Abstract Abstract
There are 63 known species of Thecaphora (Glomosporiaceae, Ustilaginomycotina), a third of which occur on Asteraceae. These smut fungi produce yellowish-brown to reddish-brown masses of spore balls in specific, mostly regenerative, plant organs. A species of Thecaphora was collected in the flower heads of Anthemischia (Anthemideae, Asteraceae) on Rhodes Island, Greece, in 2015 and 2017, which represents the first smut record of a smut fungus on a host plant species in this tribe. Based on its distinctive morphology, host species and genetic divergence, this species is described as Thecaphoraanthemidissp. nov. Molecular barcodes of the ITS region are provided for this and several other species of Thecaphora. A phylogenetic and morphological comparison to closely related species showed that Th.anthemidis differed from other species of Thecaphora. Thecaphoraanthemidis produced loose spore balls in the flower heads and peduncles of Anthemischia unlike other flower-infecting species.
Keywords: Glomosporiaceae , host specificity, internal transcribed spacer, molecular phylogenetics, smut fungi
Introduction
Thecaphora species belong to the Glomosporiaceae (Urocystidales, Ustilaginomycotina). The type species is Th.seminis-convolvuli described from Convolvulusarvensis (Convolvulaceae) collected in France (Desmazièrs 1827). Until now, 63 species of Thecaphora have been recognised (Vánky 2012), infecting host plant species in 16 different eudicot families (Vánky and Lutz 2007, Roets et al. 2008, Vánky et al. 2008, Vánky 2012). Species of Thecaphora produce sori in flowers, fruits, seeds, stems, leaves or roots, often in galls or pustules. The granular to powdery spore balls are yellowish-brown to reddish-brown, but never black. The majority of Thecaphora species produce loose or permanent spore balls without sterile cells. An exception to this is Th.smallanthi, which was reported to have large spore balls with outer spores and an internal layer of hyaline (sterile) cells (Piepenbring 2001). Three species have single spores (not united in spore balls), namely, Th.thlaspeos, Th.oxalidis (Vánky et al. 2008) and Th.capensis (Roets et al. 2008).
The Asteraceae is the largest family of eudicots with an estimated number of 30,000 species (Funk et al. 2009). The Asteraceae is divided into 13 subfamilies, including four (Asteroideae, Cichorioideae, Carduoideae and Mutisioideae) that contain about 99% of all taxa. Anthemis is a large genus in the tribe Anthemideae (subfamily Asteroideae), along with Cota, Gonospermum (including Lugoa), Nananthea, Tanacetum and Tripleurospermum (Bremer and Humphries 1993, Oberprieler et al. 2009, Presti et al. 2010). Species of Anthemis are distributed in western Eurasia, including the Mediterranean region, northern Africa and a small part of eastern Africa (Oberprieler 1998, 2001, Oberprieler et al. 2009, Presti et al. 2010). There are 62 species of Anthemis in Europe. Anthemischia belongs to the section Chiae and is a Mediterranean species common on Rhodes Island, Greece.
About 20 species of Thecaphora infect host plant species in six tribes of the Asteraceae. Taxa of the tribes Astereae and Heliantheae in the subfamily Asteroideae are often hosts of several Thecaphora species. Some less species-rich tribes, e.g. Coreopsideae, Millerieae, Polymnieae and Cynareae (subfamily Carduoideae) are also hosts of Thecaphora species. The species of Thecaphora on Asteraceae have not been studied by molecular phylogenetic methods, in contrast to species of Thecaphora on Caryophyllaceae (Vánky and Lutz 2007), Polygonaceae (Vasighzadeh et al. 2014) and Oxalidaceae (Roets et al. 2008, 2012).
Plants of Anthemischia with distorted flower heads containing mostly ligulate (ray) florets and swollen peduncles were collected near Tsambika, Rhodes Island, Greece, in 2015 and 2017. The swollen flower heads contained reddish-brown granular to powdery spore ball masses, typical of species of Thecaphora. The aim of this study was to identify the fungus and to determine its taxonomic assignment based on morphological and phylogenetic analyses of the internal transcribed spacer (ITS, barcoding locus) sequence data.
Materials and methods
Specimens
Herbarium specimens (23) of Thecaphora on a range of host plant species from across Europe and North America were examined (Tables 1, 2). The ITS sequences of specimens available on GenBank (19) and published in previous studies (Table 2) were included in the phylogenetic analysis. The nomenclature of the host plant species follows Euro+Med PlantBase (http://www.emplantbase.org/home.html) and the nomenclature of the fungi is according to Vánky (2012).
