Abstract
Methanotrophic bacteria convert methane to methanol using methane monooxygenase (MMO) enzymes. In many strains, either an iron-containing soluble (sMMO) or a copper-containing particulate (pMMO) enzyme can be produced depending on copper availability; the mechanism of this copper switch has not been elucidated. A key player in methanotroph copper homeostasis is methanobactin (Mbn), a ribosomally produced, post-translationally modified natural product with a high affinity for copper. The Mbn precursor peptide is encoded within an operon that contains a range of putative transporters, regulators, and biosynthetic proteins, but the involvement of these genes in Mbn-related processes remains unclear. Extensive time-dependent qRT-PCR studies of Methylosinus trichosporium OB3b and the constitutive sMMO-producing mutant M. trichosporium OB3b PP358 show that the Mbn operon is indeed copper-regulated, providing experimental support for its bioinformatics-based identification. Moreover, the Mbn operon is co-regulated with the sMMO operon and reciprocally regulated with the pMMO operon. Within the Mbn and sMMO operons, a subset of regulatory genes exhibits a distinct and shared pattern of expression, consistent with their proposed functions as internal regulators. In addition, genome sequencing of the M. trichosporium OB3b PP358 mutant provides new evidence for the involvement of genes adjacent to the pMMO operon in methanotroph copper homeostasis.
Introduction
Methanotrophs, bacteria that are capable of subsisting on methane (CH4) as their sole carbon and energy source,1 play a key role in the global carbon cycle, and have attracted much attention for their potential applications in bioremediation and biological gas-to-liquids conversion processes.2–4 The first step in methanotroph metabolism, the oxidation of methane to methanol, can be carried out using two entirely different enzymatic systems.1 The more common enzyme is particulate methane monooxygenase (pMMO), a membrane protein complex that is located in intracytoplasmic and/or inner membranes5 and contains a copper active site.6,7 An alternate soluble methane monooxygenase (sMMO), located in the cytoplasm of some methanotrophs, utilizes a catalytic diiron active site.8 The large subset of methanotrophs that can express both MMOs exhibits a striking copper-dependent reciprocal regulation of the two operons.9
Previous studies broadly supported a model in which the pMMO operon, encoding the three subunits PmoA, PmoB, and PmoC, is controlled by a σ70-dependent promoter (Fig. 1A), resulting in constitutive expression at an intermediate level.10–13 The operon is mildly (< 10-fold) up-regulated in the presence of copper, but the regulators involved have not been identified.14–16 The main sMMO operon, by contrast, is controlled by a σN-dependent promoter, and transcription depends on the presence of two regulatory proteins, MmoR (a σN-dependent transcriptional activator) and MmoG (a GroEL homologue) (Fig. 1B); unlike pMMO, it is markedly down-regulated in the presence of copper, with a fold change of ten to a hundred times greater than the change observed for pMMO.10,13–15,17 The unknown mechanism by which these two operons are reciprocally regulated is referred to as the “copper switch,” but no evidence exists for the interaction of known copper-dependent transcriptional regulation systems with either operon.18 Moreover, it is not known how copper triggers formation of the intracytoplasmic membranes that house pMMO.4
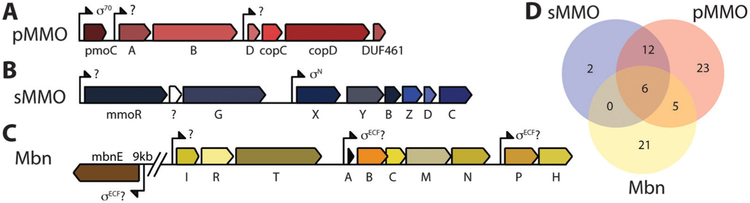
Fig. 1.
The M. trichosporium OB3b MMO and Mbn operons. (A) The pMMO operon. The order of genes in proteobacterial particulate methane monooxygenase and ammonia monooxygenase is p/amoCAB; other gene orders occur in some related monooxygenase systems. (B) The sMMO operon. The core sMMO operon (mmoXYBZDC) is found adjacent to mmoR and mmoG, but the order of these genes may vary, and additional operon members are present in some species. (C) The Mbn operon, including a core comprising biosynthesis and export genes (mbnABCMN), a group of import genes (mbn(E)IRT), and two genes of unknown function (mbnPH). (D) Overlap between sMMO, pMMO/AMO, and Mbn operons in organisms present in the JGI/IMG database. All three operons occur independently; no one operon appears to be necessary for either of the others to function.
One important agent of methanotroph copper homeostasis is methanobactin (Mbn), a small peptidic copper-binding natural product (Fig. S1, ESI†).19 Mbn is produced and secreted under conditions of copper starvation (<0.8 μM soluble copper),20 and is re-internalized after copper binding via an active process.21 A putative Mbn-related operon was recently discovered, including a small gene thought to encode an Mbn precursor peptide, suggesting that the compound is a ribosomally produced, post-translationally modified natural product.22 This small gene is repressed by copper and Mbn production is abolished when the gene is eliminated.23 Genome mining has enabled tentative assignment of other Mbn operon components in a range of species to transport, regulatory, and biosynthetic functions (Fig. 1C and Fig. S1, ESI†).24 However, direct evidence for involvement of the additional genes in copper metabolism and Mbn-related processes has not been reported.
Although the copper switch was first observed more than thirty years ago,25,26 the details of copper-dependent MMO regulation and methanotroph copper metabolism, including the role of Mbn, have not been elucidated.27 None of the three major operons appear to rely on the presence of either of the others; species possessing only Mbn, pMMO, or sMMO all exist (Fig. 1D). However, when present within the same organism, a complex copper homeostasis system appears to regulate all three operons. In this study, we report extensive time-dependent qRT-PCR analyses of Methylosinus (M.) trichosporium OB3b showing that the entire putative Mbn operon is copper-regulated and that the sMMO and Mbn genes are co-regulated. These data provide strong support for our identification of the Mbn operons. Moreover, genome sequencing of a constitutive sMMO-producing mutant reveals a multi-nucleotide frameshift deletion in a putative inner membrane copper importer adjacent to the pMMO operon, thus identifying components of an additional overlapping copper homeostasis system in methanotrophs.
