Abstract Abstract
A new species of Phaeoacremonium, P.ovale (Togniniaceae), was isolated during a diversity study of freshwater fungi from Yunnan Province in China. Morphological and cultural studies of the fungus were carried out and its sexual and asexual morphs (holomorph) are introduced herein. This species is characterised by peculiar long-necked, semi-immersed ascomata with oval to ellipsoid ascospores and ellipsoid to ovoid conidia. Phylogenetic analyses of a combined TUB and ACT gene dataset revealed that strains of P.ovale constitute a strongly supported independent lineage and are related to P.griseo-olivaceum and P.africanum. The number of nucleotide differences, across the genes analysed, also supports establishment of P.ovale as a new species.
Keywords: 1 new species, Togniniales , Sordariomycetes , Morphology, Phylogeny
Introduction
Lignicolous freshwater fungi are important in nutrient recycling (Hyde et al. 2016). A number of taxonomic studies have focused on the diversity of such fungi in the South East Asian region and these investigations have reported a number of novel species (e.g. Jeewon et al. 2003; Cabanela et al. 2007; Zhang et al. 2008; Luo et al. 2018). In this study, we report a new species of Phaeoacremonium isolated from decaying wood from a stream in Yunnan Province, China.
Phaeoacremonium (= Togninia), introduced by Crous et al. (1996), is typified by P.parasiticum and it belongs to Togniniaceae (Gramaje et al. 2015). Phaeoacremonium was reported to be the asexual morph of Togninia (Mostert et al. 2003, 2006a; Pascoe et al. 2004). Gramaje et al. (2015) proposed Phaeoacremonium over Togninia as the correct name based on common usage and this has been listed in Réblová et al. (2016) and followed in Wijayawardene et al. (2018). The species are basically characterised by black ascomata with a long neck and clavate to cylindrical asci with oval to ellipsoid, hyaline ascospores and straight or flexuous mononematous conidiophores with oval to reniform phialo-conidia (Marin-Felix et al. 2018; Spies et al. 2018).
Most species of Phaeoacremonium are plant or/and human pathogens and some have been recorded on arthropods or in soil (Groenewald et al. 2001; Guarro et al. 2003; Hemashettar et al. 2006; Mostert et al. 2006a; Damm et al. 2008; Gramaje et al. 2015) while others are causal agents of Petri disease and esca of grapevines (Pascoe et al. 2004; Rooney-Latham et al. 2005a; Mostert et al. 2006b). Phaeoacremonium species can also infect a wide range of woody hosts, such as cherry, apricot, olive and peach trees (Rumbos 1986; Di Marco et al. 2004; Kubátová et al. 2004). Recent studies have reported the importance of Phaeoacremonium species in causing brown wood streaking of Olea spp. and Prunus spp. (Mostert et al. 2006b; Damm et al. 2008; Gramaje et al. 2012; Nigro et al. 2013; Olmo et al. 2014; Carlucci et al. 2015). Rooney-Latham et al. (2004, 2005a, b) reported that, in the presence of water, spores in some Phaeoacremonium species are forcibly discharged from perithecia through the long neck and exit the ostiole to be dispersed by wind, rain or insects in order to colonise other substrates. Recently Hu et al. (2012) introduced a freshwater inhabiting species, Phaeoacremoniumaquaticum (= Togniniaaquatica).
Species of Togniniaceae have been reported to colonise substrates in different types of habitats and recent taxonomic studies have revealed additional new species (Gramaje et al. 2015). We have been studying fungi along a north-south gradient in the Asian region (Hyde et al. 2016) and, in this study, we report on two collections of Phaeoacremonium from China. The aim here is to characterise these two strains as one novel species based on morphology as well as to investigate their phylogenetic affinities with previously known Togniniaceae species based on partial TUB and ACT genes.
Materials and methods
Sample collection, morphological studies and isolation
Submerged dead wood was collected from Baoshan, Yunnan Province in China in October 2016, brought to the laboratory in zip lock plastic bags and treated in the laboratory following procedures detailed in Luo et al. (2018). Fruiting bodies were found growing on decaying wood in a sterile plastic box after two weeks of incubation and the fungus was subsequently isolated based on the method of Chomnunti et al. (2014). Specimens were examined by a Motic SMZ 168 stereomicroscope. Micromorphological characters were examined using a Nikon ECLIPSE 80i compound microscope and images were captured with a Canon EOS 600D digital camera. Identification of colours was based on Ridgway (1912). The Taro soft Image Framework programme version 0.9.0.7 was used for measurements. Single spores were isolated and grown on water agar (WA) and potato dextrose agar (PDA) media. Ascospores germinated on PDA within 1 week. The colonies were transferred to WA and PDA to promote sporulation (sporulation occurred after 30 days in PDA). The cultures were checked 2 to 3 times per week and all procedures were performed in a sterile environment and at room temperature. The morphological characters of the asexual morph were examined after sporulation. Specimens are deposited in the Kunming Institute of Botany, Academia Sinica (KUN) and duplicated in Mae Fah Luang University (MFLU) Herbarium, Chiang Rai, Thailand. Facesoffungi numbers (FoF) (http://www.facesoffungi.org/) were obtained as stated in Jayasiri et al. (2015) and Index Fungorum numbers (IF) (http://www.indexfungorum.org/names/IndexFungorumRegisterName.asp).