Table 1.
Collection records for specimens of Thecaphora examined in this study.
| Species | Host | Country | Location | Date | Collector | Herbarium accession no.* |
|---|---|---|---|---|---|---|
| Thecaphora affinis | Astragalus glycyphyllos | Slovenia | Lower Styria, region Savinjska, N of Ljubno ob Savinjii, trail to Mt. Greben Smrekovec-Komen from Primož pri Ljubnem, wayside, 46°24'21"N, 14°49'54"E, 1150 m asl | 14 July 2015 | J. Kruse | GLM F112522 |
| A. glycyphyllos | Germany | Saxony-Anhalt, SW of Zschornewitz, forestry trail nearby SW-shore of „Gürke“ (Zschornewitzer Lake) | 26 June 2007 | H. Jage | GLM F094059 | |
| Th. anthemidis | Anthemis chia | Greece | Island Rhodes, 3.5 km NE Archangelos, Tsambika, way up to monastery, northeastslope, 36°14'03"N, 28°09'19"E, 90 m asl | 26 April 2017 | V. Kummer | GLM F112531 |
| Th. haumanii | Iresine diffusa | Costa Rica | Prov. Guanacaste, 6 km NW de la barrada de la Laguna de Arenal | 1 April 1992 | R. Berndt, M. Piepenbring | M 0236177 |
| Th. leptideum | Chenopodium album | France | Lotharingia, Forbach, Kreuzberg Mt. | Aug.-Oct. 1912/1913 | A. Ludwig | M 0230099 |
| Th. molluginis | Mollugo cerviana | Romania | Bratovesti, Oltenia | 15 July 1963 | K. Lug. Eliart | M 0236178 |
| M. cerviana | Romania | Oltenia, Timburesti | 19 Sept. 1958 | L. Pop | M 0236180 | |
| Th. oxalidis | Oxalis stricta | Austria | Upper Austria, Braunau at Inn, Hagenau Inncounty, Hagenauer Street, wayside, 48°16'24"N, 13°06'03"E, 340 m asl | 18 Aug. 2014 | J. Kruse | GLM F112523 |
| O. stricta | Germany | Bavaria, Upper Franconia, Fichtelmountains, Fichtelberg, Sandgrubenway, cemetery, 605 m asl | 17 Sept. 2012 | J. Kruse | GLM F112524 | |
| O. stricta | Germany | Saxony-Anhalt, county Anhalt-Bitterfeld, Bitterfeld-Wolfen, Mühlstreet, allotment garden area „Kühler Grund“, 51°37'23"N, 12°20'08"E | 13 July 2014 | J. Kruse & H. Jage | GLM F112525 | |
| Th. pustulata | Bidens pilosa | Puerto Rico, USA | Mayagüez | 13 Mar. 1920 | H. H. Whetzel, E. W. Olive | CUP PR000458 |
| Th. seminis-convolvuli | Convolvulus arvensis | Germany | Saxony, Middlesaxony, Freiberg, Halsbrücker Street, roadside, 50°55'31"N, 13°20'56"E, 400 m asl | 11 Aug. 2017 | J. Kruse | GLM F112527 |
| C. arvensis | Germany | Hesse, c. 8.5 km SE Eschwege, Weißenborn, Sandhöfe, path, 51°07'35"N, 10°07'25"E, 250 m asl | 22 July 2017 | J. Kruse | GLM F112528 | |
| C. arvensis | Germany | Saxony-Anhalt, SSE Seeben, at Franzosenstein, wayside | 26 Aug. 2002 | H. Jage | GLM F065278 | |
| Calystegia sepium | Germany | Mecklenburg-Western Pomerania, county Vorpommern-Rügen, 1,5 km NE of Barth, Glöwitz, rest area, 54°22'15"N, 12°45'38"E, 0 m asl | 24 Aug. 2014 | J. Kruse | GLM F112526 | |
| C. sepium | Germany | North Rhine-Westphalia, county Steinfurt, Rheine, castle grounds Bentlage, between parking area and Gradierwerk, 52°17'49"N, 07°25'11"E, 35 m asl | 14 July 2017 | J. Kruse | GLM F112529 | |
| Th. seminis-convolvuli | C. sepium | Germany | Schleswig-Holstein, county Schleswig-Flensburg, Schaalby, W of Winningmay, parking area at „Reesholm“, wayside, 54°31'44"N, 09°37'53"E, 2 m asl | 30 Aug. 2014 | J. Kruse | GLM F112530 |
| Th. thlaspeos | Arabis ciliata | Austria | Tyrol, district Kufstein, county Walchsee, Kaiserwinkel, track from hickinghut towards Niederkaseralm, over Hintere Abendpoit, eastslope Mt. Hochköpfl, 47°41'25"N, 12°19'37"E, 1300 m asl | 21 July 2014 | J. Kruse | GLM F112533 |
