Abstract
Human Immunodeficiency Virus (HIV) infection continues to have major adverse public health and clinical consequences despite the effectiveness of combination Antiretroviral Therapy (cART) in reducing HIV viral load and improving immune function. As successfully treated individuals with HIV infection age, their cognition declines faster than reported for normal aging. This phenomenon underlines the importance of improving long-term care, which requires better understanding of the impact of HIV on the brain. In this paper, automated identification of patients and brain regions affected by HIV infection are modeled as a classification problem, whose solution is determined in two steps within our proposed Chained-Regularization framework. The first step focuses on selecting the HIV pattern (i.e., the most informative constellation of brain region measurements for distinguishing HIV infected subjects from healthy controls) by constraining the search for the optimal parameter setting of the classifier via group sparsity (ℓ2,1-norm). The second step improves classification accuracy by constraining the parameterization with respect to the selected measurements and the Euclidean regularization (ℓ2-norm). When applied to the cortical and subcortical structural Magnetic Resonance Images (MRI) measurements of 65 controls and 65 HIV infected individuals, this approach is more accurate in distinguishing the two cohorts than more common models. Finally, the brain regions of the identified HIV pattern concur with the HIV literature that uses traditional group analysis models.
Keywords: Computational neuroscience, Human immunodeficiency virus (HIV), MRI brain image analysis, multiple kernel learning, group sparsity
1. Introduction
Despite the success of highly active antiretroviral therapy (HAART) and combination antiretroviral therapy (cART) in extending longevity of individuals infected with the Human Immunodeficiency Virus (HIV), neurocognitive impairments still commonly occur [1, 2, 3]. Structural Magnetic Resonance Imaging (MRI) has often been used to determine the neural correlates of cognitive and motor deficits in HIV infection, indicating, for example, specific relationships between regional brain volume deficits [4, 5], memory compromise [6], and accelerated brain aging in HIV infected adults [7]. Neurocognitive and motor impairments in HIV infection, however, are similar to those reported in other age-related diagnoses [8]. To improve diagnostic specificity of MRI in HIV, this manuscript proposes a novel machine learning method and applies it to the morphometric measurements extracted from structural MRI scans collected from HIV infected and healthy control (CTRL) participants.
Conventional HIV MRI studies typically test for group differences (with respect to the CTRL cohort) by separately analyzing each image measurement for the impact of HIV [4, 9, 6, 10, 2, 1]. Separate analysis of measurements may lead to contradicting or inconclusive findings [11]. By contrast, our proposed analysis is a type of machine learning framework that considers all image measurements together to identify the subset of measurements (called patterns) specific to HIV and then relates the significance of the pattern to its accuracy in distinguishing individuals with HIV from CTRLs. A popular approach for identifying patterns uses sparse classifiers [12, 13, 14, 15, 16, 8], which assume that only a few measurements are informative for distinguishing cohorts. After identifying a pattern, the corresponding measures are often applied to a second (non-sparse) approach, which focuses only on improving classification accuracy [17, 18, 19, 20, 21, 22]. This two-step regularization procedure assumes that measurements selected by the sparse classifier define the unique, optimal pattern for distinguishing the two cohorts [23, 24, 17, 25]. This assumption, however, is generally not true because the redundancy in information across image measurements allows for multiple solutions [19]. As the two steps are based on different classification approaches, the pattern identified by the sparse classifier of the first step are generally not optimal for the non-sparse approach of the second step.
Herein, we propose an approach (denoted as Chained-Regularization) that uses the same classifier first to identify a pattern and then, using the pattern, to distinguish individuals; however, different constraints guide the parameterization of the classifier in each step. Our proposed algorithm models the selection of the most informative image measurements in the first step by confining parameterization of the classifier through group sparsity (ℓ2,1-norm) regularization [26, 8]. Group sparsity extends the concept of the ℓ1-norm [27, 28, 16] of identifying a few informative measurements for combining measurements into groups and then identifying a small number of groups [27]. The grouping can be used for explicit modeling of relationships between measurements [29]. In this work, each measurement from the regions of interest (ROIs) is grouped with its counterpart in the other brain hemisphere given our assumption that HIV infection affects the brain bilaterally. In the second step of Chained-Regularization, the classifier is trained on just the selected individual measurements with the search for the optimal parameter setting being constrained via Euclidean (ℓ2-norm) regularization. The logic of this approach is that the ℓ2,1 regularization generally improves the accuracy of classifiers in the presence of a large number of uninformative or redundant image measurements (as it is often the case of neuroimaging studies), while the ℓ2 regularization improves the accuracy of classifiers in the event that all provided image measurements are informative [17, 18]. Our chained-regularization scheme, which uses a sequential dependency approach to identify a pattern to be applied for determining group membership of individuals, is different from chain-regularization [30], a concept used in physics to describe group of objects interacting with each other in a chain.
We implement Chained-Regularization within a multiple kernel learning (MKL) framework [31, 18]. MKL is based on the assumption that samples (i.e., individual participants) that are similar to each other should be assigned to the same cohort (e.g., HIV). Similarity between two samples is measured through a pairwise comparison of the corresponding image measurements. This comparison is defined by a set of metrics (i.e., linear and nonlinear kernel functions), each capturing a unique characteristic across image measurements. The MKL algorithm now determines the combination of metrics and image measurements [18] that lead to the highest classification accuracy (see Figure 1). It thus omits the simplifying assumption of most other classifiers that the discriminating characteristics of all image measurements are best captured by a single metric (as in [18, 32, 33, 31, 34]).
Figure 1:
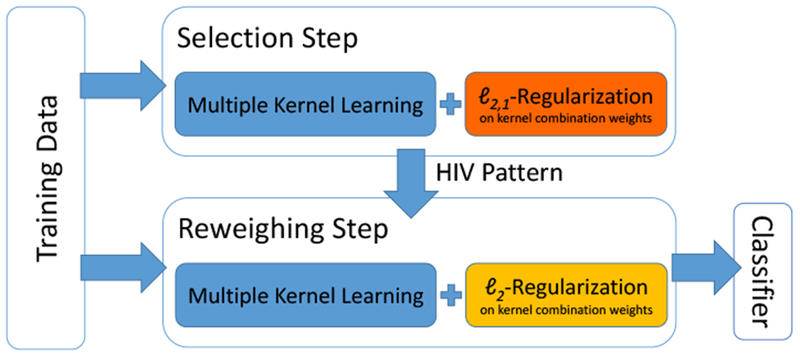
Training of the Chained-Regularization approach: The first step (top, denoted as Selection Step) selects the image measurements informative for distinguishing HIV from controls, while the second step (bottom, denoted as Reweighing Step) focuses on improving the accuracy by reweighing the selected measures for classifying the samples. Note, both steps are based on the same classifier but differ in regularizing (or constraining) its parameterization.
In summary, our analysis makes two novel contributions: (1) We propose Chained-Regularization within the MKL framework, which, in our experiments, is significantly more accurate than single-step and other two-step approaches. (2) To the best of our knowledge, this is the first study to examine both linear and non-linear supervised learning approaches to identify patterns that discriminate HIV infected from healthy control brains.
The rest of the paper is organized as follows: Section 2 introduces the materials (the data set), preprocessing, the proposed chained regularization and the experimental setup. Appendix A provides additional technical details of the proposed method. Section 3 compares our approach to other implementations on the HIV data set and reports on its identified pattern specific to HIV. Section 4 provides an in depth discussion about the findings of the previous section and their relevance with respect to the HIV literature. The paper concludes with Section 5.
2. Materials and Methods
2.1. Participant Information
Data used in this study are from 65 HIV infected individuals and 245 CTRL subjects. For classification, we match 65 CTRLs to the 65 HIV cohort. Specifically, for each HIV subject, one subject is selected from the CTRL cohort, such that they have the same sex and a minimal difference in their ages. We refer to the matched samples as ‘matched CTRL group’. The remaining 180 CTRL subjects, referred to as Confounding Factors CTRL group (CF CTRL group), are used for analysis of the confounding factors and minimizing their effects. Table 1 shows the demographic information of participants in all groups, and Figure 2 plots their age distributions. All 310 participants are tested for HIV, viral load, and CD4 T-cell count. HIV infected individuals had a CD4 count and a Karnofsky score > 70 [35]. Data from these subjects were used in previous studies [10, 4, 9].
Table 1:
Demographic information (‘svol’ = supratentorial volume).
| Total | sex |
Age (years) | svol(×106) | ||
|---|---|---|---|---|---|
| F | M | ||||
| HIV | 65 | 20 | 45 | 51.81 ± 8.44 | 1.26 ± 0.12 |
| Matched CTRL | 65 | 20 | 45 | 51.76 ± 8.44 | 1.26 ± 0.13 |
| CF CTRL | 180 | 102 | 78 | 43.36 ± 18.92 | 1.26 ± 0.13 |
Figure 2:
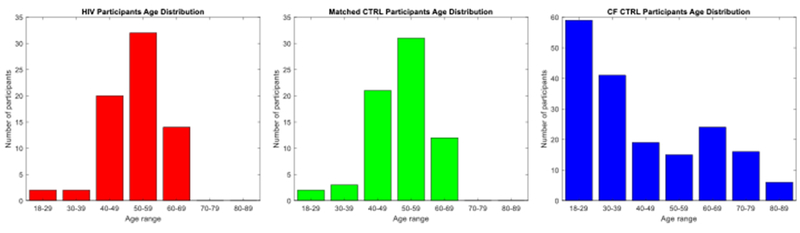
Age distribution of the participants: HIV (left), Matched CTRL (middle), and CF CTRL (right).
