Significance
Genetic studies have shown that mutations of conserved Asp residues in three analogous heterogeneous ribonucleoproteins are causative of three neurological diseases. All three Asp residues map to domains of low complexity (LC) or intrinsic disorder. These domains form labile self-associated polymers as normal functional states, and the mutations abnormally enhance the stability of the polymers via heretofore unknown mechanisms. The present study gives evidence that the charged Asp residues are closely aligned in the polymer core and removal of electrostatic repulsion enhances polymer stability. These results may provide insight into other neurodegenerative diseases also caused by mutations in LC domains.
Keywords: low-complexity sequence, hnRNPA2, neurodegenerative disease, cross-beta polymer, solid-state NMR
Abstract
Human genetic studies have given evidence of familial, disease-causing mutations in the analogous amino acid residue shared by three related RNA binding proteins causative of three neurological diseases. Alteration of aspartic acid residue 290 of hnRNPA2 to valine is believed to predispose patients to multisystem proteinopathy. Mutation of aspartic acid 262 of hnRNPA1 to either valine or asparagine has been linked to either amyotrophic lateral sclerosis or multisystem proteinopathy. Mutation of aspartic acid 378 of hnRNPDL to either asparagine or histidine has been associated with limb girdle muscular dystrophy. All three of these aspartic acid residues map to evolutionarily conserved regions of low-complexity (LC) sequence that may function in states of either intrinsic disorder or labile self-association. Here, we present a combination of solid-state NMR spectroscopy with segmental isotope labeling and electron microscopy on the LC domain of the hnRNPA2 protein. We show that, for both the wild-type protein and the aspartic acid 290-to-valine mutant, labile polymers are formed in which the LC domain associates into an in-register cross-β conformation. Aspartic acid 290 is shown to be charged at physiological pH and immobilized within the polymer core. Polymers of the aspartic acid 290-to-valine mutant are thermodynamically more stable than wild-type polymers. These observations give evidence that removal of destabilizing electrostatic interactions may be responsible for the increased propensity of the mutated LC domains to self-associate in disease-causing conformations.
The proteomes of all life forms consist predominantly of self-folding proteins. Exceptions to this norm include polypeptide regions typified by a skewed distribution of amino acids. Instead of deploying a normal sampling of the 20 aa that guide proteins to adopt distinct molecular folds, regions of attenuated sequence complexity are found in an enigmatic class of proteins believed to function in the absence of molecular order. These low-complexity (LC) domains are composed of a gibberish-like distribution of only one or a few types of amino acids. Genetic studies of familial neurodegenerative disease have identified mutations that often map to regions of low sequence complexity or intrinsic disorder (1, 2). Numerous studies have commonly concluded that disease-causing mutations favor aberrant polymerization of LC domains, prompting a transition from the state of intrinsic disorder to pathogenic polymers (3–7).
Among many examples of disease-causing mutations that map to LC regions, a perplexing, recurrent mutation has been found in the analogous amino acid residue of three related heterogeneous ribonucleoproteins (hnRNPs) (Table 1 and SI Appendix, Fig. S1). Taylor and coworkers (3) first described two unrelated families suffering from a heritable form of multisystem proteinopathy with symptoms indistinguishable from ALS and frontotemporal dementia (FTD). Surprisingly, the same mutation was found in both pedigrees, a missense mutation causing aspartic acid residue 290 to be changed to valine within the LC domain of the hnRNPA2 RNA-binding protein. Resequencing of hnRNP genes in other pedigrees of families suffering from either ALS or FTD led to the discovery of the analogous residue mutated in a distinct gene. In the latter cases, the lesion changed aspartic acid residue 262 within the LC domain of the hnRNPA1 RNA-binding protein to either valine or asparagine. Kunkel and coworkers (8) later reported pedigree studies of two unrelated South American families suffering from limb-girdle muscular dystrophy. Whole exome sequencing led to the identification of the causative mutation in both of the unrelated families as independent lesions of the analogous aspartic acid residue, in this case residue 378 within the LC domain of the hnRNPDL RNA-binding protein. Why is it that, over and over, missense mutations altering this same aspartic acid residue are causative of neurological disease?
Table 1.
Disease mutations in hnRNP proteins
| Gene product | Disease | Causative mutation |
| hnRNPA1 | ALS/multisystem proteinopathy | D262 → V(3), D262 → N(3) |
| hnRNPA2 | Multisystem proteinopathy | D290 → V(3) |
| hnRNPDL | Limb-girdle muscular dystrophy | D378 → N(8), D378 → H(8) |
Taylor and coworkers (3) found that the D-to-V mutation in hnRNPA2 prompts enhanced formation of amyloid-like aggregates, and the same observation was recently reported by Fawzi and coworkers (9). Subsequent, corroborative observations gave evidence that the aspartic acid mutations in all three proteins, hnRNPA2, hnRNPA1 and hnRNPDL, cause their respective LC domains to oligomerize into cross-β polymers having enhanced stability as measured by semidenaturing detergent agarose gel electrophoresis (10). Observations that the D290V mutation of hnRNPA2 favors cross-β polymerization as its basis of enhancing disease susceptibility can be interpreted in at least two ways. One interpretation is that the LC domain of hnRNPA2 only adopts polymeric interactions under pathologic conditions, prompted in this case by the D290V mutation. Alternatively, the LC domain of hnRNPA2 may normally adopt labile, cross-β interactions as part of its biological function. The latter interpretation offers that disposition of the aspartic acid residue commonly mutated in all three forms of neurological disease normally serves to balance the kinetics and thermodynamic stability of formation of labile, cross-β interactions.
N-Acetylimidazole (NAI) footprinting studies of the hnRNPA2 protein described the location of the cross-β core of the hnRNPA2 LC domain, allowing recognition that the disease-causing, D290V missense mutation maps within the polymer-forming core (11). NAI footprinting was further used to probe the conformation of the hnRNPA2 LC domain in the nuclei of mammalian cells, giving evidence that the native form of the protein is assembled in the same polymeric conformation as characterized for purified, recombinant protein.
Here, we report measurements by solid-state NMR spectroscopy with segmental isotope labeling and by electron microscopy to obtain a more refined understanding of the structural disposition of D290 within the LC domain of hnRNPA2. We show that D290 resides within the rigid core of hnRNPA2 LC domain polymers as part of an in-register parallel cross-β structure. We further show that this aspartic acid residue is negatively charged at physiological pH. Polymers bearing the D290V mutation are more thermodynamically stable than wild type polymer and are also organized into an in-register parallel cross-β structure. Our observations are consistent with the D290V mutation stabilizing polymers via the removal of electrostatic repulsion within the polymer core. We offer that human genetic studies have thus provided valuable help in understanding the physical forces that have evolved to properly balance a widely used category of labile, protein–protein interactions important for basic cellular function.
Results
Segmental Labeling of the hnRNPA2 LC Domain.
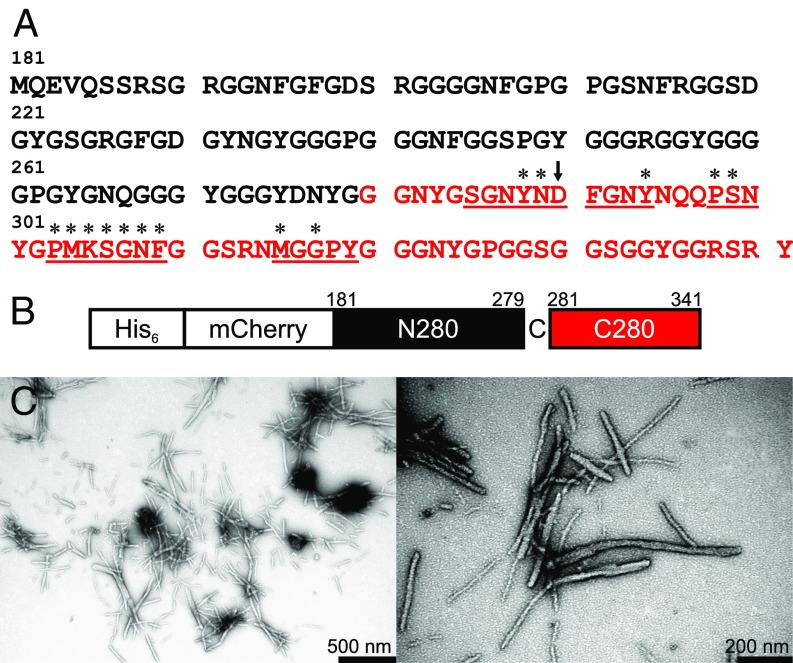
Fig. 1A shows the amino acid sequence of the hnRNPA2-LC domain, comprised of residues 181–341 of full-length hnRNPA2. Only 13 out of the 20 standard amino acids are present in hnRNPA2-LC, with Asn, Gly, Ser, and Tyr residues making up 74% of the sequence. The complete amino acid composition is listed in SI Appendix, Table S1.
Fig. 1.
