Abstract
Once pathogens have breached the mechanical barriers to infection, survived extracellular immunity and successfully invaded host cells, cell-intrinsic immunity becomes the last line of defense to protect the mammalian host against viruses, bacteria, fungi and protozoa. Many cell-intrinsic defense programs act as high-precision weapons that specifically target intracellular microbes or cytoplasmic sites of microbial replication while leaving endogenous organelles unharmed. Critical executioners of cell-autonomous immunity include interferon-inducible dynamin-like GTPases and autophagy proteins, which often act cooperatively in locating and antagonizing intracellular pathogens. Here, we discuss possible mechanistic models to account for the functional interactions that occur between these two distinct classes of host defense proteins.
Introduction
Macroautophagy (hereafter simply referred to as autophagy) is a highly conserved homeostatic process by which eukaryotic cells recycle portions of their own cytoplasm through sequestration into double-membraned autophagosomes and subsequent delivery into degradative lysosomes. This process not only removes obsolete or damaged organelles and promotes cell survival during nutrient starvation [1] but can also capture and destroy intracellular pathogens through a process known as xenophagy [2]. Multiple steps of autophagy and xenophagy, including autophagosome formation and fusion with lysosomes, require an ancient ubiquitin-like conjugation system conserved from yeast to mammals. This system is composed of several autophagy proteins (ATGs) that covalently conjugate members of the ubiquitin-like protein ATG8 protein family, such as mammalian microtubule associated protein 1 light chain 3 (LC3), to the headgroup of the membrane lipid phosphatidylethanolamine (Table 1). Similar to protein ubiquitination, ATG8 conjugation to phosphatidylethanolamine requires the function of E1-, E2- and E3-like enzymes. Following cleavage of its C-terminal arginine by the cysteine protease ATG4, the exposed penultimate glycine of ATG8 is activated by the E1-like ATG7 enzyme, then transferred to the E2-like ATG3 enzyme, before ATG8 is conjugated to phosphatidylethanolamine by the E3-like enzymatic complex consisting of ATG12–ATG5-ATG16 [1]. In addition to its essential role in degradative autophagy, the ATG8 conjugation system also regulates LC3-associated phagocytosis of pathogens and dead cells [3], the unconventional secretion of host proteins [4], exocytosis of viruses [5], and the localization of interferon (IFN)-inducible GTPases to subcellular sites of microbial colonization [6,7].
Table 1.
Glossary of host proteins discussed in the text and their functions
| Host Factors | Functions |
|---|---|
| ATG3 | An E2-like conjugating enzyme required for the lipidation of ATG8-like proteins. |
| ATG5 | As a complex with ATG12 and ATG16, it serves as an E3-like ligase for the lipidation of ATG8-like proteins. |
| ATG7 | An E1-like activating enzyme that activates ATG12 for its conjugation to ATG5 as well as ATG8 proteins for their conjugation to the lipid phosphatidylethanolamine. |
| ATG8 | A family of ubiquitin-like proteins consisting of the LC3 and GABARAP subfamilies. ATG8 proteins can be conjugated to phosphatidylethanolamine. |
| ATG12 | An ubiquitin-like protein that forms a conjugate with ATG5. |
| GBPs | A family of IFN-inducible GTPases involved in cell-intrinsic immunity and the regulation of inflammation. |
| GKS proteins | An ‘effector’ IRG subfamily that exists in mice but not in humans. |
| IRGs | A vertebrate family of IFN-inducible GTPases controlling cell-intrinsic host defense to bacterial and protozoan pathogens. |
| IRGM proteins | An IRG subfamily controlling autophagy-related processes. IRGM proteins exist both in mice and humans. |
| MX proteins | A family of IFN-inducible GTPases with antiviral functions. |
| NOD2 | A cytoplasmic sensor, which detects the bacterial peptidoglycan sub-fragment muramyl dipeptide and activates the autophagy pathway. |
| Syntaxin-17 | A membrane integrated protein that controls the fusion of autophagosomes with lysosomes. |
| ULK1 | A serine/ threonine kinase controlling autophagy initiation. |
| VLIGs | A family of very large IFN-inducible GTPases with poorly defined functions. |
IFN-inducible GTPases are grouped into four families (Table 1): Immunity Related GTPases (IRGs), Guanylate Binding Proteins (GBPs), Myxovirus-resistance (Mx) proteins and Very Large Inducible GTPases (VLIGs). While the physiological functions of VLIG proteins are poorly described, the pivotal roles for IRGs, GBPs and Mx proteins in cell-intrinsic host defense have been widely reported [7]. Current evidence suggests that Mx proteins provide resistance exclusively to viruses [8]. IRGs and GBPs on the other hand execute host defenses against a diverse group of pathogens that includes viruses, bacteria, microsporidia and protozoa [7]. In order to perform many of their antimicrobial functions, IRGs and GBPs specifically associate with intracellular microbes residing in the host cell cytosol or at pathogen-occupied supramolecular structures [9], which include viral replication compartments [10] and pathogen-containing vacuoles [7]. While these pathogen-occupied supramolecular structures bear distinct molecular signatures, some common principles that underlie their recognition by IRGs and GBPs have emerged. We will discuss the role of ATG proteins in coordinating the targeting of IRGs and GBPs to pathogen-containing vacuoles and viral replication compartments as well as the regulatory functions of IRGs and GBPs in antimicrobial autophagy.
