Abstract
Magnetic resonance (MR) imaging provides a method to obtain anatomical information from the brain in vivo not amenable to optical imaging because of its opacity. MR is non-destructive and obtains deep tissue contrast with 100 µm3 voxel resolution or better. Manganese-enhanced MRI (MEMRI) may be used to observe axonal transport and localized neural activity in the living rodent and avian brain. Such enhancement enables researchers to investigate differences in functional circuitry or neuronal activity in images of brains of different animals. Moreover, once MR images of a number of animals are aligned into a single matrix, statistical analysis can be done comparing MR intensities between different multi-animal cohorts, comprised of individuals from different mouse strains, different transgenic animals, or between different time points after an experimental manipulation. Although preprocessing steps for such comparisons (including skull-stripping and alignment) are automated for human imaging, no such automated processing has previously been readily available for mouse or other widely used experimental animals and most investigators use in-house custom processing. This protocol describes a step-wise method to perform such preprocessing for mouse.
Keywords: Magnetic Resonance Imaging (MRI), Manganese-enhanced magnetic resonance imaging (MEMRI), Computational analysis and preprocessing, Mouse and rodent brain, phenotyping brain anatomy
Introduction
Magnetic resonance imaging (MRI) is a non-invasive technology applicable to living animals that provides detailed information about brain anatomy, including volumetric information, location of fiber tracts, and circuitry responsible for functional relationships between brain regions. Challenges in generating such images from small animals include low signal-to-noise ratios since standard MR imaging detects nuclear magnetic resonance from the protons in water, and thus image detection is dependent on sample volume (Henkelman, 2010; Hoyer et al., 2014; Nieman et al., 2007; Nieman et al., 2005). Techniques to overcome this challenge are to use a high magnetic field, long imaging times, and contrast agents such as gadolinium (Fornasiero et al., 1987; Geraldes et al., 1986), chromium (Zhang et al., 2010b), and manganese (Pautler, 2004; Pautler and Koretsky, 2002; Pautler et al., 1998). Post-capture image averaging using computational tools also holds the promise of increasing detection of structural and functional detail. MR offers the possibility of obtaining anatomical information more quickly than histologic analysis and can be performed in the living animal, where changes over time and functional information can be directly imaged. However, MR images require considerable computational processing before data can be analyzed with the powerful statistical parametric mapping tools that have emerged for human imaging over the past dozen years.
Statistical parametric mapping is among the more powerful techniques developed for human brain imaging analysis (Friston, 2005). This computational approach depends on alignment of stacks of 3D MR images, often of the same subject imaged over time. For human images there are automated methods to align images. But for small animal brain images, there is no established protocol for the preparation of images for subsequent statistical analysis. The current state of the field is to use in-house custom processing unique to each lab. Most processing procedures require two major processes: an initial step, colloquially known as “skull stripping”, that masks the non-brain tissue in whole head images for subsequent alignments. This first step, skull stripping, is particularly time-consuming. For the second step, alignment of multiple extracted brains into a single matrix, several sub-steps are required. For each of these, automation programs are separately available. A pipeline to assemble these steps into a continuous process is available online from the Laboratory of Neuro Imaging (LONI) at The University of Southern California (formerly at the University of California at Los Angeles) that can perform this function for animal models (Bearer et al., 2007a; Dinov et al., 2010; Dinov et al., 2009; Medina et al., 2017; Rex et al., 2003). Use of this pipeline relies on uploading images to the LONI Pipeline server which are then processed remotely. Such distance processing poses the problems of multiple users competing for processing time, and requires the exchange of data through security checkpoints on Internet communication portals between institutions, which are especially restricted for researchers located at medical centers.
Here we present our stand-alone, self-reliant protocol for both preprocessing steps for small animal brain images (Gallagher et al., 2013; Gallagher et al., 2012; Medina et al., 2017; Zhang et al., 2010a). Our method uses software developed at several other institutions—it is not our intention here to describe the processing algorithms, which appear in publications cited below and in the websites indicated. Here we explain how to apply this software, in detail, at each step of our preprocessing protocol which takes images generated by the scanner and preprocesses them for statistical analysis. We give the rationale for each step and how it contributes to creation of aligned image stacks for comparative statistical analysis of mouse brain MR images, the ultimate goal. The overall rationale for the steps explained below is shown in Figure 1.
Figure 1. Overview of the processing events.

(A) shows a diagram of the steps required to take raw MR images to a stage where statistical analysis can be performed. Processing begins with a dataset of 3D MR images in Nifti (.nii) format with identical headers (Steps A1–3). After correcting the whole head image for magnetic field inhomogeneities (N3 correction) and scaling for intensity (modal scaling) (Steps A5–6), a mask is created and applied (Steps A7–8). Extracted brain images then undergo linear alignment and a minimal deformation atlas (MDA) of non-contrast enhanced images created. Each animal’s non-contrast enhanced image is warped to this MDA to obtain a warp field/control point grid that is then applied to the contrast-enhanced image of the same animal. This prepares a data set of aligned images for statistical analysis. (B) shows a diagram demonstrating non-linear alignment (warping) and the respective warp-field (R.E. Jacobs, by permission). An original image is warped to a template image, which changes the original grid. This new grid is the warp field. The geometric changes to the grid obtained by producing the warp field for a non-contrast-enhanced image can be applied to contrast-enhanced images from the same animal, producing a warped image via an alignment procedure that is independent of intensity values in the contrast-enhanced image. (C) shows an example of alignments performed this way(Medina et al., 2017). Pre-injection image represents the minimal deformation atlas (MDA) created from non-enhanced images, and 0.5 hr post, 6 hr post an 25 hr post are aligned contrast-enhanced images of the same cohort of animals after application of the control point grid obtained by warping their respective pre-injection image to the post-injection contrast-enhanced images.
In this protocol, we define preemptive quality control measures that include: (1) ensuring that all images have the same header parameters; and (2) aligning all images that need to be averaged together via rigid-body transformation, in instances where the position of the mouse is shifted between scans in experiments with multiple scans at one time point. We also correct for bias-field inhomogeneity through an N3 correction process (Ellegood et al., 2015; Larsen, 2014; Sled et al., 1998). Due to non-uniformities in the radio frequency pulse and the magnetic field in MR scanners, as well as anatomical issues in the test subjects, variations in intensity (also known as a bias-field) arise throughout image capture, and the true signal of the image loses some of its high-frequency components. Therefore, non-parametric non-uniform intensity normalization, also known as N3 correction, must be performed. N3 correction takes the distorted signal and sharpens its distribution to create an estimation of the true value. It then changes the distribution of the intensity of the distorted image in a way that best fits the estimated model. Finally we adjust variance in the intensity scaling due to inconsistency in the Fourier transformation of the radiofrequency signal by scaling the intensity histogram with a modal scaling process (Gallagher et al., 2013; Gallagher et al., 2012; Medina et al., 2017; Zhang et al., 2010a). The MATLAB code for that process is included here as a text file in Supplemental Materials. These steps are performed before automated removal of extra-meningeal tissue voxels on the whole-head image, and then repeated on the extracted brain image.
We call this automated masking of extra-meningeal tissues “skull-stripping”. Performing skull stripping allows us to “extract” the brain image from the whole head image as captured in intact living animals. We then create a minimal deformation atlas (MDA) image by averaging all pre-treatment images within an experimental or control group of mouse images. This allows all images to be aligned and warped to the same template, so that their size and orientation are precisely co-registered for voxel-wise statistical analysis (Kochunov et al., 2001). Flow diagrams of these two essential steps, skull stripping and brain alignment, are shown in Figure 2.
Figure 2. Flow charts of the two multi-step processes described in this protocol.

In the series of steps lettered A1–8, we check image parameters and prepare a mask to extract the brain image from non-brain tissues (left column). In the second series of steps (right column) we align the brain images into a single matrix where the anatomical features of each image are in the coordinates as every other image in the dataset.
Our protocol was developed in part to perform analyses of manganese-enhanced MR images (MEMRI). MEMRI is an emerging imaging technique that uses the paramagnetic ion, manganese, to enhance the T1-weighted signal and thereby increase signal-to-noise ratio in the MR image, improving detection of anatomical structures. This is especially useful when imaging living animals because imaging time must be short (Pautler, 2004; Pautler and Koretsky, 2002; Pautler et al., 2003; Pautler et al., 1998; Silva and Bock, 2008; Silva et al., 2004). MEMRI has been used to observe functional circuitry and axonal transport dynamics in the living brain (Bearer et al., 2007a; Bearer et al., 2007b; Bearer et al., 2009; Gallagher et al., 2013; Gallagher et al., 2012; Medina et al., 2017; Pautler, 2004; Van der Linden et al., 2004; Zhang et al., 2010a), and localized neuronal activity (Malkova et al., 2014; Yu et al., 2005). Multiple studies have been done to determine a non-toxic dose of Mn2+ (Bearer et al., 2007a; Eschenko et al., 2012; Lindsey et al., 2013).
While several papers have been published on how to perform stereotactic injection of Mn2+ for MEMRI tract-tracing in mice, this imaging approach poses particular challenges for image registration due to the hyper-intense signal from the injection site of the manganese since alignment algorithms depend on intensity patterns, the hyperintense injection site is not present in pre-injection images and its location and size may differ slightly from animal to animal. We developed some specific sub-routines in this protocol to obviate those complications. We thus arrived at a protocol that will be useful to automate the alignment of many other types of MR imaging, including of other animals, with and without contrast agents, and captured with differing pulse sequences.
Protocol
Materials
Instructions to download and install all of these recommended programs are available online at the links provided in this section. The programs listed in this section are all free, open source software. However, two. SPM and FSL, require the proprietary softwaree, “MATLAB”.
Unix-based computer
(to run SkullStrip) Mac OS system with Xcode installed (Xcode, X11 or Xquartz are Apple’s versions of the X server component of X windows system, for Mac OS X). Xcode has become the Xquartz project, an open source effort to develop an X.org X Windows for Mac OS X. Xcode is required to run MATLAB, and is available for free download at https://xquartz.macosforge.org/trac/.A virtual Linux machine running on a PC-Type Windows computer also is compatible as long as it has a 64-bit processor.
3D MR Image Viewing Program.
This is needed to review images as they move through the processing environment. We do not recommend any particular 3D program viewer, as preference of the interface should serve as a good indicator of which program(s) to choose. Examples of such programs include Statistical Parametric Mapping (SPM), FMRIB Software Library (FSL) (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki; (Jenkinson et al., 2012; Smith et al., 2004; Woolrich et al., 2009)), Fiji (or ImageJ) (http://imagej.nih.gov/ij; (Paletzki and Gerfen, 2015; Schindelin et al., 2012)), and MRIcron (http://people.cas.sc.edu/rorden/mricron/index.html; (Rorden et al., 2007)).
MATLAB
MATLAB is necessary for various processing steps and for function of various other processing programs MATLAB is an abbreviation for “matrix laboratory.” While other programming languages usually work with numbers one at a time, MATLAB® operates on whole matrices and arrays. Language fundamentals include basic operations, such as creating variables, array indexing, arithmetic, and data types. MATLAB is a matrix-based language from Mathworks for science and engineering applications, developed at University of New Mexico. It is usually available for researchers on University campuses, although it may require purchase of a license, and can be downloaded at http://www.mathworks.com/products/matlab/
FMRIB Software Library (FSL) is a comprehensive library of analytical tools for fMRI, MRI, and DTI brain imaging data that was created by the Analysis Group, FMRIB, Oxford, UK. FSL. It runs on Apple and PCs (both Linux, and Windows via a Virtual Machine), and is very easy to install. Some processing components of FSL rely on MATLAB (see below in Methods). FSL is useful as a 3D viewer, and for various processing steps, and is available for free download at http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/
Medical Image Processing, Analysis and Visualization (MIPAV) was developed to facilitate human brain MR imaging and is a product of the US National Institutes of Health. Some features of the program are also useful for rodent brain imaging. MIPAV is useful for N3 correction, and when necessary, image transformations. It is available for free download at https://mipav.cit.nih.gov
NiftyReg is an alignment tool co-created at University College London. NiftyReg also requires CMake (an open-source cross-platform build system; https://cmake.org/download/), and a compiler, such as XCode in MacOS (https://cmake.org/download/). Currently NiftyReg can use an accelerated graphics processing unit (GPU) with CUDA from NVIDIA, CUDA® is a parallel computing platform and programming module developed by NVIDIA. CUDA speeds up image processing and can be downloaded from https://developer.nvidia.com/cuda-downloads. NiftyReg is necessary for our in-house SkullStrip programs (Delora et al., 2016), and as an option for alignments later in this protocol. NiftyReg is available for free download at http://cmictig.cs.ucl.ac.uk/wiki/index.php/NiftyReg_install.
modal_scale_nii.m is a MATLAB routine developed in the Jacobs’ lab, and has been applied in a variety of papers (Bearer et al., 2007a; Bearer et al., 2007b; Delora et al., 2016; Gallagher et al., 2013; Gallagher et al., 2012; Malkova et al., 2014; Zhang et al., 2010a) although the actual processing code is previously unpublished. The “modal_scale” code is provided here with this protocol for the first time (see Supplemental Materials). This short program normalizes the gray scale between MR images in NIFTI format (.nii) based on a proportional scaling of the mode of the intensity histogram to a user-specified template image. It also produces a graph of the adjusted histogram for visual validation. This routine is useful for intensity scaling, and is provided here in the supplemental materials (see Supplemental Materials).
skullstripper.py is a program we created (Delora et al., 2016). It can be downloaded from our publication (Delora et al., 2016) or from our STMC website (http://stmc.health.unm.edu/tools-and-data/index.html). The computer code, installation instructions, and application methods are found in the Supplemental Materials for that paper. This script is dependent on NiftyReg. Instructions to set up NiftyReg are included in the application from the journal website. skullstripper.py is useful for automated masking of non-brain tissue and can be applied in batch so that multiple images are stripped in one speedy step.
