Summary
Chronic infection with Helicobacter pylori can lead to the development of gastric ulcers and stomach cancers. The helical cell shape of H. pylori promotes stomach colonization. Screens for loss of helical shape have identified several periplasmic peptidoglycan (PG) hydrolases and non-enzymatic putative scaffolding proteins, including Csd5. Both over and under expression of the PG hydrolases perturb helical shape, but the mechanism used to coordinate and localize their enzymatic activities is not known. Using immunoprecipitation and mass spectrometry we identified Csd5 interactions with cytosolic proteins CcmA, a bactofilin required for helical shape, and MurF, a PG precursor synthase, as well as the inner membrane spanning ATP synthase. A combination of Csd5 domain deletions, point mutations, and transmembrane domain chimeras, revealed that the N-terminal transmembrane domain promotes MurF, CcmA, and ATP synthase interactions while the C-terminal SH3 domain mediates PG binding. We conclude that Csd5 promotes helical shape as part of a membrane associated, multi-protein shape-complex that includes interactions with the periplasmic cell wall, a PG precursor synthesis enzyme, the bacterial cytoskeleton, and ATP synthase.
Keywords: Helicobacter pylori; Peptidoglycan; ATP Synthase; Csd5, Bactofilin; MurF
Graphical Abstract
Abbreviated Summary
Csd5 is a non-enzymatic protein that promotes helical cell shape and thus efficient colonization of the human host. We performed a structure-function analysis coupled to immunoprecipitation and mass spectrometry to identify Csd5 interaction partners and interaction domains. Here we show that Csd5 is part of multi-protein “shapeosome” complex that spans the cytoplasmic membrane and mediates interactions with the periplasmic peptidoglycan (PG) cell wall, a cytoplasmic peptidoglycan precursor synthase, a putative bactofilin, and membrane embedded ATP synthase.
Introduction
Helicobacter pylori infection accounts for a significant global cancer burden as the primary cause of stomach cancer, the third leading cause of cancer deaths worldwide (Ferlay et al., 2015). The helical cell shape of H. pylori is required for efficient stomach colonization (Sycuro et al., 2010). Thus understanding the mechanisms by which H. pylori achieves helical shape may inform novel therapeutic strategies.
The shape of most bacteria is determined by the peptidoglycan (PG) cell wall, a single macromolecule encasing the cell that determines cell shape and provides protection from osmolysis (Höltje, 1998; Typas et al., 2012). The PG cell wall consists of a meshwork of glycan chains composed of alternating N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) crosslinked by short peptides attached to NAM (Vollmer and Bertsche, 2008). For bacterial cells to grow without compromising the structural integrity of the cell wall, they must coordinate the action of PG hydrolases that cleave PG with the action of PG synthases that insert newly synthesized PG precursors into the growing cell wall. PG biosynthesis begins in the cytoplasm where precursors consisting of NAG-NAM-pentapeptides that are assembled onto an undecaprenol (UDP) lipid carrier through a series of cytoplasmic and membrane associated steps. The final lipid-linked precursor (lipidII) is then flipped into the periplasm where PG synthases assimilate the precursors into the existing wall by transglycosylation of the disaccharide (Sham et al., 2014; Scheffers and Tol, 2015). Nascent PG strands are then crosslinked to existing PG by transpeptidation to uncrosslinked PG peptides.
Genetic screens in H. pylori have identified several proteins that when deleted give rise to distinctly non-helical cell shapes. The majority of these proteins function as PG hydrolases that either break crosslinks (Csd1, Csd3) (Sycuro et al., 2010; Bonis et al., 2010) or trim uncrosslinked peptide stems (Csd6, Csd4) (Sycuro et al., 2012; Sycuro et al., 2013), giving rise to either curved-rod or straight-rod shaped cells, respectively. Two non-enzymatic proteins, CcmA and Csd5, were also identified in these screens. H. pylori CcmA has homology to bactofilins, bacterial cytoskeletal elements capable of self-assembly without a cofactor that are widely distributed in bacteria (Kühn et al., 2010) and often present as multiple (1–5) non-redundant paralogs indicating divergent cellular roles (Bulyha et al., 2013; Lin et al., 2017). In Caulobacter crescentus, the bactofilins BacA and BacB form membrane associated cytoplasmic filaments that associate with the inner membrane to direct stalk PG synthesis (Kühn et al., 2010). In Myxococcus xanthus, the bactofilin BacM forms filaments and is required for normal cell morphology (Koch et al., 2011; Zuckerman et al., 2015) while in Bacillus subtilis, the bactofilins BacE and BacF are required for bacterial motility (Andari et al., 2015). In Proteus mirabilis and Vibrio parahaemolyticus, ccmA gene expression increases in swarmer cells and overexpression in P. mirabilis induces morphological changes (Hay et al., 1999; Gode-Potratz et al., 2010). Recently, it was shown that the bactofilin LbbD from the spirochete Leptospira biflexa influences the helical pitch of the bacteria (Jackson et al., 2018). H. pylori ccmA mutants have altered global PG profiles similar to csd1 and csd3 suggesting it may regulate one or both of these enzymes (Sycuro et al., 2010). Csd5 is unique to H. pylori and Helicobacter acinonychis (Eppinger et al., 2006). Deletion of csd5 gives rise to straight cells yet global PG content analysis indicates no significant alterations in PG composition compared to wild-type (Sycuro et al., 2012). Thus, the mechanism by which Csd5 promotes helical shape is unclear.
Here we performed a structure-function analysis of H. pylori Csd5 to gain a mechanistic understanding of how this protein promotes helical cell shape. We show that N-terminal cytoplasmic (NT) and transmembrane (TM) domains, plus a C-terminal SH3 domain are each required for helical shape. Csd5 interacts directly with peptidoglycan via its C-terminal SH3 domain and the N-terminal transmembrane domain promotes interactions with CcmA, the PG precursor synthesis enzyme MurF, and F1FO ATP synthase. This work expands our understanding of the biochemical activities required to support helical shape generation and provides new insights as to how these activities may be spatially organized.
Results
Csd5 has conserved N- and C-terminal domains that each contribute to helical shape
Iterative searches using Jackhmmer (Finn et al., 2015) with the Csd5-encoding open reading frames HPG27_1195 (strain G27) and jhp1171 (strain J99) were used to identify and predict Csd5 functional domains, sequence motifs, and protein topology (Fig. 1A). We mapped predicted secondary structure features derived from these queries onto a multiple sequence alignment of diverse Csd5 variants (Fig. S1) from a hand-curated list of fully sequenced H. pylori genomes downloaded from the PATRIC database (Wattam et al., 2017).This analysis revealed distinct regions of high sequence conservation in the N and C-terminal regions of Csd5. The less conserved middle domain is predicted to be disordered, and contains within it a repetitive, glutamine rich coiled-coil (CC) motif with variable length across diverse strains of H. pylori (Fig. S1). The N-terminal region has a predicted transmembrane helix that may anchor Csd5 to the inner membrane while the C-terminus contains a bacterial SH3 domain (Kamitori and Yoshida, 2015). Bacterial SH3 domains can mediate protein-protein (Leon et al., 2010; Xu et al., 2015), protein-PG (Baba and Schneewind, 1996; Lu et al., 2006; Xu et al., 2009; Rolain et al., 2013), protein-metal (Pohl et al., 1999), or protein-nucleic acid (Wilkinson et al., 2016) interactions.
Fig. 1.
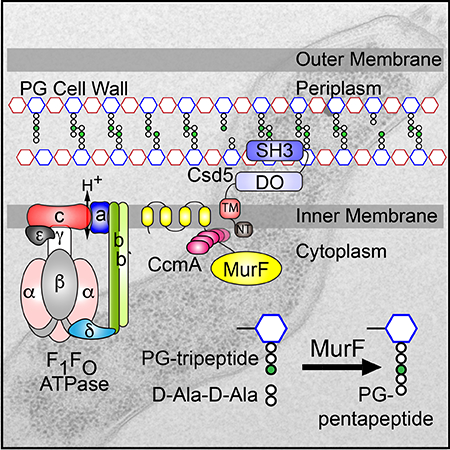
The N and C terminal domains of Csd5 are required for helical cell shape.
A. Schematic of the Gram (−) cell envelope and Csd5 predicted topology (top) and Csd5–2X-FLAG deletion variants expressed at the native locus (bottom). Numbers indicate amino acid residues of predicted domain boundaries for Csd5.
B. Phase contrast micrographs of parent (LSH100), Δcsd5 (LSH36), Csd5–2X-FLAG (WT, KBH127), ΔNT (KBH128), ΔTM (KBH129), ΔCC (KBH131), ΔDO (KBH132), ΔSH3 (KBH133). Scale bar = 5 μm.