2.2. Structural MRI Data Acquisition
Imaging data are acquired from each participant on a 3T General Electric (GE) SIGNA HDx system using an 8-channel Array Spatial Sensitivity Encoding Technique (ASSET) coil for parallel and accelerated imaging. Furthermore, Inversion Recovery-SPoiled Gradient Recalled (IR-SPGR) echo sequence (TR=7.068ms, TI=300ms, TE = 2.208ms, flip angle=15°, matrix=256 × 256, slice dimensions=1.2 × 0.9375 × 0.9375mm, 124 slices) are collected in the sagittal plane.
2.3. MRI Data Preprocessing and Feature Extraction
Preprocessing of the T1-weighted (T1w) MR images involves noise removal [36], computing signal-to-noise ratio (SNR) [37] and correcting field inhomogeneity via N4ITK (Version 2.1.0) [38]. Next, the brain mask is segmented by majority voting [39] across maps extracted by FSL BET (Version 5.0.6) [40], AFNI 3dSkullStrip (Version AFNI_011_12_21_1014) [41], FreeSurfer mri-gcut (Version 5.3.0) [42], and the Robust Brain Extraction (ROBEX) method (Version 1.2) [43], applied to bias and non-bias corrected T1w images. The refined mask is then used to repeat image inhomogeneity correction.
We further apply the cross-sectional approach of FreeSurfer (Version 5.3.0) [44, 45] to the skull-stripped T1w MRI of each subject in order to measure the mean curvature (MeanCurv), surface area (SurfArea), gray matter volume (GrayVol), and average thickness (ThickAvg) of 34 bilateral cortical Regions Of Interest (ROIs) [2 hemispheres × 4 measurement types × 34 ROIs = 272], the volumes of 8 bilateral sub-cortical ROIs (i.e., thalamus, caudate, putamen, pallidum, hippocampus, amygdala, accumbens, cerebellar cortex) [2 × 8 = 16], the volumes of 5 subregions of the corpus callosum (posterior, mid-posterior, central, mid-central and anterior), and the combined volume of all white matter hypointensities [5 + 1 = 6]. White matter hypointensities are defined according to Freesurfer as voxels inside the white matter with signal intensities lower than a threshold level [46]. Finally, volumes of the left and right lateral and third ventricles [2 × 2 = 4] are measured by non-rigidly aligning the SRI24 atlas [47] to the T1w MRI of the subject via ANTS (Version: 2.1.0) [48]. This procedure thus extracts 298 measures from each brain MRI.
For the entire matched data set, each of these 298 brain measures are normalized using their z-scores [49]. To avoid using any data for testing the model, the z-scores are parameterized by computing the mean and standard deviations of measurements across the CF CTRL cohort. Based on this distribution, the z-scores are then computed for each subject of the matched CTRL and HIV groups. Furthermore, the segmentations are used to compute the supratentorial volume (svol) for each subject. As in [50], svol is used to approximate brain size.
2.4. Confounding Factors
For each of the 298 measures, we compute the Pearson correlation between the corresponding z-scores of the 180 subjects of the CF CTRL group and the factors, i.e., age, sex, svol, race, and SNR. Some of the measures are significantly correlated with age, sex, and svol (p-value < 0.05). For each measurement, a general linear model (GLM) [51] is parameterized with respect to corresponding z-scores to omit the effect of the confounding factors. Specifically, for each image measure m ∈ {1,…, 298}, the following GLM is fit across the subjects i ∈ {1,…, 180} of the CF CTRL group with the corresponding z-score as the observation and age (), sex (), and svol () as the confounding factors:
| (1) |
After obtaining the optimal regression coefficients () across all subjects, the model is applied to the HIV and matched CTRL dataset. Specifically, the residual explained by each subject’s individual confounding factors multiplied by the regression coefficients is removed from the initial observation, i.e., the residual scores defined as
| (2) |
2.5. Pattern Extraction and Classification
In this section, the proposed Chained-Regularization technique is outlined. For the interested reader, Appendix A derives the Chained-Regularization approach in detail. Based on the residual scores of the matched data set, the accuracy of the proposed Chained-Regularization framework (denoted as ℓ2,1-ℓ2-reg; see also Figure 1) in correctly labeling HIV infected and health control subjects is measured via 10-fold (nested) cross-validation (see Figure 4). With respect to each (testing) fold, the training of ℓ2,1-ℓ2-reg on the remaining data starts with the Selection Step, i.e., extract the informative pattern for classifying samples. The training then proceeds with the Reweighing Step, i.e., finding the optimal parameterization of the classifier based on that pattern. On the testing fold, we record the labeling of subjects according to the trained ℓ2,1-ℓ2-reg. This procedure between training and testing is repeated until the labeling across all 10 testing folds are generated. Based on those labelings, we compute the Accuracy of prediction (i.e., the percentage of the testing subjects that are classified correctly into their respective classes), specificity (SPE), sensitivity (SEN) and area under the receiver operating characteristic (ROC) curve (AUC). Note, our MKL-based mapping function outputs a continuous value (more details in Appendix A) from which a binary class label is derived via thresholding. By changing the threshold, we can create the ROC curve and hence calculate the AUCs. In addition, we apply the Fisher’s exact test [52] to ensure that implementation is significantly better than chance (p-value < 0.01). The remainder of this section provides further details about training of ℓ2,1-ℓ2-reg.
Figure 4:
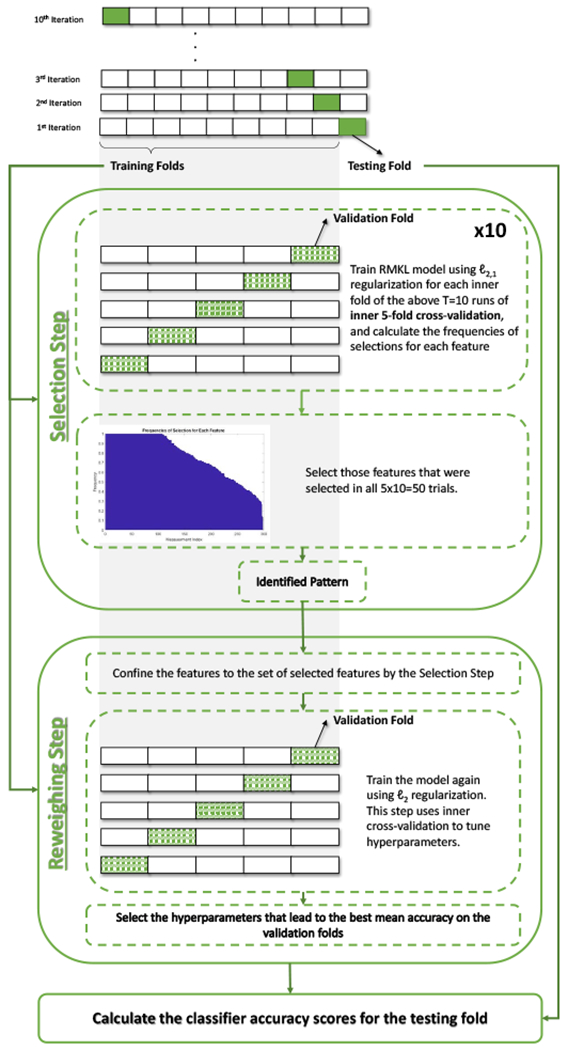
Illustration of the nested cross-validation strategy used in Chained-Regularization (ℓ2,1-ℓ2-reg). On the ith training iteration, the Selection Step selects the most informative measurements(i.e., the pattern) using ℓ2,1-regularization, and then the Reweighing Step uses that pattern to build the classifier with ℓ2-regularization. In the second step, inner cross-validation is used to choose the model hyperparameters. Next, the built classifier is used to calculate the accuracy scores on the corresponding testing fold (say Acci). The average accuracy for all folds is then reported (i.e., ).
Inspired by [18, 13], the HIV specific pattern, identified in the Selection Step during training, is defined by the optimal ‘weight’ vector specifying a linear multivariate model defined by image measurements that correctly label subjects according to the MKL model. MKL classifies samples by learning the optimal pairings between kernels and image measurements. Finding the optimal pairing is described as a minimization problem with respect to a weight vector, sparsity of which specifies the importance of pairings for class separation. We use 7 different kernels to build our multiple kernel learning model, including 3 kernel types [linear, histogram intersection kernel (HIK), and redial basis function kernel (RBF)] with different settings of their hyperparameters. These 7 kernels are defined in detail in Appendix A. Specific to our implementation, the optimal weight vector minimizes a cost function measuring classification accuracy and ‘group-sparsity’ associated with those weights. As also shown in Figure 3, group-sparsity is measured by first transforming the weight vector into a matrix so that each column represents a group and each group combines the weights associated with measurements from the same type and region (regardless of hemisphere). ℓ2,1-norm is then applied to the matrix, i.e., the ℓ2-norm is applied to each column resulting in the column being reduced to a scalar value and then the ℓ1-norm is applied to the vector of those scalar values resulting in the entire matrix being reduced to a scalar value. Note, this computation generally penalizes weight vectors that select a larger number of groups, i.e., are not sparse on a group level.
