Abstract
Morphological abnormalities of the bounding membranes of the nucleus have long been associated with human diseases from cancer to premature aging to neurodegeneration. Studies over the past few decades support that there are both cell intrinsic and extrinsic factors (e.g. mechanical force) that can lead to nuclear envelope “herniations”, a broad catch-all term that reveals little about the underlying molecular mechanisms that contribute to these morphological defects. While there are many genetic perturbations that could ultimately change nuclear shape, here, we focus on a subset of nuclear envelope herniations that likely arise as a consequence of disrupting physiological nuclear membrane remodeling pathways required to maintain nuclear envelope homeostasis. For example, stalling of the interphase nuclear pore complex (NPC) biogenesis pathway and/or triggering of NPC quality control mechanisms can lead to herniations in budding yeast, which are remarkably similar to those observed in human disease models of early-onset dystonia. By also examining the provenance of nuclear envelope herniations associated with emerging nuclear autophagy and nuclear egress pathways, we will provide a framework to help understand the molecular pathways that contribute to nuclear deformation.
Keywords: Nuclear envelope, nuclear pore complex, herniation, ESCRT, Torsin, nucleophagy, nuclear egress
Introduction
Alterations to the shape of the nucleus is a hallmark pathognomonic cellular feature of many human diseases, but it is often challenging to determine the underlying causality of these abnormalities and whether they ultimately directly contribute to disease progression[1,2]. Such is the case with the majority of examples where nuclear envelope (NE) protrusions or “herniations” have been observed extending into the cytosol. As these herniations show a continuum of size (from nano to microscale), they are challenging to categorize by morphology alone and could thus benefit from a greater understanding of the underlying molecular mechanisms that cause them. Interestingly, recent work has helped to define how physiological nuclear membrane remodeling might, when perturbed, contribute to the biogenesis of some NE herniations; they are providing a retrospective framework to help interpret the many herniations observed over the past few decades in diverse model systems. Here, we will focus on the studies that primarily explore herniations visible at the nanoscale (see Table 1) while also touching on those visible by light microscopy (often also termed NE “blebbing” or “ruffling”) observed in other contexts and more comprehensively covered elsewhere [2–5].
Table 1.
Genetic backgrounds with NE evagination and herniations
| Gene (Genetic perturbation) |
Function | Phenotype | Organism | Ref. | |
|---|---|---|---|---|---|
| NPC biogenesis/quality control | |||||
|
NUP170(repression) NUP157(deletion) |
Inner ring scaffold nucleoporins |
INM-evaginations | S. cerevisiae | [16] | INM-evagination |
| POM33 (deletion) | Transmembrane nucleoporin |
INM-evaginations | S. cerevisiae | [17] | |
|
RTN1/YOP1 (double deletion) |
Reticulon like protein (RTN1), DP1 like protein (YOP1) |
INM-evaginations | S. cerevisiae | [19] | |
| STH1 (ts allele) | RSC-Chromatin Remodeling Complex |
INM-evaginations, expanded PNS/NE |
S. cerevisiae | [20] | |
|
KAP95 (ts point mutation) |
Karyopherin/Importin | INM-evaginations, ONM invagination, NE membrane rings with stacked NPCs |
S. cerevisiae | [21] | |
| NTF2 (ts allele) | Nuclear transport, RanGDP binding protein |
INM-evaginations, nup mis-localization, flattened/loosely stacked membranes |
S. cerevisiae | [23] | |
|
GLE2 (deletion or ts allele) |
RNA export platform nucleoporin |
NE herniations, intranuclear annulate lamellae, mRNA export defect |
S. cerevisiae | [51] | NE herniation |
|
NUP116 (deletion, or truncation) |
Cytoplasmic/mRNA export platform, GLFG repeat nucleoporin |
NE herniations, RNA export defect, intranuclear annulate lamellae. (C-terminal deletion results INM evaginations only) |
S. cerevisiae | [52–54] | |
| NUP145N (deletion) | GLFG repeat nucleoporin |
NE herniations, “grape- like” clustering of NPCs |
S. cerevisiae | [46,55] | |
|
NUP145N/NUP100
(double deletion) |
GLFG repeat nucleoporins |
NE herniations, “grape- like” clustering |
S. cerevisiae | [56] | |
| NUP85 (deletion) | Y-complex/outer ring nucleoporin |
clustering of NPCs, herniations, RNA export defect |
S. cerevisiae | [38] | |
| NUP120 (deletion) | Y-complex/outer ring nucleoporin |
NE herniations, clustering of NPCs, RNA export defect |
S. cerevisiae | [42,43] | |
|
NUP145C
(truncation, ts allele) |
Y-complex/outer ring nucleoporin |
NE herniations, clustering of NPCs, RNA export defect |
S. cerevisiae | [44] | |
| SEC13 (ts alleles) | Y-complex/outer ring nucleoporin, COPII coat protein |
NE herniations, “grape- like” clustering of NPCs, disruption of ER network |
S. cerevisiae | [47,48] | |
|
NUP188 (ts allele or combination with NUP116 GLFG deletion) |
Inner ring scaffold nucleoporin |
NE herniations | S. cerevisiae | [39–41,49] | |
| NUP154 (allele) | Inner ring scaffold nucleoporin |
NE herniations, mis- localization of nucleoporins |
Drosophila | [50] | |
| NUP57 (ts allele) | FG repeat containing central channel nucleoporin |
NE herniations | S. cerevisiae | [37] | |
| MTR10/KAP111 (ts allele) | Karyopherin/Importin | NE herniations, NPC clustering, RNA export defect, expansion of PNS |
S. cerevisiae | [22] | |
| ACC1 (ts allele) | Acetyl-coA carboxylase | NE herniations, expansion of PNS, RNA export defect |
S. cerevisiae | [22,57] | |
| APQ12 (deletion) | Transmembrane NE protein, NE lipid composition regulator |
NE herniations at 23 °C and 37 °C. Hypersensitivity to membrane fluidizers. Mis-localization of cytoplasmic filaments/RNA export platform and outer ring components at 23 °C, RNA export defect |