DNA extraction, PCR amplification and sequencing
Total genomic DNA was extracted from mycelium using a Trelief Plant Genomic DNA Kit following the instructions of the manufacturer. The genomic DNA was amplified by using polymerase chain reaction (PCR) in a 25 μl reaction mixture. Partial regions of the beta-tubulin (TUB) and Actin (ACT) gene were amplified using the primer pairs T1 (O’Donnell and Cigelnik 1997) and Bt2b (Glass and Donaldson 1995), ACT-513F and ACT-783R (Carbone and Kohn 1999), respectively. The internal transcribed spacers (ITS) regions of the rDNA (ITS1-5.8S-ITS2) were also amplified using primer pairs ITS5 and ITS4 (White et al. 1990) but no further analyses were done on these due to lack of sequence data. The PCR conditions for these regions were as follows: an initial denaturation at 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 30 sec, annealing at 51 °C (TUB) or 60 °C (ACT) or 55 °C (ITS) for 50 sec and extension at 72 °C for 1 min, with a final extension at 72 °C for 10 min. PCR products were then sequenced with the primers mentioned above by a commercial sequencing provider (Tsingke Company, Beijing, P.R. China).
Phylogenetic analysis
The quality of the amplified nucleotide sequences was checked and combined by SeqMan version 7.1.0 (44.1) and Finch TV version 1.4.0 (www.geospiza.com). Sequences used by Marin-Felix et al. (2018), Spies et al. (2018) and the closest matches for our strains were retrieved from the National Center for Biotechnology Information (NCBI) by nucleotide BLAST. Sequences were aligned in MAFFT v. 7.310 (http://mafft.cbrc.jp/alignment/server/index.html) (Katoh and Standley 2016) and manually corrected in Bioedit 7.0.9.0 (Hall 1999).
The phylogenetic analyses of combined gene regions (TUB and ACT) were performed using maximum-likelihood (ML) and Bayesian Inference (BI) methods. The best-fit model (GTR+G+I) was obtained using jModelTest 2.1.10 under the Akaike Information Criterion (AIC) calculations (Darriba et al. 2012). The ML analysis was enforced with RAxML-HPC v.8 on XSEDE (Stamatakis 2014; Miller et al. 2015) with 1000 rapid bootstrap replicates. Bayesian inference was implemented by MrBayes v. 3.0b4 (Ronquist and Huelsenbeck 2003). Four simultaneous Markov chains were run for 5,000,000 generations sampling one tree every 1000th generations and other criteria as outlined by Hongsanan et al. (2017). The temperature value was lowered to 0.15, burn-in was set to 0.25. Gaps were treated as missing data with no differential weighting of transitions against transversions and the partition homogeneity test was performed to assess whether datasets from different genes were congruent. Phylogenetic trees were viewed with FigTree v1.4.0 (http:// tree.bio.ed.ac.uk/software/figtree/) and processed by Adobe Illustrator CS5. Alignment and trees were deposited in TreeBASE (submission ID: 22810). The nucleotide sequence data of the new taxon have been deposited in GenBank (Table 1).
Table 1.
Strains and GenBank accession numbers of the isolates used in this study. Isolates from this study are marked with asterisk (*) and the type strains are in bold.
| Species | Voucher/Culture | GenBank accession number | |
|---|---|---|---|
| TUB | ACT | ||
| Phaeoacremonium africanum | CBS 120863 | EU128100 | EU128142 |
| Phaeoacremonium album | CBS 142688 | KY906885 | KY906884 |
| Phaeoacremonium alvesii | CBS 110034 | AY579301 | AY579234 |
| Phaeoacremonium alvesii | CBS 729.97 | AY579302 | AY579235 |
| Phaeoacremonium amstelodamense | CBS 110627 | AY579295 | AY579228 |
| Phaeoacremonium amygdalinum | CBS 128570 | JN191307 | JN191303 |
| Phaeoacremonium amygdalinum | CBS H-20507 | JN191305 | JN191301 |
| Phaeoacremonium amygdalinum | CBS H-20508 | JN191306 | JN191302 |
| Phaeoacremonium angustius | CBS 114992 | DQ173104 | DQ173127 |
| Phaeoacremonium angustius | CBS 114991 | DQ173103 | DQ173126 |
| Phaeoacremonium argentinense | CBS 777.83 | DQ173108 | DQ173135 |
| Phaeoacremonium armeniacum | ICMP 17421 | EU596526 | EU595463 |
| Phaeoacremonium aureum | CBS 142691 | KY906657 | KY906656 |
| Phaeoacremonium australiense | CBS 113589 | AY579296 | AY579229 |
| Phaeoacremonium australiense | CBS 113592 | AY579297 | AY579230 |
| Phaeoacremonium austroafricanum | CBS 112949 | DQ173099 | DQ173122 |
| Phaeoacremonium austroafricanum | CBS 114994 | DQ173102 | DQ173125 |
| Phaeoacremonium austroafricanum | CBS 114993 | DQ173101 | DQ173124 |
| Phaeoacremonium bibendum | CBS 142694 | KY906759 | KY906758 |
| Phaeoacremoniumcanadens e | PARC327 | KF764651 | KF764499 |
| Phaeoacremonium cf. mortoniae | ICMP 18088 | HM116767 | HM116773 |
| Phaeoacremonium cinereum | CBS 123909 | FJ517161 | FJ517153 |
| Phaeoacremonium cinereum | CBS H-20215 | FJ517160 | FJ517152 |
| Phaeoacremonium cinereum | CBS H-20213 | FJ517158 | FJ517150 |
| Phaeoacremonium croatiense | CBS 123037 | EU863482 | EU863514 |
| Phaeoacremonium fraxinopennsylvanicum | CBS 101585 | AF246809 | DQ173137 |
| Phaeoacremonium fraxinopennsylvanicum | CBS 110212 | DQ173109 | DQ173136 |
| Phaeoacremonium fuscum | CBS 120856 | EU128098 | EU128141 |
| Phaeoacremonium gamsii | CBS 142712 | KY906741 | KY906740 |
