Abstract
Context
Fos null mice failed to ovulate and form a corpus luteum (CL) even when given exogenous gonadotropins, suggesting that ovarian Fos expression is critical for successful ovulation and CL formation. However, little is known about FOS in the human ovary.
Objectives
To determine the expression, regulation, and function of FOS in human periovulatory follicles.
Design/Participants
Timed periovulatory follicles were obtained from normally cycling women. Granulosa/lutein cells were collected from in vitro fertilization patients.
Main Outcome Measures
The in vivo expression after human chorionic gonadotropin (hCG) administration and in vitro regulation of FOS, JUN, JUNB, and JUND was evaluated at the mRNA and protein level. Binding of progesterone receptor (PGR) and FOS to their target genes was assessed by chromatin immunoprecipitation analyses. Prostaglandin E2 (PGE2) and progesterone were measured.
Results
The expression of FOS, JUNB, and JUND drastically increased in ovulatory follicles after hCG administration. In human granulosa/lutein cell cultures, hCG increased the expression of FOS and JUN proteins. Inhibitors of PGR and epidermal growth factor (EGF) receptors reduced hCG-induced increases in the expression and phosphorylation of FOS. PGR bound to the FOS gene. A selective FOS inhibitor blocked hCG-induced increases in PGE2 and the expression of prostaglandin (PG) synthases and transporters (PTGES, SLCO2A1, and ABCC1). FOS bound to the promoter regions of these genes.
Conclusions
The increase of FOS/activator protein 1 in human periovulatory follicles after hCG administration is mediated by collaborative actions of PGR and EGF signaling and critical for the upregulated expression of key ovulatory genes required for the rise in ovulatory PG in human granulosa cells.
The LH surge increases FOS and JUNs in ovulatory follicles, and these factors are necessary for the expression of specific ovulatory genes required for the rise in prostaglandins in the human ovary.
Fos (also known as c-Fos) belongs to the activator protein 1 (AP-1) superfamily of transcription factors. AP-1 is a dimeric protein complex composed of members of Fos (Fos, FosB, Fosl1, Fosl2), Jun (Jun, Junb, Jund), ATF, and JDP families (1). FOS contains a conserved basic leucine zipper domain, which is responsible for dimerization and DNA binding, and a transactivation domain that provides binding sites for other transcription coregulators (1). To be functional, FOS needs to form a heterodimeric complex with another basic leucine zipper domain transcription factor, Jun family proteins, resulting in the formation of the active AP-1 complex. The Fos/Jun complexes bind to DNA at a specific consensus sequence on the promoter or enhancer regions of its target genes to modulate the transcriptional activity (2). Unlike FOS, Jun family proteins can form both homo-and heterodimers. However, the FOS/JUN complex is more stable and has stronger DNA binding activity than the JUN/JUN complex (3).
The expression of Fos is rapidly and transiently induced by various external stimuli, such as growth factors, hormones, cytokines, neurotransmitters, and oncogenic stimuli in diverse tissues [reviewed in Piechaczyk and Blanchard (4)]. The cellular accumulation of FOS is also controlled by posttranslational modification. For instance, the phosphorylation of FOS by various kinases, including mitogen-activated protein kinase, protein kinase A, and protein kinase C, increased the transcriptional activity (5, 6).
In the ovary, FSH induced a rapid and transient increase in the expression of Fos, Jun, Junb, and Jund in rat granulosa cell cultures (7, 8). Similarly, LH transiently increased the levels of mRNA for Fos and Jun in granulosa cell cultures and JunB and JunD protein in rat ovaries (7, 8). In the porcine ovary, immunoreactive staining for FOS, JUN, JUNB, and JUND was detected in antral follicles and corpus luteum (CL), with a varying degree of expression for each protein (9). In cattle, FOS mRNA was detected in granulosa cells of dominant follicles collected 24 hours after LH administration (10). In humans, recent microarray data by Wissing et al. (11) listed FOS and JUN as differentially upregulated genes in granulosa cells collected 36 hours after human chorionic gonadotropin (hCG) from in vitro fertilization (IVF) patients compared with those isolated before hCG administration. In addition to the gonadotropins, prostaglandin F2a induced a transient increase in Fos mRNA in the rat ovary (12) and several members of Fos and Jun families in porcine CL (13). Together, these findings indicate that the members of Fos and Jun families are expressed in the ovary, and their expression is regulated by hormones during the different stages of follicular and luteal development.
In response to external stimuli, the AP-1 protein has been shown to control a wide range of cellular processes, including proliferation, differentiation, hypoxia, angiogenesis, steroidogenesis, and prostaglandin production (12, 14, 15). These processes are also critical for follicular development, ovulation, and luteal development (16, 17). In Fos knockout mice, follicular development was arrested at the secondary follicle stage; this was explained by reduced levels of FSH and LH (18). However, Fos-deficient mice failed to ovulate and form CL even when exogenous gonadotropins were administered (18), indicating that the ovarian expression of Fos is necessary for ovulation and luteinization.
Despite accumulating evidence showing the expression of Fos and Jun family members in the ovary and their potential impact on normal ovarian function, the specific role of the FOS/AP-1 transcription factor in the ovary has not been identified. Particularly, there is a paucity of research on the expression, regulation, and function of AP-1 proteins in the human ovary. Therefore, the objectives of this study are to (1) determine when and where FOS and JUN family members are expressed across the periovulatory period in the human ovary, (2) establish a primary human granulosa cell culture model where the in vivo expression of FOS and JUNs can be examined, (3) dissect the mechanism by which FOS and JUN expression is regulated, and (4) determine the specific role of FOS/AP-1 in regulating the expression of key ovulatory genes using a human granulosa/lutein cell (hGLC) culture model.
Materials and Methods
Materials
Unless otherwise noted, all reagents were purchased from Sigma Chemical Co. or Thermo Fisher Scientific. Both RU486 and prostaglandin E2 (PGE2) enzyme immunoassay kit were purchased from Cayman Chemical. AG1478 and T-5224 were purchased from Calbiochem and ApexBio Technology, respectively.
