Summary
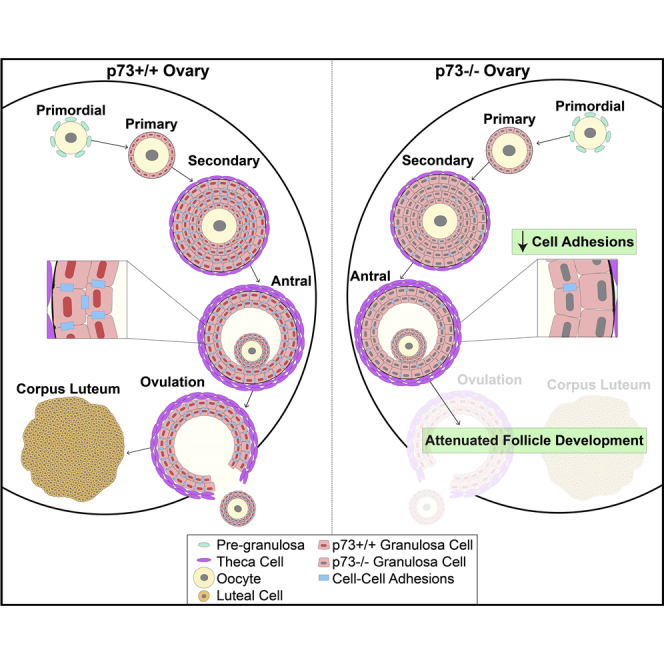
We report that p73 is expressed in ovarian granulosa cells and that loss of p73 leads to attenuated follicle development, ovulation, and corpus luteum formation, resulting in decreased levels of circulating progesterone and defects in mammary gland branching. Ectopic progesterone in p73-deficient mice completely rescued the mammary branching and partially rescued the ovarian follicle development defects. Performing RNA sequencing (RNA-seq) on transcripts from murine wild-type and p73-deficient antral follicles, we discovered differentially expressed genes that regulate biological adhesion programs. Through modulation of p73 expression in murine granulosa cells and transformed cell lines, followed by RNA-seq and chromatin immunoprecipitation sequencing, we discovered p73-dependent regulation of a gene set necessary for cell adhesion and migration and components of the focimatrix (focal intra-epithelial matrix), a basal lamina between granulosa cells that promotes follicle maturation. In summary, p73 is essential for ovarian folliculogenesis and functions as a key regulator of a gene network involved in cell-to-cell adhesion and migration.
Subject Areas: Molecular Network, Functional Aspects of Cell Biology, Developmental Biology, Omics
Graphical Abstract
Highlights
-
•
p73 is required for murine ovarian folliculogenesis and proper corpus luteum formation
-
•
p73 loss leads to defects in progesterone signaling and mammary gland branching
-
•
In murine ovaries, p73 is expressed specifically in granulosa cells
-
•
p73 regulates components of the granulosa cell focimatrix and migration
Molecular Network; Functional Aspects of Cell Biology; Developmental Biology; Omics
Introduction
The p53 family of proteins, p53, p63, and p73, are sequence-specific transcription factors that are required for cell cycle control, DNA repair, apoptosis, adhesion, organ development, and cell differentiation (Kaghad et al., 1997, Schmale and Bamberger, 1997, Yang et al., 1998, Osada et al., 1998, Trink et al., 1998, Holembowski et al., 2014). All three p53 family members share a high degree of structural and amino acid sequence similarities within their transactivation domains, DNA binding domains, and oligomerization domains (Harms and Chen, 2006, Dotsch et al., 2010), which accounts for similar genomic binding sites and regulation of overlapping target genes. Unlike p53, p63 and p73 are transcribed from two separate promoters that encode functionally divergent variants. The transcriptionally active (TA) isoform encodes the full-length protein, whereas the alternative transcript (ΔN) encodes an isoform lacking the amino-terminal transactivation domain (Kaghad et al., 1997, Yang et al., 1998). Thus, ΔNp63 and ΔNp73 isoforms act as dominant-negative regulators of TAp63 and TAp73 (Yang et al., 1998, Stiewe et al., 2002, Grob et al., 2001).
p63 and p73 play important roles in cell differentiation and tissue development. p63 is a key regulator of ectodermal differentiation and stratification of the epidermis. Mice lacking p63 fail to develop stratified epithelia, exhibit defective limb and glandular epithelial development, and die shortly after birth due to dehydration (Mills et al., 1999, Yang et al., 1999). Mice deficient for all isoforms of p73 exhibit runting, sterility, hippocampal dysgenesis and hydrocephalus, as well as chronic infection and inflammation in the lungs, sinus, and ears (Yang et al., 2000). The development of p73-isoform-specific knockout mouse models provided significant insight into the roles of select p73 isoforms. TAp73-deficient mice exhibit sterility, hippocampal dysgenesis, hydrocephalus, premature aging, genomic instability, and increased frequency of tumors (Tomasini et al., 2008). In contrast, mice that lack ΔNp73 are fertile and display signs of neurodegeneration, including hippocampal dysgenesis and hydrocephalus (Wilhelm et al., 2010). Thus the sterility defects observed in the global p73-deficient animals are due to a deficiency in the TAp73 isoform. Recently, our laboratory (Marshall et al., 2016) and others (Nemajerova et al., 2016) discovered that TAp73 is required for multiciliated cell differentiation and acts as a transcriptional regulator of a gene network required for ciliogenesis. The discovery provided mechanistic insight into the diverse phenotypes observed in p73-deficient mouse models. Impaired cilia formation in p73-deficient mice leads to insufficient clearance of pathogens from the lungs and sinuses causing chronic inflammation. Furthermore, loss of cilia in reproductive tissues decreases transport of the sperm and ova through epididymis and fallopian tubes, respectively, leading to infertility.
Other phenotypes of p73-deficient mice have been reported that are likely to originate from processes unrelated to ciliogenesis. Male mice that lack TAp73 exhibit increased DNA damage and apoptosis in spermatogonial cells within the testes, which results in defective germ cell maturation and differentiation, required for proper spermatogenesis (Inoue et al., 2014, Holembowski et al., 2014). TAp73-deficient female mice exhibit meiotic spindle formation abnormalities during oocyte maturation and impaired ovulation (Tomasini et al., 2008). We report herein that p73 expression in the ovarian follicle, the structure in which the oocyte develops, is critical for oocyte development, ovulation, and fertility. Specifically, p73 is required in granulosa cells for the expression of a p73-dependent gene set that regulates cell adhesion and migration, including genes that encode key components of granulosa-cell-associated focimatrix.
Results
p73 Is Required for Murine Ovarian Follicle Maturation
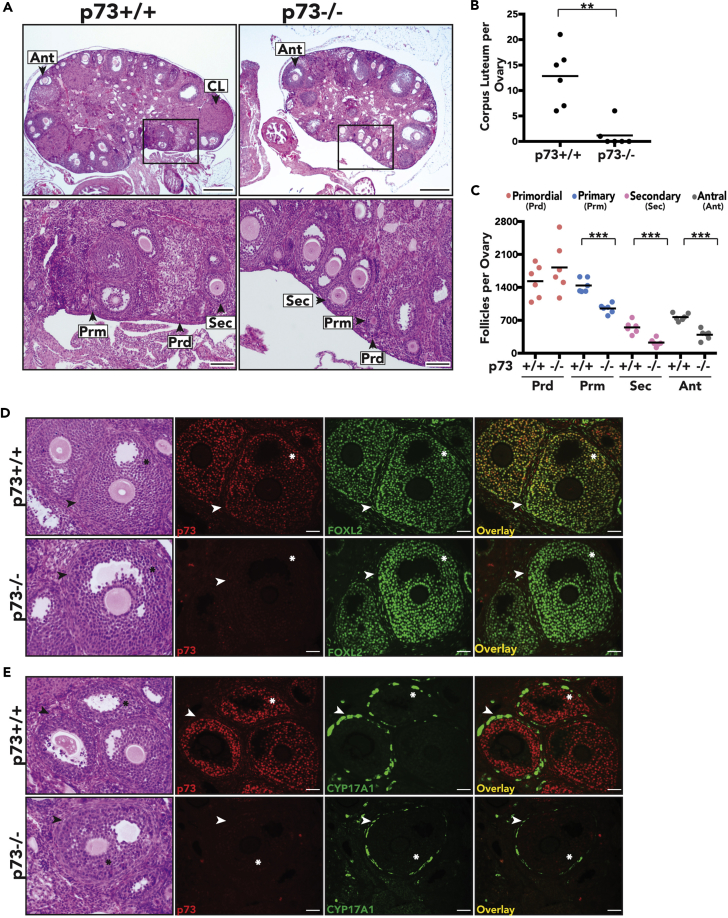
We analyzed ovaries from age-matched nulliparous female mice genetically engineered to lack functional p73 in all tissues (referred to as p73−/− hereafter and described in [Marshall et al., 2016]). Histological analysis of ovaries in 12-week-old p73+/+ female mice confirmed the presence of corpus luteum, the final stage of the mature ovarian follicle, suggesting that follicle maturation and ovulation were ongoing in mice at this age. p73+/+ ovaries harbored an average of 12 luteal structures per ovary, whereas age-matched p73−/− ovaries had an average of 1 luteal structure per ovary (p value <0.01) (Figures 1A and 1B). We quantified the total number of follicles per ovary at various stages of development (primordial, primary, secondary, and antral stages) using methodology as previously described (Kim et al., 2015) (Figures S1A and S1B). We observed a 35% decrease in the number of primary follicles (p value <0.001), a 59% decrease in the number of secondary follicles (p value <0.001), and a 49% decrease in the number of antral follicles (p value <0.001), but no significant difference in the number of primordial follicles in p73−/− mice compared with p73+/+ littermates (Figure 1C).