Figure 3:
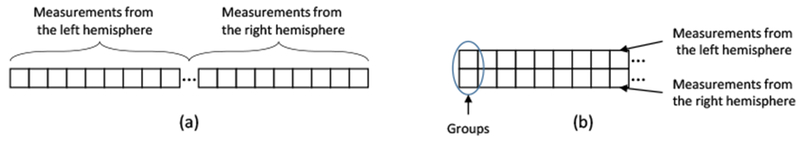
Illustration of feature grouping for group sparsity. (a) Regular sparsity (ℓ1-norm) operates on a vector that concatenates the measurements from the left and right hemispheres. (b) Group sparsity operates on the matrix formed by putting the features from the same ROIs of the left and right hemispheres in its columns.
The optimal ‘weight’ vector now depends on the weight C of the term measuring classification accuracy and the weight λ of the term measuring group sparsity within the MKL cost function (refer to Appendix A for more details). As in [13, 53], the search space for those two hyperparameters is {10−3, 10−2, 10−1, 1, 101, 102, 103}. To identify the best hyperparameter setting, we perform 5-fold inner cross-validation 10 times. Each time, we randomly divide the training data into 5 validation folds. For each validation fold, we first train our implementation of MKL with respect to a specific hyperparameter setting on the remaining training data. For that setting, we then record the accuracy of the implementation on the validation fold and the identified pattern, i.e., regional scores associated with non-zero weights. We repeat this process for each hyperparameter setting and then only keep the pattern that is associated with the highest validation accuracy across all parameter settings. Repeating this process for the remaining 4 validation folds and 9 more inner-cross validations then results in a total of 50 ‘trials’. The Selection Step then defines the HIV specific pattern as the set of residual scores that were part of all 50 trial patterns. This multi-trial selection process is considered more robust than only relying on single run of a sparse classifier [18, 23].
The Reweighing Step focuses on improving MKL’s classification accuracy when only relying on the residual scores of the HIV specific pattern. As training of the classifier is now confined to only informative image measurements, classification accuracy is generally improved by replacing the ℓ2,1-norm with the ℓ2-norm in the cost function of the MKL implementation. The Reweighing Step then performs parameter exploration of this MKL implementation via 5 fold inner cross-validation, i.e., it records the hyperparameter setting that leads to the highest average validation accuracy across the 5 inner folds. The training of ℓ2,1-ℓ2-reg is completed by training MKL with the selected hyperparameter setting on the complete training data. Note, choosing the optimal hyperparameters without including any data from the testing fold yields more reliable and reproducible results [54] than tuning the hyperparameters without any inner validation folds.
The group sparsity (in the Selection Step) guarantees ‘bilateral selection’ of each type of ROI-specific measurement (i.e., measurements on both left and right hemispheres are selected or neither one of them). The Reweighing Step then builds the final classifier relying on all selected individual measurements and the ℓ2-norm, which generates non-sparse classifiers that generalize well to unseen testing data [17, 55]. For the interested reader, Appendix A derives the Chained-Regularization approach in detail. Specifically, we first generalize the MKL approach of [13], which was specific to ℓ1-norm regularization, to regularizers that are convex and differentiable in . We then embed that approach into the proposed Chained-Regularization framework.
2.6. Alternative Implementations
To motivate the specific implementation of the Chained-Regularization approach, the nested cross-validation of Chained-Regularization is repeated with different combinations of regularizers, i.e., using ℓ1-norm in the Selection Step and ℓ2-norm in the Reweighing Step (denoted by ℓ1-ℓ2-reg), using ℓ2,1-norm in the Selection Step and ℓ1-norm in the Reweighing Step (denoted by ℓ2,1-ℓ1-reg), and using ℓ2-norm in both steps (denoted by ℓ2-ℓ2-reg). In addition, the comparison includes an implicit model for the grouping of the ROI measurements by computing the average value of each group and then using the ℓ1-norm in the Selection Step and ℓ2-norm in the Reweighing Step (denoted as Avg ℓ1-ℓ2-reg). To demonstrate the advantages of Chained-Regularization, only the MKL approach is cross-validated, i.e., omitting the Reweighing Step as well as the repeated selection procedure. The corresponding Single-Step Regularization approaches are denoted as ℓ1-reg, ℓ2-reg and ℓ2,1-reg. Note, we omitted certain alternative implementations from the experimental setup to keep the comparison concise and informative. For example, one could implement Chained-Regularization using the ℓ1-norm in both steps. While this implementation produces similar accuracy score as ℓ2,1-ℓ1-reg, the approach most likely underestimates the impact of the disease on a small number of brain regions; a risk generally associated with sparse classifiers based on the ℓ1-norm [56]. Furthermore, note that training a MKL without regularization, constraint or a penalty term (in the reweighing step) is not feasible as the underlying minimization problem is then underdetermined [18], i.e., results in an unstable classifier.
In addition to variations of Chained-Regularization, the comparison includes conventional support vector machine (SVM) classifiers widely used in neuroimaging applications to highlight the benefits of Chained-Regularization in the context of MKL. The class of alternative SVM classifiers include linear SVM, SparseSVM [57], and sparse feature selection [20] followed by a linear SVM (SFS+SVM). In addition, t-test [20], elastic-net [24], and the mutual information based feature selector minimum-redundancy maximum-relevancy (mRMR) [58] are coupled with a linear SVM classifier to further evaluate the performance of the proposed feature selection technique.
For each implementation, the accuracy scores of the previous section are computed. We also apply the DeLong test [59] to mark implementations that are significantly worse (p-value < 0.01) than the proposed ℓ2,1-ℓ1-reg.
3. Results
3.1. Comparison
Classification results of the proposed and alternative methods are summarized in Table 2. The proposed Chained-Regularization technique (ℓ2,1-ℓ2-reg) achieves the highest Accuracy (82.3%), SEN (0.84), and AUC (0.87). The SPE (0.82) is equivalent to ‘ℓ1-ℓ2-reg’ and ‘Avg ℓ1-ℓ2-reg’. All other implementations of the comparison (including ℓ2-ℓ2-reg and ℓ2,1-ℓ1-reg) not only received lower scores, but were also significantly worse than the proposed chained ℓ2,1-ℓ2 regularization. The single step regularizers received higher scores in all four performance measures than the conventional approaches with the exception of SFS+SVM. The performance scores of SFS+SVM (Accuracy: 0.69%, SPE: 0.69, SEN: 0.70 and AUC: 0.73) were higher than those of ℓ2-reg and ℓ2,1-reg but lower than ℓ1-reg (Accuracy: 70.3%, SPE: 0.70, SEN: 0.70, AUC: 0.73), the single step regularization with the highest Accuracy and AUC. Finally, only conventional methods (i.e., t-test+SVM, mRMR+SVM, SparseSVM and SVM) produced classification results that were not significantly better than chance.
Table 2:
Classification results of different approaches summarized by Accuracy, specificity (SPE), sensitivity (SEN) and area under the ROC curve (AUC). The best score in each category is in bold. Methods are marked with †, if they were significantly worse than the proposed approach (p < 0.01 according to Delong’s Test [59]). Methods marked with ‡ are significantly better than chance (p < 0.01 according to the Fisher exact test [52]).
| Method | Accuracy (%) | SPE | SEN | AUC | |
|---|---|---|---|---|---|
| Proposed | ℓ2,1-ℓ2-reg‡ | 82.3 | 0.82 | 0.84 | 0.87 |
| Chained (Baseline) | ℓ1-ℓ2-reg‡ | 81.9 | 0.82 | 0.79 | 0.86 |
| Avg ℓ1-ℓ2-reg‡ | 79.7 | 0.82 | 0.77 | 0.85 | |
| ℓ2-ℓ2-reg†‡ | 73.1 | 0.74 | 0.73 | 0.76 | |
| ℓ2,1ℓ1-reg†‡ | 72.5 | 0.72 | 0.73 | 0.76 | |
| Single Step Regularization | ℓ1-reg†‡ | 70.3 | 0.70 | 0.70 | 0.75 |
| ℓ2,1-reg†‡ | 69.7 | 0.70 | 0.68 | 0.73 | |
| ℓ2-reg†‡ | 68.7 | 0.64 | 0.70 | 0.71 | |
| Conventional Methods | SFS [20]+SVM†‡ | 69.9 | 0.69 | 0.70 | 0.73 |
| elastic-net [24]+SVM†‡ | 65.1 | 0.64 | 0.64 | 0.69 | |
| t-test [20]+SVM† | 59.1 | 0.61 | 0.56 | 0.65 | |
| mRMR [58]+SVM† | 59.6 | 0.56 | 0.61 | 0.64 | |
| SparseSVM [57]† | 57.9 | 0.55 | 0.60 | 0.64 | |
| SVM† | 56.7 | 0.57 | 0.56 | 0.60 | |
3.2. The HIV Pattern
For ℓ2,1-ℓ2-reg (the most accurate approach in the comparison), Figure 5 shows the frequencies (normalized in the range [1]) of the 298 image measurements selected by the Selection Step across the 10 runs of cross-validation on the whole matched data set considered for identifying the pattern. This figure shows the measurements with a selection frequency of 1 (selected all times), i.e., those that are actually used in the Reweighing Step of our method, with colors based on their measurement types. Note, the ordering of measurement types is arbitrary. We refer to this set of measurements as the HIV pattern. The remaining measurements are displayed in gray regardless of the type of measurement. Approximately 39% of all image measurements are selected in all runs. These measures define the HIV pattern.