Results and discussion
Copper-dependent regulation of the sMMO, Mbn, and pMMO operons
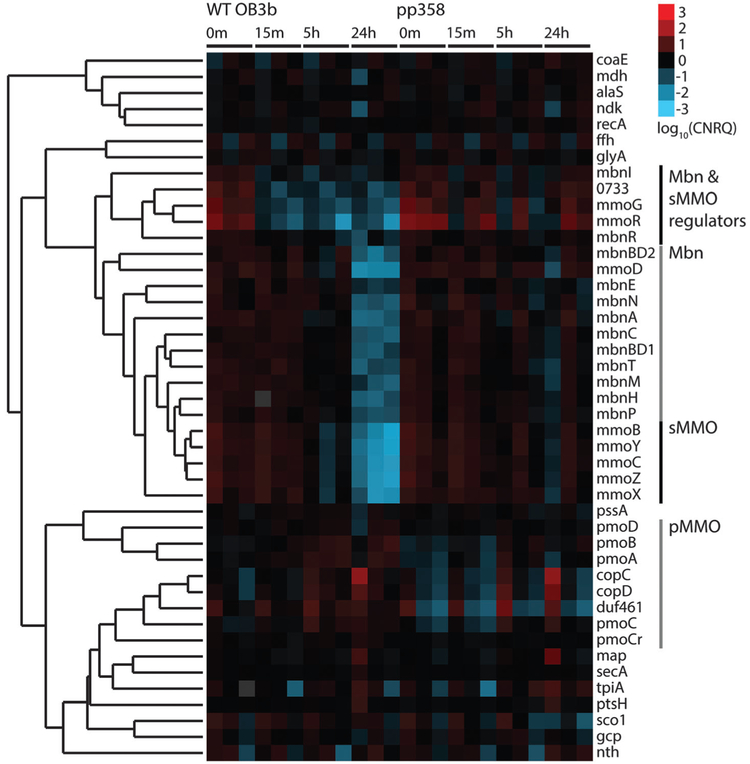
Previous investigations of the copper switch have relied on analysis at specific static copper concentrations. We chose to approach the copper switch as a dynamic and time-dependent system, relying on a set of timecourse experiments to track the transition from a copper-starved to a copper-replete state. Towards this end, samples of wildtype M. trichosporium OB3b were taken prior to the addition of copper to a final concentration of 12.5 μM and at 15 min, 5 h, and 24 h after copper addition. Normalized relative expression levels show several clear trends. Transcript levels of the main sMMO operon (mmoRGXYBZDC) drop significantly by 5 h, and are down-regulated by 2–3 orders of magnitude over the 24 h time period (Fig. 2 and Fig. S2, ESI†). This downregulation has been observed previously via proteomics15 as well as detection of the mmoX mRNA.10,14 Most important, copper-responsive regulation of the Mbn operon parallels that of sMMO. Transcript levels for the entire putative Mbn operon, mbnIRTABCMNPH, as well as mbnE, located approximately 9 kb upstream of mbnIRT (Fig. 1C), also drop significantly by 5 h and are down-regulated by 1–2 orders of magnitude over 24 h (Fig. 2 and Fig. S2, ESI†). The down-regulation of mbnA as well as all the other genes by copper is consistent with MbnA being the Mbn precursor peptide and with the entire operon functioning in copper acquisition: as copper concentrations increase, Mbn production decreases,28 and accordingly its regulatory, transport, and biosynthetic machinery is no longer highly expressed. Given that some or all of these genes are conserved in a range of Mbn operons,24 it is likely that these proteins have Mbn-related functions in other organisms as well.
Fig. 2.
Heatmap of log10-transformed calibrated normalized relative quantities (CNRQs) for genes obtained via qRT-PCR; downregulation is indicated in blue and upregulation in red. Genes were clustered via hierarchical clustering, using Kendall’s tau correlation as a similarity metric and complete linkage. Genes from each monitored operon cluster with other members of the operon, with the exception of regulatory genes from the sMMO and Mbn operons, which cluster together as a separate group. Experiments were performed with three biological replicates at four timepoints (0 min at 0 μM CuSO4,15 min at 12.5 μM CuSO4, 5 h at 12.5 μM CuSO4, and 24 h at 12.5 μM CuSO4) for both wildtype M. trichosporium OB3b and M. trichosporium OB3b PP358.
In contrast to the sMMO operon, copper-responsive regulation of pMMO has been somewhat controversial. Unlike sMMO, there is a considerable basal level of constitutive expression, mediated by a σ70 promoter.11–13 Some studies show little change in response to copper,10,29 some show a mild increase,9,15,16 and several suggest a significant increase.23,30 Our data show that genes in the pMMO operon are mildly up-regulated by less than an order of magnitude over 24 h (Fig. 2 and Fig. S2, ESI†). These results are consistent with a consensus model in which mild up-regulation of the pMMO operon does occur in the presence of copper, with the possibility of a post-transcriptional regulatory step still open.
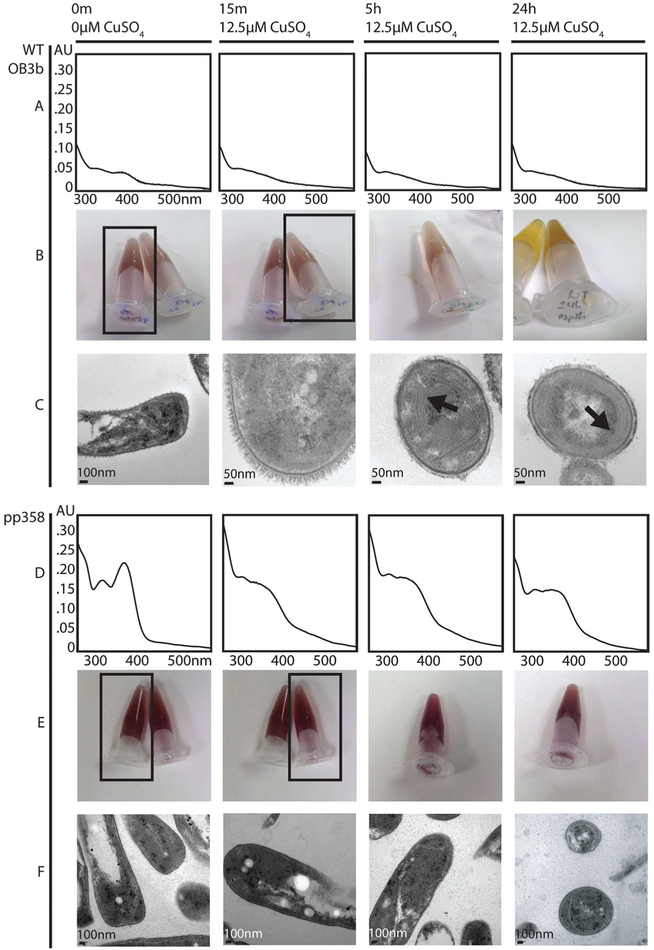
In other studies, a range of phenotypes have been used as proxies for identifying whether a given methanotroph cell population is in low- or high-copper mode, including the presence of intracytoplasmic membranes (ICMs),5,31 the naphthalene activity assay (limited to sMMO-producing strains),32 and secretion of Mbn into the spent medium (in species that produce Mbn).28 In our timecourse experiments, these phenotypes clearly track with the mRNA levels observed in the qRT-PCR analysis (Fig. 3). Metal free (apo) Mbn is visible in the spent medium at the 0 min timepoint, but has shifted to copper-loaded Mbn 15 min after copper addition (Fig. 3 and Fig. S3, ESI†). Intracellular membranes are detected primarily at the 5 h and 24 h timepoints, and naphthalene oxidation by sMMO clearly decreases at the later timepoints (Fig. 3).
