Abstract
Background
Epidermal growth factor receptor (EGFR) signalling is critical in epithelial cancer development. Human rhomboid family-1 (RHBDF1) facilitates the secretion of TGFα, an EGFR ligand, in breast cancer; however, the underlying mechanism remains unclear. We evaluated the role for RHBDF1 in clathrin-coated vesicle (CCV)-dependent pro-TGFα membrane trafficking in breast cancer cells upon stimulation by G-protein coupled receptor (GPCR) agonists.
Methods
RHBDF1 was silenced in various breast cancer cells using shRNA. TGFα levels, subcellular localization, and secretion were evaluated using ELISA, immunofluorescent staining, and coimmunoprecipitation. Phosphorylation and expression of relevant proteins were measured by western blotting. RHBDF1-dependent cell viability and invasion were measured.
Findings
RHBDF1 mediates GPCR agonist-induced EGFR phosphorylation by promoting TGFα secretion in various types of breast cancer cells. RHBDF1 not only mediates ADAM17-dependent shedding of TGFα, but is essential in membrane trafficking of pro-TGFα. RHBDF1 silencing results in blocking of clathrin uncoating from CCV, a crucial step for the plasma membrane release of pro-TGFα. Interaction of RHBDF1 with auxilin-2, a CCV protein, determines the recruitment of HSC70 to CCV to facilitate clathrin uncoating. RHBDF1 function is required for the proliferation and mobility of breast cancer cells upon stimulation by Sphingosine 1 Phosphate (S1P), a GPCR agonist. We demonstrate a significant correlation between RHBDF1 overexpression and EGFR activation in breast cancer tissues.
Interpretation
RHBDF1 is an indispensable component of the protein trafficking machinery involved in GPCR-mediated EGFR transactivation, and is an attractive therapeutic target for cancer.
Fund
National Natural Science Foundation of China (81,672,740 to ZSZ, 81,272,356 and 81,330,029 to LYL).
Keywords: RHBDF1, EGFR, TGFα, Membrane trafficking, Clathrin-coated vesicle, Breast cancer, Progression
Abbreviations: EGFR, Epidermal growth factor receptor; RHBDF1, Human rhomboid family-1 gene; CCVs, Clathrin-coated vesicles; GPCR, G protein-coupled receptors; TGFα, Transforming growth factor-α; LPA, Lysophosphatidic acid; S1P, Sphingosine-1-phosphate; ADAM17, ADAM metallopeptidase domain 17; AP1, Clathrin-associated adaptor protein complex-1; GRASP55, Golgi reassembly-stacking protein; HSC70, Heat shock cognate protein-70; H&E, Haematoxylin and Eosin staining; RT-qPCR, Reverse transcription quantitative PCR; IF, Immunofluorescence; IP, Immunoprecipitation; shRNA, Short hairpin RNA; ELISA, Enzyme-linked immuno sorbent assay
Research in context.
Evidence before this study
EGFR is frequently overexpressed in breast cancer and is critically correlated with poor prognosis in this patient population. Recent studies show that GPCR agonists induce EGFR transactivation by promoting membrane trafficking and proteolytic processing of EGFR pro-ligands. Human rhomboid family-1 (RHBDF1), a member of the rhomboid gene family, was reported to be highly expressed in cancer cells and involved in the mediation of GPCR agonist-stimulated secretion of an EGFR ligand TGFα. Moreover, RHBDF1 was reported to regulate the trafficking and maturation of A disintegrin and metalloprotease 17 (ADAM17), which is responsible for the proteolytic release of pro-TGFα on the cell surface. However, to date, the mechanisms underlying the association of RHBDF1 and TGFα remain unclear. In the study, we aimed to explore the mechanism of action of RHBDF1 to explore its potential as a novel therapeutic target.
Added value of this study
Findings from this study indicate that RHBDF1 is an essential component of protein trafficking machinery involving clathrin-coated vesicle- and ADAM17-dependent ectodomain shedding of pro-TGFα in breast cancer cells in response to G-protein coupled receptor activation. Mechanistically, the interaction of RHBDF1 with auxilin-2, a CCV protein, is required for the recruitment of HSC70 to CCV in order for clathrin uncoating to take place, which is a crucial step prior to the delivery of pro-TGFα to the plasma membrane. In addition, RHBDF1 expression is required for the proliferation and mobility of breast cancer cells in response to stimulation by S1P, a GPCR agonist. Furthermore, we show that there is a significant correlation between RHBDF1 overexpression and EGFR phosphorylation in clinical specimens of breast cancer.
Implications of all the available evidence
Our data reveal the missing link between GPCR and EGFR activation, and highlight novel relationships between RHBDF1 expression, EGFR signalling, and breast cancer development. These findings indicate that RHBDF1 is an indispensable component of the protein trafficking machinery and may act as a vital onco-modulator by mediating the clinically important EGFR transactivation signalling pathway. The findings indicate a strong potential of RHBDF1 as a novel anti-cancer drug target that may represent an improvement over current therapies.
Alt-text: Unlabelled Box
1. Introduction
The human epidermal growth factor receptor (EGFR) plays critical physiological roles in the regulation of tissue development and homeostasis in epithelial cells [1,2]. However, EGFR is also considered an oncogene, and has been frequently implicated in oncogenic transformation of many human cancers [3], including breast cancer, non-small cell lung cancer, colorectal cancer, and gastric cancer [4,5]. In addition to activation directly by its own ligands such as epidermal growth factor (EGF) and transforming growth factor-α (TGFα), EGFR may be indirectly activated by a number of G-protein coupled receptor (GPCR) agonists, such as lysophosphatidic acid (LPA) [6] or sphingosine-1-phosphate (S1P) [7]. GPCR agonist-initiated transactivation of EGFR involves a “triple-membrane-passing-signal” pathway [8], which begins with the activation of a GPCR, followed by membrane trafficking and proteolytic processing of a number of pro-ligands of EGFR such as EGF, TGFα [9], amphiregulin (AREG) [10], and heparin-binding EGF-like growth factor (HB-EGF) [11]. The spatial and temporal distribution of pro-ligands in these signalling pathways is exquisitely controlled [12]. However, the molecular mechanisms underlying GPCR-mediated EGFR transactivation remain unclear.
Human rhomboid family-1 (RHBDF1) belongs to a large family of rhomboid proteins [13]. Unlike the prototypic rhomboid-1 in Drosophila, which is a protease that processes pro-EGF, RHBDF1 apparently lacks protease activity, thus, the name “inactive rhomboid” [14]. RHBDF1 has been shown to be a key mediator of GPCR agonist-induced EGFR phosphorylation and TGFα secretion [15,16]. RHBDF1 gene silencing leads to cancer cell apoptosis or autophagy, as well as inhibition of xenograft tumour growth [17]. Additionally, RHBDF1 was found to be a critical component of a molecular switch that regulates HIF1α stability under hypoxic conditions in breast cancer cells [18]. RHBDF1 expression is also involved in the activation of EGFR signalling during mouse embryonic development [19], in cancer predisposition syndrome [20], and in the regulation of proteasome activity under endoplasmic reticulum stress [21]. Moreover, the activity of RHBDF1 and its homolog RHBDF2 (iRhom2) has been reported to be important in promoting membrane trafficking and maturation of ADAM17, which is responsible for the proteolytic release and shedding of precursor proteins, including TGFα, on the cell surface [22,23].
In the cellular context, pro-TGFα exhibits a dynamic equilibrium between the intracellular compartments and the cell surface [24]. Clathrin-coated vesicles (CCV) are involved in the trafficking of pro-TGFα and a number of plasma membrane proteins [25]. Biosynthesis and CCV-mediated trafficking of membrane proteins are regulated by a variety of signals [26]. Briefly, the proteins are tethered to the Golgi reassembly-stacking protein (GRASP55) on the trans-Golgi network [27], and are recognizable by the μ-subunit of clathrin-associated adaptor protein complex-1 (AP1μ1) prior to trafficking via CCV [28]. Then, the β1 subunit of adaptor protein complex-1 (AP1β1) is recruited to the trans-Golgi network to serve as binding sites for the polymerization of clathrin into a polyhedral coating [29]. After CCV formation and budding, heat shock cognate protein-70 (HSC70) is recruited to initiate clathrin uncoating and protein trafficking to the early endosomes or plasma membrane [30]. Dysregulation of pro-TGFα trafficking under pathological conditions has an impact on EGFR signal activation [31,32].
In this study, we focus on the molecular mechanism by which RHBDF1 expression affects GPCR-mediated EGFR transactivation in breast cancer cells. We show here that RHBDF1 is a critical component of GPCR agonist-responsive, CCV-dependent trafficking machinery of pro-TGFα. Our findings not only provide new insights into the mechanism of GPCR-mediated EGFR transactivation, but also indicate that RHBDF1 may serve as an attractive target for breast cancer therapy.