Human tissue collection
The protocol using human tissues was approved by the Human Ethics Committee of the Sahlgrenska Academy at the University of Gothenburg, and all patients had given their informed written consent before participating. Whole follicles were collected from patients across the periovulatory period as previously described (19). Women (aged 30 to 38 years) who had exhibited regular menstrual cycles and had not taken hormonal contraceptives for at least 3 months prior to their enrollment in the study underwent laparoscopic sterilization. Women were monitored by transvaginal ultrasound for two to three menstrual cycles before surgery to ascertain cycle regularity and to monitor the growth of a dominant follicle during the follicular phase. These patients were divided into three groups: preovulatory, early ovulatory, and late-ovulatory phases. In the preovulatory group, surgery was performed when the follicle reached >14 mm and ≤17.5 mm in diameter prior to the endogenous LH surge. These patients were not given hCG. The remaining women were given recombinant hCG (Ovitrelle, 250 µg) and were divided into two groups: early ovulatory (surgery between 12 and 18 hours post-hCG) and late ovulatory (surgery between 18 and 34 hours post-hCG). To confirm that these patients followed a normal hormonal pattern before the LH surge or after hCG administration, blood samples were taken at surgery and measured for serum progesterone and estradiol (the patient characteristics in Supplemental Table 1).
The whole intact follicle was removed using laparoscopic scissors and processed for either immunohistochemical or gene expression analysis. The follicle was bisected, and mural granulosa cells were gently scraped off from the interior of the follicle by small tissue forceps. The follicular fluid and cell suspension were combined and centrifuged at 500g to pellet and collect granulosa cells.
Human granulosa/lutein cell cultures
hGLCs were obtained from aspirates of IVF patients. The collection protocol was approved by the Institutional Review Board of the University of Kentucky Office of Research Integrity. Ovarian hyperstimulation was induced by the administration of recombinant human FSH in individualized doses to patients at the Bluegrass Fertility Center (Lexington, KY). IVF patients were then administered with hCG (10,000 U) on days 9 to 11, and dominant follicles were aspirated 36 hours later. The experiments with hGLCs were carried out as described previously (19, 20). Briefly, immediately after retrieval of cumulus-oocyte complexes, the remaining cells in aspirates were subjected to Percoll gradient centrifugations to remove red blood cells. The isolated cells were first examined under the microscope for their morphology and counted. If the cells isolated were of poor quality and insufficient numbers, these cells were discarded. Only the cells with normal morphology were then resuspended with OptiMEM media (Gibco) supplemented with 10% fetal bovine serum and antibiotic-antimycotic and then placed onto culture plates (2.5 × 105 cells/mL). The cells were acclimatized for 6 days, changing media every 24 hours. At the end of acclimation, the hGLCs were treated with or without various reagents ± hCG (1 IU/mL) in OptiMEM media supplemented with antibiotic-antimycotic and further cultured for stated hours.
For gene silencing experiments, the hGLCs were serum-starved for 1 hour and treated with PGR-siRNA (assay 143387; Thermo Fisher Scientific) or Stealth RNAi siRNA Negative Control GC Duplex (12935112; Thermo Fisher Scientific) in Lipofectamine RNAiMAX Transfection Reagent according to the manufacturer’s instruction. Transfected cells were incubated for 3 hours before stimulating with hCG (1 IU/mL) and cultured for 12 hours.
Immunohistochemistry
Follicles were fixed in 4% formaldehyde, embedded in paraffin, sectioned (7 μm), and processed for immunostaining. Heat-induced epitope retrieval was performed in a Biocare Medical Decloaking chamber using Dako low pH Target Retrieval Solution. Primary antibody incubation was carried out at 4°C overnight for FOS, JUN, JUNB, and JUND. Rabbit immunoglobulin G (IgG) was used in place of primary antibodies as a negative control (antibody information; Supplemental Table 2). The antibody was detected using an appropriate Immpress alkaline phosphatase kit and Vector Red AP chromogen (Vector Laboratories).
Gene expression analysis
Total RNA was isolated from granulosa cells using an RNeasy mini kit. The synthesis of first-strand cDNA was performed by reverse transcription of 500 ng total RNA using superscript III with Oligo(dT)20 primer. The levels of mRNA for genes examined were measured by quantitative PCR using Brilliant 3 Ultra-Fast SYBR green (Stratagene). Oligonucleotide primers corresponding to each gene were designed using Primer3 software (Supplemental Table 3). The relative abundance of the target transcript was normalized to GAPDH mRNA for in vivo samples and RNA18S5 for in vitro samples as previously described (19, 20) and calculated according to the 2−ΔΔCT method (21).
Western blot analysis
Whole-cell extracts were isolated from cultured cells, denatured, run on a 10% polyacrylamide gel, and then transferred onto a nitrocellulose membrane as described previously (21). The membrane was incubated overnight at 4°C in 5% skim milk/Tris-buffered saline including 0.1% Tween-20 solution containing primary antibodies against FOS, JUN, JUNB, JUND, or β-actin. The blots were incubated with the respective secondary horseradish peroxidase–conjugated antibody for 1 hour. Peroxidase activity was visualized using the SuperSignal West Pico Chemiluminescent Substrate (Pierce Chemical).
Hormone assay
The concentration of progesterone and PGE2 was measured using an Immulite kit and PGE2 ELISA kit (19). Assay sensitivity for progesterone and PGE2 was 0.02 ng/mL and 8.7 pg/mL, respectively. The intra-assay coefficients of variation for progesterone and PGE2 were 7% and 3.7%, respectively. The interassay coefficients of variation assay for progesterone and PGE2 were 12% and 8.3%, respectively.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed on progesterone receptor (PGR) binding sites in the putative FOS promoter region and FOS binding sites in the putative PTGES, SLCO2A1, and ABCC4 promoter regions using a ChIP-IT kit (Active Motif). hGLCs were fixed and the isolated nuclei were sonicated and sheared as described previously (19). Sheared chromatin was immunoprecipitated overnight at 4°C with anti-PGR or anti-FOS IgG (3 µg/mL). Normal rabbit IgG and RNA polymerase II antibody were used for negative and positive controls, respectively (Supplemental Table 2). The immunoprecipitated chromatin and input chromatin were analyzed by PCR using the primers designed to amplify fragments of the PGR responsive element or the AP-1 responsive element in respective promoter regions (Supplemental Table 3). Amplified PCR products were run on a 2% agarose gel, stained with ethidium bromide, and visualized under ultraviolet light.
Statistical analyses
All data are presented as means ± SEMs. Data were tested for homogeneity of variance by the Levene test, and log transformations were performed as appropriate. Student t test or one-way ANOVA was used to test differences in levels of mRNA for each gene across time of tissue collection, time of culture, or among treatments in vitro. If ANOVA revealed significant effects, the means were compared by the Duncan test, with P < 0.05 considered significant.