Fig. 3.
Phenotypic analysis of qRT-PCR samples. (A) Wildtype (WT) M. trichosporium OB3b spent medium UV-visible spectra. Apo Mbn is visible at 0 min (as shown by the peaks at 340 and 390 nm); addition of 12.5 μM CuSO4 to the medium results in chelation and a shift to a typical CuMbn spectrum. (B) Wildtype M. trichosporium OB3b naphthalene assay for sMMO activity. Oxidation of naphthalene by sMMO results in a pink/purple color in the presence of Fast Blue B; without sMMO activity, addition of Fast Blue B results in a primarily yellowish color. The relevant tubes for the 0 and 15 min timepoints are indicated by black boxes. (C) Transmission electron micrographs of wildtype M. trichosporium OB3b cells. By 5 h and 24 h, intracytoplasmic membranes are visible (indicated by arrows). (D) PP358 spent medium UV-visible spectra. Mbn is visible at 0 m and is more abundant than in wildtype M. trichosporium OB3b; addition of CuSO4 to the medium results in chelation and a shift to a typical CuMbn spectrum, but by 24 h features at 340 nm and 390 nm are reappearing, indicating that production of apo Mbn continues. (E) PP358 naphthalene assay for sMMO activity. Active sMMO remains present at all timepoints despite the addition of copper. (F) Transmission electron micrographs of PP358 cells. Significant numbers of intracytoplasmic membranes are not observed even at 5 h and 24 h.
The sMMO and Mbn regulatory genes
Further examination of the qRT-PCR data indicates that genes within the sMMO and Mbn operons exhibit two distinct regulatory patterns. Both operons are clearly down-regulated by copper, but the genes encoding regulatory proteins within the sMMO operon, mmoR, an uncharacterized and previously unnoted but well-conserved ORF with the current locus tag MettrDRAFT_0733, and mmoG,16 along with the proposed regulatory protein genes mbnI and mbnR in the Mbn operon24 are down-regulated within 15 min (Fig. 2 and Fig. S2, ESI†). By contrast, the rest of the operon components show no significant response over this time range, with expression levels dropping only by the 5 h and 24 h timepoints. This observation is consistent with a model in which the main operons are controlled by their internal regulators, which are the primary targets of copper or other copper-dependent regulatory proteins.
MbnI is an extracytoplasmic function (ECF) sigma factor that has already been proposed as a potential regulator for the Mbn operon, and possibly for the copper switch as well.24 Analogous ECF sigma factors in siderophore systems are partnered with inner-membrane anti-sigma factors that interact with TonB-dependent transporters to form a cell surface to cytoplasm signal transduction cascade.33–36 The MbnIRT triad comprises a potentially similar system, which would couple Mbn import to Mbn regulation. In this model, MbnI would not bind copper or Mbn, but would rather interact directly with the inner membrane anti-sigma factor MbnR and its target regulatory region in response to Mbn uptake by MbnT.24 To further assess the regulatory role of MbnI, MbnI was cloned, heterologously expressed, and purified. MbnI with a C-terminal Strep-Tactin tag expresses as a monomer and binds no metal ions as measured by ICP-MS, which is consistent with its role as part of a sigma/anti-sigma factor system. However, it does bind to a heparin column, a frequently used proxy for DNA binding (Fig. S4, ESI†).37 This initial characterization is consistent with its predicted role as an ECF sigma factor. Bioinformatic analyses and gel-shift trials involving the regions preceding MbnA, MbnI, and potentially other Mbn-related genes across methanotroph species have been inconclusive; identification of the MbnI recognition site and regulon will require further investigation.
Besides MmoR and MmoG, an additional component of the sMMO operon, MmoD, has been proposed as a regulator for the copper switch.23 Early studies indicated that MmoD interacts directly with the sMMO hydroxylase, inhibiting activity in vitro,38 but based on recent studies of an M. trichosporium OB3b Δmmo-XYBZDC1-3 mutant, it was proposed to be a copper-dependent regulatory protein23 However, mmoD is not down-regulated with the other sMMO and Mbn regulatory genes, and methanotroph MmoD proteins lack well-conserved and characterized DNA-binding and metal-binding motifs. To further probe this proposed regulatory role, we heterologously expressed and purified MmoD with a C-terminal Strep-Tactin tag. We observe more of the monomeric form than reported previously,38 possibly due to a swifter purification process and a different set of affinity tags. Whether produced without metal supplementation or isolated from cells grown with 5 mM CuSO4,no significant copper binding is observed via ICP-MS, and under no conditions does MmoD appear to bind a heparin column (Fig. S4, ESI†). These combined data support the preliminary identification of MbnI, but not MmoD, as a DNA binding protein with potential relevance to copper homeostasis. However, understanding the roles of both proteins will require extensive further investigation.
The constitutive sMMO-expressing mutant M. trichosporium OB3b PP358
In 1992, a series of constitutively sMMO-expressing strains of M. trichosporium OB3b were generated via treatment with dichloromethane, a suicide inhibitor of pMMO as well as a mutagen.39 The five final strains (PP311, PP319, PP323, PP333 and PP358) have certain shared phenotypes: they express sMMO even at copper concentrations >5 μM, and exhibit decreased levels of total cellular copper.40 A comparison of copper levels in the particulate and soluble fractions of lysed cells suggests that much of that decrease derives from the lack of pMMO, which as a cuproenzyme comprising a large portion of the cellular protein mass represents a significant fraction of the copper pool.25,26,40 However, copper levels in the soluble fractions, including cytoplasmic and periplasmic proteins, are also decreased.