C. Western blotting of whole cell extracts probed with α-FLAG M2 to detect Csd5 variants and a polyclonal antibody to Cag3 as a periplasmic loading control. WT (wild-type), IM (Inner membrane), PG (Peptidoglycan), OM (Outer membrane), CC (Coiled-coil), NT (N-terminal) TM (Transmembrane), DO (Disordered).
To assess the contribution of each of these domains or motifs to the overall function of Csd5, we generated a series of deletion variants, each encoding a C-terminal 2X-FLAG tag integrated in single-copy at the native locus (Fig. 1A). Deletion of the N-terminal tail (ΔNT), the transmembrane helix (ΔTM), or the SH3 domain resulted in predominantly rod-shaped bacteria similar to csd5 null bacteria (Fig. 1B and Fig. S2). A small population of cells in the ΔSH3 background exhibit a small degree of curvature reminiscent to the ΔccmA mutant, but overall the phenotypes between these two mutants are distinct (data not shown). To ensure that our Csd5–2X-FLAG variants were expressed we probed for Csd5 protein by immunoblotting using α-FLAG M2 antibody (Fig. 1C). The Csd5 ΔNT and ΔTM deletion variants exhibited reduced protein accumulation indicating that these proteins may be unstable, while the SH3 deletion variant was produced at similar levels to wild-type protein. Deletion of either the disordered (DO) domains or predicted coiled-coil (CC) motif in the glutamine rich linker resulted in helical cells with reduced axis length (ΔCC) or reduced sided curvature (ΔDO) and have wild-type levels of protein accumulation (Fig. 1B and Fig. S2). These data indicate functional roles for the DO, CC, and SH3 domains in cell shape generation, with the SH3 domain being required for helical shape generation. Furthermore, the N-terminal domains may be required to maintain protein stability, and/or localization of the SH3 domain.
The Csd5 transmembrane domain is required for protein function
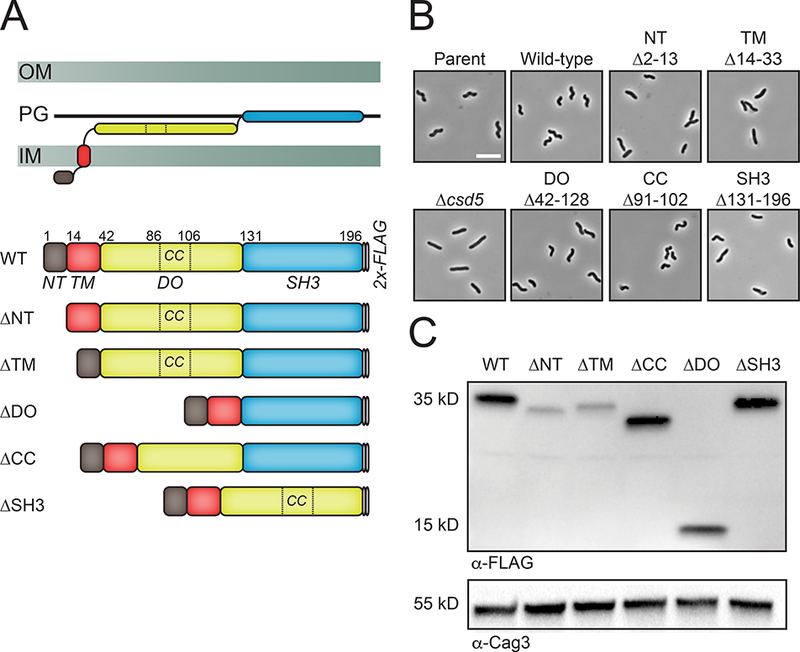
To determine if we could restore Csd5 function by replacement of the native TM sequence with a transmembrane domain from an unrelated protein, we generated chimeric Csd5 proteins containing single transmembrane domains from the H. pylori chemoreceptor TlpC (Machuca et al., 2017) and assessed their affect on cell shape, protein production, and membrane association. Replacement of the native TM sequence with either TM1 or TM2 from TlpC resulted in straight-rod bacteria (Fig. 2A) despite an improvement in steady state expression compared to the ΔNT or ΔTM variants (Fig. 2B). In addition, the chimeric Csd5 proteins exhibited membrane associations similar to that of wild-type or the ΔSH3 deletion proteins in the absence of detergent (Fig. 2C). While the ΔNT retains membrane association, the ΔTM is substantially present in the soluble fraction in the absence of detergent. Taken together, we conclude that the Csd5 N-terminal and TM domains are required for protein stability, correct subcellular localization, and protein function in vivo.
Fig. 2.
Csd5 transmembrane chimeras are non-functional but associate with membranes.
A. Top: Phase-contrast microscopy of parent (LSH100), Csd5-TlpC-TM1 (KBH169) and Csd5-TlpC-TM2 (KBH170) transmembrane (TM) chimeric variants. Bottom: Schematic of the H. pylori TlpC protein and each of the Csd5 chimeric proteins. Scale bar is 5μm. TM1 (TlpC-TM 1 domain), TM2 (TlpC –TM 2 domain), d1 (dCACH_1 domain), H1 (HAMP1 domain), H2 (HAMP2 domain), MACP (methyl-accepting chemotaxis like domain) (Machuca et al., 2017).
B. Western blotting of whole cell extracts probed with α-FLAG M2 to detect Csd5 N-terminal variants and a polyclonal antibody to Cag3 (Pinto-Santini and Salama, 2009) as a periplasmic loading control. WT (KBH127), ΔNT (KBH128), ΔTM (KBH129), ΔSH3 (KBH133)
C. Immunoblot of subcellular fractionation of Csd5 N-terminal variant baring strains showing whole-cell extracts (WC), soluble (S) and pellet (P) fractions probed with α-FLAG M2. Results representative of two independent experiments.
Csd5 interacts with CcmA, MurF, and ATP synthase
We investigated Csd5 protein interaction partners using immunoprecipitation (IP) and mass spectrometry with two independent Csd5 protein fusions, Csd5-VSV-G (vesicular stomatitis virus glycoprotein) (Table 1, Table S3, Table S4, and Fig. S3) and Csd5–2X-FLAG. We first performed IP and mass spectrometry using Csd5-VSV-G alongside control IPs of functional (helical cell shape) VSV-G fusions to MinD (cytoplasmic and and monotopic inner-membrane associated) (Szeto et al., 2002; Vecchiarelli et al., 2016), Slt (periplasmic) (Chaput et al., 2006), and a wild-type strain bearing no epitope fusion for controls. None of the top scoring proteins presented in Table 1 was present in unrelated bait, or no epitope tag control IP’s (Fig. S3). The top scoring proteins (Csd5-VSV-G IP) are known or predicted to function in the bacterial cytoplasm (Table 1, Table S3) and cytoplasmic membrane. The top unique hit in our proteomic screen was MurF, a cytoplasmic PG precursor synthesis enzyme that catalyzes the addition of D-Ala-D-Ala dipeptide to the terminal meso-diaminopimelic acid (m-DAP) residue of the UDP-linked NAM-tripeptide during cytoplasmic PG precursor biosynthesis (Smith, 2006). We also identified interactions with CcmA, a known H. pylori cell shape protein and bactofilin homolog that when mutated gives rise to curved-rod cells (Sycuro et al., 2010). Finally, our data showed robust enrichment for multiple components of the essential F1FO ATP synthase suggesting that Csd5 may be associated with this molecular machine (McGowan et al., 1997).
Table 1.
Top 10 proteins identified from Csd5-VSV-G immunoprecipitation and mass spectrometry.
| Ranka |
Protein Name |
H. pylori G27 Locus Tag |
% Coverage (± SD )b |
AVG PSM (± SD)b,c |
Mass (kD) | Description |
|---|---|---|---|---|---|---|
| 1 | MurF | HPG27_696 | 64 ± 1 | 122 ± 7 | 56 | Cytoplasmic PG precursor synthase |
| 2 | AtpA | HPG27_1079 | 62 ± 1 | 85 ± 9 | 55 | α subunit: F1ATP Synthase |
| 3 | AtpD | HPG27_1077 | 76 ± 5 | 78 ± 3 | 51 | β subunit: F1ATP Synthase |
| 4 | Csd5 | HPG27_1195 | 52 ± 3 | 60 ± 6 | 22 | Cell shape protein |
| 5 | AtpG | HPG27_1078 | 52 ± 3 | 28 ± 2 | 34 | λ subunit: FOATP Synthase |
| 6 | CcmA | HPG27_1480 | 67 ± 9 | 27 ± 1 | 15 | Cell shape protein; Putative bactofilin |
| 7 | AtpH | HPG27_1080 | 57 ± 5 | 19 ± 3 | 20 | δ subunit: F1ATP Synthase |
| 8 | AtpF | HPG27_1081 | 34 ± 1 | 14 ± 1 | 20 | b subunit: FOATP Synthase |
| 9 | AtpC | HPG27_1076 | 53 ± 5 | 14 ± 2 | 13 | ε subunit: F1ATP Synthase |
| 10 | AtpX | HPG27_1082 | 49 ± 0 | 12 ± 1 | 16 | b’ subunit: FOATP Synthase |
Uniquely present in Csd5-VSV-G IP vs. LSH100 (no tag) negative control, except AtpD.