Segmental labeling of hnRNPA2-LC polymers. (A) Wild-type amino acid sequence of the LC domain of hnRNPA2, residues 181–314. Isotopic labeling for solid-state NMR measurements on segmentally labeled hnRNPA2-LC polymers was restricted to the C-terminal segment shown in red. Residues with assigned solid-state NMR signals are underlined. Residues with β-strand backbone torsion angles determined by the experiments in this paper have an asterisk above them. (B) Schematic of the segmental labeling scheme. The N-terminal hnRNPA2-LC segment has 6x His and mCherry tags. The Gly-to-Cys mutation at residue 280 is expressed with the C-terminal segment. (C) TEM images of segmentally 13C,15N-labeled mCherry-hnRNPA2-LC polymers, negatively stained with uranyl acetate.
Fig. 1B and SI Appendix, Fig. S2 illustrate the segmental isotopic labeling scheme used in the solid-state NMR experiments presented below, which results in 15N and 13C labeling of residues 280–341, with a single Gly-to-Cys substitution at residue 280. The hnRNPA2 LC domain is expressed in two parts and then joined through chemical ligation. 13C and 15N labeling of only the C-terminal segment reduces the number of NMR-visible carboxylate sites from seven (one Glu, five Asp, and the C terminus) to two (the D290 Cγ site and the C-terminal carboxylate at Y341). The total number of isotopically labeled residues is reduced from 161 to 62. The amino acid composition of the C-terminal segment (residues 280–341) is highly similar to that of the full hnRNPA2-LC sequence (SI Appendix, Table S1) and contains all residues identified in NAI footprinting experiments as important for hnRNPA2-LC assembly into cross-β protein polymers (11). The ligated construct contains N-terminal 6x His and mCherry tags and is referred to as mCherry-hnRNPA2-LC throughout the remainder of this paper.
SI Appendix, Fig. S3A shows a hydrogel binding assay for N- and C-terminal truncated hnRNPA2-LC tagged with GFP, which reports on the ability of the hnRNPA2-LC protein to self-associate. Residues 181–260 do not bind to hydrogels of the mCherry-tagged full-length hnRNPA2-LC and do not form polymers under the same conditions as the full-length LC domain. Conversely, residues 261–341, containing the residues protected in the NAI footprinting assay in hydrogels and nuclear structures (11), bind to hydrogels of the mCherry-tagged full-length hnRNPA2-LC and form polymers that are visually similar to polymers of full-length hnRNPA2-LC in electron micrographs (SI Appendix, Fig. S3B). Fig. 1C shows transmission electron microscopy (TEM) images of negatively stained protein polymers formed by mCherry-hnRNPA2-LC. Throughout this paper we have employed the term “polymer” to refer to amyloid-like fibrils assembled from mCherry fusion proteins linked to the LC domain of hnRNPA2. The polymers are straight, unbranched, and a few nanometers wide, visually similar to polymers formed by Aβ (12), α-synuclein (13), full-length hnRNPA2 (3), mCherry-tagged fused in sarcoma (FUS) (14), and the His-tagged FUS LC domain (14). SI Appendix, Fig. S3 C and D show thioflavin T fluorescence assays for the wild-type and D290V mutant mCherry-hnRNPA2-LC polymers. Both polymers exhibit increased fluorescence at ∼500 nm, a common property of cross-β structures.
Carbonyl and Carboxylate NMR Signals from Segmentally Labeled mCherry-hnRNPA2-LC Polymers.
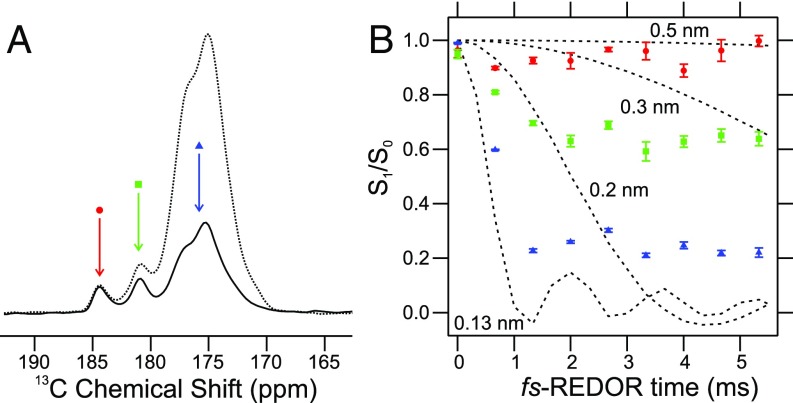
Fig. 2A shows the carbonyl/carboxylate region of a 1D solid-state 13C NMR spectrum of segmentally labeled polymers. Spectra were recorded with and without a 15N-13C dipolar recoupling period, which attenuates signals from 13C-labeled sites that have significant magnetic dipole–dipole couplings to 15N-labeled sites (i.e., that are close in space to 15N-labeled sites). The 15N-13C recoupling was implemented with the frequency-selective rotational echo double resonance (fs-REDOR) technique (15, 16). Signals at 180.9 ppm and 184.2 ppm in Fig. 2A are attributable to carboxylate carbons, while signals at 170–178 ppm are attributable to carbonyl carbons, as carboxylate carbons have larger 13C chemical shifts than carbonyl carbons (17). Both carboxylate signals exhibit little or no decay with 5.3 ms of fs-REDOR 15N-13C recoupling, while the backbone carbonyl signals and Asn/Gln side-chain carbonyls signals at 170–178 ppm decay significantly due to strong couplings to directly bonded backbone or side-chain amide 15N sites.
Fig. 2.
Identification of the carbonyl and carboxylate NMR signals from mCherry-hnRNPA2-LC polymers. (A) Carbonyl/carboxylate region of the 1D 13C solid-state NMR spectrum of segmentally labeled mCherry-hnRNPA2-LC polymers, with and without 5.3 ms of 15N-13C fs-REDOR recoupling (solid and dashed lines, respectively). Red, green, and blue arrows indicate signals arising from three distinct populations of 13C-labeled carbonyl/carboxylate sites in the polymers. (B) Normalized 15N-13C fs-REDOR decay curves for the three carbonyl/carboxylate signals. Error bars represent the rms noise in each 1D NMR spectrum. Dashed lines are numerical simulations of ideal fs-REDOR data for 15N-13C spin pairs with indicated internuclear distances. The coloring scheme and symbols are the same as in A.
Fig. 2B shows the dependence of the carbonyl and carboxylate 13C NMR signals on the fs-REDOR evolution time. As is commonly done for REDOR and fs-REDOR data (15, 18), the data are plotted as S1/S0, where S1 is the signal with the 15N-13C dipolar interaction recoupled and S0 is a reference signal without recoupling. The carbonyl signals at 170–178 ppm decay as expected for a 15N-13C bond length of 0.13 nm. The observed decay of S1/S0 to 0.2 instead of 0.0 is attributable to incomplete 1H decoupling during the fs-REDOR mixing period (15).
Since segmentally labeled mCherry-hnRNPA2-LC contains only two 13C-labeled carboxylate carbons, the signals at 180.9 ppm and 184.2 ppm can be assigned to Cγ of D290 and CO of Y341. The fact that both signals appear in Fig. 2A indicates that both D290 and Y341 are sufficiently immobilized in the polymers to permit efficient 1H-13C cross-polarization (CP), driven by 1H-13C magnetic dipole–dipole couplings (19, 20). Thus, D290 is contained within an immobilized, structurally ordered segment of the LC domain of hnRNPA2, as also indicated by previous NAI footprinting data (11). The decays of both carboxylate signals in Fig. 2B suggest 15N-13C distances greater than 0.3 nm, indicating that these sites do not have directly bonded 15N atoms, as expected. The expected intraresidue 15N-13C distances for D290 and Y341 are 0.27–0.38 and 0.24 nm, respectively. The absence of quantitative agreement between experimental and simulated decays for the carboxylate signals is attributable to motions of the D290 side chain and/or motions of the C terminus that partially average the 15N-13C couplings to smaller values, incomplete 1H decoupling, and signal overlap in the 1D spectra (discussed below).
Two-Dimensional Solid-State NMR Spectra of Segmentally Labeled mCherry-hnRNPA2-LC Polymers.