Autophagy proteins control the translocation of IFN-inducible GTPases to microbial replication compartments
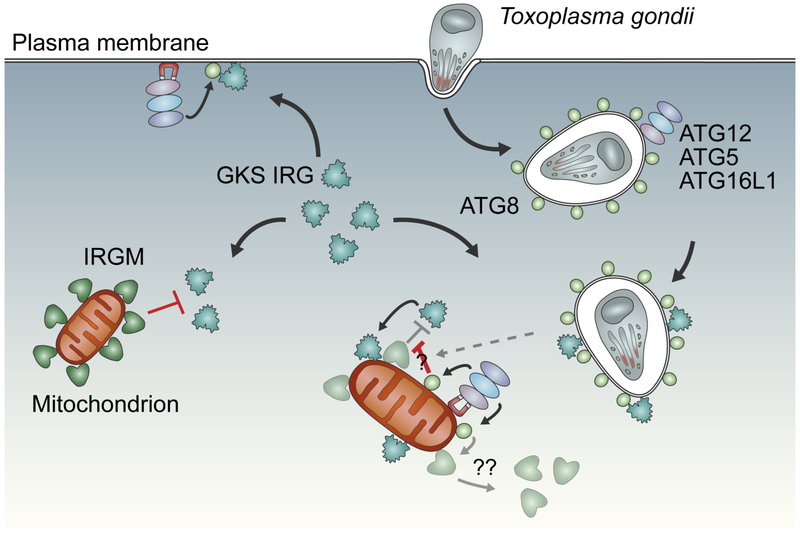
The IFN-inducible IRG resistance system of mice is highly polymorphic and is typically encoded by 10 to 20 paralogous genes in the genome of any given mouse strain [11]. Mouse IRG proteins are grouped into GKS and IRGM protein subfamilies [7,12,13]. The GKS ‘effector’ proteins are defined by a canonical GKS sequence in the nucleotide-binding pocket, which is changed to a non-canonical GMS sequence in IRGM proteins [12]. GKS proteins translocate to pathogen-containing vacuoles formed by pathogens such as Toxoplasma gondii or Chlamydia trachomatis [13-16] and promote the lytic destruction of pathogen-containing vacuoles and their occupants [17-19]. IRGM proteins regulate this translocation process, possibly through multiple mechanisms. Endomembrane-bound IRGM proteins undergo transient interactions with cytoplasmic GKS proteins to inhibit the exchange of GDP for GTP by GKS proteins [13]. Through these interactions IRGM proteins not only maintain a cytoplasmic pool of GDP-bound deployable GKS proteins [13] but also guard self-structures against off-target attacks by activated GTP-bound GKS proteins [16,20]. Whereas most endomembranes are IRGM-decorated, the paucity of IRGM on pathogen-containing vacuoles marks these microbe-occupied structures with a ‘missing-self’ pattern that permits the transition of GKS proteins into the GTP-bound, ‘membranophilic’ state and their consequential binding to pathogen-containing vacuoles [9,16] (Figure 1). However, the absence of IRGM proteins per se is insufficient to drive GKS translocation to IRGM-devoid membranes [13,20], indicating that additional ‘second’ or ‘third’ signals are required to accurately deliver GKS proteins to pathogen-containing vacuoles.
Figure 1. Multilayered regulation of GKS protein localization to pathogen-containing vacuoles.
In interferon-activated cells, the majority of endomembranes are shielded from GKS proteins by the presence of IRGM proteins. The sparsity of IRGMs at pathogen-containing vacuoles formed by T. gondii or C. trachomatis provides a ‘missing-self’ signal that permits the recruitment of GKS to these vacuoles; however, additional signals in the form of lipidated ATG8 are required to efficiently recruit GKS proteins to pathogen-containing vacuoles. In support of this ‘two-signal model’ forced localization of the ATG8 conjugation machinery (ATG12–ATG5-ATG16L1 complex) to the IRGM-free plasma membrane results in GKS recruitment at that site. In contrast, artificial placement of the ATG8 conjugation machinery at IRGM-decorated mitochondria is insufficient to recruit GKS proteins unless additional cues, like T. gondii infections, are provided. How ATG8 overrides the inhibitory function of IRGM at the mitochondria upon infection is unknown; potential mechanisms may include posttranslational modifications that block IRGM function or increase GKS affinity for ATG8-decoraetd membranes (denoted by “?”) or lead to the removal of IRGMs from mitochondrial membranes (denoted by “??”).
The first evidence for the existence of such additional signals came from studies demonstrating that ATG5 was required to deliver GKS proteins to T. gondii-containing vacuoles, resulting in gamma-interferon (IFNγ)-mediated control of T. gondii infection in tissue culture and in vivo mouse models [21]. Genetic dissection of the autophagy pathway demonstrated that GKS targeting to pathogen-containing vacuoles is dependent on the ATG8 conjugation system but independent of factors that promote autophagosome initiation and lysosomal degradation [22-26], thus indicating a noncanonical function of the ATG8 conjugation system in the delivery of GKS proteins to pathogen-containing vacuoles (Figure 1). This model was further substantiated by findings that several ATG8 homologs occupy T. gondii-containing vacuolar membranes and control GKS and GBP recruitment to pathogen-containing vacuoles [22,27,28], with a notable role for the gamma-aminobutyric acid receptor associated protein (GABARAP) subfamily of ATG8 proteins in this process [28]. Analogously, decoration of murine norovirus replication compartments with ATG8 proteins prompts GKS and GBP recruitment to replication compartments and blocks replication of murine norovirus [10], highlighting the central role for the ATG8 conjugation system in the recruitment of IFN-inducible GTPases to diverse types of microbe-occupied supramolecular structures inside infected cells. The presence of putative LC3-interacting regions in some members of the GKS and GBP protein families suggests that a subset of IFN-inducible GTPases directly interact with ATG8-decorated membranes [6] and subsequently recruit additional GKS and GBP family members to targeted membranes in a hierarchical manner. In support of this model the forced delivery of the ATG12–ATG5-ATG16L1 complex to the IRGM-devoid inner leaflet of the plasma membrane is sufficient to trigger GBP and GKS recruitment to the plasma membrane [27] (Figure 1). The engineered placement of the ATG8 conjugation machinery to IRGM-decorated mitochondria on the other hand is insufficient to mark these organelles as targets for GBPs or GKS effector GTPases, unless concomitant T. gondii infections override the inhibitory function of mitochondrial IRGM proteins [27,29]. Thus, infections may provide a third signal that strengthens the interaction of GBPs and GKS proteins with ATG8-decorated membranes. The nature and consequence of this signal is unknown but may involve posttranslational modifications that increase binding affinities between ATG8-positive membranes and its protein interaction partners [29].