2. Materials and methods
2.1. Reagents and antibodies
Anti-phospho-EGFR (Tyr1068) (D7A5) and EGFR (D38B1) antibody were purchased from Cell Signalling Technology (Danvers, MA). Cell Mask Plasma Membrane Stains was purchased from Thermo Scientific (Rockford, IL). Anti-pro-TGFα (TG86) Na+-K+-ATPase (EPR15460B), clathrin (X22), AP1β1 and μ1, HSC-70 (1B5), auxilin-2(GAK), β-actin, and inhibitor Pitstop 1 were purchased from Abcam (Cambridge, MA). Anti-GRASP55 (E-11) was purchased from Santa Cruz Biotechnology (Dallas, TX). hEGF (E9644), human transforming growth factor-α (T7924), sphingosine 1-phosphate (73914), and oleoyl-L-α-lysophosphatidic acid sodium salt (L7260) were purchased from Sigma-Aldrich (St. Louis, MO). Human TGFα DuoSet ELISA Kit was purchased from R&D Systems (Minneapolis, MN). Brefeldin A (BFA, S7046) inhibitor was purchased from Selleck (Houston, TX).
2.2. Cell culture and transfection
MDA-MB-231, MDA-MB-468, MCF-10A, MCF-7, and T-47D cell lines were purchased from the American Type Culture Collection (ATCC, USA). MCF-10A cells were cultured in Medium Dulbecco's Modified Eagle Medium (DMEM)/F12 medium supplemented with 20% horse serum; MCF-7 and T47D cells were cultured in DMEM supplemented with 0.01 mg/mL Human insulin; MDA-MB-231, MDA-MB-468 cells were cultured in DMEM supplemented with 10% foetal bovine serum at 37 °C with 5% CO2. RHBDF1 knocked-down (shRHB) and scrambled control (shScr) cell lines were generated, respectively (Fig. S1A), by using shRNA constructs in retroviral vector or scrambled shRNA cassette in pRS Vector (Origene), and selected with puromycin for 2 months. RHBDF1-overexpressing (RHB) and control (vector) cell lines were generated by transient transfection with human RHBDF1 cDNA and vector plasmids, respectively, using X-treme GENE Transfection Reagents according to the manufacturer's instructions. The cell lines used in this study were authenticated by DNA profiling using short tandem repeat (STR) analysis on an ABI 3730XL Genetic Analyzer System at Talen-bio Technology Co. LTD (China).
2.3. Condition media stimulation assay
The shScr- and shRHB-treated MDA-MB-231 cells were cultured in 6-cm plates, incubated in the presence or absence of S1P (10 nM) for 30 min, and then grown in serum-free media for 2 h. The conditioned media were harvested, centrifuged to remove cell debris, and added to sub-confluent 24-h pre-starved MDA-MB-231 cells for another 30 min. Then, the supernatants were aspirated and cell lysates were analysed by western blotting to detect total EGFR and activated EGFR levels.
2.4. ELISA assays
Quantitation of TGFα secretion levels was performed using ELISA kits (R&D Systems, USA) according to the manufacturer's instructions. The conditioned medium (100 μL) was collected from shScr- and shRHB-treated cells following stimulation with or without S1P (10 nM). Analysis was performed in triplicate for each independent experiment.
2.5. Con A enrichment of glycoprotein
To improve the detection of ADAM17, cells were lysed in TX-100 lysis buffer (1% Triton X-100, 150 mM NaCl, 50 mM Tris-HCl, pH 7.4) supplemented with complete protease inhibitor cocktail (Roche) and 10 mM 1,10-phenanthroline to prevent autocatalysis of ADAM17, and centrifuged at 20,000 ×g for 20 min. Lysates were then mixed with 50 μL of washed concanavalin A (Con A) Agarose beads and incubated for 2–3 h at 4 °C on a rotor. Beads were washed twice in the same buffer and eluted by heating for 10 min at 75 °C in sample buffer. Samples were resolved on SDS-PAGE gels for subsequent western blotting.
2.6. Biotinylation of cell plasma membrane proteins
Cells were washed with ice-cold PBS and then incubated with sulpho-NHS-SS-biotin–based Cell Surface Protein Isolation Kit (Pierce, USA) for 15 min at 4 °C. The reaction was terminated with 100 mM glycine in PBS. The cells were rinsed with PBS and lysed in RIPA lysis buffer (Millipore, GER) with added protease inhibitor cocktail (Roche, USA). The lysates were centrifuged for 10 min at 13,000 rpm. Biotinylated cell membrane proteins were precipitated with Streptavidin Sepharose solutions (GE Healthcare, USA). The pellets were gently rinsed with PBS, dissolved in 4 × SDS sample buffer, incubated at 65 °C for 10 min, and subjected to western blotting analysis.
2.7. Quantitative real-time PCR
Total RNA was extracted using the RNeasy RNA kit (Qiagen, GER), cDNA was synthesized using the EasyScript First-Strand cDNA Synthesis SuperMix (TransGen, China), and qPCR was performed using a Mastercycler ep realplex2 qPCR System (Eppendorf, GER). Results were analysed using the relative quantity (ΔΔCt) method. The expression level of each mRNA was normalized to that of β-actin mRNA and presented as a fold increase relative to values for shRHB-treated cells. All experiments were performed in triplicate. The sense and antisense primer sequences for human TGFα, AREG, HB-EGF, and β-actin were, respectively: 5′ -AAT GAC TGC CCA GAT TCC CAC-3′ and 5′ -CAA CGT ACC CAG AAT GGC AGA-3′; 5′-AGC TGC CTT TAT GTC TGC TGT G-3′ and 5′ -CGT TCC TCA GCT TCT CCT TCA T-3′; 5′ -TAT CCT CCA AGC CAC AAG CA-3′ and 5′ -TGC AGA AGT CCT TGT ATT TCC G-3′; 5′ -CCA TCA TGA AGT GTG ACG TGG A-3′ and 5′-TTC TGC ATC CTG TCG GCA A-3′.
2.8. Immunoblot analysis
Cells were lysed by sonication in lysis buffer containing 2 mM Na3VO4, 1 mM PMSF, 1 mM EDTA, and protease inhibitor cocktail (Roche), and then subjected to 12% SDS-PAGE. The proteins were transferred to a PVDF membrane (Roche, USA). The membranes were blocked and probed with appropriate antibodies. Using goat anti-mouse or rabbit IgG HRP conjugate antibodies (Thermo, USA) as secondary antibodies, the proteins were visualized using a chemiluminescent HRP substrate (Millipore, USA). Protein band densitometry was performed using ImageJ software (NIH).
2.9. Immunofluorescence analysis
Serum-starved shSrc- and shRHB-treated cells grown on chamber slides were stimulated with S1P (10 nM) for indicated durations. Cells were washed twice with phosphate-buffered saline (PBS), and then fixed with 4% paraformaldehyde at 4 °C for 15 min. After permeabilization with 0.02% Triton X-100 for 2 min, the cells were blocked with 10% goat serum at 25 °C for 1 h. The cells were incubated with a membrane dye (CellMask, ThermoFisher) and primary antibodies (1:50) in blocking solution overnight at 4 °C. After rinsing with PBS, cells were incubated with goat anti-mouse Alexa Fluor 488 conjugate (1:2000) or goat anti-rabbit Alexa Fluor 555 conjugate (1: 2000) antibodies (Invitrogen, USA) for 30 min at 25 °C in the dark, rinsed and mounted with ProLong Gold Antifade Mountant with DAPI (Invitrogen, USA), and then examined with a confocal microscope (Leica TCS SP8, GER) and processed using Image Pro Plus software.
2.10. Co-immunoprecipitation
Cells were homogenized in lysis buffer (0.5% Triton X-100, 150 mM NaCl and 5 mM EDTA) and centrifuged. The supernatants were incubated with an appropriate antibody against pro-TGFα, AP1β1, clathrin, Auxilin-2, HSC70, or Flag at 4 °C overnight. Pierce protein G-Agarose (Thermo Fisher Scientific, USA) beads were then added and incubated on a rotator at 4 °C for 2 h. The beads were collected by centrifugation, rinsed with lysis buffer, heated for 6 min at 100 °C in a 2× loading buffer, and subjected to 12% SDS-PAGE. The proteins were transferred to PVDF membranes (Millipore, USA), probed with specific antibodies and HRP-conjugated secondary antibodies, and visualized using Immobilon Western Chemiluminescent HRP substrate (Millipore, USA).