Figure 1.
p73 Is Required for Murine Ovarian Follicle Development
(A) Representative H&E images of p73+/+ and p73−/− ovaries (scale bars, 400 μm and 100 μm, respectively). Arrowheads and labels represent corpora lutea (CL) and different stages of follicle development: Prd, primordial; Prm, primary; Sec, secondary; Ant, antral follicles.
(B and C) Each data point represents the average of two independent manual quantifications of six ovaries per genotype. Bars represent the mean. (B) Total number of corpus luteum per ovary (C) Total number of follicles per ovary. **p value< 0.01, ***p value < 0.001.
(D and E) Representative H&E (asterisks represent granulosa cells and arrowheads represent theca cells) and IF images of p73+/+ and p73−/− ovaries show that (D) p73 (red) co-localizes with granulosa cell marker FOXL2 (green) in the follicles and (E) p73 (red) is not expressed in theca cells, which are stained by CYP17A1 (green) (scale bar, 50 μm).
See also Figure S1.
Analysis of p73 protein levels in murine ovaries, at 6, 9, 12, and 16 weeks of age, confirmed that full-length p73 protein was expressed in ovaries collected at all time points in p73+/+ animals and was not expressed in p73−/− animals (Figure S1C). To determine the localization of p73 protein within the ovary, we used well-established cell markers to stain follicular granulosa cells (FOXL2) (Schmidt et al., 2004) and follicular theca cells (CYP17A1) (Park et al., 2010). Through immunofluorescence (IF) staining, p73 was expressed in FOXL2-positive granulosa cells (Figure 1D, top panel) but not in CYP17A1-positive theca cells (Figure 1E, top panel). Although the number of follicles were reduced in the absence of p73, we found that, in cases in which follicles were able to form in p73−/− ovaries, the lack of p73 did not block the formation of the granulosa or theca cell layers in the follicle (Figures 1D and 1E, lower panels). We further analyzed p73 expression in human ovaries using data obtained from the Genotype-Tissue Expression (GTEx) Project (analysis date: January 19, 2018) and found that p73 is expressed at 0.6 transcripts per million (TPM). Specifically, TAp73α and TAp73β are the predominant N-terminal isoforms expressed in human ovaries (Figure S1D).
Given the potential for hetero-oligomerization between p73 and its family member p63 (Chan et al., 2004, Rocco et al., 2006, Harms and Chen, 2006), we determined if p63 and p73 were co-expressed during follicular development using dual IF detection of the proteins. p63 expression was restricted to oocytes (arrowhead) of primordial follicles in p73+/+ (Figure S1E, top panel), consistent with previously published data showing that p63 expression in primordial follicles promotes genome integrity during meiotic arrest (Suh et al., 2006). The oocyte-restricted expression pattern of p63 was unaltered in p73−/− ovaries (Figure S1E, lower panel).
The ovarian phenotypes observed in our p73−/− female mice are consistent with the fertility defects reported in TAp73-deficient animals, but not ΔNp73-deficient mice, given that both females and males are fertile in the latter (Tomasini et al., 2008, Wilhelm et al., 2010). TAp73-deficient females showed defective follicle development and significantly decreased ovulation rate relative to wild-type animals in response to exogenous hormone stimulus, and the few ovulated oocytes observed were trapped under the bursa and unable to reach the fallopian tube for implantation (Tomasini et al., 2008), possibly due to the lack of ciliated cells lining the oviduct (Marshall et al., 2016). The corpora lutea are the primary sites of progesterone production after ovulation in mice (Allen, 1941, Rothchild, 1981). Therefore, the lack of corpora lutea observed in our p73−/− female mice (Figure 1B) led us to compare the levels of circulating hormones in the p73+/+ and p73−/− female mice.
Loss of p73 Leads to a Significant Decrease in Circulating Progesterone
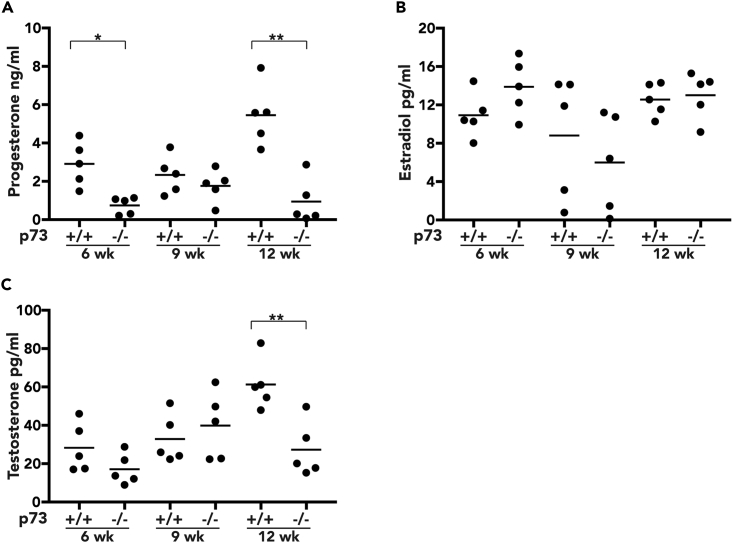
Given the impact of p73 loss on folliculogenesis and the number of corpora lutea, we measured circulating progesterone in nulliparous p73+/+ and p73−/− mice at 6, 9, and 12 weeks of age. Progesterone levels fluctuate throughout the stages of the estrous cycle (Byers et al., 2012). We assessed the estrous cycle through vaginal cytology and observed that p73−/− mice exhibit aberrant estrous cycle with prolonged diestrus stage compared with p73+/+ (Figure S2A). Given the acyclic nature of p73−/− female mice, all circulating hormones were analyzed from the only shared stage (diestrus) between both phenotypes. Progesterone levels decreased nearly 75% in p73−/− samples collected at 6 weeks of age (p value < 0.05) and 82% (p value < 0.01) at 12 weeks of age relative to p73+/+ mice (Figure 2A). Circulating estradiol levels in p73−/− mice were similar to those in p73+/+ mice at each time point analyzed (Figure 2B), although testosterone was reduced 55% in p73−/− samples harvested at 12 weeks of age (p value <0.01) (Figure 2C).
Figure 2.
Analysis of Circulating Progesterone in p73+/+ and p73−/− Female Mice
Plasma levels measure by ELISA of (A) progesterone, (B) estradiol, and (C) testosterone from five female mice per genotype at 6, 9, and 12 weeks of age; assay sensitivity range 0.2–50 ng/mL, 1–100 pg/mL, and 5–100 pg/mL, respectively. Bars represent the mean. *p value < 0.05, **p value < 0.01.
See also Figure S2.