Results
Expression of FOS and JUN family members in human periovulatory follicles
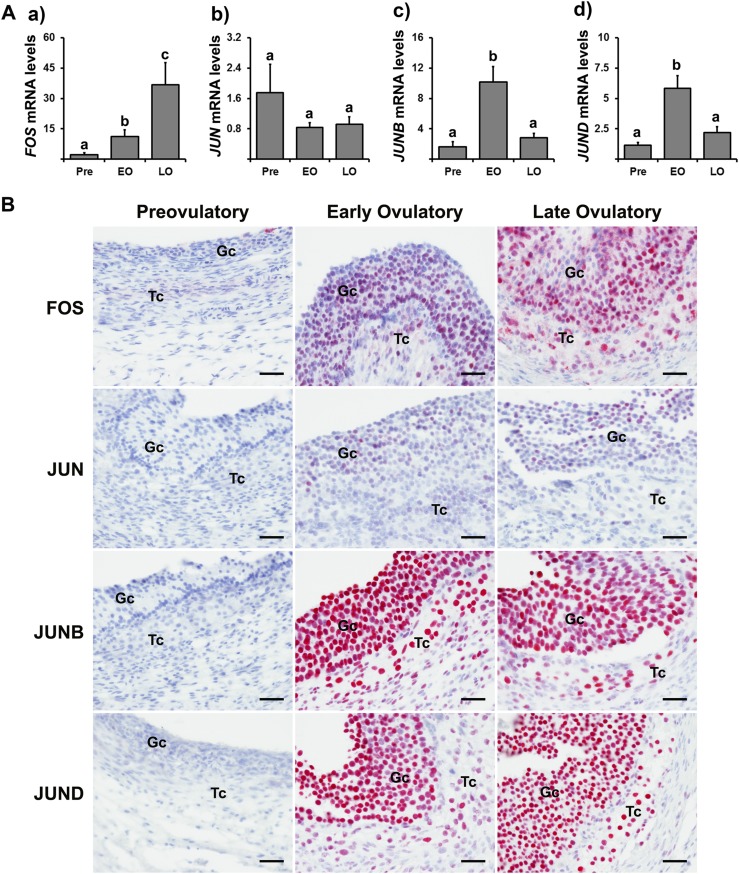
In granulosa cells of dominant follicles obtained from normally cycling women, the levels of mRNA for FOS were elevated during the early ovulatory phase compared with prior to hCG and further elevated during the late ovulatory phase [Fig. 1A(a)], whereas JUN mRNA levels were not changed throughout these ovulatory phases [Fig. 1A(b)]. In contrast, the levels of mRNA for JUNB and JUND were elevated at the early ovulatory phase but then decreased during the late ovulatory phase to the levels of the preovulatory phase [Fig. 1A(c) and 1A(d)].
Figure 1.
The expression of FOS and its binding partners, JUN, JUNB, and JUND, in periovulatory follicles after hCG administration. Dominant follicles were retrieved from the ovaries of women undergoing laparoscopic tubal sterilization before the LH surge or at defined h after recombinant hCG administration and divided into three phases: preovulatory (Pre, n = 6), early ovulatory (EO, n = 5), and late-ovulatory (LO, n = 6) phases as described in Materials and Methods. (A) The levels of mRNA for each transcript were measured by quantitative PCR in granulosa cells and normalized to the levels of GAPDH mRNA in each sample. The levels of transcript were presented as fold change to Pre levels. Bars with no common superscripts are significantly different (P < 0.05). (B) Paraffin-embedded sections of dominant follicles (n = 2/Pre, n = 4/EO, n = 4/LO) were subjected to immunohistochemical analyses to detect FOS, JUN, JUNB, and JUND. Pink/purple staining represents positive signals for these proteins. All sections were lightly stained with hematoxylin (blue) for nuclear staining. Gc, granulosa cells; Tc, theca cells. Scale bar, 100 µm for all the images. The relative abundance of immunoreactive signals for these proteins ranked by the qualitative scoring system is presented in Supplemental Table 4. A representative negative control section treated with rabbit IgG is presented in Supplemental Fig. 2.
Immunopositive staining for FOS was localized to granulosa and theca cells of dominant follicles during both early and late ovulatory phases, and the intensity of FOS staining appeared to be stronger in follicles of the late ovulatory phase (Fig. 1B). The staining for JUN was also detected in granulosa and theca cells of dominant follicles collected after recombinant hCG administration, albeit the intensity and number of immunopositive cells appeared to be weaker and sporadic compared with those of JUNB and JUND. In contrast, JUNB and JUND were localized to all granulosa and theca cells of dominant follicles collected during early and late ovulatory phases (Fig. 1B).
Effect of hCG on the expression of FOS and JUN family members in vitro
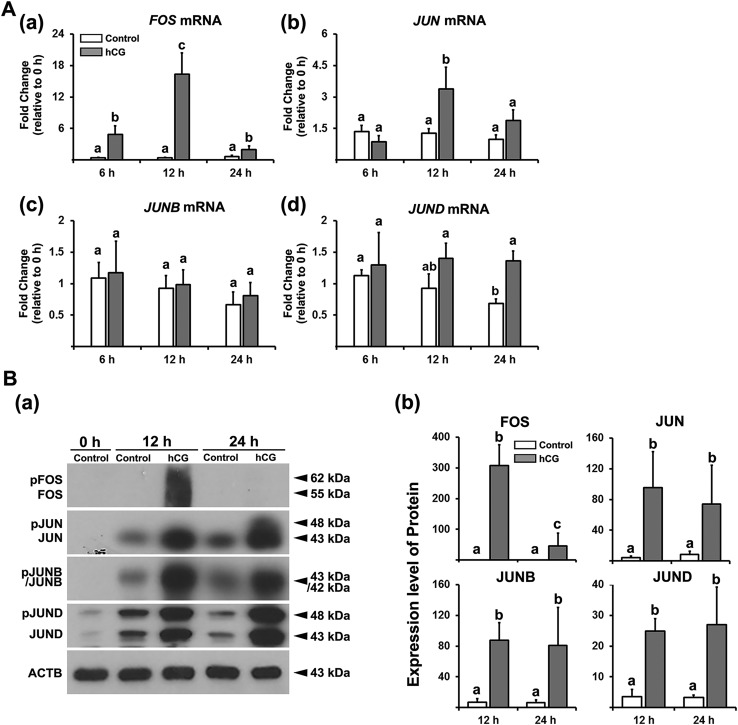
To determine whether the in vivo induction of FOS and JUN proteins during the periovulatory period can be mimicked by hCG in vitro, we used hGLCs. hCG induced transient increases in levels of mRNA and proteins for FOS [Fig. 2A(a) and 2B]. JUN mRNA and protein were also increased by hCG [Fig. 2A(b) and 2B]. Meanwhile, the levels of mRNA for JUNB and JUND were not altered by hCG [Fig. 2A(c) and 2A(d)], yet hCG increased JUNB and JUND proteins at 12 and 24 hours (Fig. 2B).
