Abstract
There are several challenges towards the development and clinical use of small molecule inhibitors, which are currently the main type of targeted therapies towards intracellular proteins. PROteolysis-TArgeting Chimeras (PROTACs) exploit the intracellular ubiquitin-proteasome system to selectively degrade target proteins. Recently, small-molecule PROTACs with high potency have been frequently reported. In this review, we summarize the emerging characteristics of small-molecule PROTACs, such as inducing a rapid, profound and sustained degradation, inducing a robust inhibition of downstream signals, displaying enhanced target selectivity, and overcoming resistance to small molecule inhibitors. In tumor xenografts, small-molecule PROTACs can significantly attenuate tumor progression. In addition, we also introduce recent developments of the PROTAC technology such as homo-PROTACs. The outstanding advantages over traditional small-molecule drugs and the promising preclinical data suggest that small-molecule PROTAC technology has the potential to greatly promote the development of targeted therapy drugs.
Keywords: PROTAC, Induced protein degradation, E3 ligases, Ubiquitin-proteasome system, Targeted therapy drugs
1. Introduction
Monoclonal antibodies and small molecule inhibitors are the two major types of targeted therapies. While monoclonal antibodies block the extracellular components of target proteins, small molecule inhibitors are highly cell permeable and can enter cells, thereby blocking the activities of intracellular target proteins and interfering with the downstream signaling pathways. In addition, the size of small molecule inhibitors usually conforms to Lipinski's rule of five [1,2], which generally ensures small molecule inhibitors being orally administered. Currently, small molecule inhibitors are the main targeted treatment towards intracellular proteins.
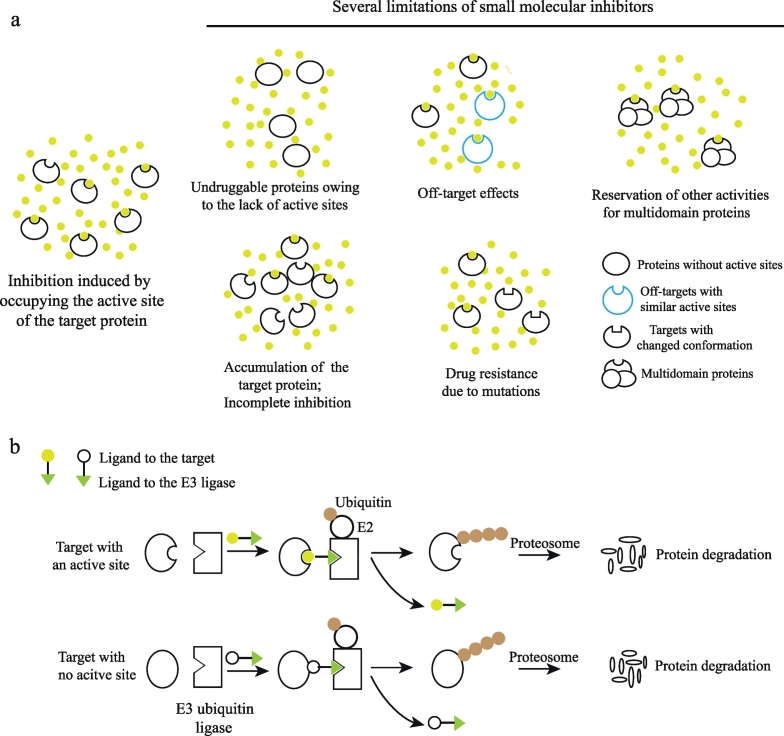
However, small molecule inhibitors have manifested several limitations (Fig. 1a). First, the target proteins of small molecular inhibitors are usually enzymes and receptors that have pockets or active sites. About 75% of the human proteome lack active sites (e.g., transcription factors, scaffolding proteins, and non-enzymatic proteins) and are thus undruggable in this strategy [3]. Second, sustainably high systemic drug levels are needed to maintain the adequate intracellular concentrations for therapeutic efficacy, which often cause off-target effects and side effects due to the competitive nature of small molecular inhibitors. Third, small molecules typically only disrupt the activity of one domain of multidomain scaffolding proteins. Functional activities of other domains and their interactions with other proteins are preserved. In cancer cells, the inhibition of multidomain kinases may lead to compensatory feedback activation of their downstream signaling cascades via other alternative kinases [4]. When treating cells with bromodomain inhibitors of multidomain and bromo-containing proteins (e.g., TRIM24), effective anti-proliferative responses were not displayed, which suggests that the inhibition of bromodomain is insufficient as an anti-cancer strategy [5,6]. Four, inhibitors may cause compensatory protein overexpression and protein accumulation [7,8], which result in incomplete depletion of target proteins and incomplete suppression of downstream signaling pathways. Finally, many cancer genes (e.g., epidermal growth factor receptor and androgen receptor) are highly mutated [9,10]. Nonsynonymous mutations on these genes may lead to the conformational changes of protein productions and thus lead to drug resistance. These limitations severely impede the extensive discovery of small molecule inhibitors and the significant and durable clinical benefits of taking small molecule inhibitors.
Fig. 1.
Overview of the mechanisms of small molecule inhibitors and PROTACs (a) In order to inhibit the activities of target proteins, small molecule inhibitors competitively bind to active sites on the target proteins. The limitations on developing and taking small molecule drugs are shown in this figure. (b) Heterobifunctional PROTAC molecules harness the ubiquitin proteasome system to selectively degrade target proteins. Currently, the generation of PROTACs relies on available small molecular inhibitors to be used as target binding ligands. Alternatively, PROTACs can bind to any crevice on the surface of the target proteins to induce their degradation.
Heterobifunctional PROTAC molecules consist of a ligand to the target protein, a ligand to the E3 ubiquitin ligase, and a linker connecting the two ligands (Fig. 1b). Once the target:PROTAC:E3 ternary complex is formed, E2 ubiquitin-conjugating enzymes transfer ubiquitin to lysine residues on the surface of the target protein. The recognition of lysine polyubiquitination signal by proteasome facilitates the degradation of the target protein. In recent years, small-molecule PROTACs with good pharmaceutical properties have been reported.
In this review, we briefly introduce the development history of the PROTAC technology. We focus on summarizing the new progress and emerging characteristics of small-molecule PROTACs particularly those reported since 2015. We emphasize the advantages of small-molecule PROTACs over small molecule inhibitors, which are currently the main type of targeted therapies towards intracellular proteins. In addition, we also introduce other modalities of PROTAC molecules, which represent the recent developments of the PROTAC technology.
2. PROTACs
The concept of PROTACs was raised by Crews and his colleagues in 2001 [11]. In this study, the synthesized chimeric compound Protac-1 contains a SCFβ-TRCP binding phosphorpeptide and small-molecule ovalicin that covalently binds to MetAP-2. In cell-free Xenopus extracts, MetAP-2 was degraded in a Protac-1-dependent manner. The first cell-permeable PROTAC was reported in 2004 [12]. The ligand to Von Hippel Lindau (VHL) E3 ligase was a peptide derived from HIF-1α, which is a substrate of VHL [13]. The membrane permeability of this peptide was accomplished by adding a poly-D-arginine tag. In cultured cells, FKBP12F36V and androgen receptor were degraded after the treatment of VHL-based PROTACs [12]. Subsequently, other proteins (e.g., MetAP-2, estrogen receptor, and aryl hydrocarbon receptor) were shown to be efficiently depleted by VHL-binding peptide-based PROTACs in cell lines [[14], [15], [16]]. Peptide-based PROTACs that induced the degradation of androgen receptor or estrogen receptor also inhibited the proliferation of androgen/estrogen-dependent cancer cells [17].
Peptide-based PROTACs have disadvantages on their high molecular weight, labile peptide bonds, poor cell penetration, and low potency which was typically in the micromolar range [18]. These shortcomings make peptide-based PROTACs poor pharmaceutical candidates.
2.1. Small-molecule PROTACs
To avoid the weaknesses of peptide-based PROTACs, all small-molecule-based PROTACs, in which E3 binding ligands are also small molecules, were created. Until now, four E3 ligases (i.e., MDM2, IAP, VHL, and cereblon) have been used for all small-molecule-based selective degradation of target proteins.