These constitutive sMMO-producing mutants were thus suggested to be defective in either copper acquisition or copper regulation of the two MMOs.28 Notably, secretion of a copperbinding compound, later characterized and identified as Mbn, was first observed in these strains, although it was later also identified in the wildtype species.40 We extended our gene expression studies to the mutant strain M. trichosporium OB3b PP358 with the goal of detecting differential gene regulation that could be linked to altered pathways. Although the continued presence of high levels of the mmoXYBZDC portion of the sMMO operon is expected from the phenotype, Mbn was uncharacterized at the time of PP358 development, and few experiments were carried out above 5 μM copper, meaning that a copper response differing from the response patterns of sMMO could not be ruled out.
The striking down-regulation of the full sMMO and Mbn operons was not observed in PP358 (Fig. 2 and Fig. S2, ESI†), although a significantly dampened copper response is eventually visible in some samples; this provides further support for the co-regulation of the sMMO and Mbn operons. As expected, sMMO activity and Mbn production are detected at high copper concentrations, and Mbn from PP358 had the same mass as the compound produced by the wildtype organism (Fig. 3 and Fig. S3, ESI†). Similarly dampened responses were observed for the pMMO operon, and the formation of significant numbers of intracytoplasmic membranes was not observed (Fig. 2 and 3). These trends verified and further characterized the PP358 phenotype while eliminating some possible causes, but its origin remained unclear.
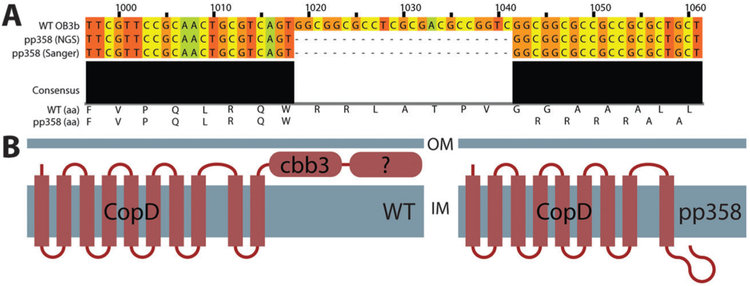
We then used next-generation sequencing to compare the PP358 genome to the published M. trichosporium OB3b genome.41 High levels of coverage (130–170×) were obtained across the entire genome, which enabled detailed identification of variants including insertions, deletions, and single nucleotide polymorphisms (Supplemental File S1, ESI†). Of high-likelihood variants that did not represent silent mutations, a significant number were in genes and genomic regions unlikely to be related to methane or copper metabolism (particularly genes related to mobile genetic elements, including transposases and related machinery). A smaller subset retained some plausible relevance to copper homeostasis (Table S1, ESI†). One variant in particular stood out: a multi-nucleotide frameshift deletion in a gene encoding CopD, an inner membrane protein thought to be responsible for importing copper into the cytoplasm in many bacterial species.42–45 Sanger sequencing of a PCR-amplified fragment containing this region confirmed the presence of this deletion (Fig. 4A). TopCons46 predicts that wildtype CopD contains 10 transmembrane helices and a C-terminal periplasmic domain. The deletion causes a frame shift in the gene, resulting in the eradication of the final transmembrane helix and the C-terminal periplasmic domains (Fig. 4B).
Fig. 4.
Mutation of CopD in the constitutive sMMO-producing PP358 strain. (A) The copD gene has undergone a multiple-nucleotide frameshift/deletion event. (B) Predicted architectures of wildtype and mutated CopD. The last transmembrane helix and the final soluble C-terminal domain are predicted to be absent as a result of the deletion.
Strikingly, the copD gene in M. trichosporium OB3b is located two genes downstream of one copy of the pMMO pmoCAB operon, preceded by the pmoD and copC genes and followed by a gene denoted DUF461 (Fig. 1A). The presence of additional genes has been noted previously in some operons encoding ammonia monooxygenase (AMO), a pMMO homolog, in which genes of unknown function follow amoCAB: amoD, which is a pmoD homolog, and in some cases amoE, a more distant pmoD homolog.47,48 In beta-proteobacterial ammonia oxidizers and alpha-proteobacterial methanotrophs, including M. trichosporium OB3b, the amoD/pmoD genes are followed by copC, which encodes a periplasmic copper binding protein CopC,49–51 copD,42,43,50 and DUF461, which encodes a second putative periplasmic copper chaperone related to PCuAC, ECuC, and other proteins thought to be involved in cytochrome c oxidase copper loading.52–54 Like pMMO, all four of these genes are mildly up-regulated in the presence of copper (Fig. 2 and Fig. S2, ESI†); interestingly, a pmoD homologue (MCA2130) was also recently identified as part of the copper-upregulated transcriptome in microarray studies of Methylococcus capsulatus (Bath).14
The copD genes from the few studied proteobacterial copCD operons42,43,50,55 are predicted by TopCons and TMHMM to encode proteins that include eight transmembrane helices. Several potential copper-binding residues, including histidines, are conserved in predicted periplasmic loop regions (Fig. S5, Supplemental Results and discussion, ESI†). By contrast, copD genes found following alpha-proteobacterial pMMO and beta-proteobacterial AMO operons contain an extra two transmembrane helices and a C-terminal periplasmic domain with a predicted heme binding site annotated as a cytochrome cbb3-like domain (PF13442, corresponding to the CcoP/FixP subunit of the enzyme (Fig. S5, ESI†)). Of the 10887 genes containing copD domains found in the JGI/IMG database at the time of analysis, only 42 genes include the cytochrome cbb3-like domain, and of those, 27 are from alpha-proteobacterial ammonia oxidizers or beta-proteobacterial methanotrophs. Gamma-proteobacterial methanotrophs and ammonia oxidizers do not have a copD gene in the immediate proximity of their AMO/pMMO operons, but they almost invariably possess copCD pairs elsewhere in their genomes. These gamma-proteobacterial copD homologues are predicted to have 14 transmembrane helices, and the extra 6 helices are part of a C-terminal domain with some homology to the cytochrome c oxidase caa3 assembly factor (CtaG), which is thought to be involved in the correct assembly of the CuA site of Bacillus subtilis cytochrome c oxidase caa3 (not to be confused with the Cox11 homologue known as CtaG that is found in Rhodobacter sphaeroides and Paracoccus denitrificans).56,57 As with the C-terminal cbb3-like domain, this domain is particularly well represented in methanotrophs and ammonia oxidizers, and interestingly, a variant with 16 helices is also common in Gram positive bacteria (Fig. S5, ESI†). Many species possess multiple copCD pairs in their genomes, potentially consistent with these proteins playing a range of roles.