| Phaeoacremonium geminum | CBS 142713 | KY906649 | KY906648 |
| Phaeoacremonium globosum | ICMP 16988 | EU596525 | EU595466 |
| Phaeoacremonium globosum | ICMP 17038 | EU596521 | EU595465 |
| Phaeoacremonium globosum | ICMP 16987 | EU596527 | EU595459 |
| Phaeoacremonium griseo-olivaceum | CBS 120857 | EU128097 | EU128139 |
| Phaeoacremonium griseorubrum | CBS 111657 | AY579294 | AY579227 |
| Phaeoacremonium griseorubrum | CBS 566.97 | AF246801 | AY579226 |
| Phaeoacremonium hispanicum | CBS 123910 | FJ517164 | FJ517156 |
| Phaeoacremonium hungaricum | CBS 123036 | EU863483 | EU863515 |
| Phaeoacremonium inflatipes | CBS 391.71 | AF246805 | AY579259 |
| Phaeoacremonium inflatipes | CBS 113273 | AY579323 | AY579260 |
| Phaeoacremonium iranianum | CBS 101357 | DQ173097 | DQ173120 |
| Phaeoacremonium iranianum | CBS 117114 | DQ173098 | DQ173121 |
| Phaeoacremonium italicum | CBS 137763 | KJ534074 | KJ534046 |
| Phaeoacremonium italicum | CBS 137764 | KJ534075 | KJ534047 |
| Phaeoacremonium italicum | CBS H-21638 | KJ534076 | KJ534048 |
| Phaeoacremonium junior | CBS 142697 | KY906709 | KY906708 |
| Phaeoacremonium krajdenii | CBS 110118 | AY579324 | AY579261 |
| Phaeoacremonium krajdenii | CBS 109479 | AY579330 | AY579267 |
| Phaeoacremonium longicollarum | CBS 142699 | KY906689 | KY906688 |
| Phaeoacremonium luteum | CBS 137497 | KF823800 | KF835406 |
| Phaeoacremonium meliae | CBS 142710 | KY906825 | KY906824 |
| Phaeoacremonium minimum | CBS 246.91 | AF246811 | AY735497 |
| Phaeoacremonium minimum | CBS 100397 | AF246806 | AY735498 |
| Phaeoacremonium mortoniae | CBS 211.97 | AF246810 | |
| Phaeoacremonium nordesticola | CMM4312 | KY030807 | KY030803 |
| Phaeoacremonium novae-zealandiae | CBS 110156 | DQ173110 | DQ173139 |
| Phaeoacremonium novae-zealandiae | CBS 110157 | DQ173111 | DQ173140 |
| Phaeoacremonium occidentale | ICMP 17037 | EU596524 | EU595460 |
| Phaeoacremonium oleae | CBS 142704 | KY906937 | KY906936 |
| *Phaeoacremoniumovale | KUMCC 17-0145 | MH395327 | MH395325 |
| *Phaeoacremoniumovale | KUMCC 18-0018 | MH395328 | MH395326 |
| Phaeoacremonium pallidum | CBS 120862 | EU128103 | EU128144 |
| Phaeoacremonium parasiticum | CBS 860.73 | AF246803 | AY579253 |
| Phaeoacremonium parasiticum | CBS 113585 | AY579307 | AY579241 |
| Phaeoacremonium parasiticum | CBS 514.82 | AY579306 | AY579240 |
| Phaeoacremonium paululum | CBS 142705 | KY906881 | KY906880 |
| Phaeoacremonium pravum | CBS 142686 | KY084246 | KY084248 |
| Phaeoacremonium proliferatum | CBS 142706 | KY906903 | KY906902 |
| Phaeoacremonium prunicola | CBS 120858 | EU128095 | EU128137 |
| Phaeoacremonium prunicola | CBS 120858 | EU128096 | EU128138 |
| Phaeoacremonium pseudopanacis | CPC 28694 | KY173609 | KY173569 |
| Phaeoacremonium roseum | PARC273 | KF764658 | KF764506 |
| Phaeoacremonium rosicola | CBS 142708 | KY906831 | KY906830 |
| Phaeoacremonium rubrigenum | CBS 498.94 | AF246802 | AY579238 |
| Phaeoacremonium rubrigenum | CBS 112046 | AY579305 | AY579239 |
| Phaeoacremonium santali | CBS 137498 | KF823797 | KF835403 |
| Phaeoacremonium scolyti | CBS 113597 | AF246800 | AY579224 |
| Phaeoacremonium scolyti | CBS 113593 | AY579293 | AY579225 |
| Phaeoacremonium scolyti | CBS 112585 | AY579292 | AY579223 |
| Phaeoacremonium sicilianum | CBS 123034 | EU863488 | EU863520 |
| Phaeoacremonium sicilianum | CBS 123035 | EU863489 | EU863521 |
| Phaeoacremonium sp. | KMU 8592 | AB986584 | AB986583 |
| Phaeoacremonium spadicum | CBS 142711 | KY906839 | KY906838 |
| Phaeoacremonium sphinctrophorum | CBS 337.90 | DQ173113 | DQ173142 |
| Phaeoacremonium sphinctrophorum | CBS 694.88 | DQ173114 | DQ173143 |
| Phaeoacremonium subulatum | CBS 113584 | AY579298 | AY579231 |
| Phaeoacremonium subulatum | CBS 113587 | AY579299 | AY579232 |
| Phaeoacremonium tardicrescens | CBS 110573 | AY579300 | AY579233 |
| Phaeoacremonium tectonae | MFLUCC 13-0707 | KT285563 | KT285555 |
| Phaeoacremonium tectonae | MFLUCC 14-1131 | KT285570 | KT285562 |
| Phaeoacremonium theobromatis | CBS 111586 | DQ173106 | DQ173132 |
| Phaeoacremonium tuscanicum | CBS 123033 | EU863458 | EU863490 |
| Phaeoacremonium venezuelense | CBS 651.85 | AY579320 | AY579256 |
| Phaeoacremonium venezuelense | CBS 110119 | AY579318 | AY579254 |
| Phaeoacremonium venezuelense | CBS 113595 | AY579319 | AY579255 |
| Phaeoacremonium vibratile | CBS 117115 | DQ649063 | DQ649064 |
| Phaeoacremonium viticola | CBS 113065 | DQ173105 | DQ173128 |
| Phaeoacremonium viticola | CBS 101737 | AF246817 | DQ173129 |
| Pleurostomophora richardsiae | CBS 270.33 | AY579334 | AY579271 |
| Wuestneia molokaiensis | CBS 114877 | AY579335 | AY579272 |
Abbreviations: CBS: CBS-KNAW Collections, Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CMM: Culture Collection of Phytopathogenic fungi “Prof. Maria Menezes”; CPC: Culture collection of Pedro Crous, housed at CBS; HKUCC: The University of Hong Kong Culture Collection; ICMP: The International Collection of Microorganisms from Plants; KMU: Kanazawa Medical University herbarium; MFLU: Mae Fah Luang University herbarium, MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; PARC: Pacific Agri-Food Research Centre.