| A. ciliata | Germany | Bavaria, Chiemgauer Alps, county Rosenheim, Priener Hut, track 8,20, way up towards Kampenwand, alpine meadow, 47°42'29"N, 12°19'27"E, 1570 m asl | 18 July 2014 | J. Kruse | GLM F112536 | |
| A. ciliata | Germany | Bavaria, Chiemgauer Alps, county Traunstein, Priener Hut, track 8,20 towards Priener Hut, alpine meadow, 47°42'07"N, 12°20'36"E, 1310 m asl | 19 July 2014 | J. Kruse | GLM F112537 | |
| A. hirsuta | Germany | Hesse, Meißnerfoothills, Werra-Meißner-county, Großalmerode, S of Weißenbach, “Bühlchen”, calcareous grassland, 51°14'55"N, 09°51'08"E, 500 m asl | 13 June 2015 | J. Kruse | GLM F112532 | |
| A. hirsuta | Germany | Bavaria, county Donau-Ries, Harburg, N of Ronheim, dry grassland, 435 m asl | 20 June 2013 | J. Kruse | GLM F112534 | |
| A. hirsuta | Germany | Bavaria, Upper Bavaria, county Weilheim, N of Pähl, E at Hartschimmelhof, N „Goaslweide“, wayside, 720 m asl | 20 July 2013 | J. Kruse | GLM F112535 |
* Acronyms: GLM = Herbarium Senckenbergianum, Görlitz, Germany; CUP = Plant Pathology Herbarium, Cornell University, New York, USA; M = Botanische Staatssammlung, Munich, Germany.
Table 2.
Specimens and GenBank sequences used for phylogenetic analyses. Sequences generated in this study are shown in bold.
| Thecaphora species | Host | Herbarium accession no. 1 | ITS GenBank accession no. | Reference |
| Th. affinis | Astragalus glycyphyllos | GLM F112522 | MH399748 | this paper |
| GLM F094059 | MH399749 | this paper | ||
| Th. alsinearum | Stellaria holostea | HUV 10535 | EF200032 | Vánky and Lutz 2007 |
| Th. amaranthi | Amaranthus hybridus | HUV 20727 | EF200013 | Vánky and Lutz 2007 |
| Th. anthemidis | Anthemis chia | GLM F112531 | MH399758 | this paper |
| Th. frezii | Arachis hypogaea | Sa-EM1* | KP994420 | Cazón et al. 2016 |
| Cba-GD2* | KP994419 | Cazón et al. 2016 | ||
| Th. haumanii | Iresine diffusa | M 0236177 | MH399764 | this paper |
| Th. hennenea | Melampodium divaricatum | HUV 14434 | EF200014 | Vánky and Lutz 2007 |
| Th. italica | Silene italica | HUV 20345 | EF200026 | Vánky and Lutz 2007 |
| HUV 20344 | EF200025 | Vánky and Lutz 2007 | ||
| Th. leptideum | Chenopodium album | M 0230099 | MH399756 | this paper |
| Th. melandrii | Silene alba | HUV 12677 | EF200024 | Vánky and Lutz 2007 |
| Th. molluginis | Mollugo cerviana | M 0236178 | MH399762 | this paper |
| M 0236180 | MH399763 | this paper | ||
| Th. oxalidis | Oxalis stricta | GLM F112524 | MH399759 | this paper |
| GLM F112523 | MH399760 | this paper | ||
| GLM F112525 | MH399761 | this paper | ||
| Th. oxytropis | Oxytropis pilosa | Kummer P 1146/3* | KF640685 | Kummer et al. 2014 |
| Kummer P 1146/2* | KF640684 | Kummer et al. 2014 | ||
| Th. pustulata | Bidens pilosa | CUP PR000458 | MH399757 | this paper |
| Th. saponariae | Saponaria officinalis | TUB 012796 | EF200022 | Vánky and Lutz 2007 |
| Th. schwarzmaniana | Rheum ribes | BASU 4242 | JX006079 | Vasighzadeh et al. 2014 |
| KRAM F-49788 | KF297811 | Vasighzadeh et al. 2014 | ||
| Th. seminis-convolvuli | Calystegia sepium | GLM F112529 | MH399742 | this paper |
| GLM F112526 | MH399743 | this paper | ||
| GLM F112530 | MH399744 | this paper | ||
| Convolvulus arvensis | GLM F112527 | MH399745 | this paper | |
| GLM F112528 | MH399746 | this paper | ||
| GLM F065278 | MH399747 | this paper | ||