The involvement of CopD in the copper switch would have several important implications for methanotroph copper regulation. First, impaired copper import into the cytoplasm will affect cytoplasmic metal-binding regulatory proteins, but not regulatory mechanisms that involve periplasmic metal sensors, ruling out two-component regulatory systems58 as likely copper switch regulators. Second, increased periplasmic copper due to reduced cytoplasmic uptake might result in up-regulation of export systems; for characterized copper resistance systems in Gram-negative bacteria, regulation frequently depends on periplasmic copper sensing via two-component regulators such as CusRS, PcoRS, and CopRS.59,60 There are no copABCDRS operons in M. trichosporium OB3b,41 but cus-like RND export systems of unknown metal specificity are present,41 and PP358 has been previously observed to have a low copper content compared to wildtype M. trichosporium OB3b at the same external copper level.40 Finally, cytoplasmic copper import may not only impact copper regulation, but may also play a role in assembly of the pMMO active site analogous to what has been proposed for cytochrome c oxidase.61–63
Conclusions
These data fill in a number of gaps in our current understanding of methanotroph copper homeostasis (Fig. 5). First, the full putative Mbn operon is coregulated with the precursor peptide, MbnA, and both are significantly down-regulated in the presence of copper, providing strong support for the bioinformatics-based identification of the Mbn operons as well as the assignment of the broader class of Mbn operons, present not only in methanotrophs but in a range of other bacterial species.24 Second, the Mbn and sMMO operons are co-regulated, suggesting that they are indeed ultimately controlled by the same copper-responsive transcription factor. In M. trichosporium OB3b, the regulatory genes in each operon are swiftly down-regulated after the addition of copper, and the main operons follow more slowly, consistent with a regulatory mode in which the in-operon regulators activate sMMO expression and Mbn biosynthesis, and are themselves repressed immediately upon copper addition, resulting in a slower decline in sMMO and Mbn as available regulator protein levels fall. Third, the mutation of the copD gene in the constitutive sMMO-producing strain PP358 provides new evidence for the involvement in a second set of copper-related genes located downstream of the pMMO operon in methanotroph copper homeostasis. The specific regulator(s) responsible for the copper switch has not yet been identified, but these combined findings provide a strong framework and many new directions for future study.
Fig. 5.
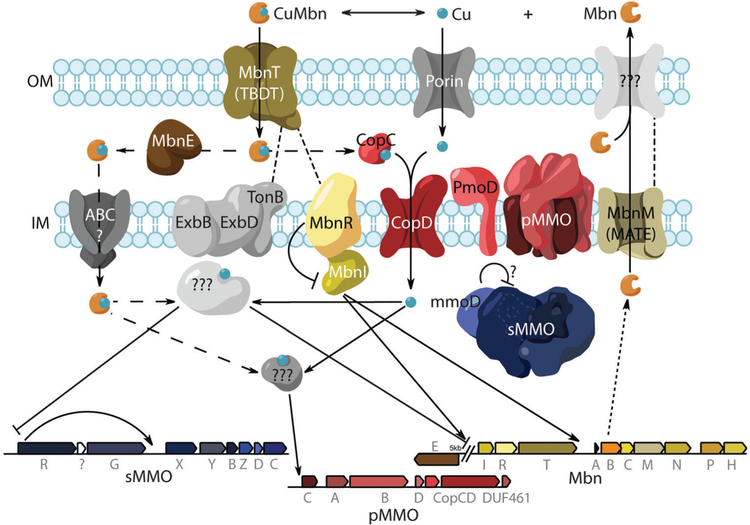
Proposed model for copper homeostasis in M. trichosporium OB3b.
Materials and methods
Additional experimental procedures are provided in ESI,† Materials and methods.
Bacterial strains and medium
Copper-starved wildtype M. trichosporium OB3b as well as the mutant strain PP358 (ATCC 55314)39 were grown in 50 mL cultures as described previously.21 Larger cultures were grown in spinner flasks specially adapted for methanotroph growth. Glassware, including spinner flasks, was treated overnight with 5% trace metal-grade nitric acid, rinsed thoroughly and autoclaved once with MilliQ-grade H2O, and a second time dry. Standard and low-copper growth media and gas mixtures were prepared as described previously.21
Timecourse experimental design
All experiments were performed with three independent biological replicates. Cultures were monitored for transition into mid-logarithmic phase growth (0.6 < O.D. < 1.0) and assayed for Mbn production and sMMO activity to confirm the correct starting phenotype. Once cultures achieved logarithmic growth, cells were harvested from spinner flasks prior to addition of CuSO4 to a final concentration of 12.5 μM, as well as 15 min, 5 h, and 24 h after copper addition. At each timepoint, cells were harvested for RNA isolation (see ESI†), OD measurement, UV-visible spectroscopy of spent medium, and transmission electron microscopy (TEM).
qRT-PCR
All qRT-PCR primers (IDT) were designed using the IDT PrimerQuest tool and were selected using constraints including 60 °C≤Tm ≤ 64°C, ΔGhomodimer ≥−7.5, ΔGheterodimer ≥−7.5, and ΔGhairpin ≥ −3, amplicon length between 70 and 150 bp, and no products with fewer than 5 mismatches and 1–2 gaps identified in the target genome using e-PCR (NCBI) (Table S3, ESI†). Each set of qRT-PCR reactions was prepared in white Bio-Rad 384-well PCR plates, and each reaction contained 1.2 μL SYBR GreenER Express Universal Master Mix (Invitrogen) and 1.2 μL of varying amounts of cDNA (1 ng for standard reactions and fourfold dilutions from 2 ng to 488 fg for primer efficiency runs), high-quality RNA (Table S4, ESI†), and/or water, dispensed via a Mosquito HTS liquid handling device (TTP), as well as 90 nL pooled forward and reverse primers (5 μM each), dispensed via an Echo liquid handler (LabCyte). Each reaction was prepared with on-plate technical triplicates, as well as no-template controls (NTC) and no-reverse-transcriptase (NRT) controls, also performed in triplicate for each primer set (NTC) or sample/primer combination (NRT). Reactions were incubated at 4 °C for 1 h, and then were run on a CFX384 (Bio-Rad), with an initial 2 m UDG inactivation step at 50 °C, followed by 2 m at 95 °C. This was followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 m, with plate reading occurring during the combined annealing/extension step. After qRT-PCR was complete, samples were allowed to anneal at 50 °C for 1 m, and melt curves were obtained by performing a plate reading step for 5 s every 0.5° from 50 °C to 95 °C. Cq values were determined in the Bio-RAD CFX software using regression for each trace. Outliers that had no Cq (in replicates where other samples had a Cq) or a melting curve not corresponding to the peak of the product were removed in the Bio-Rad software; other outliers were addressed during analysis with qBASEPLUS. In rare cases where significant gDNA contamination was observed (Cq values and products with the correct melting curve for multiple replicates and multiple genes in a given sample), the remaining RNA was treated again with Turbo DNAFree (Ambion), re-quantified, reverse transcribed again, and re-analyzed.
