Abstract
Background
The goal of this study is to identify and characterize treatment resistant tumor initiating cells (TRTICs) using orthotopic xenografts.
Methods
TRTICs were enriched from GBM cell lines using mouse xenografts treated with fractionated doses of radiation and temozolomide. TRTICs were characterized by neurosphere clonogenicity and self-renewal, serial xenotransplantation, differentiation potential, and mRNA & miRNA transcriptomic profiling. We use an unbiased approach to identify antigens encoding TRTIC and glioma stem cells (GSC) populations. Co-culture experiments of TRTIC and differentiated cells were conducted to evaluate the reliance of TRTIC differentiation on the secretome of differentiated cells.
Findings
TRTICs acquire stem-like gene expression signatures and increased side population staining resulting from the activation of multi-drug resistance genes. Genetic and functional characterization of TRTICs shows a striking resemblance with GSCs. TRTICs can differentiate towards specific progeny in the neural stem cell lineage. TRTIC-derived tumors display all the histological hallmarks of glioblastoma (GBM) and exhibit a miRNA-transcript and mRNA-transcriptomic profile associated with aggressiveness. We report that CD24+/CD44+ antigens are expressed in TRTICs and patient-derived GSCs. Double positive CD24+/CD44+ exhibit treatment resistance and enhanced tumorigenicity. Interestingly, co-culture experiments with TRTICs and differentiated cells indicated that the regulation of TRTIC differentiation could rely on the secretome in the tumor niche.
Interpretation
Radiation and temozolomide treatment enriches a population of cells that have increased iPSC gene expression. As few as 500 cells produced aggressive intracranial tumors resembling patient GBM. CD24+/CD44+ antigens are increased in TRTICs and patient-derived GSCs. The enrichment for TRTICs may result in part from the secretome of differentiated cells.
Fund
NIH/NCI 1RC2CA148190, 1R01CA108633, 1R01CA188228, and The Ohio State University Comprehensive Cancer Center.
Keywords: Treatment-resistance, Tumor-initiating, Glioma stem cell, CD24high/CD44high, Transcriptome, Neural lineage
Research in context.
Evidence before this study
Recapitulating glioblastoma (GBM) tumors similar to humans in mice for therapeutic intervention requires tumor derived glioma stem cells (GSC).
Added value of this study
We report the previously unexplored subset of treatment resistant tumor initiating cell (TRTIC) expressing iPSC genes resembling GSCs using long-term cultured GBM cell lines. TRTICs express CD24+ and CD44+ and exhibits cellular plasticity. TRTIC tumors exhibit all the histological hallmarks of GBM. Secretome of cells in the tumor niche maintains TRTIC.
Implications of all the available evidence
The intriguing implication here is the possibility of enriching TRTICs orthotopically to study the molecular basis of cellular plasticity and the possibility of developing therapeutics.
Alt-text: Unlabelled Box
1. Introduction
Glioblastoma (GBM) is the most common and aggressive type of primary brain tumor. Resistance to treatment represents a major obstacle to improving outcomes in patients with GBM. Despite major improvements in treatment modalities, the median survival time of patients following standard-of-care remains 12–15 months. A major contributor to this dismal prognosis is the near universal recurrence rate of GBM. Glioma stem cells (GSCs) are the subpopulation of cells believed to drive recurrence [[1], [2], [3], [4]]. It has been established that GSCs express genes associated with pluripotent neural stem cells (NSCs), can differentiate towards neural lineages, and have tumor-initiating ability. However, the treatment-resistant tumor-initiating cells (TRTICs) within GBM remain largely uncharacterized.
A major obstacle in studying TRTICs has been the inability to successfully isolate and maintain this subpopulation due to the lack of unique antigenic markers encoded in these cells and tumor heterogeneity. Here, we enriched and isolated the TRTIC subpopulation that survived current standard-of-care treatment, temozolomide (TMZ) and radiation treatment (RT) in NOD-SCID mice and cultured the cells in vitro. TRTICs were isolated from the residual NOD-SCID tumor after treatment and propagated as non-adherent clusters of cells, referred to as neurospheres, in growth factor-defined (bFGF and EGF) serum-free selection media originally developed for NSCs. We show that TRTICs, similar to neurospheres, have the capacity for self-renewal and the potential to differentiate to all of the principal cell types of the brain, such as neurons, astrocytes, and oligodendrocytes [[5], [6], [7], [8], [9]]. TRTICs generated at clonal density reform neurospheres after induction of differentiation and have genetic aberrations typical of brain tumors; a point that distinguishes cancer stem cells from normal stem cells. TRTICs isolated from GBM cell lines resemble GSCs isolated from patient biopsies and differ from their parental cell lines based on miRNA, mRNA profiles, and tumor forming ability. We demonstrate that TRTICs are self-renewing, proliferative, and able to reproduce the complexity of the original tumor faithfully while maintaining genetic integrity in vivo. In addition, we show that TRTICs form tumors exhibiting all of the histological hallmarks and aggressive phenotype of patient GBM. Lastly, we show that the maintenance and differentiation of the TRTIC subpopulation may rely on the secretome in the tumor niche.
2. Materials and methods
Study methodology used the Institutional guidelines and approved protocols IACUC (2009A012&-R1), IBC (2009R0169), and IRB (2009C0065 & 2014C0115). The use of human biospecimens in this study was carried out in accordance with our Institutional IRB Ethics Committee Requirements.
2.1. Statistical and data analysis
All the experiments repeated at least three times to test their validity and the results analyzed for statistical significance using the t-test and analysis of variance (ANOVA) for experiments with multiple groups. Rigorous statistical considerations applied to eliminate false positives in gene expression data and analyzed using the Partek Genomic Suite (Partek Inc., Saint Louis, MO). Ingenuity pathway analysis identified novel pathways from the sub-set of differentially regulated genes (IPA, http://www.ingenuity.com). Analyzed data displayed as mean ± s.e.m. using SAS (SAS Institute Inc., Cary, NC) and Graph Pad Prism (Graph Pad Software Inc., La Jolla, CA).
2.2. Cell culture
The primary GSCs (OSU2, OSU11, OSU16, OSU17, OSU20, OSU23, OSU53, OSU68, GBM8, PKAC2, PKAC6, & PKAC7) used in this study were isolated from GBM patient tissues and authenticated by neuro-pathologist. The commercially available cell lines (U87, U118, U251, SNB19, & T98G) obtained from ATCC. All the cells authenticated by STR analysis. DMEM (GIBCO) medium with 10% FBS (GIBCO) and penicillin/streptomycin (100 U/mL) (GIBCO) was used to grow the differentiated cells. All neurospheres were grown in DMEM F-12 (GIBCO) medium with 10% B27 supplement (GIBCO), human-basic fibroblast growth factor (20 ng/mL) (Invitrogen), human-epidermal growth factor (20 ng/mL) (Invitrogen), penicillin/streptomycin (100 U/mL) (GIBCO), and Amphotericin B (5 ng/mL) (Sigma). All the experiments reported was conducted within 10 passages.
2.3. Enriching TRTICs
Three to four week old NOD-SCID mice were orthotopically implanted with GBM cells. A small anterior-posterior incision through scalp was made down to the level of the bone. The bur hole was made exactly 2 mm from the bregma at the right frontal lobe. Cells in a 3 μL volume of medium were injected intracranially using a stereotactic frame. Initiation of treatment regimen occurred 3–4 weeks after tumor implantation. At day 1, 100 mg/kg of temozolomide was administered by p.o. gavage. The next day the mice were radiated (1 Gy) at the tumor site while sparing the exposure to other organs by housing them in a lead shield. The third day mice were monitored for their health condition, and on the fourth day temozolomide administration occurs followed by 1 gy radiation on the fifth day and monitored on the sixth day. This cycle of temozolomide administration, radiation treatment, and monitoring occured for three more cycles resulting with a cumulative dose of 500 mg/kg and a radiation dose of 5Gy. Tumor formation and tumor burden were evaluated using sequential magnetic resonance imaging (MRI), weight loss, and neurological symptoms such as lethargy, poor feeding, paralysis, appearance of distress such as poor mobility, self mutilation, hunched posture, dehydrations and skin ulcerations, and weight loss exceeding 20%. Observation of these symptoms resulted in euthanization. The brain was recovered for further studies and generation of GSCs. Tumors obtained from NOD-SCID mice was minced and enzymatically dissociated using Tryple (Invitrogen) and mechanically dissociated using a G20 10 mL syringe to obtain a cell suspension. The cell suspension was passed through cell strainer (BD) to obtain a single cell suspension. Neurosphere initiation medium was added to the cells consisting of the following: DMEM-F12 (GIBCO) medium, B27 supplement (GIBCO), human-basic fibroblast growth factor (20 ng/mL) (Invitrogen), human-epidermal growth factor (20 ng/mL) (Invitrogen), penicillin / streptomycin (100 U/mL) (GIBCO), and Amphotericin B (5 ng/mL) (Sigma). Cultures were fed every two days until they reach confluency. Cultures were passaged every week by collecting the floating spheres by centrifugation at 1000 rpm for 5 min at 4 °C. The spent media was aspirated and the pellet was mechanically dissociated and seeded in fresh medium. Additional details are reported elsewhere [53].