| Th. solani | Solanum lycopersicum | HUV 11180 | EF200037 | Vánky and Lutz 2007 |
| Th. sp. | Rheum palmatum | S. Wang 1991* | KJ579177 | Piątek et al. unpublished |
| Y. Wang 2013* | KJ579176 | Piątek et al. unpublished | ||
| HUV 21117 | KF297812 | Vasighzadeh et al. 2014 | ||
| Th. spilanthis | Acmella sp. | AFTOL 1913 | DQ832243 | Matheny et al. 2006 |
| Th. thlaspeos | Arabis hirsuta | GLM F112532 | MH399752 | this paper |
| TUB 015857 | KJ579178 | Vasighzadeh et al. 2014 | ||
| GLM F112534 | MH399750 | this paper | ||
| GLM F112535 | MH399751 | this paper | ||
| Arabis ciliata | GLM F112537 | MH399753 | this paper | |
| GLM F112533 | MH399754 | this paper | ||
| GLM F112536 | MH399755 | this paper |
1 Acronyms: AFTOL = Assembling the Fungal Tree Of Life, http://aftol.org; BASU: Herbarium of Bu-Ali Sina University, Iran; CUP = Plant Pathology Herbarium, Cornell University, New York, USA; GLM = Herbarium Senckenbergianum, Görlitz, Germany; HUV = Herbarium Ustilaginales Vánky, deposited in BRIP = Queensland Plant Pathology Herbarium, Brisbane, Australia; KRAM F = Mycological Collection of the W. Szafer Institute of Botany, Polish Academy of Sciences, Kraków, Poland; M = Botanische Staatssammlung, Munich, Germany; TUB = Herbarium Tubingense, Eberhard-Karls-Universität Tübingen, Germany; * not deposited in any public herbaria.
The morphology of the spore balls and spores of one specimen (GLM-F112531) of Thecaphora on Anthemischia was microscopically examined at 1000× in 80% lactic acid heated to the boiling point on a glass slide. Measurements of 30 spore balls and 100 spores were made with the Zeiss AxioVision software and micrographs were taken with an Olympus FE-120 camera on a Seben SBX-5 compound microscope (Seben GmbH, Berlin). The measurements are reported as maxima and minima in parentheses and the means are placed in italics.
DNA extraction, amplification and sequencing
Genomic DNA was extracted from 23 herbarium specimens of Thecaphora (Table 1) using the methods reported by Kruse et al. (2017). The ITS nrDNA was amplified by PCR as reported in Kruse et al. (2018), using M-ITS1 (Stoll et al. 2003) as forward primer and either smITS-R1 or smITS-R2 (Kruse et al. 2017) as reverse primer. The ITS of host plants was amplified using primer pair ITS1P/ITS4 (Ridgway et al. 2003) with an annealing temperature of 53 °C. The resulting amplicons were sequenced at the Senckenberg Biodiversity and Climate Research Centre (BiK-F, Senckenberg) using the ITS4 primer (White et al. 1990). Sequences were deposited in GenBank (Table 2).
Phylogenetic analysis
In total, 42 ITS sequences from 21 Thecaphora species were used in the phylogenetic analyses. Sequences were aligned with MAFFT v.7 (Katoh and Standley 2013) employing the G-INS-I algorithm and leading and trailing gaps were trimmed. The resulting alignment length was 534 bp. The methods of phylogenetic analysis were according to Kruse et al. (2018) using Minimum Evolution (ME), Maximum Likelihood (ML) and Bayesian Inference (BA). Thecaphoraitalica and allied species were selected as an outgroup, on the basis of the phylogeny presented by Vánky and Lutz (2007). Host plant species determination was verified by comparison with published sequences from Asteraceae deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) using BLASTN (Altschul et al. 1997).