2.11. Cell proliferation assay
Breast cancer cells of each type were seeded in six replicates into 96-well plate (2000 cells per well), and incubated at 37 °C in 5% CO2 with or without S1P (10 nM) in phenol-red free media for the desired time period. CellTiter 96 AQueous One Solution Reagent (Promega, Madison, WI) was used according to manufacturer's instructions to determine cell proliferation capacity in vitro. Briefly, after CellTiter 96 AQueous One Solution Reagent was added at the indicated time points to each well, cells were incubated for 4 h at 37 °C. Then, the optical density of the MTS product was measured at 490 nm using a microplate reader (Bio-Rad, Hercules, CA). Total cell numbers were calculated based on calibration curves.
2.12. Cell transwell migration/invasion assay
Transwell migration/invasion assays were performed following the manufacturer's instructions. shScr-and shRHB-treated cells were serum-starved for 24 h. For the migration assay, 1 × 105 cells were seeded in the upper chamber (Corning, USA) in a serum-free medium with/without S1P (10 nM) treatment; for the invasion assay, a layer of Matrigel was applied to the upper chamber. The lower chambers of both assays were supplemented with 10% serum-containing medium; then, the inserts were incubated in a CO2 incubator at 37 °C for 24 h. Next, cells in the upper chamber were removed using a cotton swab, and Transwell membranes were fixed with methanol for 1 h and stained with 1% crystal violet. The number of cells that migrated to the lower surface of the membrane was observed under a microscope (Nikon, JPN), and counted using Image Pro Plus software. Three replicates were obtained for each assay.
2.13. Tissue microarray and immunohistochemistry
Human breast carcinoma tissue microarray, containing 60 samples of invasive ductal carcinoma, 9 samples of medullary carcinoma, and 3 samples of cancer adjacent normal tissue, was obtained from US Biomax Corporation (BC08013a); all tissue cores were obtained from diagnostic or surgical samples as serial sections according to the supplier (Table S1). RHBDF1 expression and EGFR phosphorylation status were determined by immunohistochemical staining with DAB kit (Invitrogen, USA) according to the manufacturer's instructions. The sections were incubated with anti-phospho-EGFR antibody (Tyr1068) (#3777, 1: 50 dilution, CST) or anti-RHBDF1 antibody (LS-C81438, 1: 100 dilution, LSBio) at 4 °C overnight, followed by incubation with biotinylated secondary antibody and streptavidin-peroxidase. Sections were then visualized with DAB Chromogen Solution, counterstained with haematoxylin, and dehydrated in ethanol. Images were acquired at 20× magnification using an Aperio Digital Pathology Microsystems scanner (Leica, GER). Staining intensities of each section were scored (grades 0–3) using Image Pro Plus. Statistical analysis was performed using SPSS 19.0 software.
2.14. Statistical analysis
Correlations between RHBDF1 expression and EGFR phosphorylation were determined by calculating the Pearson correlation coefficient. All experiments were repeated at least three times, and the results are presented as the means ± SEM. Data were subjected to variance analysis (ANOVA), and 2-tailed, unpaired Student's t-tests. Differences with p-values smaller than 0.05 were considered statistically significant.
3. Results
3.1. RHBDF1 facilitates GPCR ligand-dependent EGFR transactivation by modulating secretion of TGFα
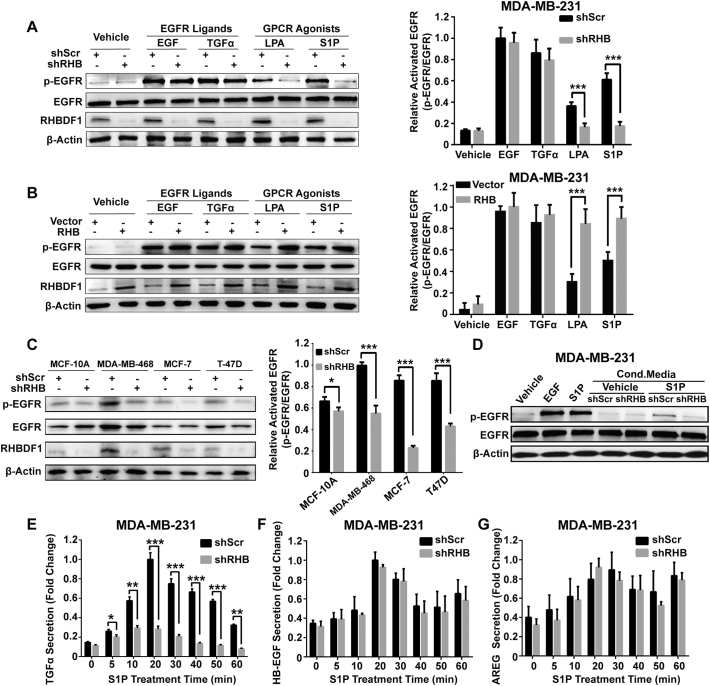
We examined EGFR phosphorylation in response to EGFR ligands EGF or TGFα, and GPCR agonists LPA or S1P, in basal breast cancer MDA-MB-231 cells. EGF or TGFα and the GPCR agonists elicit the same response of EGFR phosphorylation, whereas cells treated with shRNA against RHBDF1 (shRHB) became irresponsive to stimulation by LPA or S1P in terms of EGFR phosphorylation (Fig. 1A). Additionally, we studied the effect of RHBDF1 overexpression by transiently transfecting the cells with a plasmid containing Flag-RHBDF1 cDNA (RHB), using an empty vector (Vector) as a control (Fig. S1B). Consistently, RHBDF1 overexpression led to a marked increase in EGFR phosphorylation in response to LPA or S1P treatment (Fig. 1B). To further assess the potential causal effect of RHBDF1 on EGFR transactivation, a panel of breast cell lines were examined. RHBDF1 silencing significantly attenuated EGFR phosphorylation stimulated by S1P in another triple negative breast cancer (TNBC) cell line MDA-MB-468, and two non-TNBC breast cancer cell lines (T-47D and MCF-7) (Fig. 1C).
Fig. 1.
RHBDF1 facilitates ligand-dependent EGFR transactivation by modulating TGFα secretion. (A) RHBDF shRNA (shRHB)-transfected MDA-MB-231 cells treated with EGF (10 ng/mL) or TGFα (10 ng/mL) for 5 min, or with LPA (10 nM) or S1P (10 nM) for 30 min, and subjected to western blotting analysis; shScr, scrambled RNA control. (B) Western blotting analysis of similarly treated RHBDF1-overexpressing MDA-MB-231 cells (RHB) and empty vector-transfected control (Vector). (C) Analysis of S1P (10 nM)-triggered EGFR phosphorylation in shScr- and shRHB-treated MCF-10A, MDA-MB-468, MCF7, and T-47D cells. (D) Cultured shScr- and shRHB-treated MDA-MB-231 cells were treated with S1P (10 nM) for 30 min and maintained in serum-free media for 2 h. The collected conditioned media were added to 24 h pre-starved parental MDA-MB-231 cells. Cell lysates were subjected to immunoblot analysis to detect EGFR phosphorylation levels. Changes in the concentration of TGFα (E), HB-EGF (F) or AREG (G) in conditioned media of shScr- or shRHB-treated MDA-MB-231 cells as a function of time of S1P (10 nM) stimulation. Bar graphs represent densitometric analysis of the results of independent western blotting experiments. In all panels, data are means ± SEM (n = 3); *p < 0.05, **p < 0.01 and *** p < 0.001.
We then stimulated control shRNA (shScr)- or shRHB-treated cells with GPCR ligand S1P, collected the conditioned media, and used the conditioned media to treat parental MDA-MB-231 cells. Conditioned media from S1P-stimulated shRHB-treated cells did not induce EGFR phosphorylation as much as that from S1P-stimulated shScr-treated cells (Fig. 1D), suggesting that RHBDF1 silencing hinder the secretion of EGF-like ligands. To identify the EGFR ligand whose secretion was affected, we analysed the concentration of TGFα, AREG, or HB-EGF in the conditioned media by using ELISA. Interestingly, we found that the amount of TGFα in the conditioned media of shRHB-treated cells was less than that in shScr-treated cells following S1P stimulation (Fig. 1E), whereas RHBDF1 silencing had little effect on AREG and HB-EGF production (Figs. 1F, G and S2). These results show that RHBDF1 is a critical component of the molecular machinery involved in the transmission of GPCR ligand-initiated signals that lead to TGFα secretion and EGFR activation.