Circulating hormones originating from the pituitary gland, including follicle-stimulating hormone (FSH), luteinizing hormone (LH), and growth hormone (GH), are key regulators of the estrous cycle and thus could exert a marked influence in ovarian progesterone production. Therefore, we measured circulating FSH levels in 12-week-old nulliparous mice, finding a 50% decrease in FSH levels in p73−/− compared with age-matched p73+/+ mice (p value <0.01) (Figure S2B). These findings are consistent with previous studies showing that FSH promotes granulosa-theca cell interactions that drive the production of ovarian testosterone (Smyth et al., 1993), thus constituting a possible mechanism to explain the decrease in testosterone observed in p73−/− female mice. LH levels were modestly decreased in p73−/− mice, albeit not to a statistically significant degree (Figure S2C), and no difference in circulating GH was observed between p73+/+ and p73−/− samples (Figure S2D).
To determine if the hormonal differences in the p73−/− mice were linked to gross abnormalities in pituitary gland morphology due to hippocampal dysgenesis and hydrocephalus (Yang et al., 2000, Talos et al., 2010), we analyzed pituitary gland tissue sections from p73+/+ and p73−/− female mice. The murine pituitary gland is composed of anterior (pars distalis, D; and intermedia, I) and posterior (pars nervosa, N) lobes. Despite diminished FSH production in p73−/− mice, we did not observe any overt histological differences in the pars distalis (asterisk) (Figure S2E), where gonadotropin hormones (FSH, LH, and GH) are produced and secreted. In addition, we evaluated p73 expression in the pituitary gland of p73+/+ mice through IF. We observed p73-positive cells in pars intermedia (solid-line box), but not in pars distalis (dashed-line box) (Figure S2F). Previous studies have shown that TAp73-deficient male mice have normal levels of FSH, LH, and gonadotropin-releasing hormone (Holembowski et al., 2014). In addition, immature TAp73-deficient ovaries fail to respond to exogenous hormone stimulation and show a significantly decreased ovulation rate compared with age-matched wild-type mice (Tomasini et al., 2008), further supporting normal pituitary gland signaling. Granulosa cells in growing follicles produce activin and inhibin as a feedback mechanism to regulate FSH secretion (Knight and Glister, 2006). Thus it is possible that the loss of p73 in granulosa cells, and the decreased number of growing follicles observed in p73−/− females, may negatively interfere with ovarian-pituitary feedback signaling required for proper FSH secretion.
Ectopic Progesterone Partially Rescues Ovarian Follicle Development in p73-Deficient Mice
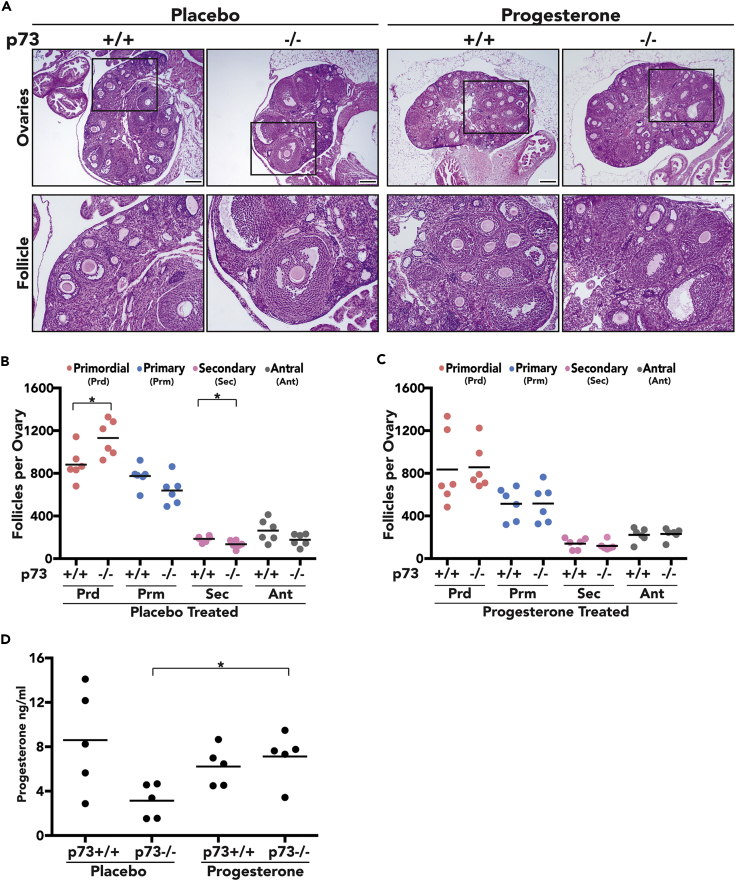
To determine if ectopic progesterone could rescue ovarian follicle maturation, 60-day-release progesterone pellets (15 mg) were implanted subcutaneously in 5-week-old nulliparous p73+/+ and p73−/− female mice and ovaries were analyzed 21 days after implantation. The ovaries from p73−/− mice with the placebo pellet harbored fewer primary, secondary, and antral follicles and were substantially smaller than placebo-treated p73+/+ mice (Figures 3A and 3B). However, ovaries from p73−/− mice treated with ectopic progesterone were similar in size to ovaries from placebo- or progesterone-treated p73+/+ mice (Figure 3A) and contained a greater number of primary, secondary, and antral follicles than placebo-treated p73−/− ovaries, similar to what was seen in progesterone- or placebo-treated p73+/+ mice (Figure 3C). As a control, circulating progesterone was measured at the time when the mice were killed to confirm that ectopic progesterone pellet increased circulating progesterone in p73−/− mice to levels comparable with both placebo- and progesterone-treated p73+/+ mice (Figure 3D).
Figure 3.
Progesterone Rescues Follicle Development in p73-Deficient Ovaries
Progesterone slow-release pellet or placebo control (15 mg/pellet, 60-day extended release) were implanted in five female mice per genotype at 5 weeks of age.
(A) Representative H&E images of placebo control and ectopic progesterone-treated ovaries of p73+/+ and p73−/− mice (scale bar, 200 μm).
(B and C) (B) Follicle quantification of placebo control and (C) progesterone-treated p73+/+ and p73−/− ovaries, respectively. Data are shown as number of follicles per ovary; Prd, primordial; Prm, primary; Sec, secondary; Ant, antral.
(D) Plasma levels of progesterone were measured through ELISA from placebo control and progesterone-treated p73+/+ and p73−/− female mice. *p value < 0.05.
See also Figure S3.
To assess the effect of ectopic progesterone on female sterility in our mouse model (Marshall et al., 2016), we determined the reproductive ability of placebo- or progesterone-treated p73+/+ and p73−/− female mice through mating trials. Twelve-week-old p73+/+ and p73−/− female mice were implanted with placebo or progesterone pellets and housed with p73+/+ males for a period of 14 days. At day 15, pregnancy status was determined and tissue was harvested for histological analysis. In p73+/+, two of two placebo-treated and two of three progesterone-treated female mice became pregnant, indicating that the level of ectopic progesterone administered did not inhibit their reproductive ability. In contrast, none of the five p73−/− mice, either placebo- (two mice) or progesterone-treated (three mice), became pregnant (data not shown). These results were not surprising given the lack of ovulation, corpus luteum formation, and ciliated cells in the p73-deficient mice (Marshall et al., 2016). Ciliated cells are required for transport of the oocyte through the oviduct and to the uterus (Critoph and Dennis, 1977, Halbert et al., 1976). Also, we analyzed the ability of ectopic progesterone to rescue corpus luteum formation by implanting placebo or progesterone pellets (5 mg) in 12-week-old nulliparous p73+/+ and p73−/− female mice. After 21 days, corpora lutea (arrowhead) were observed in p73+/+ with placebo or progesterone pellet. However, administration of ectopic progesterone in p73−/− mice was not able to rescue the formation of corpora lutea (Figure S3, arrowhead).