Statistical Parametric Mapping (SPM) is an amazing software tool that allows voxel-wise unbiased comprehensive analysis of the intensity pattern over the whole brain. SPM requires MATLAB to run. A new toolkit in SPM for mouse brain processing was developed in 2009 and can be loaded into earlier versions of SPM as an extension and was presented at the ISMRM conference in Hawaii in 2009 (http://www.spmmouse.org (Sawiak et al., 2009). With this plugin SPM allows statistical analysis to be performed on brains with different voxels sizes. It. SPM is useful for reviewing alignments, viewing intensity histograms, checking image headers, linear and non-linear warping and smoothing images before application of statistical analysis. SPM is available for free download at http://www.fil.ion.ucl.ac.uk/spm/software/spm12/. SPM12 takes Nifti-format images (*.nii), whereas an earlier version, SPM8, uses Analyze format (*.img with *.hdr).
Example MR Images to verify that the protocol works are provided in Supplemental Materials. These images are from datasets used to generate figures in this paper. They are in NIFTI (.nii) format in little-endian byte order and 16 bit signed integer type. Capture of these images is described in detail in our previous publications (Bearer et al., 2007b; Bearer et al., 2009; Gallagher et al., 2013; Gallagher et al., 2012; Manifold-Wheeler, 2016; Medina et al., 2017; Zhang et al., 2010a). Images were captured at 24h after stereotaxic injection of Mn2+ into the CA3 of the right hippocampus with a 11.7 T Bruker biospin vertical bore MR scanner.. These images are 160 × 128 × 88 voxels in dimension with voxel size of 0.1 mm, isotropic. All experimental procedures received prior approval by Caltech and UNM IACUCs.
Methods
First we provide details about the way we have written the preprocessing protocol, including command lines, user input, general information about how the commands provided can be applied, for-loops and output file directories. The following protocol steps are written with NIFTI file format (.nii) in mind.
Command lines:
“Terminal” is an application in Mac OS X that allows access to the Linux-based operating system and uses command lines to specify processing steps. Because the Mac Terminal application is based on UNIX, command lines provided below also work on a Windows-Linux computer or other computer using Linux. To open Terminal on OS X, open your Applications folder, then open the Utilities folder. Double click on the Terminal application icon. You may want to add this to your dock. Once open, Terminal will give you a “prompt”, after which you will type the command. To navigate to folders, use UNIX commands, such as “cd” to change directories, “ls” to list contents within a directory, and “/” to go up to a higher folder.
In this protocol Terminal command lines are denoted by different margins and text font. Terminal commands should be entered into the terminal as typed, with any sections of the protocol written in red text and flanked by “< >“ indicating to enter user-set file or directory names or values, which must be edited by the user according to their file and directory names on their own computer.
User input.
Our protocol requires that the user enter information. The user does so by entering command lines in Terminal application on Mac, or after the prompt in a Windows-Linux PC machine. In the next two paragraphs we describe how the commands are presented in this protocol. The user may need to have some knowledge of UNIX commands, although we provide all text needed except the specific directory and file names describing new data that the user wishes to process. We recommend that while reading this, the user has Terminal or Linux window with the prompt open on their computer, to see how those systems work.
General information about the terms and style of the protocol:
User-input portions, including file names, of each terminal command in this protocol have an “_” underscore between each word instead of spaces. This “_” is to indicate that filenames cannot have spaces in them to function properly in the given commands. If users have files with spaces in their names, the alternative is to include quotation marks at either end of a directory file path and file name when entering user’s path and file names. Terminal commands that specify directory paths can be truncated to not include the full directory path information by first navigating to the directory containing the images to be processed. For this to work, all files referred to must be located in the live directory when entering the command. For all within-program menu navigations, the nomenclature is “Parent-menu>Sub-menu>sub-sub-menu>etc.” Any command line can be modified to navigate up or down to specify files in parent or sub-directories by using UNIX commands.
About for-loop commands:
For-loop commands (for f in *) are command lines which repeat Terminal commands in batch, for all files specified. This allows the same processing protocol to be applied to multiple images and is a major component for automated processing of large datasets, vastly speeding pre-processing time. As an example, a for-loop command may have the following format:
In Terminal Enter:
for f in *.nii; do <command_function> <input_image_file _directory_path>/$f <output_image_file_directory_path>/${f}_<user_entered_filename_suffix_extension>; done
This for-loop indicates an arbitrary variable (f) to include all filenames with the suffix “.nii” (all NIFTI file extensions), and says to do a user specified “command function” to all files ($f) in the specified directory path. Similarly, any output files for the command line function can have user edited prefix or suffix additions, but keep the entire text of the original input filename (${f}) within those user defined changes. All for-loops must also contain the command “done” in order to end the for-loop function.
For-loops can be extremely useful, particularly in batch processing of data, and can be custom written in such ways as to fit the file-naming schemes for specific sets of data. For example, if one wanted to perform only the function in the for-loop for files with the suffix “_mask.nii”, rather than all “.nii” file extension files in the directory, it would only require a change of the “*.nii” portion to be written as “*_mask.nii”, or in the case where the user only wanted to perform the function on all files that started with the letter “P”, they could change the “*.nii” to be “P*”. These specifications can also be done in combination (e.g. “P*_mask.nii”).
For the purposes of the following protocols, for-loop command examples will be given only in cases where batch processing can be easily done. Any FSL command line function can be executed in a for-loop, and writing for-loops for any function is dependent on specific needs for individual data sets and file-naming schemes.
Output files.
It is very important that for all terminal commands that require user specified output directories, the specified directory has already been created. Otherwise the output files will not be written, because the specified directory will not exist. This becomes extremely important for time-intensive calculation steps done in large batches (with for-loops), where hours of computer processing time can be for naught.
Protocol
We have divided the protocol into two main multi-step processes: A. Preparing the raw images and skull stripping them; and B. Aligning the stripped images. See Figure 2 for flow charts of these two procedures. In some instances we provide instructions to perform the same step in three different ways, using different software for each alternative process.
Steps A1-A8: Preparing the images and skull stripping
This is the first procedure in this two-fold protocol, automated skull stripping.
Step A1: Check input images by viewing them in the 3D viewer.
A very important first step is to examine critically all images in the dataset visualizing each image in a 3D viewer (Figure 3). Things to watch for include the general quality of the image, whether image gray-scales are similar, and if there are any MR capture glitches, such as lines of data with obvious abnormalities or blurring. It is also imperative when working with a large set of images that there are no duplicate images, or mislabeled image filenames. Usually visual inspection is sufficient to detect these issues. Figure 3 shows visualization of MR images in FIJI prior to alignment. An example of an imaging defect is indicated in this figure. Using FIJI this glitch can be corrected, as described in the Troubleshooting section below. Any of the Image Viewing software described above is useful for this important quality control step.
Figure 3. Visualization of MR images in FIJI/ImageJ 3D image viewer.
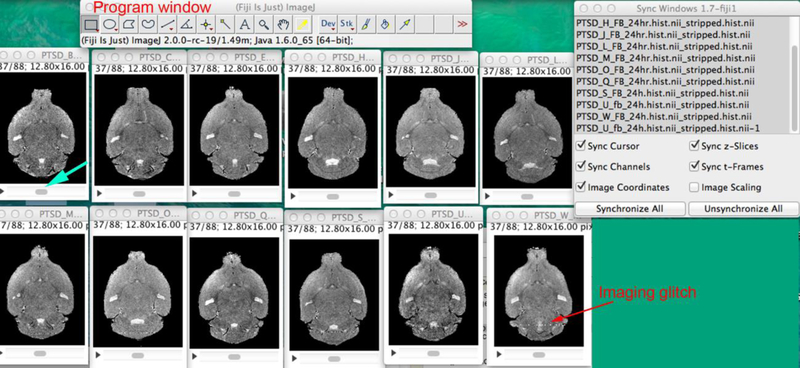
Open FIIJI and drag your set of MR images into the program window (top of screenshot above). Under the menu bar, pull down “Window” and select “Tile”. Then arrange the images on the screen. To synchronize all images so that you can scroll through the 3D stacks quickly, pull down the menu under “Analyze” select Tools>Synchronize windows. The Sync window will open (top right). You can now scroll any one image by moving the gray scroll button left or right under one of the images (turquoise arrow) and all will move synchronously through their respective stack. By watching the images, defects are apparent (red arrow).
Here we describe using FIJI as an example. Drag and drop image icons into the FIJI program window. This can be done one at a time or as a batch by selecting all and dragging them together into the program window. Go to “Window>Tile” and the images will be laid out on your screen in the order that they were opened or that they are listed in the directory. To set the gray-scale for all images equivalently at this stage (for viewing purposes) in FIJI we select Image>Adjust>Brightness/Contrast and then select Auto for each image. This function displays the image histogram and helps determine if images are signed (positive and negative intensity values) or unsigned (all positive values). All images in any particular dataset should be either “signed” or “unsigned”. This signed and unsigned refers to the intensity values being either only on the positive side of the intensity histogram horizontal axis (unsigned) or on both positive and negative sides (signed). A more detailed version of the histogram can be observed in FIJI by selecting Analyze>Histogram, or in other programs (covered in other sections).
At every step in the protocol, images should be viewed and checked to ensure that the processing has not changed their type, dimensions, and intensity scaling. Checking the histogram is particularly necessary for verifying that the modal scaling procedure, described in detail below) has produced the expected result. All processing steps should be checked to ensure proper performance by reviewing the headers of each individual image, which can be opened in MIPAV, and by viewing the output in a 3D viewer such as FIJI. More details for these quality checks is provided further along in this protocol.
Step A2: Check header to ensure all images have the same image parameters
The header file contains information about the parameters of the images. Figure 4 shows the header in MIPAV. After visual inspection, we want to ensure that all images in the dataset have the same dimensions (number of voxels in each dimension), voxel size (in mm), file type (char, short, int, float, double, etc.), byte order (Little or Big Endian), signed or unsigned, and bit type (typically either 8, 16, or 32) (Delora et al., 2016; Woods et al., 1998). Settings for each of these parameters can differ between images, and prior to further processing they must be made uniform across all images in a particular dataset. The bit size is important as it determines how many possible intensity values can be recorded in the image (Gillespy et al., 1994; Gillespy and Rowberg, 1993).
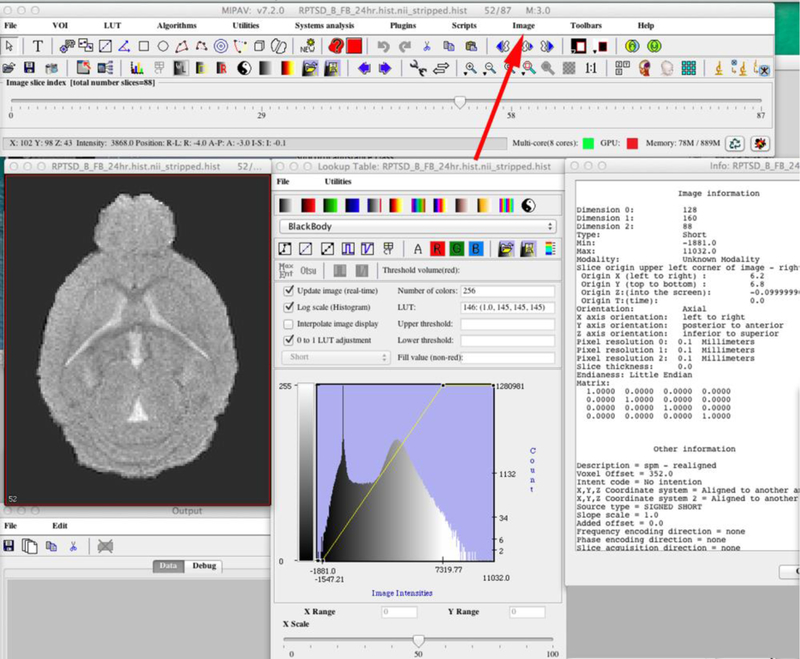
Figure 4. Reviewing the Header in MIPAV.

Open each image in MIPAV via File>Load image. By selecting the indicated icon (red arrow), the image information will open in the box adjacent to the image. MIPAV can transform images, which is sometimes necessary. See http://mipav.cit.nih.gov/pubwiki/index.php/Transform.
The header file contains information on all of the aforementioned parameters and will either be embedded within the image file (NIFTI .nii) or will be contained in a separate file alongside the image (Analyze format, .hdr) (Rajeswari and Rajesh, 2009). The header can be viewed and edited in a 3D image viewer program. These viewers will also allow changes and transformations of these parameters in order to create uniform header information across all images in the group to be processed. If you plan to use a standard template for later processing steps instead of an image from within your dataset, it is best to include that template in all steps throughout the protocol. We have provided a template in the Supplemental Materials for this paper. The template image must have the same header information as the images in your dataset.
We use MIPAV, FSL, or FIJI/ImageJ to check the header. Below are instructions for viewing header file information in FSL or FIJI/ImageJ. Figure 4 shows the MIPAV window for reviewing the header.
To view a header file in FSL:
Open Terminal
Change to directory containing image(s)
Enter the command:
fslhd <input_image_file_directory_path>/<input_image_file_name>
For batch processing of many images, enter the for-loop command:
for f in *.nii; do fslhd <input_image_directory_path>/$f; done
This command will create an in-terminal printout of the header file(s). For larger sets of files, you can use the for-loop to create printouts for every file in sequence in the terminal, and then copy the terminal printout into an easier viewing format (such as any word processor), and compare the header files in batch. To view a header file in FIJI/ImageJ:
Open FIJI/ImageJ
Drag image to FIJI/ImageJ window to open
Select Image Show Info
Repeat for all images and compare header information
Step A3: Edit header (if necessary)
If header information is not identical, the images cannot be aligned or even batch processed for these pre-alignment steps. We prefer MIPAV as the best method to adjust headers.
If the headers are not identical, the next step is to edit the header file. Again, the reason for this is so that all images have the same parameters. Attempts made to align images of different sizes and intensity histograms will fail. See Figure 4 and MIPAV website for more instructions to fix this in MIPAV. Differences may be simple and quickly fixed by typing in new header information, or more difficult requiring image adjustments, such as voxel interpolations or zero-filling. It is best to capture images with identical scan parameters to avoid the need for adjustments.