Results
Phylogenetic analyses
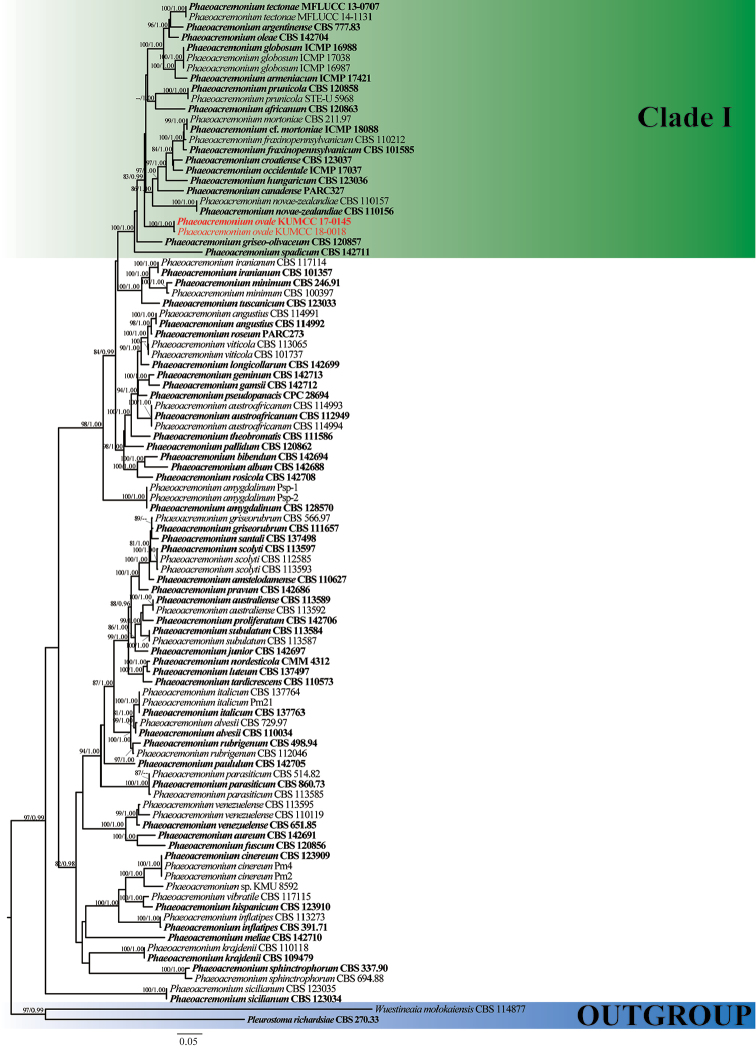
The combined TUB and ACT sequence dataset comprised 98 strains of Phaeoacremonium. The tree was rooted with Pleurostomarichardsiae (CBS 270.33) and Wuestineaiamolokaiensis (CBS 114877). The alignment comprised 947 total characters including gaps (TUB: 646bp; ACT: 301bp). ML and BI analyses yielded trees which were topologically congruent in terms of species groupings. RAxML analysis yielded a best scoring tree with a final optimisation likelihood value of -15310.399369 (Fig. 1). In the phylogenetic tree, two strains of Phaeoacremoniumovale forms a well-supported independent subclade (100%, ML/1.00, PP) and closely related to other Phaeoacremonium species in Clade I (83%, ML/0.99, PP).
Figure 1.
Maximum likelihood phylogenetic tree generated from analysis of a combined TUB and ACT sequences dataset for 98 taxa of Togniniaceae. Pleurostomarichardsiae (CBS 270.33) and Wuestineaiamolokaiensis (CBS 114877) are the outgroup taxa. ML support values greater than 70% (BSML, left) and Bayesian posterior probabilities greater than 0.90 (BYPP, right) are indicated above the nodes. The strain numbers are noted after the species names. Ex-type strains are indicated in bold. Isolates from this study are indicated in red.
Taxonomy
Phaeoacremonium ovale
S.K. Huang, R. Jeewon & K.D. Hyde sp. nov.
Figure 2.
Phaeoacremoniumovale (HKAS99550, holotype). a Substrate b, c Ascoma on host d Squashed neck e Ascoma in vertical section fPeridiumg Asci surrounded by paraphyses h Asci i Septate paraphyses j–m Asci with ascospores n Germinating ascospores. Note: Fig. i stained in Congo red reagent, fig l stained in Melzer’s reagent. Scale bars: 500 µm (c); 200 µm (d); 100 µm (e); 50 µm (f, i); 30 µm (n); 20 µm (g–h); 10 µm (j–m)
Type.