The first small-molecule PROTAC was reported in 2008 [19]. This PROTAC includes a non-steroidal androgen receptor ligand which is a selective androgen receptor modulator (SARM), a MDM2 ligand known as nutlin, and a PEG-based linker [20]. The SARM-nutlin PROTAC triggered the ubiquitination and degradation of androgen receptor. The second class of E3 ligase exploited by small-molecule PROTACs was cellular inhibitor of apoptosis protein 1 (cIAP1). Small-molecule PROTACs with cIAP1 binding ligands were also named SNIPERs (specific and nongenetic IAP-dependent protein erasers). Bestatin-based SNIPERs have shown their efficacy in the degradation of CRABP-I [21], CRABP-II [21,22], ERα [23,24], TACC3 [25], and BCR-ABL [26]. To overcome the self-degradation of cIAP1 and the low potency observed when treating cells with bestatin-based SNIPERs, an IAP antagonist LCL161 was utilized to generate SNIPERs. SNIPERs incorporating an LCL161 derivative which primarily recruit XIAP instead of cIAP1 showed nanomolar potency against ERα, BRD4, PDE4, and BCR-ABL [27]. LCL161-based SNIPERs for androgen receptor were also generated [28].
Since 2015, VHL and cereblon (CRBN) E3 ligases have been widely exploited to develop small-molecule PROTACs. Promoted by the discovery of small-molecule replacements for the HIF1α peptide fragment [[29], [30], [31]], VHL-based small-molecule PROTACs have been generated and shown to effectively degrade GFP-HaloTag fusions [32], ERRα [33], RIPK2 [33], BCR-ABL [34], BRD4 [[35], [36], [37]], TBK1 [38], several transmembrane receptor tyrosine kinases (EGFR, HER2, and c-Met) [39], and TRIM24 [5]. Immunomodulatory drugs (IMiDs) thalidomide, lenalidomide, and pomalidomide have been found to bind the CRL family E3 component CRBN [40,41]. Small-molecule PROTACs with IMiD-based CRBN binding ligands that target the Bromodomain and Extra-Terminal (BET) proteins (BRD2/3/4) [[42], [43], [44]], FKBP12 [42], BCR-ABL [34], BRD9 [45], Sirt2 [46], CDK9 [47,48], FLT3 [49], BTK [49,50], and ALK [51] have been developed.
Since 2015, more than thirty small-molecule PROTACs have been reported, and many of these reported PROTACs showed nanomolar potency (Table 1). Moreover, in vivo functional effects of several PROTACs were also studied. Next, we introduce the emerging advantages and characteristics of small-molecule PROTACs discovered from in vitro and in vivo studies of these variable PROTAC molecules.
Table 1.
Components and properties of most small-molecule PROTACs reported since 2015.
| Compound name | Target | Target ligand | E3 ligand | E3 ligase | Degradation in cell lines |
In vivo experiments on mice | Ref (Year) | ||
|---|---|---|---|---|---|---|---|---|---|
| DC50 | Dmax | Other evidences | |||||||
| SNIPER(ER)-87 | ERα | 4-OHT | An LCL161 derivative | IAP | >1 nM & <3 nM | >70% | Reduction of ERα; Suppression of tumor growth | 27 (2017) | |
| SNIPER(ABL)-38 | BCR-ABL | Dasatinib | An LCL161 derivative | IAP | >3 nM & <10 nM | >90% | 27 (2017) | ||
| SNIPER(BRD4)-1 | BRD4 | JQ1 | An LCL161 derivative | IAP | >3 nM & <10 nM | >70% | 27 (2017) | ||
| SNIPER(PDE4)-9 | PDE4 | A PDE4 inhibitor | An LCL161 derivative | IAP | ~1 nM | ~60% | 27 (2017) | ||
| MZ1 | BRD4 | JQ1 | VHL-1 | VHL | <100 nM for BRD4 | >90% | 36 (2015) | ||
| HaloPROTAC3 | GFP-HaloTag7 | Chloroalkane | A hydroxyproline derivative | VHL | 19 ± 1 nM | 90 ± 1% | 32 (2015) | ||
| PROTAC_ERRα | ERRα | A thiazolidinedione-based ligand | A hydroxyproline derivative | VHL | ~100 nM | 86% | Knockdown of ERRα | 33 (2016) | |
| PROTAC_RIPK2 | RIPK2 | A RIPK2 inhibitor | A hydroxyproline derivative | VHL | 1.4 nM | >95% | 33 (2016) | ||
| DAS-6-2-2-6-VHL | c-ABL | Dasatinib | A hydroxyproline derivative | VHL | NA | NA | >65% decrease at 1 μM | 34 (2016) | |
| DAS-6-2-2-6-CRBN | c-ABL & BCR-ABL | Dasatinib | Pomalidomide | CRBN | NA | NA | c-ABL: >85% depletion at 1 μM; BCR-ABL: >60% depletion at 1 μM | 34 (2016) | |
| BOS-6-2-2-6-CRBN | c-ABL & BCR-ABL | Bosutinib | Pomalidomide | CRBN | NA | NA | c-ABL: >90% depletion at 2.5 μM; BCR-ABL: >80% depletion at 2.5 μM | 34 (2016) | |
| dBET1 | BRD2/3/4 | JQ1 | Thalidomide | CRBN | 430 nM for BRD4 | >95% | Degradation of BRD4 and MYC; Attenuation of tumor progression | 42 (2015) | |
| ARV-771 | BRD2/3/4 | A JQ1 derivative | A HIF-1α-derived (R)-hydroxyproline | VHL | <5 nM/<1 nM for BRD2/3/4 | >90% | Degradation of the target protein; Tumor regression | 35 (2016) | |
| NA | >90% | Inhibition of the in vivo growth; Improved survival | 53 (2018) | ||||||
| NA | >90% | Reduction in leukemia burden; Improved survival | 52 (2017) | ||||||
| ARV-825 | BRD2/3/4 | OTX015 | Pomalidomide | CRBN | <1 nM for BRD4 | Near-complete depletion for BRD4 | 43 (2015) | ||
| dFKBP-1; dFKBP-2 | FKBP12 | Steel factor | Thalidomide | CRBN | 10 nM for dFKBP-1; <10 nM for dFKBP-2 | >90% | 42 (2015) | ||
| 3i | TBK1 | A TBK1 inhibitor | VHL ligand 2 | VHL | 12 nM | 96% | 38 (2017) | ||
| AT1 | BRD4 | JQ1 | A VH032 derivative | VHL | >10 nM & <100 nM for BRD4 short | >90% | 37 (2017) | ||
| dBRD9 | BRD9 | BI-7273 | Pomalidomide | CRBN | NA | NA | Marked depletion at <50 nM | 45 (2017) | |
| PROTAC 1 | Wild-type EGFR | Lapatinib | A hydroxyproline-based ligand | VHL | 39.2 nM | 97.6% | 39 (2018) | ||
| Exon 20 in. EGFR | 736.1 nM | 68.8% | 39 (2018) | ||||||
| HER2 | <100 nM | Near-complete depletion | 39 (2018) | ||||||
| PROTAC 3 | Exon 19 del EGFR | Gefitinib | A hydroxyproline-based ligand | VHL | 11.7 nM | 98.9% | 39 (2018) | ||
| L858R EGFR | 22.3 nM | 96.6% | 39 (2018) | ||||||
| PROTAC 4 | EGFR | Afatinib | A hydroxyproline-based ligand | VHL | 215.8 nM | 79.1% | 39 (2018) | ||
| PROTAC 7 | c-Met | Foretinib | A hydroxyproline-based ligand | VHL | NA | NA | Marked depletion at 500 nM | 39 (2018) | |
| PROTAC 12 | Sirt2 | Sirt2 inhibitor 3b | Thalidomide | CRBN | >0.2 μM & <1 μM | ~90% | 46 (2018) | ||
| Compound 23 | BRD2/3/4 | HJB97 | Lenalidomide | CRBN | <0.03 nM for BRD4 | Near-complete depletion | Rapid tumor regression | 44 (2018) | |
| THAL-SNS-032 | CDK9 | SNS-032 | A thalidomide derivative | CRBN | <250 nM | Near-complete depletion | 47 (2018) | ||
| PROTAC 3 | CDK9 | An aminopyrazole analog | Thalidomide | CRBN | NA | NA | ~56% depletion at 10 μM | 48 (2017) | |
| TL13-117; TL13-149 | FLT3 | AC220 | Pomalidomide | CRBN | NA | NA | Marked deletion at 10 and 100 nM | 49 (2018) | |
| DD-04-015 | BTK | RN486 | Pomalidomide | CRBN | NA | NA | Most efficient at 100 nM | 49 (2018) | |
| MS4077 (5) | ALK | Ceritinib | Pomalidomide | CRBN | 3 ± 1 nM for NPM-ALK; 34 ± 9 nM for EML4-ALK | >90% | 51 (2018) | ||
| MS4078 (6) | ALK | Ceritinib | Pomalidomide | CRBN | 11 ± 2 nM for NPM-ALK; 59 ± 16 nM for EML4-ALK | >90% | 51 (2018) | ||
| Compound 42a | AR | An AR antagonist | An LCL161 derivative | IAP | >1 μM & < 3 μM | NA | 28 (2018) | ||
| dTRIM24 | TRIM24 | IACS-7e | VL-269 | VHL | >2.5 μM & < 5 μM | ~70% | 5 (2018) | ||
| MT-802 | Wild-type BTK | An ibrutinib derivative | Pomalidomide | CRBN | 14.6 nM | >99% | 50 (2018) | ||
| C481S BTK | 14.9 nM | >99% | 50 (2018) | ||||||
Only the most potent/well-studied PROTAC molecules reported in each study are listed in this table. DC50: the concentration at which 50% degradation was observed. Dmax: the maximal level of degradation. NA: not available. Near-complete depletion: no apparent band was detected at a given concentration of the PROTAC in western blotting analysis. 4-OHT: 4-hydroxytamoxifen.