S. cerevisiae | [59,60] | |
| BRR6 (ts allele) | Transmembrane NE protein, ill-defined function, possible NE lipid composition regulator |
NE herniations at 18 °C, RNA export defect |
S. cerevisiae | [60,74] | |
|
BRR6, BRL1
(tandem depletion) |
Transmembrane NE proteins, mostly unknown function, possible NE lipid composition regulator |
NE herniations, NPC clustering |
S. cerevisiae | [73] | |
|
MGA2 (deletion) SPT23 (ts allele) |
Regulator of OLE1
transcription |
NE herniations, expansion of PNS, intralumenal vesicles |
S. cerevisiae | [58] | |
| NPL4 (ts alleles) | Component of Ufd- Cdc48 complex, ERAD, and NE reformation in mammalian cells |
NE herniations filled with electron dense material (poly-A RNA), RNA export defect |
S. cerevisiae | [116] | |
| POM152/VPS4
(double deletion) |
(POM152) Transmembrane nucleoporin, (VPS4) AAA-ATPase for ESCRT based membrane fission |
NE herniations, NPC clustering |
S. cerevisiae | [14] | |
|
TOR1A/B/3A/2A
(deletion) |
Ill-defined function | NE herniations containing K48 ubiquitin labling |
HeLa cells | [63] | |
| Torsin (knockdown) | Ill-defined function | NE herniations, abnomal DFz2C foci |
Drosophila | [70] | |
| ooc-5 (knockdown) | TorA homolog, ill- defined function |
NE herniations, mislocalization of nups and clustering of NPCs |
C. elegans | [69] | |
|
TOR1A (conditional deletion) |
Ill-defined function | NE herniations, abnormal LaminA/C and Sun1 localization |
M. musculus | [65,67,71] | |
| TOR1A ∆E | Ill-defined function, single amino acid deletion at aa302/303 Tor1A found in dystonia patients |
NE herniations | M. musculus | [64] | |
|
TOR1A EQ (overexpression) |
Ill-defined function, construct with point mutation in Walker B domain at aaE171Q, abolishing ATPase activity |
NE herniations, regions of abnormally narrow PNS |
Chinese hamster ovary cells |
[117] | |
|
LAP1/LULL1
(knockdown) |
Transmembrane Torsin co-factors, ill-defined function |
NE herniations | HeLa cells | [63] | |
ts = temperature sensitive
The nuclear envelope
The NE is contiguous with the endoplasmic reticulum (ER) and thus surrounds the genome in two biochemically distinct membranes, the outer and inner nuclear membrane (ONM and INM); the ONM and INM are separated by the perinuclear space (PNS; Figure 1). While the ONM is generally considered to be near-compositionally identical to the ER (with some exceptions, e.g. KASH-domain proteins[6]), the INM has a unique complement of integral and peripheral membrane-associated proteins that directly interface with the genome[6]. The most well described of these are the nuclear lamins, that while absent from yeasts, form a filamentous scaffold that provides mechanical support to the nucleus and helps maintain its integrity[5].
Figure 1.
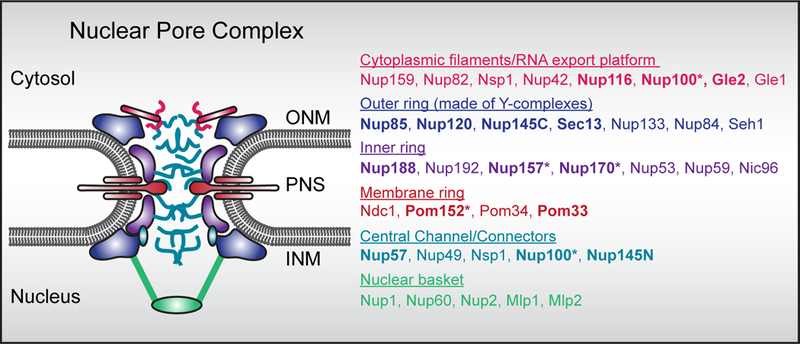
A. Schematic of the nuclear pore complex with color coded nup subcomplexes. Only budding yeast nup names are listed. Loss of function of bolded nups are associated with NE herniations either alone or upon loss of function of an additional gene (indicated by *). Also see Table 1.
Molecular traffic across the NE is controlled by nuclear pore complexes (NPCs), ~100 MD protein channels in Metazoans (~50 MD in budding yeast) that are formed by concentric inner, outer and membrane ring assemblies that scaffold a central transport channel, cytosolic filaments and nuclear basket (Figure 1)[7,8]. These architectural units are themselves constructed from modular subcomplexes of proteins termed nucleoporins or nups that are assembled step-wise into the NE, with an emerging role for unstructured motifs that connect subcomplexes together to help build the NPC[8–10]. There remains considerable interest in understanding the biochemical and morphological differences that define post-mitotic and interphase modes of NPC assembly[11–13]. Of note, recent evidence supports that post-mitotic NPC assembly begins within small (<50 nm) pores in the reforming NE, whereas interphase NPC assembly requires an INM-ONM fusion event[12,13]. Interestingly, the molecular fusogen that drives INM-ONM fusion has eluded genetic (or biochemical) identification. Identifying the fusion mechanism is a priority as it is becoming clear that some NE herniations likely arise due to defects in NPC biogenesis, and/or, surveillance mechanisms that seal off defective NPCs[14,15].
Blockade of early interphase NPC biogenesis steps
A commonly observed ultrastructural feature of the NE are small INM evaginations with a base diameter of ~40–60 nm that do not impact ONM morphology[16–23](Figure 2A, B, Table 1, blue rows).Thus, these structures fall short of “herniating” the NE, but they are likely progenitors to the more elaborate NE herniations described below. In virtually all cases where these structures have been observed, they are associated with genetic perturbation of genes encoding components of the NPC[16], NPC assembly factors[17,19], or the soluble nuclear transport apparatus including nuclear transport receptors[21,22] and the Ran GTPase[23]. This has led to the concept that these evaginations are stalled biochemical/structural intermediates in early NPC assembly. Consistent with this idea, conditional depletion of the inner ring nup gene, NUP170 in a genetic background where its paralogue NUP157 is also deleted, leads to the INM-accumulation of a distinct set of nups including components of the nuclear basket[16]. These data, in addition to other genetic and biochemical links between the INM and specific nup genes[24–29], support that key assembly events occur at the INM. Indeed, the collation of EM tomograms into a morphological timeline of interphase NPC assembly is consistent with the idea that assembly proceeds inside-out with the earliest discernable morphological feature being INM evaginations[12](Figure 2A).
Figure 2.