Ectopic Progesterone Rescues Lobulo-Alveolar Budding in p73-Deficient Mice
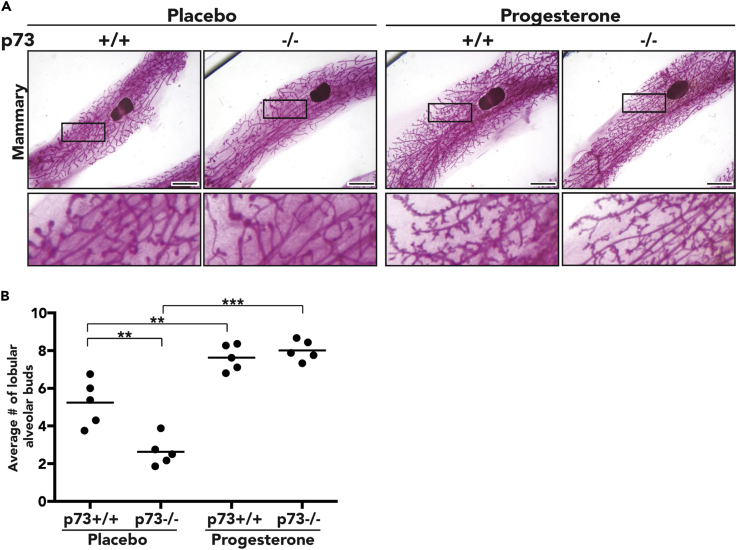
Mice that lack progesterone receptor exhibit defects in follicle rupture, causing impaired ovulation, absence of corpora lutea, and female infertility (Lydon et al., 1995, Lydon et al., 1996), similar to the phenotypes observed in our p73−/− female mice. Furthermore, impaired progesterone signaling leads to defects in proper mammary gland development by impeding the formation of lobulo-alveolar buds, small grape-like epithelial protrusions that, in the event of pregnancy, will generate the milk-producing alveoli (Hennighausen and Robinson, 2005, Bocchinfuso et al., 2000, Lydon et al., 1995). Given the similarities between the progesterone receptor-deficient mice and our p73−/− female mice, we assessed the hormonally responsive function of the mammary glands from mice treated with slow-release progesterone pellets, as described above. Using whole-mount carmine staining to visualize the mammary epithelium, we noted a 50% reduction (p value <0.01) in lobulo-alveolar budding in placebo-treated p73−/− mice compared with p73+/+ mice (Figures 4A and 4B). Progesterone-treated p73−/− female mice exhibited a complete rescue of lobular-alveolar budding and comparable mammary gland morphology to progesterone-treated p73+/+ control (Figure 4A). Ectopic progesterone significantly increased the number of lobulo-alveolar buds in p73−/− mammary gland by 200% (p value <0.001) compared with the mammary glands of placebo-treated p73−/− mice (Figure 4B).
Figure 4.
Ectopic Progesterone Rescues Lobulo-Alveolar Budding Defect in p73-Deficient Mammary Gland
(A) Whole mammary mount stained with carmine alum shows that ectopic progesterone rescues lobular-alveolar budding in p73−/− female mice (scale bar, 200 mm).
(B) Lobulo-alveolar budding quantification of placebo control or progesterone from p73+/+ and p73−/−; values shown represent the average number of side branches per primary branch. **p value< 0.01, ***p value < 0.001.
See also Figure S4.
Of note, the average number of lobular-alveolar buds was reduced in p73−/− mice by 45% at 6 weeks (p value <0.05), 60% at 9 weeks (p value <0.001), and 66% at 12 weeks (p value <0.001) of age compared with p73+/+ littermates (Figures S4A and S4B). Despite decreased lobulo-alveolar budding, the histological architecture of mammary glands was unaffected by loss of p73 (Figure S4C), with a well-organized luminal cell layer displaying apico-basal polarization (asterisks) and basally oriented myoepithelial cell layer (arrowheads), similar to what was seen in the mammary ductal epithelium of p73+/+ littermates. IF staining in p73+/+ mammary glands revealed nuclear p73 localization in basally located cells that stained positive for the myoepithelial/basal cell marker keratin 14 (Figure S4C). As expected, we did not observe p73 expression in p73−/− mammary glands (Figure S4C). Similar to our analyses in the ovaries, IF staining of p73+/+ mammary glands with antibodies against p63 confirmed myoepithelial/basal localization of p63 and p73 (Figure S4D) (Yang et al., 1999, Barbareschi et al., 2001). Interestingly, p63 co-localized with a subset of p73-expressing cells in the myoepithelial layer suggesting that these two proteins interact in specific myoepithelial cells of the mammary gland. Furthermore, we found that p63 expression was retained in the myoepithelial layer of p73−/− mammary glands, suggesting that decreased lobulo-alveolar budding in p73−/− is not due to impaired p63 expression (Figure S4D).
p73 Regulates a Biological Adhesion Gene Network in Murine Granulosa Cells
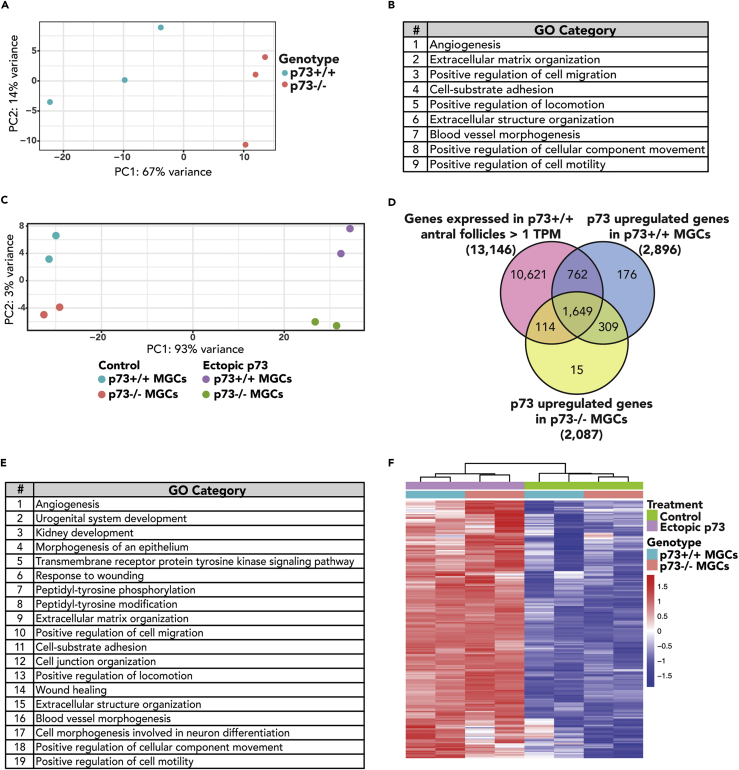
To gain mechanistic insight into the defects in follicle development and ovulation in p73−/− ovaries, laser capture microdissection was used to isolate granulosa cells from p73+/+ and p73−/− antral follicles of age-matched nulliparous mice (three mice per genotype). Principal component analysis revealed a clear separation between p73+/+ and p73−/− transcriptional changes in antral follicle samples in principal component 1 (Figure 5A). Accordingly, we identified 3,209 protein-coding genes differentially expressed between p73+/+ and p73−/− antral follicles (Table S1), of which 1,603 were enriched in p73+/+ antral follicles and 1,606 were enriched in p73−/− antral follicles. Gene Ontology (GO) enrichment (maximum false discovery rate [FDR < 2.22 × 10−16]) identified nine GO categories related to biological adhesion, including extracellular matrix organization, positive regulation of cell adhesion, cell-substrate adhesion, positive regulation of locomotion, extracellular structure organization, positive regulation of cellular component movement, and positive regulation of cell motility (Figure 5B; Table S2). These findings are consistent with previous reports of p73-mediated regulation of cell-cell adhesion and migration through integrin-β4 (Xie et al., 2018), vascular endothelial growth factor , and transforming growth factor β signaling (Fernandez-Alonso et al., 2015, Martin-Lopez et al., 2017, Bae et al., 2018).
Figure 5.
p73 Regulates a Gene Network Involved in Biological Adhesions in Antral Follicles
(A) Principal component analysis (PCA) plot of RNA sequencing (RNA-seq) analysis from LCM-isolated p73+/+ and p73−/− antral follicles (n = 3 mice/genotype).
(B) Table shows top nine GO categories enriched in p73+/+ versus p73−/− antral follicles (FDR < 2.22 × 10−16).
(C) PCA plot of RNA-seq analysis from p73+/+ and p73−/− MGCs after ectopic p73 or control.
(D) Venn diagram showing the overlap between genes expressed in p73+/+ antral follicles (TPM > 1) and upregulated after ectopic p73 expression in p73+/+ and p73−/− MGCs.
(E) Table listing the top 19 GO categories enriched in 1,649 overlapping genes from (D).
(F) Heatmap of expression for core 208 p73-upregulated granulosa cell genes. These genes were selected by identifying the 1,649 genes in (D) that were present in three or more of the enriched GO categories from (E) (FDR p value < 0.1).
