present study examined the interplay between interspecies and intraspecific interactions, and their effect on facultative mutualism over time. Mutualism is defined as reciprocal positive interactions between species resulting in a net positive effect for all participants [24, 25]. In facultative mutualistic interactions, the partners can exist and divide individually, whereas organisms in an obligate mutualistic interaction cannot grow without each other [26]. A recent study has suggested that mutualism is ubiquitous in natural environments and is an important driver of evolution [27].
In the present study, co-cultures of Xanthomonas retroflexus and Paenibacillus amylolyticus were used, originally co-isolated from the surface of a decomposing leaf [28]. The two species were cocultivated to examine whether the relationship would remain stable over a period of time and whether cocultivation would affect diversification. We found that cocultivation reinforced the mutualistic interaction, as productivity of both strains was enhanced; This was linked to an evolved wrinkled phenotype of X. retroflexus and the biofilm setting, allowing close association between the two species.
Materials and methods
See supplementary information for an extended description of the experimental procedures.
Bacterial cultivation
All bacterial isolates were stored at −80 °C in ∼20% glycerol. Strains from freeze-stock were streaked on plates of Tryptic Soy Broth (TSB; 17 g pancreatic digest of casein, 3 g papaic digest of soybean meal, 5 g sodium chloride, 2.5 g dextrose, 2.5 g dibasic potassium phosphate in 1 L distilled water, pH 7.3) complemented with 1.5 % agar (TSA) when needed. One colony was used per biological replicate.
Evolution experiments
Cultures in the exponential phase were diluted to an optical density (OD) of 0.15 at 600 nm prior to usage. Experiments were conducted in 24-well plates, which were incubated at 24 °C without shaking. X. retroflexus and P. amylolyticus were grown as both monocultures and cocultures. Every other day the content of each well was homogenised and a small fraction of the volume (1%) was transferred to a new well (Suppl. Fig. 1). On selected days the well content was plated on TSA plates complemented with 40 µg/mL Congo Red and 20 µg/mL Coomassie Brilliant Blue G250. Plates were incubated for 48 h at 24 °C before colony-forming units (CFU) were counted.
Swimming assay
Swimming plates were prepared 1 h before inoculation of the strains from half TSB complemented with 0.3% agar (Teknova). Plates were inoculated with 2 µL of the 5 mL overnight cultures, adjusted to OD600 of 0.5 and incubated at 24 °C at high humidity.
Full genome sequencing
DNA was extracted from overnight cultures using the Qiagen DNeasy Blood & Tissue kit (Qiagen, cat. no. 69504). The whole-genome sequencing library was prepared using the Nextera XT DNA library preparation kit (Illumina, USA) and subsequently quantified by fragment analyser™ (Advanced Analytical Technologies Co.). Sequencing was performed as 2 × 250-base paired-end reads using the Illumina MiSeq platform (Illumina). Data were analysed using CLC Genomic Workbench V7.5.1 (CLCBio). Obtained reads were trimmed and normalised. Raw reads from sequenced phenotypic variants were uploaded to The European Nucleotide Archive (ENA) X. retroflexus PRJEB18990 and P. amylolyticus PRJEB18991. The de novo assembly of the ancestral X. retroflexus was annotated by Rapid Annotation using Subsystem Technology (RAST) service and the annotated genome can be accessed via ENA PRJEB18431 [29–31].
Construction of strains
mCherry was introduced into the X. retroflexus strains using the mini-Tn7 system, following the general procedures described by Choi and Schweizer [32]. Insertion of PlppmCherry-TcR (via pUC18T-miniTn7-PlppmCherry-TcR) into the chromosome of the X. retroflexus strains was verified by flow cytometry, florescence microscopy, and the absence of plasmid DNA (Plasmid mini AX kit, A&A Biotechnology). gfp was introduced into P. amylolyticus via plasmid pCM20 (kindly provided by Prof. Søren Molin) [33]. P. amylolyticus was made electrocompetent according to the procedure described by Rosado et al. [34] with a few modifications. Selection was performed on TSA plates containing 10 µg/ml ERY for 48 h at 30 °C. Successful introduction of pCM20 into P. amylolyticus was verified by flow cytometry and florescence microscopy.
pUC18T-miniTn7-PlppmCherry-TcR was generated as follows: PlppmCherry was amplified by PCR and pUC18T-miniTn7-lacIq-PlppmCherry-KmR as the template. This PCR product and a pUC18T-miniTn7-KmR [32] derivative were digested with SacI and HindIII. The two fragments were fused together by T4 ligation, generating pUC18T-miniTn7-PlppmCherry-KmR, which was transformed into electrocompetent Escherichia coli Genehogs (Invitrogen) cells and selected. The TcR gene was amplified by PCR and pBR322 [35] as the template. This PCR product and pUC18T-miniTn7-PlppmCherry-KmR were digested with SpeI and SacI and subsequently fused by T4 ligation, generating pUC18T-miniTn7-PlppmCherry-TcR, which was transformed into electrocompetent E. coli Genehog cells and selected.
Construction of CDS2260 knockout mutant
A mutant of XR-Wa lacking CDS2260 was constructed (XR-Wa-ΔCDS2260::TcR) by allelic exchange utilising the pEX18 based system as described by Hmelo et al. [36]. Flanking regions of CDS2260 were amplified by PCR using X. retroflexus as template (5ʹ-CTATCTGAGCTCTGCATCGCTTGTTCTCCAAC-3ʹ in combination with 5ʹ-GGAGAACTGTGAATGCGCAAGGTCCACACGTCGATGG-3′ and 5′-TCTTGAAGACGAAAGGGCCT-GTTGTACCGTGCGAAGAGTG-3′ in combination with 5′-CTACTATCTAGAGTGGTGGATGTTGAAACCG-3ʹ). TcR was amplified using pBR322 [35] as template (5′-CACTCTTCGCACGGTACAACAGGCCCTTTCGTCTTCAAGA-3′ in combination with 5′-CCATCGACGTGTGGACCTTGCGCATTCACAGTTCTCC-3′). The three PCR fragments were fused by second-round SOE PCR using primers with overhangs, introducing SacI and Xbal restriction sites (5′-CTATCTGAGCTCTGCATCGCTTGTTCTCCAAC-3′ in combination with 5′-CTACTATCTAGAGTGGTGGATGTTGAAACCG-3ʹ). The SOE PCR product was introduced into pEX18ApGw digested with SacI and Xbal [37]. The pEX18Ap-∆CDS2260::TcR plasmid was then transformed into E. coli GeneHogs® (Thermo Fisher Scientific) and subsequently introduced into XR-Wa. The resulting deletion mutants were selected on LB medium supplemented with 60 μg/mL tetracycline and sucrose 5%.