Results
Molecular phylogenetic reconstruction
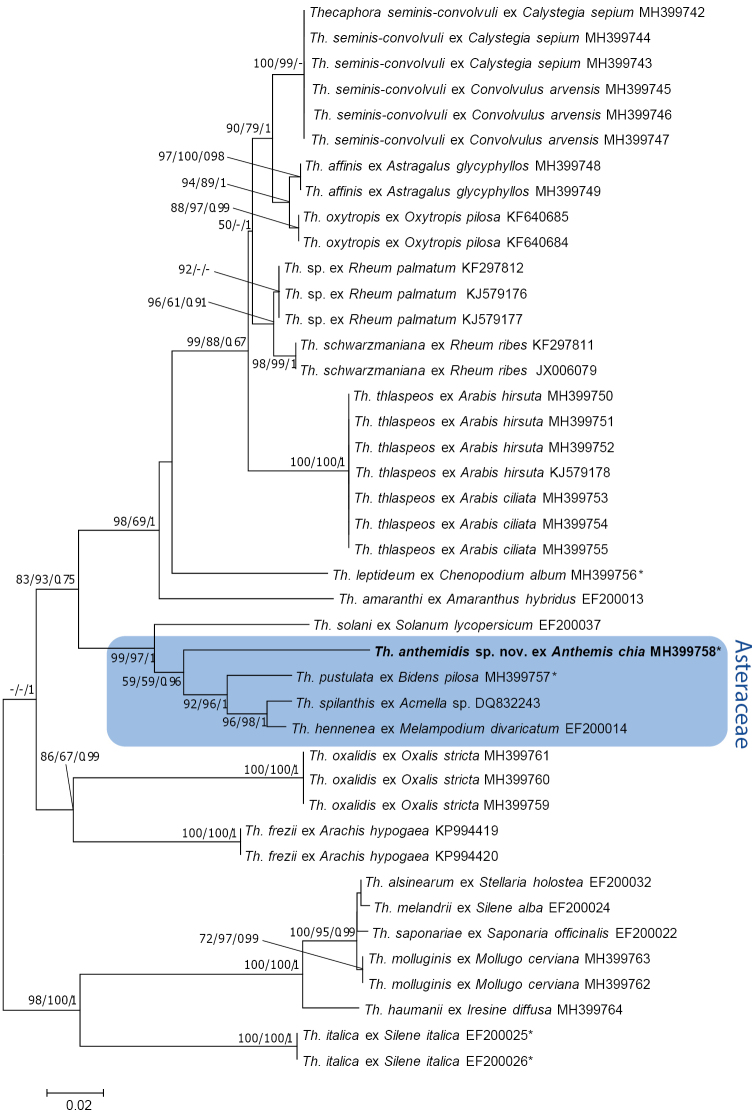
The ML and BA trees yielded consistent topologies with the ME tree (Fig. 1). The Thecaphora sp. on Anthemischia, together with three Asteracious species (Th.pustulata, Th.hennenea and Th.spilanthis) and Th.solani from Solanumlycopersicum (Solanaceae), formed a sister clade to the species on other host plant families with strong to intermediate bootstrap support (83% in ME, 93% in ML). The Thecaphora sp. on Anthemischia was sister to the other Asteracious species with low bootstrap support (59% in ME, 59% in ML), but high Bayesian posterior probability (96%). The Thecaphora species on Fabaceae were polyphyletic, with Th.frezii on Arachishypogaea sister to Th.oxalidis on Oxalisstricta (Oxalidaceae). Thecaphorafrezii was distant to a monophyletic lineage on Oxytropispilosa and Astragalusglycyphyllos, which was sister to Th.seminis-convolvuli, the type of the genus. All specimens of Th.seminis-convolvuli collected on Calystegiasepium and Convolvulusarvensis (Convolvulaceae) had identical ITS sequences, as was the case with Thecaphorathlaspeos on Arabishirsuta and A.ciliata (Brassicaceae). Within the clade of mostly Caryophyllaceae-infecting species, two species of Thecaphora infected other families of the Caryophyllales, namely Th.molluginis on Mollugocerviana (Molluginaceae) and Th.haumanii on Iresinediffusa (Amaranthaceae).
Figure 1.