Figure 2.
The effect of hCG on the expression of FOS and Jun family members in hGLCs. Granulosa/lutein cells obtained from IVF patients were cultured for 6 d and then treated without (Control) or with hCG (1 IU/mL) for 6, 12, or 24 h. (A) The levels of mRNA for FOS and Jun family members were measured by quantitative PCR and normalized to the levels of RNA18S5 in each sample (n ≥ 4 independent experiments). Bars with no common superscripts are significantly different (P < 0.05). (B) (a) FOS, JUN, JUNB, and JUND (intact and phosphorylated forms) were detected by Western blots. The membrane was reprobed with a monoclonal antibody against ACTB to assess the loading of protein in each lane. (b) The band intensities for each protein were measured by ImageJ and normalized to the intensity of β-actin (ACTB) in the corresponding sample. The experiments were repeated three times with independent samples. Bars with no common superscripts are significantly different (P < 0.05).
Regulation of FOS expression by PGR and epidermal growth factor signaling
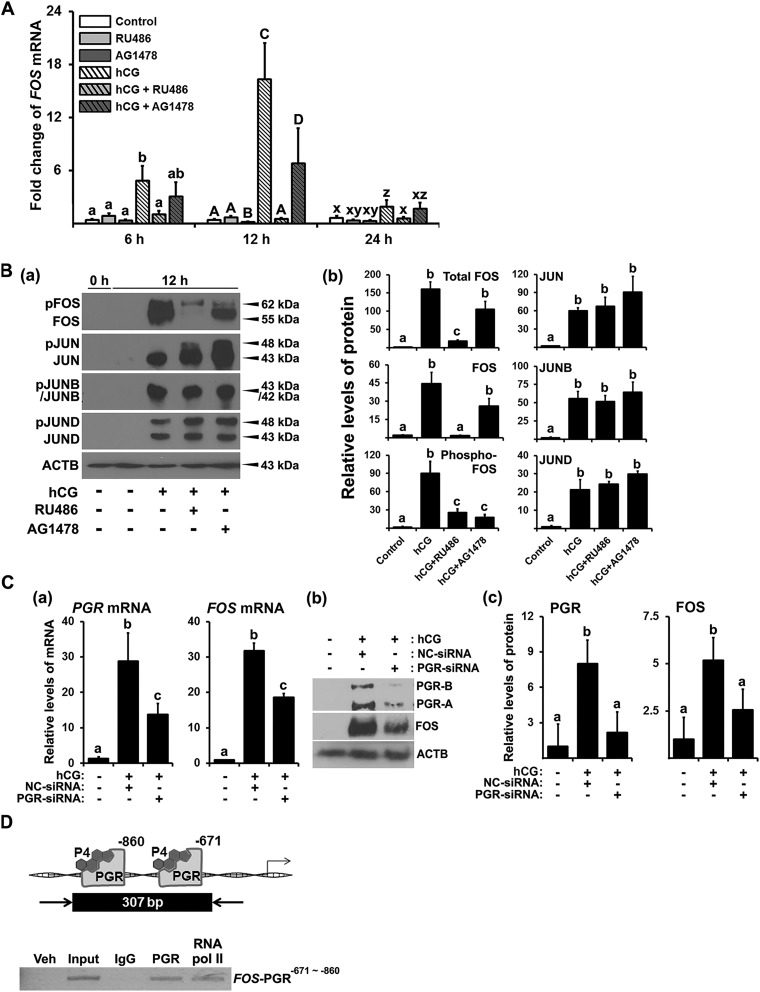
A previous study demonstrated that hCG increased progesterone/PGR and epidermal growth factor (EGF)–like peptides in cultured hGLCs, mimicking the in vivo expression of these factors in the human ovary (19). To determine whether progesterone/PGR and epidermal growth factor receptor (EGFR) signaling pathways are involved in the expression of FOS, the cells were treated with the PGR antagonist RU486 (20 µM) or the EGFR tyrosine kinase inhibitor AG1478 (10 µM). The dosage of inhibitors was chosen based on previous studies (19, 22). The hCG-induced increases in FOS mRNA were abolished by RU486 and partially inhibited by AG1478 (Fig. 3A). FOS and phosphorylated FOS were also drastically reduced in cells treated with RU486 (Fig. 3B). AG1478 reduced hCG-induced increases in FOS proteins, with the stronger reduction observed in phosphorylated FOS. In contrast, both RU486 and AG1478 had no significant effect on JUN proteins (Fig. 3B).
Figure 3.
The effects of antiprogestin, EGFR tyrosine kinase inhibitor, and PGR silencing RNA on the expression of FOS and Jun family members and binding of PGR to the FOS promoter region in hGLCs. (A) Primary hGLCs were treated with or without RU486 (progesterone receptor antagonist, 20 µM) or AG1478 (EGFR tyrosine kinase inhibitor, 10 µM) in the absence or presence of hCG (1 IU/mL) for 6, 12, or 24 h. The levels of mRNA for FOS were measured by quantitative PCR (qPCR) and normalized to those of RNA18S5 mRNA in each sample (n = 4 to 6 independent experiments). Bars with no common superscripts in each time point are significantly different (P < 0.05). (B) FOS, JUN, JUNB, and JUND were detected by Western blots. The membrane was reprobed with a monoclonal antibody against ACTB to assess the loading of protein in each lane. The band intensities for each protein were measured by ImageJ and normalized to the intensity of β-actin (ACTB) in the corresponding sample. The experiments were repeated three times with independent samples. (C) (a) The hGLCs were treated with or without hCG (1 IU/mL) in the presence of negative control (NC) or PGR silencing RNA for 10 h. The levels of mRNA for PGR and FOS were measured by qPCR and normalized to those of RNA18S5 mRNA in each sample (n = 4 independent experiments). (b) Proteins for PGR, FOS, and ACTB were detected by Western blots. (c) The band intensities for each protein were measured by ImageJ and normalized to the intensity of β-actin (ACTB) in the corresponding sample. Bars with no common superscripts in each time point are significantly different (P < 0.05). (D) Binding of PGR to the FOS promoter region was assessed by ChIP assay using hGLCs treated with hCG (1 IU/mL) for 12 h. Sheared DNA fragments immunoprecipitated with PGR antibody, RNA polymerase II antibody, or IgG and input DNA were analyzed using primer pairs specific to promoter regions of the FOS gene and represented as arrows. Amplified DNA fragments containing PGR responsive elements are represented as a black box with the indicated PCR product size. Experiments were repeated three times, each with independent patient samples. bp, base pair; veh, vehicle.