3.2. RHBDF1 modulates ADAM17-dependent shedding and membrane trafficking of pro-TGFα
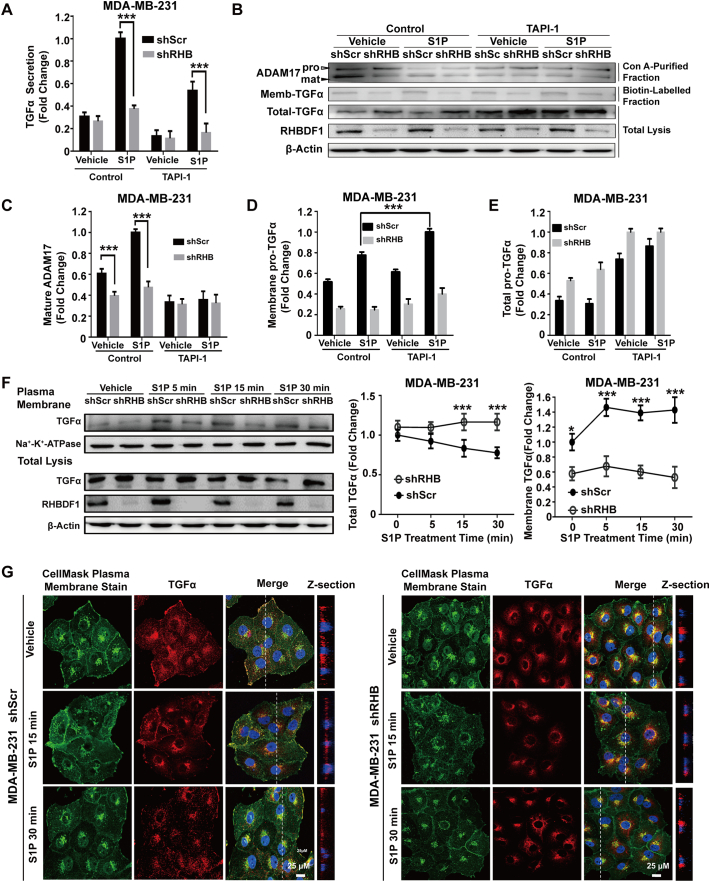
We investigated whether RHBDF1 participates in EGFR transactivation by regulating ADAM17-dependent TGFα shedding. We pre-incubated shScr- and shRHB-treated cells with or without ADAM17 inhibitor TAPI-1 (20 μM) for 1 h, and then stimulated the cells with S1P for 20 min. Measuring TGFα secretion, we found that TAPI-1 partially blocked S1P-induced TGFα secretion, compared with RHBDF1 silencing alone (Fig. 2A). Using concanavalin A (Con A) enrichment of glycoprotein and Western blot assay, we confirmed that RHBDF1 silence markedly reduced the maturation and activity of ADAM17 (Figs. 2B, C). We then analysed the content of TGFα in cell membranes and in total cell lysates, and found that TAPI-1 resulted in the accumulation of higher amounts of membrane-bound TGFα in shScr-treated cells than in shRHB-treated cells (Fig. 2D). In the meantime, TAPI-1 treatment did not significantly affect the production of TGFα in cells pre-incubated in the presence or absence of S1P (Fig. 2E). These findings indicate that RHBDF1 is involved in ADAM17-dependent shedding and distribution of pro-TGFα to the cell surface, but not in pro-TGFα production.
Fig. 2.
RHBDF1 not only modulates ADAM17-dependent shedding but also alters membrane trafficking of pro-TGFα. (A) shScr- or shRHB-treated cells were preincubated in the presence or absence of ADAM17 inhibitors TAPI-1 (20 μM) for 1 h before S1P stimulation for another 20 min; then, the TGFα in supernatant was detected by ELISA. (B) The above lysates were enriched for glycoproteins with concanavalin A (Con A) and immunoblotted for ADAM17 to detect its maturation. The cell membrane was biotin-labelled and enriched to enable detection of the level of TGFα on the cell plasma membrane. The empty arrowhead indicates immature form, whereas the black arrowhead indicates mature forms of ADAM17. (C) The percentage of mature ADAM17 was calculated by normalizing the amount of immature ADAM17. The relative levels of TGFα on the cell membrane (D) and in total lysis (E) was quantified based on densitometry. (F) Plasma membrane-bound or cell-associated total TGFα in shScr- or shRHB-treated cells were detected by western blotting analysis at indicated time intervals following S1P treatment; Na-K-ATPase, control for plasma membrane proteins. (G). Representative images of plasma membrane staining with CellMask Plasma Membrane Stain (green) and immune-fluorescence staining of pro-TGFα (red). Z-section shows the transition of pro-TGFα from locations near the nucleus to the plasma membrane. Data are means ± SEM. Student t-test, *p < 0.05, **p < 0.01 and *** p < 0.001.
We next examined the time-dependent distribution of pro-TGFα to elucidate the action of RHBDF1 in membrane trafficking. We found that the levels of plasma membrane-associated pro-TGFα were much lower in shRHB-treated cells than in shScr-treated cells upon S1P stimulation, while there was little difference in total pro-TGFα levels in the cells between the experimental groups (Fig. 2F). We then used immunofluorescence staining to determine sub-cellular localization of pro-TGFα after S1P stimulation. We found that pro-TGFα was localized in both the Golgi apparatus and plasma membrane in shScr-treated cells upon S1P stimulation; however, in shRHB-treated cells, pro-TGFα mostly remained near the nucleus 30 min after S1P stimulation (Fig. 2G). These findings suggest that RHBDF1 function be required for pro-TGFα trafficking to the cell surface in response to GPCR activation.
3.3. Clathrin uncoating, not CCV formation, in pro-TGFα trafficking process requires RHBDF1
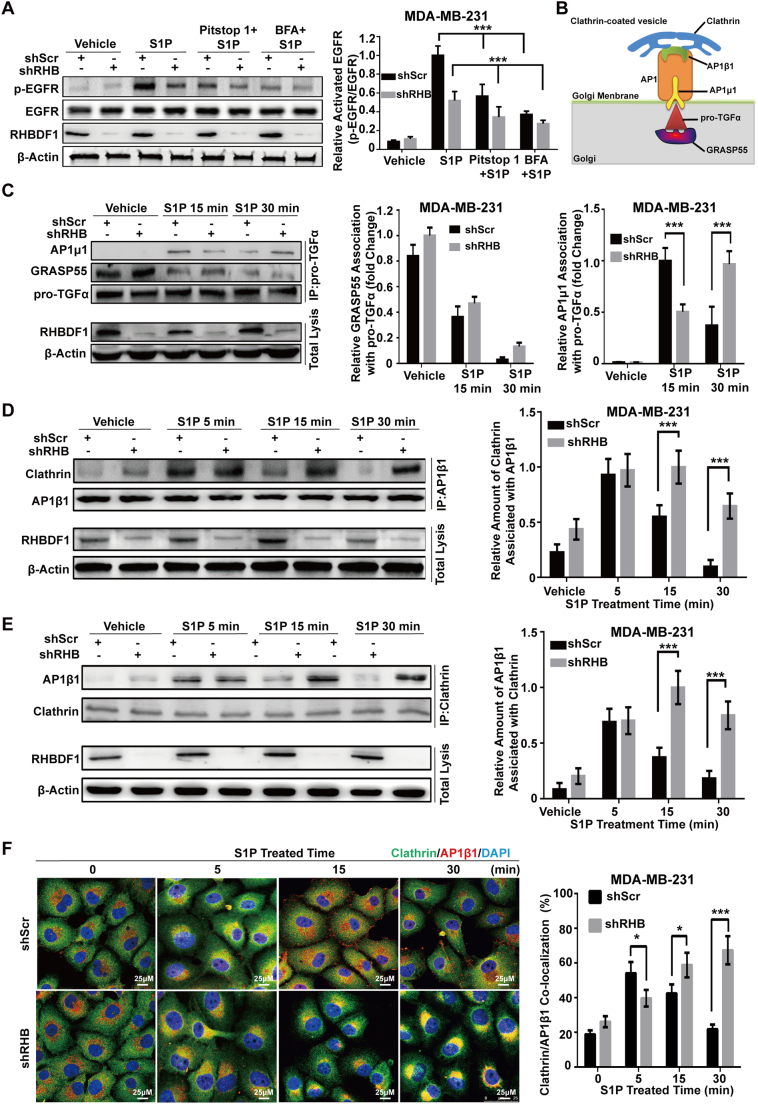
Since clathrin-coated vesicles (CCV) are known to be associated with a variety of protein trafficking processes from the trans-Golgi network to cell plasma membrane, especially those of precursor ligand proteins [25], we determined whether RHBDF1 takes part in the initiation of CCV-dependent pro-TGFα trafficking. We first compared the effect of RHBDF1 silencing with that of the clathrin terminal domain inhibitor Pitstop 1 or vesicular trafficking inhibitor Brefeldin A (BFA) on S1P-induced EGFR transactivation. We found that shRHB treatment of MDA-MB-231 cells had an inhibitory effect on EGFR phosphorylation, similar to that of each of the inhibitors. Moreover, the suppressive effect of shRHB treatment was additive with either inhibitor (Fig. 3A), suggesting that CCV plays a coordinating role with RHBDF1.
Fig. 3.