Cultivation in a planktonic environment
Cultures in the exponential phase were diluted to an OD600 of 0.15 and used to inoculate glass tubes, which were then incubated for 24 h while shaking at 250 RPM at 24 °C. Following incubation, the content was plated on TSA plates complemented with 40 µg/mL Congo Red and 20 µg/mL Coomassie Blue to allow for cell counting of both morphotypes. Plates were incubated for 48 h at 24 °C before counting CFU.
Crystal violet biofilm formation
A Calgary Biofilm Device was used to quantify biofilm formation under static conditions [38] based on the protocol previously used for these strains [39]. Cultures in the exponential phase were adjusted to an OD600 of 0.15 in TSB and then inoculated into 96-well plates. For the supernatant experiment, supernatant from P. amylolyticus was used (see supplementary information for details). The plates were then covered with a lid with pegs extending into the wells (Nunc-TSP, Termo Scientific). After 24 h of incubation at 24 °C, the pegs were washed in phosphate-buffered saline before the biofilm was stained in 1% crystal violet for 20 min followed by washing. The peg lids were then placed in a fresh microtitre plate with 96% ethanol in the wells and the absorbance of Crystal violet at OD590 nm was measured.
Biofilm cultivation using the BioFlux device
The ancestral X. retroflexus and the wrinkled phenotypic variant were tested individually and in cocultures with P. amylolyticus to observe biofilm formation in the BioFlux™ 1000 device (Fluxion Biosciences, South San Francisco, CA). A BioFlux 48-well plate was inoculated with overnight culture adjusted to 0.1 OD600 and allowed to attach for 30 min. The flow was started at 0.15 dyn cm2, yielding a flow rate of 13 µl/h, and running for 48 h at 24 °C in TSB. Image acquisition of the biofilm was performed with a CLSM (LSM800, Zeiss, Germany) equipped with a 20 × objective.
Image processing and analysis
The acquired CLSM images were segmented in Fiji and the background signal defined by a threshold set, before the image was saved as a binary mask for analysis of the spatial localisation of each species [40]. Biomass quantification, 3D aggregate estimation and co-localisation calculations were performed using the “R” statistical programming language [41]. The algorithms were included in the RCon3D package published on GitHub [42] (www.github.com/Russel88/RCon3D version 1.1). Simulated fluorescence projections and vertical cross-sections for the biofilm were performed with IMARIS software (Bitplane AG, Switzerland).
Statistical analysis
All statistical analyses were performed using R 3.4.3 [41] in RStudio [43]. CFU counting was undertaken in triplicates that were averaged prior to analysis. Unless otherwise stated, all the experiments were analysed with linear mixed-effect models [44], in which the replicas were incorporated as random intercepts. Growth curves were analysed with standard linear regression by subsetting data to only the exponential phase (manually), and afterward testing slopes of log-transformed OD measurements with an interaction between hours of growth and the isolate. ANOVA with post-hoc Tukey test was used to test whether mono-cultures were different in the crystal violet assay. To test whether co-cultures in the crystal violet assay differed from the sum of the mono-cultures, a linear model was used with the intercept set to zero and the log ratio of co-culture/mono-culture as the response.
Results
The impact of continuous co-cultivation on mutualistic interactions
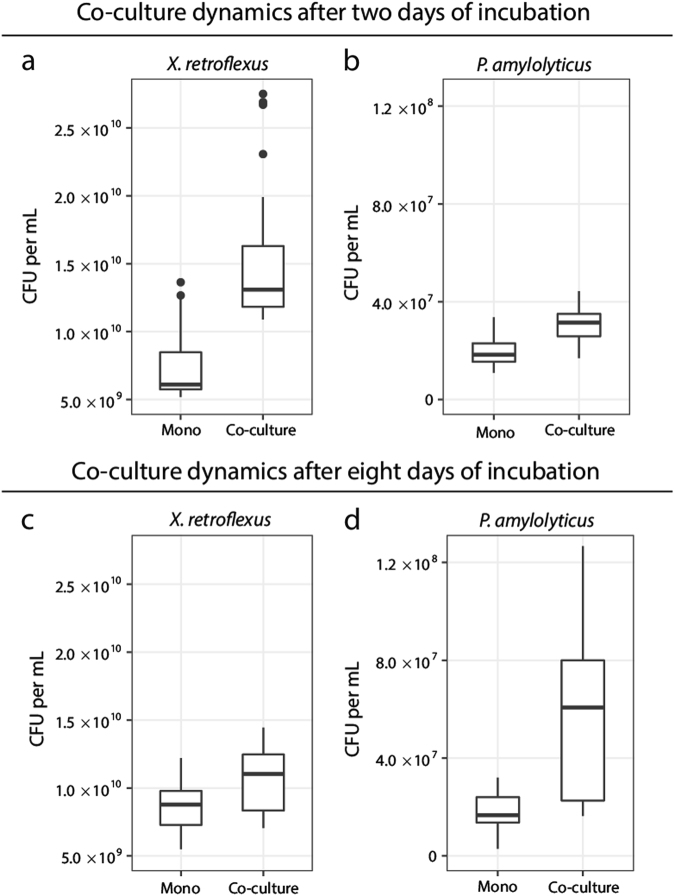
When X. retroflexus (denoted XR) and P. amylolyticus (denoted PA) were cocultured, they formed a mutualistic relationship (Fig. 1) (see Suppl. Table 1 for denotation of strains used): The cell counts of each of the two strains were significantly increased in cocultures compared with monocultures, namely by 105% for XR and 65% for PA (XR P < 0.001 and PA P < 0.001) (Fig. 1a, b). This was not the case when grown as shaken cultures (Suppl. Fig. 2). Fewer CFUs of XR were found in samples from shaken cocultures with PA compared with shaken monocultures of XR. Only PA benefited from coculturing with XR in shaken cultures (P = 0.0493 for PA with XR). Thus, the observed mutualism between the two species only occurred under static conditions (Fig. 1). Next, the stability of the mutualistic interaction was examined over time. We chose an experimental design, in which we did not select for either planktonic or the biofilm mode of growth, as all cells in the wells (planktonic, surface associated and pellicle) were mixed and a fraction was transferred. This design was chosen in order to ensure that there was no selection for a specific phenotype directly associated with a sessile or motile lifestyle. The timeframe between transfers corresponded to approximately seven generations. After approximately 26 generations for XR and 21 generations for PA (day 8), the mutualistic relationship in the co-cultured community was still observed, with respectively 24% and 28% higher cell numbers of XR and PA in coculture (Fig. 1c, d) (XR P < 0.001 and PA P < 0.001). This shows that the facultative mutualistic interaction between XR and PA persisted during a prolonged period of coexistence.