Phylogenetic tree of Thecaphora species based on ME analysis of the ITS locus. Numbers on branches denote support in ME, ML and BA, respectively. Values below 50% are denoted by ‘–‘. The bar indicates the number of substitutions per site. Ex-type sequences are highlighted with an asterisk.
Taxonomy
Thecaphora anthemidis
J. Kruse, V. Kumm. & Thines sp. nov.
827067
Figure 2.
Sori, spore balls and spores of Thecaphoraanthemidis on Anthemischia (GLM-F112531) (A–H), A habit B–C swollen flower heads and peduncles D dissected flower head with reddish granular powdery spore ball mass E young spore balls F mature spore balls G–H single spores. Scale bars: 10 µm.
Type.
Greece, Rhodes Island, 3.5 km NE Archangelos, Tsambika, on path to monastery, northeast slope, 36°14'03"N, 28°09'19"E, 90 m a.s.l, on Anthemischia, 26 Apr. 2017, V. Kummer. Holotype GLM-F112531, isotype Herbarium V. Kummer P 1971/chia4; ITS sequence GenBank MH399758.
Etymology.
From the host plant genus Anthemis.
Description.
Sori in swollen and distorted flower heads and peduncles; spore ball mass initially white, later reddish-brown, granular to powdery; spore balls subglobose to ellipsoidal, rarely ovoid, mostly regular in shape, (31–) 36–41–47 (–52) × (28–) 31–38–44 (–50) µm, length/width ratio 0.9–1.1–1.2 (n=30), under light microscopy yellowish-brown to pale yellowish-brown, composed of 2–10 (–12) loosely united spores that separate easily; spores ellipsoidal, subglobose, ovoid or cuneiform, (18–) 20–21–23 (–25) × (14–) 17–18–20 (–23) µm, length/width ratio of 1.1–1.2–1.4 (n=100), with flattened contact surfaces and rounded exposed surfaces; wall at contact surface up to 0.5 µm thick, wall at free surface up to 3 µm thick, densely verrucose with warts 0.5–1 µm high, often confluent and sometimes irregular.
Host range.
Anthemischia.
Distribution.
Greece.
Notes.
Thecaphoraanthemidis has sori in the flower heads and the peduncles, which differentiates it from the following species that produce pustules, galls or swellings on the stems of Asteraceae: Th.ambrosiae, Th.denticulata, Th.heliopsidis, Th.hennenea, Th.melampodii, Th.mexicana, Th.neomexicana, Th.piluliformis, Th.polymniae, Th.pulcherrima, Th.pustulata, Th.smallanthi and Th.spilanthis. Four of the seven previously known species of Thecaphora that infect the flower heads of Asteraceae, namely Th.arnicae, Th.burkartii, Th.californica and Th.cuneata have firmly united spores that only separate after considerable pressure, which differentiate them from Th.anthemidis that has loose spore balls. Further, Th.arnicae (spore balls comprised of up to 25 spores), Th.californica (6–20 spores) and Th.solidaginis (8 to 50 or more spores) have larger spore balls with larger numbers of spores than Th.anthemidis. The spores of Th.cuneata are radially arranged within the spore balls and Th.burkartii has spores with an outer wall 5–9 µm thick, which is more than three times thicker than in Th.anthemidis. Thecaphoralagenophorae and Th.trailii are morphologically most similar to Th.anthemidis. Thecaphoralagenophorae is only known to infect Solenogynegunnii (tribe Astereae) in Australia (Vánky 2012). Thecaphoratrailii infects species of Carduus, Cirsium and Saussurea (Asteraceae, tribe Cynareae, Carduoideae) (Vánky 2012) and further differs from Th.anthemidis by having smaller spore balls (12–30 µm) and fewer spores (2–8) per spore ball.
Discussion
The present study is the first to identify a species of Thecaphora on a host plant species in the tribe Anthemideae (Asteraceae) (see Vánky 2012). Thecaphoraanthemidis was recovered in a monophyletic group of Thecaphora species on Asteraceae, sister to Thecaphorasolani on Solanumlycopersicum (Solanaceae). Our phylogenetic hypothesis, based on the ITS region, was similar to the analyses of the LSU locus of these taxa in Vánky and Lutz (2007) and Roets et al. (2008). In the latter study, Thecaphorapolymniae, which is known only from the type collection on Polymniariparia (Polymnieae, Asteroideae, Asteraceae) from South America (Vánky 2012), clustered within a clade of taxa that infect Fabaceae, Caryophyllaceae and Amaranthaceae (Roets et al. 2008). Thecaphorapolymniae has spores with a reticulate ornamentation and this may be evidence of a host jump from one of these plant families to Asteraceae. Host jumps have been reported before in the Ustilaginomycotina (e.g. Begerow et al. 2002, Piątek et al. 2017) and are thought to be a driver of plant pathogen diversification (Choi and Thines 2015).