Average (AVG) and standard deviation (SD) from 3 technical replicates (3 injections per sample).
vs. 0 positive spectral matches (PSM) in LSH100 (except AtpD with 2 ± 2).
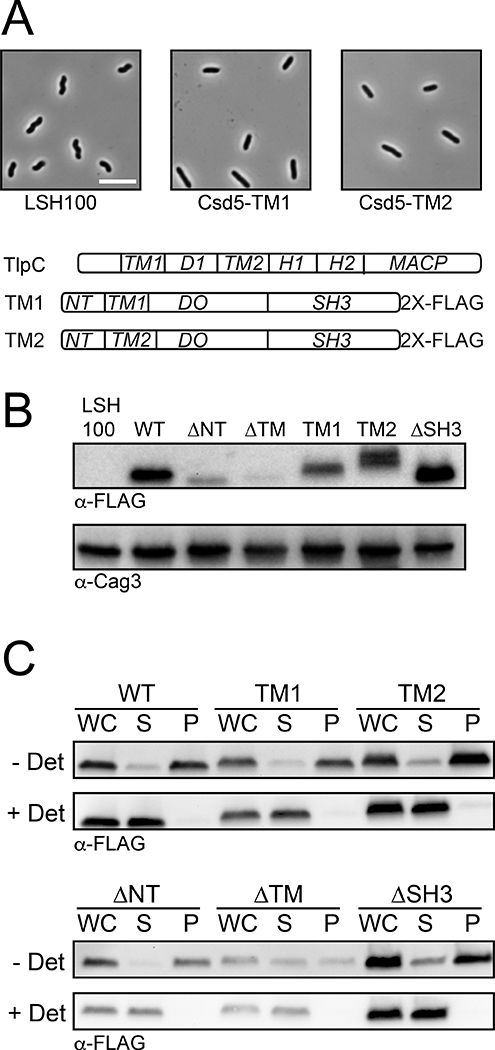
To validate these candidate interactions we performed reciprocal IPs using tagged strains followed by western blotting with polyclonal (CcmA, Csd5, AtpD) and monoclonal (FLAG, VSV-G) antibodies. We generated individual strains bearing CcmA-, MurF-3X-VSV-G, or AtpF-3X-FLAG expressed at the native locus and showed these fusions are functional (Fig. S5). CcmA-2X-FLAG cells have normal helical shape and MurF-3X-VSV-G and AtpF-3X-FLAG cells are both viable (murF and atpF are essential) and helical. AtpF (b-subunit) in the F1FO ATP synthase is part of the peripheral stalk that connects the membrane embedded FO complex to the cytoplasmic F1 ATPase (Weber, 2006; Rühle and Leister, 2015) (Fig. S4). For CcmA, MurF, and AtpF we demonstrate reciprocal Co-IP of Csd5 or Csd5–2X-FLAG (Fig. 3A and 3B). In addition, we show that with MurF, we also Co-IP CcmA and AtpD (β-subunit) of ATP synthase (Fig. 3B). Taken together, these results suggest that Csd5, MurF, CcmA, and ATP synthase interact as part of one or more multi-protein complexes.
Fig. 3.
Csd5 interacts with MurF, CcmA, and ATP synthase.
H. pylori strains bearing the indicated protein fusions were subject to immunoprecipitation (IP) using detergent solubilized whole cell extracts and α-FLAG (A) or α-VSV-G (B) antibodies to pull-down CcmA-2X-FLAG (JTH3), AtpF-3X-FLAG (KBH159), MurF-3X-VSV-G (KBH157) or a no tag-control (A, LSH100; B, KBH127).
A. CcmA (15 kD) and AtpF (20 kD) were detected in the input and IP fractions with α-FLAG while Csd5 (35 kD) was detected with α-SH3 antibody. A closed triangle denotes CcmA and closed circle denotes AtpF.
B. MurF, Csd5, CcmA, and the β-subunit of ATP synthase (AtpD) were detected in the input and IP fractions with α-VSV-G, α-SH3, α-CcmA, and α-AtpD antibodies, respectively. Representative experiments shown from a minimum of 2 biological replicates.
The Csd5 N-terminal domains are required for interactions with MurF and CcmA
We next used the domain deletions to begin to localize Csd5 interactions with MurF, CcmA, and ATP synthase. With the exception of the ΔNT strain, we could uniformly IP each of the variant bait proteins (Fig. 4A) with comparable yields despite lower levels of protein accumulation for the ΔNT and ΔTM domain deletion strains in whole cell extracts (Fig. 1C). Deletion of either the Csd5 DO domain or C-terminal SH3 domain had no effect on our ability to Co-IP MurF, CcmA, or AtpD (Fig. 4A and Fig. 4B). However, loss of the TM domain resulted in a complete loss of MurF and CcmA positive spectra (Fig. 4A) and loss of CcmA and AtpD protein by western blotting (Fig. 4B). In addition, we observed an overall decrease in the abundance of each component of the ATP synthase (Fig. S4). The NT domain also appears to be important for these interactions as we observed a significant decrease in MurF spectra and a complete loss of CcmA (Fig. 4A). These data suggest that the NT and TM domain of Csd5 are required for interactions with CcmA, MurF, and ATP synthase.
Fig. 4.
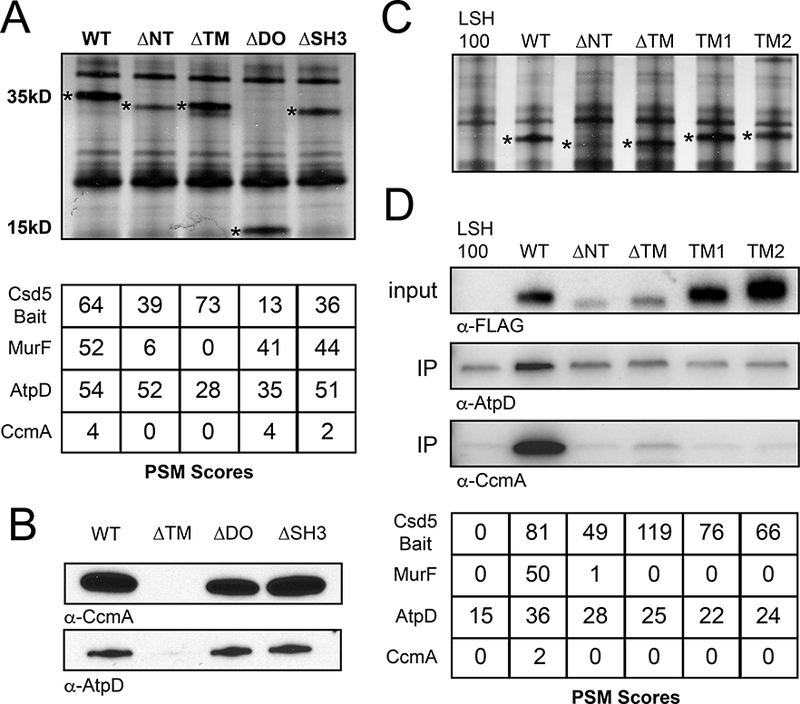
Csd5 N-terminal and transmembrane domains promote protein-protein interactions with MurF, CcmA, and ATP Synthase.
A. Top: Silver stain of Csd5 domain deletion immunoprecipitation (IP) fractions. Bait proteins indicated with an asterisk. Bottom: Mass spectrometry positive spectral match (PSM) counts for indicated Csd5–2X-FLAG domain deletion bait proteins, CcmA, and MurF from Csd5 domain deletion IPs; WT (KBH127), ΔNT (KBH128), ΔTM (KBH129), ΔDO (KBH132) and ΔSH3 (KBH133).
B. Immunoblot detection of CcmA and AtpD in IPs of indicated Csd5 deletion variants using α-CcmA and α-AtpD antibodies respectively.