Clathrin uncoating, not formation, in pro-TGFα trafficking process requires RHBDF1. (A) Western blotting analysis of EGFR phosphorylation induced by S1P in shRHB- or shScr-transfected MDA-MB-231 cells in the presence or absence of CCV inhibitor Pitstop 1 or BFA for 1 h. (B) Schema depicting RHBDF1 as a signal integrator for the clathrin-coated vesicle signalling pathway. (C) Co-immunoprecipitation of pro-TGFα-associated proteins GRASP55 and AP1μ1 in shScr- and shRHB-treated cells with S1P (10 nM) stimulation at indicated time intervals. (D, E) Reciprocal co-immunoprecipitation of AP1β1-associated clathrin in shScr- or shRHB-treated cells incubated with S1P (10 nM) for the indicated time intervals. (F) Representative immunofluorescence images showing changes in subcellular locations of clathrin (green) and AP1β1 (red) as a function of S1P treatment time; merged colour (yellow) indicates co-localization of the two proteins. Quantitative analysis of changes of the intensity of the yellow colour as a function of duration of S1P treatment. Data are means ± SEM of three independent experiments. Student t-test; * p < 0.05; *** p < 0.001.
Pro-TGFα is known to be tethered to the Golgi apparatus through interaction with Golgi reassembly-stacking protein of 55 kDa (GRASP55) prior to being collected by secretion vesicles [33], and that the cargo molecules in CCV interact with clathrin-associated protein AP1μ1 [34] (Fig. 3B). We carried out co-immunoprecipitation experiments to determine the effect of RHBDF1 silencing on the association of pro-TGFα with GRASP55 and AP1μ1; these represent two steps that mark the subcellular localization of pro-TGFα before and after vesicle recognition, respectively. We found in a co-immunoprecipitation experiment that S1P stimulation led to dissociation of pro-TGFα and GRASP55 regardless of the presence or absence of RHBDF1 in the cells; the association of AP1μ1 with pro-TGFα first increased and then decreased within 30 min in shScr-treated cells in response to S1P stimulation, indicating that pro-TGFα was being taken up by CCV and then released (Fig. 3C). However, the association of pro-TGFα with AP1μ1 continued to increase throughout the experiment in shRHB-treated cells, indicating the accumulation of pro-TGFα in CCV in the absence of RHBDF1 (Fig. 3C). These findings indicate that RHBDF1 activity is required for the release of pro-TGFα from CCV during secretion vesicle-assisted trafficking.
Since the co-localization status of clathrin with AP1β1 is a measure of CCV integrity, we performed reciprocal co-immunoprecipitation experiments to study the effect of RHBDF1 silencing on the clathrin-AP1β1 interaction. We found that S1P induced a significant increase in the association of clathrin with AP1β1 within 5 min of S1P treatment in shSrc-treated cells; this association then returned to baseline in 15 to 30 min; however, the S1P-induced association of clathrin with AP1β1 continued to increase to a high level in shRHB-treated cells, indicating that RHBDF1 activity is required for CCV uncoating (Figs. 3D, E). To corroborate this finding, we carried out immunofluorescence staining to determine the subcellular co-positioning of clathrin and AP1β1 in response to S1P treatment in the presence or absence of RHBDF1. Using antibodies specific to either clathrin or AP1β1, we found that co-localization of clathrin and AP1β1 near the nucleus became apparent within 5 min of S1P treatment, then diminished within 15 min in shScr-treated cells, whereas in sharp contrast, clathrin and AP1β1 were co-localize in shRHB-treated cells throughout the course of the experiments (Fig. 3F). These data indicate that RHBDF1 activity is a prerequisite for the uncoating of clathrin from CCV in the pro-TGFα trafficking process.
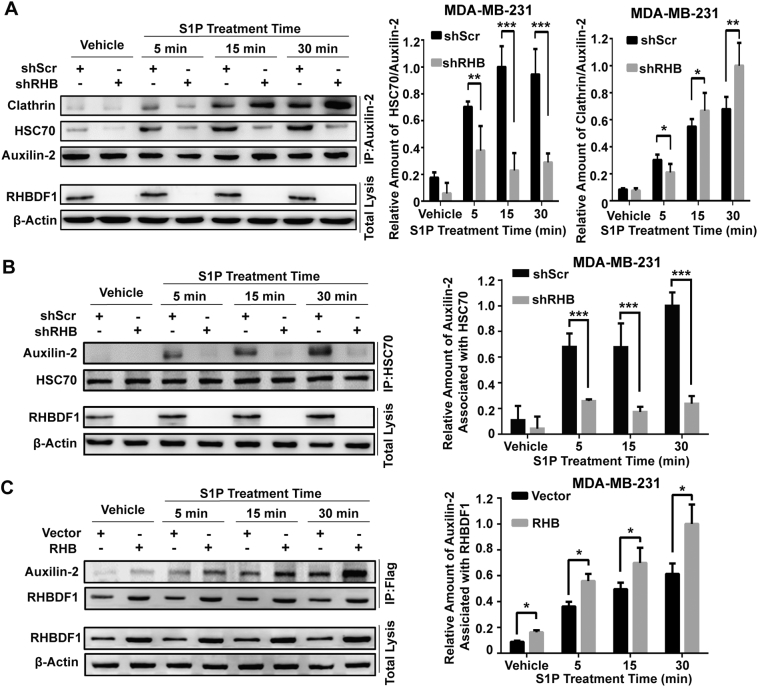
3.4. RHBDF1 enhances the recruitment of HSC70 with auxilin-2 to initiate CCV disassembly
We next analysed the interaction between auxilin-2 and HSC70 to further investigate the role of RHBDF1 in assisting clathrin uncoating, which is an initial step that leads to CCV disassembly. Using reciprocal co-immunoprecipitation, we found that, in shScr-treated cells the association of HSC70 with auxilin-2 continuously increased during the course of the experiment in response to S1P stimulation, whereas the HSC70/auxilin-2 association remained at low levels upon S1P stimulation in shRHB-treated cells, suggesting that RHBDF1 silencing leads to an inhibition of the S1P-induced interaction of HSC70 with auxilin-2 (Figs. 4A, B). Moreover, the interaction between clathrin and auxilin-2 was increasingly more apparent, during the course of the experiment, on S1P stimulation in shRHB-treated cells than in shScr-treated cells, suggesting that shRHB treatment prevented clathrin disassembly and recycling. These findings are consistent with the view that, on S1P stimulation, RHBDF1 activity is required for the recruitment of HSC70 to auxilin-2, and that the lack of this activity leads to failure of clathrin disassembly.
Fig. 4.
RHBDF1 modulates the recruitment of HSC70 with auxilin-2 to initiate CCV disassembly. (A) Co-immunoprecipitation of auxilin-2-associated clathrin and HSC70 in shScr- or shRHB-treated MDA-MB-231 cells incubated with S1P (10 nM) for the indicated time intervals. (B) Co-immunoprecipitation of HSC70-associated auxilin-2 in shScr- or shRHB-treated MDA-MB-231 cells incubated with S1P (10 nM) for the indicated time intervals. (C) Co-immunoprecipitation of RHBDF1 and auxilin-2 from RHBDF1-overexpressing MDA-MB-231 cells (RHB) and mock-transfected cell (Vector) as control. Data are means ± SEM from three independent experiments. Student's t-test; * p < 0.05; *** p < 0.001.
To verify our hypothesis, we treated Flag-RHBDF1 over-expressing and mock-transfected MDA-MB-231 cells with S1P, and then carried out co-immunoprecipitation to determine whether there was a direct interaction between Flag-RHBDF1 and auxilin-2. Remarkably, a prominent extent of interaction of Flag-RHBDF1 with auxilin-2 took place in Flag-RHBDF1-overexpressing cells in response to S1P-treatment (Fig. 4C; p < 0.05), indicating that RHBDF1 interacts directly with auxilin-2 proteins. Together, these findings support the view that binding of RHBDF1 to auxilin-2 is necessary for the recruitment of HSC70 to the latter, which would in turn lead to CCV disassembly by triggering dynamic uncoating of clathrin.
3.5. RHBDF1-enhanced proliferation and mobility of breast cancer cells correlates with EGFR activation
We next investigated whether RHBDF1 could endow breast cancer cells with proliferative advantages in tumours. We found that shRHB-treated MDA-MB-231 cells displayed a slower growth rate than shScr-treated cells, and that stimulation with S1P resulted in a much more profound increase of the growth rate of shScr-treated cells than that of shRHB-treated cells (Fig. 5A). Similarly, RHBDF1 silencing also resulted in a significant reduction in cell proliferation rates in MCF-7 or T-47D breast cancer cells in response to S1P stimulation (Figs. 5B, C). The results indicate that RHBDF1 function is required in mediating GPCR agonist-conferred growth advantage to these cancer cells, likely through EGFR transactivation.