2.4. Generating treatment resistant cells under in vitro conditions
Temozolomide (100 μM) was added to LN18, LN229, U87, U118, and T98G and irradiated with 2 Gy after two hours of TMZ addition. After 48 h of TMZ + RT, the cell growth medium was replaced to remove the dead cells and the cells were again treated with 100 μM TMZ followed by 2 Gy radiation. This step repeated for three more times, resulting in a total dose of 500 μM TMZ and 10 Gy. The cells surviving this total dose are considered treatment resistant.
2.5. Serial clonogenic analysis
To determine the self-renewal ability of TRTICs, a single-cell suspension was sorted onto a 96 well plate using a flow cytometer and cultured in serum-free growth factor-defined medium. Wells containing cells were checked daily under a microscope to count the number of cell clones. After 2 weeks, the clones were dissociated and cultured similarly in new 96-well plates to generate sub-clones.
2.6. Differentiation assay of tumor spheres
Two days after primary culture, cells were plated onto glass coverslips coated in poly-l-lysine and poly-L-ornithine (Sigma) in medium with 10% FBS in coverslips. Cells were fed with FBS-supplemented medium every 2 days, and coverslips were processed 5 days after plating using immunocytochemistry.
2.7. Radiation and chemotherapeutic sensitivity assay
Radiation was delivered using the GAMMA CELL 40 Extractor irradiator and RS 2000 Biological Irradiator. At a predetermined time after treatment, the cells were analyzed using flow cytometry after staining with AnnexinV-PE (Life Technologies) and PI (Sigma). Drug solvent DMSO was added to the control cells, MTS and/or AlmarBlue proliferation assays were used to assess viable cells after drug treatment by following manufacturers' protocol. About 5 × 103 cells plated in a 96-well plate and treated with one of the following chemotherapeutic agents at 100 μM: Temozolomide, 10 μM Doxorubicin, 10 μM Imatinib, 10 μM Paclitaxel, or 10 μM Etoposide. The cultures were incubated at 37 °C for a predetermined time (24, 48, and 72 h), 20 μL of MTS solution was added and cells were incubated for 3 h before measuring the absorbance at 490 nm.
2.8. Differentiation assay to drive TRTICs towards specific progeny
TRTICs were plated on a glass coverslips coated with poly-l-lysine and poly-L-ornithine (Sigma). Cells were fed every two days with differentiation medium, which consisted of one of the following growth factors (Invitrogen): brain-derived neurotrophic factor (BDNF), glial-derived neurotrophic factor (GDNF), bone morphogenetic protein (BMP4), ciliary neurotrophic factor (CNTF), and sonic hedgehog (SHH), at a concentration of 10 ng/mL. After 1–2 weeks, cells in the chamber slides were fixed with paraformaldehyde (4% in PBS) to stain for mAbs against mouse anti-Tubulin β-III, mouse anti-Nestin, rabbit anti-Sox2, mouse anti-MAP2, rabbit anti-GFAP, and mouse anti-Olig1 (Millipore). The staining was detected by using Alexa Fluor conjugated secondary antibodies (Molecular Probes).
2.9. Gene expression profiling
The MIAME-complaint microarray data are available in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE54660. The sample labels KPAC-U87-1 to 3 and KPAC-MGH87GSC-1 to 4 refers to the RNA samples obtained from U87 and TRTIC tumors respectively.
Additional detailed experimental procedures for magnetic resonance imaging, RNA isolation/QC, qRT-PCR, TaqMan low density array, microRNA profiling, Immunofluorescence, serial clonogenic assays, proliferation assay, flow cytometry analysis, co-culture studies, Bioplex 200, gene expression and pathway analysis are provided as supplemental material.
2.10. Immunofluorescence
The spheres were grown on poly-l-lysine poly-L-ornithine coated cover slips and fixed with 4% paraformaldehyde for 30 min at 25 °C. The fixative was aspirated and rinsed three times in PBS (10 min each). The cells were further incubated with permeable (0.3% Triton X-100 in 5% normal donkey serum in 1× PBS) or non-permeable blocking buffer (5% normal donkey serum in 1× PBS). The fixed and blocked cells were incubated with primary antibodies in permeable or non-permeable blocking buffer at 4 °C for 10 h. For dual-labeling experiments, primary antibodies generated from different species were added together. Following incubation coverslips were rinsed three times (10 min each) with PBS and the cells were overlaid with appropriate donkey-anti-mouse and/or donkey-anti-rabbit secondary antibody conjugated to Alexa Fluor for 2 h at 25 °C in dark. For double labeling, appropriate secondary antibodies were added together. After three final washes in PBS, coverslips were rinsed with double distilled water and mounted using mounting medium containing DAPI on glass slides. Fluorescence was detected and photographed with both a Leica and Zeiss ApoTome microscopes. The following antibodies were used for immunostaining: rabbit-anti-GFAP(1:250) (Chemicon), mouse-anti-nestin (1:250) (Chemicon), rabbit-anti-sox2 (1:1000) (Chemicon), mouse-anti-MAP2 (1:250) (Chemicon), mouse-anti-O1(1:500) (Chemicon), and secondary antibodies raised in goat against mouse and rabbit immunoglobulins, conjugated to the fluorophore Alexa Fluor (Invitrogen). Additionally, cells were immune-stained with 4,6-diamidino-2-phenylindole (DAPI) (Sigma), to permit counting of cell nuclei in at least 5 microscopic fields per specimen. Quantification of cells stained with each antibody could then be averaged and estimated as a percentage of total nuclei counted.
2.11. Immunohistochemistry of tumor sections
Formalin-fixed, paraffin embedded tissue sections were baked overnight at 60 °C, deparaffinized, rehydrated using xylene and ethanol graded mixture, treated with epitope retrieval agent (Dako-TRS pH 6.1) and blocked for endogenous peroxidase and biotin before the application of the primary antibody. Incubation of primary antibodies at a 1:100 dilution was performed overnight at 4 °C in a humidity chamber. Subsequent immune-detection was performed using the IgG matched secondary antibody by incubating at room temperature for 1 h in a humidity chamber, treated with chromagen, counterstained with hematoxylin, and mounted after dehydration. Thorough washing with PBS (3×) was ensured after all incubation steps.