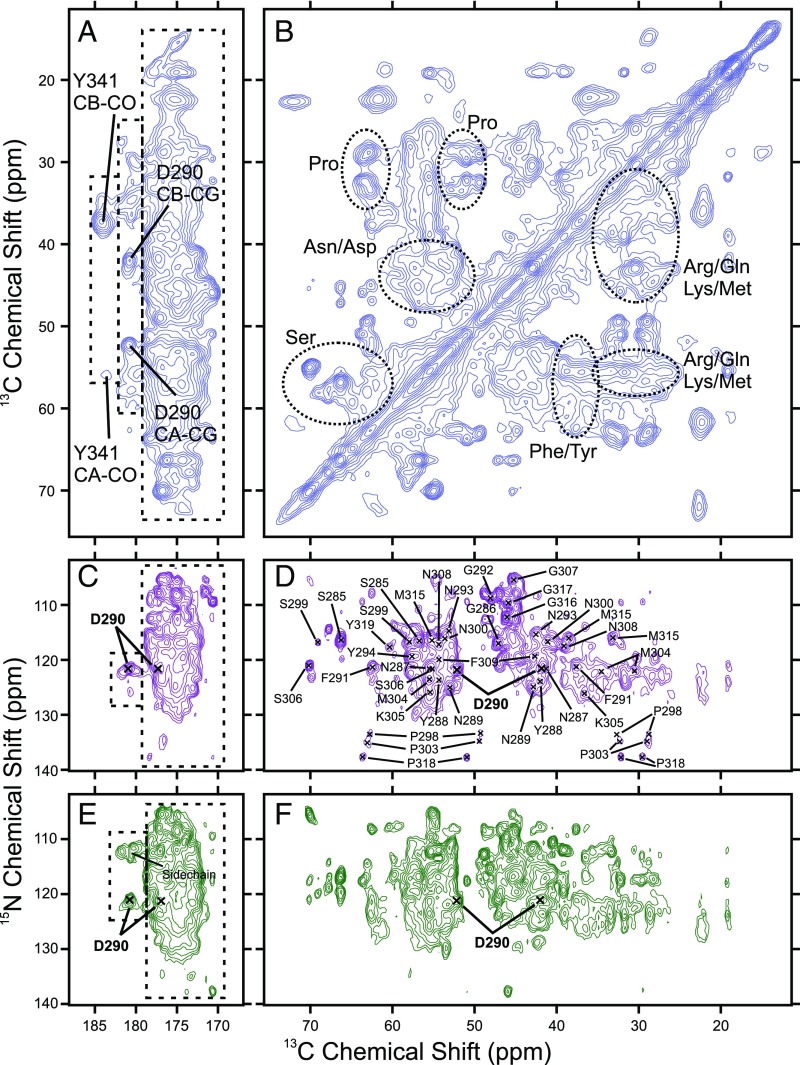
Fig. 3 shows 2D solid-state NMR spectra of the segmentally labeled polymers. The carbonyl/carboxylate-to-aliphatic cross-peak region of the 2D 13C-13C dipolar-assisted rotational resonance (DARR) spectrum (Fig. 3A) shows three distinct regions of signal intensity, delineated by dashed boxes. The carboxylate signal at 184.2 ppm shows two aliphatic cross-peaks, at 56.0 ppm and 37.4 ppm. These 13C chemical shift values are appropriate for the Cα and Cβ atoms of a Tyr residue. The carboxylate signal at 180.9 ppm shows two strong aliphatic cross-peaks, at 52.2 ppm and 41.8 ppm. These chemical shifts are within the expected ranges for the Cα and Cβ atoms of an Asp residue in a β-strand conformation (21). Thus, we assign the 184.2-ppm signal in Fig. 2A to the C-terminal carboxylate of Y341 and the 180.9 ppm signal to Cγ of D290. Additional weak cross-peaks are also present at 180.9 ppm in Fig. 3A, with aliphatic 13C chemical shifts of 57.9 ppm, 55.9 ppm, 36.4 ppm, 34.7 ppm, and 30.0 ppm. These chemical shifts are consistent with Cα, Cγ, and Cβ atoms of Gln residues. In Fig. 2B, contributions to fs-REDOR signals from these residues at 180.9 ppm explain the rapidly decaying minor component (green squares).
Fig. 3.
Two-dimensional solid-state NMR spectra of mCherry-hnRNPA2-LC polymers. (A and B) Two-dimensional 13C-13C spectrum of mCherry-hnRNPA2-LC polymers. (C and D) Two-dimensional 15N-13C NCACX spectrum of mCherry-hnRNPA2-LC polymers. (E and F) Two-dimensional 15N-13C NCOCX spectrum of the mCherry-hnRNPA2-LC polymers. The three populations of carbonyl and carboxylate signals from Fig. 2 are indicated with dashed boxes in A, C, and E. Residue-type assignments in B are based on the typical chemical shifts for each residue type. Residue-specific assignments in C–F are derived from 3D solid-state NMR experiments. Contour levels increase by successive factors of 1.4 in A–D and factors of 1.3 in E and F.
The aliphatic-to-aliphatic cross-peak region of the 2D 13C-13C DARR spectrum in Fig. 3B shows signals from most of the amino acid types present in the isotopically labeled segment of the mCherry-hnRNPA2-LC polymers (SI Appendix, Table S1). The spectrum does not allow for individual resonances to be resolved. However, based on the distributions of experimentally observed NMR chemical shifts (17), Cα-Cβ cross-peaks of Ser, Asn, Asp, Phe, and Tyr residues are identified, as are Cα-Cβ, Cα-Cγ, Cδ-Cβ, Cδ-Cγ cross-peaks of Pro residues. The Arg, Gln, Lys, and Met region of the spectrum is congested, but there is signal intensity that can be attributed to all these amino acid types.
Fig. 3 C and D show the 2D 15N-13C NCACX spectrum of the mCherry-hnRNPA2-LC polymers. Cross-peak signals in this spectrum correlate 15N chemical shifts of backbone amide sites with 13C chemical shifts within individual residues. The 180.9-ppm Cγ signal of D290 is observed in Fig. 3C at a 15N frequency of 121.5 ppm. In Fig. 3D, the D290 Cα and Cβ signals at 13C frequencies of 52.2 and 41.8 ppm are present. Three other weak 13C signals at ∼180.9 ppm with 15N frequencies of 121.9, 122.6, and 126.0 are consistent with the observation of weak Gln Cα, Cβ, and Cγ cross-peaks in Fig. 3A. No cross-peaks occur at 184.2 ppm in Fig. 3C, consistent with motional averaging of nuclear magnetic dipole–dipole couplings at the C terminus of hnRNPA2-LC.
Note that 13C NMR signals in Fig. 2A arise from magnetization transfers involving strong 1H-13C dipolar couplings, while signals in Fig. 3C arise from magnetization transfers involving weaker 15N-13C and 13C-13C couplings. Thus, signals in Fig. 3C are more sensitive to molecular motions with limited amplitudes, explaining the absence of signals from Y341 in Fig. 3C.
Fig. 3 E and F show the 15N-13C NCOCX spectrum of the mCherry-hnRNPA2-LC polymers. Most cross-peak signals in this spectrum correlate 15N chemical shifts of backbone amide sites with 13C chemical shifts of preceding residues. There is a single strong signal for Cγ at 180.9 ppm with a 15N frequency of 120.8 ppm and the corresponding Cα and Cβ cross-peaks at 52.2 and 41.8 ppm. Additionally, there are three cross-peaks near 178.1 ppm in Fig. 3E with 15N frequencies around 112 ppm. These cross-peaks represent correlations of 15N chemical shifts of Asn or Gln side-chain amide nitrogens with 13C chemical shifts of their directly bonded Cδ carbons. There is no evidence of a cross-peak at 184.2 ppm in Fig. 3E, in agreement with this site’s being the C terminus of the protein.
SI Appendix, Fig. S4 shows the 2D 13C-13C DARR, 15N-13C NCACX, and 15N-13C NCOCX spectra of mCherry-hnRNPA2-LC polymers with the D290V mutation. Cross-peak patterns in these spectra are different from cross-peak patterns in Fig. 3. This is not unexpected, since the D290V mutation removes negative charge from the polymer core structure and can produce conformational changes, thereby changing NMR chemical shifts. Importantly, spectra in SI Appendix, Fig. S4 show cross-peak signals from a valine residue, which must be V290, with chemical shifts consistent with a β-strand conformation. SI Appendix, Fig. S8C shows a TEM image of negatively stained mCherry-hnRNPA2-LC D290V mutant polymers, which are visually similar to wild-type polymers (Fig. 1C).
Assignment of NMR Signals from mCherry-hnRNPA2-LC Polymers.
To obtain additional site-specific assignments of 13C and 15N chemical shifts, 3D NCACX, NCOCX, and CONCA spectra of the segmentally labeled polymers were recorded (SI Appendix, Fig. S5). Signals were observed from 37, 38, and 42 residues in the NCACX, NCOCX, and CONCA spectra, respectively. These signals arise from residues that are strongly immobilized and structurally ordered within the polymers. Remaining 13C,15N-labeled residues that do not contribute signals to the 3D spectra are more dynamic and/or disordered.
SI Appendix, Fig. S6 shows the result of computationally assisted assignments for signals observed in the 3D NCACX, NCOCX, and CONCA spectra, obtained with the MCASSIGN Monte Carlo/simulated annealing algorithm (22). Unambiguous site-specific 13C chemical shift assignments were obtained for residues 285–294, 298–300, 303–309, and 315–319 (solid bars in SI Appendix, Fig. S6 and underlined in Fig. 1A). Assigned signals are indicated in the 2D NCACX spectrum in Fig. 3 C and D. Strip plots illustrating assignments for residues 288–292 are shown in SI Appendix, Fig. S5. SI Appendix, Table S2 lists the assigned signals. SI Appendix, Table S3 lists the unassigned signals from the NCACX, NCOCX, and CONCA spectra with possible residue-type assignments. The remaining signals (14 in the NCACX, 15 in the NCOCX, and 19 in the CONCA experiments) could not be assigned in part due to the multiple GGX and GGS motifs in residues 280–341 of the hnRNPA2-LC domain (“X” is Asn or Tyr) Altogether, signals were observed from 44 aa between residues 280 and 341, indicating that 72% of this region is immobilized.