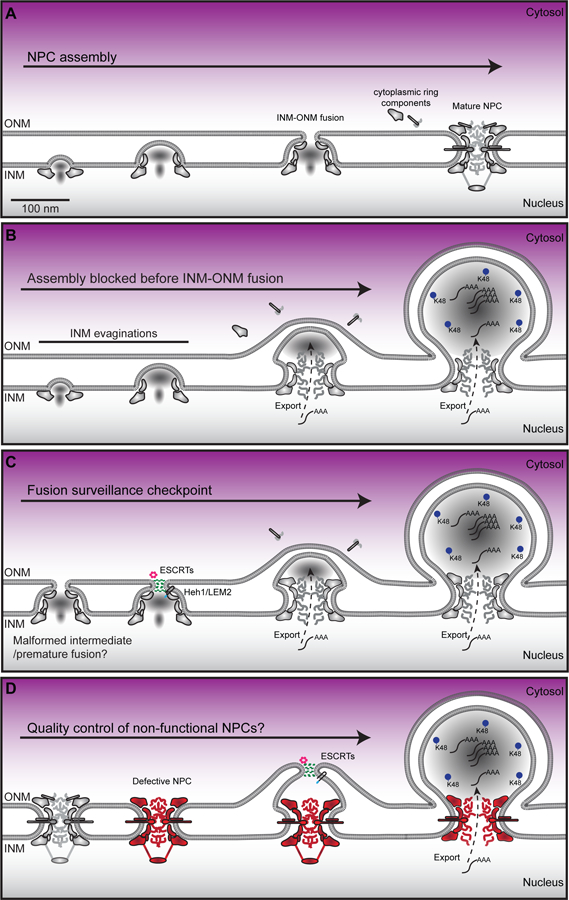
Potential mechanisms that lead to NE herniations associated with NPC biogenesis and/or quality control. A. De novo interphase NPC assembly begins by recruitment of nups to the INM followed by INM evagination. As assembly progresses fusion occurs between the INM and ONM, which is followed by the assembly of the cytosolic filaments/mRNA export platform. B. Model of herniation formation due to a block in NPC assembly before INM-ONM fusion. Poly-A (AAA) RNA has been found in herniations as has K48-linked ubiquitin. C. Model of herniation formation due to NPC assembly surveillance by the ESCRT complex (green) and the AAA ATPase Vps4 (pink) recruited by Heh1/LEM2. D. Model of herniation formation due to loss of function (e.g. oxidative damage to NPCs/degradation of nups) of NPCs. Note this model would require expansion of the pore membrane before sealing by the ESCRT (or other?) pathway.
Blockade in late interphase NPC assembly
It is likely that these small INM-evaginations can also lead to a much larger NE herniation where one can discern a neck at the INM of ~80–100 nm, often with considerable electron density enclosed within the protrusion (Figure 2B). While such a structure is not visible during “normal” NPC biogenesis, it is likely that it too is caused by a delay or abrogation of NPC assembly, but at a later step than the small INM evaginations that precede it (Table 1, teal rows). Indeed, as NPC assembly progresses, a “mushroom” like density appears to evaginate the INM before its fusion with the ONM[12](Figure 2A). These events likely coincide with the recruitment of additional nup subcomplexes capable of forming an 8-fold radially-symmetric structure (likely the nucleoplasmic outer ring), which can be visualized in en face sections of the INM[12]. Moreover, at least a subset of Phe-Gly (FG) repeat rich nups, the key proteins that establish the diffusion barrier and transport selectivity of NPCs, are also assembled before INM-ONM fusion. These observations, in the context of several other studies[19,24,30–34], have contributed to a rough ordering of nup subcomplex recruitment to a nascent NPC assembly site, with a key transition point occurring at or just after INM-ONM fusion. At this critical step, the cytosolic-facing mRNA export platform is assembled alongside other cytosolic-facing nups like Nup358, which anchors two concentric outer ring “Y-complexes” together on the cytosolic side of the NPC (at least in Metazoans[35], budding yeast only have one Y-complex ring and lack Nup358[8,36]), effectively completing NPC assembly. We suggest that genetic perturbations that prevent INM-ONM fusion lead to the formation of NE herniations (Figure 2B).
Consistent with the idea that NE herniations might arise due to a blockage of late steps in NPC assembly, they have been observed upon perturbation of several scaffold nups in yeast[37–49] and in flys[50]. In addition, due to the lack of assembly of the mRNA export platform, these alleles are often associated with an mRNA export block. For example, in budding yeast, this includes knockouts or conditional alleles of GLE2[51], NUP116[52–54] and its paralogues[46,55,56] but also extends to proteins that impact lipid homeostasis[57,58] including a membrane protein that alters membrane fluidity encoded by APQ12[59–61](first identified in a screen to uncover mRNA-export mechanisms[62]).
Interestingly, morphologically analogous herniations have also been observed in human[63], mouse[64–68], worm[69], and fly[60] cells upon disruption of the function of the ER-lumenal AAA+ ATPase Torsin A and its membrane-spanning co-factors LAP1 and LULL1[63]. Recent high resolution EM tomography studies of Torsin knockout HeLa cells further supports the conclusion that these herniations arise due to a disruption in NPC assembly, as their bases have NPC-like structures[63]. In addition, similar to the yeast counterparts, the cytosolic-facing nup, Nup358, might not be properly assembled[71]. A key challenge going forward will be to define the mechanism of Torsin A function in NPC biogenesis as it will also inform how an in-frame deletion of a glutamate residue in the Torsin A gene causes early onset dystonia[72]. But, as it is absent from yeasts, it is most likely that Torsin A plays an indirect role in NPC assembly, perhaps by controlling access of a yet-to-be defined fusogen to a nascent pore assembly site. In yeast, such a role could be performed by the essential integral NE proteins Brl1 and Brr6, which localize at INM evaginations and physically interact with nups[73]. Consistent with the interpretation that they are required for NPC assembly, NE herniations build up in the absence Brr6[60,74] or Brl1/Brr6 function[73]. While it remains to be determined how Brl1 or Brr6 contribute to INM-ONM fusion, it is remarkable that overexpression of BRL1 prevents the formation of herniations (or perhaps drives their resolution) in nup116Δ strains[73].
Taken together, the consistency in the appearance of morphologically similar NE herniations from yeast to human suggests that the INM-ONM fusion step during interphase NPC assembly is susceptible to perturbation and could be under control of regulatory mechanisms that might (for example) trigger fusion at a point where the nascent NPC is “ready” to receive cytosolic-facing components (fusion/surveillance checkpoint, see Figure 2C). Consistent with this idea, disruption of newly discovered interactions between the FG-repeats (of the GLFG-type) that fill the central transport channel and key scaffold nups, like Nup188, play important roles in NPC biogenesis, perhaps by helping to glue the scaffold together; conditional disruption of these interactions leads to herniations[41]. As the GLFG nups also play important roles in establishing the NPC diffusion barrier[75–77], it makes considerable sense to couple their assembly to the NPC scaffold to ensure that once INM-ONM fusion occurs, there will be no concomitant loss of nuclear-cytosolic compartmentalization.