To confirm the existence of a single, correctly-sized product, larger 5 μL qRT-PCR reactions were performed under the same conditions and run on a 4% NuSieve 3 : 1 agarose gel in TBE (Lonza) (Fig. S6, ESI†). Similarly, since all qRT-PCR reactions were conducted using SYBR Green to visualize product accumulation, melting curves were determined for all reactions. Primers (and, ultimately, reference gene candidates) that exhibited multiple melting peaks or multiple bands on the analytical agarose gel (Fig. S6, ESI†) were removed from candidacy. The sequences, product sizes, and locus tags for genes of interest and reference gene candidates are provided in Table S2 (ESI†).
Quantification of relative gene expression levels
Initial data screening, including visualization of melt curves, was performed in the CFX Manager software 3.1 (Bio-Rad); at this stage, reactions with no signal or a signal over 10 Cq from those of its technical replicates were removed from further calculations if the remaining technical replicates were acceptable. Primer efficiency calculation (Table S5, ESI†), identification of acceptable reference genes (Fig. S7, ESI†), and calculation of CNRQ for all genes was performed in GeNormPLUS/qBasePLUS 3.0 (Biogazelle)64,65 (Supplemental File S2, ESI†). Hierarchical clustering of log10-transformed CNRQ data was performed as described in Cluster 3.0,66 and dendrograms and heatmaps were generated from this data using JavaTreeView.67
Phenotype analysis
At each time point, 1 mL methanotroph culture was subjected to centrifugation for 15 min at 8000 × g at room temperature, and a UV-visible light spectrum was obtained from the supernatant using an Agilent 8453 UV-visible spectrophotometer. The sMMO activity assay was performed as previously described, except that cells were washed three times in salt solution prior to naphthalene incubation, and a saturating amount of naphthalene was added directly to cell suspensions.32,68 For TEM, approximately 3 × 109 cells (calculated via culture optical density) were collected via centrifugation for 20 min at 6000 × g and 4 °C. The pellet was resuspended in growth medium without metals but containing 5% EM grade glutaraldehyde (Sigma) and incubated for 4 h at 4 °C while rocking. At that time, the cells were re-pelleted and re-suspended in fresh phosphate/glutaraldehyde buffer and incubated overnight at 4 °C while rocking. The cells were centrifuged again and the resulting pellet was further fixed with 2 exchanges of a modified Karnovsky’s formula (2.5% glutaraldehyde, 2% paraformeldehyde in a 0.05 M PBS). The fixed bacteria were rinsed 3 × in 0.05 M PBS. Enrobed bacteria were embedded in resin, sectioned, and imaged using Gatan Orius camera on a JEOL 1230 TEM (80 kV accelerating voltage) (additional methods in ESI†).
Genomic DNA isolation, library preparation, and sequencing
Cells of copper-starved M. trichosporium OB3b PP358 were harvested, and the MasterPure Complete DNA Isolation kit (Epicentre) was used as described to isolate genomic DNA. dsDNA was quantified on a QuBit. When excess RNA was detected, a second RNase A (Epicentre) incubation was performed, followed by a phenol-chloroform extraction to remove the enzyme and a final clean-up step using a Genomic DNA Clean & Concentrator kit (Zymo Research). dsDNA was quantified using a QuBit prior to library construction.
Three separate gDNA libraries were prepared and sequenced. A 500 bp short insert library and a 2 kb mate pair library were prepared using TruSeq and Nextera Mate Pair kits respectively (Illumina) and were sequenced on an Illumina HiSeq2000 using 100 bp paired-end reads (Beijing Genomics Institute). The reads were filtered to remove reads with more than 10% uncalled bases and reads in which at least 40% of the bases had phred scores under 20. Reads with adapter contamination were also removed.
Reads from all three datasets were aligned to M. trichosporium OB3b using BWA MEM with default parameters. The resulting BAM file was sorted and variants were discovered using the Samtools mpileup function (version 1.2) followed by BCFtools (version 1.2) using standard parameters. Variants having a Quality score less than 20 or a read depth greater than 350 were flagged as low quality, and identified variants were manually confirmed using IGV 2.3.55.69 To confirm regions of particular interest, Sanger sequencing was employed on PCR-generated fragments (sequences in Table S6, ESI†). The resulting nucleotide sequences were aligned against the wildtype DNA sequence using MAFFT (L-INS-I mode).70
CopD bioinformatics
All ORFs containing the CopD PFAM (PF05425) were obtained from the JGI-IMG database (Supplemental Files S3 and S4, ESI†).The predicted number of helices was calculated using TMHMM.71 For genes of interest, including CopD from wildtype and mutant M. trichosporium OB3b PP358, transmembrane homology was confirmed via TopCons,46 and the JGI-IMG annotation of additional domains was supplemented by analysis with HHPred72 (SI textfile 2, ESI†).
Supplementary Material
Significance to metallomics.
Methanotrophs, bacteria that convert methane gas to methanol, have attracted intense attention for their potential applications in bioremediation and gas-to-liquids conversion processes. How these bacteria obtain copper for their key metabolic enzyme, particulate methane monooxygenase (pMMO), is not well understood. One means of copper uptake involves a copper-binding natural product called methanobactin. Analysis of copper-responsive gene expression provides evidence for the involvement of specific genes in methanobactin biosynthesis, regulation, and transport as well as in the copper-dependent expression of pMMO and an alternative iron-containing, soluble MMO (sMMO). Several other genes likely involved in cytoplasmic copper uptake have also been identified.