2.1.1. Potent and profound degradation of target proteins
Small molecule inhibitors modulate protein function through stoichiometrically occupying the active sites of target proteins. However, small-molecule PROTACs catalytically involve in multiple rounds of target protein degradation [33,43,47]. Therefore, the degradation induced by PROTACs is sub-stoichiometric.
MDM2-based PROTACs and IAP-based PROTACs with bestatin-based ligands induced remarkable degradation of target proteins at micromolar concentrations [19,[21], [22], [23], [24], [25], [26]]. PROTACs that used an LCL161 derivative as the IAP binding ligand effectively degraded target proteins at low nanomolar [27] or low micromolar [28] concentrations. In addition, the treatment of LCL161-based SNIPERs targeting ERα induced remarkable reduction of ERα and suppression of tumor growth in mice [27].
Most of small-molecule PROTACs reported from 2015 exploited VHL and CRBN E3 ligases (Table 1). The information on their degradation potency and efficacy in cultured cells are shown in Table 1. Among a series of GFP-HaloTag7-targeting PROTACs that used a small molecule to recruit VHL E3 ligase, the most potent GFP-HaloTag7-targeting PROTAC had a DC50 value of 19 nM and could degrade >90% of GFP-HaloTag7 [32]. Except the VHL-based PROTACs for c-ABL, TRIM24, and mutant isoforms of EGFR, other VHL-based PROTACs showed nanomolar potency and could degrade >85% or > 90% of the target proteins. Particularly, DC50 values of VHL-based small-molecule PROTACs for BET protein (ARV-771) and RIPK2 (PROTAC_RIPK2) were <2 nM in some cell lines [33,35]. For most of CRBN-based PROTACs, target proteins were significantly degraded at concentrations ranging from ultra-low nanomolar to hundreds of nanomolar [[42], [43], [44],46,47,49,51,52]. Several CRBN-based PROTACs (i.e., DAS-6-2-2-6-CRBN and BOS-6-2-2–6-CRBN for c-ABL/BCR-ABL, and PROTAC 3 for CDK9) dramatically depleted target proteins at low micromolar concentrations [34,48]. Six VHL/CRBN-based PROTACs (dBET1, ARV-771, ARV-825, MZ1, Compound 23, and AT1) for BET proteins depleted >90% of target proteins at nanomolar concentrations [36,37,42,52] or even picomolar concentrations [43,44].
Degradation potency and efficacy data presented in Table 1 are from in vitro experiments on cultured cell lines. In addition, small-molecule PROTACs efficiently reduced the levels of target proteins in patient-derived cells [42,52,53] and in tumor xenografts [27,33,35,42,44].
Many target proteins listed in Table 1 are associated with cancers. Several PROTACs for cancer-related proteins (e.g., ARV-771, ARV-825, and dBET1 for BRD4, THAL-SNS-032 for CDK9, PROTAC 7 for c-Met) remarkably promoted antiproliferation and/or apoptosis at nanomolar concentrations [35,39,[42], [43], [44],47,51,52]. However, some PROTACs for other cancer-related proteins (e.g., TL13-117 and TL13-149 for FLT3) showed little anti-proliferative or pro-apoptotic effects [49].
It has been observed that the half-maximal values for degrading target proteins by small-molecule PROTACs (DC50 values) could be lower than that for inhibiting or binding target proteins (IC50 values) [27,50]. For example, while the IC50 value for competitively binding wild-type BTK was 46.9 nM for MT-802, the DC50 value for degrading wild-type BTK was 14.6 nM for MT-802 [50]. However, this phenomenon was not displayed in several other studies [5,46]. Moreover, PROTACs could be more potent than their corresponding small molecule inhibitors at inhibiting the proliferation or promoting the apoptosis of cancer cells [39,47,52,53]. For example, while the IC50 value for inhibition of cell proliferation in GTL16 cells was 66.7 nM for c-Met-depleting PROTAC 7, it was 156 nM for diastereomer 8 [39]; while the IC50 value for induction of apoptosis in Mino cells was 16 ± 3 nM for ARV-825, it was 398 ± 15 nM for the BET inhibitor OTX015 [53].
2.1.2. Rapid and sustained depletion of target proteins
Induced protein degradation usually began soon after the treatment of small-molecule PROTACs, and the remarkable depletion occurred within just 1 or 2 h [22,27,33,42,43,45,47], or several hours [5,28,32,39,51]. The maximal depletion of target proteins (e.g., RIPK2, ERα, BRD4) could be achieved within several hours [27,33,36,42,43,45,47]. Notably, the reduction of target proteins was sustained [5,22,27,32,36,39,42,43,47,51]. Significant degradation could still exist after 24 or even 48 h. For example, when treating HEK 293 cells with HaloPROTAC3, 50% of GFP-HaloTag7 was degraded between 4 and 8 h, and the level of GFP-HaloTag7 at 24 h was kept as low as about 10% [32]. The remarkable degradation could maintain for several hours [51] or even longer (>10 or 24 or 48 h) [33,36,39,52,53] after the removal of PROTAC molecules. In mice, the degradation of target proteins was observed soon after a single dose of PROTACs [42,44], and the in vivo depletion persisted for >24 h [44].
2.1.3. Rapid, sustained, and robust inhibition of downstream signaling cascades
Besides target proteins, their downstream signaling cascades were also rapidly [39,51,54] and sustainably inhibited [39,43,[51], [52], [53], [54]]. For example, ALK downstream markers p-ALK and p-STAT3 were significantly inhibited after the 2-h treatment of ALK-depleting PROTACs, and the inhibition of ALK downstream signaling lasted for >10 h after the washout of ALK-depleting PROTACs [51].
When treating with small molecule inhibitors, the compensatory increase of target proteins can cause the incomplete inhibition of downstream signaling pathways. Several studies have shown that small-molecule BRD4 inhibitors (e.g., JQ1 and OTX015) led to robust BRD4 protein accumulation [43,52,53], particular with the increase of concentration and time [43]. The accumulation of BRD4 led to limited suppression of MYC and cell proliferation [43], and led to limited induction of apoptotic cell death [43,52]. While the inhibition of BRD4 caused the accumulation of BRD4, the degradation of BRD4 induced by BRD4-targeting PROTACs was nearly unaffected by treatment concentrations and treatment times [43,52]. Compared with BRD4 inhibitors, small-molecule PROTACs for BRD4 (ARV-771 and/or ARV-825) resulted in more significant and prolonged suppression of downstream proteins including MYC, more dramatic cell proliferation suppression/apoptotic cell death, and greater in vivo cancer regression [42,43,52,53]. Upon the treatment of BRD4-targeting PROTACs, more downstream proteins were perturbed than BET inhibitors [52,53]. In addition, the perturbations of protein levels were robust when changing treatment concentrations, and could even sustain for 24 h after the washout of BRD4-targeting PROTACs [52,53].