Headers can also be edited in FSL. To edit the header file information in FSL you can individually change most information using the following in FSL;
1. Open Terminal and Enter the command:
fsledithd<input_image_file_directory_path>/<input_image_filename>
This command opens up an in-terminal header file editor for the specified image, which should be self-explanatory. By comparing across your set of images you can change all header file information to be identical.
In addition, FSL provides a shortcut to copy header file geometry jn batch which is particularly useful with the following for-loop command:
for f in *.nii; do fslcpgeom <file_whose_geometry_is_to_be_copied> $f; done
This will copy all header file geometry (used by FSL) for NIFTI format files (typically the most troublesome issue when trying to process images in batch) from one file onto all target files in that same directory. This can also be done without a for-loop for a single image by starting at the “fslcpgeom” portion, and replacing the “$f” with the directory path and filename of the single file. Be aware that if images have even slightly different dimensions from each other, this can sometimes distort or stretch the images in strange ways. Before changing header file information, we suggest saving all images prior to this processing step in a different directory and making sure that all images are of the same dimensions. If your images are in different dimensions (voxel size, or x/y/z measurements), these must first be corrected in another program. The procedure for doing so will not be covered here.
Again, quality control at this point is indispensable. You must visualize all images that have had header information changed in your 3D viewer, and also check their histograms. Confirm that the edits have been saved in the file by opening each edited image in FIJI, selecting Image>Show info and checking all numbers.
Step A4: Average whole head images and align them (if necessary)
Some image capturing/experimental designs may require that several image be taken for each animal at each of the time points in a series (Figures 5 and 6) (Kovacevic et al., 2005). These images may need to be averaged together to produce a higher-quality master image for each animal at each designated time point. To average multiple images of the whole head of the same animal captured during the same imaging session (i.e., not moved in and out of the MR scanner) we use fslmaths function in FSL (Step A4.1). Often such images do not need to be aligned. However, sometimes images may have shifted in the scanner during the imaging session, which produces a fuzzy averaged image (Figure 5). If fuzziness appears in the averaged image, then backtrack and align the input images (Step A4.2).
Figure 5. Averaging images and detecting movement artifact.
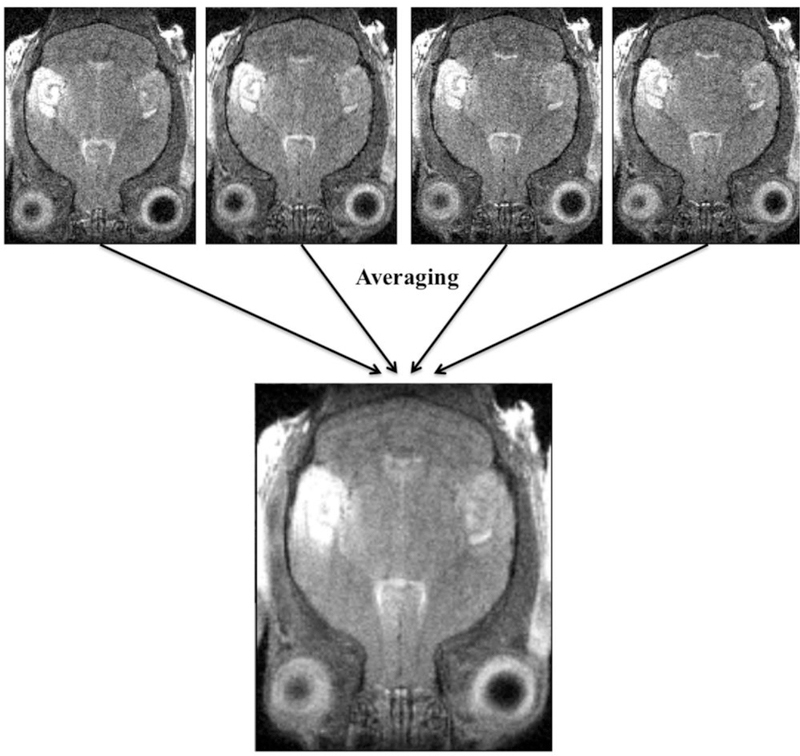
Four offset scans taken at successive times during a single imaging session of one particular mouse are offset due to the mouse moving between images. We show that through averaging these images together with “fslmaths”, a blurred average image results.
Figure 6. Correcting misalignment artifacts artifact for averaging whole head images using FLIRT in FSL.
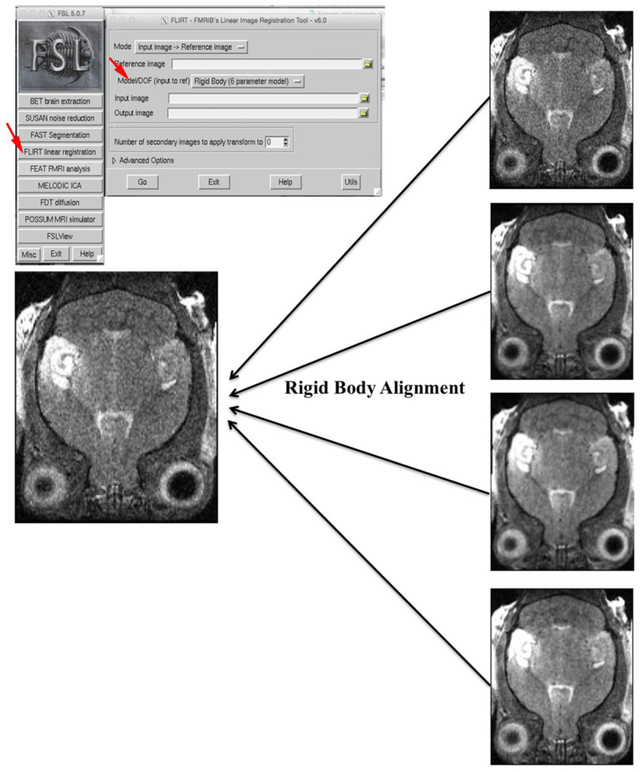
FSL FLIRT is a rigid body alignment tool that can correct movement artifacts (Step A4 and Troubleshooting). One of four offset images was randomly chosen to serve as a reference to perform an affine rigid body rotation with FSL FLIRT on the other three images as well as to itself (Step A4 and Troubleshooting). Show in the box at top left is the GUI and the window that is activated when selecting “FLIRT linear registration” in the GUI (indicated by red arrow). IN the pop-up window, select “Rigid Body”(6 parameter model). This procedure aligned all of the images together. This tool can also be used for alignments leading to the MDA (Step B3), but we prefer to do that step in NiftiReg.
Step A4.1 To average multiple whole head images when captured at the same time-point.
Open Terminal
Navigate to the directory that contains the image(s) to be averaged
Enter the following command:
fslmaths -dt <input_image_file_datatype> <input_image_file_directory_path>/<first_input_image_filename> -add<second_input_image_file_directory_path>/<second_input_image_filename> -add<third_input_image_file_directory_path>/<third_input_image_filename> -div <number_of_images_being_averaged><output_image_file_directory_path>/<output_image_filename> -odt<output_image_file_datatype>
An averaged image will be placed in the current directory. Note that “-dt” signifies the data-type used in internal calculations. These can include char, short, int, float, or double. Also note that “-odt” signifies the output data-type. Selecting “-odt input” will create an output image that is the same data-type as the input images. If nothing is selected for “-dt” or “-odt” then 32 bit float will be the default. This can be used to average as many images into a single image as one would like, just by continuing to “-add <input_image_directory_path>/<n-th_input_image_filename>“ for each desired image in sequence into the command line.
Inspect the averaged image in your 3D viewer. If it is blurry, as shown in Figure 5, the animal may have moved during the scans. Minor movements of the stereotactic device or movement of the animal being imaged can lead to the resulting images being off-centered from each other.
Step A4.2 Align images if necessary
If the averaged image appears blurry, use FSL FLIRT to correct (Figure 6). In the FSL GUI, select “FLIRT”. In the pop-up window type in the path to the first one of your images as “Image input”. This image will serve as the reference image to which the others are aligned. IN the pull down menu for “Model/DOF (input to ref) select “Rigid Body (6 parameter mode)”. Set the “Number of secondary images to be transformed” at the number of scans in dataset, including the one you are also using as a reference image. Enter the names of all these images in the input image window. In the output image, we recommend giving output images the same name as the input, but with an added suffix that helps know that they were processed, such a _FL.nii. Always remember to perform any processing step on copies of the original images in a new directory exclusively created for that step to avoid overwriting any previous manipulations or the raw image. Average the output images as described above and visually inspect the result for crispness.
Step A5: N3 correction to balance field inhomogeneities
As described above, high B fields in the MR scanner may not be homogeneous, and thus inhomogeneities in this magnetic field may produce varying intensities in the image. These variations can be equalized by an “N3 correction” (Sled et al., 1998). N3 correction can be run in SPM or MIPAV. Instructions and information about N3 are explored at length by Sled et al. (Sled et al., 1998) and Larsen et al. (Larsen, 2014) and the MIPAV website:
http://mipav.cit.nih.gov/pubwiki/index.php/Shading_Correction:_Inhomogeneity_N3_Correction
You will need to create a new folder for N3 corrected images as otherwise the program will overwrite your original images. Name this folder “N3_corrected”. Place copies of the images you wish to correct in this folder.
1. Open MIPAV
2. Open all images to be corrected that were placed in the new N3_corrected folder in MIPAV.
2. Go to Algorithms>Shading correction>Inhomogeneity N3 correction in MIPAV.
3. Type appropriate parameters and press ok.
For parameters, we typically use the following settings: Signal threshold 1.0, 50 iterations, end tolerance 0.0001, field distance in mm (this is generated by the image, for our typical mouse brain it is about 2.785 mm), subsampling factor 4.0, kernel 0.15, and Wiener filter noise 0.01. We select “whole brain” and no options.
N3 corrected images will be saved in the N3_corrected folder. These new images will overwrite the images you copied into that folder. So be sure that originals are saved somewhere else.
To do N3 correction processing in batch, writing a MIPAV script in the internal script editor (accessed via Scripts>Record script) can be very useful. Note that this is not a command, it is a script, i.e. it is a sub-program that runs inside MIPAV. For our above-specified parameters, the following script can be used. In this case “image 1” refers to the input image and “image 2” its corrected output, which are user defined filenames.
IHN3Correction(“input_image_1 ext_image $image1”, “do_process_whole_image boolean true”, “signal_threshold float 1.0”, “end_tolerance float 0.0001”, “max_iterations int 50”, “field_distance_mm float 2.785”, “subsampling_factor float 4.0”, “kernel_fwhm float 0.15”, “wiener_noise_filter float 0.01”, “do_auto_histo_thresholding boolean false”, “do_create_field_image boolean false”)
CloseFrame(“input_image_1 image $image1”)
SaveImage(“input_image_1 image $image2”)
CloseFrame(“input_image_1 image $image2”)
Step A6: Modal scaling of intensity maps
In addition to the fact that the image can have non-uniformities in intensity for the reasons described above, the gray scale of every image can vary. This means that the intensity range of one image may not be the same as that of the next image taken. To correct for this, we scale the mode of the intensity histogram of each image to that of a reference image (Bearer et al., 2007b; Kovacevic et al., 2005). We call this step “modal scaling”. The reference image for whole head modal scaling can be a whole head image selected at random from the dataset to be processed.
We created a MATLAB script to do this (see Supplemental Materials for a MATLAB based script to perform this function, and a template mouse brain image in NIFTI format).
Open MATLAB
Create a new directory for the images you plan to scale. Place copies of the images to be scaled, along with “modal_scale_nii.m” script file, into the new directory and migrate to that directory in the main MATLAB window.
At the prompt in the MATLAB command window type “modal_scale_nii”, which should bring up a GUI prompt asking for the user to enter number of bins to display the histogram, and the delta value to find the peaks of signal (defaults are 1000 for both, which we find sufficient).
Select the reference image you would like to use to scale all other images and whether you would like the histograms plotted for you.
After running the program, check by visualizing them and clicking on the histogram plots in your 3D Viewer (See Figure 7 for how to visualize the histogram in MIPAV, and Figure 8 to visualize in MATLAB “model_scaling” module). Histograms and headers can also be checked in FIJI. This will give useful information as to whether your images were properly scaled. Some header file discrepancies can affect the scaling calculations. If the images are not scaled appropriately as determined by coincident peaks in the histogram, re-check their pre-scaling headers to find discrepancies and correct them before running the program again.
Figure 7. MIPAV example of an intensity histogram.
The intensity histogram can be obtained under Image>histogram. Once an image is scaled to the mode of the histogram, the histogram should have the same peak and extent in all images in the dataset.
Figure 8. Histogram before and after modal scaling.
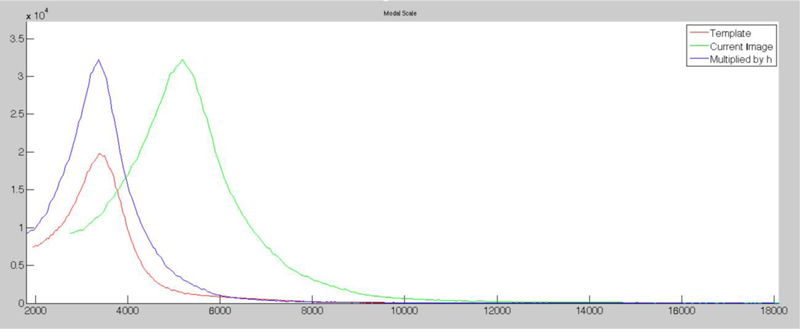
Graph showing the histograms of images before and after modal scaling using a template (red) to adjust the intensity scale of a current image (green) giving a final adjusted histogram for the current image matching the intensity distribution of the template (blue). Note that these are unsigned images (all intensity values are positive), and that the peaks of the histograms for the template (blue) and modal-scaled (red) images are similar, whereas prior to modal scaling the peak of the intensity histogram for the new image (green) was not coincident with the template histogram. (The graph is generated in MATLAB after running our modal scaling (see Supplemental data for code).