Previously, only two ITS sequences of Thecaphora species infecting Asteraceae (Th.spilanthis and Th.hennenea) were available on GenBank, which together with the new sequences reported in this study, represents only 20% of all Thecaphora species known to occur on Asteraceae. In addition to the sequence of Th.anthemidis, we have provided barcode sequences of the ITS region for eight other taxa not previously available on GenBank (Table I). Future studies should address whether species of Thecaphora that infect the flower heads of Asteraceae form a monophyletic group.
Supplementary Material
Acknowledgements
The authors are grateful to Ulrike Damm and Michaela Schwager of the Herbarium Senckenbergianum Görlitz (GLM) for providing us with herbarium numbers for private collections and to the curators Dagmar Triebel of the Herbarium Munich (M), Ulrike Damm of the Herbarium Senckenbergianum Görlitz (GLM) and Scott LaGreca of the Cornell University New York (CUP) for loaning specimens from their keeping. We also thank all private collectors of the specimens investigated in this study for giving material to public herbaria. Furthermore, we want to thank the Ministry for Environment and Energy, Directorate for Forest Management for the collection permission in Rhodes (Greece). Alistair McTaggart is thanked for proofreading the paper and helpful advice.
Citation
Kruse J, Kummer V, Shivas RG, Thines M (2018) The first smut fungus, Thecaphora anthemidis sp. nov. (Glomosporiaceae), described from Anthemis (Asteraceae). MycoKeys 41: 39–50. https://doi.org/10.3897/mycokeys.41.28454
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begerow D, Lutz M, Oberwinkler F. (2002) Implications of molecular characters for the phylogeny of the genus Entyloma. Mycological Research 106: 1392–1399. 10.1017/S0953756202006962 [DOI] [Google Scholar]
- Bremer K, Humphries CJ. (1993) Generic monograph of the Asteraceae-Anthemideae. Bulletin of the British Museum (Natural History) Botany series 23: 71–177. [Google Scholar]
- Cazón I, Conforto C, Fernández FD, Paredes JA, Rago AM. (2016) Molecular detection of Thecaphorafrezii in peanut (Arachishypogaea L.) seeds. Journal of Plant Pathology 98: 327–330. http://www.jstor.org/stable/44280452 [Google Scholar]
- Choi YJ, Thines M. (2015) Host jumps and radiation, not co‐divergence drives diversification of obligate pathogens. A case study in downy mildews and Asteraceae PLoS One. 10: 10.1371/journal.pone.0133655 [DOI] [PMC free article] [PubMed]
- Desmazières JBHJ. (1827) Plantes cryptogames du Nord de la France (Cryptogamic plants of France). Edn 1: No. 274. Exsiccata, 44 fascicles.
- Funk VA, Susanna A, Stuessy TF, Robinson HE. (2009) Classification of Compositae. In: Funk VA, Susanna A, Stuessy T, Bayer R. (Eds) Systematics, Evolution, and Biogeography of Compositae.International Association for Plant Taxonomy, Vienna, 171–189.
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse J, Choi YJ, Thines M. (2017) New smut-specific primers for the ITS barcoding of Ustilaginomycotina. Mycological Progress 16: 213–221. 10.1007/s11557-016-1265-x [DOI] [Google Scholar]
- Kruse J, Dietrich W, Zimmermann H, Klenke F, Richter U, Richter H, Thines M. (2018) Ustilago species causing leaf-stripe smut revisited. IMA Fungus 9: 49–73. 10.5598/imafungus.2018.09.01.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummer V, Lutz M, Richter U, Ristow M, Zimmermann H. (2014) Thecaphoraoxytropis – erste Nachweise in Europa. Boletus 35: 5–15. [Google Scholar]
- Matheny PB, Gossmann JA, Zalar P, Kumar TA, Hibbett DS. (2006) Resolving the phylogenetic position of the Wallemiomycetes: an enigmatic major lineage of Basidiomycota. Botany 84: 1794–1805. 10.1139/b06-128 [DOI] [Google Scholar]
- Oberprieler C. (1998) The systematics of Anthemis L. (Compositae, Anthemideae) in W and C North Africa. Bocconea 9: 1–328. [Google Scholar]
- Oberprieler C. (2001) Phylogenetic relationships in Anthemis L. (Compositae, Anthemideae) based on nrDNA ITS sequence variation. Taxon 50: 745–762. 10.2307/1223705 [DOI] [Google Scholar]
- Oberprieler C, Himmelreich S, Källersjö M, Valles J, Vogt R. (2009) Anthemideae. In: Funk VA, Susanna A, Stuessy TF, Bayer R. (Eds) Systematics, evolution, and biogeography of Compositae.International Association for Plant Taxonomy,Vienna, 631–666.