RU486 can block both PGR and glucocorticoid receptor (23). To verify that FOS expression is mediated by PGR, PGR small interfering RNA (siRNA) was used. hCG-induced PGR expression was suppressed by PGR siRNA, which in turn reduced the expression of FOS in hGLCs (Fig. 3C).
To determine whether PGR acts directly on the FOS gene, we performed ChIP analyses on hGLCs cultured with hCG for 12 hours. PCR data demonstrated the enrichment of chromatin fragments containing two PGR response elements (Supplemental Fig. 1A) in the 5′-flanking region of the human FOS gene in samples immune-precipitated with PGR antibody, indicating that PGR bound to the promoter region of the FOS gene (Fig. 3D).
Effect of FOS inhibition on prostaglandin synthesis and progesterone production in vitro
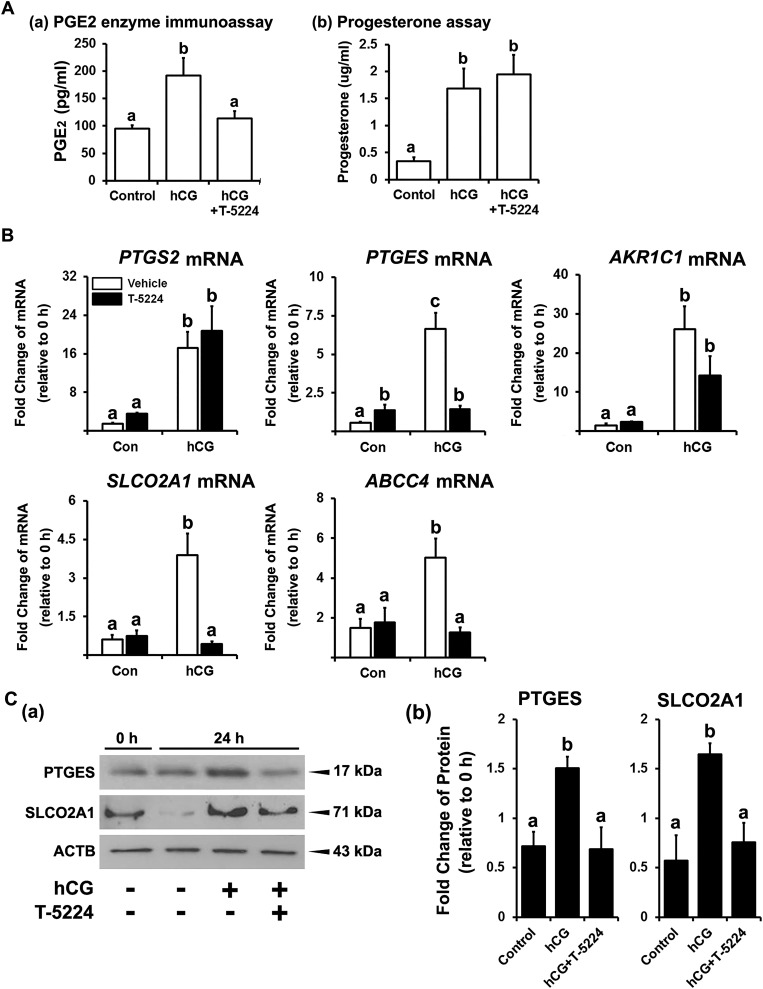
To investigate the function of FOS in human granulosa cells, we used a specific FOS inhibitor, T-5224, which blocks the binding of FOS to its specific binding site in the DNA (24). T-5224 inhibited hCG-stimulated increases in the levels of PGE2 [Fig. 4A(a)] but had no effect on progesterone production in hGLCs [Fig. 4A(b)].
Figure 4.
Effect of FOS inhibitor on the production of progesterone and PGE2 and on the expression of mRNA for PG synthases and transporters in hGLCs. Primary hGLCs were treated with or without T-5224 (FOS inhibitor, 20 µM) in the absence or presence of hCG (1 IU/mL). (A) The levels of PGE2 and progesterone in media collected at 24 h were measured by PGE2 (a) enzyme immunoassay and (b) progesterone assay, respectively. (B) The levels of mRNA for PG synthases (PTGS2, PTGES, and AKR1C1) and transporters (SLCO2A1 and ABCC4) at 12 h were measured by quantitative PCR. The levels of transcripts were normalized to those of RNA18S5 mRNA in each sample (n = 4 to 6 independent experiments). (C) (a) PTGES and SLCO2A1 were detected by Western blots. The membrane was reprobed with a monoclonal antibody against ACTB to assess the loading of protein in each lane. (b) The band intensities for each protein were measured by ImageJ and normalized to the intensity of β-actin (ACTB) in the corresponding sample. The experiments were repeated three times with an independent sample. Bars with no common superscripts in each time point are significantly different (P < 0.05). Con, control.
To determine the mechanisms by which FOS regulates PGE2 levels in hGLCs, we assessed the effect of T-5224 on the expression of several downstream target genes associated with prostaglandin (PG) synthesis and transport. T-5224 inhibited the hCG-induced increases in the levels of mRNA for PGE2 synthase, PTGES, and PG transporters, SLCO2A1 and ABCC4, but had no effects on the levels of mRNA for PTGS2 and prostaglandin F2α synthase, AKR1C1(Fig. 4B).