This script should automatically create a sub-directory within the new one directory you created for this step. This directory will be called “output” and it will now contain the scaled images. These processed images will be renamed with “.hist” in the suffix of the filename by the script.
Step A7: Skull-stripping (also known as “Masking non-brain tissue”, or “brain extraction”)
Extra-meningeal tissues, such as the skull, mouth, nares, ears, etc., must be masked out of the images, so that only the brain remains. We call this process “skull-stripping,” also known as masking or extraction. For human images, there are well-known programs that perform this task using edge detection methods, such as BrainSuite (http://brainsuite.org/processing/surfaceextraction/bse/; (Shattuck and Leahy, 2001; Shattuck and Leahy, 2002) or Brain Extraction Toolkit (BET) (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/BET/UserGuide; (Smith, 2002)). However, the automations of these programs do not work well on mouse images because mouse brains are not spherical in shape, as in humans, and mice have a narrower space of cerebral spinal fluid separating the brain and the skull, giving less distinct edges of the brain. This means that masks created with these programs are poor in quality. For mice, we therefore developed a “SkullStrip” program, skullstripper.py (Delora et al., 2016), to automate mask production for rodent brain images for MEMRI studies (Figure 9). The code for this program together with instructions in a “readme.txt” file, are available as Supplemental Materials to Delora et al. (Delora et al., 2016), and on our website: http://stmc.health.unm.edu/tools-and-data/ as described above.
Figure 9. Diagram of automated brain extraction: Skullstripper.
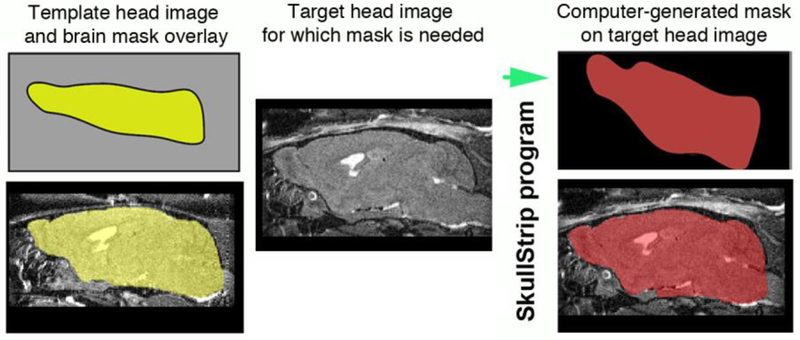
Our SkullStrip program, skullstripper.py, takes a reference template image of a whole head together with and its respective mask (yellow, left panels) and warps the template mask to fit a new, target head image (gray, middle panel). Output of the computer-generated mask and its overlay on the new target image shows the precise fit this process produces (red, right panels).
Our SkullStrip program first requires that a refined mask be created for one of the images in the dataset to serve as the “reference mask.” For MEMRI datasets we use a non-contrast-enhanced whole head image, typically the pre-injection image. The mask is first made with the automation in Brainsuite or the Brain Extraction Toolkit (BET) and the resultant mask is further hand-refined in FSLView (Figure 10) (see (Delora et al., 2016) for more detailed instructions). This produces the reference mask for a reference whole head image, that are used together to generate masks for all images in the dataset.
Figure 10. Example of creating the reference mask or refining a mask manually in FSL.

Skullstripper.py requires input of a head image and its respective mask. To prepare a reference mask, or to refine masks (Step A7 and Troubleshooting), we use FSLView. View each mask overlaid onto the whole head image and check the accuracy. Masks must be checked in each coordinate (coronal, axial and sagittal) at each slice in the 3D dataset of each individual brain. Once the skull-stripping processing has been performed, some masks in the batch may have errors. The Transparency Bar, Lock, Pencil Icon, and Editing Tools are highlighted.
Our program, skullstripper.py, will warp the reference whole head non-contrast-enhanced image to fit over new whole head images in the dataset (see diagram in Figure 9 and an example of a mask in Figure 11) (Delora et al., 2016)). The program then applies the same warp field used to fit the reference image to the new images, takes the resultant control point grid (warp field) and applies it to warp the reference mask. This process warps the reference mask to fit the new input images. The program automatically repeats these steps for any images indicated in a for-loop batch. Thus the reference mask is warped to fit the original matrices of the new images, which themselves are not warped or otherwise distorted by this process. In the case of MEMRI, the post-injection image cannot be warped directly, as the hyper-intense Mn2+ signal distorts computational alignment calculations based on gray-scale, and thus would produce a distorted image. Thus, by warping a template mask to fit the contrast-enhanced image, distortions produced by hyper-intense signal are avoided.
Figure 11. Application of a thresholded mask to create the brain-stripped image for alignment.
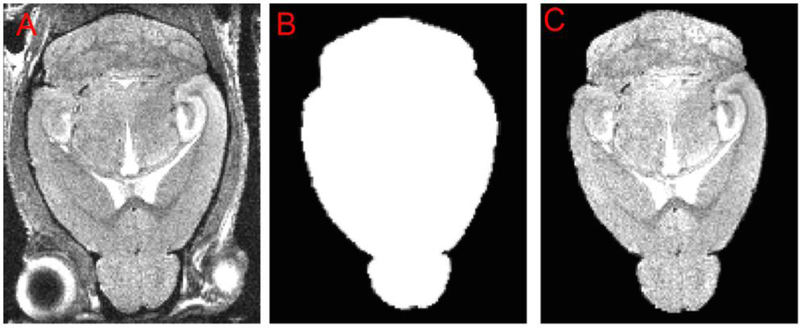
This shows the results of the SkullStrip program, skullstripper.py, to mask the extra-meningeal tissue in the whole head image. This is the final results after following steps A1-A8. (A) A new whole-head image. (B) The mask generated by skullstripper.py and thresholded to produce a black-white image derived from the reference image and reference mask and warped to fit the image in (A). (C) Application of the mask to the whole head image cause the non-brain tissues to disappear from the image because they are masked out. This resultant image is shown here as a sincle axial slice. However, the mask must fit in all three orthogonal dimensions and must be checked in a 3D viewer in each coronal, axial, and sagittal slice across the whole brain.
Delora et al. (Delora et al., 2016) describe the SkullStrip program at length and provide a protocol in the Supplemental Materials accompanying that paper. Note that the open source program, NiftyReg (Ma et al., 2014; Modat et al., 2010), must be installed to run skullstripper.py. Installation and compilation of NiftiReg routines prior to processing is required, otherwise the commands below will not work. All instructions for download, compilation and creation of command paths, are presented in (Delora et al., 2016). Below are brief step-wise instructions to apply skullstripper.py using NiftyReg dependencies:
Open Terminal and change the terminal directory to the directory containing all image(s) referred to in the following command line, as well as making sure that the skullstripper.py program is copied into the same directory. (Note this step is unique to the stripping command line, because the skullstripping.py program is written to refer to files within the command terminal directory. One could modify the skullstripping.py code to refer to any specified directory path; however, the details of those modifications will not be discussed here).
In Terminal Enter the for-loop command:
for f in *.nii; do python skullstripper.py –in $f -ref <pre-injection_reference_image_filename> -refmask <pre-injection_reference_mask_filename> -out ${f}_mask; done
Warped-mask files for each corresponding image will now be placed in a new automatically created output sub-directory called “masks”.
Thresholding the masks:
In order to filter out noise in the mask that may have been caused from warping, we set a threshold, meaning that anything with an intensity value below that threshold is removed from the mask. Thus a black-white mask is created with no fuzzy gray borders. This can be done in SPM or using the “fslmaths” function in FSL. The threshold number can be changed, higher or lower, according to the intensity scale of the image.
We like to enter “${f}_thrmask” as the output name. This signifies that the name of the image to be masked will be kept with the suffix “_thrmask.nii” added to the end of its name to denote that this mask has had a threshold applied. For the purposes of this protocol, subsequent command lines will assume this nomenclature.
1. To threshold the mask using FSL, in Terminal Enter:
fslmaths<input_image_file_directory_path>/<input_image_filename> -thr <value_to_threshold_at><output_image_file_directory_path>/<output_image_filename>_thrmask –odt <output_image_file_datatype>
2. To run a batch of masks on many images in your dataset, use this for-loop command:
for f in *_mask.nii; do fslmaths<mask_image_file_directory_path>/$f -thr <value_to_threshold_at><mask_output_file_directory_path>/${f}_thrmask -odt<output_image_file_datatype>; done
Masks with the threshold applied will be placed in the indicated output directory.
The most important aspect of applying a threshold to your masks is finding the right amount of threshold without creating masking artifacts (often these happen on the border of masked images, which we refer to as “edging artifacts”). Since the output mask files generally have values that mostly fall between 0 and 255, your threshold value will fall somewhere between there. We suggest opening masks in an image viewer and applying a threshold based on any strange edging artifacts created by the mask (if you apply the masks in the steps below, you can get a better idea of the edging artifacts we are referring to). Essentially, choosing a mask threshold value may require some observational predictions or trial and error to best fit your images.
The mask should appear as a black-white image with no fuzzy borders after masking. Visually inspect masks for imperfections. We prefer to perform this action in FSL view (Figure 10). If the mask does not have any imperfections, continue to the next step. If it does have imperfections, the mask may need hand-refining, as you did when creating the reference mask.
Step A8: Apply mask
The next step is to apply the thresholded masks to their respective whole head images (Figure 11). We prefer to use the “fslmaths” function in FSL. Again, create a new folder for the output files. This avoids over-writing your input and allows you to follow the processing step-by-step.
We like to enter “${f}_masked” as the output image name. This signifies that the original name of the image to be masked will be kept with the suffix “_masked.nii” added to the end of its name to denote that this image has been masked. Note the “_mask.nii_thrmask.nii” is based on our previous nomenclature that generates threshold masks with the following extensions added to the “<input_image_filename>“.
Note that “-dt” signifies the data-type used in internal calculations. These can include “char,” “short,” “int,” “float,” “double” etc. Also note that “-odt” signifies the output data-type. Selecting “-odt input” will create an output image that is the same data-type as the input images. If nothing is selected for “-dt” or “-odt” 32-bit float will be the default image type.
1. Open Terminal and enter the following command to create a single mask:
fslmaths -dt <input_image_file_datatype><input_image_file_directory_path>/<input_image_filename> -mas<mask_file_directory_path>/<mask_filename><masked_image_output_directory_path>/<masked_image_output_image_filename>_masked -odt <output_image_file_datatype>
2. To run a batch applying masks to a series of images, use this command:
for f in *.nii; do fslmaths -dt <input_image_file_datatype><input_image_file_directory_path>/$f -mas${f}_mask.nii_thrmask.nii<masked_image_output_directory_path>/${f}_masked -odt<output_datatype>; done
Masked images of the brain will be placed in the indicated output director. These images will be used for all subsequent steps.
Steps B1-B8. Aligning skull-stripped images
The goal of this second series of pre-processing steps is to create a stack of aligned 3D images for use in subsequent statistical analysis, with all brain structures at the same coordinate position throughout the stack and normalized to the same peak intensity. Here we describe how to perform the steps to align the brains after they have been skull-stripped (masked), as shown on the right side of the flow chart, Figure 2. Some of these steps repeat the processing describe in the above section, A1–8, but on the extracted brain images. When a step is similar to those previously described, the reader will be referred to those commands and Figures that appeared earlier in this protocol. While command lines remain the same, input and output filenaes and directories will differ between Steps A1–8 and Steps B 1–8.
Step B1. Bias-field correction (Second N3 correction -- if necessary)
A second N3 correction may be necessary on the extracted brain images because extra-meningeal tissues have intensity values that differ from that of the brain, and thus may have interfered with the N3 processing of the brain regions in the whole head image. For example the skull is very light-shaded at low intensity compared to the brain. In order to minimize the influence of extra-meningeal tissue, brain images may also need to be N3 corrected. See Step A5 above for how to perform N3 correction in MIPAV to repeat this processing step.
Depending on the MR scanner and the images being processed, this second inhomogeneity correction (post-skull-stripping) may not be required. We recommend users determine this based on inspection of their own images looking for intensity variations that do not appear correlated with brain anatomy. If users are concerned about being able to determine this, we recommend applying a second N3 correction to the brain-extracted images.
Step B2: Second modal scaling (if needed)
The extracted brain has less intensity variation than the whole head image. Hence repeat of the modal scaling is likely to refine the scale of the intensity histogram. To perform modal scaling on the extracted brain images, follow the instructions in Step A6 above, replacing the whole head images with the stripped brains as input.
The histogram of intensity should be checked after this procedure to ensure that modal scaling has brought the mode and scale of the histogram for all images to similar intensity values. Again, signed images have both negative and positive values whereas unsigned are only positive. The histogram reflects this. All images must be the same signed or unsigned type to proceed (Review Figures 7 and 8 for examples).
We also recommend checking image type before and after scaling. If images in the same dataset scale improperly based on their sign or unsigned designation, one option is to convert all files to an unsigned format, such as 32-bit float, which can be done in Fiji, MIPAV or MRicron. Histograms can be viewed in FIJI/ImageJ, MIPAV, or SPM as described above in Step A1 and 2. Refer to Figures 3 and 4 for examples of visualizing the images in FIJI and the histogram in MIPAV respectively. In FIJI/ImageJ the histogram may also be checked in the window that pops up when selecting Image>Adjust Brightness/Contrast.
For the same reasons as described in Step B1 above for N3 correction, modal scaling may also not need to be repeated on the stripped brain images. Check the histograms before and after attempting to perform a repeated modal scaling on the extracted brain. If no change is visible in either the image or the histograms after running the modal scaling, the step may be eliminated in subsequent processing of images in the dataset. If uncertain as to whether this second modal scaling changes the histogram, be conservative and apply it.