Although ATG-independent mechanisms of pathogen-containing vacuolar targeting by IFN-inducible GTPases also exist [30], compelling evidence demonstrates that ATG8 lipidation of pathogen-containing vacuoles and viral replication compartments promotes the recruitment of GBPs and GKS proteins to these intracellular centers of microbial replication. However, the mechanism by which the ATG8 conjugation machinery is deployed to pathogen-containing vacuoles or replication compartments is unknown. In vitro reconstitution experiments demonstrated direct binding of recombinant ATG5 protein to liposomes in a negative charge-dependent manner [31]. Therefore, the transient association of ATG5 with T. gondii-containing vacuoles following shortly after infection [27] may similarly depend on interactions between ATG5 and negatively charged lipids present in nascent T. gondii-containing vacuoles. Alternatively, ATG5 or an ATG5 adaptor protein could directly interact with microbial molecules present on pathogen-containing vacuoles or viral replication compartments, paralleling the interactions described for ATG5 and the Shigella protein IcsA [32]. Lastly, microbe-mediated activities such as the insertion of protein secretion channels into pathogen-containing vacuolar membranes could induce aberrant-self signals that trigger the recruitment of ATG5 and IFN-inducible GTPases to pathogen-containing vacuoles [30,33]. The latter two models would account for the specificity with which the ATG8 conjugation machinery is delivered to vesicles containing pathogenic microbes.
IFN-inducible GTPases promote ubiquitination of pathogen-containing vacuoles and trigger autophagy-related defenses
GBPs are recruited to sites of intracellular microbial colonization, where they promote antimicrobial responses that eliminate the pathogen. Current evidence indicates that these antimicrobial responses are diverse, vary with the particular GBP, and may be host species-specific. They include: (i) lysis of pathogen-containing vacuoles to extricate vacuolar pathogens from their safe intracellular niches [34,35]; (ii) recruitment of antimicrobial effectors such as the NADPH oxidase to bacteria-containing phagosomes [36]; (iii) promotion of vacuole maturation and elimination through lysosomal fusion [36]; (iv) lysis of cytosolic bacteria [37-39] and (v) suppression of the actin-based motility used by some cytosolic bacteria for cell-to-cell spread [40,41]. Knowledge of GBP functioning at the molecular level is still insufficient to explain such a broad array of activities.
There is now considerable evidence that some of the antimicrobial functions of IFN-inducible GTPases are linked to their ability to regulate cellular ubiquitination events. Studies in mouse cells have shown that a sentinel event controlled by IRG proteins is the recruitment of ubiquitin E3 ligases such as TRAF6 and Trim21 to the C. trachomatis- and T. gondii-containing vacuoles [17,42,43]. The ubiquitin moieties then serve as binding sites for the ubiquitin-binding protein and autophagy receptor p62/SQSTM1, which in turn attracts mouse GBPs to pathogen-containing vacuoles [17,44]. In human cells, IFNγ cell priming similarly promotes ubiquitination of C. trachomatis- and T. gondii-containing vacuoles but in a seemingly IRG- and GBP-independent manner [45-48]. There is also evidence that GBPs may interact with autophagy proteins to drive downstream responses that occur subsequent to GBP loading on the vacuole. For instance, mouse GBP7 facilitates ATG4 recruitment to mycobacterial phagosomes [36], and GBP1 and GBP2 mediate xenophagic destruction of C. trachomatis in human macrophages [49]. However, such studies are few at this point, underscoring the lack of broad knowledge concerning the interphase between GBPs and autophagy proteins, and the manner in which this impacts their antimicrobial functions.
Mouse and human IRGM proteins regulate autophagy induction and autophagosome maturation in response to infections
As alluded to above, IRGM proteins are biochemically and functionally distinct from the GKS proteins in possessing a methionine substitution for the canonical lysine residue in the G1 GTP-binding motif [12]. Humans possess one IRGM gene that is expressed as 5 differentially spliced mRNAs (IRGMa-e) [12,50], while mice possess three distinct genes, named Irgml (previously known as LRG-47), Irgm2 (GTPI) and Irgm3 (IGTP) [12]. Interest in the IRGMs has been piqued by a large body of literature associating human IRGM gene variants with inflammatory and infectious diseases, including Crohn’s disease [51-53], non-alcoholic fatty liver disease [54], ankylosing spondylitis [55], sepsis [56], and mycobacterial infection [57,58]. Increased risks for the development of inflammatory diseases is linked to carriage of the IRGM minor allele, which encompasses several polymorphisms including a 20-kb deletion within the IRGM promoter region [52]. The various polymorphisms associated with the IRGM minor allele are in nearly complete linkage disequilibrium [59,60], making it difficult to assess the functional consequences of individual polymorphisms and their relevance to disease in the absence of more detailed studies. Because the genetic changes common to the IRGM minor allele are predominantly found outside the protein-coding region, we can nonetheless presume that these genetic alterations affect gene expression rather than protein function. This is supported by studies showing that mRNA expression of the minor allele is diminished relative to that of the major allele [56,59]; though conversely, one study proffers that the minor allele mRNA is impervious to negative regulation by mir-196, a microRNA expressed in colonic epithelial cells during inflammation [61]. Collectively, these studies suggest then that either decreases or increases in IRGM levels may promote disease dependent on the cellular context.
The degree to which human and mouse IRGMs share cellular functions has been debated [60,62,63]. Mouse and human IRGMs differ from each other in some notable ways: mouse IRGMs do possess the aforementioned GKS IRG regulatory function (described above) that human IRGM lacks. Further, expression of mouse IRGMs but not human IRGM is induced by IFN signaling [13,16,64]. In spite of these differences, a growing body of evidence suggests shared functions, particularly in modulating antimicrobial autophagy (Figure 2). Absence of either mouse IRGM1 or human IRGM leads to impaired xenophagic clearance of pathogens, especially in macrophages [20,44,50,65-67]. For human IRGM, the underlying mechanism involves the ability of the protein to associate with autophagy proteins including the Unc-51-like autophagy activating kinase 1 (ULK1) of the autophagy initiation complex and to mediate activation of this complex via phosphorylation [68-70]. Human IRGM also interacts with the bacterial peptidoglycan sensor nucleotide-binding oligomerization domain-containing protein 2 (NOD2) and ATG16L1 [68], known mediators of infection-induced autophagy [71]. Whereas these observations suggest that human IRGM plays a role in the nucleation or elongation of autophagosomes, a recent study demonstrates a role for human IRGM in the recruitment of the membrane fusion protein Syntaxin-17 to the autophagosome, with Syntaxin-17 then stimulating autophagosome-lysosome fusion [72]. In sum, these studies indicate that human IRGM controls several stages of the autophagic process in response to infectious triggers such as cell invasion-mediated NOD2 activation. The mechanism through which mouse IRGM1 or its paralogs regulate autophagy is less well studied.