C. Silver stain of Csd5 N-terminal deletions and Csd5-TlpC transmembrane chimera IP fractions. TM1 (KBH169), TM2 (KBH170). Bait proteins indicated by an asterisk.
D. Top: Immunoblot detection of AtpD and CcmA in input and IP fractions of indicated Csd5 variants using α-CcmA and α-AtpD antibodies; Csd5 variants detected in the input fractions with α-FLAG antibody. Bottom: Mass spectrometry PSM counts for Csd5 bait proteins, MurF, AtpD and CcmA in the indicated IP fractions.
To further probe the role and specificity of the TM domain for interactions with CcmA, MurF, and ATP synthase, we performed IP-MS using our Csd5 chimeric proteins containing either TM1 or TM2 from TlpC. Despite robust IP of both the TM1 or TM2 chimeric variants of Csd5 (Fig. 4C and 4D, Table S5), we are unable to demonstrate IP of MurF or CcmA using these strains, and we observed no enrichment for AtpD above the background levels of ATP synthase routinely observed in these experiments (Fig. 4D).
To probe whether Csd5, MurF, CcmA, and ATP synthase exist in a single complex, we performed IP and mass spectrometry using CcmA-2X-FLAG as bait in the presence or absence of formaldehyde crosslinking. We observed a crosslinking specific increase in spectral counts for both MurF and Csd5 (Fig. 5A) and also for components of the ATP synthase (Table S6). We next tested whether Csd5 could pull down MurF in the absence of CcmA using a ccmA deletion strain. Deletion of ccmA had no effect on immunoprecipitation of MurF by Csd5 (Fig. 5B) suggesting that the Csd5 and MurF interaction is direct. Taken together we conclude that Csd5, MurF, and CcmA interact individually and/or together with ATP synthase to promote maintenance of cell shape.
Fig. 5.
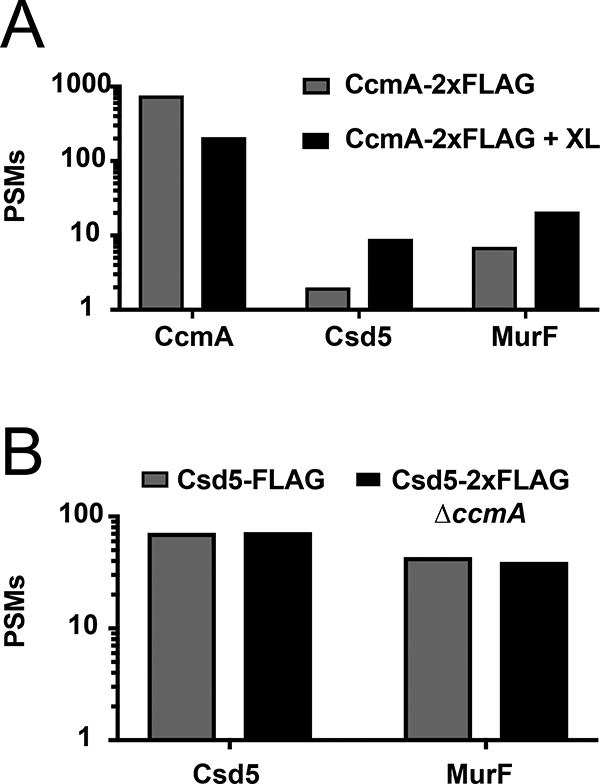
Csd5 interacts directly with MurF and CcmA.
A. Mass spectrometry positive spectral match (PSM) counts for CcmA, Csd5, and MurF from CcmA-2X-FLAG immunoprecipitation (IP) with and without formaldehyde crosslinking (XL).
B. PSM counts for Csd5 and MurF from Csd5–2X-FLAG (KBH127) and Csd5–2X-FLAG ΔccmA (KBH126) IPs.
The SH3 domain RT-loop is required for SH3 domain stability and promotes interaction with PG
None of the observed Csd5 protein-protein interactions required the SH3 domain. To further probe the function of the SH3 domain we investigated a putative interaction with the PG cell wall. Bacterial SH3 domains have been implicated as cell wall targeting domains that bind directly to PG (Baba and Schneewind, 1996; Lu et al., 2006). We identified an amino acid (R146) in the RT-loop motif that is strictly conserved in H. pylori and also conserved with the cell wall targeting SH3 domain of Lystostaphin (ALE-1, PDB ID 1R77) from Staphylococcus simulans (Fig. 6A and Fig. S1) (Lu et al., 2006). We identified a second amino acid (T151) that is slightly less conserved in both Csd5 and ALE-1 and is positioned at the tip of the surface-accessible RT-loop in our homology model (Fig. 6A). We generated strains expressing single copy mutants of csd5-R146A or csd5-T151A at the native locus. Mutation of the conserved arginine resulted in a complete loss of Csd5 protein in vivo and a complete loss of helical cell shape due to an apparent loss of protein stability (Fig. 6B). In contrast, the Csd5-T151A variant strain resulted in no visible change in protein accumulation but cell shape was affected, yielding cells with reduced side curvature indicating this loop region is important for maintaining helical cell shape (Fig. 6B).
Fig. 6.
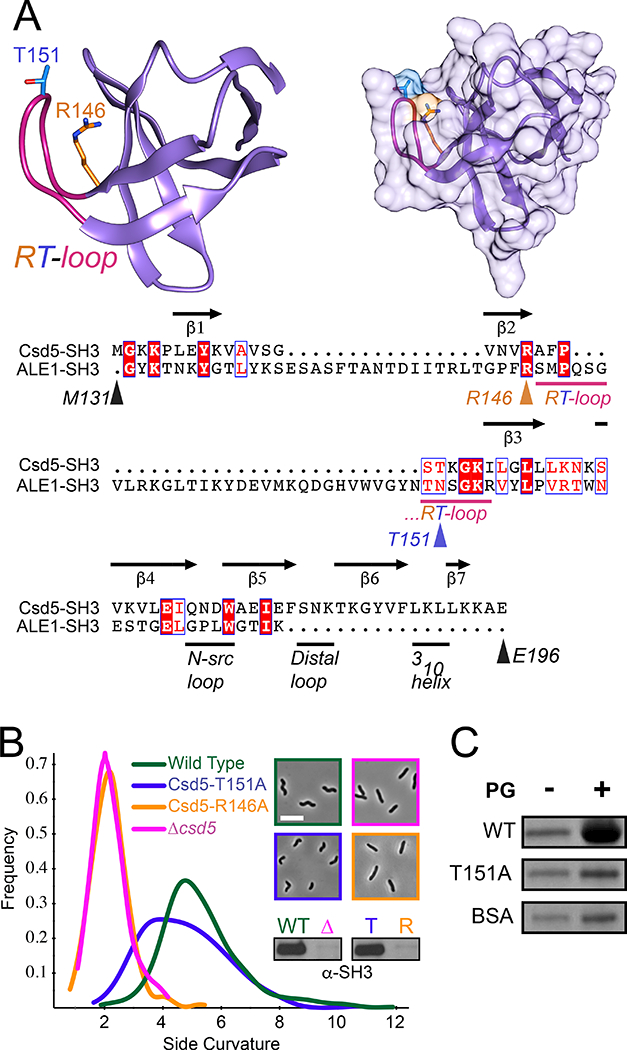
The Csd5-SH3 domain interacts with peptidoglycan.
A. Phyre2 (Kelley et al., 2015) homology model of the Csd5 SH3 domain highlighting the RT-loop (pink) and mutated sites R146 (Orange) and T151 (Blue) as sticks (Left). Surface representation of SH3 domain model (Right). Graphics generated using Chimera (Pettersen et al., 2004). Sequence alignment of H. pylori Csd5 SH3 domain with Staphyloccocus simulans ALE-1 cell wall binding domain (PDB ID 1R77) and Csd5 secondary structure and loop domain assignments (RT-loop, N-src loop, distal loop, and 310 helix) based on established SH3 domain loop nomenclature (Noble et al., 1993; Kamitori and Yoshida, 2015). Colored arrows or bars indicate mutated sites and loops, respectively (Bottom). Alignment graphic generated using Clustal Omega (Sievers et al., 2011) and ESPRIT 3.0 (Robert and Gouet, 2014).
B. Phase-contrast microscopy and quantitative cell shape analysis using CellTool software (Sycuro et al., 2010) of wild-type (LSH100), Δcsd5 (LSH36) and complemented full length Csd5-SH3 variants R146A (KMH1) and T151A (KMH2). Smooth histogram showing cell population side curvature (x-axis) as a density function (188–382 cells analyzed per strain). Western blots performed using whole cell extracts with an α-Csd5-SH3 domain antibody.