For multidomain proteins, the binding of small molecule inhibitors disrupts the function of only one domain. Because of the dynamic nature of the kinome, the inhibition of kinase activity can cause compensatory feedback activation of downstream signaling pathways via alternative kinases [4,55]. Burslem et al. [39] generated small-molecule PROTACs capable of degrading transmembrane receptor tyrosine kinases (RTKs). They found that the PROTAC-induced degradation led to a rapid inhibition of downstream signaling. The inhibition induced by PROTACs still maintained while that induced by the corresponding inhibitor was reversed. In addition, PROTACs caused a greater anti-proliferative efficacy than did inhibitors. Therefore, the degradation of RTKs can abrogate their scaffolding roles and thus suppress kinome rewiring caused by the compensatory feedback activation.
2.1.4. Profound in vivo tumor suppression
In several studies, a marked attenuation of cancer progression was observed in immune-depleted mice engrafted with cancer cells [27,35,42,44,52,53]. PROTACs for estrogen receptor [27] and BET protein [35,42,44,52,53] were intraperitoneally [27,42], subcutaneously [35,52,53], or intravenously [44] administered. It is worth to note that, compared with small molecule inhibitors, PROTACs for the same proteins induced stronger in vitro proliferation suppression/apoptotic cell death [5,28,35,43,52,53] and stronger in vivo growth inhibition [35,52], and also induced greater survival improvements [52,53]. For example, the subcutaneous injection of BET-targeting PROTAC ARV-771 caused a greater reduction in leukemia burden and/or improved survival of mice compared to the oral treatment of BET inhibitor OTX015 [52,53].
2.1.5. Overcome drug resistance due to mutations
Nonsynonymous mutations at active sites can cause resistance to small molecule inhibitors. Notably, such mutations can frequently occur in some cancer-associated genes. For example, EGFR exon 20 insertion accounts for at least 9% of all EGFR-mutated cases [56], which is associated with the resistance to EGFR-tyrosine kinase inhibitors; >80% of chronic lymphocytic leukemia patients develop resistance to the BTK inhibitor ibrutinib due to the C481S mutation in the BTK gene. Because the degradation of target proteins induced by PROTAC molecules only relies on transient and reversible associations with their substrates, PROTACs have the potential to effectively degrade mutant target proteins.
PROTACs exploiting small molecule inhibitors, which are designed for wild-type proteins, to recruit target proteins have been shown to effectively degrade mutant proteins [50]. BTK-depleting PROTAC MT-802 which used an ibrutinib derivative as the target ligand showed equivalent potency against wild-type and C481S BTK [50]. Specifically, in X-linked agammaglobulinemia cells the DC50 values of MT-802 for wild-type and C481S BTK were, respectively, 14.6 nM and 14.9 nM, and >99% of wild-type and C481S BTK could be degraded. Another BTK-depleting PROTAC P13I consisting of ibrutinib and pomalidomide also efferently induced the degradation of wild-type and C481S BTK (DC50 for wild-type BTK = ~10 nM, DC50 for C481S BTK = 30 nM) [57]. Furthermore, both of these two BTK degraders showed enhanced kinase selectivity over ibrutinib. EGFR with an exon 20 insertion was moderately degraded (DC50 = 736.2 nM, Dmax = 68.8%) by EGFR-depleting PROTAC incorporating lapatinib [39]. However, this PROTAC induced a profound degradation of wild-type EGFR at low-nanomolar concentrations (DC50 = 39.2 nM, Dmax = 97.6%). Further optimizations on the lapatinib-based EGFR-depleting PROTAC are needed.
2.1.6. Enhanced target selectivity
Based on proteome-wide approaches, several studies have discovered that small-molecule PROTACs displayed high specificity for their target proteins [5,33,37,42,45,47]. Winter et al. [42] compared the proteomic changes of dBET1 treatment with JQ1 and vehicle controls using isobaric tagging which allowed the detection of 7429 proteins. MYC and PIM1 were down regulated in both of the JQ1 and dBET1 treatment conditions. Notably, only BRD2, BRD3, and BRD4 were significantly and markedly depleted in dBET1-treated cells. Bondeson et al. showed that only two (RIPK2 and the unrelated kinase MAPKAPK3) of the ~7000 quantified proteins were significantly degraded upon the treatment of PROTAC_RIPK2 [33]. Other kinases (such as RIPK3, ABL and TESK) that can bind to PROTAC_RIPK2 were not degraded. Based on an unbiased, multiplexed quantitative mass-spectrometry-based proteomics approach, Olson et al. [47] found that CDK9 was the most depleted protein after the treatment of THAL-SNS-032. Meanwhile, CDK9 was the only CDK that exhibited more than two-log-fold significant downregulation although THAL-SNS-032 have high affinities to other CDK such as CDK1, CDK2, and CDK7.
Some proteome-wide studies and several other studies suggest that the selectivity of PROTACs can be beyond the intrinsic binding specificity of the target binding ligands [33,[36], [37], [38],45,47,58]. Notably, although the BET inhibitor JQ1 lacks selectivity towards BRD2, BRD3, and BRD4, JQ1-based PROTACs MZ1 and AT1 induced selective depletion of BRD4 [36,37]. Although the TBK1 binding ligand of compound 3i exhibits poor selectivity for TBK1 over IKKε, compound 3i displayed excellent selectivity against IKKε [38].
2.1.7. Modifying the potency and selectivity
When generating PROTACs, their selectivity towards particular target proteins and their potency can be modified through adjusting the length and composition of linkers [21,23,27,32,34,36,38,44,59], altering the binding ligands to the target proteins and/or E3 ligases [27,32,34,36,38,39,44,46,59], and altering the choice of recruited E3 ligases [34,46]. HaloPROTAC7 with three ethylene glycol units showed the highest degradation efficacy among a series of HaloPROTACs with variable linker lengths [32]. Lai et al. showed that degradation profiles (c-ABL and BCR-ABL) could be changed through varying the inhibitor warheads (imatinib, bosutinib, and dasatinib) and the recruited E3 ligases (VHL and CRBN) [34]. Moreover, a more potent inhibitor warhead towards the target protein does not necessarily generate a more potent PROTAC [59]. Although the BET inhibitor that is used as target binding ligand lacks selectivity for any of BRD2/3/4, the preferential removal of BRD4 over BRD2 and BRD3 was fulfilled by modifying the chemical structure of linker [36]. Recently, researchers based on the available crystal structures to generate highly potent and selective PROTAC molecules [37,49]. For example, Huang et al. utilized the available target:inhibitor co-crystal structures to choose the site for installing a linker [49].
Through adjusting the stereochemistry of functional PROTACs [5,33,36,39,42] or modifying the E3-recruiting ligands (e.g., adding a methyl group on the glutarimide nitrogen of pomalidomide or lenalidomide) [43,44,50,51], inactive PROTACs that can bind target proteins but not E3 ligases were generated and were used as good control molecules with nearly identical physicochemical properties.