We isolated and cultured primary mouse granulosa cells (MGCs) from p73+/+ mice and transduced the cells with lentivirus expressing TAp73β for 48 hr. We did not observe any difference in cell morphology or the levels of cleaved poly (ADP-ribose) polymerase 1 (PARP1) between control MGCs and those expressing ectopic p73 (Figures S5A and S5B). The rationale for using TAp73β was based on previously published data showing that TAp73β exhibits the highest level of transcriptional activity among p73 isoforms (Ueda et al., 1999, Lee and La Thangue, 1999) as well as the fact that the TAp73β isoform is highly expressed in human ovaries (Figure S1D). We measured global gene expression changes by RNA sequencing after ectopic p73 in MGCs isolated from both p73+/+ and p73−/− female mice and identified clear separation of samples after ectopic p73 (Figure 5C). Differential expression of 5,178 genes was identified in p73+/+ MGCs after ectopic expression of TAp73β, including 2,896 upregulated genes (Table S3). Similarly, we identified 3,391 differentially expressed genes after TAp73β expression in p73−/− MGCs (2,087 upregulated genes) (Table S4). We identified 1,649 genes commonly upregulated in TAp73β-expressing MGCs and in p73+/+ antral follicles (>1 TPM) (Figure 5D, Table S5). GO pathway enrichment analysis identified 19 GO categories (FDR < 2.22 × 10−16), including biological adhesion and migration (Figure 5E and Table S6). A core set of 208 genes overlapped in at least 3 of the 19 enriched GO categories (Figures 5F and S5C), including Adam10, Adamts12, Icam1, Pxn, and Mmp14. Importantly, we identified multiple genes required for the formation of the follicular focimatrix (focal intra-epithelial matrix), which is the extracellular matrix that aggregates between granulosa cells and increases as follicles progress to pre-ovulatory stage (Irving-Rodgers et al., 2004, Irving-Rodgers et al., 2009, Irving-Rodgers et al., 2010). Key focimatrix genes identified include Lama1, Lama2, Lama5, Lamb1, Hspg2, Nid1, and Nid2. These data suggest that p73 is a key regulator of genes involved in cell adhesion and migration.
p73 Regulates Cell Adhesion and Migration
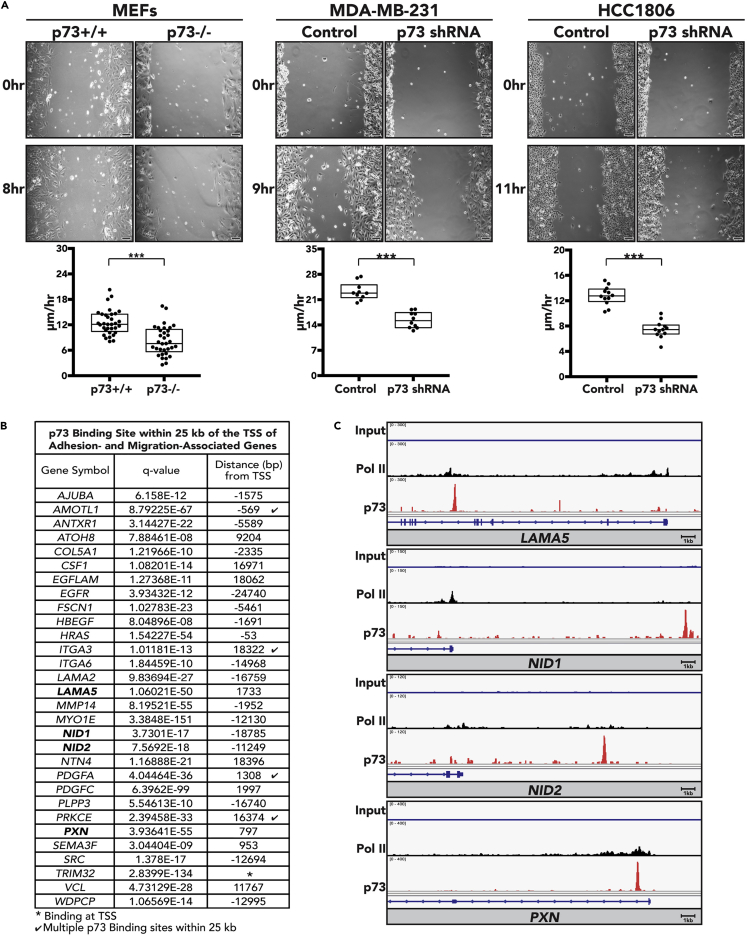
We used a well-established in vitro migration assay to analyze the effect of p73 on cell adhesion and migration. To set up the assay, magnetically attachable stencils (MAtS) were placed in culture vessels to create “gaps” in monolayer cell cultures (Ashby et al., 2012). The measurement of cell movement into the gaps over time provided a reporter assay for cell adhesion and migration. Due to technical constraints of large-scale MGC isolation and culture, we used mouse embryonic fibroblasts (MEFs) isolated from p73+/+ and p73−/− embryos as well as transformed epithelial cell lines that readily grow in culture. Lentivirally delivered short hairpin RNA sequences against p73 were used to “knockdown” p73 in transformed cell lines (MDA-MB-231 and HCC1806). Cells were plated near confluency surrounding the MAtS and cultured in serum-free media for 14 hr. MAtS were then removed (T = 0 hr) and migration was monitored for 8–11 hr, revealing that loss of p73 significantly decreased cellular migration rate in p73−/− MEFs, MDA-MB-231, and HCC1806 cells when compared with p73+/+ MEFs or vector control (Figure 6A). Loss of p73 expression was confirmed through qRT-PCR and western blot analysis (Figures S6A and S6B).
Figure 6.
p73 Regulates Cell Migration
(A) MEFs isolated from p73+/+ and p73−/− mice, MDA-MB-231, and HCC1806 cells stably expressing control short hairpin RNA (shRNA) and p73 shRNA were plated in culture dishes containing magnetic stencils and grown to confluency (scale bar, 100 μm). Each dot represents the percentage gap closure per field of view. ***p value < 0.001.
(B) Table listing adhesion- and migration-associated genes from the core set of 208 genes bound by p73 within 25 kb of their TSS (in HCC1806 cells). For each gene, the q value of the nearby p73 peak and its distance from the TSS of the gene are included.
(C) Integrative Genomics Viewer images for selected genes from (B) with tracks for input, p73, and Pol II ChIP-seq in HCC1806 cells. Each sample was normalized to 1X depth of coverage. Individual tracks within a gene are scaled equally. RefSeq gene annotations are in blue schematics at the bottom of each panel on the same scale as the ChIP-seq tracks.
See also Figures S6 and S7 and Table S7.
Given that the most significant difference in cell migration was observed in p73-deficient HCC1806 cells, we performed chromatin immunoprecipitation sequencing (ChIP-seq) on parental HCC1806 cells to determine if p73 directly binds near the transcriptional start site (TSS) of genes involved in cell migration. HCC1806 cells were formaldehyde cross-linked and processed for p73 and RNA polymerase II (Pol II) ChIP-seq, as described in the Transparent Methods. Quality control analysis of these data demonstrated clear separation between ChIP and input signal for p73 and Pol II (Figure S7A). Because the p73 ChIP replicates were highly correlated (Figure S7B), we pooled the samples for peak calling to increase peak detection sensitivity. We identified 3,555 p73 and 19,696 Pol II genomic binding sites (Table S7). Motif analysis showed strong enrichment for the p53 family binding motif (Figure S7C) (Rosenbluth et al., 2008, el-Deiry et al., 1992, Lokshin et al., 2007, Smeenk et al., 2008). We identified known binding sites in p73 target genes MDM2 and CDKN1A (Figures S7D and S7E) (Barak et al., 1993, Juven et al., 1993, Espinosa and Emerson, 2001) (Robinson et al., 2011, Thorvaldsdottir et al., 2013) as well as a binding site in the newly reported p73 target gene ITGB4 (integrin-β4) (Xie et al., 2018). Since we were comparing murine gene expression data with human ChIP data, we focused our analysis on genes that were increased after p73 expression in MGCS and for which the binding of p73 occurred within 25 kb of the TSS in HCC1806 ChIP. From the 208 p73-regulated core gene set, we found 30 adhesion- and migration-associated genes with a p73 binding site within 25 kb of the TSS of the human gene homolog (Figure 6B). Of immediate interest were p73 binding sites near genes encoding adhesion PXN and focimatrix components LAMA5, NID1, and NID2 (Figure 6C). Paxillin is a scaffolding protein that regulates cytoskeleton remodeling, cell migration, and focal adhesions (Huang et al., 2003, Hu et al., 2014, Deramaudt et al., 2014).
p73 is necessary for cell migration in transformed epithelial cell line models. Through ChIP-seq, we identified p73 binding within 25 kb of the TSS of genes involved in cell-to-cell adhesion and migration, including NID1, NID2, LAMA5, and PXN. Furthermore, ectopic p73 expression is sufficient to upregulate the expression of these genes in MGCs. Collectively, these data support the conclusion that p73 regulates ovarian folliculogenesis and ovulation, in part through regulated expression of adhesion and focimatrix genes necessary for proper follicle maturation.