Steps B3-B4: Affine alignments images and creation of the minimal deformation atlas (MDA)
In order to align all images in a dataset into the same space for statistical analysis, all the images need to be co-registered to either a minimal deformation atlas (MDA) created from the non-enhanced images in the same dataset, or to a standardized mouse brain template (Figure 12 and Supplemental materials for a template). We often create our MDA from non-contrast-enhanced images (Figures 13 and 14 for an example and a diagram of these procedures, respectively). The steps below can be applied to all images, or to only the non-contrast enhanced images. If applied to all images, the MDA should still only be created from non-contrasted-enhanced images. Contrast-enhanced images can subsequently be separately aligned using steps described next in Step B5.
Figure 12. Image of an axial slice from a template 3D MR image as possible reference for brain image alignments.
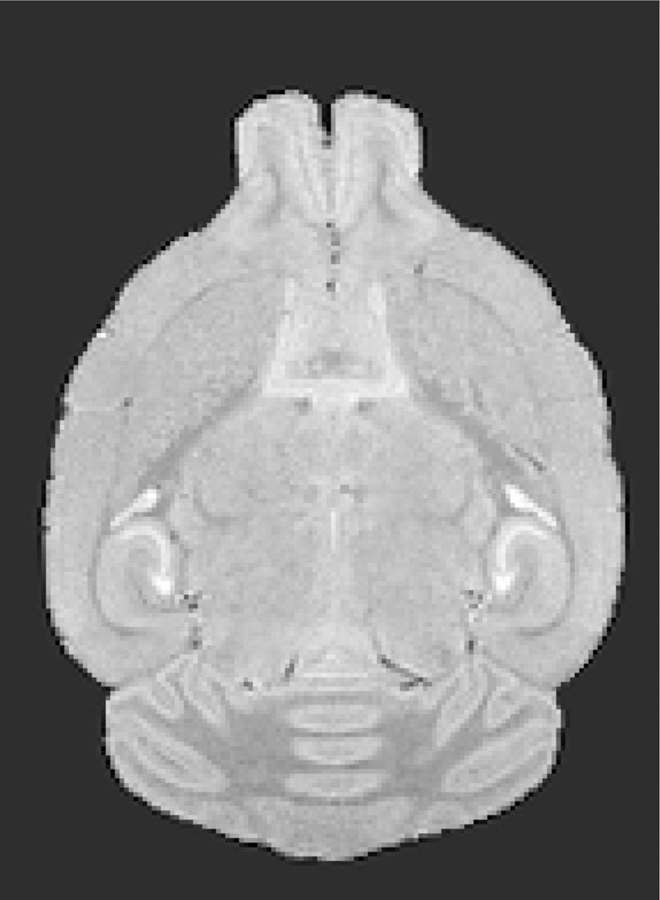
Shown is a single axial slice from our 3D template MR image showing intensity scaling and skull-stripping, both useful for modal scaling and alignments of other images (see Supplemental Materials for this image in NIFTI format).
Figure 13: Example of a Minimal Deformation Atlas (MDA).

(A) shows a single pre-injection image from a data set of 6 non-contrast enhanced images. (B) is the averaged image from all 6 animals in this dataset, the minimal deformation atlas (MDA). Notice that the MDA has finer detail than the pre-injection image. Application of this preprocessing described in this protocol produces more consistent MDAs and finer detail than when images are manually stripped of non-brain tissue and processed through other preprocessing procedures. Also see (Delora et al., 2016).
Figure 14: Diagram of Steps B3–6 for aligning non-contrast and contrast-enhanced brain images.

A diagrammatic representation of the alignment procedures described in Steps B3-B6. In Step B3, non-contrast enhanced brain images from Step B2 are affine transformed (NC1, NC2, NC3 etc). In Step B4 these aligned non-contrast-enhanced images are averaged, and then re-aligned against their averaged image. These re-aligned images are then averaged to create the MDA. In Step B5, each set of contrast-enhanced images from Step B2 (C1a-x, C2a-x, C3a-x, etc., where 1,2,3,…n are the mice numbers, and a,b,c,…x are experimental time points) are aligned to their respective non-contrast-enhanced image from the same animal using a rigid body rotation. In Step B6, the re-aligned non-contrast-enhanced images for each mouse (NC1r, NC2r, etc.) representing the “step 3 images” from the output of Step B4.2, are warped to the MDA, to obtain a Warp field for each mouse (WF1, WF2, etc.). The warp field is then applied to all rigid body aligned contrast images for each mouse from Step B5, one mouse at a time, to produce the output with all images from the dataset aligned-warped (NC1.w, NC2.w, etc and C1a.w, C1b.w etc.; C2a.w, C2b.w, etc) where *.w indicates that images that have been warped.
The process of creating an MDA has two steps: 1) Affine linear alignment to a respective target (reference or template image) (Step B3); 2) Obtain an average image of non-contrast-enhanced brain-stripped images (Step B4). For MEMRI, because the Mn2+ signal can cause miscalculations in the alignment using a warping process, the MDA is comprised only of baseline, non-contrast enhanced images (pre-injection with Mn2+). Non-linear warping alignment is done for both non-enhanced and contrast-enhanced images based on the warp-field generated by warping the non-enhanced image for each individual to the MDA and applying the warp-field to all the images in the dataset for that animal. This will be described later in Steps B5 and 6.
Step B3. Affine Transformations (linear alignment) of either all images, or only contrast-enhanced images.
The decision whether or not to align all images or only the non-contrast-enhanced images at this steps depends on the intensity of the contrast enhancement. For images with dramatic hyper-intense injection sites, we find it best to separate the two transformations, performing a full affine transformation on the non-contrast enhanced images (Step B3), and separately performing only a 6-parameter rigid body rotation only on non-contrast enhanced images, described further below in Step B5.
For all of these processing approaches, you first must choose the reference image. This can be one of the “non-contrast-enhanced original pre-injection images” in your dataset, or a standard template image that has been brought to the same dimensions and type as your dataset, such the one provided with this protocol (See Figure 12 and Supplementary material). At the conclusion of this processing step, all images will be aligned to the chosen reference image.
If a reference image is created from the dataset, a multi-step process is required, wherein first a random image is selected from within the dataset (output from Step B2) to serve as the first round alignment target as described in Step B3. Next the first MDA is created by averaging those aligned images (Step B4.1). Then the same pre-alignment images from Step B2 are aligned again to this first MDA (Step B4.2) and the resulting re-aligned images averaged to create the final MDA (Step B4.3). These aligned images will be used in Step B6. This is to avoid a bias towards the randomly selected reference image in the first round of alignments.
For full affine transformation, there are three alternative computational approaches presented here: FSL FLIRT (Step B3.1), NiftyReg (Step B3.2) or MATHLAB/SPM (Step B3.3)
It is important to note that each of these alignment methods uses different mathematical algorithms to align the images (the details of which are not described here), and thus we recommend using the same alignment method for all images in a batch. An affine transformation preserves the points, straight lines, and planes, and sets of parallel lines remain parallel in the image. Affine transformations can be a simple rotation of the image, such as we performed in Step A4 above. Such rotation is called “rigid body transformation” and is a 6 parameter rotation. With more parameters, the image is more distorted while lines in the original grid remain parallel. The number of parameters to choose will depend on the degree of differences between the images in your dataset. For brains that appear highly similar, rigid body rotations (6 parameters) are often sufficient. For transgenic animals that have large anatomical differences, a full affine with 12 parameters may be necessary. Always review the images side-by-side on the screen in your 3D visualization program to determine how much to transform your dataset. The best transformation will use the least number of parameters to achieve images that appear identically oriented.
Step B3.1 FSL FLIRT affine transformation (linear alignment)
Step B3.1 describes application of FLIRT (FMRIB’s Linear Image Registration Tool - http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FLIRT; FMRIB is the Oxford Centre for Functional MRI of the Brain – (https://www.ndcn.ox.ac.uk/divisions/fmrib) in FSL. This is the same program you used in the optional step of aligning whole head images (Step A4), but now we use a full affine transformation with command lines using the default settings in FSL, whereas for Step A4 we used the FSL GUI for correcting movement artifacts with a 6 parameter rigid body rotation. Since this protocol was created for MEMRI tract tracing experiments, we use “non-contrast enhanced” and “pre-injection” interchangeably. Pre-injection refers to the images collected for each animal before injection of the contrast agent, Mn2+. For more help in determining FSL options for alignment transformations, we suggest looking at the FSL online user guide and wiki at http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FLIRT/UserGuide. There will be various options for alignment within FSL.
Step B3.1. Perform affine transformation, using FSL FLIRT
1. Open Terminal and enter the following command:
flirt –in <input_image_file_directory_path>/<input_image_filename>-ref <reference_pre-injection_image_directory_path>/<reference_pre-injection_image_filename> –out<output_image_file_directory>/<output_image_filename>
2. To run a batch, enter the following for-loop command:
for f in *.nii; do flirt –in $f -ref <reference_pre-injection_image_directory_path>/<reference_pre-injection_image_filename> –out<output_image_file_directory_path>/${f}_flirt; done
Alternative Step B3.2 To perform affine transformation using Reg Alladin in NiftyReg
Reg-Alladin is one of the alignment tools in NiftyReg (Ourselin, 2001). For more information about setting parameters (options) for your specific dataset, consult this website: https://sourceforge.net/p/niftyreg/git/ci/dev/tree/. The complete code can be found here: http://cmictig.cs.ucl.ac.uk/wiki/index.php/Reg_aladin.
Alternative Step B3.2 To perform affine transformation with reg_aladin
1. Open Terminal and Enter:
reg_aladin –ref <reference_pre-injection_image_filedirectory_path>/<reference_pre-injection_image_filename>-flo<input_image_file_directory_path>/<input_image_filename> -res<output_image_file_directory_path>/<output_image_filename>
2. To run a batch, enter the following for-loop command:
for f in *.nii; do reg_aladin –ref <reference_pre-injection_image_file_directory_path>/<reference_pre-injection_image_filename> -flo $f -res<output_image_file_directory_path>/<output_image_filename>; done
Alternative Step B3.3. To perform affine transformation in SPM/MATLAB:
Open MATLAB, and navigate to the directory with images, then open SPM in MATLAB. Select “PET & VBM,” and in the main SPM window, select Realign>Estimate & Reslice and enter reference/target images and desired settings. (See the SPM manual for more details). Run batch, and these images will be placed in current directory with the prefix “r” (for default settings).
For subsequent directions in this section, these images will be referred to as “step 1 images.”
Step B4: To obtain an averaged image of the non-contrast-enhanced affine-transformed images-- creation of the minimal deformation atlas (MDA).
We create the MDA in several steps. First, in Step B4.1, we average the non-contrast-enhanced images created in Step B3 (“first step images”). Then in Step 4.2 we re-align the same input images again, this time to the averaged image created in Step 4.1, following the same commands/steps as described in Step B3 except using the average image from Step 4.1 as the reference image. We perform this alignment step this second time in order to avoid having any particular reference image drive the averaging, or biasing the averaged image to any particular image in the dataset, when a reference image is selected from within the dataset. In Step 4.3, we then average only the non-contrast enhanced images from Step 4.2. This becomes the MDA to be used in Steps B5 and B6.
Step B4. To obtain an average image (minimal deformation atlas, MDA) of all non-contrast-enhanced brain images, perform the following steps:
Step B4.1. Average affine transformed images, “step 1 images”.
With FSL, average the “step 1 images” from Step B3 as described in Step A4.1 using fslmaths. For subsequent directions in this section, these images will be referred to as “step 2 averaged image.”
Step B4.2. Second affine alignment
Repeat the affine transformation on the “step 1 images” using the commands you followed in Step 3, with reference image being the “step 2 averaged image”, the average image, created in Step B4.1. This will correct for bias for the particular pre-injection image that was originally chosen for affine alignments. For subsequent directions in this section, these images will be referred to as “step 3 images.”
Step B4.3. Obtain MDA
Average only the non-contrast enhanced images from Step B4.2, “step 3 images,” together using the same averaging commands as in Steps A4.1 and Step B4.1.
The resulting averaged image of the non-contrast enhanced images will be the “MDA” (Figure 13 for an example comparing a single image from a dataset of non-contrast-enhanced images that were aligned and averaged). Additional examples of single images and their averaged aligned image can be found in our papers as cited throughout this protocol.
Step B5. Rigid body alignment of contrast-enhanced images.
This step is only performed when the contrast-enhanced images were not included in Step B3. If the dataset includes both non-enhanced and contrast-enhanced images, each animal’s contrast-enhanced images can now be aligned to its non-contrast enhanced image individually. If all images in the dataset are not enhanced or if the contrast did not produce dramatic localized hyper-intensity, this step can be skipped and you can proceed directly to the warping step (Step B6).
First, create a new directory with the images you plan to process and perform functions within that directory.
To perform a rigid body affine transformation with six degrees of freedom of each modally scaled post-injection image (input image from Step B2) to its respective “non-contrast-enhanced original pre-injection images” that was affine-transformed to the MDA (reference image from Step B4.2 “step 3 images). We recommend that the non-contrast enhanced original pre-injection image for each animal be run in parallel through the rigid body affine transformation to insure uniformity in preprocessing environment. There are 3 alternative computational approaches to perform this step. Instructions for each are provided above in Step B3: 1) To use the methods for FSL, follow commands in Step B3.1; for Reg_aladin in Step B3.2, where rigid body specifications must be entered to the respective command lines during processing; and MATLAB/SPM follow Step B3.3. Note that the default settings in MATLAB/SPM do an affine transformation with six degrees of freedom (6DoF).
To perform affine in FSL, follow steps B5.1
Step B5.1. To perform affine transformation of each contrast enhanced images to its respective per-injection image with FSL,
1. Open Terminal and enter the command:
flirt –in <input_image_file_directory_path>/<input_image_filename> -ref <reference_pre-injection_image_directory_path>/<reference_pre-injection_image_filename> -dof 6 –out<output_image_file_directory>/<output_image_filename>
2. To run in batch, use this for-loop command:
for f in *.nii; do flirt –in $f -ref <reference_pre-injection_image_directory_path>/<reference_pre-injection_image_filename> -dof 6 –out<output_image_file_directory_path>/${f}_flirt; done
Alternative Step B5.2.