Secondary Structure in mCherry-hnRNPA2-LC Polymers.
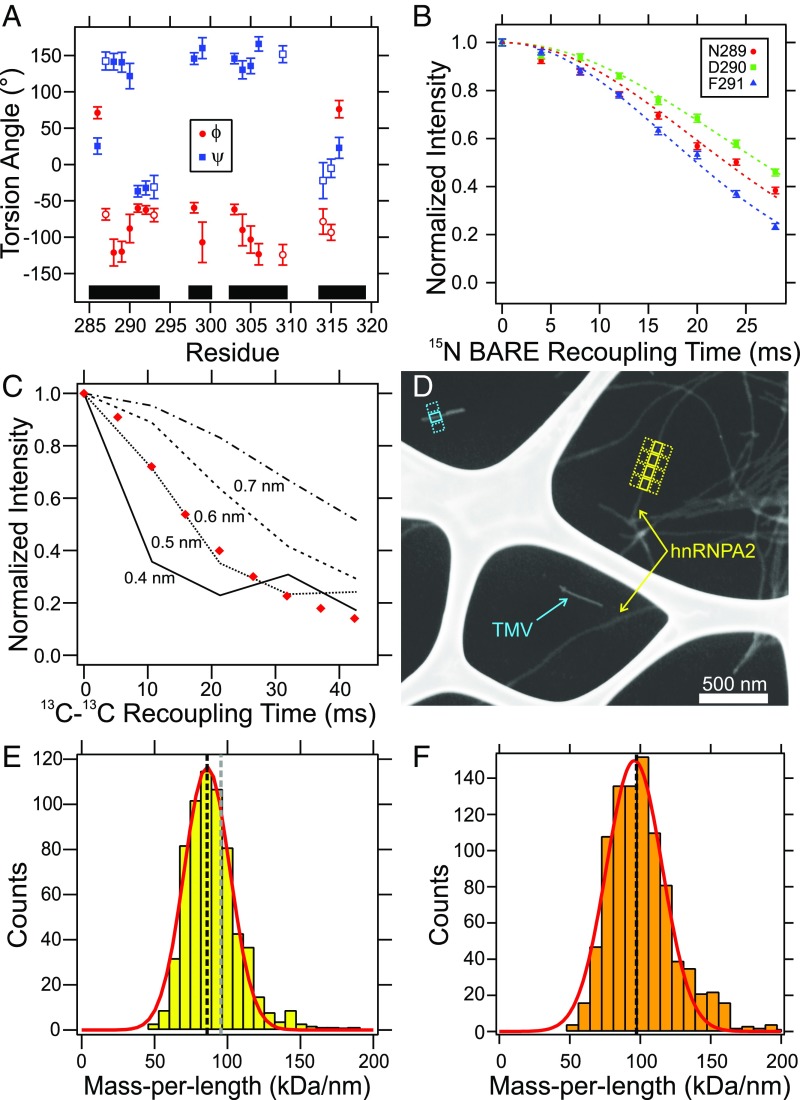
Fig. 4A shows predictions of backbone ϕ/ψ torsion angles in mCherry-hnRNPA2-LC polymers by the TALOS-N program (23), based on the assigned solid-state NMR chemical shifts. Residues 288–290, 298–299, 303–306, and 309 are predicted to have the characteristic torsion angles of a β-strand conformation. Residues 291–293 and 314–316 are predicted to have characteristic torsion angles of non-β-strand structures. SI Appendix, Fig. S7F shows a 2D NCA solid-state NMR spectrum of the mCherry-hnRNPA2-LC polymers collected with a 15N BAckbone REcoupling (15N-BARE) constant-time homonuclear dipole–dipole recoupling block (24). Resonances with unique assignments are indicated with arrows in SI Appendix, Fig. S7F and overlapping resonances are shown in gray. The decay of the signal intensities as a function of the 15N-BARE recoupling time is plotted in Fig. 4C and SI Appendix, Fig. S7 A–E. Signal decay in these measurements depends on backbone ϕ/ψ torsion angles and, equivalently, amide 15N-15N distances between successive residues. In experiments on globular microcrystalline proteins under the same experimental conditions at 28-ms recoupling time, residues in β-strand conformations had signal decays to 0.5 or greater, residues in turn structures or at the end of β-strands had signal decays between 0.2 and 0.5, and residues in helical structures had signal decays to 0.2 or smaller (24). Normalized signals from residues 288, 294, 299, 307–309, 315, and 317 exhibit limited decay (to final values of 0.5 or greater) after 28 ms, which is consistent with fully extended β-strand conformations. The decay of signal intensity for residues 291–293 is substantially greater (to final values of ∼0.2), indicating that these residues may form a turn with relatively short sequential amide 15N-15N distances. The remaining sites (285–286, 289–290, 300, 305, 316, and 319) have intermediate decay (to final values of 0.2–0.5), indicating these sites have backbone conformations that are not fully extended.
Fig. 4.
Structural measurements on mCherry-hnRNPA2-LC polymers. (A) Predictions of backbone ϕ/ψ torsion angles by TALOS-N, based on assigned solid-state NMR chemical shifts. Error bars are uncertainties reported by TALOS-N. Filled and open symbols represent “strong” and “generous” predictions, respectively. Black bars indicate regions where solid-state NMR signals are assigned. (B) Normalized 15N-BARE decay curves for residues N289–F291. Error bars represent the rms noise in the NMR spectrum and dashed lines are Gaussian fits to the data. (C) 13C-13C PITHIRDS-CT dipolar recoupling data for mCherry-hnRNPA2-LC polymers that were 13C-labeled only at backbone carbonyl sites of Tyr residues within residues 280–341. Red diamonds are normalized peak intensities. Lines are simulated data for linear chains of 13C labels with the indicated interatomic spacings. Error due to noise is smaller than the symbol size. (D) Dark-field TEM image of unstained mCherry-hnRNPA2-LC polymers. Yellow and cyan arrows indicate mCherry-hnRNPA2-LC polymers and TMV rods, respectively. Examples of 60- × 80-nm integration boxes used in the MPL measurement are shown. (E) Histogram of apparent MPL values. The red line is a single Gaussian fit to the data, centered at 86.0 ± 0.6 kDa/nm with a full-width half-maximum value of 22.0 ± 0.9 kDa/nm. The vertical gray dashed line is the expected MPL value for cross-β polymers with an intermolecular spacing of 0.48 nm and with the mCherry-hnRNPA2-LC monomer molecular weight of 45.8 kDa. The vertical black dashed line is for an average molecular weight of 40.9 kDa, corresponding to partial cleavage of the mCherry tag for 45% of the monomers. (F) Histogram of apparent MPL values for mCherry-hnRNPA2-LC D290V mutant polymers. The red line is a Gaussian fit to the data, centered at 95.8 ± 1.0 kDa/nm with a full-width half-maximum value of 28.1 ± 1.4 kDa/nm. The vertical black dashed line is the expected value for cross-β polymers with an intermolecular spacing of 0.48 nm and the mCherry-hnRNPA2-LC D290V molecular weight of 46.2 kDa.
Although residues 289–290 are predicted by TALOS-N to have fully extended β-strand conformations but have intermediate 15N-BARE signal decay values, the predictions and 15N-BARE measurements are consistent with D290’s being contained at the C-terminal end of a β-strand in the structurally ordered core of mCherry-hnRNPA2-LC polymers. TALOS-N predicts small values of |ψ| for residues 314–316, which would imply relative short 15N-15N distances, but the 15N-BARE data for residues 315–317 indicate relatively long 15N-15N distances. We attribute this disagreement to uncertainties in the TALOS-N predictions. Residues with characteristic β-strand φ/ψ torsion angles are indicated with an asterisk in Fig. 1A.
Intermolecular 13C-13C Distances in mCherry-hnRNPA2-LC Polymers.
Fig. 4C shows measurements of 13C-13C dipole–dipole couplings in mCherry-hnRNPA2-LC polymers that were 13C-labeled only at backbone carbonyl sites of Tyr residues 283, 288, 294, 301, 319, 324, 335, and 341, using the 13C-13C constant-time, one-third-rotor-period π pulse (PITHIRDS-CT) dipolar recoupling technique (25). This Tyr-labeled sample was prepared by ligating an unlabeled N-terminal construct to a 13CO-Tyr-labeled C-terminal construct, as in Fig. 1B and SI Appendix, Fig. S2. After subtraction of a nondecaying 10% contribution attributable to natural-abundance 13C NMR signals, the decay of experimental 13C NMR signal amplitudes (red diamonds) with increasing recoupling time is in good agreement with simulated 13C-13C PITHIRDS-CT data for a linear chain of 13C labels with a 0.5-nm interatomic distance (dotted line). Thus, these 13C-13C PITHIRDS-CT data are consistent with an in-register parallel β-sheet structure that contains all or most of the labeled Tyr residues (more specifically, labeled Tyr residues that are strongly immobilized in the polymers). The 13C-13C PITHIRDS-CT data for segmentally 13CO-Tyr–labeled mCherry-hnRNPA2-LC D290V mutant polymers are also consistent with a 0.5 nm distance between labeled sites (SI Appendix, Fig. S8B). In TEM images, mCherry-hnRNPA2-LC D290V mutant polymers were morphologically similar to mCherry-hnRNPA2-LC polymers (Fig. 1C with SI Appendix, Fig. S8A). Together, the 13C-13C PITHIRDS-CT measurements show that the hnRNPA2-LC domain is organized in an in-register parallel cross-β structure for both wild-type and D290V mutant polymers.