3. Results
3.1. TRTICs possess an induced pluripotent stem cell (iPSC) signature
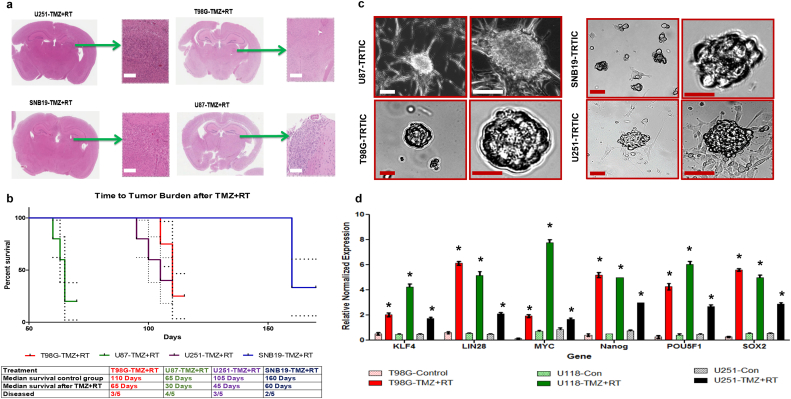
U251, T98G, SNB19, and U87 cells were implanted into the right frontal lobe of NOD-SCID mice. Upon tumor formation, we administered temozolomide (TMZ) and radiation treatment (RT) (Fig. 1a). The time to tumor burden after TMZ + RT varied across cell lines shows their different treatment sensitivity, U87 being more resistant compared to other tumors (Fig. 1b). In each cell line, a subset of xenograft mice exhibited residual tumors after TMZ + RT in the following numbers: U87 (80%), U251 (60%), T98G (60%), and SNB19 (40%) (Fig. 1b). H&E staining of coronal section of mice bearing U87 tumors exhibited a TMZ + RT dose dependent effect (Fig. S1a). Among the three treatment arms TMZ, RT, and TMZ + RT, the mice treated with TMZ + RT survived longer than other treatment arms (Fig. S1b). We cultured the cells isolated from the residual tumors after TMZ + RT in vitro as neurospheres (Figs. 1c & S1c). The morphology of these neurospheres closely resembled the neurospheres derived from induced pluripotent stem cells in earlier reports [[10], [11], [12]]. The TRTICs derived from the residual tumors surviving TMZ + RT demonstrated a higher expression level of induced pluripotent stem cell (iPSC) reprograming factors (Figs. 1d & S1d) compared to the respective parental cells. We were able to serial passage these TRTICs as neurospheres. The first pass single cell neurosphere assay consisted of 87–93% clones and the second pass consisted of 90–95% clones. This capacity for repeated generation of neurospheres from single cells is generally viewed as evidence of self-renewal [8,9]. Taken together, these data suggest that these isolated TRTICs possess a stem cell phenotype.
Fig. 1.
TRTICs were enriched from residual tumors following temozolomide and radiation treatment. a. Hematoxylin and Eosin stained coronal sections of NOD-SCID mice bearing U251, T98G, SNB19, and U87 residual tumors after temozolomide and radiation treatment (n = 5). b. Kaplan-Meier survival plots of treated tumors (n = 5) (log-rank (Mantel-Cox) test) c. Microscopic images of TRTICs derived from residual tumors (n = 5). d. Relative normalized expression of iPSC factors and stem cell genes in parental cell lines and TRTICs (n = 6). (Two-way ANOVA: * p < 0.001). Scale bar = 50 μm.
To determine whether the upregulation of iPSC gene expression in TRTICs was an artifact of culturing the residual tumors in growth factor defined medium, we conducted in vitro studies using U87 cells and treating them with TMZ, RT, and TMZ + RT for 7 days. We used pluripotency expression profiling arrays to analyze changes in stem cell phenotype. Cells surviving TMZ + RT had significant upregulation of Nanog, LIN28, and SOX2 (Fig. S1e) (Supplemental Table 1). Interestingly, Gamma-aminobutyric acid receptor subunit beta-3 (GABRB3), a gene encoding a member of the ligand-gated ion channel family, is highly expressed in the TMZ + RT arm. In undifferentiated cells, GABRB3 serves as the receptor for gamma-aminobutyric acid and acts as the major inhibitory transmitter of the nervous system. The GABRB3 gene has been associated with mental disability. The upregulation of GABRB3 could potentially connect the neurocognitive decline in GBM patients after TMZ + RT treatments, which will require further detailed investigation.
3.2. TRTIC tumors recapitulate the histological hallmarks of GBM, tumor growth kinetics and enhanced tumorigenicity
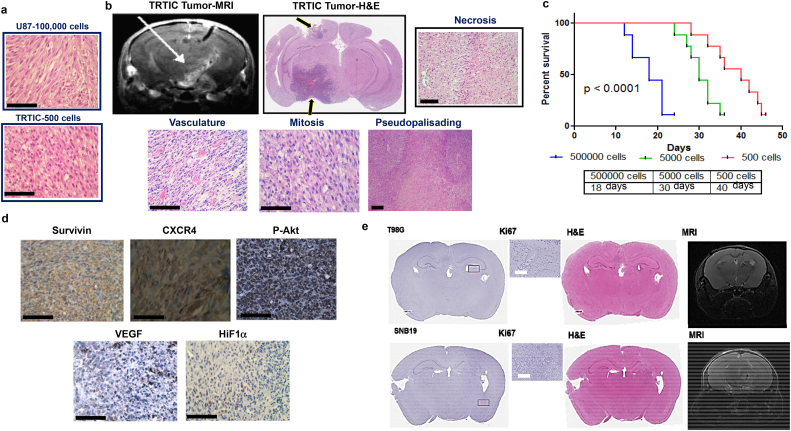
TRTICs produced a significantly denser tumor compared to parental U87 cells (Fig. 2a). TRTIC-derived tumors resembled the key histological hallmarks of patient GBM (Fig. 2b). MRI imaging showed significant migration of the TRTICs away from the needle track crossing the midline recapitulating the invasive and migratory behavior of GBM (Fig. 2b). Growth kinetics measured by implanting 500,000, 5000, and 500 TRTICs into NOD-SCID mice exhibited an increase in median survival following each serial dilution (Fig. 2c). Immunostaining of the TRTIC-derived tumors detected high expression levels of proteins linked to GBM aggressiveness and pro-survival (Fig. 2d). The high expression of VEGF and HIF1α along the periphery of the pseudopalisadic regions of the TRTIC-derived tumors is consistent with pseudopalisading in patient GBM as the periphery of necrotic regions are severely hypoxic, overexpress HIF1α, and secrete pro-angiogenic factor VEGF. As would also be expected in patient GBM, analysis of the pseudopalisadic regions of the TRTIC-derived tumors showed increased expression of CXCR4, a protein involved in metastasis, angiogenesis, and the upregulation of HIF1α. The treatment responsive mice that did not show symptoms due to tumor burden underwent MRIs. Coronal sections isolated from these mice stained with H&E and Ki-67 further confirmed the absence of tumors (Fig. 2e). Next, we investigated whether we could generate TRTIC like cells without tumor stroma under cell culture conditions. Indeed LN229, LN18, U118 and T98G produced clonal cells surviving TMZ + RT that exhibited an increased iPSC factors and stem cell gene signature (Fig. S2a–c). Overall, these data indicate that TRTICs can be enriched by treating GBM cell lines and xenograft tumors with TMZ + RT. However, TRTICs obtained from xenografts are superior due to in vitro enrichment since in vivo TRTICs recapitulated GSC properties better than in vitro TRTICs and possessed an extensive self-renewal capacity. Additionally, this could be due to the interaction with stroma under in vivo conditions.
Fig. 2.
TRTIC-derived tumors recapitulate hallmarks of patient GBM. a. Hematoxylin and Eosin stained sections of tumors formed from 100,000 U87 parental cells and 500 U87 derived TRTIC cells (n = 5). b. MRI scan showing invasive nature and Hematoxylin and Eosin stained sections of U87 TRTIC-derived tumors exhibiting histological hallmarks of GBM (n = 3). c. Kaplan-Meier survival plots of serial xenotransplantation of U87 TRTICs (n = 10) (log-rank (Mantel-Cox) test). d. Immunohistochemical staining of tumor sections with proteins associated with aggressive nature (n = 3). e. MRI scans and Hematoxylin and Eosin stained coronal sections of temozolomide and radiation responsive NOD-SCID mouse bearing T98G and SNB19 tumors (n = 3). Scale bar = 50 μm.
3.3. TRTICs resemble GSCs
U87-derived TRTICs and GSCs exhibited similar levels of lineage marker gene expression, with the exception of ABCG2 (Fig. S3a). The viability of TRTICs and GSCs following RT + TMZ were similar with no significant differences in the clonogenic survival (Fig. S3b, c & d). The multidrug resistance of GSCs conferred by ABC transporters is the basis for cells exhibiting side population (SP) [[13], [14], [15]]. TRTICs demonstrated higher expression of the ABC transporters (Fig. S3a). We determined that about 0.1% of the TRTIC population were SP cells (Fig. S3e), which are verapamil sensitive [16]. The presence of SP cells confers drug efflux properties and can contribute to the drug resistance nature of TRTICs.