Triggering NPC quality control
But what are the consequences if INM-ONM fusion occurs prematurely, or, if an existing NPC loses key components due to degradation because of oxidative (or other) damage? For example, while the NPC scaffold is extremely long lived, particularly in post-mitotic cells like neurons[78–80], the GLFG-rich Nup116 is lost from replicatively-old NPCs in budding yeast[81,82]. This observation is telling as nup116Δ NPCs have also been shown to acquire double-membrane seals after a shift to a non-permissive growth temperature resulting in morphologically-identical herniations as those associated with defective NPC assembly[52]. Thus, loss of function of Nup116 results in herniations that could arise either due to a lack of INM-ONM fusion, or, through surveillance mechanisms that seal off defective NPCs and/or NPC assembly intermediates[52] (Figure 2C, D).
A mechanism for how cells could re-seal a nuclear pore formed through premature INM-ONM fusion, or, one capable of sealing off a defective NPC likely requires the function of the endosomal complexes required for transport (ESCRT). This assertion is based on our work demonstrating the NE recruitment of the ESCRT component Chm7 (the yeast orthologue of CHMP7) by the integral INM protein Heh1 (the orthologue of human LEM2) under conditions in which NPC assembly is blocked[15]; Chm7 recruitment is most striking in nup116Δ and apq12Δ cells, strongly correlating NE herniations with Chm7 function. Moreover, the viability and the maintenance of nuclear-cytosolic compartmentalization of nup116Δ[15] and apq12Δ[83] cells requires CHM7. A role for the ESCRT machinery in NPC quality control is attractive as they are well established to form spiraling polymers capable of stitching membranes together at multiple subcellular locations[84]. In this way, they might also help seal the NE at the end of mitosis, or after NE rupture events[85–88]. Indeed, ESCRTs are emerging as a key molecular machinery that surveils the integrity of many membrane-bound compartments including endo-lysosomes[89].
In all of these scenarios, the precise mechanism of Chm7 and downstream ESCRT components like Snf7 (CHMP4 in humans), function at the NE remains ill defined. Specifically, while one can imagine a mechanism in which Chm7 counteracts premature INM-ONM fusion by, for example, re-sealing a small (<100 nm diameter) hole (Figure 2C), it is more challenging to contemplate how an existing defective NPC is sealed over, which would likely require local membrane remodeling to expand the pore membrane (Figure 2D). Likewise, how a large (>100 nm diameter) NE rupture could be repaired solely by the ESCRT machinery is also difficult to imagine. An attractive model for both of these examples, however, might be the local delivery and NE-incorporation of ER-membrane sheets, in analogy to how Drosophila embryos expand their NE during rapid early cell divisions[90].
Herniations associated with nuclear egress and nucleophagy
Interestingly, NE herniations that are morphologically similar to those associated with NPC biogenesis have also been observed as intermediates in pathways of nuclear egress. For example, herpesvirus exits the nucleus by budding through the NE lumen/PNS[91,92](Figure 3). While the molecular machinery that drives herpesvirus egress is encoded by the viral genome (the nuclear egress complex/NEC) and is thus distinct from NPC biogenesis, there is nonetheless evidence for the involvement of ESCRTs[93,94] and Torsin A[95] (and LULL1[96]) in the viral lifecycle hinting at a functional relationship with host nuclear membrane remodeling pathways. Further, Torsin A has been implicated in an egress pathway for so-called “Mega” RNPs that functions in the cells of Drosophila neuromuscular junctions[70,97] and in sea urchin embryos[98]. A key challenge, however, will be to distinguish if the NE herniations observed in these cells are functionally distinct from those linked with NPC biogenesis in mammalian cell culture[63]. Regardless, the concept that proteins/RNAs might exit the nucleus through a vesicular intermediate remains attractive and has been hypothesized to function as a clearance mechanism for nuclear aggregates[99]. While there is no direct evidence to yet support this idea, such a pathway could be used to remove defective NPC assembly intermediates from the INM[14,100]; interestingly, K48-linked ubiquitin can be found concentrated within the herniations of Torsin knockout cell lines[63](Figure 2B–D), but the identity of the ubiquitylated proteins remains unknown.
Figure 3.
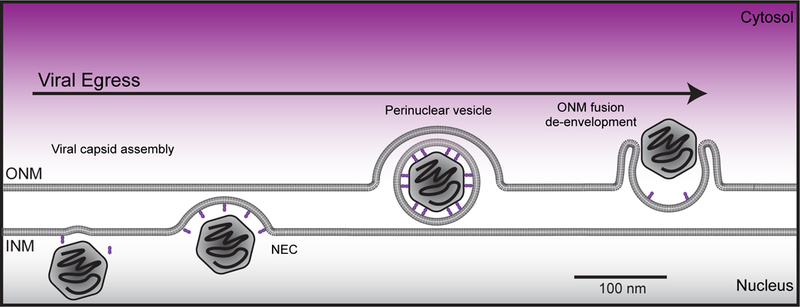
Schematic of herpesvirus nuclear egress pathway. A mature viral capsid (rounded hexagon), filled with viral DNA, is recruited to the INM by the virally encoded, nuclear egress complex (NEC), which is shown in purple. The NEC deforms the INM, encapsulating the viral capsid in an intermediate perinuclear vesicle. The vesicle fuses with the ONM, in conjunction with disassembly of the NEC and release of the viral capsid into cytosol, where it further matures.
That herniations of the NE could be associated with quality control pathways that clear defective components from the INM and/or nucleus can also be gleaned from work on nuclear autophagy or “nucleophagy” pathways. For example, NE herniations can be visualized in the vacuoles of budding yeast as an intermediate in piece meal microautophagy of the nucleus (PMN)[101].The mechanisms that drive herniation formation remain ill-defined, but clearly depend on direct connections between the vacuole membrane and the ONM[101] that likely also extend to the INM[102]. While a direct analog of this pathway has yet to be discovered in mammalian systems, it is not coincidental that the clearance of Lamin B1 (observed upon overexpression of an oncogenic HRasV12 protein) in mammalian cell culture might also proceed through a NE herniation intermediate[103]. Likewise, the overexpression of an Epstein virus protein, BFRF1 might trigger a nucleophagy-type pathway that incorporates the resolution of a NE herniation into a cytosolic vesicle targeted by the autophagy pathway[104]. It remains conceptually challenging, however, to contemplate the molecular mechanisms behind how a NE herniation could be pinched off from the nucleus while maintaining nuclear integrity, let alone how it engages the autophagy machinery.