CHINA, Yunnan Province, Baoshan, stream along the roadside; saprobic on dead wood, 21 December 2016; Huang S.K. (KUN HKAS99550, holotype; MFLU MFLU18-1076, isotype); ex-type living culture (KUMCC 17-0145; KUMCC 18-0018). GenBank no. (ITS: MH399732, TUB: MH395327, ACT: MH395325; ITS: MH399733, TUB: MH395328, ACT: MH395326)
Etymology.
The name ovale refers to the oval shaped ascospores.
Description.
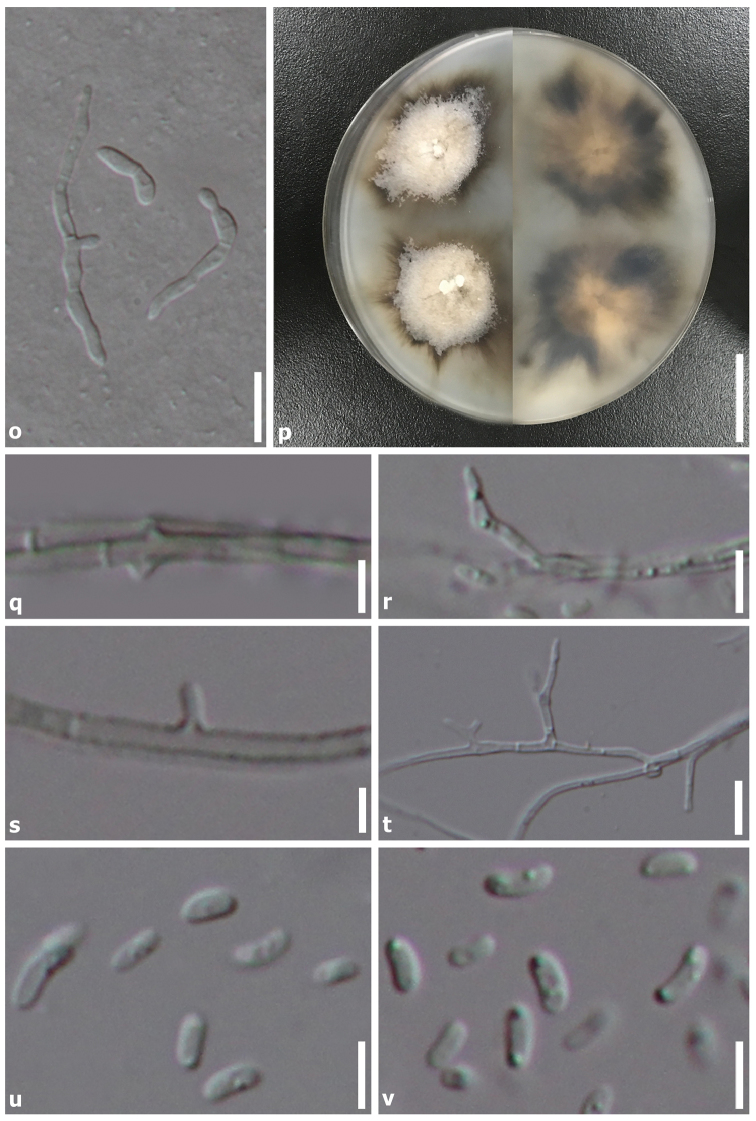
Sexual morph: Ascomata 225–300 μm (n = 5), on wood, perithecial, solitary, semi-immersed, unilocular, subglobose to globose, black, ostiolate, with ostiolar neck erumpent through bark of host when mature. Neck 445–645 × 35–45 μm (x̄ = 530 × 40 μm, n = 5), centrally ostiolate, contorted, lined with hyaline periphyses. Peridium 17–40 μm diam., membranous, composed of dark brown to hyaline cells of textura angularis. Hamathecium composed of 2–6 μm wide, hyaline, septate paraphyses, slightly constricted at septa and gradually narrowed towards apex. Asci 11–20 × 3–6 μm (x̄ = 15.5 × 5 μm, n = 30), 8-spored, unitunicate, clavate, with short pedicel, apically rounded. Ascospores 3–5 × 1.5–3 μm (x̄ = 3.5 × 2 μm, n = 50), bi-seriate, hyaline, oval to ellipsoid, aseptate, smooth-walled, rounded at the ends. Asexual morph: Mycelium on culture, partly superficial, composed of septate, branched, hyaline, rarely verrucose, hyphae 1.5–3 μm diam., rarely with adelophialides. Conidiophores usually arising from hyaline hyphae, mononematous, unbranched, occasionally constricted at basal septum, hyaline. Phialides 8–15 × 2–4 μm (x̄ = 9.5 × 3 μm, n = 20), terminal, monophialidic, elongate-ampulliform and attenuated at base. Conidia 2.5–6 × 1–2.5 µm (x̄ = 4 × 2 μm, n = 30), hyaline, ellipsoid to ovoid, aseptate.
Culture characteristics.
Ascospore germinating on PDA within 1 week at 23°C, germ tubes produced from ends. Colonies growing on PDA, reaching 2 cm diam. and sporulating after 30 days. Colonies semi-immersed to superficial, irregular in shape, flat, slightly raised, with undulate edge, slightly rough on surface, cottony to fairly fluffy, colony from above, greyish-brown (5F3–5, Ridgway 1912) at the margin, initially write to cream (5A1–3) in the centre, becoming dark brown (5F7–8) at the margin, orange-white (5B1–3) at the centre; from below, initially, greyish-brown at the margin, white at the centre, becoming dark brown at the margin, orange-white at the centre, producing brown pigmentation in agar.
Discussion
Phaeoacremonium is currently accommodated in the monogeneric family Togniniaceae (Wijayawardene et al. 2018). To date, 65 species are accepted in this genus (Mostert et al. 2006b; Gramaje et al. 2015; Marin-Felix et al. 2018; Spies et al. 2018). While most of the species are commonly isolated as asexual morphs, some taxa have been recovered in their sexual morph state, viz. Phaeoacremoniumaquaticum (= Togniniaaquatica), P.viticola (= T.viticola), P.novae-zealandiae (= T.novae-zealandiae) (Hausner et al. 1992; Mostert et al. 2006a; Hu et al. 2012).