C. SDS-PAGE of PG sedimentation assay in the presence (+) or absence (−) of PG cell walls with purified GST-SH3 fusions to wild-type, mutant (T151A) or BSA as a negative control, stained with Coomasie Blue. Representative of 2 independent experiments.
To determine if the SH3 domain can interact with PG directly, we purified wild type and mutant (T151A) SH3 domains as recombinant fusions to GST (Fig. S6). Each protein (WT and T151A variant or controls) was individually mixed and incubated in the presence or absence of purified PG from wild-type H. pylori bacteria. We show that the wild-type, but not the T151A variant, is preferentially pulled down in a PG dependent manner (Fig. 6C and Fig. S7A). Cleavage and separation of the GST domain revealed that the WT SH3 domain in isolation binds PG sacculi (Fig. S7B) while GST alone does not (Fig. S7C). These combined data establish the RT-loop as a functional determinant for direct SH3 domain interaction with the bacterial cell wall.
Discussion
While curvature generation in curved-rod organisms such as Caulobacter crescentus and Vibrio cholera is thought to result from biased PG insertion along the outer curve of the cell, opposite the inner curvature localized cytoskeletal or periskeletal cell spanning filaments (Cabeen et al., 2009; Bartlett et al., 2017), helical curvature of H. pylori has been suggested to result from modulation of PG crosslinking (Sycuro et al., 2010; Bonis et al., 2010). Shared phenotypes (straight-rod cell shape) between Δcsd5 and the periplasmic PG carboxypeptidases Δcsd6 and Δcsd4 led to a hypothesis that Csd5 may localize these enzymes to either promote or inhibit the activity of periplasmic PG hydrolases that successively trim uncrosslinked PG-tetrapeptide to PG-dipeptide that can no longer participate in crosslinking (Sycuro et al., 2012; Sycuro et al., 2013; Kim et al., 2014; Chan et al., 2015). Despite significant efforts and a report that recombinant Csd5 and Csd4 interact directly (Kim et al., 2014), we have not been able to reproduce this finding with our in vivo IP’s and formaldehyde crosslinking experiments. Instead, in this work we have identified and localized Csd5 interactions with a cell shape associated putative bactofilin (CcmA), a PG precursor synthase (MurF) and to the PG cell wall. In addition, we have identified an association between these factors and ATP synthase. These results suggest the existence of a shape promoting protein complex that spans the cytoplasmic membrane and may connect the cell wall in the periplasm with the cytoplasmic cytoskeleton (Fig. 7). Future studies to investigate both the role of the ATP synthase as well as the membrane association and filament formation activities of CcmA would lend further support to this model.
Fig. 7.
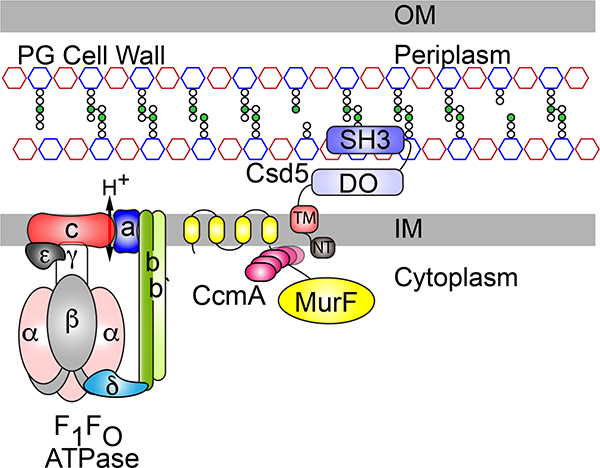
Model of the Csd5 H. pylori cell shape promoting protein complex.
Csd5 protein is anchored in the inner membrane where it mediates interactions between a putative cytoskeletal bactofilin CcmA, the PG precursor synthase MurF (D-Ala-D-Ala ligase), and the rotary F1FO ATP synthase. The C-terminal SH3 domain interacts with peptidoglycan in the periplasm and links Csd5 to cytoplasmic cell shape and cell elongation factors essential for helical shape (CcmA) and growth (MurF). OM (outer membrane), IM (inner membrane), PG (peptidoglycan), NT (N-terminal), TM (transmembrane), DO (disordered). The PG cell wall is depicted with alternating red (N-acetylmuramic acid) and blue (N-acetylglucosamine) octagons. meso-diaminopimelic acid (m-DAP;green circles) in the PG peptide stems (white circles).
The SH3 domain of Csd5 could serve as a sensor for recognition of specific PG ligands for proper positioning of the protein complex in the membrane. The similarity of the H. pylori Csd5 SH3 domain to NlpC/P60 SH3b2 domain (Xu et al., 2015) and to the Gram (+) ALE-1 cell wall targeting domain (Lu et al., 2006) suggests it may recognize either free stem peptides (e.g. di-, tri-, tetra-, or penta-) or specific crosslink species present in the cell wall sacculus (Gründling and Schneewind, 2006). Crystal structures of ALE-1 with (PDB ID 5LEO) and without (PDB ID 1R77) a penta-glycine PG fragment are deposited in the RCSB (PDB; http://www.rcsb.org/pdb/) protein databank (Berman et al., 2000). Our finding that mutations in conserved residues of the RT-loop can affect cell shape and disrupt binding to PG in vitro, indicates a role for the SH3 RT-loop in PG substrate recognition.
The ATP-dependent synthesis of PG precursors is carried out in the cytoplasm and is divided into cytosolic (MurA-F) and membrane associated (MurG, MraY, MurJ) steps (Laddomada et al., 2016). In H. pylori it appears that MurF may deviate from this model as iterative searching using Jackhmmer seeded with the H. pylori MurF sequence identified 4 predicted N-terminal transmembrane domains preceding the enzymatic domain of this highly conserved and essential protein. A PSI-BLAST search (excluding the Epsilonproteobacteria) seeded with the H. pylori MurF sequence was limited to orthologs with greater than 80% query coverage. This analysis revealed widespread prevalence of N-terminal transmembrane extensions in MurF from Gram (−), Gram (+), and organisms with minimal or reduced genomes (Nelson and Stegen, 2015; Brown et al., 2015) (data not shown). That our Csd5 ΔTM deletion mutants and TM chimeras fail to IP MurF, CcmA, and ATP synthase may suggest intra-membrane protein interactions that maintain these proteins within a single complex. MurF has been shown to associate directly with the elongasome and cytoskeleton through interactions with MurG (Favini-Stabile et al., 2013), MraY (White et al., 2010), and MreB (Mohammadi et al., 2007). Consistent with this, we observe a low level of MurG spectral hits in our Csd5-VSV-G IP (Table S3). Csd5 may thus be localized to sites of new PG synthesis through its association with MurF to modulate cell wall growth.
The H. pylori ATP synthase subunit stoichiometry varies from the prototypical bacterial synthase, where instead of a single dimeric b-subunit, there exist two distinct b (AtpF) and b’ (AtpX) subunits that form the peripheral stalk dimer (Fig. 7 and Fig. S4) (Weber, 2006). In both mitochondria and chloroplasts, the formation of ATP synthase dimers and even larger superassemblies have been associated with regions of high membrane curvature (Rexroth et al., 2004; Seelert and Dencher, 2011; Hahn et al., 2016). In mitochondria, the dimer rows of ATP synthase are essential for the formation of highly curved lamellar cristae (Paumard et al., 2002; Davies et al., 2012). Specialized subunits promote dimerization at the interface between the peripheral stalks of adjacent ATP synthases (Arnold et al., 1998). To date, ATP synthase dimers have not been observed in bacteria and future work will explore this possibility. Alternatively, association between MurF and ATP synthase could be involved in modulating local rates of PG synthesis or ATP turnover (Lee et al., 2013).
The identification of Csd5 protein interactions with CcmA, a known cell shape protein and putative cytoskeletal bactofilin, and with MurF, a known cell elongation factor, were unexpected but not surprising given the connections between PG synthesis, cell shape and intermediate filament proteins in curved organisms (Ausmees et al., 2003; Koch et al., 2011). We have discovered the first example of a Gram (−) SH3 domain interaction with peptidoglycan and provide evidence of a cell shape promoting protein complex in H. pylori. Future work characterizing these interactions in more detail will expand our understanding of the diversity of mechanisms cells use to regulate and control cell shape.