Mechanically-induced NE herniations
While a nucleophagy mechanism would likely require maintenance of NE integrity, there are many other NE herniations that can lead to NE rupture. These herniations are sufficiently large that they can be visualized by light microscopy and might contribute to the often dramatic nuclear deformations that have been associated with cancers and other human diseases, including the laminopathies (extensively reviewed elsewhere[2,4,105]). While the provenance of these herniations are not always understood, there is evidence to support that at least a subset might be caused by direct mechanical force imposed by extrinsic environmental factors. For example, during cell migration through constrictions that are smaller than the nucleus, herniations of the NE arise and often lead to NE rupture[86,87]. These herniations are also often associated with a break or discontinuity within the lamin network that underlies the INM[86,87,106,107].
Herniations associated with mechanical strain on the nucleus have also been observed in cell culture, primarily of cancer cells[106–109]. In this scenario, mechanical force that drives herniation growth is likely also imposed by nuclear confinement, but in this case through the perinuclear actin network that could directly connect to the NE through the linker of nucleoskeleton and cytoskeleton (LINC) complexes, which directly connect the cytoskeleton to nuclear factors through a translumenal bridge[109]. As is with the case of NE herniations caused by cell migration through constrictions, these herniations are chromatin-filled and occur at sites of lamina discontinuity[107,109,110]. It remains enigmatic how mechanical force is translated into the formation of a herniation but it is clear that this process is directly impacted by extracellular cues some of which trigger elaborate feedback mechanisms that modulate the output of both nuclear and cytosolic cytoskeletons[3,111–113]. It is likely that a combination of misregulation of these feedback mechanisms with defects in the mechanical networks themselves (e.g. the lamina) increase the propensity of NE herniations in vivo and might directly contribute to disease mechanisms.
Outlook
While we have focused on NE herniations that, by their very definition, protrude outward from the nucleus, there are also interesting intranuclear membrane intrusions[114] that might contribute to forming the “nucleoplasmic reticulum”[115]. Like the herniations discussed here, the nucleoplasmic reticulum has also been associated with pathology, even as it is likely a product of physiological processes that remain to be fully understood. Thus, a key challenge for the future is to identify the molecular mechanisms driving specific nuclear membrane remodeling events that can lead to often remarkable morphological changes to the NE. The introduction of membrane-remodeling proteins[17,19] and ATP-utilizing machineries[14,63,116] to the NE is clearly a step in this direction, however, it might be that their local regulation is the most important factor for defining whether membrane remodeling is productive or not. Once these regulatory processes are understood, perhaps there is a chance to define how they might contribute to disease.
References
- 1.Dey P (2010) Cancer nucleus: morphology and beyond. Diagn. Cytopathol 38, 382–90. [DOI] [PubMed] [Google Scholar]
- 2.Burke B and Stewart CL (2014) Functional architecture of the cell’s nucleus in development, aging, and disease. Curr. Top. Dev. Biol 109, 1–52. [DOI] [PubMed] [Google Scholar]
- 3.Kirby TJ and Lammerding J (2018) Emerging views of the nucleus as a cellular mechanosensor. Nat. Cell Biol 20, 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatch E and Hetzer M (2014) Breaching the nuclear envelope in development and disease. J. Cell Biol 205, 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke B and Stewart CL (2013) The nuclear lamins: flexibility in function. Nat. Rev. Mol. Cell Biol 14, 13–24. [DOI] [PubMed] [Google Scholar]
- 6.Ungricht R and Kutay U (2017) Mechanisms and functions of nuclear envelope remodelling. Nat. Rev. Mol. Cell Biol 18, 229–245. [DOI] [PubMed] [Google Scholar]
- 7.Beck M and Hurt E (2016) The nuclear pore complex: understanding its function through structural insight. Nat. Rev. Mol. Cell Biol 18, 73–89. [DOI] [PubMed] [Google Scholar]
- 8.Kim SJ, Fernandez-Martinez J, Nudelman I, Shi Y, Zhang W, Raveh B, Herricks T, Slaughter BD, Hogan JA, Upla P, et al. (2018) Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P and Hurt E (2011) Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 146, 277–89. [DOI] [PubMed] [Google Scholar]
- 10.Teimer R, Kosinski J, Von Appen A, Beck M and Hurt E (2017) A short linear motif in scaffold Nup145C connects Y-complex with pre-Assembled outer ring Nup82 complex. Nat. Commun 8, 1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doucet CM, Talamas JA and Hetzer MW (2010) Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell 141, 1030–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otsuka S, Bui KH, Schorb M, Hossain MJ, Politi AZ, Koch B, Eltsov M, Beck M and Ellenberg J (2016) Nuclear pore assembly proceeds by an inside-out extrusion of the nuclear envelope. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otsuka S, Steyer AM, Schorb M, Hériché J-K, Hossain MJ, Sethi S, Kueblbeck M, Schwab Y, Beck M and Ellenberg J (2018) Postmitotic nuclear pore assembly proceeds by radial dilation of small membrane openings. Nat. Struct. Mol. Biol 25, 21–28. [DOI] [PubMed] [Google Scholar]
- 14.Webster BM, Colombi P, Jäger J and Lusk CP (2014) Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell 159, 388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webster BM, Thaller DJ, Jäger J, Ochmann SE, Borah S and Lusk CP (2016) Chm7 and Heh1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing. EMBO J 35, 2447–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makio T, Stanton LH, Lin CC, Goldfarb DS, Weis K and Wozniak RW (2009) The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly. J. Cell Biol 185, 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chadrin A, Hess B, San Roman M, Gatti X, Lombard B, Loew D, Barral Y, Palancade B and Doye V (2010) Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J. Cell Biol 189, 795–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belgareh N, Snay-Hodge C, Pasteau F, Dagher S, Cole CN and Doye V (1998) Functional characterization of a Nup159p-containing nuclear pore subcomplex. Mol. Biol. Cell 9, 3475–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson TR, Lazarus MD, Hetzer MW and Wente SR (2009) ER membrane - bending proteins are necessary for de novo nuclear pore formation. J Cell Biol 184, 659–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Titus LC, Dawson TR, Rexer DJ, Ryan KJ and Wente SR (2010) Members of the RSC chromatin-remodeling complex are required for maintaining proper nuclear envelope structure and pore complex localization. Mol. Biol. Cell 21, 1072–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan KJKJ, Zhou YY and Wente SR (2007) The karyopherin Kap95 regulates nuclear pore complex assembly into intact nuclear envelopes in vivo. Mol. Biol. Cell 18, 886–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadowaki T (1994) Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J. Cell Biol 126, 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan KJ, McCaffery JM and Wente SR (2003) The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J. Cell Biol 160, 1041–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vollmer B, Lorenz M, Moreno-Andrés D, Bodenhöfer M, De Magistris P, Astrinidis SA, Schooley A, Flötenmeyer M, Leptihn S and Antonin W (2015) Nup153 Recruits the Nup107–160 Complex to the Inner Nuclear Membrane for Interphasic Nuclear Pore Complex Assembly. Dev. Cell 33, 717–728. [DOI] [PubMed] [Google Scholar]