Fig. 1.
Co-cultivation of two facultative mutualists. Cell numbers of X. retroflexus in monocultures and cocultures with P. amylolyticus. a, b The facultative mutualism between the two species after two days. There was a significant increase of X. retroflexus in cocultures compared with monocultures (P < 0.001) and also a significant increase of P. amylolyticus in cocultures compared with monocultures (P < 0.001). c, d The facultative mutualism between the two species was maintained after 8 days of cocultivation in the static environment (X. retroflexus P < 0.001 and P. amylolyticus P < 0.001). Boxplots are Tukey style and P-values are from a linear mixed-effect model. Data are based on 25 biological replicates from three separate experiments (n = 25)
Emergence of wrinkled colony variants
The colony morphology was examined at day 8 and day 16 to identify phenotypic (and possibly genotypic) variants and to compare monocultures and cocultures. Twenty-five biological replicates of monocultures and cocultures were screened for new phenotypic variants (75 biological replicates in total). After 8 days of cocultivation, a wrinkled phenotypic colony variant of X. retroflexus (denoted XR-W) was observed. The phenotypic wrinkled variant emerged in both monocultures and cocultures with P. amylolyticus, indicating that the appearance of this variant was not dependent on the presence of PA. The wrinkled phenotype was observed in 20 of the 25 biological monoculture replicates and in 21 of the 25 coculture replicates. Yet at day 16 the cell counts of the wrinkled X. retroflexus variant had significantly increased in the cocultures (>10-fold more at day 16 compared with day 8), whereas no change from day 8 to day 16 was observed in the monocultures (P = 0.001 and P = 0.748, respectively). All the biological replicates of the cocultures at day 16 (25 of 25) contained the wrinkled variant, whereas only 18 of the 25 monocultures did so (Suppl. Fig. 3), indicating that the presence of PA positively selected for the wrinkled variant of X. retroflexus between day 8 and day 16.
Identification and interactions of the wrinkled colony variant
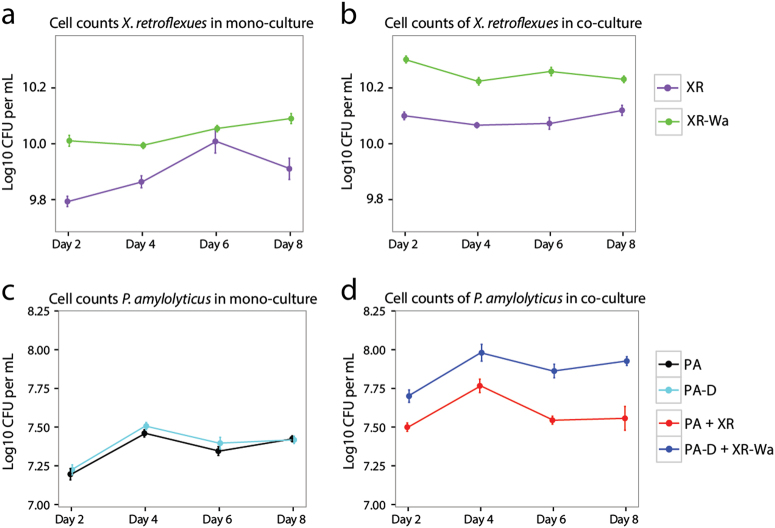
The higher frequencies of wrinkled colony variants observed in cocultures at day 16 led to an examination of the interaction between this phenotypic variant and PA, since the new phenotypic variant could be expected to destabilise the interaction by ensuring a better spatial position for XR in a pellicle; a biofilm formed at the air–liquid interface in static cultures. A wrinkled X. retroflexus colony variant (denoted XR-Wa) that was found in particularly high frequencies in one of the coculture replicates was isolated for further studies. We found that monocultures of XR-Wa exhibited an increase in cell numbers compared with the ancestral XR after 2 days of growth (P < 0.001) (Fig. 2a).
Fig. 2.