Experimental Procedures
Bacterial Strains and Growth Conditions
Strains used in this work, as well as primers and plasmids used in strain construction are described in Table S1 and S2. H. pylori was grown in Brucella broth supplemented with 10% fetal bovine serum (Invitrogen) without antimicrobials or on horse blood agar plates with antimicrobials as described (Chan et al., 2015) Bacteria were incubated at 37°C under micro-aerobic conditions in a tri-gas incubator (10% CO2, 10% O2 , and 80% N2). For resistance marker selection, horse blood plates were supplemented with 15 μg/ml chloramphenicol; 25 μg/ml kanamycin; or 60 mg/ml sucrose. For plasmid selection and maintenance in Escherichia coli, cultures were routinely grown in Luria broth (LB) or agar at 37°C supplemented with 100 μg/ml ampicillin or 25 μg/ml kanamycin.
Generating polyclonal antisera against the Csd5 SH3 domain and CcmA
We generated clones of GST-CcmA (pJF-1480) and GST-Csd5-SH3 (pKB7) for recombinant expression and purification of CcmA (genomic position:1,607,196–1,607,510) and Csd5 (genomic position: 1,313,682–1,313,888) antigens respectively. Each protein was expressed in E. coli BL21 DE3, and purified on glutathione sepharose 4B (GE Healthcare) resin by gravity. For Csd5 but not CcmA, the GST tag was subsequently removed by Thrombin CleanCleave™ protease (Sigma) with the uncut fusion protein and cleaved GST tag removed in a subsequent purification step. The resulting protein preparations were submitted for antibody production (R & R Research LLC). Approximately 0.5 mg of protein antigen was used for primary immunization and 0.25 mg of antigen was used for 3 boosts prior to the final bleed. Resulting anti-sera were used for western blotting against H. pylori whole cell lysates from wild-type and mutant (ΔccmA or Δcsd5) bacteria to ensure specific detection of CcmA or Csd5.
Construction of H. pylori epitope-tagged strains
Strains containing single or multi-copy FLAG or VSV-G epitope gene fusions were generated by PCR SOEing (Horton, 1995) to target integration at the native locus after natural transformation (Clayton and Mobley, 2010). All strains (Table S1) and plasmids (Table S2) were validated by diagnostic PCR, Sanger sequencing and western blotting as appropriate. Refer to supplemental experimental methods for detailed descriptions of H. pylori strain construction.
H. pylori quantitative cell shape analysis
Phase-contrast microscopy was performed and resulting images were thresholded using the ImageJ software package. Quantitative analysis of thresholded images of ~200 bacteria per strain was used to measure both side curvature and central axis length with the CellTool software package as described previously (Sycuro et al., 2010).
H. pylori cell fractionation
2 optical density at 600 nm units (OD600) of H. pylori bacteria expressing functional FLAG fusions to Csd5 variants grown to mid-log phase (0.3–0 7 OD600) were harvested by centrifugation for 10 minutes at 8,000 × g at room temperature (RT). Frozen cell pellets were resuspended in 0.9ml of Buffer A (20 mM Tris pH 8.0, 150 mM NaCl, 2% Glycerol, 1.0%) with or without 1.0% Triton X-100. Each suspension was uniformly sonicated at 10% power with a microtip (Sonic Dismembrator Model 500, Branson) for 6–8 seconds and 100 μl volumes of normalized whole cell extracts, soluble and pellet fractions were collected by centrifugation at 20,000 × g for 10 minutes at 4°C and analyzed by SDS-PAGE followed by western blotting using α-FLAG (Sigma) primary antibody as described below.
Immunoprecipitation and crosslinking of H. pylori cell shape proteins
25 OD’s of H. pylori bacteria expressing functional FLAG or VSV-G fusions to cell shape proteins (Csd5, CcmA, MurF and AtpF) grown to mid-log phase were harvested by centrifugation for 10 minutes at 8000 × g at RT. Fresh or frozen cell pellets were detergent solubilized in 1.9 ml of chilled Buffer A (20 mM Tris pH 8.0, 150 mM NaCl, 2% Glycerol) supplemented with 1.0% Triton X-100, 10 units of Benzonase nuclease (EMD) and EDTA-free protease inhibitors (Pierce). The cells were sonicated for 5–10 seconds using short pulses at 10% power until visibly cleared, then centrifuged at 20,000 × g for 30 minutes at 4°C. The soluble fraction was then incubated with 40 μl of equilibrated (with Buffer A) α-FLAG M2 (magnetic or agarose) or VSV-G mouse monoclonal agarose beads (Sigma) and incubated for 90 minutes at 4°C. The beads are then subjected to 3 × 15 ml washes (room temp) with cold Buffer B (20 mM Tris pH 8.0, 150 mM NaCl, 2% Glycerol and 0.1% Triton X-100). Beads were then boiled in 2x bead volumes of protein sample buffer (66 mM Tris-HCl, pH 6.8 26% (w/v) glycerol, 2.1% SDS, 0.01% Bromophenol blue) without beta-mercaptoethanol and subjected to SDS-PAGE, silver staining (SilverQuest™, Invitrogen), western blotting and/or mass spectrometry analysis as appropriate. For crosslinking experiments, 25 OD’s of whole cells were incubated on ice with 1.0% formaldehyde in 2.0 ml of cold 1X phosphate buffered saline (PBS) pH7.2 for 30 minutes and then quenched with 0.83M glycine pH 2.2 for 10 minutes. Cells were harvested by centrifugation and frozen at −80°C prior to immunoprecipitation as described above.
Immunoblotting H. pylori whole-cell extracts and immunoprecipitates
Whole cell exGtracts were prepared by harvesting 1.0 OD600 of log phase (0.3–0.7 OD600) H. pylori by centrifugation for 2 minutes at max speed in a microcentrifuge and resuspending in 2x protein sample buffer at 10.0 OD600 per ml and boiled for 10 min. Whole cell extracts or input and IP fractions were separated on 4–15% gradient (BioRad TGX) gels by SDS-PAGE and transferred onto PVDF membranes using the BioRad Turbo-transfer system according to the manufacturer’s instructions. Membranes were blocked for 1 hour at RT with 5% non-fat milk in TBS-T (0.5 M Tris,1.5 M NaCl, pH 7.6, plus 0.05% Tween 20). Membranes were incubated with primary antibody overnight at 4°C (or 1.5 hours at RT) with primary antibodies; 1:5000 dilution for α-Flag M2 (Sigma), 1:5000 for α-VSV-G (Sigma), 1:5000 for α-SH3, 1:10,000 for α-CcmA, 1:10,000 for AtpD (Agrisera, Sweden), 1:20,000 dilution for α-Cag3 (Pinto-Santini and Salama, 2009), in TBS-T. Four 10 minute washes with TBS-T were followed by a 1.25 hour incubation at RT with appropriate horseradish peroxidase-conjugated α-immunoglobulin G (Santa Cruz Biotechnology) antibody at 1:20,000 dilution in TBS-T (α-rabbit for Cag3, Csd5, CcmA; α-mouse for FLAG and VSV-G). After four more TBS-T washes, antibody detection was performed with ECL Plus (Pierce) (Cag3, CcmA, CcmA-2X-FLAG, SH3, AtpF-3X-FLAG) or Immobilon (Millipore) (SH3, Csd5–2X-FLAG, MurF-3X-VSV-G, AtpF-3X-FLAG, AtpD) detection kits, following the manufacturer’s protocol and imaged directly on film or with the BioRad Gel Documentation System.
Mass spectrometry analysis of H. pylori immunoprecipitates
Immunoprecipitated and washed α-VSV-G monoclonal agarose beads (Table 1 and Fig. S3) were stored at −80°C following three washes with 20 mM ammonium bicarbonate and treated with 20 μg/ml trypsin for 5.25 hours at 37°C. For mass spectrometry, trypsinized bead suspensions were thawed and supernatant collected. The beads were washed twice with 20 μl of 20 mM ammonium bicarbonate and combined with the supernatant followed by reduction with 5mM TCEP (Pierce) for 1 hour at 37°C. Samples were next alkylated with 10 mM iodoacetamide for 30 minutes in the dark at RT. Reactions were quenched by addition of 16 mM N-acetyl-L-Cysteine (Sigma). Samples were then purified and concentrated on C18 reverse spin columns (Pierce) according to the manufacturers directions. Samples were quantified by Absorbance (A280) and submitted to Northwestern Proteomics (Chicago, IL) for protein identification as previously described (Whitney et al., 2015).
For α-FLAG IP’s (Fig. 3A and Fig. S4), samples were run ~ 1cm into the wells of a 4–15% SDS-PAGE (Bio-Rad TGX) 10-well 50μl gels. After electrophoresis a 1cm gel slice containing the entire IP was excised and submitted for total protein identification to the Fred Hutch Proteomics Facility, as further described in supplemental experimental methods.