Fig. 5.
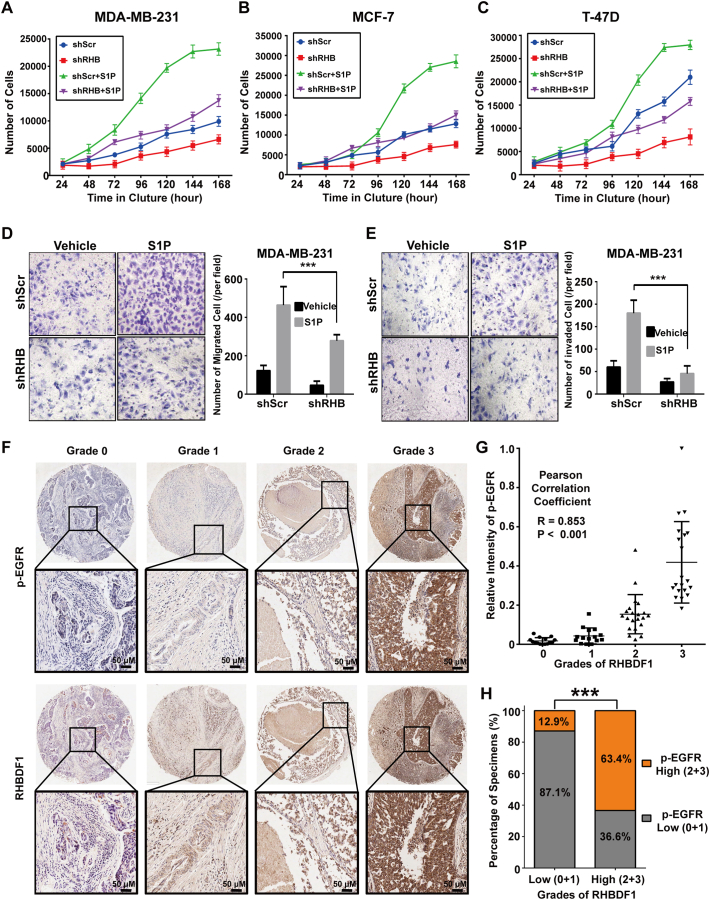
RHBDF1 enhances proliferation and mobility of breast cancer cells and correlates with EGFR activation in breast cancer patients. (A) Cell proliferation assay for MDA-MB-231 cells upon S1P stimulation, investigating the effects of RHBDF1 silencing in vitro. The number of shScr- or shRHB-treated MDA-MB-231 cells was monitored in the presence or absence of S1P using the CellTiter 96 AQueous one-solution cell proliferation assay with averages from sextuplicate wells. (B) Cell proliferation assay for shScr- or shRHB-treated MCF-7 cells with or without S1P stimulation. (C) Cell proliferation assay for shScr- or shRHB-treated T-47D cells with or without S1P stimulation. (D) Representative images of shScr- or shRHB-treated cells that crossed the polycarbonate membrane of the Transwell chamber, with or without S1P stimulation, to detect the migration of cells (magnification ×100). (E) Representative images of shScr- or shRHB-treated cells that crossed the Matrigel-coated polycarbonate membrane of the Transwell chamber for detection of the invasion of cells (magnification ×100). (F) Representative images of immunohistochemical analysis of breast cancer specimens showing four grades (0–3) of RHBDF1 and p-EGFR protein staining (magnification, ×200). (G) Dot plots of the grades of RHBDF1 and the relative intensity of p-EGFR in the specimens (R = 0.853, p < 0.001). (H) Percentages of specimens displaying p-EGFR levels in relation to grades of RHBDF1. Student's t-test; *** p < 0.001.
We then investigated RHBDF1 actions on breast cancer cell migration and invasion by using transwell assays. We found that S1P stimulation elicits a more pronounced increase in cell migration in shScr-treated cells than in shRHB-treated cells (Fig. 5D). We then set a Matrigel basement membrane on the transwells to investigate cell invasion behaviour in vitro. We found that cell invasion from the Matrigel was significantly increased upon S1P treatment of the shSrc-treated cells; however, this effect was not so apparent with shRHB-treated cells (Fig. 5E). These results suggest that RHBDF1 silencing significantly suppress basal and S1P-induced migration and invasion in breast cancer cells.
To establish further clinical relevance of RHBDF1 in breast cancer, we assessed the correlation between EGFR activation status and RHBDF1 expression by immunohistochemical analysis of clinical specimens of breast cancer (Table S1). We categorized the staining patterns of the sections into 4 intensity grades (0–3 for negative or marginal, low, medium, and high, respectively) and found that RHBDF1 expression levels in the specimens were significantly and positively correlated with EGFR phosphorylation levels (Pearson correlation statistical analysis, R = 0.853, p < 0.001; n = 72) (Fig. 5G). Specifically, 87.1% of the specimens with low RHBDF1 expression (grades 0–1; 31 specimens) exhibited low levels of EGFR activation (grades 0–1), whereas 63.4% of the specimens with high RHBDF1 expression (grades 2–3; 41 specimens) showed high levels of EGFR activation (grades 2–3; χ2-test, p < 0.001) (Fig. 5H). Collectively, our findings indicate that RHBDF1 supports pro-tumorigenic functions in breast cancer cells, including proliferation, invasion, and migration, and that RHBDF1 expression levels are significantly correlates with EGFR activation in breast cancer in clinical settings.
4. Discussion
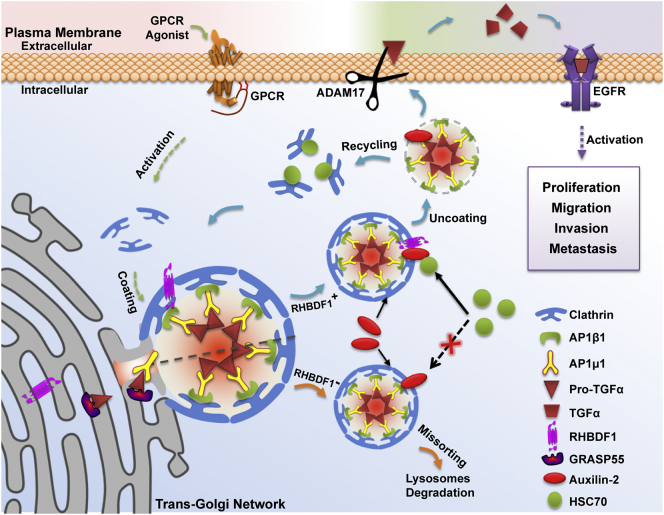
Our data highlight the important regulatory role of RHBDF1 in protein trafficking machinery, which is present on the ER-Golgi membrane and is also an important signalling hub in CCV-dependent pro-TGFα trafficking in response to GPCR agonist stimulation (Fig. 6). Based on our findings we propose that the formation of a RHBDF1/auxilin-2 protein complex enables the recruitment of HSC70 to CCV, which initiates a process leading to the disassembly of clathrin and release of pro-TGFα for ADAM17-dependent shedding and EGFR activation. Elimination of RHBDF1 by gene silencing disables the recruitment of HSC70 to auxilin-2, subjecting pro-TGFα-containing CCV to failure of clathrin uncoating and subsequent delivery of pro-TGFα to the plasma membrane. Since an increment in the levels of the cell surface growth factor results in augmented levels of soluble growth factor, this paradigm is in agreement with our previous findings that elimination of RHBDF1 with siRNA lead to greatly diminished secretion of TGFα, and that the engineered overexpression of RHBDF1 results in facilitated TGFα export by epithelial cancer cells in response to GPCR ligand stimulation. Our findings thus reveal a missing step that connects GPCR activation and membrane distribution of the EGFR ligand TGFα.
Fig. 6.
Schematic representation of RHBDF1-facilitated membrane trafficking of pro-TGFα via CCV pathway. The diagram depicts the formation of a RHBDF1/auxilin-2 protein complex, after CCV coating and budding in response to GPCR stimulation enables the recruitment of HSC70, which initiates clathrin uncoating from CCV to release pro-TGFα for shedding and, consequently, EGFR activation. Removal of RHBDF1 abolishes the recruitment of HSC70 to auxilin-2, resulting in failure of clathrin uncoating in pro-TGFα-containing CCV.