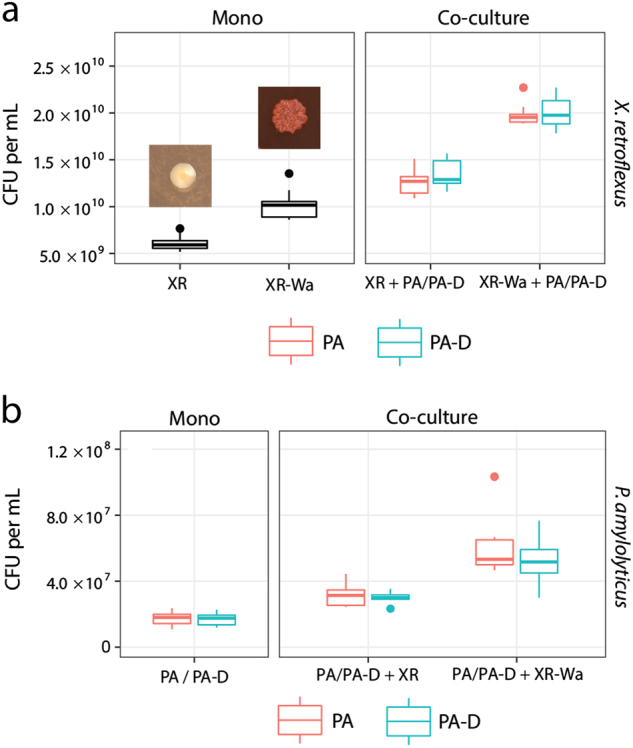
Ancestral vs. wrinkled X. retroflexus: The wrinkled variant reinforced the mutualistic interaction. a Cell numbers of X. retroflexus in mono and coculture with P. amylolyticus after 2 days. XR was found in lower numbers in monocultures compared with the wrinkled XR-Wa (P < 0.001). Colony phenotypes of XR (right) on TSA and wrinkled XR-Wa on Congo Red plates (left). b In the coculture with P. amylolyticus both XR and XR-Wa benefited significantly from cocultivation with P. amylolyticus (Both P < 0.001), resulting in significantly higher CFU for XR-Wa than XR (P < 0.001). b Cell numbers of P. amylolyticus in monoculture and coculture with X. retroflexus. Ancestral P. amylolyticus reached the same cell numbers as the evolved P. amylolyticus (P = 1.0) in monoculture. The coculture with derived P. amylolyticus and XR-Wa reached a higher biomass than the coculture with both ancestral strains (P < 0.001). Boxplots are Tukey style. P-values are from a linear mixed-effect model. Data based on nine biological replicates from two separate experiments (n = 9)
As an additional control, a derived X. retroflexus isolate (denoted XR-D) and a derived P. amylolyticus isolate (denoted PA-D), defined by having a similar colony morphology to the ancestral strains, were isolated from the coculture at day 8 to examine if they retained their original characteristics. Moreover, XR-D and PA-D were isolated in order to dismiss potential adaptation and optimisation towards growth in the media. Thus ancestral XR and PA strains were compared with XR-D and PA-D: No differences in growth rates were found for either XA-D (P = 0.645) or PA-D (P = 0.471) (Suppl. Table 2). The mutualistic relationship was also confirmed for the derived variants, as both XR-D and PA-D benefitted from cocultivation with PA and XR, respectively (Suppl. Fig. 4). In addition, it was confirmed that PA and PA-D did not differ in terms of productivity (P = 1.0). Cell counts of wrinkled XR-Wa in cocultures with PA were significantly higher than the cell counts of XR in the cocultures containing XR and PA, showing a 58% increase (Fig. 2a) (P < 0.001). The same was true when using PA-D in the coculture with XR-Wa, yielding a 49% increase (P < 0.001). Both PA-D and PA also increased additionally in cell numbers when cocultured with XR-Wa, by 96% for PA and by 69% for PA-D compared with the ancestral community with PA and XR (PA-D P < 0.001 and PA P < 0.001) (Fig. 2b). These results indicate that the facultative mutualistic relationship was not only retained in the derived coculture composed of wrinkled XR-Wa and PA-D, but was in fact reinforced as the productivity of both species was enhanced.
To confirm whether the increased productivity would remain stable for multiple generations, the different combinations were cultivated for 8 days (Fig. 3). This showed that the increased productivity of cocultures with XR-Wa (P < 0.001) and PA-D (P < 0.001), compared with cocultures with XR and PA, remained stable for the duration of these experiments. The percentage increase was 37% for XR-Wa with PA-D and 90% for PA-D with XR-Wa compared with XR with PA during the same time period.
Fig. 3.
Ancestral vs. wrinkled X. retroflexus: The facultative mutualistic interaction is stable over time. a + b Cell numbers of X. retroflexus in monoculture and coculture with P. amylolyticus after 8 days of growth. a XR was found in lower numbers in monocultures compared with the wrinkled XR-Wa (P < 0.001). b In the coculture with P. amylolyticus both XR and XR-Wa benefited significantly from cocultivation with P. amylolyticus (Both P < 0.001), resulting in significantly higher CFU for XR-Wa than XR (P < 0.001). c + d Cell numbers of P. amylolyticus in monoculture and coculture with X. retroflexus after 8 days of growth. c PA reached the same cell numbers as the derived PA-D (P = 0.934) in the monoculture. d The coculture with PA-D and XR-Wa reached higher biomass than the coculture with both ancestral strains (P < 1 × 10−5). Means and SEM are shown. P-values are from linear mixed-effect model. Data based on nine biological replicates from two separate experiments (n = 9)
Characterisation of wrinkled X. retroflexus colony variants
A high degree of variance was found in the proportions of the wrinkled X. retroflexus phenotypes among the biological replicates after 16 days of cocultivation (Suppl. Fig. 3). It was speculated that this could indicate difference in abilities of the individual wrinkled strains to establish in the coculture. Therefore, the growth and fitness of isolates with the wrinkled phenotype from two additional biological replicates were examined (denoted XR-Wb and XR-Wc). XR-Wb and XR-Wc were chosen because they were both present in smaller proportions than the efficient XR-Wa strain in the individual biological replicates. In addition, the colony morphology of XR-Wc was similar to XR-Wa, while the wrinkledness of XR-Wb was less pronounced (Suppl. Fig. 5). We speculated that this difference might influence the interaction in the coculture. The two phenotypic variants were, therefore, cocultured with both PA and the co-isolated derived P. amylolyticus isolates (denoted PA-Db and PA-Dc). The cell counts from cocultivating XR-Wc with either PA-Dc or PA were similar to those of XR-Wa in coculture, as these increased in coculture (with PA-Dc P = 0.006 and PA P < 0.001) (Suppl. Fig. 6a). Cell numbers of both P. amylolyticus variants were also higher when cocultured with XR-Wc (Suppl. Fig. 6b). Cell numbers of XR-Wb meanwhile were similar to those of the ancestral X. retroflexus (PA-Db P = 1.0 and PA P = 0.372) (Suppl. Fig. 6a). However, P. amylolyticus still benefited more from cocultivation with XR-Wb compared with the ancestral X. retroflexus (with PA-Db P = 0.007 or with PA P = 0.016) (Suppl. Fig. 6b). This shows that the different wrinkled variants (XR-Wa, XR-Wb and XR-Wc) did not benefit equally from interacting with P. amylolyticus (Fig. 3, Suppl. Fig. 6), which may explain the observed variance in the ratios of ancestral and wrinkled variants in the experimental replicates.