Binding of FOS to promoter regions of PTEGS, SLCO2A1, and ABCC4
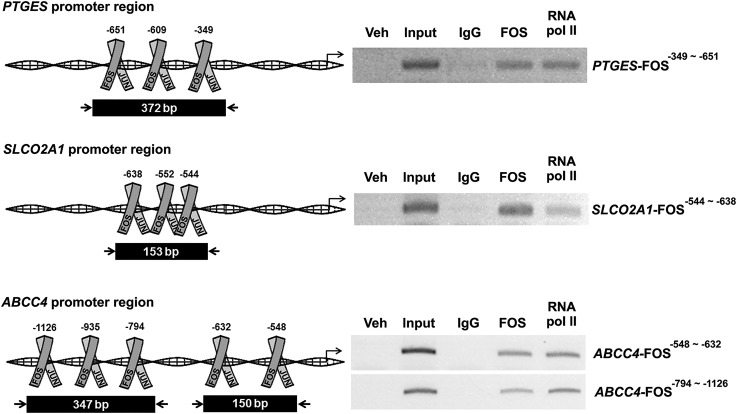
To assess how FOS regulates the transcription of PTGES, SLCO2A1, and ABCC4 in hGLCs, we screened for potential FOS/AP-1 binding sites in the 5′-flanking region of the human PTGES, SLCO2A1, and ABCC4 genes (Supplemental Fig. 1B–1D). ChIP analysis was conducted using hGLCs collected at 12 hours after hCG stimulation. Immunoprecipitation with FOS antibody, not control IgG, yielded the enriched chromatin fragments containing the FOS/AP-1 specific sites (Fig. 5), demonstrating that FOS bound to the promoter region of the human PTGES, SLCO2A1, and ABCC4 genes.
Figure 5.
Binding of FOS to promoter regions of PTGS2, PTGES, and SLCO2A1. ChIP assay was performed using hGLCs treated with hCG (1 IU/mL) for 12 h. DNA fragments immunoprecipitated with FOS antibody, RNA polymerase II antibody, or IgG and input DNA were analyzed using primer pairs specific to promoter regions of the PTGES, SLCO2A1, and ABCC4 genes and represented as arrows. Amplified DNA fragments containing FOS responsive elements are represented as black boxes with the indicated PCR product size. Experiments were repeated three times, each with independent patient samples. bp, base pair; veh, vehicle.
Discussion
Ovulation is a complex process that requires precisely coordinated actions of diverse intracellular and extracellular factors induced by the preovulatory LH surge. Specific transcription factors induced by the LH surge play a critical role in this process by regulating the transcription of genes that exert targeted and regulated actions in ovulatory follicles. Accumulating evidence has demonstrated the necessity of several transcription factors for successful ovulation and luteal formation such as CCAAT/enhancer binding protein α/β (25), PGR (26), Nr5a2 (27), and core binding factors (28). Adding to the list of these transcription factors, the current study unveiled for the first time, to our knowledge, that the expression of FOS and Jun family members is induced in human ovulatory follicles, and FOS plays an important role in the expression of key ovulatory genes critical for successful ovulation in the human ovary.
Using dominant follicles collected from normally cycling women, we observed that the expression of FOS, JUNB, and JUND increases after hCG administration during the periovulatory period. Interestingly, the levels of mRNA for JUN were not changed throughout ovulatory phases, whereas the immunopositive staining for JUN protein was detected in dominant follicles obtained only after hCG administration. Therefore, it remains to be determined how JUN protein is accumulated, without transcriptional upregulation in ovulatory follicles. Nevertheless, these data together demonstrated that hCG increases FOS and all Jun family members, JUN, JUNB, and JUND, in human periovulatory follicles. To be functional, FOS needs to form a heterodimeric complex with one of the Jun family members, whereas JUN proteins can dimerize with the same or different Jun members to form the AP-1 complex (1, 29). Therefore, our findings indicate that multiple combinations of heterodimers and homodimers of the AP-1 complex may exist in human ovulatory follicles and function to differentially regulate the expression of genes involved in the periovulatory process.
In the light of our findings demonstrating the periovulatory upregulation of FOS and JUN proteins, we have investigated the regulatory mechanisms by which hCG increases the expression of these genes. Previously, we have shown that hCG increases the expression of key ovulatory genes, including PGR, PTGS2, and AREG in our hGLC cultures, similar to the in vivo induction of these genes (19). Similarly, hCG increased the expression of FOS, yet the expression was transient and peaked at 12 hours. Meanwhile, hCG increased the levels of mRNA for JUN but not JUNB and JUND, exhibiting a discrepancy in transcriptional regulation of these genes between the in vivo and in vitro models. However, the levels of JUN, JUNB, and JUND proteins were increased by hCG in hGLC cultures, suggesting that the accumulation of JUN proteins by hCG is likely mediated at the posttranscriptional level such as phosphorylation and protein stabilization in vitro. Importantly, our in vitro study showed that hCG increased the expression of FOS and the levels of JUN proteins during the first 12 hours of cultures, similar to those observed in the in vivo model.
Among the cellular pathways activated by the LH surge or hCG administration, progesterone/PGR and EGF signaling pathways are essential for ovulation (26, 30). Using our in vitro model where hCG activates key ovulatory pathways such as progesterone/PGR, EGF signaling, and prostaglandin production (19), we have investigated whether these pathways are involved in hCG-induced increases in FOS expression and accumulation of JUNs. Indeed, the progesterone/PGR pathway played a critical role in mediating hCG-induced increases in FOS expression. Support for this concept was the finding that blocking the action of PGR with RU486 and knocking down the expression of PGR by siRNA inhibited hCG-induced increases in FOS expression. Moreover, ChIP data suggest that PGR could act as a transcriptional activator mediating the increases in FOS expression in human granulosa cells. The regulation of progesterone/PGR on FOS expression is not unique to granulosa cells. In breast cancer cell lines, progestins increased the expression of FOS (31), whereas in neurons, a progestin antagonist blocked Fos expression (32). In addition to progesterone/PGR, the present data indicated that the activation of EGF signaling is involved in the transcription and phosphorylation of FOS. For instance, the inhibition of EGFR activation by AG1478 reduced the levels of FOS mRNA and phosphorylated FOS protein. This is consistent with the data from previous studies showing that EGF treatment increased the levels of FOS mRNA in bovine luteal cells (33) and nonovarian cells (34). Besides the transcriptional regulation, the phosphorylation of FOS by various kinases has been reported to be one of the mechanisms controlling the cellular levels and activity of FOS (5, 6, 35). ERK1/2 has been shown to stabilize FOS through direct phosphorylation (36, 37), thereby allowing heterodimerization with JUN proteins to form transcriptionally active AP-1 complexes. In hGLC cultures, the stimulation of EGFR by its ligands was shown to activate ERK1/2 (22). Therefore, it needs to be further determined whether EGF signaling increases the expression and phosphorylation of FOS through the activation of ERK1/2 in hGLC cultures. Interestingly, none of the JUN proteins was reduced by inhibition of PGR or EGFR, indicating that hCG increases the accumulation of JUN proteins using posttranscriptional mechanisms independent of progesterone/PGR and EGF signaling pathways.