- 25.Mészáros N, Cibulka J, Mendiburo MJ, Romanauska A, Schneider M and Köhler A (2015) Nuclear Pore Basket Proteins Are Tethered to the Nuclear Envelope and Can Regulate Membrane Curvature. Dev. Cell 33, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yewdell WT, Colombi P, Makhnevych T and Lusk CP (2011) Lumenal interactions in nuclear pore complex assembly and stability. Mol. Biol. Cell 22, 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Q, Pante N, Misteli T, Elsagga M, Crisp M, Hodzic D, Burke B and Roux KJ (2007) Functional association of Sun1 with nuclear pore complexes. J. Cell Biol 178, 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD and Hodzic D (2006) Coupling of the nucleus and cytoplasm: Role of the LINC complex. J. Cell Biol 172, 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talamas JA and Hetzer MW (2011) POM121 and Sun1 play a role in early steps of interphase NPC assembly. J. Cell Biol 194, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madrid AS, Mancuso J, Cande WZ and Weis K (2006) The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J. Cell Biol 173, 361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onischenko E, Stanton LH, Madrid AS, Kieselbach T and Weis K (2009) Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J. Cell Biol 185, 475–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dultz E and Ellenberg J (2010) Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J. Cell Biol 191, 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaik M, Flemming D, Von Appen A, Kastritis P, Mücke N, Fischer J, Stelter P, Ori A, Bui KH, Baßler J, et al. (2015) Structural basis for assembly and function of the Nup82 complex in the nuclear pore scaffold. J. Cell Biol 208, 283–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez-Martinez J, Kim SJ, Shi Y, Upla P, Pellarin R, Gagnon M, Chemmama IE, Wang J, Nudelman I, Zhang W, et al. (2016) Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform. Cell 167, 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull M-T, Banterle N, Parca L, Kastritis P, et al. (2015) In situ structural analysis of the human nuclear pore complex. Nature 526, 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. (2007) The molecular architecture of the nuclear pore complex. Nature 450, 695–701. [DOI] [PubMed] [Google Scholar]
- 37.Bucci M and Wente SR (1998) A Novel Fluorescence-based Genetic Strategy Identifies Mutants of Saccharomyces cerevisiae Defective for Nuclear Pore Complex Assembly. Mol. Biol. Cell 9, 2439–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A and Hurt EC (1996) A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 84, 265–275. [DOI] [PubMed] [Google Scholar]
- 39.Zabel U, Doye VV, Tekotte H, Wepf R, Grandi P, Hurtll EC, Hurt EC, Hurtll EC and Hurt EC (1996) Nic96p is required for nuclear pore formation and fuctionally interacts with a novel nucleoporin, Nup188p. J. Cell Biol 133, 1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teixeira MT, Siniossoglou S, Podtelejnikov S, Bénichou JC, Mann M, Dujon B, Hurt E and Fabre E (1997) Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins. EMBO J 16, 5086–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Onischenko E, Tang JH, Andersen KR, Knockenhauer KE, Vallotton P, Derrer CP, Kralt A, Mugler CF, Chan LY, Schwartz TU, et al. (2017) Natively Unfolded FG Repeats Stabilize the Structure of the Nuclear Pore Complex. Cell 171, 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aitchison JD, Blobel G and Rout MP (1995) Nup120p : A Yeast Nucleoporin Required for NPC Distribution and mRNA Transport. J. Cell Biol 131, 1659–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heath CV, Copeland CS, Amberg DC, Priore V Del, Snyder M and Cole CN (1995) Nuclear Pore Complex Clustering and Nuclear Accumulation of Poly(A) RNA Associated with Mutation of the Saccharomyces cerevisiae RAT2 / NUP120 Gene. J. Cell Biol 131, 1677–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dockendorff TC, Heath CV, Goldstein AL, Snay CA and Cole CN (1997) C-terminal truncations of the yeast nucleoporin Nup145p produce a rapid temperature-conditional mRNA export defect and alterations to nuclear structure. Mol. Cell. Biol 17, 906–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wente SR and Blobel G (1994) NUP145 Encodes a Novel Yeast Glycine-Leucine-Phenylalanine-Glycine (GLFG) Nucleoporin Required for Nuclear Envelope Structure. J. Cell Biol 125, 955–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emtage JL, Bucci M, Watkins JL and Wente SR (1997) Defining the essential functional regions of the nucleoporin Nup145p. J. Cell Sci 110, 911–925. [DOI] [PubMed] [Google Scholar]
- 47.Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U and Hurt E (2000) Structure and assembly of the Nup84p complex. J. Cell Biol 149, 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryan KJ and Wente SR (2002) Isolation and characterization of new Saccharomyces cerevisiaemutants perturbed in nuclear pore complex assembly. BMC Genet 5, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nehrbass U, Rout MP, Maguire S, Blobel G and Wozniak RW (1996) The yeast nucleoporin Nup188p interacts genetically and physically with the core structures of the nuclear pore complex. J. Cell Biol 133, 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gigliotti S, Callaini G, Andone S, Riparbelli MG, Pernas-Alonso R, Hoffmann G, Graziani F and Malva C (1998) Nup154, a new Drosophila gene essential for male and female gametogenesis is related to the Nup155 vertebrate nucleoporin gene. J. Cell Biol 142, 1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy R, Watkins J and Wente SR (1996) GLE2, a saccharomyces cerevisiae homologue of the schizosaccharomyces pomble export factor RAE1, is required for nuclear pore complex structure in function. Mol. Biol. Cell 7, 1921–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wente SR (1993) A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J. Cell Biol 123, 