Acknowledgements
This work was supported by NSF grant MCB0842366 and the Institute for Sustainability and Energy at Northwestern (ISEN). G. E. K. was supported by American Heart Association predoctoral fellowship 14PRE20460104 and the Northwestern University Presidential Fellowship. Imaging work was performed at the Biological Imaging Facility generously supported by the Office for Research (Northwestern University). The qRT-PCR experiments were performed at the High Throughput Analysis Facility, supported by the Robert H. Lurie Cancer Center and the Office for Research (Northwestern University). Nucleic acid quality control and genome sequencing were performed with the help of the NUSeq Core Facility (Northwestern University.) Mass spectrometry was performed at the Integrated Molecular Structure Education and Research Center. We additionally thank Dr. George Georgiou for permission to use the PP358 strain (sourced from ATCC) and Dr. Stefan Green (University of Illinois, Chicago) for assistance with the PP358 genome.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c5mt00289c
References
- 1.Sirajuddin S and Rosenzweig AC, Biochemistry, 2015, 54, 2283–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Semrau JD, Front. Microbiol, 2011, 2, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haynes CA and Gonzalez R, Nat. Chem. Biol, 2014, 10, 331–339. [DOI] [PubMed] [Google Scholar]
- 4.Hanson RS and Hanson TE, Microbiol. Rev, 1996, 60, 439–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brantner CA, Remsen CC, Owen HA, Buchholz LA and Perille Collins ML, Arch. Microbiol, 2002, 178, 59–64. [DOI] [PubMed] [Google Scholar]
- 6.Lieberman RL and Rosenzweig AC, Nature, 2005, 434, 177–182. [DOI] [PubMed] [Google Scholar]
- 7.Balasubramanian R, Smith SM, Rawat S, Yatsunyk LA, Stemmler TL and Rosenzweig AC, Nature, 2010, 465, 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenzweig AC, Frederick CA, Lippard SJ and Nordlund P, Nature, 1993, 366, 537–543. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen AK, Gerdes K and Murrell JC, Mol. Microbiol, 1997, 25, 399–409. [DOI] [PubMed] [Google Scholar]
- 10.Ali MH and Murrell JC, Microbiology, 2009, 155, 761–771. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert B, McDonald IR, Finch R, Stafford GP, Nielsen AK and Murrell JC, Appl. Environ. Microbiol, 2000, 66, 966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stolyar S, Franke M and Lidstrom ME, J. Bacteriol, 2001, 183, 1810–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stafford GP, Scanlan J, McDonald IR and Murrell JC, Microbiology, 2003, 149, 1771–1784. [DOI] [PubMed] [Google Scholar]
- 14.Larsen Ø and Karlsen OA, MicrobiologyOpen, 2015, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kao W-C, Chen Y-R, Yi EC, Lee H, Tian Q, Wu K-M, Tsai S-F, Yu SS-F, Chen Y-J, Aebersold R and Chan SI, J. Biol. Chem, 2004, 279, 51554–51560. [DOI] [PubMed] [Google Scholar]
- 16.Choi DW, Kunz RC, Boyd ES, Semrau JD, Antholine WE, Han J-I, Zahn JA, Boyd JM, de La Mora AM and DiSpirito AA, J. Bacteriol, 2003, 185, 5755–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iguchi H, Yurimoto H and Sakai Y, FEMS Microbiol. Lett, 2010, 312, 71–76. [DOI] [PubMed] [Google Scholar]
- 18.Stanley SH, Prior SD, Leak DJ and Dalton H, Biotechnol. Lett, 1983, 5, 487–492. [Google Scholar]
- 19.Kim HJ, Graham DW, DiSpirito AA, Alterman MA, Galeva N, Larive CK, Asunskis D and Sherwood PMA, Science, 2004, 305, 1612–1615. [DOI] [PubMed] [Google Scholar]
- 20.Bandow NL, Gallagher WH, Behling LA, Choi DW, Semrau JD, Hartsel SC, Gilles VS and DiSpirito AA, Methods Enzymol, 2011, 495, 259–269. [DOI] [PubMed] [Google Scholar]
- 21.Balasubramanian R, Kenney GE and Rosenzweig AC, J. Biol. Chem, 2011, 286, 37313–37319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krentz BD, Mulheron HJ, Semrau JD, DiSpirito AA, Bandow NL, Haft DH, Vuilleumier S, Murrell JC, McEllistrem MT, Hartsel SC and Gallagher WH, Biochemistry, 2010, 49, 10117–10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Semrau JD, Jagadevan S, DiSpirito AA, Khalifa A, Scanlan J, Bergman BH, Freemeier BC, Baral BS, Bandow NL, Vorobev A, Haft DH, Vuilleumier S and Murrell JC, Environ. Microbiol, 2013, 15, 3077–3086. [DOI] [PubMed] [Google Scholar]
- 24.Kenney GE and Rosenzweig AC, BMC Biol, 2013, 11, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeda K and Tanaka K, Anton. Leeuw. Int. J. G, 1980, 46, 15–25. [DOI] [PubMed] [Google Scholar]
- 26.Prior SD and Dalton H, Microbiology, 1985, 131, 155–163. [Google Scholar]
- 27.Semrau JD, DiSpirito AA and Yoon S, FEMS Microbiol. Rev, 2010, 34, 496–531. [DOI] [PubMed] [Google Scholar]
- 28.DiSpirito AA, Zahn JA, Graham DW, Kim HJ, Larive CK, Derrick TS, Cox CD and Taylor A, J. Bacteriol, 1998, 180, 3606–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vorobev A, Jagadevan S, Baral BS, DiSpirito AA, Freemeier BC, Bergman BH, Bandow NL and Semrau JD, Appl. Environ. Microbiol, 2013, 79, 5918–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knapp CW, Fowle DA, Kulczycki E, Roberts JA and Graham DW, Proc. Natl. Acad. Sci. U. S. A, 2007, 104, 12040–12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott DC, Brannan J and Higgins IJ, J. Gen. Microbiol, 1981, 125, 63–72. [Google Scholar]
- 32.Brusseau GA, Tsien HC, Hanson RS and Wackett LP, Biodegradation, 1990, 1, 19–29. [DOI] [PubMed] [Google Scholar]