Figure 5:
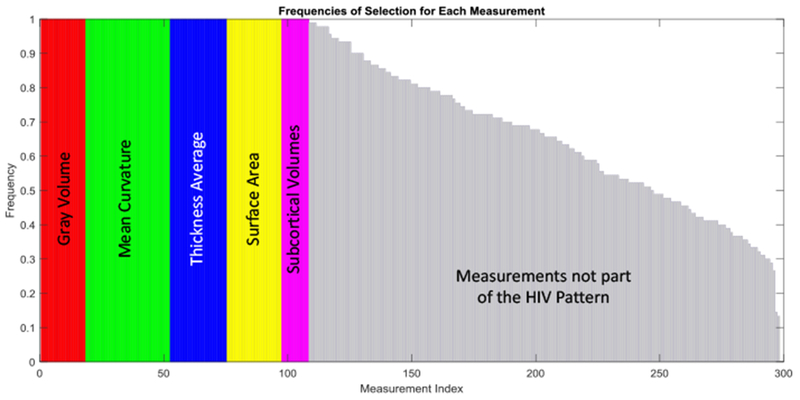
Frequencies of selection for each of the 298 features. Colors encode measurement types. The measurements in gray are those ignored in the Reweighing step.
To analyze the significance of each type of measurement (Mean Curvature, Surface Area, Gray Matter Volume, Average Thickness, and Subcortical ROI Volumes), we first create a baseline for comparison by performing 10-fold cross validation just on the Reweighing Step with the scores being confined to the HIV pattern, recording the testing accuracy for each fold, and then computing the mean and standard deviation in the accuracy score across all 10 folds. The results in an accuracy of 87.69% ± 1.69 (mean ± standard deviation), which we refer to as ‘All Measurements’ in Table 3. For each measurement type, we then omit the corresponding measures from the data, perform 10-fold cross-validation of the Selection Step on this subset of data, record the pattern, and repeat the previous cross-validation of the Reweighing Step with respect to that pattern.
Table 3:
The mean±standard of the classification Accuracy and area under the ROC curve (AUC) of the proposed method with different subsets of the 298 measurements. Entries marked with f are significantly worse (p < 0.01; Delong’s Test) compared to ‘All Measurements’.
| Method | Accuracy (%) | AUC |
|---|---|---|
| All Measurements | 87.7 ± 1.69 | 0.87 ± 0.03 |
| No Average Thickness | 79.6 ± 1.96 | 0.78 ± 0.07 |
| No Mean Curvature† | 77.2 ± 1.92 | 0.84 ± 0.05 |
| No Surface Area† | 73.9 ± 1.77 | 0.79 ± 0.03 |
| No Gray Matter Volume† | 66.8 ± 1.49 | 0.74 ± 0.06 |
| Only Cortical Measurements† | 69.2 ± 2.10 | 0.75 ± 0.02 |
With respect to using subsets of the measurements, omitting Average Thickness from the data resulted in the pattern with the highest mean accuracy score (79.6% ± 1.96). Omitting Mean Curvature, Surface Area, or Volume from the HIV pattern resulted in accuracy scores that were significantly lower than those produced by All Measurements (or the HIV pattern). The same was true when confining classification to cortical gray matter volumes.
Beyond the type of measurements, Table 4 lists and Figure 6 visualizes the selected cortical regions. 35% of all cortical measurements are selected by our method. Furthermore, a total of 52% of the subcortical measurements are selected. Figure 7 shows the selected subcortical regions (i.e., hippocampus, amygdala, accumbens and cerebellar cortex) along with the white matter structures (i.e., corpus callosum posterior and mid-posterior) selected by our approach. In addition to these ROIs, hypointensity lesion volumes are also selected. Note, as also argued in [8], the coefficients computed by sparse classifiers simply parameterize a linear multivariate model (explained in detail in Appendix A), which predicts the class labels. Thus, coefficients are informative with respect to feature selection but are not good indicators for differentiating the importance among the selected features with respect to identifying cohorts.
Table 4:
Cortical surface ROIs and their measurement types selected by our method.
| ROI | GrayVol | MeanCurv | ThickAvg | SurfArea |
|---|---|---|---|---|
| Bankssts | ✓ | ✓ | ||
| Caudalanteriorcingulate | ✓ | ✓ | ✓ | |
| Caudalmiddlefrontal | ✓ | ✓ | ✓ | |
| Cuneus | ||||
| Entorhinal | ✓ | |||
| Fusiform | ✓ | ✓ | ✓ | |
| Inferiorparietal | ✓ | |||
| Inferiortemporal | ✓ | |||
| Isthmuscingulate | ✓ | ✓ | ✓ | |
| Lateraloccipital | ✓ | |||
| Lateralorbitofrontal | ✓ | ✓ | ||
| Lingual | ||||
| Medialorbitofrontal | ||||
| Middletemporal | ✓ | ✓ | ||
| Parahippocampal | ✓ | ✓ | ✓ | |
| Paracentral | ✓ | ✓ | ✓ | ✓ |
| Parsopercularis | ✓ | ✓ | ||
| Parsorbitalis | ✓ | |||
| Parstriangularis | ✓ | |||
| Pericalcarine | ✓ | ✓ | ||
| Postcentral | ✓ | |||
| Posteriorcingulate | ✓ | ✓ | ✓ | |
| Precentral | ✓ | ✓ | ✓ | |
| Precuneus | ✓ | ✓ | ||
| Rostralanteriorcingulate | ✓ | ✓ | ||
| Rostralmiddlefrontal | ✓ | ✓ | ✓ | |
| Superiorfrontal | ✓ | ✓ | ||
| Superiorparietal | ✓ | ✓ | ✓ | |
| Superiortemporal | ✓ | |||
| Supramarginal | ||||
| Frontalpole | ✓ | |||
| Temporalpole | ✓ | |||
| Transversetemporal | ✓ | ✓ | ✓ | |
| Insula | ✓ | ✓ | ✓ |
Figure 6:
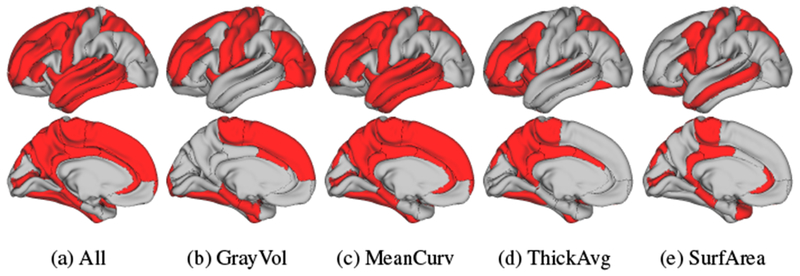
cortical ROIs selected by our proposed approach. (b-e) show the selected ROIs for each measurement type separately, while (a) visualizes the union of the four types.
Figure 7:
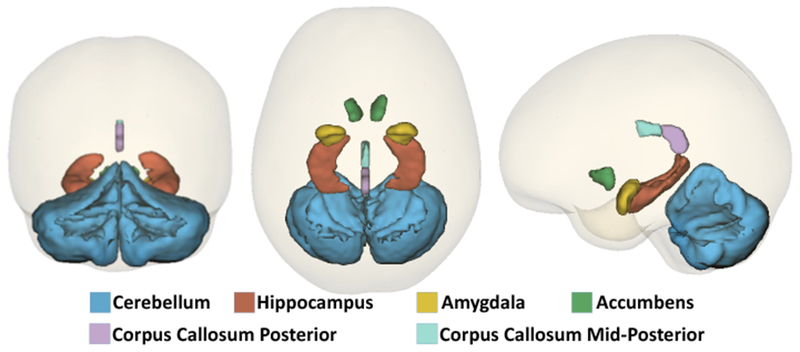
Subcortical ROIs and white matter structures selected by our method.
The comparison of different approaches (see Table 2) revealed that our Chained-Regularization approach was significantly better than confining analysis to any one single step (i.e., ℓ2,1-reg or ℓ2-reg). Our approach was also significantly better than alternative implementations of the Chained-Regularization that used the same type of regularizer for both steps (i.e., sparse regularizer (ℓ2,1-ℓ1-reg) or Euclidean regularizer (ℓ2-ℓ2-reg)). This finding underlines the importance of selecting two regularizers that complement each other for our approach, i.e., the first regularizer models the selection of the measurements, while the second one reweighs the influence of the selected measurements in order to improve the classification accuracy.
Choosing alternative complementary regularizers by replacing the ℓ2,1-norm in the Selection Step with other sparse regularizers (while leaving the Reweighing Step unchanged) results in non-significantly lower accuracy scores compared with the proposed approach. Unlike other implementations of the Chained-Regularization, our proposed ℓ2,1 regularizer was the only one that explicitly modeled the bilateral effect of HIV on the brain by grouping measurements across hemispheres. This additional modelling constraint simplifies the classification task and results in higher performance scores.