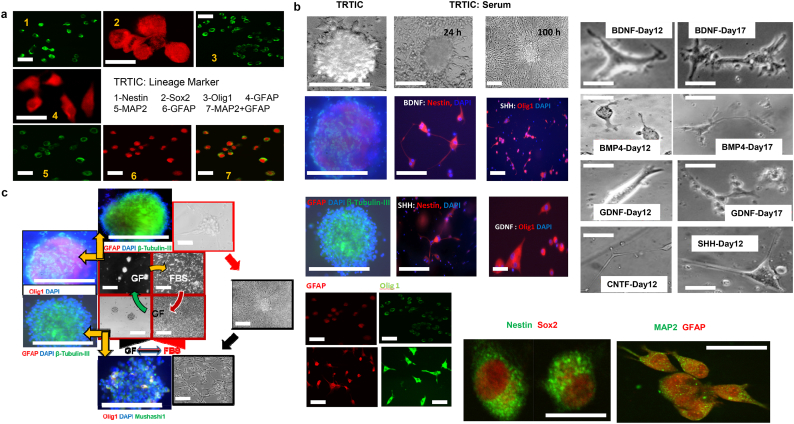
3.4. Harnessing TRTICs by driving differentiation to a specific progeny
The observation that TRTICs express genes associated with the plasticity of neural stem cells (NSCs) [17] fueled our hypothesis that perhaps these TRTICs could similarly differentiate towards specific progeny. The TRTIC spheres grown in serum free culture conditions express neural lineage markers (Fig. 3a). TRTICs were differentiated into adherent cells by replacing the media with serum containing media. Upon differentiation, TRTICs upregulated the expression of neural lineage markers (Fig. 3b). This is not surprising as the stem cell phenotype is associated with plasticity. Next, we investigated the ability of TRTICs to differentiate to specific progeny within the neural lineage using specific growth factors (i.e. BDNF, SHH, BMP4, GDNF or CNTF). After a week of culturing TRTICs in neurosphere growth medium with specific growth factor, the change in cell-type specific markers showed that these growth factors were sufficient to drive differentiation of TRTICs to the neuronal (β-tubulin), astroglial (GFAP), and oligodendroglial (Olig1) progenitor cell types (Fig. 3b). The change in phenotype was confirmed by the approximate 3- to 5- fold increase in the transcriptional level of the lineage markers following addition of the growth factors (Supplemental Table 2). Thus, we demonstrated that BDNF, CNTF, and GDNF drive TRTIC differentiation to neuronal progenitors, while SHH drives TRTICs to proliferate and maintain neuronal and oligodendroglial lineages. An interesting observation is that serum differentiation was robust and yielded an adherent phenotype within 12 h, whereas differentiation under non-serum conditions by using specific growth factors: BDNF, CNTF, GDNF, and SHH decreased the proliferative potential of TRTICs and the transformation to a specific phenotype took more than a week of culturing. The ability of growth factors to control the differentiation of TRTICs poses the possibility that they may play a key role in regulating the maintenance and differentiation of TRTICs. Interestingly, withdrawal of growth factors and supplementation with serum induced differentiation of the TRTIC-neurosphere to adherent cells. Similarly, substituting serum medium with serum free growth factor defined medium reversed this process changing the adherent phenotype to neurospheres (Fig. 3c). This plasticity supports the notion of interconversion from neurosphere to the differentiated adherent phenotype based on the availability of growth factors in the tumor secretome.
Fig. 3.
TRTICs are capable of differentiation to specific progeny in the neural lineage and dedifferentiation. a. Immunofluorescence staining of neural progenitor lineage markers in U87 TRTICs (n = 3). b. Immunofluorescence images of U87 TRTICs differentiated to specific progeny using the indicated growth factors (n = 3). Specific progeny are marked as follows: astroglial (GFAP), oligodendroglial (Olig1), neuronal (β-tubulin III) and neural progenitors (Nestin). Microscopic images on the right demonstrate the morphology of the U87-TRTICs treated with the indicated growth factors. c. Immunofluorescence imaging of TRTICs plasticity between differentiated and dedifferentiated states (n = 3). Brain-derived neurotrophic factor (BDNF); Bone morphogenetic protein 4 (BMP4); Glial cell derived neurotrophic factor (GDNF); Ciliary neurotrophic factor (CNTF); Sonic hedgehog (SHH). Scale bar = 50 μm.
3.5. TRTICs maintain genetic integrity throughout serial xenotransplantation
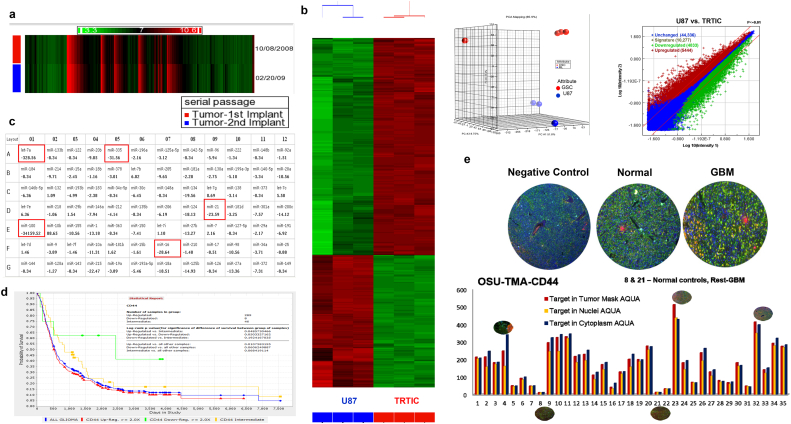
RNA isolated from pooled tumor cells following the first and second implants of TRTICs yielded a small list of differentially regulated genes (Fig. 4a). There were 4 genes which exhibited a 5-fold change (Supplemental Table 3). The differentially regulated gene list subjected to pathway analysis indicates that there were no hits for any biologically relevant processes, showing enrichment scores of zero. Therefore, the genome harbored genetic abnormalities are consistent with the parental infiltrative tumor and suggestive of TRTICs maintaining genetic integrity upon serial implantation.
Fig. 4.
TRTICs maintain genetic integrity and have increased expression of CD44. a. Heat map of differentially regulated genes of serially passaged U87 TRTIC-derived tumors (n = 3). b. Heat map, principal component analysis (PCA), and volcanic plot of the transcriptomic profiles of U87 parental- and U87 TRTIC-derived tumors (n = 3). c. microRNA expression fold change of U87 parental- and TRTIC- derived tumors (n = 3). d. Kaplan Meier plots of REMBRANDT data showing the poor prognosis of CD44 expression in GBM. e. Representative immunofluorescence images of AQUA platform and fluorescence intensity analysis of CD44 in tumor microarrays comprising normal and GBM tissues (n = 3).
3.6. Differential regulation of transcriptomic profiles
The transcriptomic profiles were generated for both U87-derived and TRTIC-derived tumors per MIAMI standards. Analysis of the data clustered samples into two groups and exhibited a contrasting set of differentially regulated transcripts (Fig. 4b). Pathway analysis of these profiles showed the regulation of transcripts associated with DNA replication, cell cycle control, DNA damage response, and immune response (Supplemental Table 4). Out of the statistically significant differentially regulated top 50 genes, CD44 was the only surface antigen. This suggests that CD44 antigen expression may be used to identify the TRTIC subpopulation.
The microRNA (miRNA) expression profiles for the U87-derived and TRTIC-derived tumors showed significantly different miRNA profiles (Fig. 4c). Out of the total miRNAs profiled, TRTICs expressed a higher number of miRNAs (76%) than U87 cells (62%). TRTIC-derived tumors showed increased expression of oncogenic miRNA mir-10b and decreased expression of miRNAs targeting the tumor suppressors: let-7a, mir-335, mir-16, mir-21 and mir-100. This result clearly delineates the cells based on their tumorigenic potential and treatment resistance phenotypes.
3.7. Validation of CD44 expression in GBM patient cohorts
The cell surface antigen CD44 was one of the top 50 upregulated genes (ranking at 32) whose expression levels are low in NSCs. Recurrent GBM tumors tend to shift to a phenotype with a higher expression of CD44 [18]. Increased CD44 expression correlates (p = 0.05) with decreased patient survival in 343 patients from Rembrandt data (http://rembrandt.nci.nih.gov) (Fig. 4d). CD44 immunofluorescent staining patient tissue microarrays comprising tissues from GBM patients and normal autopsy brain tissue showed higher expression in patient cohorts compared to normal samples (Fig. 4e). Further, patients that underwent concomitant and adjuvant temozolomide following radiotherapy showed an association between CD44 expression and resistance to therapy [19]. The heatmap of the data clearly separates the normal versus tumor samples (Fig. S4). Interestingly, the TMZ + RT treated recurrent cases had higher expression levels of CD44. Collectively, these observations suggest CD44 is one of the best prognostic markers for GBM patients.