As a component of the AP-1 transcription factor, FOS has been shown to regulate the expression of diverse genes in a variety of cell types (38, 39). Of particular interest is a report showing that FOS regulates the expression of Ptgs2 and PGE2 production in the amnion during pregnancy and labor in mice (14). In the ovary, the LH surge induces a rapid increase in PG production in ovulatory follicles, which is essential for successful ovulation in various species, including nonhuman primates (40). Therefore, we have hypothesized that FOS is involved in the rise of PG in human periovulatory follicles. This hypothesis was tested by inhibiting the action of FOS by using T-5224 in a hGLC model in which hCG can increase FOS expression and PG production (19). T-5224 is a selective FOS inhibitor whose mechanism of action is to block the DNA binding activity of FOS (41). The present data indicated that T-5224 reduced hCG-induced increases in PGE2 and levels of mRNA for PGE2 synthase, PTGES, and PG transporters, SLCO2A1 and ABCC4. ChIP data also revealed that FOS bound to the promoter regions of these genes, suggesting the direct transcriptional action of FOS on these genes. Together, these data indicated that FOS plays an important role in the hCG-induced PG synthesis and transport by acting as a transcriptional regulator in human periovulatory granulosa cells. In addition to PG, FOS has been shown to transcriptionally repress StAR expression in rat luteal cells (12). However, we found that T-5224 has no effect on hCG-induced increases in STAR expression (Supplemental Fig. 3) and progesterone production in human granulosa cells.
In summary, the current study provides novel findings, to our knowledge, that document spatiotemporally regulated expression of FOS and Jun family members by hCG in dominant follicles during the ovulatory period in humans. We discovered that the hCG-induced increase in FOS expression is dependent on collaborative actions of progesterone/PGR and EGF signaling pathways as a direct transcriptional activator and posttranscriptional enhancer, respectively. As a transcription factor, FOS/AP-1 upregulated the expression of genes involved in PG production and transport in hGLCs. On the basis of these findings, we conclude that FOS/AP-1 plays an important role in ovulation by acting as a key downstream mediator of the progesterone/PGR and EGF signaling pathways necessary for PG production and transport in human ovulatory follicles. Because a phase II clinical trial is under way for T-5224, this study provides fundamental information that might help in dissecting mechanisms of the ovulatory process in humans and for the development of novel/improved contraceptives.
Supplementary Material
Acknowledgments
Financial Support: This research was supported by the Lalor Foundation Postdoctoral Fellowship (to Y.C.), P01HD71875 (to M.J., T.E.C., and M.B.) and R03HD095098 (to M.J.) from the Foundation for the National Institutes of Health, and the BTPSRF of the University of Kentucky Markey Cancer Center (P30CA177558).
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- AP-1
activator protein 1
- ChIP
chromatin immunoprecipitation
- CL
corpus luteum
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- hCG
human chorionic gonadotropin
- hGLC
human granulosa/lutein cell
- IgG
immunoglobulin G
- IVF
in vitro fertilization
- PG
progesterone
- PGE2
prostaglandin E2
- PGR
progesterone receptor
- siRNA
small interfering RNA
References
- 1. Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117(Pt 25):5965–5973. [DOI] [PubMed] [Google Scholar]
- 2. Chiu R, Boyle WJ, Meek J, Smeal T, Hunter T, Karin M. The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes. Cell. 1988;54(4):541–552. [DOI] [PubMed] [Google Scholar]
- 3. Halazonetis TD, Georgopoulos K, Greenberg ME, Leder P. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell. 1988;55(5):917–924. [DOI] [PubMed] [Google Scholar]
- 4. Piechaczyk M, Blanchard JM. c-fos proto-oncogene regulation and function. Crit Rev Oncol Hematol. 1994;17(2):93–131. [DOI] [PubMed] [Google Scholar]
- 5. Tanos T, Marinissen MJ, Leskow FC, Hochbaum D, Martinetto H, Gutkind JS, Coso OA. Phosphorylation of c-Fos by members of the p38 MAPK family: role in the AP-1 response to UV light. J Biol Chem. 2005;280(19):18842–18852. [DOI] [PubMed] [Google Scholar]
- 6. Abate C, Marshak DR, Curran T. Fos is phosphorylated by p34cdc2, cAMP-dependent protein kinase and protein kinase C at multiple sites clustered within regulatory regions. Oncogene. 1991;6(12):2179–2185. [PubMed] [Google Scholar]
- 7. Ness JM, Kasson BG. Gonadotropin regulation of c-fos and c-jun messenger ribonucleic acids in cultured rat granulosa cells. Mol Cell Endocrinol. 1992;90(1):17–25. [DOI] [PubMed] [Google Scholar]
- 8. Sharma SC, Richards JS. Regulation of AP1 (Jun/Fos) factor expression and activation in ovarian granulosa cells: relation of JunD and Fra2 to terminal differentiation. J Biol Chem. 2000;275(43):33718–33728. [DOI] [PubMed] [Google Scholar]
- 9. Rusovici R, LaVoie HA. Expression and distribution of AP-1 transcription factors in the porcine ovary. Biol Reprod. 2003;69(1):64–74. [DOI] [PubMed] [Google Scholar]
- 10. Dias FC, Khan MI, Sirard MA, Adams GP, Singh J. Differential gene expression of granulosa cells after ovarian superstimulation in beef cattle. Reproduction. 2013;146(2):181–191. [DOI] [PubMed] [Google Scholar]
- 11. Wissing ML, Kristensen SG, Andersen CY, Mikkelsen AL, Høst T, Borup R, Grøndahl ML. Identification of new ovulation-related genes in humans by comparing the transcriptome of granulosa cells before and after ovulation triggering in the same controlled ovarian stimulation cycle. Hum Reprod. 2014;29(5):997–1010. [DOI] [PubMed] [Google Scholar]
- 12. Shea-Eaton W, Sandhoff TW, Lopez D, Hales DB, McLean MP. Transcriptional repression of the rat steroidogenic acute regulatory (StAR) protein gene by the AP-1 family member c-Fos. Mol Cell Endocrinol. 2002;188(1-2):161–170. [DOI] [PubMed] [Google Scholar]