With the exception of SFS+SVM (which is also significantly less accurate than our proposed method), the worst performing methods are common (two-step) approaches that used different methods for feature selection and classification. Such approaches view pattern identification and classification as two disconnected machine learning tasks [20, 60, 61]. Thus, the optimal pattern identified in the first step is generally not optimal for the classifier in the second step, which would explain the low accuracy scores.
All implementations led to SPE, SEN, and AUC values similar to their Accuracy score (which is being maximized). This concurrence emerged because our data set is well balanced between the two cohorts. The residual scores of the imaging measures further minimizes the risk of biasing analysis towards one cohort.
Note that ℓ2,1-ℓ2-reg iteratively runs several trials of nested cross-validation in the Selection Step for reliable selection of the relevant features (i.e., the HIV pattern). However, training based on this procedure is computational expensive, the training time is insignificant in comparison with the years it took to acquire the data. With the implementations done only on a single computing core of a machine with an Intel® Core™ i7-4712HQ CPU @ 2.30 GHz with 16 Gigabytes of memory, using Matlab R2017a, it took approximately 6 hours to train the model and search all possible settings for the hyperparameters and tune them. Note, our implementation was not optimized and therefore computation times may be improved. Furthermore, the training is done only once, after which the model parameters are saved and run on test data. The testing time of ℓ2,1-ℓ2-reg is less than 0.01 second, which is similar to the other implementations of this comparison. Note that the increase in the running time of our method (compared to single-step methods) is mainly due to the constant number of trials that we repeat the method to get a more robust pattern selected, i.e., the increase in the running time is not exponential to the number of subjects or measurements. Therefore, the method is scalable for larger number of inputs. However, if the number of measurements dramatically increases, the approach faces the so-called ‘Small-Sample-Size’ problem, a common issue in machine learning [62]. This problem arises when the number available samples (N) is far fewer than the number of features (d) extracted from them (i.e., N << d). Under these settings, all machine learning and pattern recognition methods fail to identify the intrinsic space of the samples.
The HIV pattern identified by the proposed Chained-Regularization technique composes of approximately 39% of the 298 measurements that were selected in all 50 training runs (see Figure 5). This frequent selection of such a large number of measurements does not contradict the sparsity constraint of MKL but rather is due to grouping of measurement and the accounting done by the Chained-Regularization approach. Classifiers relying on group sparsity (ℓ2,1) tend to select more measurements than those relying solely on sparsity (ℓ1 [27]). Furthermore, our method marks a measurement as informative if it is selected by MKL at least once in connection with one of the 7 kernels (i.e., Linear, HIK, and RBF with 5 instances of its hyperparamter setting). That those kernel-measurement pairs are actually sparsely selected by MKL is shown in Figure 8, which lists each pair separately. By doing so, our Chained-Regularization approach avoids underestimating the impact of the disease to a small region of the brain as commonly done by sparse classifiers [56].
Figure 8:
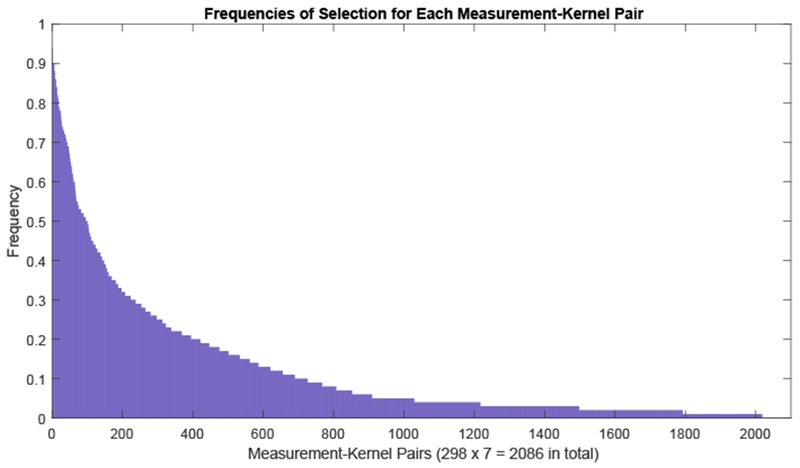
Frequencies of selection for each of the measurement-subkernel pair. Note that 298 brain measurements are used, together with 7 different subkernels resulting in 2098 total pairs.
As indicated by Table 4, combining the four different types of measurements used in our analysis is essential for creating a highly accurate HIV pattern. Of all measurement types, the Mean Curvature is the most frequent measurement type present in the pattern. However, when performing classification without the Mean Curvature (see Table 3), the drop in accuracy is less then when omitting Surface Area and Gray Matter Volume scores. The opposite is true for the regional gray matter volumes, which are least often selected, but whose omission from the pattern results in the largest drop in accuracy. This observation acknowledges that the number of times any type of measurement is part of a selected pattern does not necessarily indicate how important that type is for characterizing the disease.
4. Discussion
The measurements composing the HIV pattern identified by the most accurate approach, the proposed Chained-Regularization, are in agreement with the literature, which suggests that HIV infection is associated with volume deficits in cortical, subcortical, and white matter regions [63, 64, 65, 66, 67, 68, 69, 70, 71, 72]. As identified using our automated, machine learning method (Table 4), the literature indicates that cortical areas affected in HIV relative to healthy controls include frontal, cingulate, sensorimotor, and parietal regions [9, 73, 74, 75, 76, 77]. For other cortical regions identified herein, reports on the effects of HIV are relatively less common: [9] lists temporoparietal regions; [71, 78] include effects of HIV on thinning of the temporal cortices; [72] describes effects on insula; [79] lists parahippocampal cortex. That our methods identified regions not commonly reported in the HIV literature (e.g., Caudalanteriorcingulate, Isthmuscingulate, Lateralorbitalfrontal, Parsopercularis, and Frontalpole) may be due to our inclusion of cortical measures such as mean curvature, thickness, and surface area, which are not typical metrics used in the HIV literature. Instead, the imaging literature usually focuses on the effects of HIV on gray matter volume (see the following for exceptions [80, 81, 82]). Indeed, in studies that assess cortical thickness rather than cortical volume, HIV effects are evident in areas such as the insula, orbitofrontal, temporal, and cingulate cortices [78, 83], similar to the ones identified here.
As also confirmed by our results, white matter is notably affected by HIV infection. Damage to myelin sheathes may be reflected in lower than normal white matter volume and greater prevalence of white matter hyperintensities [84] (deemed “hypointensities” by FreeSurfer [44]). Indeed, examination of brain microstructural integrity using DTI has detected subtle HIV-related differences from controls (e.g., lower fractional anisotropy and higher mean diffusivity) in myelin and axonal integrity [85, 86, 87, 88, 89, 90], even in normal-appearing white matter [91, 92, 93].
Subcortical regions frequently reported in the literature to have significantly smaller volumes in HIV subjects relative to healthy controls include hippocampus and basal ganglia structures [64, 94, 95, 96, 4]. Regarding the basal ganglia and limbic structures, our results specifically identify the accumbens and amygdala, whereas the literature more frequently cites the caudate, putamen, and pallidum (e.g., [65, 94, 95, 96]).
Although our approach does not identify the thalamus, a structure as particularly susceptible to HIV despite other reports (e.g., [9, 83, 4, 6]), our scheme does note cerebellum as a significant contributor to diagnosis differences. This inclusion is consistent with several other studies report HIV-related gray matter volume deficits in the cerebellum [68, 69, 97]. The functional consequences of HIV effects on the cerebellum have been reported [98]; yet the cerebellum is generally underappreciated in the imaging literature as common analysis methods are designed with the neocortex in mind and may be suboptimal for the analysis of the cerebellum.
One of the main limitations of the proposed study is the large imbalance between the number of HIV patients and CTRL subjects. We addressed this issue by matching (and hence balancing) a subset of the CTRLs to the HIV group. However, this greatly reduced the number of samples used for extracting the pattern and testing, and thus the power of the analysis. To preserve the power of the provided data, expanding Chained-Regularization for explicit modeling of the imbalance between the cohorts can be a direction for future work.
5. Conclusion
We presented Chained-Regularization, a two-step-approach to identifying disease-specific patterns and performing pattern-based classification that, unlike the state-of the art, uses the same classification model for first identifying informative measures and then improving the accuracy of the classification based on the selected measures. Our choice of classification approach was a generalized version of the MKL method proposed by [13]. In the Selection Step, parameterization of MKL was confined by groups sparsity (ℓ2,1-norm) and in the Reweighing Step, the parameterization was penalized by the Euclidean (ℓ2-norm) regularization. This implementation was more accurate than alternative implementations and significantly better than common (two-step) approaches using different methods for feature selection and classification.
The Chained-Regularization approach identified a number of brain regions comporting with the literature and designated a few novel regions that (to our knowledge) have not been previously described in the HIV literature. These regions would benefit from further investigation as an improved understanding of the diseases remains critical for advancing the long-term care for the large number of HIV infected patients, who (even with suppressed viral loads) can suffer from cognitive disorders associated with HIV. Our current contribution in improving this understanding is in providing an automated, impartial approach for identifying key brain regions implicated in HIV infection.