3.8. Unbiased approach to identify antigens encoding chemo- and radio- resistance in GSCs using multiparametric flow cytometry
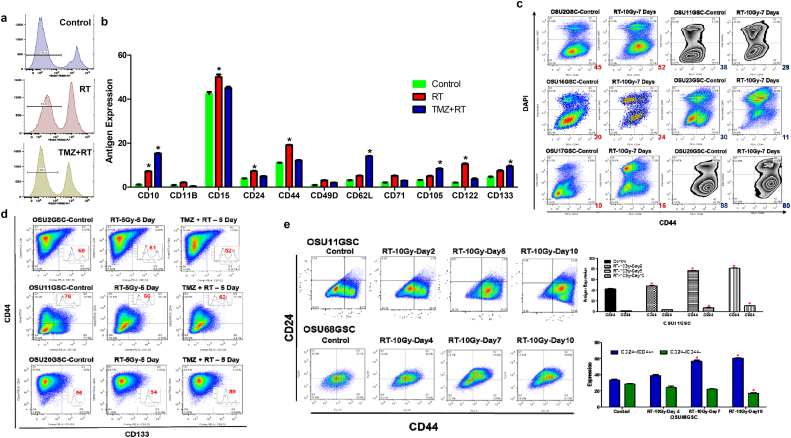
We directed our efforts to identify antigens encoding the TRTIC sub-population of GSCs using multiparametric flow cytometry analysis. Twenty-five surface antigens were selected using a gene expression data set profiled from GBM patients with poor survival benefits. Antigen expression was measured in GSCs following five days of treatment. Flow cytometry analysis demonstrated that about 70% and 85% of GSCs survived after RT and TMZ + RT, respectively (Fig. 5a). In the GSCs surviving treatment, there was an increase in the expression levels of eight antigens: CD10, CD15, CD24, CD44, CD62L, CD105, CD122, and CD133 (Figs. 5b, S5 & S6). Among these antigens, we tested the utility of CD133 and CD44 as markers for treatment resistance (RT and TMZ + RT) in our GSCs. These two antigens were selected due to their association with GSCs. Antigens CD10. CD15, CD62L, CD105, and CD122 were excluded due to their extremely low abundance in some GSCs. We selected a panel of GSCs with CD133+ and CD133− populations (Fig. S7a).
Fig. 5.
Changes in antigen expression following radiation and radiation + temozolomide treatments. a. PI staining of OSU11GSC cells following 10 Gy radiation and 10 Gy radiation +100 μM temozolomide treatment (n = 3). b. Quantified expression level of surface antigens in OSU11GSC cells after 10 Gy radiation and 10 Gy radiation +100 μM temozolomide treatments (n = 3) (Two-way ANOVA:p < 0.0001). c. Representative flow cytometry plots showing the expression level of CD44 positive cells seven days after 10 Gy radiation treatment of OSU2GSC, OSU11GSC, OSU16GSC, OSU17GSC, OSU20GSC and OSU23GSC cells (n = 3). d. Flow cytometry plots of CD44 and CD133 expressing cells after 10 Gy radiation and 10 Gy radiation +100 μM temozolomide treatment of OSU2GSC, OSU11GSC, and OSU20GSC cells (n = 3). PI staining quantifying the number of live cells is represented inside each plot. e. Expression of CD24 and CD44 after 10 Gy radiation treatment in OSU11GSC and OSU68GSC (n = 3) (Two-way ANOVA:p < 0.0001).
We measured the change in CD133 expressing cells in GSCs after treatment. While OSU11GSC and OSU20GSC exhibited a decrease in CD133 expressing cells after RT and TMZ + RT, OSU2GSC exhibited an increase in CD133 antigen presenting cells after TMZ + RT (Fig. S7b). Next, we sorted pure populations of CD133+ and CD133− cells from OSU11GSC and PKAC6GSC cells to determine whether CD133 expression dictated radiation sensitivity. In both cell lines, the CD133− subpopulation of cells exhibited an increased radiation resistance compared to CD133+ cells. (Fig. S7c & d). Therefore, we concluded that CD133 may only act as a marker for TRTICs in a subset of GSCs. We next focused on CD44+ as a marker for treatment-resistance in GSCs. The abundance of CD44 presenting in GSCs was measured 7 days following RT (Fig. 5c). Three cell lines showed an increase in CD44+ subpopulations, while the other three cells lines demonstrated decreased CD44+ subpopulations. Therefore, CD44 also identifies TRTICs in a subset of GSCs. Thus, use of a single antigen, CD133 or CD44, may not be useful to identify treatment resistance GSCs.
3.9. Two is better than one: CD24+/CD44+ subpopulation is enriched in TRTICs
Next, we investigated whether using two antigens would identify TRTIC subsets in GSCs. We tested two antigen combinations: CD133+/CD44+ and CD24+/CD44+. OSU2GSC, OSU11GSC, and OSU20GSC cells were treated with RT and TMZ + RT. CD133 and CD44 expressing cells were measured after 5 days. Among the three cell lines, only OSU11GSC showed an increase in CD133+/CD44+ cells following treatment (Fig. 5d). Thus, the CD44+/CD133+ antigen expressing cells may not be unique to TRTIC subset of GSCs. We next evaluated whether CD24+/CD44+ cells identified TRTICs. OSU11GSC and OSU68GSC cells demonstrated an increased subpopulation of CD24+/CD44+ presenting cells in a dose- and time-dependent manner following RT (Figs. 5e, S8b & c). OSU53GSC and PKAC2GSC showed a similar increase in CD24+/CD44+ antigen expression after RT (Fig. 6a). To validate these findings, we sorted pure populations of CD24+/CD44+/AAD− and CD24−/CD44−/AAD− cells to determine their radiation sensitivity. The Annexin V assay clearly shows that the CD24+/CD44+ cells have increased resistance to radiation, whereas CD24−/CD44− cells are more sensitive to radiation (Fig. S8b & c). Therefore, we conclude that the CD24+/CD44+ cells may encode a treatment resistant subpopulation of GSCs.
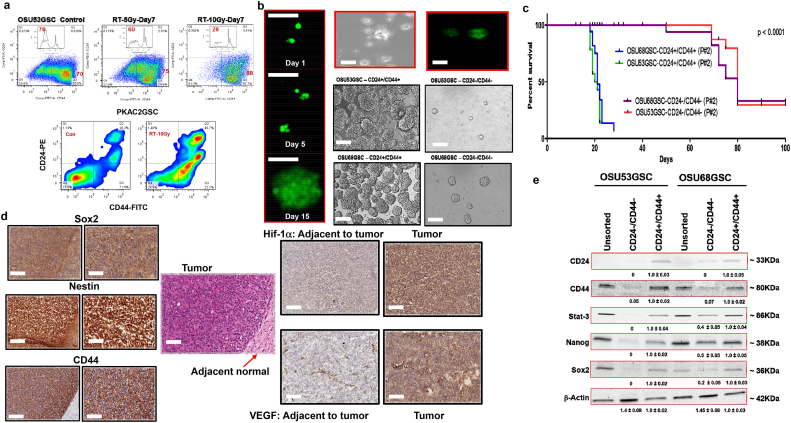
Fig. 6.
CD44+/CD24+ antigens identify TRTICs and GSCs. a. Flow cytometry plot showing the expression of CD24 and CD44 in OSU53GSC and PKAC2GSC cells after 10 Gy radiation treatment (n = 3). b. Phase contrast and GFP+/CD24+/CD44+ GFP cells showing sphere-forming ability at day1 to day15. Microscopic images of OSU53GSC and OSU68GSC CD44+/CD24+ and CD44-/CD24- TRTICs 10 days after sorting pure population (n = 3). c. Kaplan-Meier survival plots of NOD-SCID mice bearing tumors from CD24+/CD44+ and CD24-/CD44- populations of OSU53GSC and OSU68GSC cells (n = 3) (log-rank (Mantel-Cox) test). d. Immunohistochemistry of Sox2, Nestin, CD44, Hif-1α and VEGF in U87 TRTIC-derived tumors (n = 3). The Hematoxylin and Eosin stained tumor section demonstrates the adjacent normal tissue in the Sox2, Nestin and CD44 images. The Hif-1α and VEGF adjacent normal tissue is represented as individual boxes to the left of their respective tumor staining. e. Western blots showing the protein expression pattern of unsorted, CD44+/CD24+ and CD24-/CD44- OSU53GSC and OSU68GSC cells (n = 3). Scale bar = 50 μm.