To perform the affine transformation in NiftyReg, follow steps B5.2. As above, “reference pre-injection image” refers to the affine-transformed image of that same animal in Step B4.2, “step 3 images”, and “input filename” refers to the post-injection contrast-enhanced images from Step B2. To perform this in NiftyReg follow the steps below:
1. For NiftiReg, Open Terminal and enter the following commands:
reg_aladin –ref <reference_pre-injection_image_file_directory_path>/<reference_pre-injection_image_filename> -flo<input_image_file_directory_path>/<input_image_filename> -rigOnly -res <output_image_file_directory_path>/<output_image_filename>
2. To run this in batch, enter the following for-loop command:
for f in *.nii; do reg_aladin –ref <reference_pre-injection_image_file_directory_path>/<reference_pre-injection_image_filename> -flo $f -rigOnly -res<output_image_file_directory_path>/<output_image_filename>; done
Alternative Step B5.3 to perform rigid body alignment in MATLAB/SPM
Follow the procedure from Step B3.3, using the “step 3 images” as reference and Step B2 contrast images as input. Select 6 parameter rigid body rotation.
Images resulting from the work we just performed will be referred to as “affine non-enhanced images” (Step B4.2, “step 3 images”) and “affine contrast-enhanced images,” (Step B5, contrast-enhanced images) respectively.
Step B6: Nonlinear alignment (warping)
To achieve a dataset of fully aligned images, we employ a complicated procedure diagrammed in 14, using images produced in Steps B2-B5. We warp the “non-enhanced image” for each animal that was affine-transformed (“step 3 images” from Step B4.2) to the MDA to obtain a control point grid (aka “warp field”). This grid is then applied to affine-transformed contrast-enhanced image from the same animal, as obtained in Step B5. This warp-field aligns images with unusual contrast in localized areas, such as Mn2+-enhancement, without allowing the contrast to affect alignment. Since the “non-contrast-enhanced images” do not have any Mn+2 signal, we apply the same warp-field to post-injection images that we obtained from the non-contrast-enhanced image of the same animal. We also apply this warp-field to the “non-contrast-enhanced image,” used to obtain it, which ensures uniformity in preprocessing.
To do this, we can use either MATLAB/SPM in batch or our NiftyReg SkullStrip program for individual animals one at a time, in the following ways.
B6.1. Warping in MATLAB/SPM
Open MATLAB, and navigate MATLAB to directory with images, then open SPM in MATLAB. Select “PET & VBM,” and in the main SPM window, select Normalize>Estimate & Reslice and enter reference/target images and desired settings. (See the SPM manual for more details). Enter images like so:
Source Image: non-contrast-enhanced original pre-injection image 1
Images to Write: post-injection image 1 time point 1
Source Image: non-contrast-enhanced original pre-injection image 1
Images to Write: post-injection image 1 time point 2 (3, 4, etc.)
Source Image: non-contrast-enhanced original pre-injection image 1
Images to Write: non-contrast-enhanced original pre-injection image 1
Repeat these inputs for each animal and its time-points.
Run batch, and images will be placed in current MATLAB directory with the prefix “w.”
Alternative Step B.6.2. Warping individual animal images in NiftyReg, an alternative procedure using skullstripper.py to obtain the warp field and apply it.
Using skullstripper.py can only be done for each individual image separately unless a more complex for-loop command is written. Her we provide only the command for single images.
If using NiftyReg-based skullstripper.py, as in the earlier Step A7, first create a new directory containing all image(s) that you will use and refer to in the command line (i.e. if “MDA_image_filename”, is the filename that you assigned to your MDA image, ie “MDA”). Also make sure that the skullstripper.py program is copied into that same directory, since this program is written to refer to files within the command terminal directory.
Here we are warping the images based on the control point grid generated from the warp of the pre-injection images onto the MDA. The output file may be named “MDA”. Hence, after each warping each post-injection image, check the output file and edit the file name. The MDA may need to be copied each time into the directory, since the output file may overwrite the original MDA.
In the skullstripper.py program, “MDA” refers to the rigid body aligned pre-injeciton (non-contrast enhanced images), “Affine_Pre-injection” refers to the pre-injection rigid body transformed image for each individual in the dataset obtained in Step B3.3, “step 3 images”. The skulltripper.py program will warp this image to the MDA creating the warp field (control point grid) to be applied to the post-injection images of the same animal. “Affine-post-injection” refers to the same mouse’s contrast-enhanced image that was rigid body transformed in Step B5. “Output_Image_Name” should be the same as the post-injection mouse name with the suffix “w”. The processing has to be run on one image at a time. Before each new run the “-ref”, “-refmask” and “-out” filenames must be adjusted for each image pair, keeping “-in <MDA>“ the same.
Open Terminal and enter the command:
python skullstripper.py -in <MDA> -ref <Affine_Pre-Injection> -refmask <Affine-Post-Injection> -out <Output_Directory><Output_Image_Name>
Step B7. Smoothing.
Individual voxels often differ in intensity from neighboring voxels. To minimize sharp transitions from one voxel to the next, the images are smoothed. This process avoids pixilation in the subsequent statistical maps. Smoothing changes the intensity of each voxel, based on the average of its surrounding voxels. While multiple types of viable smoothing settings exist, we use a Gaussian smoothing kernel, which is the standard setting in MATLAB/SPM. We usually set the dimension three times greater than the original voxel size of the image. Hence we select Full Width at Half Maximum (FWHM) at three times the voxel dimensions in the SPM protocol. The SPM Processing Manual provides more detail on this protocol for smoothing (http://www.fil.ion.ucl.ac.uk/spm/doc/manual.pdf).
Open MATLAB, and navigate MATLAB to directory with images, then open SPM in MATLAB. Select “PET & VBM,” and in the main SPM window select “Smooth.”
Select files to smooth, and change the FWHMs (Full Width at Half Maximum; we typically use a size three times the voxel size, e.g. our voxels are usually 0.1 mm isotropic, thus we use a smoothing FWHM, of 0.3mm), and run batch.
Files will now be smoothed with a Gaussian kernel.
Step B8: Final images ready for statistical parametric mapping, region of interest analysis, or other statistical approaches for comparisons between time points, between different strains, and between transgenic animals.
At this point, you will have a dataset of skull-stripped, modally scaled and aligned images ready for statistical parametric mapping, independent component analysis, region of interest measurements or other analyses that require co-registration of images and intensity normalization. Look at these images and confirm that each aligned image looks sharp, that the header and file parameters have been preserved, and that the histograms are consistent in range and peak. Registration should be validated either by visualization in any of the 3D viewer programs discussed above, or in MATLAB/SPM “Check Reg” button in the GUI (Figure 15). You will also have a MDA and a mask template that can be applied for the automated pre-processing of many more images provided they have the same MR capture parameters. Such aligned images are readily loaded into MATLAB/SPM or other analysis programs to obtain statistical maps of intensity changes between animals or over time in the same animal.
Figure 15: MATLAB/SPM GUI for co-registration and check registration.
After opening MATLAB/SPM and clicking on the “Coregistration” button (red arrow), images can be loaded and then processed. The button for smoothing (Smooth) is also found on this GUI, on the upper right of the blue box, to the right of the red arrow.
Skull-stripped aligned brain images may be analyzed for differences between groups of mice. We have used this approach to discover how specific genetic knock-outs affect circuitry. For example, knock-out of the serotonin transporter gene (SERT KO) has been demonstrated to produce one of the four validated rodent models of PTSD based on behavior (Pitman et al., 2012). How this behavior is reflected by functional circuitry in the brain is an open question. Using MEMRI with injection of Mn2+ into the medial prefrontal cortex, a region implicated in PTSD, led us to discover that the mesocortical circuitry was profoundly altered in the SERT knock-out mouse (Bearer et al., 2009). This discovery was enabled by computational alignment of brain images from 12 wildtype and 12 SERT KO mice obtained before and at 6 different time points after injection followed by statistical parametric mapping comparing intensity patterns between time points within each group.
In studies of the important memory circuit from hippocampus to medial septum in the forebrain, we found that Down syndrome, the most common genetic cause of mental retardation, produces more robust transport than normal, likely due increased expression of APP from the triplicated chromosome 21 and consequent failure of synaptic pruning (Bearer et al., 2007b). Application of this alignment protocol has also allowed us to discover the roles of particular proteins in axonal transport, such as the microtubule motor, kinesin-1. Whole brain detection of transport dynamics in living mammals has been challenging for traditional optical imaging techniques. With MEMRI images captured over time we were able to show the role of kinesin-1 in axonal transport in the living brain for the first time (Medina et al., 2017). Using this protocol for alignment we have also shown the effect of psychoactive drugs on neuronal activation, particularly focusing on treatments in a mouse model of schizophrenia (Malkova et al., 2014).
Many questions remain to be answered that application of MRI together with the power of mouse transgenics can address. Mouse models offer the capability of manipulations under experimental control. For example, does early life trauma, known to be a risk factor for PTSD, impact circuitry in the same way as the SERT gene knock out? And how do the senile plaques of Alzheimer’s disease affect functional circuitry in the important memory tracts from hippocampus to forebrain? For these questions, mouse models exist. All that remains is to perform imaging and analyze the results. A major obstacle in such imaging experimentation has been the enormous effort involved in brain extraction and image alignment when automated processing was neither readily available nor standardized. Custom processing differs between laboratories, which may interfere with reproducibility. A standardized processing protocol avoids this caveat. It is our hope that the procedure described here will transcend this obstacle and promote more application of powerful MRI technologies to rodent animal models of human neuropsychiatric disorders.
Commentary
Background Information
Magnetic resonance imaging (MRI) is a powerful method to observe the anatomy and function in the living brain. MR has a number of advantages over more traditional anatomical approaches, such as histologic examination and computerized tomography (CT). MR can be repeatedly carried out to enable collection of longitudinal information over an animal’s lifetime, not possible with either histology, which requires sacrifice, or computerized tomography (CT) scanning, which is destructive and has less soft-tissue contrast for detection of neuronal nuclei within the brain.
However, several issues limit applications of MR. One limitation is the intense computational image analysis required to extract significant biological information other than gross anatomical abnormalities from MR brain images. The amount of imaging time required to obtain enough images from at least six animals for statistical power is costly. Typically only one mouse can be imaged at a time, although numbers of animals imaged in one session may be expanded by using special holders (Lerch et al., 2011; Scholz et al., 2016). Finally, due to the small size of the rodent brain and the low signal-to-noise ratios inherent in MR imaging, a high field magnet is necessary to capture whole brain images in a reasonable time to sustain the living animal inside the MR machine. Averaging many images computationally in a protocol such as that presented here enhances signal-to-noise ratios and identifies consistently occurring structural differences among a cohort of animals. Additionally contrast enhancement with manganese-enhanced magnetic resonance imaging (MEMRI), a contrast agent for T1-weighted MR scans, improves the detection of anatomic structures. MEMRI can be used for in vivo tracing of neuronal pathways (Pautler, 2004; Pautler and Koretsky, 2002; Pautler et al., 2003; Pautler et al., 1998; Van der Linden et al., 2004). A single image (Figure 13A) is shown next to an MDA (Figure 13B). Notice that the MDA (Minimal Deformation Atlas) has much more fine detail than the single image (Delora et al., 2016). By facilitating and standardizing image processing for rodent brain imaging analysis as described here, MR of the brains of living animals will be easier and more reliable, and therefore more accessible.
Why manganese?
The manganese ion (Mn2+) is emerging as a powerful contrast agent in T1-weighted MR imaging (Fornasiero et al., 1987; Geraldes et al., 1986; Mendonca-Dias et al., 1983; Pautler, 2004; Pautler et al., 2003; Pautler et al., 1998; Silva et al., 2004; Wadghiri et al., 2004; Watanabe et al., 2001). As a paramagnetic ion, Mn2+ causes a reduction in the relaxation time of protons in adjacent water molecules thereby producing a hyper-intense signal in T1-wieghted MR images (Mendonca-Dias et al., 1983).
How manganese is delivered
There are multiple techniques of administering manganese (II) chloride (MnCl2) so that Mn2+ can be taken up into neuronal circuits (Bearer et al., 2007b; Bearer et al., 2009; Gallagher et al., 2013; Gallagher et al., 2012; Pautler, 2004; Pautler et al., 2003; Zhang et al., 2010a): (1) Stereotaxic injection directly into a region of the brain (2) Intranasal administration (Chuang et al., 2009; Massaad and Pautler, 2011; Pautler and Koretsky, 2002; Smith et al., 2007; Smith et al., 2011); (3) Oral gavage (Wang et al., 2012); (4) Intravitreal injection, in which Mn2+ is taken up in the retinal ganglion cells and enters the brain by transport within axons of the optic nerve (Bearer et al., 2007a; Gallagher et al., 2012; Pautler et al., 1998); and (5) Intraperitoneal injection where after Mn2+ passes through the blood brain barrier via diffusion or active transport (Malkova et al., 2014; Massaad and Pautler, 2011; Van der Linden et al., 2004). The animal is anesthetized both when delivering the Mn2 and during the MRI scan. It is placed in a tube that serves as an anesthetic chamber and is also used to hold the mouse in place (Balaban and Hampshire, 2001).
Important applications of MEMRI
MEMRI has two important functions, which include (1) Contrast enhancement of anatomical structures of the brain for anatomical studies (Koretsky and Silva, 2004; Massaad and Pautler, 2011). (2) Measuring brain activity by entering activated cells through voltage-gated calcium ion (Ca+2) channels (Koretsky and Silva, 2004; Malkova et al., 2014; Massaad and Pautler, 2011; Van der Linden et al., 2004; Yu et al., 2005). MEMRI studies have discovered changes in neuronal circuitry in Down syndrome (Bearer et al., 2007b), Alzheimer’s disease (Smith et al., 2011; Wang et al., 2012), serotonin transport and other monoamine transporters (Bearer et al., 2009; Gallagher et al., 2013; Zhang et al., 2010a) and odor-induced activation in the olfactory bulbs (Chuang et al., 2009; Pautler, 2004; Smith et al., 2007; Smith et al., 2011) among other things in mice.