The wrinkled phenotypic variants of X. retroflexus were genome-sequenced to characterise their genotypes, along with three additional wrinkled X. retroflexus isolates originating from other replicates to provide a more substantial sample cohort (XR-Wd, XR-We and XR-Wf). Mutations were identified in all isolates with the wrinkled phenotype (Suppl. Table 3). All of the strains carried mutations in diguanylate cyclase (DGC)-encoding genes predicted to synthesise the second messenger cyclic di-GMP (c-di-GMP), based on the presence of GGDEF domains with conserved active sites. The mutations in c-di-GMP DGC genes were all found outside regions that encoded identified conserved protein domains and none were nonsense mutations. In all X. retroflexus XR-W strains, with the exception of XR-Wb, mutations were identified in CDS 2260, but each one was unique. The mutated c-di-GMP DGC-encoding gene of XR-Wb was CDS 300. Hence, none of the mutations were predicted to disrupt the active GGDEF site or terminate expression of the respectable DGC.
To examine if this mutation was linked to the observed phenotype, we constructed a knockout mutant in which the CDS 2260 gene was deleted from XR-Wa. This resulted in the colony morphology phenotype similar to the ancestral XR, which confirmed the linkage of the mutations to the shift in phenotype. As increased wrinkledness on Congo Red plates is often linked to increased biofilm forming ability and reduced motility, this was further assessed. When tested on swimming agar plates, XR-Wa exhibited reduced swimming activity compared with XR (Suppl. Fig. 7) as expected. Reduced motility was also observed for XR-Wc, but not for XR-Wb, further emphasising that the different mutations resulted in different phenotypes typically regulated via c-di-GMP. In accordance, we found that pellicle biofilms formed at the air–liquid interface of static cultures, were denser amongst XR-Wa compared with XR cultures (Suppl. Video). Interestingly, in the presence of P. amylolyticus, a thicker pellicle formed than when X. retroflexus was cultured alone. When coculturing both the wrinkled and ancestral X. retroflexus variants with P. amylolyticus (XR, XR-Wa, and PD), 76% of the pellicle was dominated by XR-Wa, compared with 24% for XR and only 0.1% for PA-D.
Wrinkled colony variants of X. retroflexus increased biofilm formation and surface attachment
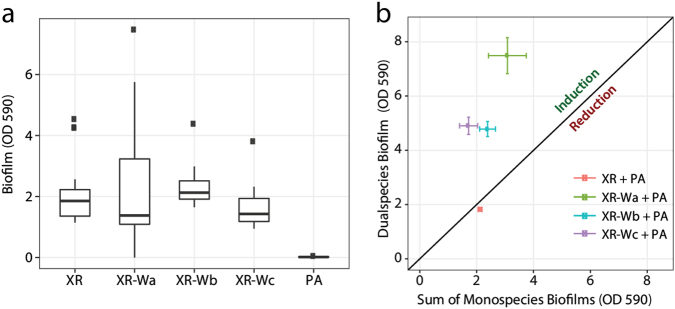
To examine the ability of the individual strain to form biofilm, the biofilm-forming capability of XR was compared with that of the wrinkled variant in a static biofilm assay where the biofilm formed on pegs and was quantified by crystal violet staining (Calgary device). Contrary to our expectations, no differences were found between monospecies biofilm biomass production of XR and the three wrinkled phenotypes (XR-Wa, XR-Wb and XR-Wc) (Fig. 4a).
Fig. 4.
Biofilm formation under static conditions. Biofilm formation under static conditions in the Calgary device using crystal violet to assess biofilm formation after 24 h of growth in TSB media. a Mono species biofilm formation. Boxplots are Tukey style. b Data points above the line represent cocultures that are performing better than the sum of their mono species biofilms. This biofilm induction was higher when XR-Wa, XR-Wb or XR-Wc was cocultured with PA than when XR was cocultured with PA (P < 0.001, P = 0.005 and P < 0.001, respectively). Data based on 20, 12, 11, and 11 (XR, XR-Wa, XR-Wb, and WR-Wc) biological replicates each with four technical replicates. Means and SEM are shown
XR and the wrinkled variants were also cocultured with PA in the Calgary device. A similar biofilm biomass was observed for the coculture with XR compared with the sum of the two monospecies cultures (P = 0.511) (Fig. 4b). However, overall, biofilm production of cocultures containing the wrinkled X. retroflexus variants was significantly enhanced, with increases ranging from 97 to 189% (P < 0.001 for XR-Wa, XR-Wb or XR-Wc with PA). We observed a biofilm inducing effect on XR-Wa, when adding spent supernatant of PA, where biofilm production equalled levels produced in the presence of PA cells (Suppl. Fig. 8). This effect was not observed when adding spent PA supernatant to the ancestral XR.
To further characterise the mutualistic interaction, cocultures of PA + XR-Wa and PA + XR-Wb were grown in shaken cultures. As pellicles do not form in shaken cultures, these functioned as biofilm-negative controls. Only PA gained from cocultivation with the wrinkled X. retroflexus strains in shaken cultures (P < 0.001 for XR-Wa and XR-Wb with PA) (Suppl. Fig. 9). These results correspond to those presented in Fig. 1 showing that mutualism between the two species only occurred under conditions that support the establishment of biofilms, either on a solid surface or at the air–liquid interface. In combination, these results suggest that the reinforced facultative mutualism observed between XR-Wa and PA was exclusively linked to the structured biofilm setting and at least partly mediated by diffusible compounds present in the PA supernatant.
Organisation of cells within biofilms
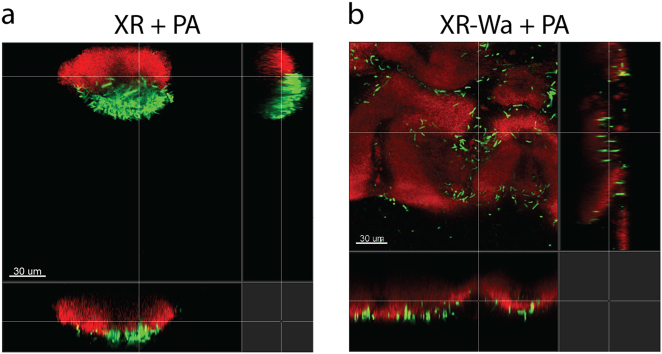
To evaluate the spatial organisation in the dual species biofilms, the BioFlux flow cell system was used to follow the organisation of cells during biofilm development. Monocultures of XR-Wa produced more biofilm compared with XR (Suppl. Fig. 10). PA was unable to form a stable biofilm on its own during the 48 h. However, PA resided stably in biofilms with XR over time by attaching onto XR as the biofilm developed (Fig. 5a). When PA was cocultivated in biofilms with XR-Wa, it established in the biofilm and positioned in the outer layers, resulting in enhanced biofilm production compared with that in cocultures with WR (Fig. 5b).