- Oberprieler C, Himmelreich S, Vogt R. (2007) A new subtribal classification of the tribe Anthemideae (Compositae). Willdenowia 37: 89–114. 10.3372/wi.37.37104 [DOI] [Google Scholar]
- Piątek M, Lutz M, Sousa FMP, Santos ARO, Félix CR, Landell MF, Gomes FCO, Rosa CA. (2017) Pattersoniomycestillandsiae gen. et comb. nov.: linking sexual and asexual morphs of the only known smut fungus associated with Bromeliaceae. Organisms Diversity & Evolution 17: 531–543. 10.1007/s13127-017-0340-8 [DOI] [Google Scholar]
- Piepenbring M. (2001) New species of smut fungi from the neotropics. Mycological Research 105: 757–767. 10.1017/S0953756200004135 [DOI] [Google Scholar]
- Presti RML, Oppolzer S, Oberprieler C. (2010) A molecular phylogeny and a revised classification of the Mediterranean genus Anthemis sl (Compositae, Anthemideae) based on three molecular markers and micromorphological characters. Taxon 59: 1441–1456. http://www.jstor.org/stable/20774040 [Google Scholar]
- Ridgway KP, Duck JM, Young JPW. (2003) Identification of roots from grass swards using PCR-RFLP and FFLP of the plastid trn L (UAA) intron. BMC Ecology 3: 8. 10.1186/1472-6785-3-8 [DOI] [PMC free article] [PubMed]
- Roets F, Dreyer LL, Wingfield MJ, Begerow D. (2008) Thecaphoracapensis sp. nov., an unusual new anther smut on Oxalis in South Africa. Persoonia 21: 147–152. 10.3767/003158508X387462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roets F, Curran H, Dreyer LL. (2012) Morphological and reproductive consequences of an anther smut fungus on Oxalis. Sydowia 64: 267–280. 10.4102/sajs.v107i3/4.653 [DOI] [Google Scholar]
- Stoll M, Piepenbring M, Begerow D, Oberwinkler F. (2003) Molecular phylogeny of Ustilago and Sporisorium species (Basidiomycota, Ustilaginales), based on internal transcribed spacer (ITS) sequences. Canadian Journal of Botany 81: 976–984. 10.1017/S0953756204002229 [DOI] [Google Scholar]
- Vánky K, Lutz M. (2007) Revision of some Thecaphora species (Ustilaginomycotina) on Caryophyllaceae. Mycological Research 111: 1207–1219. 10.1016/j.mycres.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Vánky K, Lutz M, Bauer R. (2008) About the genus Thecaphora (Glomosporiaceae) and its new synonyms. Mycological Progress 7: 31–39. 10.1007/s11557-007-0550-0 [DOI] [Google Scholar]
- Vánky K. (2012) Smut Fungi of the World. APS Press, St Paul, Minnesota, 1458 pp. [Google Scholar]
- Vasighzadeh A, Zafari D, Selçuk F, Hüseyin E, Kurşat M, Lutz M, Piątek M. (2014) Discovery of Thecaphoraschwarzmaniana on Rheumribes in Iran and Turkey: implications for the diversity and phylogeny of leaf smuts on rhubarbs. Mycological Progress 13: 881–892. 10.1007/s11557-014-0972-4 [DOI] [Google Scholar]
- White TJ, Bruns T, Lee SJWT, Taylor JL. (1990) Amplification and direct sequencing of fungal ribosomal RNA sequences for phylogenetics. In: Innis N, Gelfand D, Sninsky J, White T. (Eds) PCR Protocols: a guide to methods and applications.San Diego, Academic Press, 315–322.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.