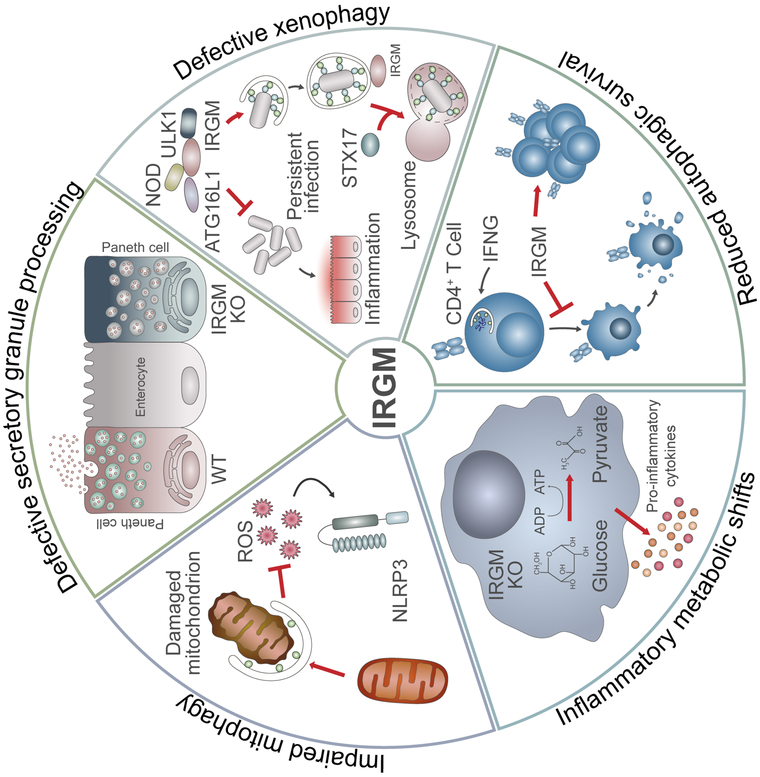
Figure 2. Functional consequences of IRGM dysregulation.
Defects in IRGM function perturb autophagy and related processes in mouse and human cells, thereby nurturing a pro-inflammatory milieu linked to Crohn’s and other diseases. The absence of IRGM results in defective processing of secretory granules in Paneth cells, defects in antimicrobial autophagy (xenophagy), disrupted T cell homeostasis and changes in mitochondrial dynamics and metabolism that lead to exacerbated cytokine production during inflammation. In many cases, defects in IRGM closely mirror defects in another autophagy gene, ATG16L1, which like IRGM is an established susceptibility factor for the development of Crohn’s disease. WT = wild type; KO = knockout; STX17 = Syntaxin-17; IFNG = gamma-interferon; ROS = radical oxygen species.
There is evidence that altered autophagy regulation as a consequence of human IRGM or mouse IRGM1 deficiencies promotes the onset of inflammatory diseases such as colitis through multiple mechanisms (Figure 2). (i) Xenophagic removal of bacterial pathogens is defective in the absence of human IRGM or mouse IRGM1 [7,65-67,73], which may lead to persistent infections with pathogens or pathobionts, thereby causing chronic intestinal inflammation. (ii) Absence of IRGM1 leads to alterations in autophagic processing of the secretory granules in intestinal Paneth cells [74], the components of which are important for homeostasis of bacterial populations in the ileum. This connection between autophagic regulation of Paneth cells and Crohn’s disease was first established by the identification of another gene, ATG16L1, as a Crohn’s disease susceptibility gene [51,75], and by the phenotype of Atg16l1 hypomorphic mice [76,77] that parallels that of Irgm1-deficient mice with respect to altered Paneth cell function and increased susceptibility to dextran sodium sulfate-induced colitis [74]. (iii) Decreased levels of human IRGM expression or disruption of Irgm1 gene function in mice also lead to altered mitochondrial dynamics [50,74,78], suggesting a possible role for IRGM proteins in the clearance of mitochondria via mitophagy. Components of damaged mitochondria that are not removed through mitophagy are known to activate the Nucleotide-binding oligomerization domain-like receptor pyrin domain-containing-3 (NLRP3) inflammasome and to trigger inflammation [79,80]. Further, diminished mitophagy has also been connected to altered Paneth cell function [81]. (iv) Irgm1 deletion prompts striking metabolic changes in mouse cells, particularly increases in glycolysis, that lead to escalated production of proinflammatory cytokines [78]. Again, a parallel exists with ATG16L1, as impaired autophagy through its deficiency also leads to increases in glycolysis and cytokine production [77,82,83]. (v) IRGM1 regulates autophagic survival of proliferating T cells during immune activation and accordingly Irgm1-deficient mice become lymphopenic in response to infections [84-87]. Crohn’s disease is well documented to be a T cell-driven disease, and further, autophagy deficiencies disrupt the functional integrity of the regulatory T cell compartment leading to increased intestinal inflammation in mouse models [82,83]. Clearly, much work is needed to clarify which of these mechanisms are relevant to inflammatory diseases influenced by IRGM proteins, as well as to determine whether phenotypes associated with IRGM1 deficiency in mice are also present in humans carrying IRGM disease variants, and vice versa.