Fig. 5.
Biofilm produced after 48 h in BioFlux in TSB media. a Biofilm formation of XR (red) and PA (green). b Biofilm formation of wrinkled XR-Wa (red) and PA (green). PA is more aggregated in cocultures with XR (red) than in those containing XR-Wa (red)
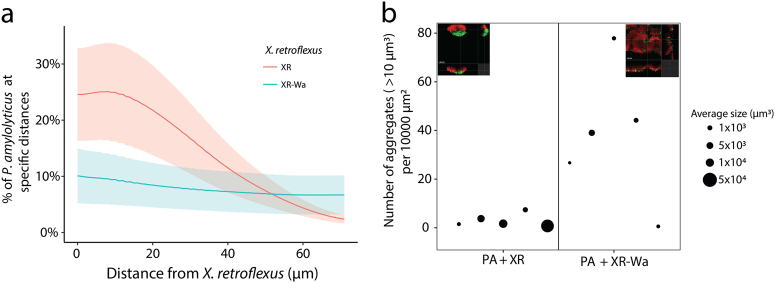
To analyse the spatial organisation of P. amylolyticus and the two versions of X. retroflexus, we identified specific parts of the biofilm where both species were present. These selected parts were then subjected to image analysis. Colocalisation analysis of the two species in biofilms revealed that PA was consistently closely associated with XR (Figs. 5a and 6a). When cocultured with XR-Wa, PA became more evenly distributed (few aggregates) in the outer layer of the biofilms (Figs. 5b and 6). With XR-Wa, PA produced small aggregates whereas with XR, PA produced larger aggregates. Interestingly, the structure of both versions of X. retroflexus remained unchanged between monospecies and dual species biofilms (Fig. 5 and Suppl. Fig. 9). Figure 6b shows the quantification of PA aggregate sizes in dual species biofilms with XR and XR-Wa. The more rugged biofilm structure of XR-Wa contained many small aggregates of PA positioned in the clefts of the structure. In contrast, few very large aggregates of PA were observed in biofilms with XR. The overall spatial structures of XR and XR-Wa biofilms were distinct, and PA exhibited changed organisation depending on the coculture partner. In both cases, PA resided in the outer layers of the biofilm. The wrinkled phenotypic variant XR-Wa did not outgrow PA (see Suppl. Fig. 11 for biovolume quantification). The increased biovolume of XR-Wa allowed for PA to settle more places compared with the relatively small amount with XR.
Fig. 6.
Biofilm formation in BioFlux. a Colocalisation of PA relative to X. retroflexus. Lines are means and shaded areas are SEM of the biological replica. b Aggregates of PA in cocultures with XR or XR-Wa in the BioFlux were estimated by joining neighbouring pixels in aggregates and then counting those larger than 10 μm3. Area of point sizes are scaled by average aggregate volume, one point corresponding to one biological replica. Data from five biological replicates, each with 1–5 (median 3) technical replicates
Discussion
Bacterial communities play various crucial roles in ecological, industrial and clinical settings. More often than not, they consist of multiple species. This study examined whether and how a facultative mutualistic relationship between two species changed as a response to prolonged coexistence (>50 generations).
The mutualistic interaction was maintained after 26 generations of cocultivation, as at this point both strains benefited from cocultivation. Furthermore, the study showed the emergence of wrinkled colony variants of X. retroflexus, which was unlikely to be due to genetic drift as a large number of cells (>10,000,000 cells) were subcultured at each transfer. As the new phenotypic variants emerged in both monocultures and cocultures, it was clear that interspecies interactions were not the direct cause of the observed intraspecific variation. However, P. amylolyticus did seem to affect the intraspecific diversification of X. retroflexus by applying an additional selective pressure, demonstrating that intraspecific variation impacts community composition.
Coexisting bacteria isolated from their natural habitats have previously been found to interact more synergistically in biofilm formation compared with isolates from different environments, indicating that coexistence affects adaptation processes in the community [45]. In line with this, interactions in complex bacterial communities have also been shown to become less competitive over time, resulting in increased productivity [46]. However, it has also been demonstrated that two species, with a one-way directed dependency, over time adapted towards an exploitative interaction since the biomass of one species was markedly reduced [47]. The present study, however, shows that adaptation can lead to reinforcement of an already positive interaction, as both the wrinkled X. retroflexus variant and P. amylolyticus benefited from the adaptation, reflected in higher biomass production of both strains (Fig. 3). In addition, P. amylolyticus in cocultures stabilised the presence of the wrinkled X. retroflexus (Fig. 4). This indicates that the presence of P. amylolyticus served to positively select for specific mutants of X. retroflexus, and P. amylolyticus itself benefited from the presence of the variants. In contrast, no selection for phenotypic changes in P. amylolyticus induced by cocultivation was observed within the timeframe of this study.
The results presented in this study emphasise the need for considering (i) intraspecific variation when examining interactions between species, and (ii) intraspecific trait variation in communities when exploring community assembly mechanisms. Further research is needed to completely understand the selection pressure maintaining intraspecific diversification in order to disclose its impact on interspecies coexistence. Previous research has shown that the diversification of microbial populations and productivity is affected by cooccurring species [23, 48]. This is in line with our observation of P. amylolyticus applying an additional selective pressure on X. retroflexus. However, these studies underline the importance of intraspecific diversification and the need to improve our understanding of this link if predictions are to be made about the species interplays in complex microbiomes. This general problem of determining the complex dynamics of species coexistence is a subject of increased attention, as it potentially predicts how species interactions impact community composition and ecosystem functioning.