Acknowledgment
This research was supported in part by the NIH grants U01 AA017347, R37-AA010723, K05-AA017168, K23-AG032872, K24-MH098759, and R01-MH113406. It was also supported by the Creative and Novel Ideas in HIV Research (CNIHR) Program through a supplement to the University of Alabama at Birmingham (UAB) Center For AIDS Research funding (NIH P30 AI027767). This funding was made possible by collaborative efforts of the Office of AIDS Research, the National Institute of Allergy and Infectious Diseases, and the International AIDS Society.
Appendix A. Multiple Kernel Learning for Feature Selection and Classification
Table A.5:
Notations: Note that throughout this paper, we refer to matrices with bold capital letters (e.g., A), vectors with small bold letters (e.g., a), and scalars or functions with all non-bold letters. is the scalar in row i and column j of A, while ai the ith row and aj the jth column of A.
| Notation | Description |
|---|---|
| N | Number of training samples |
| d | Dimensionality of the feature vectors |
| d′ | The dimensionality of the selected features set |
| Feature matrix of all samples | |
| The class labels for each of the samples | |
| The new reduced feature matrix, after feature selection | |
| k(x, xn) | Subkernel function between the two samples x and xn |
| α | Weights vector learned to aggregate subkernels into a kernel |
| k(x, xn, α) | Aggregate kernel of the two samples x and xn, using weights α |
| ∥a∥1 | The ℓ1 norm of vector a (i.e., ) |
| ∥a∥2 | The ℓ2 norm of vector a (i.e., ) |
| ∥A∥2,1 | The ℓ2,1 norm of the matrix A (i.e., ) |
| The set of non-negative real numbers |
The MKL of [13, 18] classifies samples by learning the optimal pairings between kernels and image measurements. Finding the optimal paring is described as a minimization problem with respect to a weight vector (denoted α), sparsity of which specifies the importance of pairings for class separation. To make the minimization problem tractable, the search for the optimal weight vector is constrained by a regularization term . The minimization problem is furthermore characterized by a prediction function f (x, α) that maps the image measurements x of a sample to a label or cohort (i.e., y). The max-margin term (or for short) then measures the distance between the ‘support vectors’ of the classes (i.e., HIV and CTRL) as defined by f(·, ·) (see [53, 99] for detailed definition and see Table A.5 for the notations). Introducing the loss function L(y, f(x, α)) for measuring the difference between the predicted and actual label of a sample, the final term of the minimization problem computes that difference across all training samples, i.e., . Thus, the regularized MKL approach is completely defined by
| (A.1) |
where C and λ are trade-off hyperparameters, the constraint α ≥ 0 is needed to efficiently solve the minimization problem (similar to [13, 18]), and is a Reproducing Kernel Hilbert Space (RKHS) [100]. Note that α ≥ 0 guarantees that the search for the optimal parameters is done in the space of non-negative values, in which we can define flexible (convex and smooth) regularization functions. The regularizers ℓ2,1 and ℓ1 are only smooth in the domain of non-negative values [18, 27]. For more details, refer to Section Appendix A.1.
In the above objective, function f is expressed in terms of the aggregated kernel function k(·, ·, α), the weight wn of a training samples ‘n’ in the decision process, and bias parameter b [101, 53]1:
| (A.2) |
As shown in Figure A.9 (and [13]), the aggregated kernel function k(·,·, α) applies a set of subkernels {k1 (·, ·),…, kκ(·, ·)} to each single residual score and then computes a weighted average across all subkernels and residual scores with the weight defined by α, i.e.,
| (A.3) |
An efficient solution to Eq. (A.1) requires the subkernels to be positive semidefinite (PSD), which is a common constraint for kernel methods [101]. Note, that any linear combination (with non-negative coefficients) of PSD subkernels also results in PSD kernel (as in Eq. (A.3)). For our specific application, we choose three types of subkernels. The first one is a Linear (LIN) kernel, which is one of the simplest and most widely used kernels in machine learning:
| (A.4) |
As an alternative to the linear kernel, k(·, ·, ·) also includes the histogram intersection kernel (HIK) [102], a non-linear kernel popular for non-negative features. This kernel is applied to the absolute values of the residuals in X (as in Eq. (2)), i.e.,
| (A.5) |
Finally, the implementation includes several instances of the Radial Basis Function (RBF) or the Gaussian kernel [103], a popular, non-linear kernel that depends on the kernel hyperparameter σ:
| (A.6) |
This kernel can be built by different values of its hyperparameter, σ. We build several instances of the RBF subkernel with respect to σ ∈ {10−2, 10−1, 1, 10, 102}. Doing so avoids hyperparameter tuning for this kernel, as MKL solves Eq. (A.1) with respect to α to select the pairs of subkernels and residual scores that best fit the data.
Assuming that (which is explicitly defined later) is convex and differentiable for non-negative input values (i.e., α ≥ 0), the solution to Eq. (A.1) can be efficiently determined via Optimize-RMKL(·) (see Algorithm 1). Inspired by [13], “Optimize-RMKL(·)”, iteratively solves the equation based on Block Coordinate Descent [104], i.e., by alternating between optimizing for f and α until convergence. When optimizing for f (with α being fixed), Eq. (A.1) reduces to a SVM that can be solved
Algorithm 1.
“Regularized multiple kernel learning” (RMKL), as in Eq. (A.1).
| Optimize-RMKL (y, X, , C, λ) | |
| Input: Training features X, labels y, the regularization function , and hyperparamters C and λ. | |
| 1: | t ←, α0 = 1 |
| 2: | repeat |
| 3: | ft+1 ← SVMSolver(yk(.,., αt)). |
| 4: | αt+1 ← Solve (A.1) by using f* = ft+1 and regularization , using PGD. |
| 5: | αt+1 ← max(0, α), t ← t + 1. |
| 6: | . |
| 7: | until , or , or t > 100 |
| Output: f*, αt. |
with standard approaches (e.g., LIBSVM [105]). To determine the optimal feature-kernel weights α (with f being fixed), projected gradient decent (PDG) [106] is applied to Eq. (A.1).
Appendix A.1. Chained ℓ2,1,-ℓ2 Regularization
Given the training data as well as the search space {10−3, 10−2, 10−1, 1, 101, 102, 103} for both hyperparameters λ and C of Optimize-RMKL(·), our Chained-Regularization approach (called Chained-Reg; see also Algorithm 2) now makes use of Optimize-RMKL(·) in the Selection Step and the Reweighing Step. In the Selection Step, the regularizer of Eq. (A.1) is defined by group sparsity (i.e., ℓ2,1 norm) so that Optimize-RMKL(·) identifies the kernel-measurement pairs that best distinguish the two cohorts. To explicitly define , we introduce the group matrix , in which each column represents a group according to bilateral dependencies of ROI measurements and the entries of the rows are the elements of α corresponding to those measurements. Then, the first step of the Chained-Regularization defines as
| (A.7) |
Algorithm 2.
Chained ℓ2,1-ℓ2 regularization for joint feature selection & classification.
 |
As mentioned, for the selection process to be reliable, we compute a distribution over the selected features by repeatedly solving Eq. (A.1) and then select residual scores based on that distribution. Specifically, the training data is split into 5 (inner)
Figure A.9:
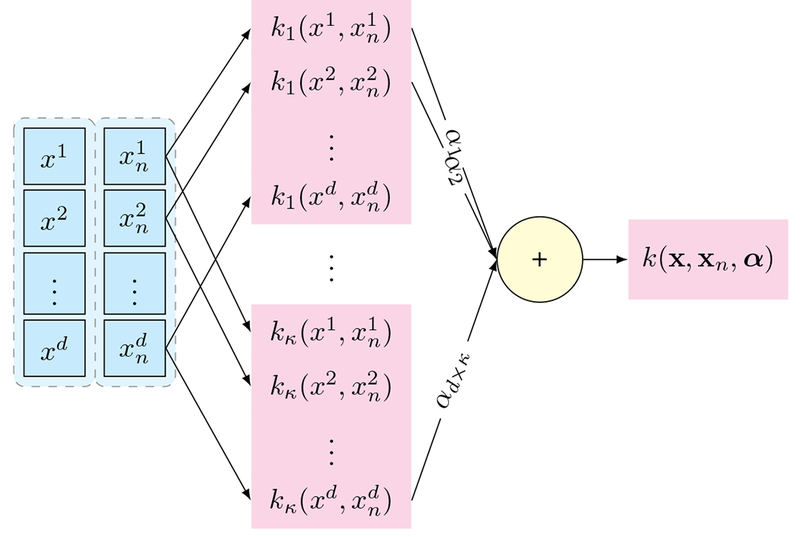
An illustration of computing the kernel for each pair of samples (x and xn), similar to what is presented in Eq. (A.3). The final kernel is computed by a weighted aggregation of κ different kernels applied on each single feature.
folds based on random sampling. For each fold, the approach records the set of selected kernel-feature pairs associated with the most accurate (hyperparameter) setting of Optimize-RMKL(·). The accuracy of a setting is determined by parameterizing Optimize-RMKL(·) accordingly, training the approach on the remaining training data, and applying the resulting implementation to the inner fold. The entire process of splitting the training data and recording the set of selected kernel-feature pairs is repeated 9 more times to produce a total of 50 sets. The features selected in all trials then define the ‘selected measurement matrix’ X’. Note, this conservative threshold minimizes the chance to introduce another hyperparameter that requires tuning. However, one can implement other selection criteria if required by the application.