Mn2+, a calcium analogue, enters neurons though voltage-gated calcium channels (Drapeau and Nachshen, 1984; Merritt et al., 1989; Narita et al., 1990; Takeda, 2003). It is then carried down the axon primarily by anterograde transport (probably via transport vesicles, mitochondria, and endoplasmic reticulum, all of which have Ca+ channels), and then crosses active synapses (Bearer et al., 2007b; Lin and Koretsky, 1997; Sloot and Gramsbergen, 1994; Takeda et al., 1998). We demonstrated that Mn2+ is carried by the kinesin-based microtubule transport system and preferentially crosses active synapses (Bearer et al., 2007a). For detection of functional circuitry, MR images are taken at multiple time points after localized stereotactic injection into the brain. Statistical parametric mapping after preprocessing of these time-lapse images, as described in this protocol, reveals functional circuits highlighted by the Mn2+ hyper-intense signal. Statistical comparisons between genetically different cohorts, when similarly preprocessed and co-registered, reveals phenotypic differences.
Critical Parameters
A number of issues can impact the quality of the images and make them difficult to analyze, including 1) differing capture parameters (such as pulse sequences, the pattern of radiofrequency impulses transmitted across the sample that activates the nuclear magnetic resonance producing the signal, thereby creating the image); 2) electronic noise interference; and 3) movement of the animal during the imaging session. It is critically important to visually inspect the images to confirm that intensity patterns of anatomical structures appear similar soon after they have been captured and prior to animal sacrifice to check that capture parameters are identical and the image has not been blurred by noise or movement ,because these may be hard, or even impossible, to correct post-capture.
Capture parameters:
Multiple pulse sequences exist in the MEMRI world, each of which gives different contrast weights. Some frequently used pulse sequences include RARE (rapid acquisition with relaxation enhancement, or fast spin echo) or FLASH (fast low angle shot, or spoiled gradient echo). These pulse sequences have different echo time and repeat time of the activating electronic current (the pulse), and differing orientation of the pulse relative to the B field inside the magnet (90o, 180o, etc.). We do not recommend a specific pulse sequence, but it is best if each image within a dataset is captured with the same pulse sequence, since the pulse sequence affects the contrast and thereby the relative intensities of various structures throughout the brain. If different images within the same dataset are accidently taken with different pulse sequences, which happens when using MR scanners in core facilities where the pulse sequences are set differently for each user, correction will be difficult. Such differences in capture parameters can be a particularly large problem if the error is not discovered until after the mice have already been sacrificed and new images cannot be re-captured. Another parameter set during capture is the dimensions, i.e. voxel size and number of slices. If these parameters differ between images in the dataset, they can be corrected post-capture by computational approaches such interpolation to reduce the size or zero filling to enlarge it. Care must be taken not to alter the data significantly when performing these corrections.
Electronic noise interference:
Another issue is that MR images may have electronic noise interference that blurs the image. Interference can be detected in the image by blurring of the whole image, or corruption of single slices. If this is not discovered in time to re-capture the image, that subject’s images may not be usable for alignment and statistical analysis. However, if only a single slice is affected, that can be computationally corrected by replacing the slice with an average slice created from the adjacent slices to either side of the corrupted slice.
Movement within the scanner:
Movement artifact can often be corrected computationally, as we described above here (See Step A4.2 and Figures 5 and 6.)
Hardware:
It is also important to have a suitable computer available for image preprocessing. Our Skullstripper.py program runs in Linux or Mac. Hence, a Mac computer with a 64-bit processor, or a virtual Linux PC machine also with 64-bit processing capabilities, are required. We recommend that the computer have at least 8 Gb of RAM and large storage space. It is critical that images be saved at each step of preprocessing to backtrack any errors that may have occurred along the way or to repeat the process from an earlier step, i.e. “Save the Game”. Saving images at each step will take up a large amount of storage space. The computer must also have the correct versions of the programs mentioned in the materials section loaded before commencing with this preprocessing protocol.
Troubleshooting
In addition to the issues described in Critical Parameters above, many of which cannot be rectified post-capture, some more minor problems may some up, such as off-center images or mask imperfections. These are readily fixed computationally.
Off Center image:
If one or more images in the dataset are off-center, the resulting averaged image will be blurred (Figures 5 and 6). As stated in Step A4, images taken simultaneously of the same subject at the same time point may need to be averaged together. To correct for off-center images, chose one of the images as a reference image and perform a rigid body affine transformation of the other image(s) to it or vice versa (Step 4.2 and Figure 6). We recommend that the reference image used to transform the others be transformed into itself as well to ensure uniformity in the preprocessing steps and their output. Furthermore, if this step must be done to one or a few sets of images, we recommend that it be done to all images within all sets to ensure uniformity. Also see Step B5 for details on rigid body transformation. The averaged image will have more distinct anatomical features after affine transformation once the off-center image is corrected (Figure 13).
Mask imperfections:
Imperfections in masks is another common troubleshooting issue. Large imperfections show up when visually inspecting the mask as described in Step A8, “Check Mask” section. Smaller imperfections may not be obvious until they produce edge artifacts after performing statistical parametric mapping. If there are mask imperfections, defective masks must be refined by hand. To do this, open the whole head image and modify the mask as described in Step A7 by using the manual editing tool, Brainsuite (Shattuck and Leahy, 2002), the Brain Extraction Toolkit (BET) (Smith, 2002) or FSLView (Figure 10). In FSLView, uncheck the lock to enable editing of the mask and click the pencil icon to show editing tools.
Anticipated Results
Following the protocol described, images will be uniform in intensity, shape, and size without extra-meningeal tissue. Notable milestones in the protocol are the “N3 Correction” and “Modal Scale” step, in which the histogram of the images will be altered to a standard for uniformity, necessary for alignments. Checking these key steps helps determine if the preprocessing is on track or not. Similarly, checking the mask helps to ensure that the skull stripping program has produced expected results. The skull-stripping step should create a series of white, binary masks which should fit over their corresponding whole-head images to create an image with the extra-meningeal tissue removed (Figure 11b). One common mistake here is when the output of the Skull Strip program is a different image type than the whole head image receiving the mask. If fuzzy borders occur, check the image type in FIJI and make sure both mask and whole head image are the same type. It is anticipated that the histogram after modal scaling will reveal co-incident peaks of the template and scaled images (Figure 8). Creation of the MDA is a notable milestone. As an average of all pre-injection images, it should have more fine detail that any individual pre-injection image (Figure 13). Check the MDA in the 3D image viewer and confirm that it is sharper, and not blurrier, than the input images. Our automated program is more consistent than manual preprocessing, and thus produces a sharper averaged image.
Time Considerations
Previous to the development of this protocol, preprocessing with manual skull-stripping could take weeks and involved a team of researchers. When following this protocol, time required is greatly reduced compared to previous manual processing to several days for one researcher, but may vary depending on the size of the dataset being preprocessed. For example, preprocessing of 12–24 images of 8–16 Mb size using a Mac computer with 16Mb of RAM can be accomplished in a few days’ time when following this automation preprocessing protocol. Delays usually occur from user error, in naming files and entering command lines, or if images are not captured with the same pulse sequence, or have other artifacts. In our experience, practiced users can perform the protocol in this number of images in less than a day when all images are captured consistently.
If all the images have identical headers, things go quickly and smoothly (Steps A1 and A2). Time and effort required to bring all images into the same dimensions is hard to predict, as this depends on the amount of editing required.
After everything is ready to run, there are no lunch breaks as processing proceeds at second to minute rates. Changing voxel sizes and headers (A1–3), image averaging (A4), intensity corrections (A5–6) creating masks (A7) and applying masks (A8) can be performed in a matter of seconds per image using this protocol. Preparing the reference mask takes considerable time, perhaps a day or two if a template whole head image and its reference mask are not available. Second N3 correction and modal scaling can take several seconds to minutes for each image (B1–2). . Performing affine transformations and warping images can take several seconds to several minutes per image (B3 and B5). The entire process of creating a MDA can take approximately an hour (B3–5), and smoothing can be done is few minutes.
Supplementary Material
Significance Statement.
Magnetic resonance imaging (MRI) provides the opportunity to determine the anatomy of the brain in living animals. Anatomical features detected by MRI, such as volumes of brain and sub-brain regions (detected with anatomical MRI), functional connections (detected using manganese-enhancement or blood-oxygen-level-dependent imaging), and fiber tracts (detected using diffusion tensor imaging), reveal phenotypic differences among different mouse strains, transgenic animals, and experimental models. To identify statistically significant differences from images of these anatomical features, multiple 3D images of different individuals must have the brain extracted from non-brain tissue and be co-registered. The protocol presented here provides detailed step-wise instructions for automation of these crucial preprocessing steps to produce datasets of aligned images suitable for subsequent statistical analysis. This automation protocol reduces the time needed for preprocessing from weeks or even months to just a few days. Use of this protocol will also standardize MR image preparation for analysis and thus promote reproducibility. By exponentially reducing the time required for preprocessing, this automation facilitates MR image analysis for phenotyping of rodent models of brain disorders, Similar methodology may be applicable to automation of analysis of MRI data from other organs and tissues.
Acknowledgments
This work was funded by The Harvey Family Endowment (ELB), NIGMS (P50GM085273) (ELB), and NIDA (DA018184) (REJ with subaward to ELB), NINDS (NS046810, and NS062184) (ELB). We are indebted to Russell E. Jacobs and the Beckman Institute Imaging Center at Caltech for access to the 11.7 MR scanner, the images, the template image, and the modal scaling code, as well as advice and coaching for over a dozen years on MR technology. We also gratefully acknowledge UNM’s BA-MD program for support of Christopher Medina. We thank Kevin P. Reagan, Sharon Wu Lin and XiaoWei Zhang for technical assistance, Krish Subramaniam for the modal scaling program, and current and former student helpers Brianna S. Mulligan, Amber Jean Zimmerman, Abdul Faheem Mohed, Adam Mitchell, Adam Delora, and Frances Chaves.
Footnotes
Conflicts of Interest
The authors report no conflicts of interest.
This paper provides the SkullStrip program and more specific dialogue on its applications and instructions on how to use it.
Internet Resources
http://fsl.fmrib.ox.ac.uk/fsl/fslwiki
Provides information on FMRIB Software Library (FSL), such as download instructions and instructions for use.
Provides information on FIJI/ImageJ, such as download instructions and instructions for use.
http://people.cas.sc.edu/rorden/mricron/index.html
Provides information on MRIcron, such as download instructions and instructions for use.
MIPAV instructions for N3 corrections.
http://brainsuite.org/processing/surfaceextraction/bse/
Provides information on BrainSuite, such as download instructions and instructions for use.
http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/BET/UserGuide
FSL user guide for Brain Extraction Toolkit (BET).
http://www.fil.ion.ucl.ac.uk/spm/doc/manual.pdf
SPM12 Manual.