Discussion
We discovered that p73 is required for proper ovarian follicle development, ovulation, and subsequent corpus luteum formation and progesterone production. Similar to prior findings made with TA-specific p73-deficient male mice (Inoue et al., 2014), we observed a significant decrease in levels of circulating progesterone in our p73−/− female mice. Furthermore, we demonstrated that the mammary branching defect observed in p73−/− female mice is secondary to decreased levels of progesterone through a complete rescue of the branching defect after ectopic progesterone administration. Through analysis of gene expression between p73+/+ and p73−/− antral follicles and modulation of p73 expression in various model systems, from primary culture of MGCs to human epithelial cells, we discovered p73-dependent regulation of genes crucial for biological adhesion (Pxn) and extracellular matrix interactions required to form proper focimatrix (Lama5, Nid1/2, and Hspg2). Focimatrix levels have been previously linked with steroidogenesis and CYP11A1 (Irving-Rodgers et al., 2009, Matti et al., 2010) during ovarian follicle development. Consistent with defective focimatrix formation in our p73-deficient ovarian follicles, we observed a significant decrease in expression of Cyp11a1, as well as other hormonally regulated genes including prolactin receptor (Prlr), luteinizing hormone/choriogonadotropin receptor (Lhcgr), oxytocin receptor (Oxtr), steroidogenic factor 1 (Nr5a1), and activin B receptor (Acvr1c) in p73−/− antral follicles (Table S1). NR5A1 is a transcriptional activator required for the formation of steroidogenic tissues, and cell-specific knockout experiments have shown that Nr5a1 is necessary for male and female fertility (Ferraz-de-Souza et al., 2011, Jeyasuria et al., 2004). Mice that lack ACVR1C expression in granulosa cells exhibit striking similarities to our p73−/− mice including defective follicle development, absence of corpora lutea, and decreased levels of circulating FSH (Sandoval-Guzman et al., 2012), providing a possible mechanism for the decreased FSH levels in our p73−/− females. Future studies are needed to determine the direct or indirect mechanism by which p73 regulates the expression of genes required for proper steroidogenesis and hormone signaling in antral follicles.
The lack of functional p73 protein in murine ovaries results in an absence of corpora lutea and an increase in the number of primordial follicles, suggesting a defect in primordial-to-primary follicle transition. We also observed a decrease in FSH levels, which supports the reduced number of developing follicles in p73−/− mice. FSH, secreted from the pituitary gland, is positively and negatively regulated by activin and inhibin, respectively, which are secreted from granulosa cells (Knight and Glister, 2006). From our analysis, p73 is expressed in the pars intermedia, and not in pars distalis where FSH, LH, and GH are produced. Previous studies have demonstrated that p73-deficient mice exhibit hippocampal dysgenesis and hydrocephalus (Yang et al., 2000, Talos et al., 2010, Marshall et al., 2016). In our p73−/− mice, we also observed hippocampal dysgenesis and hydrocephalus to varying degrees and on a mouse-to-mouse basis across our cohort of p73-deficient mice; we are not able to rule out the possible effect of these phenotypes on pituitary gland function. Future experiments are needed to determine the impact of hippocampal dysgenesis and hydrocephalus on pituitary gland signaling and hormone secretion.
p73 is necessary for multiciliated cell development (Marshall et al., 2016, Nemajerova et al., 2016), and we observed expression of p73 in ciliated cells that line the oviductal epithelium in mice (Marshall et al., 2016). A major role of these ciliated cells is to transport the oocyte to the uterus for implantation (Critoph and Dennis, 1977, Halbert et al., 1976). Mouse models that lack ciliated cells, such as gene knockouts of Foxj1 (Gomperts et al., 2004) and Gemc1 (Terre et al., 2016), have similar phenotypes to our p73-deficient mice (Marshall et al., 2016), including female and male infertility. Gemc1−/− ovaries exhibit a follicle development defect similar to our p73−/− female mice (Terre et al., 2016). We are not able to rule out a possible paracrine mechanism between the ciliated cells in the oviduct and the granulosa cells in the ovaries. It would be of interest to determine if ciliated cells have additional roles in fertility besides transport of the oocyte, in terms of a direct effect on ovarian follicle development and ovulation through cell-to-cell signaling mechanisms. We posit that p73 is required in both granulosa cells within the follicle and ciliated cells in the oviduct to promote proper follicle development, ovulation, and oocyte implantation.
In summary, we discovered that p73 is required for ovarian follicle development, ovulation, and subsequent corpus luteum formation. The mammary gland branching defect observed in our p73−/− female mice was due to decreased circulating levels of progesterone. We identified a p73-regulated core gene set involved in biological adhesion and cell migration in MGCs. Furthermore, we demonstrated that p73 is necessary for proper epithelial cell migration and identified p73 binding near the TSS of focimatrix component genes and crucial regulators of cell adhesion and migration like PXN. We conclude that p73 acts as critical regulator of cell-to-cell adhesion, extracellular matrix interactions, and cell migration and promotes proper follicle development, ovulation, and fertility.
Limitations of the Study
A limitation of the study was our inability to isolate and culture sufficient numbers of the murine granulosa cells required for cell migration and ChIP experiments, due to technical constraints associated with purification of cells from murine ovaries. To address this limitation, we used another tissue for generation of primary cultures of murine cells from p73+/+ and p73−/− mice as well as the established HCC1806 cell line to analyze the role of p73 in cell migration. For continuity, we used HCC1806 cell line to identify genes involved in migration, to which p73 directly binds through ChIP-seq.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Donna Hicks for technical assistance with progesterone pellet implantation in mice, Dr. So-Youn Kim for well-developed MGC isolation and culture protocols, Violeta Sanchez for help with histology, Andries Zijlstra for migration assay methods and protocols, and Harold Moses for assistance with histopathology analysis. Vanderbilt Hormone and Analytical Services Core (supported by NIH (USA) grants: DK059637 and DK020593) was used for progesterone, estradiol, testosterone, and growth hormone analysis, and the UVA Center for Research in Reproduction (supported NIH (USA) grant: HD28934) was used for FSH and LH analysis. This research was supported by NIH (USA) grants CA105436, CA098131, and CA068485 to J.A.P. and training grant support GM062459 to G.L.S.G.
Author Contributions
G.L.S.G. outlined and conducted the majority of the experiments and prepared manuscript drafts. J.S.B. performed ChIP-seq and RNA-seq bioinformatics analysis and assisted with experimental design. C.B.M. performed ChIP-seq and assisted in the follicle quantification. T.M.S. provided bioinformatics analysis and assisted with experimental design. Q.S. provided biostatistical and bioinformatics analyses. K.N.J. provided assistance with isolation and culture of MGCs. K.L.B. conducted pathology analysis of murine ovaries. B.J.V. provided advice and assistance on ChIP-seq experiments. R.S.C. assisted and provided guidance on progesterone pellet experiments. J.A.P was involved in all aspects of the study, including experimental design, manuscript preparation, and acquisition of funding.
Declaration of Interests
The authors declare no competing interests.
Published: October 26, 2018
Footnotes
Supplemental Information includes Transparent Methods, seven figures, and seven tables and can be found with this article online at https://doi.org/10.1016/j.isci.2018.09.018.