Conclusions
The innate immune system employs pattern recognition receptors to detect the presence of invasive microbes. While the field of immunology has made tremendous progress in our understanding of how this antimicrobial alarm system is activated [88], we know far less about how the innate immune system subsequently ‘handcuffs’ microbial intruders, many of which hide within infected host cells. The latter task requires for the host to detect the microbe’s precise location inside a host cell. IFN-inducible dynamin-like GTPases and autophagy proteins have emerged as cooperative partners in host defense that are able to localize and destroy intracellular pathogens; IRGs and GBPs can detect ‘non-self,’ ‘aberrant-self’ and ‘missing-self’ patterns associated with intracellular microbes [9,16,30,40]; autophagy proteins are also able to detect the location of pathogen-occupied supramolecular structures independent of IFN-inducible GTPases [10,27,32] and subsequently recruit GBPs and GKS proteins via ATG8-lipidated membranes. Nevertheless, a molecular and biochemical appreciation of these processes is still lacking and the underlying mechanisms need to be defined.
Once bound to their microbial targets, GBPs and mouse GKS proteins solicit a diverse repertoire of defense programs that include the ubiquitination and autophagic destruction of the captured microbes [17,36,42,49]. Human IRGM and its mouse orthologs appear to intersect with the autophagy machinery in a different manner. While dispensable for canonical autophagy [70,89], IRGM proteins promote the formation and maturation of autophagosomes in response to microbial stimuli [50,66-69,72]. Recent evidence reveals that human IRGM promotes autolysosome formation by assisting in the recruitment of the vesicle fusion protein Syntaxin-17 to immature autophagosomes [72]. Whether other human IRGM isoforms or their murine orthologs also act as adaptors for Syntaxin-17 or instead function as adaptors for other membrane remodeling proteins remains to be tested. Nonetheless, the physical and functional interactions between IRGM and several autophagy proteins set a framework for the future exploration of the role of IRGM isoforms in autophagy-related host defense programs.
Acknowledgements
This work was supported by National Institute Health grants R01AI103197 (to JC), T32AI007090 (to HMB), R01AI127518 and DP2CA225208 (to SH) and Veterans Administration grant I01 BX002369 (to GT). JC holds an Investigator in the Pathogenesis of Infectious Disease Awards from the Burroughs Wellcome Fund.
References
- 1.Gatica D, Lahiri V, Klionsky DJ: Cargo recognition and degradation by selective autophagy. Nat Cell Biol 2018, 20:233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine B: Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 2005, 120:159–162. [DOI] [PubMed] [Google Scholar]
- 3.Sil P, Muse G, Martinez J: A ravenous defense: canonical and non-canonical autophagy in immunity. Curr Opin Immunol 2018, 50:21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabouille C: Pathways of Unconventional Protein Secretion. Trends Cell Biol 2017, 27:230–240. [DOI] [PubMed] [Google Scholar]
- 5.Munz C: The Autophagic Machinery in Viral Exocytosis. Front Microbiol 2017, 8:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown HM, Biering SB, Zhu A, Choi J, Hwang S: Demarcation of Viral Shelters Results in Destruction by Membranolytic GTPases: Antiviral Function of Autophagy Proteins and Interferon-Inducible GTPases. Bioessays 2018:e1700231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pilla-Moffett D, Barber MF, Taylor GA, Coers J: Interferon-inducible GTPases in host resistance, inflammation and disease. J Mol Biol 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haller O, Staeheli P, Schwemmle M, Kochs G: Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol 2015, 23:154–163. [DOI] [PubMed] [Google Scholar]
- 9.Coers J: Self and Non-self Discrimination of Intracellular Membranes by the Innate Immune System. PLoS Pathog 2013, 9:e1003538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *10.Biering SB, Choi J, Halstrom RA, Brown HM, Beatty WL, Lee S, McCune BT, Dominici E, Williams LE, Orchard RC, et al. : Viral Replication Complexes Are Targeted by LC3-Guided Interferon-Inducible GTPases. Cell Host Microbe 2017, 22:74–85 e77. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors demonstrate that the replication compartment of viruses is targeted and disrupted by IFN-inducible GTPases in an ATG-dependent manner.
- 11.Lilue J, Muller UB, Steinfeldt T, Howard JC: Reciprocal virulence and resistance polymorphism in the relationship between Toxoplasma gondii and the house mouse. Elite 2013, 2:e01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bekpen C, Hunn JP, Rohde C, Parvanova I, Guethlein L, Dunn DM, Glowalla E, Leptin M, Howard JC: The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol 2005, 6:R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunn JP, Koenen-Waisman S, Papic N, Schroeder N, Pawlowski N, Lange R, Kaiser F, Zerrahn J, Martens S, Howard JC: Regulatory interactions between IRG resistance GTPases in the cellular response to Toxoplasma gondii. EMBO J 2008, 27:2495–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coers J, Bernstein-Hanley I, Grotsky D, Parvanova I, Howard JC, Taylor GA, Dietrich WF, Starnbach MN: Chlamydia muridarum evades growth restriction by the IFN-gamma-inducible host resistance factor Irgb10. J Immunol 2008, 180:6237–6245. [DOI] [PubMed] [Google Scholar]
- 15.Degrandi D, Konermann C, Beuter-Gunia C, Kresse A, Wurthner J, Kurig S, Beer S, Pfeffer K: Extensive characterization of IFN-induced GTPases mGBP1 to mGBP10 involved in host defense. J Immunol 2007, 179:7729–7740. [DOI] [PubMed] [Google Scholar]