In this study, we introduce a novel taxon of Phaeoacremonium from dead wood collected in a stream in the Yunnan Province, China and describe its sexual and asexual morph. Examination of morphological characters reveal that our species is sufficiently distinct from extant species to establish it as a new species. Analyses of the combined DNA sequence dataset from partial TUB and ACT genes also support that this taxon is a Phaeoacremonium species and phylogenetically distinct from other species (Fig. 1). The two strains of P.ovale constitute a strongly supported independent lineage close to other species as depicted in Clade I. Phylogeny also reveals a close relationship to P.griseo-olivaceum, but with low support. To further support P.ovale as a new species, we compared nucleotide differences with other related species as recommended by Jeewon and Hyde (2016). Comparison of the 533 nucleotides across the TUB region reveals 43 bp (10%) differences, 256 bp of the ACT region reveals 22 bp (8.5%) differences and 517 bp of the ITS region reveals 4 bp (1%) differences compared to P.griseo-olivaceum (CBS 120857). Examination of the TUB region reveals 59 bp (11%) difference compared to P.africanum (CBS 120863) while the ACT region reveals 19 bp (7%) and ITS region reveals 17 bp (3%) differences, but the latter clusters in a different subclade in our phylogeny and is therefore considered distinct. There are also some morphological similarities between P.ovale and P.africanum in terms of black ascomata with a long neck, clavate asci and small, oval to ellipsoid ascospores in sexual morph and ellipsoid to ovoid, aseptate conidia in asexual morph (Damm et al. 2008). Despite a morphological resemblance to P.africanum and close relationship to P.griseo-olivaceum, there are other differences across these species. Phaeoacremoniumovale was collected from an aquatic habitat and from dead wood in China whereas the former two species were collected from Prunus spp. in South Africa (Damm et al. 2008). In addition, conidial size of P.africanum and P.griseo-olivaceum are 5–12 × 1.5–2 µm and 5–8 × 1.5–2 µm, whereas conidia of P.ovale measure 2.5–6 × 1–2.5 µm (Damm et al. 2008; Fig. 3). No sequence data of the TUB and ACT gene are available for P.aquaticum and P.leptorrhynchum and therefore we provide ITS sequences of our strains and compare them with those two species. Comparison of ITS regions reveals 61 bp (12%) differences with P.aquaticum (IFRDCC 3035) and 11 bp (2%) differences with P.leptorrhynchum (UAMH9590). In addition, our new species is also morphologically different from them. Phaeoacremoniumovale is morphologically different as ascospores of P.aquaticum and P.leptorrhynchum are reniform (ascospores of P.ovale are oval/ellipsoid) and measure 5–6 × 1–1.5 µm and 7–10 × 1–1.5 µm, respectively. Phaeoacremoniuminconspicuum as described by Gramaje et al. (2015) also appears morphologically similar to P.ovale in terms of clavate asci and hyaline, aseptate ascospores (Eriksson and Yue 1990), but could not be included in our analyses as DNA sequences are unavailable. However, the ascospore shape and size of P.inconspicuum is different (allantoid, measuring 7–10 × 1.5–2 µm) (Eriksson and Yue 1990; Réblová 2011).
Figure 3.
Phaeoacremoniumovale (HKAS99550, holotype). o Germinating ascospores, p 7 weeks of culture plate (above, left/reverse, right), q Mycelium with adelophialides r–t Branched conidiophores u–vConidia. Scale bars: 20 mm (p); 20 µm (o); 10 µm (r, t); 5 µm (q, s, u–v).
Supplementary Material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31760014), the Science and Technology Foundation of Guizhou Province (No. [2016]2863 & [2017]5788). Dr. Qi Zhao is acknowledged for his support and thanks to Dr. Sajeewa Maharachchikumbura for his valuable help in phylogenetic analyses. Shaun Pennycook is thanked for checking and correcting the Latin name.
Citation
Huang S-K, Jeewon R, Hyde KD, Bhat DJ, Chomnunti P, Wen T-C (2018) Beta-tubulin and Actin gene phylogeny supports Phaeoacremonium ovale as a new species from freshwater habitats in China. MycoKeys 41: 1–15. https://doi.org/10.3897/mycokeys.41.27536
References
- Cabanela MV, Jeewon R, Hyde KD. (2007) Morphotaxonomy and phylogeny of Paoayensislignicola gen et sp. nov. (ascomycetes) from submerged wood in Paoay Lake, Ilocos Norte, the Philippines. Cryptogamie, Mycologie 28: 301–310. [Google Scholar]
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91(3): 553–556. 10.2307/3761358 [DOI] [Google Scholar]
- Carlucci A, Lops F, Cibelli F, Raimondo ML. (2015) Phaeoacremonium species associated with olive wilt and decline in southern Italy. European Journal of Plant Pathology 141(4): 717–729. 10.1007/s10658-014-0573-8 [DOI] [Google Scholar]
- Chomnunti P, Hongsanan S, Aguirre-Hudson B, Tian Q, Peršoh D, Dhami MK, Alias AS, Xu JC, Liu XZ, Stadler M, Hyde KD. (2014) The sooty moulds. Fungal Diversity 66: 1–36. 10.1007/s13225-014-0278-5 [DOI] [Google Scholar]