Given the selected measurements, the Reweighing Step of the Chained-Regularization again applies Optimize-RMKL(·) to solve Eq. (A.1), but now with respect to X’ and the regularizer defined by the Euclidean norm, i.e.,
| (A.8) |
To find the most accurate reweighing α2 of the selected measurements, 5-fold (inner) cross-validation coupled with hyperparameter exploration is performed. The accuracy of each hyperparameter setting is computed by first recording the classification results on the fold not used for training Optimize-RMKL(·) and then averaging those results across all folds. With respect to the most accurate setting of Optimize-RMKL(·), Chained-Regularization returns the kernel function f and corresponding weight vector α2 to define the classifier for the testing data.
We end the description of Chained-Regularization by noting that the proposed approach is not specific to the two norms discussed here. As mentioned, Optimize-RMKL(·) only requires that the chosen norms are convex and differentiable in , which, for example, ℓ1 and ℓ2,1 norms are.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Clifford DB, Ances BM, Hiv-associated neurocognitive disorder, The Lancet infectious diseases 13 (11) (2013) 976–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Heaton RK, Franklin DR, Ellis RJ, McCutchan JA, Letendre SL, LeBlanc S, Corkran SH, Duarte NA, Clifford DB, Woods SP, et al. , Hiv-associated neurocognitive disorders before and during the era of combination antiretroviral therapy: differences in rates, nature, and predictors, Journal of neurovirology 17 (1) (2011) 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fama R, Sullivan EV, Sassoon SA, Pfefferbaum A, Zahr NM, Impairments in component processes of executive function and episodic memory in alcoholism, hiv infection, and hiv infection with alcoholism comorbidity, Alcoholism: Clinical and Experimental Research 40 (12) (2016) 2656–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pfefferbaum A, Rosenbloom MJ, Sassoon SA, Kemper CA, Deresinski S, Rohlfing T, Sullivan EV, Regional brain structural dysmorphology in human immunodeficiency virus infection: effects of acquired immune deficiency syndrome, alcoholism, and age, Biological psychiatry 72 (5) (2012) 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Castelo J, Sherman S, Courtney M, Melrose R, Stern C, Altered hippocampal-prefrontal activation in hiv patients during episodic memory encoding, Neurology 66 (11) (2006) 1688–1695. [DOI] [PubMed] [Google Scholar]
- [6].Fama R, Rosenbloom MJ, Sassoon SA, Rohlfing T, Pfefferbaum A, Sullivan EV, Thalamic volume deficit contributes to procedural and explicit memory impairment in hiv infection with primary alcoholism comorbidity, Brain imaging and behavior 8 (4) (2014) 611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cole JH, Underwood J, Caan MW, De Francesco D, van Zoest RA, Leech R, Wit FW, Portegies P, Geurtsen GJ, Schmand BA, et al. , Increased brain-predicted aging in treated hiv disease, Neurology 88 (14) (2017) 1349–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang Y, Kwon D, Esmaeili-Firidouni P, Pfefferbaum A, Sullivan EV, Javitz H, Valcour V, Pohl KM, Extracting patterns of morphometry distinguishing hiv associated neurodegeneration from mild cognitive impairment via group cardinality constrained classification, Human brain mapping 37 (12) (2016) 4523–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pfefferbaum A, Rogosa DA, Rosenbloom MJ, Chu W, Sassoon SA, Kemper CA, Deresinski S, Rohlfing T, Zahr NM, Sullivan EV, Accelerated aging of selective brain structures in human immunodeficiency virus infection: a controlled, longitudinal magnetic resonance imaging study, Neurobiology of aging 35 (7) (2014) 1755–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pfefferbaum A, Rosenbloom MJ, Rohlfing T, Adalsteinsson E, Kemper CA, Deresinski S, Sullivan EV, Contribution of alcoholism to brain dysmorphology in hiv infection: effects on the ventricles and corpus callosum, Neuroimage 33 (1) (2006) 239–251. [DOI] [PubMed] [Google Scholar]
- [11].Witten IH, Frank E, Hall MA, Pal CJ, Data Mining: Practical machine learning tools and techniques, Morgan Kaufmann, 2016. [Google Scholar]
- [12].Saeys Y, Inza I, Larrañaga P, A review of feature selection techniques in bioinformatics, bioinformatics 23 (19) (2007) 2507–2517. [DOI] [PubMed] [Google Scholar]
- [13].Adeli E, Wu G, Saghafi B, An L, Shi F, Shen D, Kernel-based joint feature selection and max-margin classification for early diagnosis of parkinsons disease, Scientific reports 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Adeli-Mosabbeb E, Thung K-H, An L, Shi F, Shen D, Robust feature-sample linear discriminant analysis for brain disorders diagnosis, in: Neural Information Processing Systems (NIPS), 2015.
- [15].Bron E, Smits M, van Swieten J, Niessen W, Klein S, Feature selection based on svm significance maps for classification of dementia, in: Machine Learning in Medical Imaging, Vol. 8679, 2014, pp. 272–279. [Google Scholar]
- [16].Rosa MJ, Portugal L, Hahn T, Fallgatter AJ, Garrido MI, Shawe-Taylor J, Mourao-Miranda J, Sparse network-based models for patient classification using fmri, Neuroimage 105 (2015) 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ng AY, Feature selection, L1 vs. L2 regularization, and rotational invariance, in: Proceedings of the twenty-first international conference on Machine learning, ACM, 2004, p. 78. [Google Scholar]
- [18].Varma M, Babu BR, More generality in efficient multiple kernel learning, in: ICML, 2009, pp. 1065–1072. [Google Scholar]
- [19].Mwangi B, Tian TS, Soares JC, A review of feature reduction techniques in neuroimaging, Neuroinformatics 12 (2) (2014) 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Guyon I, Elisseeff A, An introduction to variable and feature selection, JMLR 3 (2003) 1157–1182. [Google Scholar]
- [21].Rondina JM, Hahn T, de Oliveira L, Marquand AF, Dresler T, Leitner T, Fallgatter AJ, Shawe-Taylor J, Mourao-Miranda J, SCoRSA method based on stability for feature selection and mapping in neuroimaging, IEEE transactions on medical imaging 33 (1) (2014) 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bron EE, Smits M, Niessen WJ, Klein S, Feature selection based on the svm weight vector for classification of dementia, IEEE journal of biomedical and health informatics 19 (5) (2015) 1617–1626. [DOI] [PubMed] [Google Scholar]
- [23].Nie F, Huang H, Cai X, Ding CH, Efficient and robust feature selection via joint ℓ2,1,-norms minimization, in: Neural Information Processing Systems, 2010, pp. 1813–1821. [Google Scholar]
- [24].Zou H, Hastie T, Regularization and variable selection via the elastic net, J. of Royal Statistical Society: Series B (Statistical Methodology) 67 (2) (2005) 301–320. [Google Scholar]
- [25].Bauer S, Nolte L-P, Reyes M, Fully automatic segmentation of brain tumor images using support vector machine classification in combination with hierarchical conditional random field regularization, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, 2011, pp. 354–361. [DOI] [PubMed] [Google Scholar]
- [26].Elhamifar E, Vidal R, Robust classification using structured sparse representation, in: Computer Vision and Pattern Recognition, 2011.