Supplemental Information
References
- Allen W.M. The chemical and physiological properties, and clinical uses of the corpus luteum hormone, progesterone. Bull. N. Y. Acad. Med. 1941;17:508–518. [PMC free article] [PubMed] [Google Scholar]
- Ashby W.J., Wikswo J.P., Zijlstra A. Magnetically attachable stencils and the non-destructive analysis of the contribution made by the underlying matrix to cell migration. Biomaterials. 2012;33:8189–8203. doi: 10.1016/j.biomaterials.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae W.K., Hong C.S., Park M.R., Sun E.G., Lee J.H., Kang K., Ryu K.H., Shim H.J., Hwang J.E., Cho S.H., Chung I.J. TAp73 inhibits cell invasion and migration by directly activating KAI1 expression in colorectal carcinoma. Cancer Lett. 2018;415:106–116. doi: 10.1016/j.canlet.2017.12.002. [DOI] [PubMed] [Google Scholar]
- Barak Y., Juven T., Haffner R., Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbareschi M., Pecciarini L., Cangi M.G., Macri E., Rizzo A., Viale G., Doglioni C. p63, a p53 homologue, is a selective nuclear marker of myoepithelial cells of the human breast. Am. J. Surg. Pathol. 2001;25:1054–1060. doi: 10.1097/00000478-200108000-00010. [DOI] [PubMed] [Google Scholar]
- Bocchinfuso W.P., Lindzey J.K., Hewitt S.C., Clark J.A., Myers P.H., Cooper R., Korach K.S. Induction of mammary gland development in estrogen receptor-alpha knockout mice. Endocrinology. 2000;141:2982–2994. doi: 10.1210/endo.141.8.7609. [DOI] [PubMed] [Google Scholar]
- Byers S.L., Wiles M.V., Dunn S.L., Taft R.A. Mouse estrous cycle identification tool and images. PLoS One. 2012;7:e35538. doi: 10.1371/journal.pone.0035538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W.M., Siu W.Y., Lau A., Poon R.Y. How many mutant p53 molecules are needed to inactivate a tetramer? Mol. Cell. Biol. 2004;24:3536–3551. doi: 10.1128/MCB.24.8.3536-3551.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critoph F.N., Dennis K.J. Ciliary activity in the human oviduct. Obstet. Gynecol. Surv. 1977;32:602–603. doi: 10.1097/00006254-197709000-00015. [DOI] [PubMed] [Google Scholar]
- Deramaudt T.B., Dujardin D., Noulet F., Martin S., Vauchelles R., Takeda K., Ronde P. Altering FAK-paxillin interactions reduces adhesion, migration and invasion processes. PLoS One. 2014;9:e92059. doi: 10.1371/journal.pone.0092059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotsch V., Bernassola F., Coutandin D., Candi E., Melino G. p63 and p73, the ancestors of p53. Cold Spring Harb. Perspect. Biol. 2010;2:a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry W.S., Kern S.E., Pietenpol J.A., Kinzler K.W., Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- Espinosa J.M., Emerson B.M. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell. 2001;8:57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- Fernandez-Alonso R., Martin-Lopez M., Gonzalez-Cano L., Garcia S., Castrillo F., Diez-Prieto I., Fernandez-Corona A., Lorenzo-Marcos M.E., Li X., Claesson-Welsh L. p73 is required for endothelial cell differentiation, migration and the formation of vascular networks regulating VEGF and TGFbeta signaling. Cell Death Differ. 2015;22:1287–1299. doi: 10.1038/cdd.2014.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraz-de-Souza B., Lin L., Achermann J.C. Steroidogenic factor-1 (SF-1, NR5A1) and human disease. Mol. Cell Endocrinol. 2011;336:198–205. doi: 10.1016/j.mce.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomperts B.N., Gong-Cooper X., Hackett B.P. Foxj1 regulates basal body anchoring to the cytoskeleton of ciliated pulmonary epithelial cells. J. Cell Sci. 2004;117:1329–1337. doi: 10.1242/jcs.00978. [DOI] [PubMed] [Google Scholar]
- Grob T.J., Novak U., Maisse C., Barcaroli D., Luthi A.U., Pirnia F., Hugli B., Graber H.U., De Laurenzi V., Fey M.F. Human delta Np73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ. 2001;8:1213–1223. doi: 10.1038/sj.cdd.4400962. [DOI] [PubMed] [Google Scholar]
- Halbert S.A., Tam P.Y., Blandau R.J. Egg transport in the rabbit oviduct: the roles of cilia and muscle. Science. 1976;191:1052–1053. doi: 10.1126/science.1251215. [DOI] [PubMed] [Google Scholar]
- Harms K.L., Chen X. The functional domains in p53 family proteins exhibit both common and distinct properties. Cell Death Differ. 2006;13:890–897. doi: 10.1038/sj.cdd.4401904. [DOI] [PubMed] [Google Scholar]
- Hennighausen L., Robinson G.W. Information networks in the mammary gland. Nat. Rev. Mol. Cell Biol. 2005;6:715–725. doi: 10.1038/nrm1714. [DOI] [PubMed] [Google Scholar]
- Holembowski L., Kramer D., Riedel D., Sordella R., Nemajerova A., Dobbelstein M., Moll U.M. TAp73 is essential for germ cell adhesion and maturation in testis. J. Cell Biol. 2014;204:1173–1190. doi: 10.1083/jcb.201306066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.L., Lu S., Szeto K.W., Sun J., Wang Y., Lasheras J.C., Chien S. FAK and paxillin dynamics at focal adhesions in the protrusions of migrating cells. Sci. Rep. 2014;4:6024. doi: 10.1038/srep06024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Rajfur Z., Borchers C., Schaller M.D., Jacobson K. JNK phosphorylates paxillin and regulates cell migration. Nature. 2003;424:219–223. doi: 10.1038/nature01745. [DOI] [PubMed] [Google Scholar]
- Inoue S., Tomasini R., Rufini A., Elia A.J., Agostini M., Amelio I., Cescon D., Dinsdale D., Zhou L., Harris I.S. TAp73 is required for spermatogenesis and the maintenance of male fertility. Proc. Natl. Acad. Sci. U S A. 2014;111:1843–1848. doi: 10.1073/pnas.1323416111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving-Rodgers H.F., Harland M.L., Rodgers R.J. A novel basal lamina matrix of the stratified epithelium of the ovarian follicle. Matrix Biol. 2004;23:207–217. doi: 10.1016/j.matbio.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Irving-Rodgers H.F., Harland M.L., Sullivan T.R., Rodgers R.J. Studies of granulosa cell maturation in dominant and subordinate bovine follicles: novel extracellular matrix focimatrix is co-ordinately regulated with cholesterol side-chain cleavage CYP11A1. Reproduction. 2009;137:825–834. doi: 10.1530/REP-08-0485. [DOI] [PubMed] [Google Scholar]
- Irving-Rodgers H.F., Hummitzsch K., Murdiyarso L.S., Bonner W.M., Sado Y., Ninomiya Y., Couchman J.R., Sorokin L.M., Rodgers R.J. Dynamics of extracellular matrix in ovarian follicles and corpora lutea of mice. Cell Tissue Res. 2010;339:613–624. doi: 10.1007/s00441-009-0905-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyasuria P., Ikeda Y., Jamin S.P., Zhao L., De Rooij D.G., Themmen A.P., Behringer R.R., Parker K.L. Cell-specific knockout of steroidogenic factor 1 reveals its essential roles in gonadal function. Mol. Endocrinol. 2004;18:1610–1619. doi: 10.1210/me.2003-0404. [DOI] [PubMed] [Google Scholar]
- Juven T., Barak Y., Zauberman A., George D.L., Oren M. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene. 1993;8:3411–3416. [PubMed] [Google Scholar]
- Kaghad M., Bonnet H., Yang A., Creancier L., Biscan J.C., Valent A., Minty A., Chalon P., Lelias J.M., Dumont X. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- Kim S.Y., Ebbert K., Cordeiro M.H., Romero M., Zhu J., Serna V.A., Whelan K.A., Woodruff T.K., Kurita T. Cell autonomous phosphoinositide 3-kinase activation in oocytes disrupts normal ovarian function through promoting survival and overgrowth of ovarian follicles. Endocrinology. 2015;156:1464–1476. doi: 10.1210/en.2014-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight P.G., Glister C. TGF-beta superfamily members and ovarian follicle development. Reproduction. 2006;132:191–206. doi: 10.1530/rep.1.01074. [DOI] [PubMed] [Google Scholar]