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bailer SM, Siniossoglou S, Podtelejnikov A, Hellwig A, Mann M and Hurt E (1998) Nup116p and Nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor Gle2p. EMBO J 17, 1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho AK, Raczniak GA, Ives EB and Wente SR (1998) The Integral Membrane Protein Snl1p Is Genetically Linked to Yeast Nuclear Pore Complex Function. Mol. Biol. Cell 9, 355–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wente SR and Blobel GG (1994) NUP145 encodes a novel yeast glycine-leucine-phenylalanine-glycine (GLFG) nucleoporin required for nuclear envelope structure. J. Cell Biol 125, 955–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wente SR, Rout MP and Blobel G (1992) A new family of yeast nuclear pore complex proteins. J. Cell Biol 119, 705–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneiter R, Hitomi M, Ivessa AS, Fasch E, Kohlwein SD and Tartakoff AM (1996) A Yeast Acetyl Coenzyme A Carboxylase Mutant Links Very-Long-Chain Fatty Acid Synthesis to the Structure and Function of the Nuclear Membrane-Pore Complex. Mol. Cell. Biol 16, 7161–7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang S, Skalsky Y and Garfinkel DJ (1999) MGA2 or SPT23 is required for transcription of the delta9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics 151, 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scarcelli JJ, Hodge CA and Cole CN (2007) The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. J. Cell Biol 178, 799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hodge CA, Choudhary V, Wolyniak MJ, Scarcelli JJ, Schneiter R and Cole CN (2010) Integral membrane proteins Brr6 and Apq12 link assembly of the nuclear pore complex to lipid homeostasis in the endoplasmic reticulum. J. Cell Sci 123, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lone MA, Atkinson AE, Hodge CA, Cottier S, Martínez-Montañés F, Maithel S, Mène-Saffrané L, Cole CN and Schneiter R (2015) Yeast Integral Membrane Proteins Apq12, Brl1, and Brr6 Form a Complex Important for Regulation of Membrane Homeostasis and Nuclear Pore Complex Biogenesis. Eukaryot. Cell, American Society for Microbiology 14, 1217–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baker KE, Coller J and Parker R (2004) The yeast Apq12 protein affects nucleocytoplasmic mRNA transport. RNA 10, 1352–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laudermilch E, Tsai P-L, Graham M, Turner E, Zhao C and Schlieker C (2016) Dissecting Torsin/cofactor function at the nuclear envelope: a genetic study. Mol. Biol. Cell 27, 3964–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goodchild RE, Kim CE and Dauer WT (2005) Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron 48, 923–932. [DOI] [PubMed] [Google Scholar]
- 65.Liang CC, Tanabe LM, Jou S, Chi F and Dauer WT (2014) TorsinA hypofunction causes abnormal twisting movements and sensorimotor circuit neurodegeneration. J. Clin. Invest 124, 3080–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rose AE, Zhao C, Turner EM, Steyer AM and Schlieker C (2014) Arresting a Torsin ATPase reshapes the endoplasmic reticulum. J. Biol. Chem 289, 552–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanabe LM, Liang C-C and Dauer WT (2016) Neuronal Nuclear Membrane Budding Occurs during a Developmental Window Modulated by Torsin Paralogs. Cell Rep 16, 3322–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim CE, Perez A, Perkins G, Ellisman MH and Dauer WT (2010) A molecular mechanism underlying the neural-specific defect in torsinA mutant mice. Proc. Natl. Acad. Sci 107, 9861–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.VanGompel MJW, Nguyen KCQ, Hall DH, Dauer WT and Rose LS (2015) A novel function for the Caenorhabditis elegans torsin OOC-5 in nucleoporin localization and nuclear import. Mol. Biol. Cell 26, 1752–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jokhi V, Ashley J, Nunnari J, Noma A, Ito N, Wakabayashi-Ito N, Moore MJ and Budnik V (2013) Torsin Mediates Primary Envelopment of Large Ribonucleoprotein Granules at the Nuclear Envelope. Cell Rep 3, 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pappas SS, Liang C-C, Kim S, Rivera CO and Dauer WT (2018) TorsinA dysfunction causes persistent neuronal nuclear pore defects. Hum. Mol. Genet 27, 407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ozelius LJ, Hewett JW, Page CE, Bressman SB, Kramer PL, Shalish C, De Leon D, Brin MF, Raymond D, Corey DP, et al. (1997) The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat. Genet 17, 40–48. [DOI] [PubMed] [Google Scholar]
- 73.Zhang W, Neuner A, Rüthnick D, Sachsenheimer T, Lüchtenborg C, Brügger B and Schiebel E (2018) Brr6 and Brl1 locate to nuclear pore complex assembly sites to promote their biogenesis. J. Cell Biol 217, 877–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Bruyn Kops A, Guthrie C and Kops A. d. B.; Guthrie C (2001) An essential nuclear envelope integral membrane protein, Brr6p, required for nuclear transport. EMBO J 20, 4183–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hülsmann BB, Labokha AA and Görlich D (2012) The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell 150, 738–751. [DOI] [PubMed] [Google Scholar]
- 76.Popken P, Ghavami A, Onck PR, Poolman B and Veenhoff LM (2015) Size-dependent leak of soluble and membrane proteins through the yeast nuclear pore complex. Mol. Biol. Cell 26, 1386–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Timney BL, Raveh B, Mironska R, Trivedi JM, Kim SJ, Russel D, Wente SR, Sali A and Rout MP (2016) Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol 215, 57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.D’Angelo MA, Raices M, Panowski SH and Hetzer MW (2009) Age-Dependent Deterioration of Nuclear Pore Complexes Causes a Loss of Nuclear Integrity in Postmitotic Cells. Cell 136, 284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Savas JN, Toyama BH, Xu T, Yates JR and Hetzer MW (2012) Extremely long-lived nuclear pore proteins in the rat brain. Science 335, 942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR and Hetzer MW (2013) Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 154, 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lord CL, Timney BL, Rout MP and Wente SR (2015) Altering nuclear pore complex function impacts longevity and mitochondrial function in S. cerevisiae. J. Cell Biol 208, 729–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Janssens GE, Meinema AC, González J, Wolters JC, Schmidt A, Guryev V, Bischoff R, Wit EC, Veenhoff LM and Heinemann M (2015) Protein biogenesis machinery is a driver of replicative aging in yeast. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bauer I, Brune T, Preiss R and Kölling R (2015) Evidence for a Nonendosomal Function of the Saccharomyces cerevisiae ESCRT-III-Like Protein Chm7. Genetics 201, 1439–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stoten CL and Carlton JG (2018) ESCRT-dependent control of membrane remodelling during cell division. Semin. Cell Dev. Biol 74, 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Olmos Y, Perdrix-Rosell A and Carlton JG (2016) Membrane Binding by CHMP7 Coordinates ESCRT-III-Dependent Nuclear Envelope Reformation. Curr. Biol 26, 2635–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Raab M, Gentili M, de Belly H, Thiam H-R, Vargas P, Jimenez AJ, Lautenschlaeger F, Voituriez R, Lennon-Dumenil A-M, Manel N, et al. (2016) ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352, 359–362. [DOI] [PubMed] [Google Scholar]