Our findings are consistent with the view that RHBDF1 functions as a chaperone. The RHBDF1 protein is apparently not a protease because of the absence of a catalytic centre commonly found in the rhomboid family of proteins that function as proteases [35,36]. Yet in addition to auxilin-2, RHBDF1 has been shown to directly interact with a number of proteins. For instance, we showed previously that RHBDF1 functions to maintain HIF1α stability under hypoxic conditions by interacting with RACK1, an action that disrupts RACK1 binding to HIF1α, which would otherwise leads to degradation of the latter [18]. Others have shown that RHBDF1 interacts with the 20S proteasome assembly chaperones PAC1 and PAC2 to affect ER stress-associated proteasome protein stability [21]. Similarly, RHBDF2 (iRhom2), a homologous protein of RHBDF1 and also an “inactive” rhomboid protein, can recruit the translocon-associated protein TRAPβ to stimulator of interferon genes STING complex and facilitate the trafficking of STING from the endoplasmic reticulum to peri-nuclear microsomes [37]. It has also been shown that RHBDF1 and its homolog RHBDF2 regulate the trafficking and maturation of ADAM17 [22], and are essential upstream regulators of ADAM17-dependent EGFR signalling [38], receptor tyrosine kinase MET signalling [39], and colony stimulating factor 1 receptor signalling [40]. The ability of RHBDF1 to interact with various proteins is highly supportive of our hypothesis that RHBDF1 possesses chaperone-like functions.
Of the known EGFR ligands, TGFα has been reported to be expressed more highly in triple-negative breast cancer, and is synthesized as a transmembrane precursor requiring ADAM17-dependent proteolytic release to activate its receptor [41]. It is plausible that ADAM17-dependent EGFR signalling is partly the result of RHBDF1-facilitated TGFα trafficking and secretion. Our findings are indicative that RHBDF1 participate in the trafficking and maturation of ADAM17. It is of interest to note that some members of the mammalian rhomboid family with protease activities, such as RHBDL4, can also take part in promoting membrane trafficking of several proteins in response to GPCR activation, whilst also acting as a shedding enzyme for pro-TGFα on the plasma membrane [42]. It is plausible that RHBDF1 may have evolved from rhomboid intramembrane protease ancestors, although the evolution of the rhomboid superfamily is not clear and is difficult to trace owing to the low overall sequence identity of its members. Based on the current findings, the ability to bind with a variety of proteins may have been inherited and expanded to perform biological functions involved in regulating the fate of secretory proteins, including subcellular membrane trafficking.
Interestingly, RHBDF1 silencing similarly inhibited proliferation, migration, and invasion in the absence and presence of S1P stimulation in several types of breast cancer cells, suggesting that the ability of RHBDF1 to enhance cancer development may not necessarily be connected to GPCR-mediated EGFR transactivation. It is possible that the GPCR-independent activities of RHBDF1 derive from its ability to maintain HIF-1α from undergoing degradation under hypoxic condition, as we reported previously [18].
The pathological relevance of RHBDF1 in cancer development and progression is further underscored by the significant correlations between RHBDF1 and EGFR activation in clinical specimens of breast cancer, suggesting that RHBDF1 perform an oncogenic role of promoting tumorigenesis and metastasis in the context of EGFR activation. The expression, activation, and mutation status of EGFR are pivotal in the development of cancers [43,44], Alzheimer's disease [45], viral infection [46], and neuronal diseases [47]. Therefore, the role of RHBDF1 in the membrane trafficking of proteins places this non-protease member of the rhomboid family at the centre of cancer development, and it may be similarly implicated in other disease conditions, an aspect that is worth of further investigations. It is interesting that RHBDF2 has also been described as being associated with various disease conditions[48].
That RHBDF1 plays a critical role in assisting membrane trafficking and maturation of secretory ligands of EGFR such as TGFα in response to GPCR activation makes this non-protease member of the rhomboid family a potentially valuable targets for the treatment of cancer as well as other EGFR-related diseases. Further insights from future studies of the mechanisms underlying such modulation will help to develop means to interfere with RHBDF1-assisted, GPCR-mediated EGFR transactivation.
Funding sources
This project was supported by grants from the National Natural Science Foundation of China (81672740 to ZSZ, 81272356 and 81330029 to LYL).
Declaration of interests
We declare that there are no financial or other relationships that may lead to a conflict of interest related to this study.
Author contributions
JL, ZSZ, and LYL conceived and designed the study. JL, TRB, SG, ZZ, XMP, LSZ, and DLD performed experiments and analysed the data. JL, ZSZ, and LYL wrote and revised of the manuscript. All authors approved the final version of the manuscript.
Acknowledgements
We thank Dr. Qiangzhe Zhang, Mr. Jian Shen, and Ms. Nannan Xiao of Nankai University for their skilful assistance with the experiments.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.09.038.
Contributor Information
Zhi-Song Zhang, Email: zzs@nankai.edu.cn.
Lu-Yuan Li, Email: liluyuan@nankai.edu.cn.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- 1.Schlessinger J. Receptor tyrosine kinases: legacy of the first two decades. Cold Spring Harbor perspectives in biology. 2014;6(3). [DOI] [PMC free article] [PubMed]
- 2.Overstreet J.M., Wang Y., Wang X., Niu A., Gewin L.S., Yao B. Selective activation of epidermal growth factor receptor in renal proximal tubule induces tubulointerstitial fibrosis. FASEB J. 2017;31(10):4407–4421. doi: 10.1096/fj.201601359RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sigismund S., Avanzato D., Lanzetti L. Emerging functions of the EGFR in cancer. Mol Oncol. 2018;12(1):3–20. doi: 10.1002/1878-0261.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian Y., Lin J., Tian Y., Zhang G., Zeng X., Zheng R. Efficacy and safety of anti-EGFR agents administered concurrently with standard therapies for patients with head and neck squamous cell carcinoma: a systematic review and meta-analysis of randomized controlled trials. Int J Cancer. 2017;142(11):2198–2206. doi: 10.1002/ijc.31157. [DOI] [PubMed] [Google Scholar]
- 5.Chong C.R., Janne P.A. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med. 2013;19(11):1389–1400. doi: 10.1038/nm.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harper K., Lavoie R.R., Charbonneau M., Brochu-Gaudreau K., Dubois C.M. The hypoxic tumor Microenvironment promotes invadopodia formation and metastasis through LPA1 receptor and EGFR cooperation. Mol Cancer Res. 2018:1389–1400. doi: 10.1158/1541-7786.MCR-17-0649. [DOI] [PubMed] [Google Scholar]
- 7.Wang X., Maruvada R., Morris A.J., Liu J.O., Wolfgang M.J., Baek D.J. Sphingosine 1-phosphate activation of EGFR as a novel target for meningitic escherichia coli penetration of the blood-brain barrier. PLoS Pathog. 2016;12(10) doi: 10.1371/journal.ppat.1005926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ockenga W., Kuhne S., Bocksberger S., Banning A., Tikkanen R. Epidermal growth factor receptor transactivation is required for mitogen-activated protein kinase activation by muscarinic acetylcholine receptors in HaCaT keratinocytes. Int J Mol Sci. 2014;15(11):21433–21454. doi: 10.3390/ijms151121433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rayego-Mateos S., Morgado-Pascual J.L., Sanz A.B., Ramos A.M., Eguchi S., Batlle D. TWEAK transactivation of the epidermal growth factor receptor mediates renal inflammation. J Pathol. 2013;231(4):480–494. doi: 10.1002/path.4250. [DOI] [PubMed] [Google Scholar]
- 10.Nagathihalli N.S., Beesetty Y., Lee W., Washington M.K., Chen X., Lockhart A.C. Novel mechanistic insights into ectodomain shedding of EGFR Ligands Amphiregulin and TGF-alpha: impact on gastrointestinal cancers driven by secondary bile acids. Cancer Res. 2014;74(7):2062–2072. doi: 10.1158/0008-5472.CAN-13-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X.P., Fu J.Y., Yang R.C., Liu W.T., Zhang T., Yang B. EGFR transactivation contributes to neuroinflammation in Streptococcus suis meningitis. J Neuroinflammation. 2016;13(1):274. doi: 10.1186/s12974-016-0734-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francavilla C., Papetti M., Rigbolt K.T., Pedersen A.K., Sigurdsson J.O., Cazzamali G. Multilayered proteomics reveals molecular switches dictating ligand-dependent EGFR trafficking. Nat Struct Mol Biol. 2016;23(6):608–618. doi: 10.1038/nsmb.3218. [DOI] [PubMed] [Google Scholar]