- Crous PW, Gams W, Wingfield MJ, van Wyk PS. (1996) Phaeoacremonium gen. nov. associated with wilt and decline diseases of woody hosts and human infections. Mycologia 88(5): 786–796. 10.1080/00275514.1996.12026716 [DOI] [Google Scholar]
- Damm U, Mostert L, Crous PW, Fourie PH. (2008) Novel Phaeoacremonium species associated with necrotic wood of Prunus trees. Persoonia 20: 87–102. 10.3767/003158508X324227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9(8): 772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed]
- Di Marco S, Calzarano F, Osti F, Mazzullo A. (2004) Pathogenicity of fungi associated with a decay of kiwifruit. Australasian Plant Pathology 33: 337–342. 10.1071/AP04024 [DOI] [Google Scholar]
- Eriksson O, Yue JZ. (1990) Notes on bambusicolous pyrenomycetes. No.s 1-10. Mycotaxon 38: 201–220. [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved gene from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramaje D, García-Jiménez J, Armengol J. (2012) Fungal trunk pathogens in Spanish grapevine nurseries: a survey of current nursery management practices in Spain. Phytopathologia Mediterranea 51: 411–412. [Google Scholar]
- Gramaje D, Mostert L, Groenewald JZ, Crous PW. (2015) Phaeoacremonium: From esca disease to phaeohyphomycosis. Fungal Biology 119: 759–783. 10.1016/j.funbio.2015.06.004 [DOI] [PubMed] [Google Scholar]
- Groenewald M, Kang JC, Crous PW, Gams W. (2001) ITS and β-tubulin phylogeny of Phaeoacremonium and Phaeomoniella species. Mycological Research 105: 651–657. 10.1017/S0953756201004282 [DOI] [Google Scholar]
- Guarro J, Alves SH, Gené J, Grazziotin NA, Muzzuco R, Dalmagro C, Capilla J, Zaror L, Mayayo E. (2003) Two cases of subcutaneous infection due to Phaeoacremonium spp. Journal of Clinical Microbiology 41: 1332–1336. 10.1128/JCM.41.3.1332-1336.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hausner G, Eyjólfsdóttir GG, Reid J, Klassen GR. (1992) Two additional species of the genus Togninia. Canadian Journal of Botany 70(4): 724–734. 10.1139/b92-093 [DOI] [Google Scholar]
- Hemashettar BM, Siddaramappa B, Munjunathaswamy BS, Pangi AS, Pattan J, Andrade AT, Padhye AA, Mostert L, Summerbell RC. (2006) Phaeoacremoniumkrajdenii, a cause of white grain eumycetoma. Journal of Clinical Microbiology 44: 4619–4622. 10.1128/JCM.01019-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongsanan S, Maharachchikumbura SSN, Hyde KD, Samarakoon MC, Jeewon R, Zhao Q, Al-Sadi AM, Bahkali AH. (2017) An updated phylogeny of Sordariomycetes based on phylogenetic and molecular clock evidence. Fungal Diversity 84: 25–41. 10.1007/s13225-017-0384-2 [DOI] [Google Scholar]
- Hu DM, Cai L, Hyde KD. (2012) Three new ascomycetes from freshwater in China. Mycologia 104(6): 1478–1489. 10.3852/11-430 [DOI] [PubMed] [Google Scholar]
- Hyde KD, Fryar S, Tian Q, Bahkali AH, Xu JC. (2016) Lignicolous freshwater fungi along a north-south latitudinal gradient in the Asian/Australian region; can we predict the impact of global warming on biodiversity and function? Fungal Ecology 19: 190–200. 10.1016/j.funeco.2015.07.002 [DOI]
- Jayasiri SC, Hyde KD, Ariyawansa HA, Bhat J, Buyck B, Cai L, Dai YC, Abd-Elsalam KA, Ertz D, Hidayat I, Jeewon R, Jones EBG, Bahkali AH, Karunarathna SC, Liu JK, Luangsa-ard JJ, Lumbsch HT, Maharachchikumbura SSN, McKenzie EHC, Moncalvo JM, GhobadNejhad M, Nilsson H, Pang KL, Pereira OL, Phillips AJL, Raspé O, Rollins AW, Romero AI, Etayo J, Selçuk F, Stephenson SL, Suetrong S, Taylor JE, Tsui CKM, Vizzini A, Abdel-Wahab MA, Wen TC, Boonmee S, Dai DQ, Daranagama DA, Dissanayake AJ, Ekanayaka AH, Fryar SC, Hongsanan S, Jayawardena RS, Li WJ, Perera RH, Phookamsak R, de Silva NI, Thambugala KM, Tian Q, Wijayawardene NN, Zhao RL, Zhao Q, Kang JC, Promputtha I. (2015) The Faces of fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Diversity 74: 3–18. 10.1007/s13225-015-0351-8 [DOI] [Google Scholar]
- Jeewon R, Cai L, Zhang K, Hyde KD. (2003) Dyrithiopsislakefuxianensis gen et sp. nov. from Fuxian Lake, Yunnan, China and notes on the taxonomic confusion surrounding Dyrithium. Mycologia 95: 911–920. 10.1080/15572536.2004.11833050 [DOI] [PubMed] [Google Scholar]
- Jeewon R, Hyde KD. (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7: 1669–1677. 10.5943/mycosphere/7/11/4 [DOI] [Google Scholar]
- Katoh K, Standley DM. (2016) A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32(13): 1933–1942. 10.1093/bioinformatics/btw108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubátová A, Kolarik M, Pazoutová S. (2004) Phaeoacremoniumrubrigenum–hyphomycete associated with bark beetles found in Czechia. Folia Microbiology 49: 99–104. 10.1007/BF02931381 [DOI] [PubMed] [Google Scholar]
- Luo ZL, Hyde KD, Bhat DJ, Jeewon R, Maharachchikumbura SSN, Bao DF, Li WL, Su XJ, Yang XY, Su HY. (2018) Morphological and molecular taxonomy of novel species Pleurotheciaceae from freshwater habitats in Yunnan, China. Mycological Progress 17(5): 511–530. 10.1007/s11557-018-1377-6 [DOI] [Google Scholar]