The wrinkled colony variants identified in the present study have also been observed in other bacterial species, and examination has provided further insight into the genetic determinants of biofilm matrix and formation. Moreover, these variants have been found in both natural and clinical environments [49–51]. Variants displaying wrinkled colony morphologies are generally more tolerant to antimicrobials and other stressors, indicating that this can be a beneficial trait in some environments. In the wrinkled X. retroflexus variants, isolated and characterised in the present study, mutations were localised in genes predicted to synthesise c-di-GMP. An elevated level of c-di-GMP is typically associated with reduced motility, the wrinkled colony phenotype among gamma-proteobacteria, and increased biofilm formation and aggregation [16, 52, 53]. It was therefore hypothesised that mutations in the DGC genes facilitate enhanced DGC activity, thus inducing biofilm formation and reducing motility. The ability to reduce or enhance the production of exopolysaccharides by mutational changes in the c-di-GMP regulatory network has been suggested as a means of facilitating adaptation to new environments [54]. It also seems that adaptions that optimise the biofilm phenotype are generally important for the fitness of bacteria [55]. The observations presented here suggest that changes in matrix composition can facilitate coexistence in multispecies biofilm by modifying the spatial organisation. In this case, increased matrix formation by X. retroflexus led to a reinforced positive interaction for both species in the static assay. In addition, biofilm production was uniquely stimulated in the wrinkled variant of X. retroflexus when exposed to spent P. amylolyticus supernatant, suggesting a changed response by this variant to the presence of another species. This supports the postulate of Marchal et al. [56] that beneficial interactions between organisms select for attachment, providing a spatial structure that can be conducive for the evolution of active mutualistic interactions.
This study has provided evidence that a structured environment can support mutualistic interactions between facultative partners. We observed the emergence of a new variant that enhanced its own growth, but not at the expense of the other member. In fact, the facultative mutualistic interaction was maintained and even enhanced by the prevailing variant, facilitated by close interspecies association in the structured biofilm setting.
Data availability
Data and R scripts can be obtained from the authors upon reasonable requests.
Electronic supplementary material
Acknowledgements
The authors would like to thank Anette Løth and Jonas Oftebro-Svendsen for their assistance in the laboratory. This study was funded by grants from The Danish Council for Independent Research, ref no: DFF-1110-6571, and from the Villum Foundation, project no: 10098. We would also like to thank the three anonymous reviewers for their excellent feedback and suggestions.
Author contributions
HLR designed the study in collaboration with JR, JSM, JH, SJS and MB. HLR, JH, JR, MFA and JSM performed the experiments. JR, HLR and JSM analysed the data. HLR wrote the first draft of the manuscript, and all the authors contributed substantially to revisions.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0165-2) contains supplementary material, which is available to authorised users.
References
- 1.Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, et al. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–8. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitra RK, Nandy RK, Ramamurthy T, Bhattacharya SK, Yamasaki S, Shimada T, et al. Molecular characterisation of rough variants of Vibrio cholerae isolated from hospitalised patients with diarrhoea. J Med Microbiol. 2001;50:268–76. doi: 10.1099/0022-1317-50-3-268. [DOI] [PubMed] [Google Scholar]
- 3.Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 4.Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol Microbiol. 2004;53:497–515. doi: 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- 5.Beyhan S, Bilecen K, Salama SR, Casper-Lindley C, Yildiz FH. Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J Bacteriol. 2007;189:388–402. doi: 10.1128/JB.00981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burmølle M, Webb J, Rao D, Hansen L, Sørensen S, Kjelleberg S. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol. 2006;72:3916–23. doi: 10.1128/AEM.03022-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen LB, Ren D, Burmolle M, Sorensen SJ. Distinct gene expression profile of Xanthomonas retroflexus engaged in synergistic multispecies biofilm formation. ISME J. 2017;11:300–3. doi: 10.1038/ismej.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee KW, Periasamy S, Mukherjee M, Xie C, Kjelleberg S, Rice SA. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J. 2014;8:894–907. doi: 10.1038/ismej.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwering M, Song J, Louie M, Turner RJ, Ceri H. Multi-species biofilms defined from drinking water microorganisms provide increased protection against chlorine disinfection. Biofouling. 2013;29:917–28. doi: 10.1080/08927014.2013.816298. [DOI] [PubMed] [Google Scholar]
- 10.Madsen JS, Sorensen SJ, Burmolle M. Bacterial social interactions and the emergence of community-intrinsic properties. Curr Opin Microbiol. 2017;42:104–9. doi: 10.1016/j.mib.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 11.Elias S, Banin E. Multi-species biofilms: living with friendly neighbors. FEMS Microbiol Rev. 2012;36:990–1004. doi: 10.1111/j.1574-6976.2012.00325.x. [DOI] [PubMed] [Google Scholar]
- 12.Nadell CD, Drescher K, Foster KR. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol. 2016;14:589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 13.Rendueles O, Ghigo JM. Multi-species biofilms: how to avoid unfriendly neighbors. FEMS Microbiol Rev. 2012;36:972–89. doi: 10.1111/j.1574-6976.2012.00328.x. [DOI] [PubMed] [Google Scholar]
- 14.Rendueles O, Ghigo JM. Mechanisms of competition in biofilm communities. Microbiol Spectr. 2015;3. doi:10.1128/microbiolspec.mb-0009-2014. PMID: 26185066. [DOI] [PubMed]
- 15.Foster KR, Bell T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr Biol. 2012;22:1845–50. doi: 10.1016/j.cub.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Madsen JS, Lin YC, Squyres GR, Price-Whelan A, de Santiago Torio A, Song A, et al. Facultative control of matrix production optimizes competitive fitness in Pseudomonas aeruginosa PA14 biofilm models. Appl Environ Microbiol. 2015;81:8414–26. doi: 10.1128/AEM.02628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliveira NM, Martinez-Garcia E, Xavier J, Durham WM, Kolter R, Kim W, et al. Biofilm formation as a response to ecological competition. PLOS Biol. 2015;13:e1002191. doi: 10.1371/journal.pbio.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xavier JB, Foster KR. Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci USA. 2007;104:876–81. doi: 10.1073/pnas.0607651104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aschehoug ET, Callaway RM. Morphological variability in tree root architecture indirectly affects coexistence among competitors in the understory. Ecology. 2014;95:1731–6. doi: 10.1890/13-1749.1. [DOI] [PubMed] [Google Scholar]