Mass per Length of mCherry-hnRNPA2-LC Polymers.
Fig. 4D shows a dark-field TEM image of unstained mCherry-hnRNPA2-LC polymers. From such images, mass-per-length (MPL) values for individual polymer segments are determined by integration of image intensities within rectangular regions centered on the polymers, with subtraction of average integrated intensities in rectangular regions of the same size on either side of the polymers (yellow boxes in Fig. 4D) (26). Intensities are calibrated by using tobacco mosaic virus (TMV) rods, which have a known MPL value (cyan boxes in Fig. 4D). The resulting MPL histogram can be fit with a single Gaussian peak, centered at 86.0 ± 0.6 kDa/nm and with a 22 ± 0.9 kDa/nm width. A histogram of variations in background intensities (SI Appendix, Fig. S9A) has a width of 23 ± 1.2 kDa/nm, indicating that the variations in individual MPL values in Fig. 4E are due to noise in the images, rather than real structural variations.
Given that the molecular weight of an mCherry-hnRNPA2-LC monomer is 45.8 kDa and that the spacing between β-strands along the polymer growth direction in an ideal cross-β structure is 0.475 ± 0.005 nm, mCherry-hnRNPA2-LC polymers with an in-register parallel cross-β structure are expected to have MPL ≈ 96.4 kDa/nm (gray vertical dashed line in Fig. 4E). However, liquid chromatography MS data indicate that a fraction of the mCherry tags were subject to partial cleavage, reducing the molecular weight of mCherry-hnRNPA2-LC for 45% of the mCherry-hnRNPA2-LC monomers in our polymers (Experimental Methods and SI Appendix, Fig. S10). Correction of the average molecular weight for mCherry-hnRNPA2-LC results in an expected MPL of 86.0 kDa/nm (black vertical dashed line in Fig. 4E), in excellent agreement with the Gaussian fit to the histogram in Fig. 4E.
Fig. 4F and SI Appendix, Fig. S9 C–E show MPL data for mCherry-hnRNPA2-LC D290V mutant polymers. A Gaussian fit to the MPL histogram in Fig. 4F is centered at 95.8 ± 1.0 kDa/nm, in agreement with the expected value of 97.3 kDa/nm for mCherry-hnRNPA2-LC D290V (molecular weight 46.2 kDa) in an in-register parallel cross-β structure. The 28.1 ± 1.4 kDa/nm width of the MPL histogram in Fig. 4F agrees well with variations in background intensity (30.8 ± 1.1 kDa/nm; SI Appendix, Fig. S9E).
The Carboxylate Sites in the mCherry-hnRNPA2-LC Polymers Are Negatively Charged at Physiological pH.
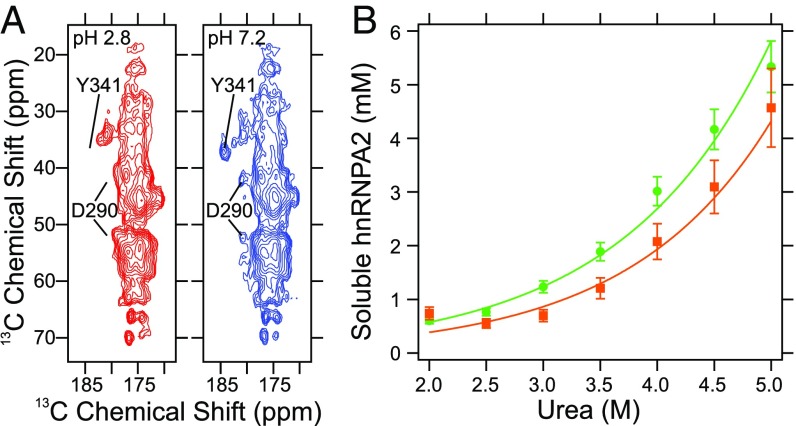
SI Appendix, Fig. S11 shows the carbonyl/carboxylate region of 1D solid-state NMR spectra of the segmentally labeled mCherry-hnRNPA2-LC polymers exchanged into buffer with pH values between 2.8 and 7.2. As the pH decreases, the signal intensities of the D290 Cγ and Y341 C-terminal carboxylate resonances decrease (at 180.9 and 184.2 ppm, respectively), shifting into the bulk carbonyl signals between 170 and 178 ppm. The shift toward lower chemical shift values is expected for carboxylate sites that become uncharged as the pH is lowered. Fig. 5A shows regions of 2D 13C-13C DARR NMR spectra of the polymers at pH 2.8 and 7.2. The aliphatic-carboxylate cross-peaks at 180.9 and 184.2 ppm are missing at pH 2.8 and present at pH 7.2, confirming that the resonances have shifted into the bulk carbonyl signal intensity at lower pH. The overall appearance of the carbonyl resonances in Fig. 5A are highly similar and indicate that decreased pH does not alter the structure of the mCherry-hnRNPA2-LC polymers. Additionally, the spectra in Fig. 5A are consistent with the spectrum in Fig. 3A, confirming that buffer exchange does not alter the mCherry-hnRNPA2-LC polymer conformation.
Fig. 5.
Charge state of the D290 side chain and polymer thermodynamic stability. (A) Carbonyl/carboxylate region of a 2D 13C-13C DARR solid-state NMR spectrum of mCherry-hnRNPA2-LC polymers. Red and blue contours are spectra of samples at pH 2.8 and 7.2, respectively. Locations of the D290 and Y340 cross-peaks at pH 7.2 are indicated in both spectra. (B) Soluble GFP-hnRNPA2-LC concentrations after wild-type (green circles) and D290V mutant (orange squares) polymers were exposed to urea for 21–22 h. Color-coded lines are a simultaneous fit to the data assuming the polymers are at equilibrium with monomeric soluble protein and a linear dependence of polymer stability on urea concentration.
Polymers with the D290V Mutation Are More Thermodynamically Stable than Wild-Type Polymers.
Polymers prepared from the D290V mutant of the hnRNPA2-LC domain display enhanced stability as measured by semidenaturing detergent agarose gel electrophoresis (10). To quantify the stability difference, we measured concentrations of soluble proteins after wild-type and D290V mutant GFP-hnRNPA2-LC polymers were incubated in buffered solutions with urea concentrations between 2 and 5 M (Fig. 5B). Under these conditions, a model where the free energy change, ΔG, of polymer dissociation depends linearly on the urea concentration (27), monomeric protein exchanges with the polymer ends, and the system has reached steady-state (28) describes the thermodynamic equilibrium. Best-fit curves to the data give free energy changes in the absence of denaturant, ΔG0, for wild-type and D290V polymers of 21.9 ± 0.3 and 23.1 ± 0.6 kJ/mol, respectively. Therefore, the aspartic acid-to-valine mutation stabilizes the polymers by 1.2 kJ/mol. The slope of the linear dependence of ΔG on denaturant concentration (typically referred to as m) for the wild-type and D290V mutant polymers is −1.88 ± 0.08 kJ/mol⋅M and −1.96 ± 0.17 kJ/mol⋅M, respectively. The ΔΔG0 ≈ 1.2 ± 0.7 kJ/mol change in polymer dissociation free energy is similar to ΔΔG0 values for effects of single-amino-acid substitutions on unfolding of globular proteins, which range from 0.3 kJ/mol to 19.8 kJ/mol (29).
Although the difference in thermodynamic stability indicated by data in Fig. 5B is small (roughly 0.5 RT, where R is the gas constant and T is temperature), it may be sufficient to overcome a delicate balance of interactions between LC domains. We do not argue that rigid β-sheets are the functional unit for the hnRNPA2 LC domain, but rather that limited interactions between protein monomers with cross-β character facilitate its function. In this instance, a small change in thermodynamic stability could significantly alter the functional activity of hnRNPA2.
Discussion
The main conclusions from experiments described above include the following. (i) The immobilized core of cross-β polymers formed by the LC domain of hnRNPA2 consists of about 44 aa within residues 280–341 (Figs. 2 and 3 and SI Appendix, Figs. S5 and S6). (ii) The core adopts an in-register parallel cross-β structure (Fig. 4 A–E and SI Appendix, Figs. S7 and S9 A and B). (iii) In wild-type polymers, D290 is part of the immobilized core, positioned near the C-terminal end of a β-strand segment (Figs. 2 and 4 A and B and SI Appendix, Fig. S7). (iv) At physiological pH, the D290 side-chain carboxylate group is negatively charged (Fig. 5A and SI Appendix, Fig. S11). (v) D290V mutant polymers also have an in-register parallel cross-β structure, with V290 being part of the immobilized core (Fig. 4F and SI Appendix, Figs. S4, S8B, and S9 C–E). (vi) D290V mutant polymers have greater thermodynamic stability, by ∼1.2 ± 0.7 kJ/mol according to urea-driven polymer dissolution experiments (Fig. 5B).