- 16.Haldar AK, Saka HA, Piro AS, Dunn JD, Henry SC, Taylor GA, Frickel EM, Valdivia RH, Coers J: IRG and GBP host resistance factors target aberrant, “non-self” vacuoles characterized by the missing of “self” IRGM proteins. PLoS Pathog 2013, 9:e1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haldar AK, Foltz C, Finethy R, Piro AS, Feeley EM, Pilla-Moffett DM, Komatsu M, Frickel EM, Coers J: Ubiquitin systems mark pathogen-containing vacuoles as targets for host defense by guanylate binding proteins. Proc Natl Acad Sci U S A 2015, 112:E5628–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ling YM, Shaw MH, Ayala C, Coppens I, Taylor GA, Ferguson DJ, Yap GS: Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J Exp Med 2006, 203:2063–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martens S, Parvanova I, Zerrahn J, Griffiths G, Schell G, Reichmann G, Howard JC: Disruption of Toxoplasma gondii parasitophorous vacuoles by the mouse p47-resistance GTPases. PLoS Pathog 2005, 1:e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maric-Biresev J, Hunn JP, Krut O, Helms JB, Martens S, Howard JC: Loss of the interferon-gamma-inducible regulatory immunity-related GTPase (IRG), Irgm1, causes activation of effector IRG proteins on lysosomes, damaging lysosomal function and predicting the dramatic susceptibility of Irgm1-deficient mice to infection. BMC Biol 2016, 14:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, et al. : Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 2008, 4:458–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi J, Park S, Biering SB, Selleck E, Liu CY, Zhang X, Fujita N, Saitoh T, Akira S, Yoshimori T, et al. : The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity 2014, 40:924–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haldar Ak, Piro AS, Pilla DM, Yamamoto M, Coers J: The E2-like conjugation enzyme Atg3 promotes binding of IRG and Gbp proteins to Chlamydia- and Toxoplasma-containing vacuoles and host resistance. PLoS One 2014, 9:e86684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khaminets A, Hunn JP, Konen-Waisman S, Zhao YO, Preukschat D, Coers J, Boyle JP, Ong YC, Boothroyd JC, Reichmann G, et al. : Coordinated loading of IRG resistance GTPases on to the Toxoplasma gondii parasitophorous vacuole. Cell Microbiol 2010, 12:939–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohshima J, Lee Y, Sasai M, Saitoh T, Su Ma J, Kamiyama N, Matsuura Y, Pann-Ghill S, Hayashi M, Ebisu S, et al. : Role of mouse and human autophagy proteins in IFN-gamma-induced cell-autonomous responses against Toxoplasma gondii. J Immunol 2014, 192:3328–3335. [DOI] [PubMed] [Google Scholar]
- 26.EM Selleck, Fentress SJ, Beatty WL, Degrandi D, Pfeffer K, Virgin HWt, Macmicking JD, Sibley LD: Guanylate-binding protein 1 (Gbp1) contributes to cell-autonomous immunity against Toxoplasma gondii. PLoS Pathog 2013, 9:e1003320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **27.Park S, Choi J, Biering SB, Dominici E, Williams LE, Hwang S: Targeting by AutophaGy proteins (TAG): Targeting of IFNG-inducible GTPases to membranes by the lC3 conjugation system of autophagy. Autophagy 2016, 12:1153–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]; By deliberate placement of the ATG8 conjugation machinery to the plasma membrane and mitochondrial outer membrane, the authors demonstrate that autophagy proteins are necessary and sufficient to control the translocation of IFN-inducible GTPases to membranes.
- *28.Sasai M, Sakaguchi N, Ma JS, Nakamura S, Kawabata T, Bando H, Lee Y, Saitoh T, Akira S, Iwasaki A, et al. : Essential role for GABARAP autophagy proteins in interferon-inducible GTPase-mediated host defense. Nat Immunol 2017, 18:899–910. [DOI] [PubMed] [Google Scholar]; Even though all known mammalian homologs of ATG8 are translocated onto the parasitophorus vacuole membrane of T. gondii, the authors demonstrate that only GABARAP proteins play an essential role in controlling the function of IFN-inducible GTPases.
- 29.Choi J, Biering SB, Hwang S: Quo vadis? Interferon-inducible GTPases go to their target membranes via the LC3-conjugation system of autophagy. Small GTPases 2016:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **30.Feeley EM, Pilla-Moffett DM, Zwack EE, Piro AS, Finethy R, Kolb JP, Martinez J, Brodsky IE, Coers J: Galectin-3 directs antimicrobial guanylate binding proteins to vacuoles furnished with bacterial secretion systems. Proc Natl Acad Sci U S A 2017, 114:E1698–E1706. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that GBPs detect the aberrant cytosolic exposure of host glycans normally confined to the luminal side of host vacuoles. This detection program is ATG-independent and induced by bacterial translocons that serve as conduits for the secretion of bacterial virulence factors across eukaryotic membranes.
- 31.Romanov J, Walczak M, Ibiricu I, Schuchner S, Ogris E, Kraft C, Martens S: Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J 2012, 31:4304–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C: Escape of intracellular Shigella from autophagy. Science 2005, 307:727–731. [DOI] [PubMed] [Google Scholar]
- 33.Coers J: Sweet host revenge: Galectins and GBPs join forces at broken membranes. Cell Microbiol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meunier E, Dick MS, Dreier RF, Schurmann N, Kenzelmann Broz D, Warming S, Roose-Girma M, Bumann D, Kayagaki N, Takeda K, et al. : Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature 2014, 509:366–370. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto M, Okuyama M, Ma JS, Kimura T, Kamiyama N, Saiga H, Ohshima J, Sasai M, Kayama H, Okamoto T, et al. : A Cluster of Interferon-gamma-Inducible p65 GTPases Plays a Critical Role in Host Defense against Toxoplasma gondii. Immunity 2012. [DOI] [PubMed] [Google Scholar]
- 36.Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD: A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science 2011, 332:717–721. [DOI] [PubMed] [Google Scholar]
- **37.Li P, Jiang W, Yu Q, Liu W, Zhou P, Li J, Xu J, Xu B, Wang F, Shao F: Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature 2017. [DOI] [PubMed] [Google Scholar]; Together with Refs. [40**,41**] this study identifies the secreted Shigella protein IpaH9.8 as an inhibitor of human GBP1 and its homologs. Li et al. and Wandel et al. [41**] further characterize IpaH9.8 as an ubiquitin E3 ligase that tags GBPs for proteasomal degradation.
- 38.Man SM, Karki R, Malireddi RK, Neale G, Vogel P, Yamamoto M, Lamkanfi M, Kanneganti TD: The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat Immunol 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Ruhl S, Dussurgey S, Dick MS, Kistner A, Rigard M, et al. : Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **40.Piro AS, Hernandez D, Luoma S, Feeley EM, Finethy R, Yirga A, Frickel EM, Lesser CF, Coers J: Detection of Cytosolic Shigella flexneri via a C-Terminal Triple-Arginine Motif of GBP1 Inhibits Actin-Based Motility. MBio 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides genetic evidence that human GBP1 through its unique C-terminal protein motif detects non-self bacterial lipopolysaccharide on the surface of cytosolic Shigella. Furthermore, this study as well as Wandel et al. [41**] demonstrate that human GBP1 bound to Shigella blocks actin-based bacterial motility and bacterial cell-to-cell spread.