- Marin-Felix Y, Hernández-Restrepo M, Wingfield MJ, Akulov A, Carnegie AJ, Cheewangkoon R, Gramaje D, Groenewald JZ, Guarnaccia V, Halleen F, Lombard L, Luangsaard J, Marincowitz S, Moslemi A, Mostert L, Quaedvlieg W, Schumacher RK, Spies CFJ, Thangavel R, Taylor PWJ, Wilson AM, Wingfield BD, Wood AR, Crous PW. (2018) Genera of phytopathogenic fungi: GOPHY 2. Studies in Mycology. 10.1016/j.simyco.2018.04.002 [DOI] [PMC free article] [PubMed]
- Miller AM, Schwartz T, Pickett BE, He S, Klem EB, Scheuermann RH, Passarotti M, Kaufman S, O’Leary MA. (2015) A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evol Bioinform 11: 43–48. 10.4137/EBO.S21501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostert L, Crous PW, Groenewald JZE, Gams W, Summerbell RC. (2003) Togninia (Calosphaeriales) is confirmed as teleomorph of Phaeoacremonium by means of morphology, sexual compatibility and DNA phylogeny. Mycologia 95(4): 646–659. 10.1080/15572536.2004.11833069 [DOI] [PubMed] [Google Scholar]
- Mostert L, Groenewald JZ, Summerbell RC, Gams W, Crous PW. (2006a) Taxonomy and pathology of Togninia (Diaporthales) and its Phaeoacremonium anamorphs. Studies in Mycology 54: 1–115. 10.3114/sim.54.1.1 [DOI] [Google Scholar]
- Mostert L, Halleen F, Fourie P, Crous PW. (2006b) A review of Phaeoacremonium species involved in Petri disease and esca of grapevines. Phytopathologia Mediterranea 45: 12–29. [Google Scholar]
- Nigro F, Boscia D, Antelmi I, Ippolito A. (2013) Fungal species associated with a severe decline of olive in Southern Italy. Journal of Plant Pathology 95(3): 668–668. [Google Scholar]
- O’Donnell K, Cigelnik E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. 10.1006/mpev.1996.0376 [DOI] [PubMed] [Google Scholar]
- Olmo D, Gramaje D, Agustí-Brisach C, Leon M, Armengol J. (2014) First report of Phaeoacremoniumvenezuelense associated with decay of apricot trees in Spain. Plant Disease 98(7): 1001. 10.1094/PDIS-12-13-1198-PDN [DOI] [PubMed]
- Pascoe IG, Edwards J, Cunnington JH, Cottral E. (2004) Detection of the Togninia teleomorph of Phaeoacremoniumaleophilum in Australia. Phytopathologia Mediterranea 43: 51–58. [Google Scholar]
- Réblová M. (2011) New insights into the systematics and phylogeny of the genus Jattaea and similar fungi of the Calosphaeriales. Fungal Diversity 49: 167–198. 10.1007/s13225-011-0099-8 [DOI] [Google Scholar]
- Réblová M, Miller AN, Rossman AY, Seifert KA, Crous PW, Hawksworth DL, Abdel-Wahab MA, Cannon PF, Daranagama DA, De Beer ZW, Huang SK, Hyde KD, Jayawardena R, Jaklitsch W, Jones EB, Ju YM, Judith C, Maharachchikumbura SS, Pang KL, Petrini LE, Raja HA, Romero AI, Shearer C, Senanayake IC, Voglmayr H, Weir BS, Wijayawarden NN. (2016) Recommendations for competing sexual-asexually typified generic names in Sordariomycetes (except Diaporthales, Hypocreales, and Magnaporthales). IMA Fungus 7(1): 131–153. 10.5598/imafungus.2016.07.01.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway R. (1912) Color Standards and Color Nomenclature. Washington, DC. 10.5962/bhl.title.144788 [DOI]
- Ronquist F, Huelsenbeck JP. (2003) Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Rooney-Latham S, Eskalen A, Gubler WD. (2004) Ascospore discharge and occurrence of Togniniaminima (anamorph = Phaeoacremoniumaleophilum) in California vineyards. (Abstr.) Phytopathology 94: S57.
- Rooney-Latham S, Escalen A, Gubler WD. (2005a) Teleomorph formation of Phaeoacremoniumaleophilum, cause of esca and grapevine decline in California. Plant Disease 89: 177–184. 10.1094/PD-89-0177 [DOI] [PubMed] [Google Scholar]
- Rooney-Latham S, Eskalen A, Gubler WD. (2005b) Ascospore release of Togniniaminima, cause of esca and grapevine decline in California. Online. Plant Health Progress. 10.1094/PHP-2005-0209-01-RS [DOI] [PubMed]
- Rumbos IC. (1986) Phialophoraparasitica, causal agent of cherry dieback. Journal of Phytopathology 117: 283–287. 10.1111/j.1439-0434.1986.tb00944.x [DOI] [Google Scholar]
- Spies CFJ, Moyo P, Halleen F, Mostert L. (2018) Phaeoacremonium species diversity on woody hosts in the Western Cape Province of South Africa. Persoonia 40: 26–62. 10.3767/persoonia.2018.40.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayawardene NN, Hyde KD, Lumbsch HT, Liu JK, Maharachchikumbura SSN, Ekanayaka AH, Tian Q, Phookamsak R. (2018) Outline of Ascomycota: 2017. Fungal Diversity 88: 167–263. 10.1007/s13225-018-0394-8 [DOI] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR Protocols: A Guide to Methods and Applications. Academic Press, San Diego, 315–322.
- Zhang Y, Jeewon R, Fournier J, Hyde KD. (2008) Multi-gene phylogeny and morphotaxonomy of Amniculicolalignicola: novel freshwater fungus from France and its relationships to the Pleosporales. Fungal Biology 112: 1186–1194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.