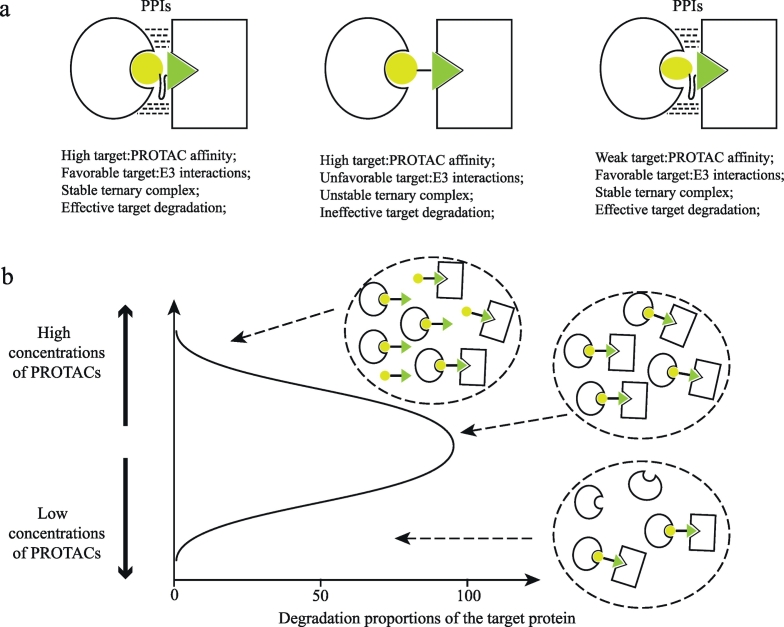
2.1.8. PPI induced stable ternary complexes
The preferential degradation of substrates bound by the target ligands has been observed in several studies [33,36,38,49]. This phenomenon leads researchers to suspect the structural basis of target selectivity. Researchers solved the crystal structure of the BRD4:MZ1:VHL ternary complex [37]. The structure reveals that MZ1 folds into itself and induces extensive protein–protein interactions (PPIs) between BRD4 and VHL E3 ligase, which promote the formation of stable and cooperative complexes (Fig. 2a). In addition, the specific de novo intermolecular PPIs drive the specificity of cooperative recognition. Bondeson et al. examined the degradation profile of a promiscuous PROTAC that can bind >50 kinases [58]. They found that only a subset of target kinases was degraded, and the formation of stable ternary complexes but not the target binding affinity correlated with degradation potency. Therefore, the efficient degradation relies on the formation of stable ternary complexes induced by the PPIs between the E3 ubiquitin ligase and the target protein (Fig. 2a). Moreover, even weak kinase:PROTAC affinity can be compensated by favorable target:E3 interactions, which results in the formation of stable ternary complexes and subsequently efficient degradation.
Fig. 2.
Characteristics of PROTACs (a) Protein-protein interactions (PPIs) between the target protein and the E3 ligase can stabilize the target:PROTAC:E3 complex even when the affinity between the target and the PROTAC is weak. The formation of stable ternary complexes is required for the induced protein degradation. (b) A hook effect shows when the systemic concentration of PROTACs is too high. High concentrations of dimeric PROTAC:E3 and PROTAC:target complexes inhibit the formation of degradation-inducing ternary complexes.
2.1.9. Hook effect
A hook effect (the reduced degradation at high concentrations of small-molecule PROTACs) was frequently observed in studies that investigated the degradation efficacy of small-molecule PROTACs [27,32,33,39,42,43,46,47] (Fig. 2b). The hook effect commonly occurs with three-component systems. When the concentration of PROTAC molecules is significantly higher than the DC50 value, autoinhibition of the formation of E3:PROTAC:target ternary complexes appears because of the high concentrations of PROTAC:E3 and PROTAC:target binary complexes.
2.2. Clinical development
Protein degradation induced by heterobifunctional molecules holds great promise in being used as a new pharmaceutical paradigm. At present, several companies (e.g., Arvinas, C4 therapeutics, Kymera Therapeutics, and Captor Therapeutics) are focusing on pushing the development of this technology and utilizing it to develop targeted therapeutics. The first company is Arvinas (New Haven, Connecticut, USA), which was launched in 2013 and founded by Crews [60]. C4 therapeutics (Cambridge, Massachusetts, USA) was launched in 2016. Kymera Therapeutics (Cambridge, Massachusetts, USA) and Captor Therapeutics (Wroclaw, Dolnoslaskie, Poland) were both founded in 2017. Arvinas is making efforts to advance its first two PROTAC drugs into clinical trials. These two oral PROTACs selectively degrade androgen receptor for prostate cancer and estrogen receptor for breast cancer, respectively.
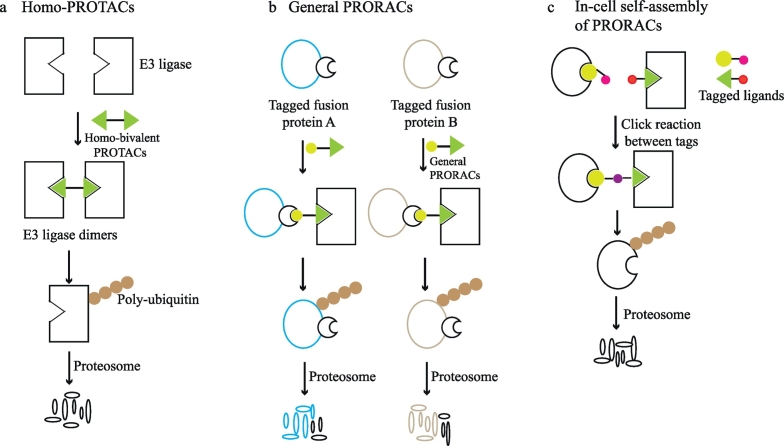
2.3. Depletion of E3 ligases through homo-PROTACs
E3 ubiquitin ligases are a huge protein family comprising >600 members. Overexpression of several E3 ligases (e.g., MDM2, IAP, and SCF) are frequently observed in human cancers and are associated with increased chemo-resistance and poor clinic prognosis [61]. Because E3 ligases lack active sites, have extensive PPIs with other proteins, and are multidomain protein, it is quite challenging to develop small molecule inhibitors that can effectively inhibit the activities of E3 ligases [62]. Given the advantages of heterobifunctional PROTACs over small molecule inhibitors, researchers adjusted the available heterobifunctional PROTACs paradigm and developed Homo-PROTACs [63]. Homo-PROTACs are bivalent small molecules and are intended to dimerize an E3 ligase and then induce its self-degradation (Fig. 3a). Utilizing VHL inhibitors, the most potent VHL degrader (CM11) was identified by modifying the length of the linker region and the stereochemistry. Symmetric trans-trans CM11 induced potent (DC99 = 10 nM for pVHL30), sustained, and isoform-selective degradation of VHL. Like heterobifunctional PROTACs, CM11 also exhibited the hook effect at high concentrations. Homo-PROTACs may be a powerful new strategy for drugging E3 ligases.
Fig. 3.
Other modalities of PROTACs (a) Homo-PROTACs are bivalent small-molecules that can trigger the dimerization of an E3 ligase and its subsequent self-degradation. (b) In contrast to typical PROTACs, general PROTACs cross-link E3 ligases and tagged fusion proteins and subsequently degrade fusion proteins. General PROTACs can be flexibly utilized to degrade variable proteins and study the functions of particular proteins. (c) Through bio-orthogonal click combination of two tagged small molecule precursors, heterobifunctional PROTACs can be formed intracellularly and successfully induce the degradation of target proteins. This approach was created to overcome the high molecular weight nature of typical PROTACs which contain two small-molecule ligands and a linker.
2.4. General PROTACs for tagged fusion proteins
PROTAC molecules listed in Table 1 require available target-selective molecules to be used as target binding ligands. General small-molecule PROTACs that target HaloTag [32], His-Tag [64], and FKBP12F36V [65] fusion proteins have been generated, which can extensively induce the selective degradation of proteins of interest (Fig. 3b). Through transgene expression and/or CRISPR-mediated locus-specific knock-in, researchers studied the kinetic properties of these general PROTACs towards variable target proteins. Particularly, heterobifunctional degraders including a FKBP12F36V-directed ligand and a CRBN binding ligand potently induced the rapid and profound degradation of FKBP12F36V-tagged proteins such as BRD4, KRASG12V, and MYC [65]. The degradation of FKBP12F36V-KRASG12V altered the levels of phosphorylated MEK and AKT and ERK-dependent transcriptional signaling, which suggests that KRASG12V is a functional oncoprotein. Though general PROTACs functional effects of proteins of interest can be evaluated, which can facilitate the selection of candidate proteins for further drug development.