- 13. Diaz FJ, Luo W, Wiltbank MC. Prostaglandin F2α regulation of mRNA for activating protein 1 transcriptional factors in porcine corpora lutea (CL): lack of induction of JUN and JUND in CL without luteolytic capacity. Domest Anim Endocrinol. 2013;44(2):98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li H, Zhou J, Wei X, Chen R, Geng J, Zheng R, Chai J, Li F, Jiang S. miR-144 and targets, c-fos and cyclooxygenase-2 (COX2), modulate synthesis of PGE2 in the amnion during pregnancy and labor. Sci Rep. 2016;6(1):27914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072(2-3):129–157. [DOI] [PubMed] [Google Scholar]
- 16. Richards JS, Russell DL, Ochsner S, Espey LL. Ovulation: new dimensions and new regulators of the inflammatory-like response. Annu Rev Physiol. 2002;64(1):69–92. [DOI] [PubMed] [Google Scholar]
- 17. Stocco C, Telleria C, Gibori G. The molecular control of corpus luteum formation, function, and regression. Endocr Rev. 2007;28(1):117–149. [DOI] [PubMed] [Google Scholar]
- 18. Xie C, Jonak CR, Kauffman AS, Coss D. Gonadotropin and kisspeptin gene expression, but not GnRH, are impaired in cFOS deficient mice. Mol Cell Endocrinol. 2015;411:223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Choi Y, Wilson K, Hannon PR, Rosewell KL, Brännström M, Akin JW, Curry TE Jr, Jo M. Coordinated regulation among progesterone, prostaglandins, and EGF-like factors in human ovulatory follicles. J Clin Endocrinol Metab. 2017;102(6):1971–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Al-Alem L, Puttabyatappa M, Rosewell K, Brännström M, Akin J, Boldt J, Muse K, Curry TE Jr. Chemokine ligand 20: a signal for leukocyte recruitment during human ovulation? Endocrinology. 2015;156(9):3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- 22. Fang L, Yu Y, Zhang R, He J, Sun YP. Amphiregulin mediates hCG-induced StAR expression and progesterone production in human granulosa cells. Sci Rep. 2016;6(1):24917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Spitz IM, Bardin CW. Mifepristone (RU 486)—a modulator of progestin and glucocorticoid action. N Engl J Med. 1993;329(6):404–412. [DOI] [PubMed] [Google Scholar]
- 24. Ye N, Ding Y, Wild C, Shen Q, Zhou J. Small molecule inhibitors targeting activator protein 1 (AP-1). J Med Chem. 2014;57(16):6930–6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fan HY, Liu Z, Johnson PF, Richards JS. CCAAT/enhancer-binding proteins (C/EBP)-α and -β are essential for ovulation, luteinization, and the expression of key target genes. Mol Endocrinol. 2011;25(2):253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robker RL, Russell DL, Espey LL, Lydon JP, O’Malley BW, Richards JS. Progesterone-regulated genes in the ovulation process: ADAMTS-1 and cathepsin L proteases. Proc Natl Acad Sci USA. 2000;97(9):4689–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bertolin K, Gossen J, Schoonjans K, Murphy BD. The orphan nuclear receptor Nr5a2 is essential for luteinization in the female mouse ovary. Endocrinology. 2014;155(5):1931–1943. [DOI] [PubMed] [Google Scholar]
- 28. Lee-Thacker S, Choi Y, Taniuchi I, Takarada T, Yoneda Y, Ko C, Jo M. Core binding factor β expression in ovarian granulosa cells is essential for female fertility. Endocrinology. 2018;159(5):2094–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Curran T, Franza BR Jr. Fos and Jun: the AP-1 connection. Cell. 1988;55(3):395–397. [DOI] [PubMed] [Google Scholar]
- 30. Park JY, Su YQ, Ariga M, Law E, Jin SL, Conti M. EGF-like growth factors as mediators of LH action in the ovulatory follicle. Science. 2004;303(5658):682–684. [DOI] [PubMed] [Google Scholar]
- 31. Musgrove EA, Lee CS, Sutherland RL. Progestins both stimulate and inhibit breast cancer cell cycle progression while increasing expression of transforming growth factor alpha, epidermal growth factor receptor, c-fos, and c-myc genes. Mol Cell Biol. 1991;11(10):5032–5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blaustein JD, Gréco B. A progestin antagonist blocks vaginocervical stimulation-induced fos expression in neurones containing progestin receptors in the rostral medial preoptic area. J Neuroendocrinol. 2002;14(2):109–115. [DOI] [PubMed] [Google Scholar]
- 33. Chen DB, Davis JS. Epidermal growth factor induces c-fos and c-jun mRNA via Raf-1/MEK1/ERK-dependent and -independent pathways in bovine luteal cells. Mol Cell Endocrinol. 2003;200(1–2):141–154. [DOI] [PubMed] [Google Scholar]
- 34. Takeuchi K, Shibamoto S, Nagamine K, Shigemori I, Omura S, Kitamura N, Ito F. Signaling pathways leading to transcription and translation cooperatively regulate the transient increase in expression of c-Fos protein. J Biol Chem. 2001;276(28):26077–26083. [DOI] [PubMed] [Google Scholar]
- 35. Abate C, Baker SJ, Lees-Miller SP, Anderson CW, Marshak DR, Curran T. Dimerization and DNA binding alter phosphorylation of Fos and Jun. Proc Natl Acad Sci USA. 1993;90(14):6766–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nat Cell Biol. 2002;4(8):556–564. [DOI] [PubMed] [Google Scholar]
- 37. Monje P, Marinissen MJ, Gutkind JS. Phosphorylation of the carboxyl-terminal transactivation domain of c-Fos by extracellular signal-regulated kinase mediates the transcriptional activation of AP-1 and cellular transformation induced by platelet-derived growth factor. Mol Cell Biol. 2003;23(19):7030–7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Braselmann S, Bergers G, Wrighton C, Graninger P, Superti-Furga G, Busslinger M. Identification of Fos target genes by the use of selective induction systems. J Cell Sci Suppl. 1992;16:97–109. [DOI] [PubMed] [Google Scholar]
- 39. Malik AN, Vierbuchen T, Hemberg M, Rubin AA, Ling E, Couch CH, Stroud H, Spiegel I, Farh KK, Harmin DA, Greenberg ME. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat Neurosci. 2014;17(10):1330–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Duffy DM. Novel contraceptive targets to inhibit ovulation: the prostaglandin E2 pathway. Hum Reprod Update. 2015;21(5):652–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Aikawa Y, Morimoto K, Yamamoto T, Chaki H, Hashiramoto A, Narita H, Hirono S, Shiozawa S. Treatment of arthritis with a selective inhibitor of c-Fos/activator protein-1. Nat Biotechnol. 2008;26(7):817–823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.