3.10. Clonogenicity and tumorigenicity of CD24+/CD44+ GSCs
To elucidate whether a single CD24+/CD44+ cancer cell can form clones, we initiated a series of single-cell-cloning experiments. Using single-cell sorting of spheroid cells by flow cytometry, we established that about 50% of cells have the capacity to induce a monoclonal culture as judged by the successful formation of spheroids. FACS deposited GFP-transduced GFP+/CD24+/CD44+ spheroid cell into different amounts of GFP−/CD24−/CD44− cells from the same culture invariably resulted in GFP+ spheres (Fig. 6b). Exclusion of the transmission of the expression vector indicated that the CD24+/CD44+ and not the CD24−/CD44− cells contain clonogenic capacity. In agreement, limiting dilution experiments from a spheroid culture showed that 1 in 2 CD24+/CD44+ cells have the capacity to generate spheroids (Fig. 6b), whereas 1 in 250 CD24−/CD44− cells have this ability. To fully ascertain that this is also true for directly isolated tumor cells, we isolated and cultured CD24+/CD44+ and CD24−/CD44− cells directly ex-patient. In this setting, the CD24+/CD44+ cell fraction generated single cell derived spheroids, whereas the CD24−/CD44− cells were incapable of doing so. This clearly confirmed that the clonogenic potential of our spheroid cultures resides in the CD24+/CD44+ cells. The single cell derived cultures obtained by FACS deposition displayed similar expansion rates as the original culture, indicating that we did not select for rapidly proliferating cells. Dramatic increase in the CD24+/CD44+ population in CSCs (40%) after high doses of radiation and a drastic decrease in CD24−/CD44− (Fig. 6a & S9) suggests an association of radiation resistance with double positive cells. Mice bearing sorted population double positive and double negative (CD24/CD44) of OSU68GSC and OSU53GSC developed tumor symptoms in three weeks compared to 10 weeks (Fig. 6c). Thus, the CD24+/CD44+ subpopulation is more aggressive in these GSCs. Coronal sections of double positive tumors express stem cell markers nestin and SOX2 within the pseudopalisadic region suggesting the region contains TRTICs and facilitates an escape from chemotherapy and RT toxicities (Fig. 6d). Thus, the microvascular hyperplasia of the TRTIC-derived tumors provides a new vasculature and promotes peripheral tumor expansion. Higher expression of stem cell proteins and STAT3 in the CD24+/CD44+ cells compared to double negative cells (Fig. 6e) further supports their aggressive nature.
3.11. Differentiated cells promote TRTIC maintenance and increase radiation resistance
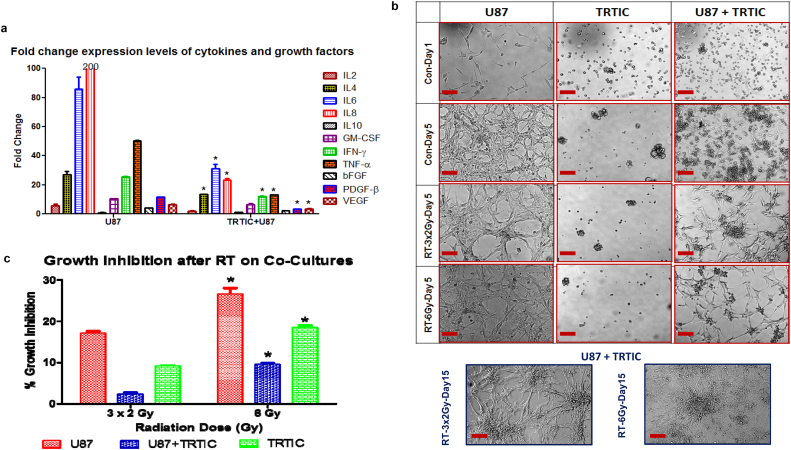
The iPSC gene expression in TRTICs suggests that these cells remain in an undifferentiated state. In the healthy adult brain, NSCs receive maintenance and differentiation signals from the surrounding glial cells, neurons, and extracellular matrix. In GBMs, it has been shown that endothelial cells interact with self-renewing brain tumor cells and secrete factors that maintain these cells in a stem-like state [[20], [21], [22], [23], [24]]. We reasoned that TRTICs might likewise remain in an undifferentiated state due to growth factors in their tumor secretome. An equal number of U87, TRTICs, and co-cultures of U87 and TRTICs were incubated for 24 h. The media was then collected and a panel of Bioplex assay was used to determine the concentration of growth factors and cytokines in the secretome (Fig. 7a). Interestingly, a significant decrease in the secretome of the U87 cells in the presence of TRTICs in co-cultures occurred. This suggests that TRTICs may utilize the secreted growth factors from the U87 cells for their survival and maintenance. Co-cultures exhibited an increased radiation resistance possibly revealing a contribution to radiation resistance due to secretome (Fig. 7b & c). Previous reports suggest that cross talk occurs between GSCs and their differentiated counterparts through brain-derived neurotrophic growth factor mediated paracrine stimuli to promote tumor growth [25]. This supports our findings that this interaction is crucial for treatment resistance and tumor recurrence,
Fig. 7.
Radioprotection of TRTICs by adjacent monolayer U87 cells and cytokine secretome for maintaining TRTICs. a. Expression level of various growth factors and cytokines from U87 parental cells and U87 parental cells co-cultured with U87 derived TRTIC (n = 3) (Two-way ANOVA:p < 0.0001). b. Microscopic images of U87 parental cells, U87 derived TRTICs and co-culture of U87 parental cells with U87 derived GSCs following 10 Gy (n = 3). c. Growth inhibition of U87 parental cells, U87 derived TRTIC and co-culture of U87 parental cells with U87 derived TRTICs following 10 Gy (n = 3) (Two-way ANOVA:p < 0.0001). Scale bar = 50 μm.
4. Discussion
We show that established cancer cell lines maintain TRTICs over long periods of time. This raises the possibility that these TRTICs may provide attractive models for studying the molecular basis that defines stem cells. For curative therapies, it is imperative to develop a model that mimics clinical observations and exhibits all histological hallmarks of the disease. We report that these TRTICs can differentiate into mature cell types, recapitulating the complexity of GBM. The cellular heterogeneity of brain tumors suggests that only a small fraction of cancer cells regenerate tumor and that targeting these cells could be an innovative approach to eradicating the tumor. Spheres generated at clonal density can reform spheres after induction of differentiation and have genetic aberrations typical of brain tumors. This is an important point that distinguishes cancer stem cells from normal stem cells. These cells express iPSC factors, such as Sox2, Oct-3/4, and Nanog, at higher levels. Oct-4 works in concert with Nanog and Sox2 to maintain stem cell identity and repress genes that promote differentiation [26]. We demonstrate that TRTICs have increased expression of stem cell markers, such as Nanog, POU5F1 (OCT4), and SOX2 as well as pluripotency markers TDGF1, DNMT3B, GABRB3, and GDF3 compared to untreated control. These markers represent gene products functionally associated with maintenance of the undifferentiated embryonic stem cell state and the stem cell markers. Up regulation of TERT is indicative of exceeding the Hayflick limit and an increased oncogenesis in the treatment resistance phenotype. The decrease in the expression level of bone morphogenetic protein 4 (BMP4), a negative regulator of stem cells, in TRTICs may be due to the capability of BMP4 in inhibiting the growth of the GSCs and abolishing their ability to establish glioblastomas after transplantation [27], which supports our findings. Enriching cancer stem cells by in vivo irradiation has been attempted earlier using breast tumors [28], which additionally supports our findings.