By use of similar methods of segmental labeling, solid-state NMR spectroscopy and electron microscopy we have recently developed a detailed molecular structural model for polymers formed by the 214-residue LC domain of the FUS RNA-binding protein (30). FUS-LC polymers contain an in-register parallel cross-β structure formed by a specific 57-residue segment that lacks purely hydrophobic amino acid residues (other than a single proline). The FUS core-forming segment contains two fully structured regions, composed of 16 and 33 aa, which are separated by a disordered loop of eight residues. Within the FUS-LC polymer core, ∼50% of the structurally ordered residues participate in β-strands, which range in length from three to six residues. The FUS-LC polymer core appears to be stabilized largely by nonhydrophobic interactions including hydrogen bonds, polar zippers (31), and electrostatic dipole–dipole interactions among polar side chains.
Whereas the data presented herein are not sufficient to determine the full structure of hnRNPA2-LC polymers, certain features appear to be in common with FUS-LC polymers. Data shown in Figs. 3 and 4 and SI Appendix, Figs. S5 and S6 give evidence that the core of hnRNPA2-LC polymers spans a segment of ∼50 residues, interspersed internally by short disordered segments. The segmental labeling strategy we have employed does not allow us to probe structure preceding residue 280 of the hnRNPA2-LC domain. NAI footprinting data have, however, given evidence that residues 181–281 are freely accessible to chemical acetylation, a strong indicator that this region lacks rigid, polymeric structure (11). Chemical shifts with residue-specific and residue-type assignments suggest that about half of the rigid core is disposed in a β-strand structure (Fig. 4A). Measurements of backbone 15N dipolar coupling interactions support the chemical shift-based secondary structure analysis (Fig. 4B and SI Appendix, Fig. S7). Additional solid-state NMR measurements on additional samples may allow unambiguous identification of all core-forming segments, eventually enabling development of a detailed structural model of hnRNPA2-LC polymers.
The primary focus of the present study has been aimed toward an understanding of why residue D290 of the hnRNPA2 LC domain, when mutated to valine, might predispose patients to neurodegenerative disease. Analysis of muscle tissue from disease-bearing patients has revealed cytoplasmic inclusions containing aggregated hnRNPA2 (3). Kinetic aggregation assays have further shown that a synthetic hexapeptide representing residues 287–292 of the hnRNPA2 LC domain was more prone to aggregation if bearing the D-to-V missense mutation (3). Subsequent studies of a chimeric hnRNPA2-Sup35 fusion protein in yeast cells revealed a marked enhancement in prion formation in derivatives bearing the D290V mutation (32). Finally, biochemical analysis of LC domains from hnRNPA2, hnRNPA1, and hnRNPDL revealed enhanced polymer stability for all three proteins upon mutation of D290 (hnRNPA2), D262 (hnRNPA1), or D378 (hnRNPDL) to valine or asparagine (10). All of these observations are readily explained by the simple idea that replacement of aspartic acid by valine or asparagine within an in-register parallel cross-β structure eliminates electrostatic repulsion among aspartate side chains of neighboring monomers, replacing these destabilizing interactions with stabilizing hydrophobic or polar zipper interactions. Data presented herein are supportive of this idea. Since the normal biological function of hnRNPA2 is believed to involve reversible cross-β self-assembly (11), increased stability of these structures within cells may enhance the probability of nucleation of pathogenic fibrillar assemblies. It may be of use to note that several recent papers have offered observations consistent with the biological utility of cross-β interactions formed from various LC-domain-containing proteins (33, 34).
A potential shortcoming of the studies described in this paper may be attributed to the use of the hnRNPA2 LC domain in isolation from its two RNA recognition motif (RRM) domains—these RRM domains readily adopt stable, well-folded conformations that facilitate direct interaction with RNA substrates. Given that NAI footprinting studies have revealed similar conformations for cross-β polymers formed from the isolated hnRNPA2 LC domain and its counterpart in nuclei of mammalian cells (11), we cautiously endorse the validity of the reductionist approach of this and earlier studies.
We close by making note of computational studies investigating the frequency of occurrence of amino acids localized to specific secondary folds within all protein structures in the protein database. According to these global calculations, the least-favored residue in β-strands is aspartic acid, and the most-favored is valine (35). This recurrent mutational change within three different hnRNP proteins, we offer, may be of significance to our understanding of both neurological disease mechanism and the normal biological function of LC domains.
Experimental Methods
Protein Expression, Labeling, and Purification.
Preparation of GFP-fused or mCherry-fused hnRNPA2-LC (residues 181–341) was performed as previously described (11, 14). The expression plasmids of the GFP-fused N-terminal half (181–260) or the C-terminal half (261–341) of hnRNPA2-LC were constructed in a pHis-parallel-GFP vector, and the proteins were overexpressed and purified as previously described (14).
To produce the segmentally labeled mCherry-hnRNPA2-LC, we used the artificially designed Cfa intein (36). First, the 3′ end of the DNA fragment coding the N-terminal region of hnRNPA2-LC (amino acids 181–279) was ligated to the 5′ end of the Cfa intein DNA fragment by using a two-step PCR technique. The resulting DNA fragment was then inserted into the BamHI/XhoI site of the pHis-parellel-mCherry vector. The DNA fragments of hnRNPA2-LC (181–341) or the C-terminal region of hnRNPA2-LC (amino acids 281–341) was also ligated into the BamHI/XhoI site of the pHis-parellel-mCherry vector with a caspase-3 cleavage site (GDEVDA or GDEVDC) inserted between mCherry and hnRNPA2-LC. The design of the N-terminal and C-terminal fragments is summarized in SI Appendix, Fig. S2. The D290V mutation was introduced by the quick-change technique. All of the sequences were confirmed by DNA sequencing.
His6-mCherry-hnRNPA2-LC(181-279)-Cfa and His6-mCherry-GDEVDC-hnRNPA2-LC(281-341) were transformed into Escherichia coli. BL21(DE3). Nonlabeled His6-mCherry-hnRNPA2-LC(181-279)-Cfa was overexpressed in LB medium with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) overnight at 16 °C as described before (30). The wild type and D290V mutant of His6-mCherry-GDEVDC-hnRNPA2-LC(281–341) were overexpressed in M9 medium with 13C-glucose and 15N-ammonium chloride for universal 13C,15N labeling as described before (30), or in M9 medium containing 13CO-Tyr and other regular amino acids for tyrosine-specific labeling as reported before (37). In both cases, inoculated M9 media with labeling materials were shaken at 37 °C until OD600 reached around 1.0. Then, culture temperature was reduced to 16 °C, and IPTG was added to a final concentration of 0.5 mM for overnight expression. Cells were harvested by centrifugation, washed with PBS, and stored at −80 °C.
For purification of the above N- and C-terminal fragments of hnRNPA2-LC, cells were resuspended in a lysis buffer containing 2 M urea, 500 mM NaCl, 50 mM Tris⋅HCl (pH 8.0), 2 mM Tris[2-carboxyethyl]phosphine hydrochloride (TCEP), and one tablet of protease inhibitor mixture (Sigma-Aldrich), and lysed by sonication (70% power for 3 min, Fisher Scientific Model FB705) on ice. The cell lysate was centrifuged at 32,000 × g for 1 h at 4 °C, and the supernatant was loaded onto an Ni-NTA (Qiagen) affinity column. Unbound impurities were removed with buffer containing 20 mM imidazole, 2 M urea, 150 mM NaCl, 25 mM Tris⋅HCl (pH 8.0), and 2 mM TCEP. The bound proteins were eluted from the Ni-NTA column with buffer containing 300 mM imidazole, 2 M urea, 150 mM NaCl, 25 mM Tris⋅HCl (pH 8.0), and 2 mM TCEP. The eluted proteins were concentrated with a 10-kDa molecular weight cut-off (MWCO) Amicon Ultra centrifugal filter device (Millipore) to a concentration of ∼2 mM, flash-frozen in liquid nitrogen, and stored at −80 °C.
Polymer Preparation.
GFP-fused half-constructs of hnRNPA2-LC (200–300 μM) were dialyzed in a gelation buffer containing 20 mM Tris⋅HCl (pH 7.5), 200 mM NaCl, 20 mM β-mercaptoethanol, 0.5 mM EDTA, and 0.1 mM PMSF overnight at room temperature. The protein solutions were centrifuged at 20,000 × g for 1 min to remove any precipitate then concentrated with a 10-kDa MWCO Amicon Ultra centrifugal filter device to a final concentration of 200 μM. The concentrated proteins were incubated at 4 °C for several days. Polymer formation was checked with TEM as described previously (10).