Purification of H. pylori peptidoglycan cell wall sacculi
Isolation of H. pylori cell wall sacculi was performed as described (Glauner, 1988). Briefly, 330 ml of H. pylori cells in shaken liquid culture were grown to 0.6–0.8 OD/ml. Cells were harvested by centrifugation in a TOMY TX-160 centrifuge with a TMA-27 rotor at 5000 rpm for 10 minutes at 4°C and subsequently resuspended in 6 ml chilled Dulbecco’s PBS (Gibco). The cell suspension was added dropwise to 6 ml boiling 8% SDS and boiled for a further 6.5 hours. Sacculi were collected the first time by ultracentrifugation in a Beckman-Coulter Optima L-90K ultracentrifuge with a SW41 Ti rotor at 28000 rpm for 60 minutes at 28°C. Sacculi were resuspended in 2.5 ml 4% SDS and boiled for four hours. Sacculi were again collected by ultracentrifugation. This and all further ultracentrifugation steps were performed in a Beckman TL-100 ultracentrifuge with a TLA 100.3 rotor at 70000 rpm for 60 minutes at 25°C. Sacculi were resuspended in 2.5 ml 1% SDS, and boiled for four hours. We then collected sacculi by ultracentrifugation, resuspended in 2.5 ml of 50mM sodium phosphate pH 7.4. We incubated sacculi at 37°C with 0.25 mg α-chymotrypsin (Sigma, C4129) for four hours, added an additional 0.25 mg α-chymotrypsin, and incubated overnight. Following incubation, we added 300 μl of 10% SDS and boiled samples for four hours. Sacculi were collected by ultracentrifugation, resuspended in 2.5 ml 1% SDS, and boiled for four hours. We then washed sacculi by repeated ultracentrifugation and resuspension in ddH2O until supernatant was demonstrated free from SDS by the Hayashi test (Hayashi, 1975). Purified SDS-free sacculi were resuspended in 500 μl ddH2O plus 0.02% sodium azide and stored at 4°C until use.
Purification of recombinant GST-SH3 domain variants
Plasmids containing N-terminal GST fusions to wild type (pKB7) and mutant SH3 (pKM4-R146A, and pKM5-T151A) were transformed into E. coli protein production host BL21 DE3. Strains were grown overnight at 37°C in the presence of LB with 0.2% glucose and 100 μg/ml ampicillin. The next day, cells were diluted 1/100 into fresh media without glucose, grown to mid-log (~0.5 – 0.8 OD) at 37°C, chilled on ice for 30 minutes, then induction for protein expression by addition of 1.0mM IPTG. Flasks were transferred to 16°C and incubated overnight with shaking. Cells were harvested by centrifugation and flash frozen in liquid nitrogen or used immediately. Cells were suspended in 1/10 culture volume of lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 10 mM MgCl2, 5% Glycerol), supplemented with 10U of Benzonase nuclease (EMD) and 1mM PMSF (Fisher) and sonicated at 20% power using 4 × 30 second pulses on ice (1 minute rest on ice between each pulse). Lysates were then cleared at 20,000 × g at 4°C. The cleared lysate was applied to equilibrated glutathione sepharose 4B affinity resin (25 μl slurry/ml culture volume) and incubated for 2 hours at RT with mixing. The protein bound GST resin was washed with 3 × 10 column volumes and proteins were eluted (5 × 1 column volume) from the beads using 20mM glutathione in lysis buffer. Fractions were analyzed by SDS-PAGE for purity and yield (Fig. S6). For some experiments, the SH3 domain was separated from the GST domain by thrombin cleavage as described above for antigen preparation.
Peptidoglycan Co-sedimentation Assay
To 35 μl of binding buffer (1X TBS + 5% Glycerol), zero or 5 μl (equivalent to ~2.4 ODs of bacteria) of purified wild-type H. pylori sacculi (peptidoglycan) and 10 μl of recombinant SH3 domain or control proteins (GST or BSA), at a concentration of 0.5 mg/ml, were mixed (input) and incubated for 30 minutes at room temperature with constant agitation. The sacculi were sedimented by centrifugation at maximum speed in a bench-top microcentrifuge for 3 minutes at room temperature. The supernatant (output) fraction was collected and the pellets were washed with 1.0 ml of binding buffer and centrifuged again for 3 minutes. The input, output and pellet samples were normalized in 2X SDS-PAGE sample buffer, analyzed by SDS-PAGE and stained with Coomasie blue.
Supplementary Material
Acknowledgments
We thank Thomas Rodriguez and Marcus Hernandez and Desiree Yang for construction of H. pylori strains csd5-VSV-G:cat, slt::cat, and ccmA::kan respectively. We thank Dr. Joseph Mougous for reagents and helpful guidance. This work was supported in part by R01 AI094839 (NRS), R01 AI054423 (NRS), T32 CA009657 (KMB), a National Science Foundation Graduate Research Fellowship Grant DGE-1256082 (KMB), and the Proteomics and Genomics Shared Resources of NIH/NCI Cancer Center Support Grant P30 CA015704. Additional proteomics services were performed by the Northwestern Proteomics Core Facility, generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center and the National Resource for Translational and Developmental Proteomics supported by P41 GM108569. We declare no conflicts of interest.
References
- Andari, El J, Altegoer F, Bange G, and Graumann PL (2015) Bacillus subtilis bactofilins are essential for flagellar hook and filament assembly and dynamically localize into structures of less than 100 nm diameter underneath the cell membrane. PloS one, 10(10), e0141546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold I, Pfeiffer K, Neupert W, Stuart RA, and Schägger H (1998) Yeast mitochondrial F1F0-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J 17: 7170–7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausmees N, Kuhn JR, and Jacobs-Wagner C (2003) The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115: 705–713. [DOI] [PubMed] [Google Scholar]
- Baba T, and Schneewind O (1996) Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J 15: 4789–4797. [PMC free article] [PubMed] [Google Scholar]
- Bartlett TM, Bratton BP, Duvshani A, Miguel A, Sheng Y, Martin NR, et al. (2017) A periplasmic polymer curves Vibrio cholerae and promotes pathogenesis. Cell 168: 172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. (2000) The protein data bank. Nucleic Acids Res 28: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonis M, Ecobichon C, Guadagnini S, Prevost MC, and Boneca IG (2010) A M23B family metallopeptidase of Helicobacter pylori required for cell shape, pole formation and virulence. Mol Microbiol 78: 809–819. [DOI] [PubMed] [Google Scholar]
- Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, et al. (2015) Unusual biology across a group comprising more than 15% of domain bacteria. Nature 523: 208–211. [DOI] [PubMed] [Google Scholar]
- Bulyha I, Lindow S, Lin L, Bolte K, Wuichet K, Kahnt J, et al. (2013) Two small GTPases act in concert with the bactofilin cytoskeleton to regulate dynamic bacterial cell polarity. Dev Cell 25: 119–131. [DOI] [PubMed] [Google Scholar]
- Cabeen MT, Charbon G, Vollmer W, Born P, Ausmees N, Weibel DB, and Jacobs-Wagner C (2009) Bacterial cell curvature through mechanical control of cell growth. EMBO J 28: 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan ACK, Blair KM, Liu Y, Frirdich E, Gaynor EC, Tanner ME, et al. (2015) Helical shape of Helicobacter pylori requires an atypical glutamine as a zinc ligand in the carboxypeptidase Csd4. J Biol Chem 290: 3622–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaput C, Labigne A, and Boneca IG (2006) Characterization of Helicobacter pylori lytic gransglycosylases Slt and MltD. J Bacteriol 189: 422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton CL, and Mobley HLT (2010) Helicobacter pylori Protocols. Humana Press. [Google Scholar]
- Davies KM, Anselmi C, Wittig I, Faraldo-Gómez JD, and Kühlbrandt W (2012) Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci USA 109: 13602–13607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppinger M, Baar C, Linz B, Raddatz G, Lanz C, Keller H, et al. (2006) Who ate whom? Adaptive Helicobacter genomic changes that accompanied a host jump from early humans to large felines. PLoS Genet 2: e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favini-Stabile S, Contreras-Martel C, Thielens N, and Dessen A (2013) MreB and MurG as scaffolds for the cytoplasmic steps of peptidoglycan biosynthesis. Environ Microbiol 15: 3218–3228. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136: E359–86. [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, et al. (2015) HMMER web server: 2015 update. Nucleic Acids Res 43: W30–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauner B (1988) Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem 172: 451–464. [DOI] [PubMed] [Google Scholar]
- Gode-Potratz CJ, Kustusch RJ, Breheny PJ, Weiss DS, and McCarter LL (2010) Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol Microbiol 79: 240–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründling A, and Schneewind O (2006) Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. J Bacteriol 188: 2463–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn A, Parey K, Bublitz M, Mills DJ, Zickermann V, Vonck J, et al. (2016) Structure of a complete ATP synthase dimer reveals the molecular basis of inner mitochondrial membrane morphology. Mol Cell 63: 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay NA, Tipper DJ, Gygi D, and Hughes C (1999) A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J Bacteriol 181: 2008–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K (1975) A rapid determination of sodium dodecyl sulfate with methylene blue. Anal Biochem 67: 503–506. [DOI] [PubMed] [Google Scholar]