2.5. In-cell assembly of PROTACs
Small-molecule PROTAC molecules usually possess relatively large size (typically 700–1100 Da), which may provide more opportunities for metabolic attack. Additionally, the size of PROTACs do not obey Lipinski's rule of five, which is a rule of thumb that evaluates if a chemical compound possesses certain pharmacological or biological properties of an orally active drug in humans. Lebraud et al. reported that heterobifunctional PROTACs can be formed intracellularly through bio-orthogonal click combination of two tagged smaller ligands [66] (Fig. 3c). In this study, BRD4 and ERK1/2 were successfully degraded when treating cultured cells with one precursor for a few hours followed by the treatment of the other precursor. However, the sequential treatment of two precursors and the potential difficulty for functional PROTACs to rapidly achieve high intracellular concentrations can dramatically slow the degradation progress.
3. Other protein degradation strategies
PROTACs utilize the intracellular ubiquitin-proteasome system to induce selective degradation. In addition to the PROTAC strategy, other protein degradation strategies (e.g., selective hormone receptor degraders and hydrophobic tagging) have also been discovered or developed. Several selective estrogen/androgen receptor modulators (e.g., tamoxifen, fulvestrant and AZD3514) have been developed [[67], [68], [69]]. They can bind estrogen/androgen receptor and cause the increased surface hydrophobicity and subsequent degradation of estrogen/androgen receptor. Among them, fulvestrant has been approved by FDA for the treatment of estrogen receptor-positive advanced breast cancer [70]. When using the hydrophobic tagging (HyT) strategy, a known ligand against a given target protein is conjugated to a hydrophobic tag (i.e., adamantane or Boc3-Arg) [71,72]. The binding of hydrophobic tags to the surface of target proteins results in the degradation of target proteins via the proteasome through causing localized conformational instability and/or unfolding of target proteins, and subsequent recruitment of chaperones. The HyT strategy has successfully utilized to induce the degradation of Her3 [73] and androgen receptor [74].
4. Challenges and directions
The relatively large size of small-molecule PROTAC molecules may severely affect their drug-like properties that would make small-molecule PROTACs becoming orally active drugs in humans. Currently, PROTACs are usually intraperitoneally, subcutaneously, or intravenously administered to mice to study their in vivo properties. The pharmaceutical properties of small-molecule PROTACs should be further optimized to make PROTACs being orally administered.
The development of PROTACs thus far relies on available small molecule inhibitors. Even target proteins that have low affinities with PROTACs can be effectively degraded if PROTACs can induce extensive PPIs between target proteins and E3 ligases [58]. Considering the unnecessary of high-affinity ligands to target proteins, proteins with no active occupation sites for small molecule inhibitors can be the potential targets of PROTACs. Screening small molecule libraries against proteins without active sites have great potentials in extending the repository of PROTAC-based targeted drugs. Meanwhile, generating PROTACs that bind to the crevices on the surface of kinases/receptors can avoid the resistance to small molecule inhibitors when drug-resistant variants emerge.
Although small-molecule PROTACs showed enhanced target selectivity compared with their corresponding small molecule inhibitors, off-targets were still detected in some studies [33,75,76]. Off-target proteins can be targets of the target ligands [75] or the IMiD-based CRBN binding ligands [65,76]. Small-molecule PROTACs may sometimes fail to degrade their potential targets. Ishoey et al. [76] synthesized a series of PROTACs consisting of promiscuous kinase inhibitors and phthalimide. However, these PROTACs were unable to degrade any of their consensus targets.
Since 2015, most of the reported small-molecule PROTACs induce the degradation of target proteins through recruiting VHL or CRBN E3 ligases. E3 ligases are a large protein family. More E3 ligases can be harnessed through discovering binding ligands to other E3 ligases, which can speed up the development of PROTACs targeting a given protein.
Search strategy and selection criteria
PubMed was used as the main search engines. References were searched using following terms: PROteolysis-TArgeting Chimeras, PROTAC, induced protein degradation, hydrophobic tagging, small molecule inhibitor, targeted drugs.
Funding
This work was supported by the National Key Research and Development Project of China [grant number 2018ZX09711002-003-011] and the Team Project of Guangdong Natural Science Foundation [grant number 2016A030312014].
Conflict of interest
The authors declared that there is no conflict of interest.
References
- 1.Lavanya V., Mohamed Adil A.A., Neesar A., Arun K.R., Shazia J. Small molecule inhibitors as emerging cancer therapeutics. Integr Cancer Sci Ther. 2014;1 [Google Scholar]
- 2.Wu P., Nielsen T.E., Clausen M.H. Small-molecule kinase inhibitors: an analysis of FDA-approved drugs. Drug Discov Today. 2016;21:5–10. doi: 10.1016/j.drudis.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Toure M., Crews C.M. Small-molecule PROTACS: new approaches to protein degradation. Angew Chem Int Ed Engl. 2016;55:1966–1973. doi: 10.1002/anie.201507978. [DOI] [PubMed] [Google Scholar]
- 4.Graves L.M., Duncan J.S., Whittle M.C., Johnson G.L. The dynamic nature of the kinome. Biochem J. 2013;450:1–8. doi: 10.1042/BJ20121456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gechijian L.N., Buckley D.L., Lawlor M.A., Reyes J.M., Paulk J., Ott C.J. Functional TRIM24 degrader via conjugation of ineffectual bromodomain and VHL ligands. Nat Chem Biol. 2018;14:405–412. doi: 10.1038/s41589-018-0010-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vangamudi B., Paul T.A., Shah P.K., Kost-Alimova M., Nottebaum L., Shi X. The SMARCA2/4 ATPase domain surpasses the bromodomain as a drug target in SWI/SNF-mutant cancers: insights from cDNA rescue and PFI-3 inhibitor studies. Cancer Res. 2015;75:3865–3878. doi: 10.1158/0008-5472.CAN-14-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leiser D., Pochon B., Blank-Liss W., Francica P., Glück A.A., Aebersold D.M. Targeting of the MET receptor tyrosine kinase by small molecule inhibitors leads to MET accumulation by impairing the receptor downregulation. FEBS Lett. 2014;588:653–658. doi: 10.1016/j.febslet.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 8.Spiegel J., Cromm P.M., Zimmermann G., Grossmann T.N., Waldmann H. Small-molecule modulation of Ras signaling. Nat Chem Biol. 2014;10:613–622. doi: 10.1038/nchembio.1560. [DOI] [PubMed] [Google Scholar]
- 9.Dogan S., Shen R., Ang D.C., Johnson M.L., D'Angelo S.P., Paik P.K. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18:6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisermann K., Wang D., Jing Y., Pascal L.E., Wang Z. Androgen receptor gene mutation, rearrangement, polymorphism. Transl Androl Urol. 2013;2:137–147. doi: 10.3978/j.issn.2223-4683.2013.09.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakamoto K.M., Kim K.B., Kumagai A., Mercurio F., Crews C.M., Deshaies R.J. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci U S A. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneekloth J.S., Jr., Fonseca F.N., Koldobskiy M., Mandal A., Deshaies R., Sakamoto K. Chemical genetic control of protein levels: selective in vivo targeted degradation. J Am Chem Soc. 2004;126:3748–3754. doi: 10.1021/ja039025z. [DOI] [PubMed] [Google Scholar]
- 13.Ohh M., Park C.W., Ivan M., Hoffman M.A., Kim T.Y., Huang L.E. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 14.Zhang D., Baek S.-H., Ho A., Kim K. Degradation of target protein in living cells by small-molecule proteolysis inducer. Bioorg Med Chem Lett. 2004;14:645–648. doi: 10.1016/j.bmcl.2003.11.042. [DOI] [PubMed] [Google Scholar]