Literature cited
- Balaban RS and Hampshire VA 2001. Challenges in small animal noninvasive imaging. ILAR journal / National Research Council, Institute of Laboratory Animal Resources 42:248–262. [DOI] [PubMed] [Google Scholar]
- Bearer EL, Falzone TL, Zhang X, Biris O, Rasin A, and Jacobs RE 2007a. Role of neuronal activity and kinesin on tract tracing by manganese-enhanced MRI (MEMRI). NeuroImage 37 Suppl 1:S37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearer EL, Zhang X, and Jacobs RE 2007b. Live imaging of neuronal connections by magnetic resonance: Robust transport in the hippocampal-septal memory circuit in a mouse model of Down syndrome. NeuroImage 37:230–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearer EL, Zhang X, Janvelyan D, Boulat B, and Jacobs RE 2009. Reward circuitry is perturbed in the absence of the serotonin transporter. NeuroImage 46:1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang KH, Lee JH, Silva AC, Belluscio L, and Koretsky AP 2009. Manganese enhanced MRI reveals functional circuitry in response to odorant stimuli. NeuroImage 44:363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delora A, Gonzales A, Medina CS, Mitchell A, Mohed AF, Jacobs RE, and Bearer EL 2016. A simple rapid process for semi-automated brain extraction from magnetic resonance images of the whole mouse head. Journal of neuroscience methods 257:185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinov I, Lozev K, Petrosyan P, Liu Z, Eggert P, Pierce J, Zamanyan A, Chakrapani S, Van Horn J, Parker DS, Magsipoc R, Leung K, Gutman B, Woods R, and Toga A 2010. Neuroimaging study designs, computational analyses and data provenance using the LONI pipeline. PloS one 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinov ID, Van Horn JD, Lozev KM, Magsipoc R, Petrosyan P, Liu Z, Mackenzie-Graham A, Eggert P, Parker DS, and Toga AW 2009. Efficient, Distributed and Interactive Neuroimaging Data Analysis Using the LONI Pipeline. Frontiers in neuroinformatics 3:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapeau P and Nachshen DA 1984. Manganese fluxes and manganese-dependent neurotransmitter release in presynaptic nerve endings isolated from rat brain. The Journal of physiology 348:493–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegood J, Anagnostou E, Babineau BA, Crawley JN, Lin L, Genestine M, DiCicco-Bloom E, Lai JK, Foster JA, Penagarikano O, Geschwind DH, Pacey LK, Hampson DR, Laliberte CL, Mills AA, Tam E, Osborne LR, Kouser M, Espinosa-Becerra F, Xuan Z, Powell CM, Raznahan A, Robins DM, Nakai N, Nakatani J, Takumi T, van Eede MC, Kerr TM, Muller C, Blakely RD, Veenstra-VanderWeele J, Henkelman RM, and Lerch JP 2015. Clustering autism: using neuroanatomical differences in 26 mouse models to gain insight into the heterogeneity. Molecular psychiatry 20:118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenko O, Evrard HC, Neves RM, Beyerlein M, Murayama Y, and Logothetis NK 2012. Tracing of noradrenergic projections using manganese-enhanced MRI. NeuroImage 59:3252–3265. [DOI] [PubMed] [Google Scholar]
- Fornasiero D, Bellen JC, Baker RJ, and Chatterton BE 1987. Paramagnetic complexes of manganese(II), iron(III), and gadolinium(III) as contrast agents for magnetic resonance imaging. The influence of stability constants on the biodistribution of radioactive aminopolycarboxylate complexes. Investigative radiology 22:322–327. [DOI] [PubMed] [Google Scholar]
- Friston KJ 2005. Models of brain function in neuroimaging. Annual review of psychology 56:57–87. [DOI] [PubMed] [Google Scholar]
- Gallagher JJ, Zhang X, Hall FS, Uhl GR, Bearer EL, and Jacobs RE 2013. Altered reward circuitry in the norepinephrine transporter knockout mouse. PloS one 8:e57597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher JJ, Zhang X, Ziomek GJ, Jacobs RE, and Bearer EL 2012. Deficits in axonal transport in hippocampal-based circuitry and the visual pathway in APP knock-out animals witnessed by manganese enhanced MRI. NeuroImage 60:1856–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes CF, Sherry AD, Brown RD 3rd, and Koenig SH 1986. Magnetic field dependence of solvent proton relaxation rates induced by Gd3+ and Mn2+ complexes of various polyaza macrocyclic ligands: implications for NMR imaging. Magnetic resonance in medicine 3:242–250. [DOI] [PubMed] [Google Scholar]
- Gillespy T 3rd, Richardson ML, and Rowberg AH 1994. Displaying radiologic images on personal computers: practical applications and uses. Journal of digital imaging 7:101–106. [DOI] [PubMed] [Google Scholar]
- Gillespy T 3rd and Rowberg AH 1993. Radiological images on personal computers: introduction and fundamental principles of digital images. Journal of digital imaging 6:81–87. [DOI] [PubMed] [Google Scholar]
- Henkelman RM 2010. Systems biology through mouse imaging centers: experience and new directions. Annual review of biomedical engineering 12:143–166. [DOI] [PubMed] [Google Scholar]
- Hoyer C, Gass N, Weber-Fahr W, and Sartorius A 2014. Advantages and challenges of small animal magnetic resonance imaging as a translational tool. Neuropsychobiology 69:187–201. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, and Smith SM 2012. Fsl. NeuroImage 62:782–790. [DOI] [PubMed] [Google Scholar]
- Kochunov P, Lancaster JL, Thompson P, Woods R, Mazziotta J, Hardies J, and Fox P 2001. Regional spatial normalization: toward an optimal target. Journal of computer assisted tomography 25:805–816. [DOI] [PubMed] [Google Scholar]
- Koretsky AP and Silva AC 2004. Manganese-enhanced magnetic resonance imaging (MEMRI). NMR in biomedicine 17:527–531. [DOI] [PubMed] [Google Scholar]
- Kovacevic N, Henderson JT, Chan E, Lifshitz N, Bishop J, Evans AC, Henkelman RM, and Chen XJ 2005. A three-dimensional MRI atlas of the mouse brain with estimates of the average and variability. Cerebral cortex 15:639–645. [DOI] [PubMed] [Google Scholar]
- Larsen CT, Iglesias JE, and Van Leemput K . 2014. N3 Bias Field Correction Explained as a Bayesian Modeling Method. . Springer International Publishing 1–12. DOI: 10.1007/978-3-319-12289-2_1.[*This reference does not have a pre-defined format and therefore uses the “Generic” format]
- Lerch JP, Sled JG, and Henkelman RM 2011. MRI phenotyping of genetically altered mice. Methods in molecular biology 711:349–361. [DOI] [PubMed] [Google Scholar]
- Lin YJ and Koretsky AP 1997. Manganese ion enhances T1-weighted MRI during brain activation: an approach to direct imaging of brain function. Magnetic resonance in medicine 38:378–388. [DOI] [PubMed] [Google Scholar]
- Lindsey JD, Grob SR, Scadeng M, Duong-Polk K, and Weinreb RN 2013. Ocular integrity following manganese labeling of the visual system for MRI. Magnetic resonance imaging 31:865–874. [DOI] [PubMed] [Google Scholar]
- Ma D, Cardoso MJ, Modat M, Powell N, Wells J, Holmes H, Wiseman F, Tybulewicz V, Fisher E, Lythgoe MF, and Ourselin S 2014. Automatic structural parcellation of mouse brain MRI using multi-atlas label fusion. PLoS One 9:e86576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova NV, Gallagher JJ, Yu CZ, Jacobs RE, and Patterson PH 2014. Manganese-enhanced magnetic resonance imaging reveals increased DOI-induced brain activity in a mouse model of schizophrenia. Proceedings of the National Academy of Sciences of the United States of America 111:E2492–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manifold-Wheeler B, Gonzales A, Chaves F, Jacobs RE, Bearer EL 2016. Alterations in the hippocampal-forebrain circuitry with aging and with plaque formation. Neurobiology of Aging MS in preparartion [Google Scholar]
- Massaad CA and Pautler RG 2011. Manganese-enhanced magnetic resonance imaging (MEMRI). Methods in molecular biology 711:145–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina CS, Biris O, Falzone TL, Zhang X, Zimmerman AJ, and Bearer EL 2017. Hippocampal to basal forebrain transport of Mn2+ is impaired by deletion of KLC1, a subunit of the conventional kinesin microtubule-based motor. NeuroImage 145:44–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonca-Dias MH, Gaggelli E, and Lauterbur PC 1983. Paramagnetic contrast agents in nuclear magnetic resonance medical imaging. Seminars in nuclear medicine 13:364–376. [DOI] [PubMed] [Google Scholar]
- Merritt JE, Jacob R, and Hallam TJ 1989. Use of manganese to discriminate between calcium influx and mobilization from internal stores in stimulated human neutrophils. The Journal of biological chemistry 264:1522–1527. [PubMed] [Google Scholar]
- Modat M, Ridgway GR, Taylor ZA, Lehmann M, Barnes J, Hawkes DJ, Fox NC, and Ourselin S 2010. Fast free-form deformation using graphics processing units. Computer methods and programs in biomedicine 98:278–284. [DOI] [PubMed] [Google Scholar]
- Narita K, Kawasaki F, and Kita H 1990. Mn and Mg influxes through Ca channels of motor nerve terminals are prevented by verapamil in frogs. Brain research 510:289–295. [DOI] [PubMed] [Google Scholar]
- Nieman BJ, Bishop J, Dazai J, Bock NA, Lerch JP, Feintuch A, Chen XJ, Sled JG, and Henkelman RM 2007. MR technology for biological studies in mice. NMR in biomedicine 20:291–303. [DOI] [PubMed] [Google Scholar]
- Nieman BJ, Bock NA, Bishop J, Chen XJ, Sled JG, Rossant J, and Henkelman RM 2005. Magnetic resonance imaging for detection and analysis of mouse phenotypes. NMR in biomedicine 18:447–468. [DOI] [PubMed] [Google Scholar]
- Ourselin S, Roche A, Subsol G, Pennec X, and Ayache N . 2001. Reconstructing a 3D structure from serial histological sections. Image and Vision Computing 19:25–31. [Google Scholar]
- Paletzki R and Gerfen CR 2015. Whole Mouse Brain Image Reconstruction from Serial Coronal Sections Using FIJI (ImageJ). Current protocols in neuroscience / editorial board, Jacqueline N. Crawley … [et al. ] 73:1 25 21–21. [DOI] [PubMed] [Google Scholar]
- Pautler RG 2004. In vivo, trans-synaptic tract-tracing utilizing manganese-enhanced magnetic resonance imaging (MEMRI). NMR in biomedicine 17:595–601. [DOI] [PubMed] [Google Scholar]
- Pautler RG and Koretsky AP 2002. Tracing odor-induced activation in the olfactory bulbs of mice using manganese-enhanced magnetic resonance imaging. NeuroImage 16:441–448. [DOI] [PubMed] [Google Scholar]
- Pautler RG, Mongeau R, and Jacobs RE 2003. In vivo trans-synaptic tract tracing from the murine striatum and amygdala utilizing manganese enhanced MRI (MEMRI). Magnetic resonance in medicine 50:33–39. [DOI] [PubMed] [Google Scholar]
- Pautler RG, Silva AC, and Koretsky AP 1998. In vivo neuronal tract tracing using manganese-enhanced magnetic resonance imaging. Magnetic resonance in medicine 40:740–748. [DOI] [PubMed] [Google Scholar]
- Pitman RK, Rasmusson AM, Koenen KC, Shin LM, Orr SP, Gilbertson MW, Milad MR, and Liberzon I 2012. Biological studies of post-traumatic stress disorder. Nature reviews. Neuroscience 13:769–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeswari R and Rajesh R 2009. On the efficient compression of FMRI data series of brain. The neuroradiology journal 21:737–743. [DOI] [PubMed] [Google Scholar]
- Rex DE, Ma JQ, and Toga AW 2003. The LONI Pipeline Processing Environment. NeuroImage 19:1033–1048. [DOI] [PubMed] [Google Scholar]
- Rorden C, Karnath HO, and Bonilha L 2007. Improving lesion-symptom mapping. Journal of cognitive neuroscience 19:1081–1088. [DOI] [PubMed] [Google Scholar]
- Sawiak SJ, Wood NI, Williams GB, Morton AJ, and Carpenter TA 2009. Voxel-based morphometry in the R6/2 transgenic mouse reveals differences between genotypes not seen with manual 2D morphometry. Neurobiol Dis 33:20–27. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, and Cardona A 2012. Fiji: an open-source platform for biological-image analysis. Nature methods 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz J, LaLiberte C, van Eede M, Lerch JP, and Henkelman M 2016. Variability of brain anatomy for three common mouse strains. NeuroImage [DOI] [PubMed] [Google Scholar]
- Shattuck DW and Leahy RM 2001. Automated graph-based analysis and correction of cortical volume topology. IEEE transactions on medical imaging 20:1167–1177. [DOI] [PubMed] [Google Scholar]
- Shattuck DW and Leahy RM 2002. BrainSuite: an automated cortical surface identification tool. Medical image analysis 6:129–142. [DOI] [PubMed] [Google Scholar]
- Silva AC and Bock NA 2008. Manganese-enhanced MRI: an exceptional tool in translational neuroimaging. Schizophrenia bulletin 34:595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AC, Lee JH, Aoki I, and Koretsky AP 2004. Manganese-enhanced magnetic resonance imaging (MEMRI): methodological and practical considerations. NMR in biomedicine 17:532–543. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, and Evans AC 1998. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE transactions on medical imaging 17:87–97. [DOI] [PubMed] [Google Scholar]
- Sloot WN and Gramsbergen JB 1994. Axonal transport of manganese and its relevance to selective neurotoxicity in the rat basal ganglia. Brain research 657:124–132. [DOI] [PubMed] [Google Scholar]
- Smith KD, Kallhoff V, Zheng H, and Pautler RG 2007. In vivo axonal transport rates decrease in a mouse model of Alzheimer’s disease. NeuroImage 35:1401–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Paylor R, and Pautler RG 2011. R-flurbiprofen improves axonal transport in the Tg2576 mouse model of Alzheimer’s disease as determined by MEMRI. Magnetic resonance in medicine 65:1423–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM 2002. Fast robust automated brain extraction. Human brain mapping 17:143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, and Matthews PM 2004. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage 23 Suppl 1:S208–219. [DOI] [PubMed] [Google Scholar]
- Takeda A 2003. Manganese action in brain function. Brain research. Brain research reviews 41:79–87. [DOI] [PubMed] [Google Scholar]
- Takeda A, Kodama Y, Ishiwatari S, and Okada S 1998. Manganese transport in the neural circuit of rat CNS. Brain research bulletin 45:149–152. [DOI] [PubMed] [Google Scholar]
- Van der Linden A, Van Meir V, Tindemans I, Verhoye M, and Balthazart J 2004. Applications of manganese-enhanced magnetic resonance imaging (MEMRI) to image brain plasticity in song birds. NMR in biomedicine 17:602–612. [DOI] [PubMed] [Google Scholar]
- Wadghiri YZ, Blind JA, Duan X, Moreno C, Yu X, Joyner AL, and Turnbull DH 2004. Manganese-enhanced magnetic resonance imaging (MEMRI) of mouse brain development. NMR in biomedicine 17:613–619. [DOI] [PubMed] [Google Scholar]
- Wang FH, Appelkvist P, Klason T, Gissberg O, Bogstedt A, Eliason K, Martinsson S, Briem S, Andersson A, Visser SA, Ivarsson M, Lindberg M, Agerman K, and Sandin J 2012. Decreased axonal transport rates in the Tg2576 APP transgenic mouse: improvement with the gamma-secretase inhibitor MRK-560 as detected by manganese-enhanced MRI. The European journal of neuroscience 36:3165–3172. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Michaelis T, and Frahm J 2001. Mapping of retinal projections in the living rat using high-resolution 3D gradient-echo MRI with Mn2+-induced contrast. Magnetic resonance in medicine 46:424–429. [DOI] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Holmes CJ, Cherry SR, and Mazziotta JC 1998. Automated image registration: I. General methods and intrasubject, intramodality validation. Journal of computer assisted tomography 22:139–152. [DOI] [PubMed] [Google Scholar]
- Woolrich MW, Jbabdi S, Patenaude B, Chappell M, Makni S, Behrens T, Beckmann C, Jenkinson M, and Smith SM 2009. Bayesian analysis of neuroimaging data in FSL. NeuroImage 45:S173–186. [DOI] [PubMed] [Google Scholar]
- Yu X, Wadghiri YZ, Sanes DH, and Turnbull DH 2005. In vivo auditory brain mapping in mice with Mn-enhanced MRI. Nature neuroscience 8:961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bearer EL, Boulat B, Hall FS, Uhl GR, and Jacobs RE 2010a. Altered neurocircuitry in the dopamine transporter knockout mouse brain. PloS one 5:e11506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bearer EL, Perles-Barbacaru AT, and Jacobs RE 2010b. Increased anatomical detail by in vitro MR microscopy with a modified Golgi impregnation method. Magnetic resonance in medicine 63:1391–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