Overexpression of multidrug resistance (MDR) genes are associated with high levels of resistance to a variety of anticancer agents due to enhanced drug efflux and has been linked to clinical drug resistance in a wide variety of hematological malignancies [15,29]. The higher expression levels of MDR genes in TRTICs compared to U87 cells correlates with the presence SP positive verapamil sensitive cells. The persistence of SP cells in cancer cell lines cultured in serum-containing medium for decades suggests that SP cells may be a general source of cancer stem cells [30]. Higher expression of MDR and iPSC genes in TRTICs expressing CD44 coincides with a previous study that has connected CD44-Nanog-Sox2-STAT3-MDR signaling. Multidrug resistance and tumor progression is associated with CD44 expressing cells. Hyaluronan induced interaction between CD44 and Nanog activates the expression of pluripotent stem cell regulator Sox2 and forms a complex with STAT3 in the nucleus leading to transcriptional activation of MDR genes resulting in the efflux of chemotherapeutic drugs [31]. Overexpression of CD24 in ovarian carcinoma effusions compared to solid lesions may be due to the acquisition of cancer stem cell characteristics [32]. CD24 expression level directly affects cisplatin sensitivity and affects the expression of critical apoptotic, stem cell, and drug resistance genes. CD24 presents a strong rationale for its utilization as a predictive indicator to stratify head and neck cancer patients for platinum-based therapy [33]. A report in nasopharyngeal carcinoma (NPC) identified CSC with co-expression of CD24 and CD44 are enriched in high-stage clinical samples. They show that CD24 and CD44 collaboratively drive the reprogramming of NPC through STAT3 mediated stemness and epithelial-mesenchymal transition in NPC [34]. All these recent reports support our findings that there may be a significant association between CD24 and CD44 expressing cells and treatment resistance and cancer stem cells in GBM tumors.
Through mRNA and miRNA profiles, we have shown GSC associative markers. We were the first to report that CD44 is a marker for GSCs [35] and establishing an association of CD44 with GSCs. Subsequently, other investigators confirmed our report by providing mechanistic insights [[36], [37], [38], [39], [40]]. Recurrent GBM tumors tend to shift to a phenotype with a higher expression of CD44 [18]. Additionally, CD44 is used as a marker to isolate cancer stem cells in breast, prostate, pancreatic, and colorectal cancers [[41], [42], [43], [44]]. A report shows that let-7 inhibition leads to reprogramming of induced pluripotent stem cells (iPSCs), which supports the role of reprogramming in GSCs [45]. LIN28, a mRNA binding protein expressed in embryonic stem cells and is a factor in iPSC generation. LIN28 binds to the let-7 pre-miRNA and blocks the production of the mature let-7 miRNA [46]. Collectively the let-7 down regulation along increased expression of iPSC factors [47,48], such as Oct4, Sox2, C-Myc, and Nanog in GSCs, are suggestive of reorganization leading to an aggressive form. TRTICs expressing double positive CD24/CD44 has heightened activation of STAT3 which is supported by previous studies showing the association with the aggressive nature of GBM and in other cancers. It has been shown that STAT3 promotes tumorigenicity of GSCs [49]. CD24+/CD44+ pancreatic cancer stem-like cells had increased activation of STAT3 [50]. It has been shown that STAT3 labels reactive astrocytes implicated in brain metastasis [51]. We show that TRTICs exhibits differentiation plasticity and can be differentiated to neural lineage subtypes by utilizing suitable growth factor. The observation that specific growth factors drive TRTIC differentiation and decrease proliferation may provide an opportunity for therapeutic targeting of TRTICs.
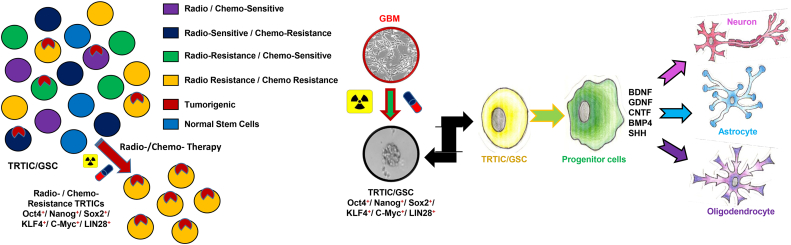
We have attempted to answer an important question as to how TRTICs exists within differentiated cells. A possibility could be due to the influence of the secretome of differentiated cells helps to maintain the TRTICs. Endothelial cells interact with self-renewing brain tumor cells and secrete factors that maintain these cells in a stem-like state [[20], [21], [22], [23], [24]]. Clinically, the tumor shrinking after treatment might result from the tumor comprising of TRTICs and differentiated cells, when the tumors are treated the differentiated cells undergo cell death leading to the shrinkage of the tumor. Through co-culture studies, we have shown that differentiated cells may provide a protective niche for TRTICs. Therefore, the small sub-population of TRTICs escapes treatment and resides within the residual shrunk tumor. Due to the lack of cytokines and growth factors, the quiescent TRTICs start to differentiate as shown by our driving differentiation studies. A portion of this left-over TRTICs then differentiate and once they are differentiated cells they can provide cytokines and growth factors to the remaining TRTICs for their survival and maintenance, leading to tumor recurrence and relapse. The interesting implication is the ability to drive the TRTICs to a specific progeny that exhibits treatment sensitivity. Thus, chemo-/radiation- therapies and/or antagonizing the cytokine and growth factors that fuel TRTICs is a strategy to target TRTIC differentiated counter parts. Graphical summary of TRTIC isolation and their differentiation characteristics are shown in Fig. 8.
Fig. 8.
Snapshot showing that GBM comprises of multiple clonal populations of cells and are heterogeneous. Chemotherapy and radiation treatments eliminate the treatment sensitive subset of cells, enriching TRTICs expressing iPSC genes. These TRTICs can be directed to differentiate to a specific neural progeny by providing appropriate growth factors.
We have begun to understand the molecular mechanisms involved in this conversion, which were previously unknown because of limited availability and abundance in tumors and differentiated cancer cells. The model we have developed to enrich TRTICs will be useful to study the molecular basis of cellular plasticity and exploit TRTICs for therapeutic benefit. In many ways, our study supports the perspectives on the properties of stem cells [52]. The limitation of the study is that TRTICs obtained after treating U87 tumors with DNA damaging agents, TMZ and RT could have led to genomic aberrations, which require exome sequencing. Furthermore, a comprehensive analysis of secretome is required to identify the critical factors required for maintenance and survival of TRTICs to eradicate TRTICs. Nevertheless, we anticipate that the current report provides a complete and comprehensive characterization of TRTICs.
Acknowledgments
Acknowledgments
The authors thank Dr. Samuel Rabkin, and Dr. Hiroaki Wakimoto, Massachusetts General Hospital and Harvard Medical School for his critical input, Dr. Kimerly Powell and Dr. Anna Bratasz, OSU-CCC small animal imaging core, and Dr. David Dombkowski, MGH-Harvard Flow core, Dr. Reddy Gali, Harvard Medical School for his help with bio-Informatics, and Ananya Kamalakannan for her help with graphical abstract.
Funding
NIH/NCI 1RC2CA148190, 1R01CA108633, 1R01CA188228, and The Ohio State University Comprehensive Cancer Center.
Disclosure of potential conflicts of interest
The authors declare no potential conflicts of interests.
Author contributions
Conceptualization: KP & AC, Investigation: Formal Analysis and Data Curation: KP, JRJ, KTL & ARC, Writing, Review and Editing: KP, JRJ & AC, Funding Acquisition: KP & AC.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.09.019.