- 15.Lee H., Puppala D., Choi E.Y., Swanson H., Kim K.B. Targeted degradation of the aryl hydrocarbon receptor by the PROTAC approach: a useful chemical genetic tool. Chembiochem. 2007;8:2058–2062. doi: 10.1002/cbic.200700438. [DOI] [PubMed] [Google Scholar]
- 16.Bargagna-Mohan P., Baek S.H., Lee H., Kim K., Mohan R. Use of PROTACS as molecular probes of angiogenesis. Bioorg Med Chem Lett. 2005;15:2724–2727. doi: 10.1016/j.bmcl.2005.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Gonzalez A., Cyrus K., Salcius M., Kim K., Crews C.M., Deshaies R.J. Targeting steroid hormone receptors for ubiquitination and degradation in breast and prostate cancer. Oncogene. 2008;27:7201–7211. doi: 10.1038/onc.2008.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai A.C., Crews C.M. Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov. 2017;16:101–114. doi: 10.1038/nrd.2016.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneekloth A.R., Pucheault M., Tae H.S., Crews C.M. Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics. Bioorg Med Chem Lett. 2008;18:5904–5908. doi: 10.1016/j.bmcl.2008.07.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marhefka C.A., Gao W., Chung K., Kim J., He Y., Yin D. Design, synthesis, and biological characterization of metabolically stable selective androgen receptor modulators. J Med Chem. 2004;47:993–998. doi: 10.1021/jm030336u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh Y., Ishikawa M., Naito M., Hashimoto Y. Protein knockdown using methyl bestatin-ligand hybrid molecules: design and synthesis of inducers of ubiquitination-mediated degradation of cellular retinoic acid-binding proteins. J Am Chem Soc. 2010;132:5820–5826. doi: 10.1021/ja100691p. [DOI] [PubMed] [Google Scholar]
- 22.Okuhira K., Ohoka N., Sai K., Nishimaki-Mogami T., Itoh Y., Ishikawa M. Specific degradation of CRABP-II via cIAP1-mediated ubiquitylation induced by hybrid molecules that crosslink cIAP1 and the target protein. FEBS Lett. 2011;585:1147–1152. doi: 10.1016/j.febslet.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 23.Okuhira K., Demizu Y., Hattori T., Ohoka N., Shibata N., Nishimaki-Mogami T. Development of hybrid small molecules that induce degradation of estrogen receptor-alpha and necrotic cell death in breast cancer cells. Cancer Sci. 2013;104:1492–1498. doi: 10.1111/cas.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demizu Y., Okuhira K., Motoi H., Ohno A., Shoda T., Fukuhara K. Design and synthesis of estrogen receptor degradation inducer based on a protein knockdown strategy. Bioorg Med Chem Lett. 2012;22:1793–1796. doi: 10.1016/j.bmcl.2011.11.086. [DOI] [PubMed] [Google Scholar]
- 25.Ohoka N., Nagai K., Hattori T., Okuhira K., Shibata N., Cho N. Cancer cell death induced by novel small molecules degrading the TACC3 protein via the ubiquitin-proteasome pathway. Cell Death Dis. 2014;5 doi: 10.1038/cddis.2014.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demizu Y., Shibata N., Hattori T., Ohoka N., Motoi H., Misawa T. Development of BCR-ABL degradation inducers via the conjugation of an imatinib derivative and a cIAP1 ligand. Bioorg Med Chem Lett. 2016;26:4865–4869. doi: 10.1016/j.bmcl.2016.09.041. [DOI] [PubMed] [Google Scholar]
- 27.Ohoka N., Okuhira K., Ito M., Nagai K., Shibata N., Hattori T. In Vivo knockdown of pathogenic proteins via specific and nongenetic inhibitor of apoptosis protein (IAP)-dependent protein erasers (SNIPERs) J Biol Chem. 2017;292:4556–4570. doi: 10.1074/jbc.M116.768853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shibata N., Nagai K., Morita Y., Ujikawa O., Ohoka N., Hattori T. Development of protein degradation inducers of androgen receptor by conjugation of androgen receptor ligands and inhibitor of apoptosis protein ligands. J Med Chem. 2018;61:543–575. doi: 10.1021/acs.jmedchem.7b00168. [DOI] [PubMed] [Google Scholar]
- 29.Buckley D.L., Van Molle I., Gareiss P.C., Tae H.S., Michel J., Noblin D.J. Targeting the von Hippel-Lindau E3 ubiquitin ligase using small molecules to disrupt the VHL/HIF-1α interaction. J Am Chem Soc. 2012;134:4465–4468. doi: 10.1021/ja209924v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buckley D.L., Gustafson J.L., Van Molle I., Roth A.G., Tae H.S., Gareiss P.C. Small-molecule inhibitors of the interaction between the E3 ligase VHL and HIF1α. Angew Chem Int Ed Engl. 2012;51:11463–11467. doi: 10.1002/anie.201206231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galdeano C., Gadd M.S., Soares P., Scaffidi S., Van Molle I., Birced I. Structure-guided design and optimization of small molecules targeting the protein-protein interaction between the von Hippel-Lindau (VHL) E3 ubiquitin ligase and the hypoxia inducible factor (HIF) alpha subunit with in vitro nanomolar affinities. J Med Chem. 2014;57:8657–8663. doi: 10.1021/jm5011258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buckley D.L., Raina K., Darricarrere N., Hines J., Gustafson J.L., Smith I.E. HaloPROTACS: use of small molecule PROTACs to induce degradation of HaloTag fusion proteins. ACS Chem Biol. 2015;10:1831–1837. doi: 10.1021/acschembio.5b00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bondeson D.P., Mares A., Smith I.E., Ko E., Campos S., Miah A.H. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol. 2015;11:611–617. doi: 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai A.C., Toure M., Hellerschmied D., Salami J., Jaime-Figueroa S., Ko E. Modular PROTAC design for the degradation of oncogenic BCR-ABL. Angew Chem Int Ed Engl. 2016;55:807–810. doi: 10.1002/anie.201507634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raina K., Lu J., Qian Y., Altieri M., Gordon D., Rossi A.M. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci U S A. 2016;113:7124–7129. doi: 10.1073/pnas.1521738113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zengerle M., Chan K.H., Ciulli A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem Biol. 2015;10:1770–1777. doi: 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gadd M.S., Testa A., Lucas X., Chan K.H., Chen W., Lamont D.J. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol. 2017;13:514–521. doi: 10.1038/nchembio.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crew A.P., Raina K., Dong H., Qian Y., Wang J., Vigil D. Identification and characterization of Von Hippel-Lindau-recruiting proteolysis targeting chimeras (PROTACs) of TANK-binding kinase 1. J Med Chem. 2018;61:583–598. doi: 10.1021/acs.jmedchem.7b00635. [DOI] [PubMed] [Google Scholar]
- 39.Burslem G.M., Smith B.E., Lai A.C., Jaime-Figueroa S., McQuaid D.C., Bondeson D.P. The advantages of targeted protein degradation over inhibition: an RTK case study. Cell Chem Biol. 2018;25:67–77. doi: 10.1016/j.chembiol.2017.09.009. [e3] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito T., Ando H., Suzuki T., Ogura T., Hotta K., Imamura Y. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 41.Lopez-Girona A., Mendy D., Ito T., Miller K., Gandhi A.K., Kang J. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia. 2012;26:2326–2335. doi: 10.1038/leu.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winter G.E., Buckley D.L., Paulk J., Roberts J.M., Souza A., Dhe-Paganon S. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu J., Qian Y., Altieri M., Dong H., Wang J., Raina K. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem Biol. 2015;22:755–763. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou B., Hu J., Xu F., Chen Z., Bai L., Fernandez-Salas E. Discovery of a small-molecule degrader of bromodomain and extra-terminal (BET) proteins with picomolar cellular potencies and capable of achieving tumor regression. J Med Chem. 2018;61:462–481. doi: 10.1021/acs.jmedchem.6b01816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Remillard D., Buckley D.L., Paulk J., Brien G.L., Sonnett M., Seo H.S. Degradation of the BAF complex factor BRD9 by heterobifunctional ligands. Angew Chem Int Ed Engl. 2017;56:5738–5743. doi: 10.1002/anie.201611281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schiedel M., Herp D., Hammelmann S., Swyter S., Lehotzky A., Robaa D. Chemically induced degradation of sirtuin 2 (Sirt2) by a proteolysis targeting chimera (PROTAC) based on sirtuin rearranging ligands (SirReals) J Med Chem. 2018;61:482–491. doi: 10.1021/acs.jmedchem.6b01872. [DOI] [PubMed] [Google Scholar]