To make the segmentally labeled mCherry-hnRNPA2-LC, unlabeled His6-mCherry-hnRNPA2-LC(181–279)-Cfa and universally 13C, 15N or 13CO Tyr specifically labeled His6-mCherry-GDEVDC-hnRNPA2-LC(281–341) were mixed to initiate an intein ligation reaction as described previously (30). The reaction is summarized in SI Appendix, Fig. S2. Briefly, the intein reaction mixtures contained 200 μM unlabeled His6-mCherry-hnRNPA2-LC(181–279)-Cfa, 200 μM labeled, wild-type or D290V mutant His6-mCherry-GDEVDC-hnRNPA2-LC(281–341), 800 mM sodium 2-mercaptoethanesulfonate, 100 mM potassium phosphate (pH 7.2), 100 mM potassium chloride, 5 mM TCEP, 1 mM EDTA, and 2 μg/mL caspase-3. The reaction mixtures were incubated at 37 °C with gentle rotation for 16 h for the wild type and for 5 h for the D290V mutant. Precipitate that formed in the first hour of the wild-type reaction was removed by centrifugation at 10,000 × g for 2 min. For the mutant reaction, precipitates were removed at 30 min and 1 h by centrifugation. After incubation, the majority of the ligated product (mCherry-hnRNPA2-LC) formed polymers. A typical yield of the product was 30–40% amount of the initial N-terminal fragment for both the wild type and the D290V mutant based on visual judgments of SDS/PAGE. The polymers were harvested by centrifugation at 6,000 × g for 5 min. The polymer pellet was washed three times with a buffer containing 25 mM Tris⋅HCl (pH 7.5) and 100 mM NaCl. Purity and morphology of the washed polymers were checked by SDS/PAGE and TEM, respectively.
For MPL measurements a small amount of the 13CO Tyr mCherry-hnRNPA2-LC polymer sample was added to 300 μL of a solution of 20 mM sodium phosphate (pH 7.4) and 1 mM TCEP using a pipette tip. The solution was placed in an ice bath and tip sonicated using a Branson Sonifier 250 at a duty cycle of 15% using an output setting of 0.15. TEM images of the solution revealed short seed polymers ∼25 nm in length or smaller. Then, 0.5 mL of purified mCherry-hnRNPA2-LC (not segmentally labeled) protein at a concentration of ∼110 μL was dialyzed against 2 L of 20 mM Tris⋅HCl, 200 mM sodium chloride, 20 mM β-mercaptoethanol, 0.1 mM phenylmethonylsulfonyl fluoride, and 0.5 mM EDTA for 24 h using a Slide-A-Lyzer 3500 MWCO dialysis cassette (Thermo Fisher Scientific). The solution was recovered from the dialysis cassette and centrifuged at 16,000 × g for 20 min to remove any precipitated material. The protein concentration of the supernatant was 70 μM based on optical absorbance at 280 nm. Five hundred microliters of the soluble protein solution was mixed with the sonicated polymers by brief vortexing and allowed to sit quiescently at room temperature for 24 h. Long, thin polymers were observed in negatively stained TEM micrographs (SI Appendix, Fig. S9B).
For the MPL measurement of the mCherry-hnRNPA2-LC D290V mutant polymers, 100 μL of the wild-type polymers was diluted with 200 μL of 20 mM sodium phosphate (pH 7.4). The dilution was tip sonicated using a Branson Sonifier 250 an output of 0.1 and a 20% duty cycle for 5 min. Forty-two microliters of pure mCherry-hnRNPA2-LC D290V in 20 mM Tris⋅HCl (pH 7.5), 500 mM sodium chloride, 20 mM β-mercaptoethanol, 0.1 mM phenylmethonylsulfonyl fluoride, 0.5 mM EDTA, and 2 M urea at 758 μM was diluted to 108 μM with 258 μL of 20 mM sodium phosphate (pH 7.4). The solution was centrifuged at 17,000 × g to remove precipitated material before adding 20 μL of the sonicated material. Polymers were allowed for form at 4 °C for 3 d. Long, thin polymers were observed in negatively stained TEM micrographs (SI Appendix, Fig. S9C).
For the pH titration solid-state NMR measurements, the mCherry-hnRNPA2-LC polymers were unpacked from the NMR rotor using centrifugation. Citric acid-sodium phosphate buffers were prepared by mixing 0.1 M citric acid with 0.2 M sodium phosphate dibasic at the following ratios (in milliliters): pH 3.0 = 8.0/2.1, pH 4.4 = 5.6/4.4, pH 6.0 = 3.7/6.3, pH 7.4 = 0.9/9.1; 71.7 mg of TCEP powder was added to 10-mL aliquots of the buffers for a final concentration of 5 mM. The measured pH values were 2.8, 4.0, 5.7, and 7.2, respectively. The pellet was resuspended in 400 μL of pH 7.3 buffer by vortexing and bath sonication. The sample was split into four parts, diluted with 900 μL of each buffer (pH 2.8, 4.0, 5.7, and 7.2), equilibrated for 1 h and 15 min, then centrifuged at 228,000 × g for 3.5 h at 4 °C. The pellets were resuspended in 200 μL of buffer of the same pH, bath-sonicated, and transferred to a new centrifuge tube. Samples were diluted with 200 μL of buffer and were equilibrated at 4 °C for 2 d. Samples were harvested by centrifugation at 336,000 × g for 4 h at 4 °C.
Solid-State NMR Measurements.
The 1D fs-REDOR, 2D DARR, 2D NCACX, 2D NCOCX, 3D NCACX, and 3D NCOCX data were collected on a 14.1-T (599.1 MHz 1H NMR frequency) magnet using a Varian InfinityPlus console and a Varian Balun solenoid 3.2-mm magic angle spinning (MAS) probe. The 3D CONCA and 15N-BARE measurements were collected on a 17.5-T (745.9 MHz 1H NMR frequency) magnet using a Varian InfinityPlus console and a Black Fox 3.2-mm MAS probe. The 13C-13C PITHIRDS-CT data were collected on a 9.4-T (400.6 MHz 1H NMR frequency) magnet using a Varian 3.2-mm T3 MAS probe and either a Bruker Avance III console or a Tecmag Redstone console. For the 13C-13C PITHIRDS-CT experiments the cooling air was set such that the sample temperature was ∼32 °C. For all other experiments, the sample temperature was ∼20 °C. Unless otherwise indicated 1H decoupling used the two-pulse phase-modulated method (38). The 2D and 3D experiments (CC, NCACX, NCOCX, and CONCA) utilized DARR (39) for 13C-13C transfers and CP for heteronuclear magnetization transfers (20). The 13C-13C PITHIRDS-CT, and 15N-BARE experiments were performed as previously described (24, 25). Acquisition parameters for all NMR experiments are listed in SI Appendix, Table S4. Data were processed using NMRPipe (40) and Spinsight (Varian) software. Integration of the 1D data was performed in the Spinsight software and all plots were prepared using Igor Pro-6.3.7.2 (WaveMetrics).
The mCherry-hnRNPA2-LC segmentally and uniformly 13C,15N-labeled polymers were packed into a Varian-style 3.2-mm thin-walled MAS rotor using a swinging-bucket centrifuge spinning at 20,000 × g for 16 h (20 min for the pH titration samples). For the 1H-13C CP-MAS/fs-REDOR recoupling measurements (15, 16), a soft Gaussian pulse was applied on resonance with the carbonyl/carboxylate signals and alternating transients were subtracted to provide a spectrum that only contained carbonyl/carboxylate signals. During the fs-REDOR mixing the 15N-13C dipolar interaction was selectively reintroduced using a hard π pulse on the 15N channel during the center of the 13C soft Gaussian pulse. fs-REDOR simulations were performed in SIMPSON (41) for an isolated 15N-13C spin pair. For the 2D and 3D NCACX, NCOCX, and CONCA experiments 15N-13C magnetization transfers used the SPECIFIC-CP method (42) using continuous wave 1H decoupling. For N-Cα transfers the 15N carrier frequency was placed at the center of the amide nitrogen signals and the 13C carrier frequency was placed at the center of the α-carbon signals. For N–CO transfers the 15N carrier frequency was placed at the center of the amide nitrogen signals and the 13C carrier frequency was placed at the center of the carbonyl carbon signals. Sample packing and experimental conditions were similar for the segmentally and uniformly 13C,15N-labeled mCherry-hnRNPA2-LC D290V mutant polymers.