Appendix A. Supplementary data
Supplementary Figures
Supplementary Materials
References
- 1.Lathia J.D., Mack S.C., Mulkearns-Hubert E.E., Valentim C.L., Rich J.N. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson M., Hassiotou F., Nowak A. Glioblastoma stem-like cells: at the root of tumor recurrence and a therapeutic target. Carcinogenesis. 2015;36(2):177–185. doi: 10.1093/carcin/bgu243. [DOI] [PubMed] [Google Scholar]
- 3.Sanai N., Alvarez-Buylla A., Berger M.S. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353(8):811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 4.Das S., Srikanth M., Kessler J.A. Cancer stem cells and glioma. Nat Clin Pract Neurol. 2008;4(8):427–435. doi: 10.1038/ncpneuro0862. [DOI] [PubMed] [Google Scholar]
- 5.Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N.M. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 6.Reynolds B.A., Tetzlaff W., Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J Neurosci. 1992;12(11):4565–4574. doi: 10.1523/JNEUROSCI.12-11-04565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reynolds B.A., Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255(5052):1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 8.Jordan C.T., Guzman M.L., Noble M. Cancer stem cells. N Engl J Med. 2006;355(12):1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 9.Seaberg R.M., van der Kooy D. Stem and progenitor cells: the premature desertion of rigorous definitions. Trends Neurosci. 2003;26(3):125–131. doi: 10.1016/S0166-2236(03)00031-6. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 12.Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448(7151):318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 13.Bleau A.M., Huse J.T., Holland E.C. The ABCG2 resistance network of glioblastoma. Cell Cycle. 2009;8(18):2936–2944. [PubMed] [Google Scholar]
- 14.Broadley K.W., Hunn M.K., Farrand K.J., Price K.M., Grasso C., Miller R.J. Side population is not necessary or sufficient for a cancer stem cell phenotype in glioblastoma multiforme. Stem Cells. 2011;29(3):452–461. doi: 10.1002/stem.582. [DOI] [PubMed] [Google Scholar]
- 15.Gottesman M.M., Fojo T., Bates S.E. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2(1):48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 16.Komuro H., Saihara R., Shinya M., Takita J., Kaneko S., Kaneko M. Identification of side population cells (stem-like cell population) in pediatric solid tumor cell lines. J Pediatr Surg. 2007;42(12):2040–2045. doi: 10.1016/j.jpedsurg.2007.08.026. [DOI] [PubMed] [Google Scholar]
- 17.Martino G., Pluchino S. The therapeutic potential of neural stem cells. Nat Rev Neurosci. 2006;7(5):395–406. doi: 10.1038/nrn1908. [DOI] [PubMed] [Google Scholar]
- 18.Phillips H.S., Kharbanda S., Chen R., Forrest W.F., Soriano R.H., Wu T.D. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 19.Murat A., Migliavacca E., Gorlia T., Lambiv W.L., Shay T., Hamou M.F. Stem cell-related "self-renewal" signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26(18):3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 20.Calabrese C., Poppleton H., Kocak M., Hogg T.L., Fuller C., Hamner B. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11(1):69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 21.Jones D.L., Wagers A.J. No place like home: anatomy and function of the stem cell niche. Nat Rev Mol Cell Biol. 2008;9(1):11–21. doi: 10.1038/nrm2319. [DOI] [PubMed] [Google Scholar]
- 22.Palmer T.D., Willhoite A.R., Gage F.H. Vascular niche for adult hippocampal neurogenesis. J Comp Neurol. 2000;425(4):479–494. doi: 10.1002/1096-9861(20001002)425:4<479::aid-cne2>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 23.Ramirez-Castillejo C., Sanchez-Sanchez F., Andreu-Agullo C., Ferron S.R., Aroca-Aguilar J.D., Sanchez P. Pigment epithelium-derived factor is a niche signal for neural stem cell renewal. Nat Neurosci. 2006;9(3):331–339. doi: 10.1038/nn1657. [DOI] [PubMed] [Google Scholar]
- 24.Shen Q., Goderie S.K., Jin L., Karanth N., Sun Y., Abramova N. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 2004;304(5675):1338–1340. doi: 10.1126/science.1095505. [DOI] [PubMed] [Google Scholar]
- 25.Wang X., Prager B.C., Wu Q., Kim L.J.Y., Gimple R.C., Shi Y. Reciprocal signaling between glioblastoma stem cells and differentiated tumor cells promotes malignant progression. Cell Stem Cell. 2018;22(4):514–528. doi: 10.1016/j.stem.2018.03.011. [e5] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piccirillo S.G., Reynolds B.A., Zanetti N., Lamorte G., Binda E., Broggi G. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444(7120):761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 28.Diehn M., Cho R.W., Lobo N.A., Kalisky T., Dorie M.J., Kulp A.N. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458(7239):780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doyle L.A., Ross D.D. Multidrug resistance mediated by the breast cancer resistance protein BCRP (ABCG2) Oncogene. 2003;22(47):7340–7358. doi: 10.1038/sj.onc.1206938. [DOI] [PubMed] [Google Scholar]
- 30.Setoguchi T., Taga T., Kondo T. Cancer stem cells persist in many cancer cell lines. Cell Cycle. 2004;3(4):414–415. doi: 10.4161/cc.3.4.799. [DOI] [PubMed] [Google Scholar]
- 31.Bourguignon L.Y., Peyrollier K., Xia W., Gilad E. Hyaluronan-CD44 interaction activates stem cell marker Nanog, Stat-3-mediated MDR1 gene expression, and ankyrin-regulated multidrug efflux in breast and ovarian tumor cells. J Biol Chem. 2008;283(25):17635–17651. doi: 10.1074/jbc.M800109200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davidson B. CD24 is highly useful in differentiating high-grade serous carcinoma from benign and malignant mesothelial cells. Hum Pathol. 2016;58:123–127. doi: 10.1016/j.humpath.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Modur V., Joshi P., Nie D., Robbins K.T., Khan A.U., Rao K. CD24 expression may play a role as a predictive indicator and a modulator of cisplatin treatment response in head and neck squamous cellular carcinoma. PLoS One. 2016;11(6) doi: 10.1371/journal.pone.0156651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen Y.A., Wang C.Y., Chuang H.Y., Hwang J.J., Chi W.H., Shu C.H. CD44 and CD24 coordinate the reprogramming of nasopharyngeal carcinoma cells towards a cancer stem cell phenotype through STAT3 activation. Oncotarget. 2016;7(36):58351–58366. doi: 10.18632/oncotarget.11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palanichamy K., Chakravarti A. Brain tumor stem cell marker - distinct populations of tumor-initiating cells expressing CD44/CD24 derived from cancer stem cells mediates chemo and radio resistance. Int J Radiat Oncol. 2009;75(3) (S545-S) [Google Scholar]
- 36.Xu Y., Stamenkovic I., Yu Q. CD44 attenuates activation of the hippo signaling pathway and is a prime therapeutic target for glioblastoma. Cancer Res. 2010;70(6):2455–2464. doi: 10.1158/0008-5472.CAN-09-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anido J., Saez-Borderias A., Gonzalez-Junca A., Rodon L., Folch G., Carmona M.A. TGF-beta receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18(6):655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 38.Stieber D., Golebiewska A., Evers L., Lenkiewicz E., Brons N.H., Nicot N. Glioblastomas are composed of genetically divergent clones with distinct tumourigenic potential and variable stem cell-associated phenotypes. Acta Neuropathol. 2014;127(2):203–219. doi: 10.1007/s00401-013-1196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Behnan J., Isakson P., Joel M. Langmoen IA; Vik-Mo EO, et al. Recruited brain tumor-derived mesenchymal stem cells contribute to brain tumor progression. Stem Cells: 2013. Cilio C. [DOI] [PubMed] [Google Scholar]
- 40.Bhat K.P., Balasubramaniyan V., Vaillant B., Ezhilarasan R., Hummelink K., Hollingsworth F. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patrawala L., Calhoun T., Schneider-Broussard R., Li H., Bhatia B., Tang S. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25(12):1696–1708. doi: 10.1038/sj.onc.1209327. [DOI] [PubMed] [Google Scholar]
- 43.Li C., Heidt D.G., Dalerba P., Burant C.F., Zhang L., Adsay V. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67(3):1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 44.Dalerba P., Dylla S.J., Park I.K., Liu R., Wang X., Cho R.W. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104(24):10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Worringer K.A., Rand T.A., Hayashi Y., Sami S., Takahashi K., Tanabe K. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell. 2014;14(1):40–52. doi: 10.1016/j.stem.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Viswanathan S.R., Daley G.Q., Gregory R.I. Selective blockade of microRNA processing by Lin28. Science. 2008;320(5872):97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 49.Kim E., Kim M., Woo D.H., Shin Y., Shin J., Chang N. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23(6):839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin L., Jou D., Wang Y., Ma H., Liu T., Fuchs J. STAT3 as a potential therapeutic target in ALDH+ and CD44+/CD24+ stem cell-like pancreatic cancer cells. Int J Oncol. 2016;49(6):2265–2274. doi: 10.3892/ijo.2016.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Priego N., Zhu L., Monteiro C., Mulders M., Wasilewski D., Bindeman W. STAT3 labels a subpopulation of reactive astrocytes required for brain metastasis. Nat Med. 2018 Jul;24(7):1024–1035. doi: 10.1038/s41591-018-0044-4. [DOI] [PubMed] [Google Scholar]
- 52.McCulloch E.A., Till J.E. Perspectives on the properties of stem cells. Nat Med. 2005;11(10):1026–1028. doi: 10.1038/nm1005-1026. [DOI] [PubMed] [Google Scholar]
- 53.Palanichamy K., Acus K., Jacob R.J., Chakravarti A. Clinically relevant brain tumor model and device development for experimental therapeutics. J Anal Oncol. 2015;4:5–12. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Materials