- 47.Olson C.M., Jiang B., Erb M.A., Liang Y., Doctor Z.M., Zhang Z. Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat Chem Biol. 2018;14:163–170. doi: 10.1038/nchembio.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robb C.M., Contreras J.I., Kour S., Taylor M.A., Abid M., Sonawane Y.A. Chemically induced degradation of CDK9 by a proteolysis targeting chimera (PROTAC) Chem Commun (Camb) 2017;53:7577–7580. doi: 10.1039/c7cc03879h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang H.T., Dobrovolsky D., Paulk J., Yang G., Weisberg E.L., Doctor Z.M. A chemoproteomic approach to query the degradable kinome using a multi-kinase degrader. Cell Chem Biol. 2018;25:88–99. doi: 10.1016/j.chembiol.2017.10.005. [e6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buhimschi A.D., Armstrong H.A., Toure M., Jaime-Figueroa S., Chen T.L., Lehman A.M. Targeting the C481S ibrutinib-resistance mutation in Bruton's tyrosine kinase using PROTAC-mediated degradation. Biochemistry. 2018;57:3564–3575. doi: 10.1021/acs.biochem.8b00391. [DOI] [PubMed] [Google Scholar]
- 51.Zhang C., Han X.R., Yang X., Jiang B., Liu J., Xiong Y. Proteolysis targeting chimeras (PROTACs) of anaplastic lymphoma kinase (ALK) Eur J Med Chem. 2018;151:304–314. doi: 10.1016/j.ejmech.2018.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saenz D.T., Fiskus W., Qian Y., Manshouri T., Rajapakshe K., Raina K. Novel BET protein proteolysis-targeting chimera exerts superior lethal activity than bromodomain inhibitor (BETi) against post-myeloproliferative neoplasm secondary (s) AML cells. Leukemia. 2017;31:1951–1961. doi: 10.1038/leu.2016.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun B., Fiskus W., Qian Y., Rajapakshe K., Raina K., Coleman K.G. BET protein proteolysis targeting chimera (PROTAC) exerts potent lethal activity against mantle cell lymphoma cells. Leukemia. 2018;32:343–352. doi: 10.1038/leu.2017.207. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X., Lee H.C., Shirazi F., Baladandayuthapani V., Lin H., Kuiatse I. Protein targeting chimeric molecules specific for bromodomain and extra-terminal motif family proteins are active against pre-clinical models of multiple myeloma. Leukemia. 2018;32:1–16. doi: 10.1038/s41375-018-0044-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jun H.J., Acquaviva J., Chi D., Lessard J., Zhu H., Woolfenden S. Acquired MET expression confers resistance to EGFR inhibition in a mouse model of glioblastoma multiforme. Oncogene. 2012;31:3039–3050. doi: 10.1038/onc.2011.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arcila M.E., Nafa K., Chaft J.E., Rekhtman N., Lau C., Reva B.A. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013;12:220–229. doi: 10.1158/1535-7163.MCT-12-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun Y., Zhao X., Ding N., Gao H., Wu Y., Yang Y. PROTAC-induced BTK degradation as a novel therapy for mutated BTK C481S induced ibrutinib-resistant B-cell malignancies. Cell Res. 2018;28:779–781. doi: 10.1038/s41422-018-0055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bondeson D.P., Smith B.E., Burslem G.M., Buhimschi A.D., Hines J., Jaime-Figueroa S. Lessons in PROTAC design from selective degradation with a Promiscuous Warhead. Cell Chem Biol. 2018;25:78–87. doi: 10.1016/j.chembiol.2017.09.010. [e5] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan K.H., Zengerle M., Testa A., Ciulli A. Impact of target Warhead and linkage vector on inducing protein degradation: comparison of bromodomain and extra-terminal (BET) degraders derived from triazolodiazepine (JQ1) and tetrahydroquinoline (I-BET726) BET inhibitor scaffolds. J Med Chem. 2018;61:504–513. doi: 10.1021/acs.jmedchem.6b01912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bouchie A., Allison M., Webb S., Defrancesco L. Nature biotechnology's academic spinouts of 2013. Nat Biotechnol. 2014;32:229–238. doi: 10.1038/nbt.2846. [DOI] [PubMed] [Google Scholar]
- 61.Sun Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia. 2006;8:645–654. doi: 10.1593/neo.06376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arkin M.R., Tang Y., Wells J.A. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol. 2014;21:1102–1114. doi: 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maniaci C., Hughes S.J., Testa A., Chen W., Lamont D.J., Rocha S. Homo-PROTACs: bivalent small-molecule dimerizers of the VHL E3 ubiquitin ligase to induce self-degradation. Nat Commun. 2017;8:830. doi: 10.1038/s41467-017-00954-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Okitsu K., Hattori T., Misawa T., Shoda T., Kurihara M., Naito M. Development of a small hybrid molecule that mediates degradation of His-Tag fused proteins. J Med Chem. 2018;61:576–582. doi: 10.1021/acs.jmedchem.7b00413. [DOI] [PubMed] [Google Scholar]
- 65.Nabet B., Roberts J.M., Buckley D.L., Paulk J., Dastjerdi S., Yang A. The dTAG system for immediate and target-specific protein degradation. Nat Chem Biol. 2018;14:431–441. doi: 10.1038/s41589-018-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lebraud H., Wright D.J., Johnson C.N., Heightman T.D. Protein degradation by in-cell self-assembly of proteolysis targeting chimeras. ACS Cent Sci. 2016;2:927–934. doi: 10.1021/acscentsci.6b00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Connor C.E., Norris J.D., Broadwater G., Willson T.M., Gottardis M.M., Dewhirst M.W. Circumventing tamoxifen resistance in breast cancers using antiestrogens that induce unique conformational changes in the estrogen receptor. Cancer Res. 2001;61:2917–2922. [PubMed] [Google Scholar]
- 68.Wittmann B.M., Sherk A., McDonnell D.P. Definition of functionally important mechanistic differences among selective estrogen receptor down-regulators. Cancer Res. 2007;67:9549–9560. doi: 10.1158/0008-5472.CAN-07-1590. [DOI] [PubMed] [Google Scholar]
- 69.Bradbury R.H., Acton D.G., Broadbent N.L., Brooks A.N., Carr G.R., Hatter G. Discovery of AZD3514, a small-molecule androgen receptor downregulator for treatment of advanced prostate cancer. Bioorg Med Chem Lett. 2013;23:1945–1948. doi: 10.1016/j.bmcl.2013.02.056. [DOI] [PubMed] [Google Scholar]
- 70.Di Leo A., Jerusalem G., Petruzelka L., Torres R., Bondarenko I.N., Khasanov R. Results of the CONFIRM phase III trial comparing fulvestrant 250 mg with fulvestrant 500 mg in postmenopausal women with estrogen receptor-positive advanced breast cancer. J Clin Oncol. 2010;28:4594–4600. doi: 10.1200/JCO.2010.28.8415. [DOI] [PubMed] [Google Scholar]
- 71.Neklesa T.K., Crews C.M. Chemical biology: greasy tags for protein removal. Nature. 2012;487:308–309. doi: 10.1038/487308a. [DOI] [PubMed] [Google Scholar]
- 72.Long M.J., Gollapalli D.R., Hedstrom L. Inhibitor mediated protein degradation. Chem Biol. 2012;19:629–637. doi: 10.1016/j.chembiol.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xie T., Lim S.M., Westover K.D., Dodge M.E., Ercan D., Ficarro S.B. Pharmacological targeting of the pseudokinase Her3. Nat Chem Biol. 2014;10:1006–1012. doi: 10.1038/nchembio.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gustafson J.L., Neklesa T.K., Cox C.S., Roth A.G., Buckley D.L., Tae H.S. Small-molecule-mediated degradation of the androgen receptor through hydrophobic tagging. Angew Chem Int Ed Engl. 2015;54:9659–9662. doi: 10.1002/anie.201503720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Savitski M.M., Zinn N., Faelth-Savitski M., Poeckel D., Gade S., Becher I. Multiplexed proteome dynamics profiling reveals mechanisms controlling protein homeostasis. Cell. 2018;173:260–274. doi: 10.1016/j.cell.2018.02.030. [e25] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ishoey M., Chorn S., Singh N., Jaeger M.G., Brand M., Paulk J. Translation termination factor GSPT1 is a phenotypically relevant off-target of heterobifunctional phthalimide degraders. ACS Chem Biol. 2018;13:553–560. doi: 10.1021/acschembio.7b00969. [DOI] [PubMed] [Google Scholar]