- 20.Forsman A. Rethinking phenotypic plasticity and its consequences for individuals, populations and species. Heredity. 2015;115:276–84. doi: 10.1038/hdy.2014.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Libberton B, Coates RE, Brockhurst MA, Horsburgh MJ. Evidence that intraspecific trait variation among nasal bacteria shapes the distribution of Staphylococcus aureus. Infect Immun. 2014;82:3811–5. doi: 10.1128/IAI.02025-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolnick DI, Amarasekare P, Araujo MS, Burger R, Levine JM, Novak M, et al. Why intraspecific trait variation matters in community ecology. Trends Ecol Evol. 2011;26:183–92. doi: 10.1016/j.tree.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence D, Fiegna F, Behrends V, Bundy JG, Phillimore AB, Bell T, et al. Species interactions alter evolutionary responses to a novel environment. PLOS Biol. 2012;10:e1001330. doi: 10.1371/journal.pbio.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bronstein JL. The exploitation of mutualisms. Ecol Lett. 2001;4:277–87. doi: 10.1046/j.1461-0248.2001.00218.x. [DOI] [Google Scholar]
- 25.Holland JN, Bronstein JL. Mutualism. In: Fath BD, Jørgensen SE, editors. Encyclopedia of Ecology. Oxford: Academic Press; 2008. pp. 2485–91. [Google Scholar]
- 26.Bronstein JL. The evolution of facilitation and mutualism. J Ecol. 2009;97:1160–70. doi: 10.1111/j.1365-2745.2009.01566.x. [DOI] [Google Scholar]
- 27.Takimoto G, Suzuki K. Global stability of obligate mutualism in community modules with facultative mutualists. Oikos. 2016;125:535–40. doi: 10.1111/oik.02741. [DOI] [Google Scholar]
- 28.de la Cruz-Perera CI, Ren D, Blanchet M, Dendooven L, Marsch R, Sorensen SJ, et al. The ability of soil bacteria to receive the conjugative IncP1 plasmid, pKJK10, is different in a mixed community compared to single strains. FEMS Microbiol Lett. 2013;338:95–100. doi: 10.1111/1574-6968.12036. [DOI] [PubMed] [Google Scholar]
- 29.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucleic Acids Res. 2014;42:D206–214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi KH, Schweizer HP. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc. 2006;1:153–61. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 33.Timmusk S, Grantcharova N, Wagner EG. Paenibacillus polymyxa invades plant roots and forms biofilms. Appl Environ Microbiol. 2005;71:7292–7300. doi: 10.1128/AEM.71.11.7292-7300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosado A, Duarte GF, Seldin L. Optimization of electroporation procedure to transform B. polymyxa SCE2 and other nitrogen-fixing Bacillus. J Microbiol Methods. 1994;19:1–11. doi: 10.1016/0167-7012(94)90020-5. [DOI] [Google Scholar]
- 35.Sutcliffe JG. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/SQB.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- 36.Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc. 2015;10:1820–41. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi KH, Schweizer HP. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 2005;5:1–11. doi: 10.1186/1471-2180-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol. 1999;37:1771–6. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren D, Madsen JS, de la Cruz-Perera CI, Bergmark L, Sorensen SJ, Burmolle M. High-throughput screening of multispecies biofilm formation and quantitative PCR-based assessment of individual species proportions, useful for exploring interspecific bacterial interactions. Microb Ecol. 2014;68:146–54. doi: 10.1007/s00248-013-0315-z. [DOI] [PubMed] [Google Scholar]
- 40.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Team RCD. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 42.Liu W, Russel J, Roder HL, Madsen JS, Burmolle M, Sorensen SJ. Low-abundant species facilitates specific spatial organization that promotes multispecies biofilm formation. Environ Microbiol. 2017;19:2893–905. doi: 10.1111/1462-2920.13816. [DOI] [PubMed] [Google Scholar]
- 43.Allaire J (2012). RStudio: Integrated development environment for R. Boston, MA: RStudio Inc.
- 44.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. arXiv preprint arXiv:1406.5823 (2014).
- 45.Madsen JS, Røder HL, Russel J, Sørensen H, Burmølle M, Sørensen SJ. Coexistence facilitates interspecific biofilm formation in complex microbial communities. Environ Microbiol. 2016;18:2565–74. doi: 10.1111/1462-2920.13335. [DOI] [PubMed] [Google Scholar]
- 46.Rivett DW, Scheuerl T, Culbert CT, Mombrikotb SB, Johnstone E, Barraclough TG, et al. Resource-dependent attenuation of species interactions during bacterial succession. ISME J. 2016;10:2259–68. doi: 10.1038/ismej.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hansen SK, Rainey PB, Haagensen JA, Molin S. Evolution of species interactions in a biofilm community. Nature. 2007;445:533–6. doi: 10.1038/nature05514. [DOI] [PubMed] [Google Scholar]
- 48.Bailey SF, Dettman JR, Rainey PB, Kassen R. Competition both drives and impedes diversification in a model adaptive radiation. Proc Biol Sci. 2013;280:20131253. doi: 10.1098/rspb.2013.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rice EW, Johnson CJ, Clark RM, Fox KR, Reasoner DJ, Dunnigan ME, et al. Chlorine and survival of “rugose” Vibrio cholerae. Lancet. 1992;340:740. doi: 10.1016/0140-6736(92)92289-R. [DOI] [PubMed] [Google Scholar]
- 50.Beyhan S, Yildiz FH. Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signalling pathway. Mol Microbiol. 2007;63:995–1007. doi: 10.1111/j.1365-2958.2006.05568.x. [DOI] [PubMed] [Google Scholar]
- 51.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol. 2010;75:827–42. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–70. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x. [DOI] [PubMed] [Google Scholar]
- 53.Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH. Amyloid adhesins are abundant in natural biofilms. Environ Microbiol. 2007;9:3077–90. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- 54.Lind PA, Farr AD, Rainey PB. Evolutionary convergence in experimental Pseudomonas populations. ISME J. 2017;11:589–600. doi: 10.1038/ismej.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flynn KM, Dowell G, Johnson TM, Koestler BJ, Waters CM, Cooper VS. Evolution of ecological diversity in biofilms of Pseudomonas aeruginosa by altered cyclic diguanylate signaling. J Bacteriol. 2016;198:2608–18. doi: 10.1128/JB.00048-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marchal M, Goldschmidt F, Derksen-Muller SN, Panke S, Ackermann M, Johnson DR. A passive mutualistic interaction promotes the evolution of spatial structure within microbial populations. BMC Evol Biol. 2017;17:106. doi: 10.1186/s12862-017-0950-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and R scripts can be obtained from the authors upon reasonable requests.