- 13.Freeman M. Rhomboids: 7 years of a new protease family. Semin Cell Dev Biol. 2009;20(2):231–239. doi: 10.1016/j.semcdb.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Freeman M. The rhomboid-like superfamily: molecular mechanisms and biological roles. Annu Rev Cell Dev Biol. 2014;30:235–254. doi: 10.1146/annurev-cellbio-100913-012944. [DOI] [PubMed] [Google Scholar]
- 15.Zou H., Thomas S.M., Yan Z.W., Grandis J.R., Vogt A., Li L.Y. Human rhomboid family-1 gene RHBDF1 participates in GPCR-mediated transactivation of EGFR growth signals in head and neck squamous cancer cells. FASEB J. 2009;23(2):425–432. doi: 10.1096/fj.08-112771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakagawa T., Guichard A., Castro C.P., Xiao Y., Rizen M., Zhang H.Z. Characterization of a human rhomboid homolog, p100hRho/RHBDF1, which interacts with TGF-alpha family ligands. Dev Dyn. 2005;233(4):1315–1331. doi: 10.1002/dvdy.20450. [DOI] [PubMed] [Google Scholar]
- 17.Yan Z., Zou H., Tian F., Grandis J.R., Mixson A.J., Lu P.Y. Human rhomboid family-1 gene silencing causes apoptosis or autophagy to epithelial cancer cells and inhibits xenograft tumor growth. Mol Cancer Ther. 2008;7(6):1355–1364. doi: 10.1158/1535-7163.MCT-08-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Z., Liu F., Zhang Z.S., Shu F., Zheng Y., Fu L. Human rhomboid family-1 suppresses oxygen-independent degradation of hypoxia-inducible factor-1alpha in breast cancer. Cancer Res. 2014;74(10):2719–2730. doi: 10.1158/0008-5472.CAN-13-1027. [DOI] [PubMed] [Google Scholar]
- 19.Maretzky T., McIlwain D.R., Issuree P.D., Li X., Malapeira J., Amin S. iRhom2 controls the substrate selectivity of stimulated ADAM17-dependent ectodomain shedding. Proc Natl Acad Sci U S A. 2013;110(28):11433–11438. doi: 10.1073/pnas.1302553110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maney S.K., McIlwain D.R., Polz R., Pandyra A.A., Sundaram B., Wolff D. Deletions in the cytoplasmic domain of iRhom1 and iRhom2 promote shedding of the TNF receptor by the protease ADAM17. Sci Signal. 2015;8(401) doi: 10.1126/scisignal.aac5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee W., Kim Y., Park J., Shim S., Lee J., Hong S.H. iRhom1 regulates proteasome activity via PAC1/2 under ER stress. Sci Rep. 2015;5:11559. doi: 10.1038/srep11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McIlwain D.R., Lang P.A., Maretzky T., Hamada K., Ohishi K., Maney S.K. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science. 2012;335(6065):229–232. doi: 10.1126/science.1214448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cavadas M., Oikonomidi I., Gaspar C.J., Burbridge E., Badenes M., Felix I. Phosphorylation of iRhom2 Controls Stimulated Proteolytic Shedding by the Metalloprotease ADAM17/TACE. Cell Rep. 2017;21(3):745–757. doi: 10.1016/j.celrep.2017.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaksonen M., Roux A. Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2018;19(5):313–326. doi: 10.1038/nrm.2017.132. [DOI] [PubMed] [Google Scholar]
- 25.Robinson M.S. Forty years of Clathrin-coated Vesicles. Traffic. 2015;16(12):1210–1238. doi: 10.1111/tra.12335. [DOI] [PubMed] [Google Scholar]
- 26.Goldenring J.R. A central role for vesicle trafficking in epithelial neoplasia: intracellular highways to carcinogenesis. Nat Rev Cancer. 2013;13(11):813–820. doi: 10.1038/nrc3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiang Y., Zhang X., Nix D.B., Katoh T., Aoki K., Tiemeyer M. Regulation of protein glycosylation and sorting by the Golgi matrix proteins GRASP55/65. Nat Commun. 2013;4:1659. doi: 10.1038/ncomms2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarro Negredo P., Edgar J.R., Wrobel A.G., Zaccai N.R., Antrobus R., Owen D.J. Contribution of the clathrin adaptor AP-1 subunit micro1 to acidic cluster protein sorting. J Cell Biol. 2017;216(9):2927–2943. doi: 10.1083/jcb.201602058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canagarajah B.J., Ren X.F., Bonifacino J.S., Hurley J.H. The clathrin adaptor complexes as a paradigm for membrane-associated allostery. Protein Sci. 2013;22(5):517–529. doi: 10.1002/pro.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMahon H.T., Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12(8):517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 31.Singh B., Coffey R.J. From wavy hair to naked proteins: the role of transforming growth factor alpha in health and disease. Semin Cell Dev Biol. 2014;28:12–21. doi: 10.1016/j.semcdb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimoda M., Horiuchi K., Sasaki A., Tsukamoto T., Okabayashi K., Hasegawa H. Epithelial cell-derived a disintegrin and metalloproteinase-17 confers resistance to colonic inflammation through EGFR activation. EBioMedicine. 2016;5:114–124. doi: 10.1016/j.ebiom.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo A., Zhong C., Lane W.S., Derynck R. Transmembrane transforming growth factor-alpha tethers to the PDZ domain-containing, Golgi membrane-associated protein p59/GRASP55. EMBO J. 2000;19(23):6427–6439. doi: 10.1093/emboj/19.23.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonifacino J.S. Adaptor proteins involved in polarized sorting. J Cell Biol. 2014;204(1):7–17. doi: 10.1083/jcb.201310021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemberg M.K., Adrain C. Inactive rhomboid proteins: new mechanisms with implications in health and disease. Semin Cell Dev Biol. 2016;60:29–37. doi: 10.1016/j.semcdb.2016.06.022. [DOI] [PubMed] [Google Scholar]
- 36.Adrain C., Freeman M. New lives for old: evolution of pseudoenzyme function illustrated by iRhoms. Nat Rev Mol Cell Biol. 2012;13(8):489–498. doi: 10.1038/nrm3392. [DOI] [PubMed] [Google Scholar]
- 37.Luo W.W., Li S., Li C., Lian H., Yang Q., Zhong B. iRhom2 is essential for innate immunity to DNA viruses by mediating trafficking and stability of the adaptor STING. Nat Immunol. 2016;17(9):1057–1066. doi: 10.1038/ni.3510. [DOI] [PubMed] [Google Scholar]
- 38.Li X., Maretzky T., Weskamp G., Monette S., Qing X., Issuree P.D. iRhoms 1 and 2 are essential upstream regulators of ADAM17-dependent EGFR signaling. Proc Natl Acad Sci U S A. 2015;112(19):6080–6085. doi: 10.1073/pnas.1505649112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cataisson C., Michalowski A.M., Shibuya K., Ryscavage A., Klosterman M., Wright L. MET signaling in keratinocytes activates EGFR and initiates squamous carcinogenesis. Sci Signal. 2016;9(433) doi: 10.1126/scisignal.aaf5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qing X., DR L., Mortha A., Lavin Y., Redecha P., Issuree P.D. iRhom2 regulates CSF1R cell surface expression and non-steady state myelopoiesis in mice. Eur J Immunol. 2016;46(12) doi: 10.1002/eji.201646482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calvo V., Giricz O., Kenny P.A. TACE-regulated TGF-alpha shedding drives GRB7-dependent invasion in triple-negative breast cancer cell lines. Cancer Res. 2013;73 doi: 10.1002/ijc.28295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wunderle L., Knopf J.D., Kuhnle N., Morle A., Hehn B., Adrain C. Rhomboid intramembrane protease RHBDL4 triggers ER-export and non-canonical secretion of membrane-anchored TGFalpha. Sci Rep. 2016;6:27342. doi: 10.1038/srep27342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z.W., Ouyang K.H., Zhao J., Chen H., Xiong L., Wang W.J. Structural characterization and hypolipidemic effect of Cyclocarya paliurus polysaccharide in rat. Int J Biol Macromol. 2016;91:1073–1080. doi: 10.1016/j.ijbiomac.2016.06.063. [DOI] [PubMed] [Google Scholar]
- 44.Karachaliou N., Chaib I., Cardona A.F., Berenguer J., Bracht J.W.P., Yang J. Common Co-activation of AXL and CDCP1 in EGFR-mutation-positive non-smallcell lung cancer associated with poor prognosis. EBioMedicine. 2018;29:112–127. doi: 10.1016/j.ebiom.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neefjes J., van der Kant R. Stuck in traffic: an emerging theme in diseases of the nervous system. Trends Neurosci. 2014;37(2):66–76. doi: 10.1016/j.tins.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 46.Soares H., Lasserre R., Alcover A. Orchestrating cytoskeleton and intracellular vesicle traffic to build functional immunological synapses. Immunol Rev. 2013;256(1):118–132. doi: 10.1111/imr.12110. [DOI] [PubMed] [Google Scholar]
- 47.Poulsen E.T., Larsen A., Zollo A., Jorgensen A.L., Sanggaard K.W., Enghild J.J. New insights to clathrin and adaptor protein 2 for the design and development of therapeutic strategies. Int J Mol Sci. 2015;16(12):29446–29453. doi: 10.3390/ijms161226181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee M.Y., Nam K.H., Choi K.C. iRhoms; its Functions and Essential Roles. Biomol Ther. 2016;24(2):109–114. doi: 10.4062/biomolther.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2