- 87.Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K and Lammerding J (2016) Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Robijns J, Molenberghs F, Sieprath T, Corne TDJ, Verschuuren M and De Vos WH (2016) In silico synchronization reveals regulators of nuclear ruptures in lamin A/C deficient model cells. Sci. Rep 6, 30325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Skowyra ML, Schlesinger PH, Naismith TV and Hanson PI (2018) Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360, 5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hampoelz B, Mackmull MT, Machado P, Ronchi P, Bui KH, Schieber N, Santarella-Mellwig R, Necakov A, Andrés-Pons A, Philippe JM, et al. (2016) Pre-assembled Nuclear Pores Insert into the Nuclear Envelope during Early Development. Cell 166, 664–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lye MF, Wilkie AR, Filman DJ, Hogle JM and Coen DM (2017) Getting to and through the inner nuclear membrane during herpesvirus nuclear egress. Curr. Opin. Cell Biol 46, 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johnson DC and Baines JD (2011) Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol 9, 382–394. [DOI] [PubMed] [Google Scholar]
- 93.Lee SC, Wu CH and Wang CW (2010) Traffic of a viral movement protein complex to the highly curved tubules of the cortical endoplasmic reticulum. Traffic 11, 912–930. [DOI] [PubMed] [Google Scholar]
- 94.Lee C-P, Liu G-T, Kung H-N, Liu P-T, Liao Y-T, Chow L-P, Chang L-S, Chang Y-H, Chang C-W, Shu W-C, et al. (2016) The Ubiquitin Ligase Itch and Ubiquitination Regulate BFRF1-Mediated Nuclear Envelope Modification for Epstein-Barr Virus Maturation. J. Virol 90, 8994–9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maric M, Shao J, Ryan RJ, Wong C-S, Gonzalez-Alegre P and Roller RJ (2011) A Functional Role for TorsinA in Herpes Simplex Virus 1 Nuclear Egress. J. Virol 85, 9667–9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Turner EM, Brown RSH, Laudermilch E, Tsai P-L and Schlieker C (2015) The Torsin Activator LULL1 Is Required for Efficient Growth of Herpes Simplex Virus 1. J. Virol 89, 8444–8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang Y-T, Li Q, et al. (2012) Nuclear Envelope Budding Enables Large Ribonucleoprotein Particle Export during Synaptic Wnt Signaling. Cell 149, 832–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.LaMassa N, Arenas-Mena C and Phillips GR (2018) Electron microscopic characterization of nuclear egress in the sea urchin gastrula. J. Morphol 279, 609–615. [DOI] [PubMed] [Google Scholar]
- 99.Rose A and Schlieker C (2012) Alternative nuclear transport for cellular protein quality control. Trends Cell Biol 22, 509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Webster BM and Lusk CP (2016) Border Safety: Quality Control at the Nuclear Envelope. Trends Cell Biol 26, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Roberts P, Moshitch-Moshkovitz S, Kvam E, O ‘toole E, Winey M, Goldfarb DS, O’Toole E, Winey M and Goldfarb DS (2003) Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Millen JI, Pierson J, Kvam E, Olsen LJ and Goldfarb DS (2008) The Luminal N-Terminus of Yeast Nvj1 is an Inner Nuclear Membrane Anchor. Traffic 9, 1653–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dou Z, Xu C, Donahue G, Shimi T, Pan JA, Zhu J, Ivanov A, Capell BC, Drake AM, Shah PP, et al. (2015) Autophagy mediates degradation of nuclear lamina. Nature 527, 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu G-T, Kung H-N, Chen C-K, Huang C, Wang Y-L, Yu C-P and Lee C-P (2018) Improving nuclear envelope dynamics by EBV BFRF1 facilitates intranuclear component clearance through autophagy. FASEB J 32. [DOI] [PubMed] [Google Scholar]
- 105.Robijns J, Houthaeve G, Braeckmans K and De Vos WH (2018) Loss of Nuclear Envelope Integrity in Aging and Disease. Int. Rev. Cell Mol. Biol 336, 205–222. [DOI] [PubMed] [Google Scholar]
- 106.De vos WH, Houben F, Kamps M, Malhas A, Verheyen F, Cox J, Manders EMM, Verstraeten VLRM, Van steensel MAM, Marcelis CLM, et al. (2011) Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum. Mol. Genet 20, 4175–4186. [DOI] [PubMed] [Google Scholar]
- 107.Vargas JD, Hatch EM, Anderson DJ and Hetzer MW (2012) Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus 3, 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hatch EM, Fischer AH, Deerinck TJ and Hetzer MW (2013) Catastrophic Nuclear Envelope Collapse in Cancer Cell Micronuclei. Cell 154, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hatch EM and Hetzer MW (2016) Nuclear envelope rupture is induced by actin-based nucleus confinement. J. Cell Biol 215, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Deviri D, Discher DE and Safran SA (2017) Rupture Dynamics and Chromatin Herniation in Deformed Nuclei. Biophys. J 113, 1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stewart RM, Zubek AE, Rosowski KA, Schreiner SM, Horsley V and King MC (2015) Nuclear-cytoskeletal linkages facilitate cross talk between the nucleus and intercellular adhesions. J. Cell Biol 209, 403–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thakar K, May CK, Rogers A and Carroll CW (2017) Opposing roles for distinct LINC complexes in regulation of the small GTPase RhoA. Mol. Biol. Cell 28, 182–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.King MC and Lusk CP (2016) A model for coordinating nuclear mechanics and membrane remodeling to support nuclear integrity. Curr. Opin. Cell Biol 41, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Goodwin EC, Motamedi N, Lipovsky A, Fernández-Busnadiego R and DiMaio D (2014) Expression of DNAJB12 or DNAJB14 causes coordinate invasion of the nucleus by membranes associated with a novel nuclear pore structure. PLoS One 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Malhas A, Goulbourne C and Vaux DJ (2011) The nucleoplasmic reticulum: form and function. Trends Cell Biol 21, 362–73. [DOI] [PubMed] [Google Scholar]
- 116.Dehoratius C and Silver PA (1996) Nuclear Transport Defects and Nuclear Envelope Alterations Are Associated with Mutation of the Saccharomyces cerevisiae NPL4 Gene. Mol. Biol. Cell 7, 1835–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Naismith TV, Heuser JE, Breakefield XO and Hanson PI (2004) TorsinA in the nuclear envelope. Proc. Natl. Acad. Sci 101, 7612–7617. [DOI] [PMC free article] [PubMed] [Google Scholar]