- 33.Van Hove B, Staudenmaier H and Braun V, J. Bacteriol, 1990, 172, 6749–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Angerer A, Enz S, Ochs M and Braun V, Mol. Microbiol, 1995, 18, 163–174. [DOI] [PubMed] [Google Scholar]
- 35.Koebnik R, Trends Microbiol, 2005, 13, 343–347. [DOI] [PubMed] [Google Scholar]
- 36.Ferguson AD, Amezcua CA, Halabi NM, Chelliah Y, Rosen MK, Ranganathan R and Deisenhofer J, Proc. Natl. Acad. Sci. U. S. A, 2007, 104, 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nawapan S, Charoenlap N, Charoenwuttitam A, Saenkham P, Mongkolsuk S and Vattanaviboon P, J. Bacteriol, 2009, 191, 5159–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merkx M and Lippard SJ, J. Biol. Chem, 2002, 277, 5858–5865. [DOI] [PubMed] [Google Scholar]
- 39.Phelps PA, Agarwal SK, Speitel GE and Georgiou G, Appl. Environ. Microbiol, 1992, 58, 3701–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fitch MW, Graham DW, Arnold RG, Agarwal SK, Phelps PA, Speitel GE and Georgiou G, Appl. Environ. Microbiol, 1993, 59, 2771–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stein LY, Yoon S, Semrau JD, DiSpirito AA, Crombie A, Murrell JC, Vuilleumier S, Kalyuzhnaya MG, Op den Camp HJM, Bringel F, Bruce D, Cheng J-F, Copeland A, Goodwin L, Han S, Hauser L, Jetten MSM, Lajus A, Land ML, Lapidus A, Lucas S, Médigue C, Pitluck S, Woyke T, Zeytun A and Klotz MG, J. Bacteriol, 2010, 192, 6497–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chillappagari S, Miethke M, Trip H, Kuipers OP and Marahiel MA, J. Bacteriol, 2009, 191, 2362–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serventi F, Youard ZA, Murset V, Huwiler S, Bühler D, Richter M, Luchsinger R, Fischer H-M, Brogioli R, Niederer M and Hennecke H, J. Biol. Chem, 2012, 287, 38812–38823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cha JS and Cooksey DA, Appl. Environ. Microbiol, 1993, 59, 1671–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X-X and Rainey PB, Environ. Microbiol, 2008, 10, 3284–3294. [DOI] [PubMed] [Google Scholar]
- 46.Tsirigos KD, Peters C, Shu N, Käll L and Elofsson A, Nucleic Acids Res, 2015, 43, W401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.El Sheikh AF, Poret-Peterson AT and Klotz MG, Appl. Environ. Microbiol, 2007, 74, 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park S and Ely RL, Arch. Microbiol, 2007, 189, 541–548. [DOI] [PubMed] [Google Scholar]
- 49.Arnesano F, Banci L, Bertini I, Mangani S and Thompsett AR, Proc. Natl. Acad. Sci. U. S. A, 2003, 100, 3814–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee SM, Grass G, Rensing C, Barrett SR, Yates CJD, Stoyanov JV and Brown NL, Biochem. Biophys. Res. Commun, 2002, 295, 616–620. [DOI] [PubMed] [Google Scholar]
- 51.Zhang L, Koay M, Maher MJ, Xiao Z and Wedd AG, J. Am. Chem. Soc, 2006, 128, 5834–5850. [DOI] [PubMed] [Google Scholar]
- 52.Abriata LA, Banci L, Bertini I, Ciofi-Baffoni S, Gkazonis P, Spyroulias GA, Vila AJ and Wang S, Nat. Chem. Biol, 2008, 4, 599–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blundell KLIM, Hough MA, Vijgenboom E and Worrall JAR, Biochem. J, 2014, 459, 525–538. [DOI] [PubMed] [Google Scholar]
- 54.Dash BP, Alles M, Bundschuh FA and Richter O, Biochim. Biophys. Acta, 2015, 1847, 202–211. [DOI] [PubMed] [Google Scholar]
- 55.Wijekoon CJK, Young TR, Wedd AG and Xiao Z, Inorg. Chem, 2015, 54, 2950–2959. [DOI] [PubMed] [Google Scholar]
- 56.Bengtsson J, von Wachenfeldt C, Winstedt L, Nygaard P and Hederstedt L, Microbiology, 2004, 150, 415–425. [DOI] [PubMed] [Google Scholar]
- 57.Greiner P, Hannappel A, Werner C and Ludwig B, Biochim. Biophys. Acta, 2008, 1777, 904–911. [DOI] [PubMed] [Google Scholar]
- 58.Ma Z, Jacobsen FE and Giedroc DP, Chem. Rev, 2009, 109, 4644–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rensing C and Grass G, FEMS Microbiol. Rev, 2006, 27, 197–213. [DOI] [PubMed] [Google Scholar]
- 60.Rademacher C and Masepohl B, Microbiology, 2012, 158, 2451–2464. [DOI] [PubMed] [Google Scholar]
- 61.Ekici S, Yang H, Koch H-G and Daldal F, mBio, 2011, 3, e00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ekici S, Pawlik G, Lohmeyer E, Koch H-G and Daldal F, Biochim. Biophys. Acta, Bioenerg, 2012, 1817, 898–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ekici S, Turkarslan S, Pawlik G, Dancis A, Baliga NS, Koch H-G and Daldal F, mBio, 2013, 5, e01055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, de Paepe A and Speleman F, Genome Biol, 2002, 3, research0034.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hellemans J, Mortier G, de Paepe A, Speleman F and Vandesompele J, Genome Biol, 2007, 8, R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eisen MB, Spellman PT, Brown PO and Botstein D, Proc. Natl. Acad. Sci. U. S. A, 1998, 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saldanha AJ, Bioinformatics, 2004, 20, 3246–3248. [DOI] [PubMed] [Google Scholar]
- 68.Bowman JP and Sayler GS, Biodegradation, 1994, 5, 1–11. [DOI] [PubMed] [Google Scholar]
- 69.Thorvaldsdottir H, Robinson JT and Mesirov JP, Briefings Bioinf, 2013, 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Katoh K and Standley DM, Mol. Biol. Evol, 2013, 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krogh A, Larsson B, von Heijne G and Sonnhammer EL, J. Mol. Biol, 2001, 305, 567–580. [DOI] [PubMed] [Google Scholar]
- 72.Soding J, Biegert A and Lupas AN, Nucleic Acids Res, 2005, 33, W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.