- **41.Wandel MP, Pathe C, Werner EI, Ellison CJ, Boyle KB, von der Malsburg A, Rohde J, Randow F: GBPs Inhibit Motility of Shigella flexneri but Are Targeted for Degradation by the Bacterial Ubiquitin Ligase IpaH9.8. Cell Host Microbe 2017, 22:507–518 e505. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Refs. [37**,40**].
- 42.Lee Y, Sasai M, Ma JS, Sakaguchi N, Ohshima J, Bando H, Saitoh T, Akira S, Yamamoto M: p62 Plays a Specific Role in Interferon-gamma-Induced Presentation of a Toxoplasma Vacuolar Antigen. Cell Rep 2015, 13:223–233. [DOI] [PubMed] [Google Scholar]
- 43.Foltz C, Napolitano A, Khan R, Clough B, Hirst EM, Frickel EM: TRIM21 is critical for survival of Toxoplasma gondii infection and localises to GBP-positive parasite vacuoles. Sci Rep 2017, 7:5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Traver MK, Henry SC, Cantillana V, Oliver T, Hunn JP, Howard JC, Beer S, Pfeffer K, Coers J, Taylor GA: Immunity-related GTPase M (IRGM) proteins influence the localization of guanylate-binding protein 2 (GBP2) by modulating macroautophagy. J Biol Chem 2011, 286:30471–30480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clough B, Wright JD, Pereira PM, Hirst EM, Johnston AC, Henriques R, Frickel EM: K63-Linked Ubiquitination Targets Toxoplasma gondii for Endo-lysosomal Destruction in IFNgamma-Stimulated Human Cells. PLoS Pathog 2016, 12:e1006027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selleck EM, Orchard RC, Lassen KG, Beatty WL, Xavier RJ, Levine B, Virgin HW, Sibley LD: A Noncanonical Autophagy Pathway Restricts Toxoplasma gondii Growth in a Strain-Specific Manner in IFN-gamma-Activated Human Cells. MBio 2015, 6:e01157–01115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coers J, Haldar AK: Ubiquitination of pathogen-containing vacuoles promotes host defense to Chlamydia trachomatis and Toxoplasma gondii. Communicative & Integrative Biology 2015, 8:e1115163–1115161–1115163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haldar AK, Piro AS, Finethy R, Espenschied ST, Brown HE, Giebel AM, Frickel EM, Nelson DE, Coers J: Chlamydia trachomatis Is Resistant to Inclusion Ubiquitination and Associated Host Defense in Gamma Interferon-Primed Human Epithelial Cells. MBio 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Al-Zeer MA, Al-Younes HM, Lauster D, Abu Lubad M, Meyer TF: Autophagy restricts Chlamydia trachomatis growth in human macrophages via IFNG-inducible guanylate binding proteins. Autophagy 2013, 9:50–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E, Ponpuak M, Master S, Pilli M, White E, et al. : Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol 2010, 12:1154–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Massey DC, Parkes M: Genome-wide association scanning highlights two autophagy genes, ATG16L1 and IRGM, as being significantly associated with Crohn’s disease. Autophagy 2007, 3:649–651. [DOI] [PubMed] [Google Scholar]
- 52.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, et al. : Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D, et al. : Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet 2007, 39:830–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin YC, Chang PF, Lin HF, Liu K, Chang MH, Ni YH: Variants in the autophagy-related gene IRGM confer susceptibility to non-alcoholic fatty liver disease by modulating lipophagy. J Hepatol 2016, 65:1209–1216. [DOI] [PubMed] [Google Scholar]
- 55.Xia Q, Wang M, Yang X, Li X, Zhang X, Xu S, Shuai Z, Xu J, Fan D, Ding C, et al. : Autophagy-related IRGM genes confer susceptibility to ankylosing spondylitis in a Chinese female population: a case-control study. Genes Immun 2017, 18:42–47. [DOI] [PubMed] [Google Scholar]
- 56.Kimura T, Watanabe E, Sakamoto T, Takasu O, Ikeda T, Ikeda K, Kotani J, Kitamura N, Sadahiro T, Tateishi Y, et al. : Autophagy-related IRGM polymorphism is associated with mortality of patients with severe sepsis. PLoS One 2014, 9:e91522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Intemann CD, Thye T, Niemann S, Browne EN, Amanua Chinbuah M, Enimil A, Gyapong J, Osei I, Owusu-Dabo E, Helm S, et al. : Autophagy gene variant IRGM −261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog 2009, 5:e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.King KY, Lew JD, Ha NP, Lin JS, Ma X, Graviss EA, Goodell MA: Polymorphic allele of human IRGM1 is associated with susceptibility to tuberculosis in African Americans. PLoS One 2011, 6:e16317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, et al. : Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet 2008. 40:1107–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bekpen C, Xavier RJ, Eichler EE: Human IRGM gene “to be or not to be”. Semin Immunopathol 2010, 32:437–444. [DOI] [PubMed] [Google Scholar]
- 61.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hebuterne X, et al. : A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet 2011, 43:242–245. [DOI] [PubMed] [Google Scholar]
- 62.Coers J, Starnbach MN, Howard JC: Modeling infectious disease in mice: co-adaptation and the role of host-specific IFNgamma responses. PLoS Pathog 2009. 5:e1000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hunn JP, Feng CG, Sher A, Howard JC: The immunity-related GTPases in mammals: a fast-evolving cell-autonomous resistance system against intracellular pathogens. Mamm Genome 2011, 22:43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Henry SC, Daniell XG, Burroughs AR, Indaram M, Howell DN, Coers J, Starnbach MN, Hunn JP, Howard JC, Feng CG, et al. : Balance of Irgm protein activities determines IFN-{gamma}-induced host defense. J Leukoc Biol 2009, 85:877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V: Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004, 119:753–766. [DOI] [PubMed] [Google Scholar]
- 66.Singh SB, Davis AS, Taylor GA, Deretic V: Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 2006, 313:1438–1441. [DOI] [PubMed] [Google Scholar]
- 67.Lapaquette P, Glasser AL, Huett A, Xavier RJ, Darfeuille-Michaud A: Crohn’s disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell Microbiol 2010, 12:99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chauhan S, Mandell MA, Deretic V: IRGM Governs the Core Autophagy Machinery to Conduct Antimicrobial Defense. Mol Cell 2015, 58:507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *69.Hansen MD, Johnsen IB, Stiberg KA, Sherstova T, Wakita T, Richard GM, Kandasamy RK, Meurs EF, Anthonsen MW: Hepatitis C virus triggers Golgi fragmentation and autophagy through the immunity-related GTPase M. Proc Natl Acad Sci U S A 2017, 114:E3462–E3471. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors establish a novel role for IRGM in autophagy-mediated membrane remodeling, showing that human IRGM localizes to the Golgi apparatus where it triggers Golgi membrane fragmentation in response to hepatitic C virus that supports viral replication.