- Lee C.W., La Thangue N.B. Promoter specificity and stability control of the p53-related protein p73. Oncogene. 1999;18:4171–4181. doi: 10.1038/sj.onc.1202793. [DOI] [PubMed] [Google Scholar]
- Lokshin M., Li Y., Gaiddon C., Prives C. p53 and p73 display common and distinct requirements for sequence specific binding to DNA. Nucleic Acids Res. 2007;35:340–352. doi: 10.1093/nar/gkl1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydon J.P., DeMayo F.J., Conneely O.M., O'Malley B.W. Reproductive phenotypes of the progesterone receptor null mutant mouse. J. Steroid Biochem. Mol. Biol. 1996;56:67–77. doi: 10.1016/0960-0760(95)00254-5. [DOI] [PubMed] [Google Scholar]
- Lydon J.P., DeMayo F.J., Funk C.R., Mani S.K., Hughes A.R., Montgomery C.A., Jr., Shyamala G., Conneely O.M., O'Malley B.W. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 1995;9:2266–2278. doi: 10.1101/gad.9.18.2266. [DOI] [PubMed] [Google Scholar]
- Marshall C.B., Mays D.J., Beeler J.S., Rosenbluth J.M., Boyd K.L., Santos Guasch G.L., Shaver T.M., Tang L.J., Liu Q., Shyr Y. p73 is required for multiciliogenesis and regulates the Foxj1-associated gene network. Cell Rep. 2016;14:2289–2300. doi: 10.1016/j.celrep.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Lopez M., Maeso-Alonso L., Fuertes-Alvarez S., Balboa D., Rodriguez-Cortez V., Weltner J., Diez-Prieto I., Davis A., Wu Y., Otonkoski T. p73 is required for appropriate BMP-induced mesenchymal-to-epithelial transition during somatic cell reprogramming. Cell Death Dis. 2017;8:e3034. doi: 10.1038/cddis.2017.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matti N., Irving-Rodgers H.F., Hatzirodos N., Sullivan T.R., Rodgers R.J. Differential expression of focimatrix and steroidogenic enzymes before size deviation during waves of follicular development in bovine ovarian follicles. Mol. Cell Endocrinol. 2010;321:207–214. doi: 10.1016/j.mce.2010.02.019. [DOI] [PubMed] [Google Scholar]
- Mills A.A., Zheng B., Wang X.J., Vogel H., Roop D.R., Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Nemajerova A., Kramer D., Siller S.S., Herr C., Shomroni O., Pena T., Gallinas Suazo C., Glaser K., Wildung M., Steffen H. TAp73 is a central transcriptional regulator of airway multiciliogenesis. Genes Dev. 2016;30:1300–1312. doi: 10.1101/gad.279836.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada M., Ohba M., Kawahara C., Ishioka C., Kanamaru R., Katoh I., Ikawa Y., Nimura Y., Nakagawara A., Obinata M., Ikawa S. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat. Med. 1998;4:839–843. doi: 10.1038/nm0798-839. [DOI] [PubMed] [Google Scholar]
- Park M., Shin E., Won M., Kim J.H., Go H., Kim H.L., Ko J.J., Lee K., Bae J. FOXL2 interacts with steroidogenic factor-1 (SF-1) and represses SF-1-induced CYP17 transcription in granulosa cells. Mol. Endocrinol. 2010;24:1024–1036. doi: 10.1210/me.2009-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco J.W., Leong C.O., Kuperwasser N., DeYoung M.P., Ellisen L.W. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9:45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Rosenbluth J.M., Mays D.J., Pino M.F., Tang L.J., Pietenpol J.A. A gene signature-based approach identifies mTOR as a regulator of p73. Mol. Cell. Biol. 2008;28:5951–5964. doi: 10.1128/MCB.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothchild I. The regulation of the mammalian corpus luteum. Recent Prog. Horm. Res. 1981;37:183–298. doi: 10.1016/b978-0-12-571137-1.50009-8. [DOI] [PubMed] [Google Scholar]
- Sandoval-Guzman T., Gongrich C., Moliner A., Guo T., Wu H., Broberger C., Ibanez C.F. Neuroendocrine control of female reproductive function by the activin receptor ALK7. FASEB J. 2012;26:4966–4976. doi: 10.1096/fj.11-199059. [DOI] [PubMed] [Google Scholar]
- Schmale H., Bamberger C. A novel protein with strong homology to the tumor suppressor p53. Oncogene. 1997;15:1363–1367. doi: 10.1038/sj.onc.1201500. [DOI] [PubMed] [Google Scholar]
- Schmidt D., Ovitt C.E., Anlag K., Fehsenfeld S., Gredsted L., Treier A.C., Treier M. The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development. 2004;131:933–942. doi: 10.1242/dev.00969. [DOI] [PubMed] [Google Scholar]
- Smeenk L., van Heeringen S.J., Koeppel M., van Driel M.A., Bartels S.J., Akkers R.C., Denissov S., Stunnenberg H.G., Lohrum M. Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res. 2008;36:3639–3654. doi: 10.1093/nar/gkn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth C.D., Miro F., Whitelaw P.F., Howles C.M., Hillier S.G. Ovarian thecal/interstitial androgen synthesis is enhanced by a follicle-stimulating hormone-stimulated paracrine mechanism. Endocrinology. 1993;133:1532–1538. doi: 10.1210/endo.133.4.8404591. [DOI] [PubMed] [Google Scholar]
- Stiewe T., Theseling C.C., Putzer B.M. Transactivation-deficient Delta TA-p73 inhibits p53 by direct competition for DNA binding: implications for tumorigenesis. J. Biol. Chem. 2002;277:14177–14185. doi: 10.1074/jbc.M200480200. [DOI] [PubMed] [Google Scholar]
- Suh E.K., Yang A., Kettenbach A., Bamberger C., Michaelis A.H., Zhu Z., Elvin J.A., Bronson R.T., Crum C.P., McKeon F. p63 protects the female germ line during meiotic arrest. Nature. 2006;444:624–628. doi: 10.1038/nature05337. [DOI] [PubMed] [Google Scholar]
- Talos F., Abraham A., Vaseva A.V., Holembowski L., Tsirka S.E., Scheel A., Bode D., Dobbelstein M., Bruck W., Moll U.M. p73 is an essential regulator of neural stem cell maintenance in embryonal and adult CNS neurogenesis. Cell Death Differ. 2010;17:1816–1829. doi: 10.1038/cdd.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terre B., Piergiovanni G., Segura-Bayona S., Gil-Gomez G., Youssef S.A., Attolini C.S., Wilsch-Brauninger M., Jung C., Rojas A.M., Marjanovic M. GEMC1 is a critical regulator of multiciliated cell differentiation. EMBO J. 2016;35:942–960. doi: 10.15252/embj.201592821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini R., Tsuchihara K., Wilhelm M., Fujitani M., Rufini A., Cheung C.C., Khan F., Itie-Youten A., Wakeham A., Tsao M.S. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22:2677–2691. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trink B., Okami K., Wu L., Sriuranpong V., Jen J., Sidransky D. A new human p53 homologue. Nat. Med. 1998;4:747–748. doi: 10.1038/nm0798-747. [DOI] [PubMed] [Google Scholar]
- Ueda Y., Hijikata M., Takagi S., Chiba T., Shimotohno K. New p73 variants with altered C-terminal structures have varied transcriptional activities. Oncogene. 1999;18:4993–4998. doi: 10.1038/sj.onc.1202817. [DOI] [PubMed] [Google Scholar]
- Wilhelm M.T., Rufini A., Wetzel M.K., Tsuchihara K., Inoue S., Tomasini R., Itie-Youten A., Wakeham A., Arsenian-Henriksson M., Melino G. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev. 2010;24:549–560. doi: 10.1101/gad.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie N., Vikhreva P., Annicchiarico-Petruzzelli M., Amelio I., Barlev N., Knight R.A., Melino G. Integrin-beta4 is a novel transcriptional target of TAp73. Cell Cycle. 2018;17:589–594. doi: 10.1080/15384101.2017.1403684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A., Kaghad M., Wang Y., Gillett E., Fleming M.D., Dotsch V., Andrews N.C., Caput D., McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R.T., Tabin C., Sharpe A., Caput D., Crum C., McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yang A., Walker N., Bronson R., Kaghad M., Oosterwegel M., Bonnin J., Vagner C., Bonnet H., Dikkes P., Sharpe A. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.