The 13CO Tyr-labeled mCherry-hnRNPA2-LC polymers were packed into a Varian style 3.2-mm medium-walled rotor using a swinging bucket centrifuge with ∼1.5 mm Teflon spacers on either side of the sample to center the polymers in the probe coil. The peak intensity was plotted as a function of recoupling time. The rms noise in each measurement was calculated from 145 baseline points that were free of spectral artifacts in all nine spectra. The rms noise was 0.001 for all sites relative to the normalized peak intensity in each data point. The noise was calculated in Excel (Microsoft). To account for signal decay in the 13C-13C PITHIRDS-CT measurement due to natural abundance signals the measurement was performed on hnRNPA2-LC polymers without isotopic enrichment of the expression media. The data were fit to the equation
where t is the 13C-13C PITHIRDS-CT recoupling time in milliseconds. The data were then normalized to put it on the same scale as the experimental data. Ten percent of the decay was subtracted from the 13C-13C PITHIRDS-CT decay curves for the isotopically labeled sample based on the relative number of natural abundance 13CO to isotopically labeled 13CO Tyr sites. Decay curves were simulated using FORTRAN programs for a linear chain of 13C atoms separated by 0.4, 0.5, 0.6, and 0.7 nm. The data presented in SI Appendix, Fig. S8B were collected on mCherry-hnRNPA2-LC D290V mutant polymer samples segmentally isotopically enriched at 13CO Tyr. The NMR rotor packing, data collection, and analysis were performed in the same way as the 13CO Tyr-labeled mCherry-hnRNPA2-LC wild-type polymer sample.
Hydrogel Binding Assay.
Preparation of hydrogels of the wild type of mCherry-hnRNPA2-LC was carried out as described before (11). GFP-fused half constructs of hnRNPA2-LC were diluted in 1 mL gelation buffer at a final concentration of 1 μM then added to dishes containing the mCherry-hnRNPA2-LC hydrogels. After incubation at 4 °C overnight, the hydrogels were analyzed by confocal microscopy for bound GFP signals as previously described (43).
Thioflavin T Fluorescence Assay.
Ten microliters of mCherry-hnRNPA2-LC wild-type and D290V mutant polymers used in the MPL measurements were added to 200-μL solutions of either water alone or 20 μM Thioflavin T (Sigma). Ten microliters of water was added to 200 μL of 20 μM thioflavin T solution for the nonpolymer control. Fluorescence spectra were recorded on a StellarNet, Inc. fluorimeter with excitation at 423 nm. The signal averaging time was 500 ms and eight scans were summed for the spectra presented in SI Appendix, Fig. S3 C and D.
Additional information on the experimental methods can be found in SI Appendix.
Supplementary Material
Acknowledgments
S.L.M. thanks an anonymous donor for the generous provision of unrestricted funds. This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health, PRAT Fellowship Fi2GM117604 (to D.T.M.) through the National Institute of General Medical Sciences, and National Institute of General Medical Sciences Grant U01GM107623 (to S.L.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806174115/-/DCSupplemental.
References
- 1.Harrison AF, Shorter J. RNA-binding proteins with prion-like domains in health and disease. Biochem J. 2017;474:1417–1438. doi: 10.1042/BCJ20160499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapeli K, Martinez FJ, Yeo GW. Genetic mutations in RNA-binding proteins and their roles in ALS. Hum Genet. 2017;136:1193–1214. doi: 10.1007/s00439-017-1830-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim HJ, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackenzie IR, et al. TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron. 2017;95:808–816.e9. doi: 10.1016/j.neuron.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo W, et al. An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nat Struct Mol Biol. 2011;18:822–830. doi: 10.1038/nsmb.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson BS, et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel A, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 8.Vieira NM, et al. A defect in the RNA-processing protein HNRPDL causes limb-girdle muscular dystrophy 1G (LGMD1G) Hum Mol Genet. 2014;23:4103–4110. doi: 10.1093/hmg/ddu127. [DOI] [PubMed] [Google Scholar]
- 9.Ryan VH, et al. Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol Cell. 2018;69:465–479.e7. doi: 10.1016/j.molcel.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin Y, et al. Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell. 2016;167:789–802.e12. doi: 10.1016/j.cell.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang S, et al. The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid-like droplets, and nuclei. Cell. 2015;163:829–839. doi: 10.1016/j.cell.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu JX, et al. Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuttle MD, et al. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat Struct Mol Biol. 2016;23:409–415. doi: 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato M, et al. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaroniec CP, Tounge BA, Herzfeld J, Griffin RG. Frequency selective heteronuclear dipolar recoupling in rotating solids: Accurate (13)C-(15)N distance measurements in uniformly (13)C,(15)N-labeled peptides. J Am Chem Soc. 2001;123:3507–3519. doi: 10.1021/ja003266e. [DOI] [PubMed] [Google Scholar]
- 16.Jaroniec CP, Tounge BA, Rienstra CM, Herzfeld J, Griffin RG. Measurement of 13C−15N distances in uniformly 13C labeled biomolecules: J-decoupled REDOR. J Am Chem Soc. 1999;121:10237–10238. [Google Scholar]
- 17.Ulrich EL, et al. BioMagResBank. Nucleic Acids Res. 2008;36(Suppl 1):D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gullion T, Schaefer J. Rotational-echo double-resonance NMR. J Magn Reson (1969) 1989;81:196–200. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Hartmann SR, Hahn EL. Nuclear double resonance in the rotating frame. Phys Rev. 1962;128:2042–2053. [Google Scholar]
- 20.Pines A, Gibby M, Waugh J. High resolution NMR of dilute spins in solids. Bull Am Phys Soc. 1971;16:1408. [Google Scholar]
- 21.Wishart DS. Interpreting protein chemical shift data. Prog Nucl Magn Reson Spectrosc. 2011;58:62–87. doi: 10.1016/j.pnmrs.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Hu K-N, Qiang W, Tycko R. A general Monte Carlo/simulated annealing algorithm for resonance assignment in NMR of uniformly labeled biopolymers. J Biomol NMR. 2011;50:267–276. doi: 10.1007/s10858-011-9517-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu KN, Qiang W, Bermejo GA, Schwieters CD, Tycko R. Restraints on backbone conformations in solid state NMR studies of uniformly labeled proteins from quantitative amide 15N-15N and carbonyl 13C-13C dipolar recoupling data. J Magn Reson. 2012;218:115–127. doi: 10.1016/j.jmr.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tycko R. Symmetry-based constant-time homonuclear dipolar recoupling in solid state NMR. J Chem Phys. 2007;126:064506. doi: 10.1063/1.2437194. [DOI] [PubMed] [Google Scholar]
- 26.Chen B, Thurber KR, Shewmaker F, Wickner RB, Tycko R. Measurement of amyloid fibril mass-per-length by tilted-beam transmission electron microscopy. Proc Natl Acad Sci USA. 2009;106:14339–14344. doi: 10.1073/pnas.0907821106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greene RF, Jr, Pace CN. Urea and guanidine hydrochloride denaturation of ribonuclease, lysozyme, α-chymotrypsin, and β-lactoglobulin. J Biol Chem. 1974;249:5388–5393. [PubMed] [Google Scholar]
- 28.O’Nuallain B, Shivaprasad S, Kheterpal I, Wetzel R. Thermodynamics of A β(1-40) amyloid fibril elongation. Biochemistry. 2005;44:12709–12718. doi: 10.1021/bi050927h. [DOI] [PubMed] [Google Scholar]
- 29.Pons J, Rajpal A, Kirsch JF. Energetic analysis of an antigen/antibody interface: Alanine scanning mutagenesis and double mutant cycles on the HyHEL-10/lysozyme interaction. Protein Sci. 1999;8:958–968. doi: 10.1110/ps.8.5.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray DT, et al. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell. 2017;171:615–627.e16. doi: 10.1016/j.cell.2017.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers: Their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paul KR, et al. Effects of mutations on the aggregation propensity of the human prion-like protein hnRNPA2B1. Mol Cell Biol. 2017;37:e00652-16. doi: 10.1128/MCB.00652-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes MP, et al. Atomic structures of low-complexity protein segments reveal kinked β sheets that assemble networks. Science. 2018;359:698–701. doi: 10.1126/science.aan6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qamar S, et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell. 2018;173:720–734.e15. doi: 10.1016/j.cell.2018.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koehl P, Levitt M. Structure-based conformational preferences of amino acids. Proc Natl Acad Sci USA. 1999;96:12524–12529. doi: 10.1073/pnas.96.22.12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens AJ, et al. Design of a split intein with exceptional protein splicing activity. J Am Chem Soc. 2016;138:2162–2165. doi: 10.1021/jacs.5b13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baxa U, et al. Characterization of beta-sheet structure in Ure2p1-89 yeast prion fibrils by solid-state nuclear magnetic resonance. Biochemistry. 2007;46:13149–13162. doi: 10.1021/bi700826b. [DOI] [PubMed] [Google Scholar]
- 38.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- 39.Takegoshi K, Nakamura S, Terao T. 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- 40.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 41.Bak M, Rasmussen JT, Nielsen NC. SIMPSON: A general simulation program for solid-state NMR spectroscopy. J Magn Reson. 2000;147:296–330. doi: 10.1006/jmre.2000.2179. [DOI] [PubMed] [Google Scholar]
- 42.Baldus M, Petkova AT, Herzfeld J, Griffin RG. Cross polarization in the tilted frame: Assignment and spectral simplification in heteronuclear spin systems. Mol Phys. 1998;95:1197–1207. [Google Scholar]
- 43.Kato M, Lin Y, McKnight SL. Cross-β polymerization and hydrogel formation by low-complexity sequence proteins. Methods. 2017;126:3–11. doi: 10.1016/j.ymeth.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.