- Horton RM (1995) PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol Biotechnol 3: 93–99. [DOI] [PubMed] [Google Scholar]
- Höltje JV (1998) Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol R 62: 181–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson KM, Schwartz C, Wachter J, Rosa PA, and Stewart PE (2018) A widely conserved bacterial cytoskeletal component influences unique helical shape and motility of the spirochete Leptospira biflexa. Mol Microbiol 108: 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitori S, & Yoshida H (2015). Structure-Function Relationship of Bacterial SH3 Domains In SH Domains (pp. 71–89). Springer, Cham. [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Kim J, Im HN, An DR, Lee M, Hesek D, et al. (2014) Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a d,l-carboxypeptidase controlling the helical cell shape. Acta Crystallogr D 70: 2800–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MK, McHugh CA, and Hoiczyk E (2011) BacM, an N-terminally processed bactofilin of Myxococcus xanthus, is crucial for proper cell shape. Mol Microbiol 80: 1031–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn J, Briegel A, Mörschel E, Kahnt J, Leser K, Wick S, et al. (2010) Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J 29: 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laddomada F, Miyachiro MM, & Dessen A (2016). Structural insights into protein-protein interactions involved in bacterial cell wall biogenesis. Antibiotics, 5(2), 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Pontes MH, and Groisman EA (2013) A bacterial virulence protein promotes pathogenicity by inhibiting the bacterium’s own F1Fo ATP synthase. Cell 154: 146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon E, Navarro-Aviles G, Santiveri CM, Flores-Flores C, Rico M, Gonzalez C, et al. (2010) A bacterial antirepressor with SH3 domain topology mimics operator DNA in sequestering the repressor DNA recognition helix. Nucleic Acids Res 38: 5226–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Valeriano MO, Harms A, Søgaard-Andersen L, & Thanbichler M (2017). Bactofilin-mediated organization of the ParABS chromosome segregation system in Myxococcus xanthus. Nat commun 8(1), 1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JZ, Fujiwara T, Komatsuzawa H, Sugai M, and Sakon J (2006) Cell wall-targeting domain of glycylglycine endopeptidase distinguishes among peptidoglycan cross-bridges. J Biol Chem 281: 549–558. [DOI] [PubMed] [Google Scholar]
- Machuca MA, Johnson KS, Liu YC, Steer DL, Ottemann KM, & Roujeinikova A (2017). Helicobacter pylori chemoreceptor TlpC mediates chemotaxis to lactate. Scientific reports, 7(1), 14089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan CC, Cover TL, and Blaser MJ (1997) Analysis of F1F0-ATPase from Helicobacter pylori. Infect Immun 65: 2640–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi T, Karczmarek A, Crouvoisier M, Bouhss A, Mengin-Lecreulx D, and Blaauwen, den T (2007) The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol Microbiol 65: 1106–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WC, and Stegen JC (2015) The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front Microbiol 6: 713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble ME, Musacchio A, Saraste M, Courtneidge SA, and Wierenga RK (1993) Crystal structure of the SH3 domain in human Fyn; comparison of the three-dimensional structures of SH3 domains in tyrosine kinases and spectrin. EMBO J 12: 2617–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, et al. (2002) The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J 21: 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004) UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Pinto-Santini DM, and Salama NR (2009) Cag3 Is a novel essential component of the Helicobacter pylori Cag type IV secretion system outer membrane subcomplex. J Bacteriol 191: 7343–7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl E, Holmes RK, and Hol WG (1999) Crystal structure of a cobalt-activated diphtheria toxin repressor-DNA complex reveals a metal-binding SH3-like domain. J Mol Biol 292: 653–667. [DOI] [PubMed] [Google Scholar]
- Rexroth S, Meyer zu Tittingdorf JMW, Schwaßmann HJ, Krause F, Seelert H, and Dencher NA (2004) Dimeric H+-ATP synthase in the chloroplast of Chlamydomonas reinhardtii. Biochimica et Biophysica Acta (BBA)-Bioenergetics, 1658(3), 202–211. [DOI] [PubMed] [Google Scholar]
- Robert X, and Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42: W320–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolain T, Bernard E, Beaussart A, Degand H, Courtin P, Egge-Jacobsen W, et al. (2013) O-glycosylation as a novel control mechanism of peptidoglycan hydrolase activity. J Biol Chem 288: 22233–22247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rühle T, & Leister D (2015). Assembly of F1F0-ATP synthases. Biochimica et Biophysica Acta (BBA)- Bioenergetics, 1847(9), 849–860. [DOI] [PubMed] [Google Scholar]
- Scheffers DJ, and Tol MB (2015) LipidII: Just another brick in the wall? PLoS Pathog 11: e1005213–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelert H, and Dencher NA (2011) ATP synthase superassemblies in animals and plants: two or more are better. Biochimica et Biophysica Acta (BBA)-Bioenergetics, 1807(9), 1185–1197. [DOI] [PubMed] [Google Scholar]
- Sham LT, Butler EK, Lebar MD, Kahne D, Bernhardt TG, and Ruiz N (2014) Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345: 220–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA (2006) Structure, function and dynamics in the Mur family of bacterial cell wall ligases. J Mol Biol 362: 640–655. [DOI] [PubMed] [Google Scholar]
- Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, and Salama NR (2010) Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell 141: 822–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sycuro LK, Rule CS, Petersen TW, Wyckoff TJ, Sessler T, Nagarkar DB, et al. (2013) Flow cytometry-based enrichment for cell shape mutants identifies multiple genes that influence Helicobacter pylori morphology. Mol Microbiol 90: 869–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sycuro LK, Wyckoff TJ, Biboy J, Born P, Pincus Z, Vollmer W, and Salama NR (2012) Multiple peptidoglycan modification networks modulate Helicobacter pylori’s cell shape, motility, and colonization potential. PLoS Pathog 8: e1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto TH, Rowland SL, Rothfield LI, and King GF (2002) Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc Natl Acad Sci USA 99: 15693–15698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, and Vollmer W (2012) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10: 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli AG, Li M, Mizuuchi M, Hwang LC, Seol Y, Neuman KC, and Mizuuchi K (2016) Membrane-bound MinDE complex acts as a toggle switch that drives Min oscillation coupled to cytoplasmic depletion of MinD. Proc Natl Acad Sci USA 113: E1479–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, and Bertsche U (2008) Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochimica et Biophysica Acta (BBA)- Biomembranes 1778(9), 1714–1734. [DOI] [PubMed] [Google Scholar]
- Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, et al. (2017) Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45: D535–D542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J (2006) ATP synthase: subunit-subunit interactions in the stator stalk. Biochim Biophys Acta 1757: 1162–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CL, Kitich A, and Gober JW (2010) Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol Microbiol 76: 616–633. [DOI] [PubMed] [Google Scholar]
- Whitney JC, Quentin D, Sawai S, LeRoux M, Harding BN, Ledvina HE, et al. (2015) An interbacterial NAD(P)+ glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell 163: 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M, Chaban Y, and Wigley DB (2016) Mechanism for nuclease regulation in RecBCD. eLife 5: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Mengin-Lecreulx D, Liu XW, Patin D, Farr CL, Grant JC, et al. (2015) Insights into substrate specificity of NlpC/P60 cell wall hydrolases containing bacterial SH3 domains. mBio 6: e02327–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Sudek S, McMullan D, Miller MD, Geierstanger B, Jones DH, et al. (2009) Structural basis of murein peptide specificity of a gamma-D-glutamyl-l-diamino acid endopeptidase. Structure 17: 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerman DM, Boucher LE, Xie K, Engelhardt H, Bosch J, and Hoiczyk E (2015) The bactofilin cytoskeleton protein BacM of Myxococcus xanthus forms an extended β-sheet structure likely mediated by hydrophobic interactions. PLoS ONE 10: e0121074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