- 70.Gregoire IP, Richetta C, Meyniel-Schicklin L, Borel S, Pradezynski F, Diaz O, Deloire A, Azocar O, Baguet J, Le Breton M, et al. : IRGM is a common target of RNA viruses that subvert the autophagy network. PLoS Pathog 2011, 7:e1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, et al. : Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol 2010, 11:55–62. [DOI] [PubMed] [Google Scholar]
- **72.Kumar S, Jain A, Farzam F, Jia J, Gu Y, Choi SW, Mudd MH, Claude-Taupin A, Wester MJ, Lidke KA, et al. : Mechanism of Stx17 recruitment to autophagosomes via IRGM and mammalian Atg8 proteins. J Cell Biol 2018, 217:997–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that human IRGM physically interacts with ATG8 and the membrane fusion protein Syntaxin-17 to form an autophagsome recognition particle required for efficient autophagosome maturation.
- 73.Henry SC, Daniell X, Indaram M, Whitesides JF, Sempowski GD, Howell D, Oliver T, Taylor GA: Impaired macrophage function underscores susceptibility to Salmonella in mice lacking Irgm1 (LRG-47). J Immunol 2007, 179:6963–6972. [DOI] [PubMed] [Google Scholar]
- 74.Liu B, Gulati AS, Cantillana V, Henry SC, Schmidt EA, Daniell X, Grossniklaus E, Schoenborn AA, Sartor RB, Taylor GA: Irgm1-deficient mice exhibit Paneth cell abnormalities and increased susceptibility to acute intestinal inflammation. Am J Physiol Gastrointest Liver Physiol 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al. : A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet 2007, 39:207–211. [DOI] [PubMed] [Google Scholar]
- 76.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. : A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008, 456:259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. : Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 2008, 456:264–268. [DOI] [PubMed] [Google Scholar]
- *78.Schmidt EA, Fee BE, Henry SC, Nicholsa AG, Shinohara ML, Rathmell JC, MacIver NJ, Coers J, Ilkayeva OR, Koves TR, et al. : Metabolic alterations contribute to enhanced inflammatory cytokine production in Irgm1-deficient macrophages. J Biol Chem 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show for the first time that perturbations in IRGM protein function leads to altered metabolism that promotes inflammation. Metabolic analyses demonstrated that Irgm1-deficient macrophages manifest increased glycolysis and accumulation of long chain acyl carnitines, both of which contribute to increased production of inflammatory cytokines in absence of infection.
- 79.Mills EL, Kelly B, O’Neill LAJ: Mitochondria are the powerhouses of immunity. Nat Immunol 2017, 18:488–498. [DOI] [PubMed] [Google Scholar]
- 80.Prochnicki T, Latz E: Inflammasomes on the Crossroads of Innate Immune Recognition and Metabolic Control. Cell Metab 2017, 26:71–93. [DOI] [PubMed] [Google Scholar]
- 81.Matsuzawa-Ishimoto Y, Shono Y, Gomez LE, Hubbard-Lucey VM, Cammer M, Neil J, Dewan MZ, Lieberman SR, Lazrak A, Marinis JM, et al. : Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J Exp Med 2017, 214:3687–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wei J, Long L, Yang K, Guy C, Shrestha S, Chen Z, Wu C, Vogel P, Neale G, Green DR, et al. : Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat Immunol 2016, 17:277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kabat AM, Harrison OJ, Riffelmacher T, Moghaddam AE, Pearson CF, Laing A, Abeler-Dorner L, Forman SP, Grencis RK, Sattentau Q, et al. : The autophagy gene Atg16l1 differentially regulates Treg and TH2 cells to control intestinal inflammation. Elite 2016, 5:e12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Feng CG, Weksberg DC, Taylor GA, Sher A, Goodell MA: The p47 GTPase Lrg-47 (Irgm1) Links Host Defense and Hematopoietic Stem Cell Proliferation. Cell Stem Cell 2008, 2:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.King KY, Baldridge MT, Weksberg DC, Chambers SM, Lukov GL, Wu S, Boles NC, Jung SY, Qin J, Liu D, et al. : Irgm1 protects hematopoietic stem cells by negative regulation of IFN signaling. Blood 2011, 118:1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Feng CG, Zheng L, Jankovic D, Bafica A, Cannons JL, Watford WT, Chaussabel D, Hieny S, Caspar P, Schwartzberg PL, et al. : The immunity-related GTPase Irgm1 promotes the expansion of activated CD4+ T cell populations by preventing interferon-gamma-induced cell death. Nat Immunol 2008, 9:1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Coers J, Gondek DC, Olive AJ, Rohlfing A, Taylor GA, Starnbach MN: Compensatory T Cell Responses in IRG-Deficient Mice Prevent Sustained Chlamydia trachomatis Infections. PLoS Pathog 2011, 7:e1001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brubaker Sw, Bonham KS, Zanoni I, Kagan JC: Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol 2015, 33:257–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Matsuzawa T, Kim BH, Shenoy AR, Kamitani S, Miyake M, Macmicking JD: IFN-gamma Elicits Macrophage Autophagy via the p38 MAPK Signaling Pathway. J Immunol 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]