- [27].Hastie T, Tibshirani R, Wainwright M, Statistical learning with sparsity: the lasso and generalizations, CRC press, 2015. [Google Scholar]
- [28].Wright J, Yang AY, Ganesh A, Sastry SS, Ma Y, Robust face recognition via sparse representation, IEEE Transactions on Pattern Analysis and Machine Intelligence 31 (2) (2009) 210–227. [DOI] [PubMed] [Google Scholar]
- [29].Yuan M, Lin Y, Model selection and estimation in regression with grouped variables, Journal of the Royal Statistical Society: Series B (Statistical Methodology) 68 (1) (2006) 49–67. [Google Scholar]
- [30].Minesaki Y, An efficient conservative integrator with a chain regularization for the few-body problem, The Astronomical Journal 150 (4) (2015) 102. [Google Scholar]
- [31].Gonen M, Alpaydin E, Multiple kernel learning algorithms, Journal of machine learning research 12 (July) (2011) 2211–2268. [Google Scholar]
- [32].Zhu X, Thung K-H, Adeli E, Zhang Y, Shen D, Maximum mean discrepancy based multiple kernel learning for incomplete multimodality neuroimaging data, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, 2017, pp. 72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sonnenburg S, Rätsch G, Schäfer C, Schölkopf B, Large scale multiple kernel learning, Journal of Machine Learning Research 7 (July) (2006) 1531–1565. [Google Scholar]
- [34].Liu F, Zhou L, Shen C, Yin J, Multiple kernel learning in the primal for multimodal alzheimers disease classification, IEEE journal of biomedical and health informatics 18 (3) (2014) 984–990. [DOI] [PubMed] [Google Scholar]
- [35].Karnofsky DA, The clinical evaluation of chemotherapeutic agents in cancer, Evaluation of chemotherapeutic agents (1949). [Google Scholar]
- [36].Coupé P, Yger P, Prima S, Hellier P, Kervrann C, Barillot C, An optimized blockwise nonlocal means denoising filter for 3-d magnetic resonance images, IEEE transactions on medical imaging 27 (4) (2008) 425–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cosman PC, Gray RM, Olshen RA, Evaluating quality of compressed medical images: Snr, subjective rating, and diagnostic accuracy, Proceedings of the IEEE 82 (6) (1994) 919–932. [Google Scholar]
- [38].Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC, N4ITK: improved n3 bias correction, IEEE transactions on medical imaging 29 (6) (2010) 1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rohlfing T, Russakoff DB, Maurer CR, Performance-based classifier combination in atlas-based image segmentation using expectation-maximization parameter estimation, IEEE transactions on medical imaging 23 (8) (2004) 983–994. [DOI] [PubMed] [Google Scholar]
- [40].Smith SM, Fast robust automated brain extraction, Human brain mapping 17 (3) (2002) 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cox RW, Afni: software for analysis and visualization of functional magnetic resonance neuroimages, Computers and Biomedical research 29 (3) (1996) 162–173. [DOI] [PubMed] [Google Scholar]
- [42].Sadananthan SA, Zheng W, Chee MW, Zagorodnov V, Skull stripping using graph cuts, Neuroimage 49 (1) (2010) 225–239. [DOI] [PubMed] [Google Scholar]
- [43].Iglesias JE, Liu C-Y, Thompson PM, Tu Z, Robust brain extraction across datasets and comparison with publicly available methods, IEEE transactions on medical imaging 30 (9) (2011) 1617–1634. [DOI] [PubMed] [Google Scholar]
- [44].Dale AM, Fischl B, Sereno MI, Cortical surface-based analysis: I. segmentation and surface reconstruction, Neuroimage 9 (2) (1999) 179–194. [DOI] [PubMed] [Google Scholar]
- [45].Reuter M, Schmansky NJ, Rosas HD, Fischl B, Within-subject template estimation for unbiased longitudinal image analysis, Neuroimage 61 (4) (2012) 1402–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mon A, Ab C, Durazzo T, Meyerhoff D, Potential effects of fat on magnetic resonance signal intensity and derived brain tissue volumes, Obesity research & clinical practice) 10 (2) (2016) 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Rohlfing T, Zahr NM, Sullivan EV, Pfefferbaum A, The SRI24 multichannel atlas of normal adult human brain structure, Human brain mapping 31 (5) (2010) 798–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Avants BB, Epstein CL, Grossman M, Gee JC, Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain, Medical image analysis 12 (1) (2008) 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zill D, Wright WS, Cullen MR, Advanced engineering mathematics, Jones & Bartlett Learning, 2011. [Google Scholar]
- [50].Pfefferbaum A, Rohlfing T, Rosenbloom MJ, Chu W, Colrain IM, Sullivan EV, Variation in longitudinal trajectories of regional brain volumes of healthy men and women (ages 10 to 85 years) measured with atlas-based parcellation of mri, Neuroimage 65 (2013) 176–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Madsen H, Thyregod P, Introduction to general and generalized linear models, CRC Press, 2010. [Google Scholar]
- [52].Fisher RA, The logic of inductive inference, Journal of the Royal Statistical Society 98 (1) (1935) 39–82. [Google Scholar]
- [53].Chapelle O, Training a support vector machine in the primal, Neural computation 19 (5) (2007) 1155–1178. [DOI] [PubMed] [Google Scholar]
- [54].Arlot S, Celisse A, et al. , A survey of cross-validation procedures for model selection, Statistics surveys 4 (2010) 40–79. [Google Scholar]
- [55].Cortes C, Mohri M, Rostamizadeh A, L 2 regularization for learning kernels, in: Proceedings of the Twenty-Fifth Conference on Uncertainty in Artificial Intelligence, AUAI Press, 2009, pp. 109–116. [Google Scholar]
- [56].Sabuncu MR, A universal and efficient method to compute maps from image-based prediction models, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, 2014, pp. 353–360. [DOI] [PubMed] [Google Scholar]
- [57].Tan M, Wang L, Tsang IW, Learning sparse SVM for feature selection on very high dimensional datasets, in: ICML, 2010, pp. 1047–1054. [Google Scholar]
- [58].Peng H, Long F, Ding C, Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy, IEEE Transactions on Pattern Analysis and Machine Intelligence 27 (8) (2005) 1226–1238. [DOI] [PubMed] [Google Scholar]
- [59].DeLong ER, DeLong DM, Clarke-Pearson DL, Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach, Biometrics (1988) 837–845. [PubMed] [Google Scholar]
- [60].Garrett D, Peterson DA, Anderson CW, Thaut MH, Comparison of linear, nonlinear, and feature selection methods for EEG signal classification, IEEE TNSRE 11 (2) (2003) 141–144. [DOI] [PubMed] [Google Scholar]
- [61].Tohka J, Moradi E, Huttunen H, ADNI, Comparison of feature selection techniques in machine learning for anatomical brain mri in dementia, Neuroinformatics (2016) 1–18. [DOI] [PubMed] [Google Scholar]
- [62].Raudys SJ, Jain AK, et al. , Small sample size effects in statistical pattern recognition: Recommendations for practitioners, IEEE Transactions on pattern analysis and machine intelligence 13 (3) (1991) 252–264. [Google Scholar]
- [63].DI SCLAFANI V, MACKAY RS, MEYERHOFF DJ, NORMAN D, WEINER MW, FEIN G, Brain atrophy in hiv infection is more strongly associated with cdc clinical stage than with cognitive impairment, Journal of the International Neuropsychological Society 3 (3) (1997) 276–287. [PMC free article] [PubMed] [Google Scholar]
- [64].Aylward EH, Henderer J, McArthur JC, Brettschneider P, Harris G, Barta P, Pearlson G, Reduced basal ganglia volume in hiv-1-associated dementia results from quantitative neuroimaging, Neurology 43 (10) (1993) 2099–2099. [DOI] [PubMed] [Google Scholar]
- [65].Stout JC, Ellis RJ, Jernigan TL, Archibald SL, Abramson I, Wolfson T, McCutchan JA, Wallace MR, Atkinson JH, Grant I, Progressive cerebral volume loss in human immunodeficiency virus infection: a longitudinal volumetric magnetic resonance imaging study, Archives of neurology 55 (2) (1998) 161–168. [DOI] [PubMed] [Google Scholar]
- [66].Ragin AB, Du H, Ochs R, Wu Y, Sammet CL, Shoukry A, Epstein LG, Structural brain alterations can be detected early in hiv infection, Neurology 79 (24) (2012) 2328–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Underwood J, Cole JH, Caan M, De Francesco D, Leech R, van Zoest RA, Su T, Geurtsen GJ, Schmand BA, Portegies P, et al. , Gray and white matter abnormalities in treated human immunodeficiency virus disease and their relationship to cognitive function, Clinical Infectious Diseases 65 (3) (2017) 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kallianpur KJ, Shikuma C, Kirk GR, Shiramizu B, Valcour V, Chow D, Souza S, Nakamoto B, Sailasuta N, Peripheral blood hiv dna is associated with atrophy of cerebellar and subcortical gray matter, Neurology 80 (19) (2013) 1792–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Tagliati M, Simpson D, Morgello S, Clifford D, Schwartz R, Berger J, Cerebellar degeneration associated with human immunodeficiency virus infection, Neurology 50 (1) (1998) 244–251. [DOI] [PubMed] [Google Scholar]
- [70].Aylward EH, Brettschneider PD, McArthur JC, Harris GJ, et al. , Magnetic resonance imaging measurement of gray matter volume reductions in hiv dementia, The American journal of psychiatry 152 (7) (1995) 987. [DOI] [PubMed] [Google Scholar]
- [71].Thompson PM, Dutton RA, Hayashi KM, Toga AW, Lopez OL, Aizenstein HJ, Becker JT, Thinning of the cerebral cortex visualized in hiv/aids reflects cd4+ t lymphocyte decline, Proceedings of the National Academy of Sciences 102 (43) (2005) 15647–15652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Zhou Y, Li R, Wang X, Miao H, Wei Y, Ali R, Qiu B, Li H, Motor-related brain abnormalities in hiv-infected patients: a multimodal mri study, Neuroradiology (2017) 1–10. [DOI] [PubMed] [Google Scholar]
- [73].Wang B, Liu Z, Liu J, Tang Z, Li H, Tian J, Gray and white matter alterations in early hiv-infected patients: Combined voxel-based morphometry and tract-based spatial statistics, Journal of Magnetic Resonance Imaging 43 (6) (2016) 1474–1483. [DOI] [PubMed] [Google Scholar]
- [74].Janssen MA, Meulenbroek O, Steens SC, Góraj B, Bosch M, Koopmans PP, Kessels RP, Cognitive functioning, wellbeing and brain correlates in hiv-1 infected patients on long-term combination antiretroviral therapy, AIDS 29 (16) (2015) 2139–2148. [DOI] [PubMed] [Google Scholar]
- [75].Clark US, Walker KA, Cohen RA, Devlin KN, Folkers AM, Pina MJ, Tashima KT, Facial emotion recognition impairments are associated with brain volume abnormalities in individuals with hiv, Neuropsychologia 70 (2015) 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Li Y, Li H, Gao Q, Yuan D, Zhao J, Structural gray matter change early in male patients with hiv, International journal of clinical and experimental medicine 7 (10) (2014) 3362. [PMC free article] [PubMed] [Google Scholar]
- [77].Heaps JM, Joska J, Hoare J, Ortega M, Agrawal A, Seedat S, Ances BM, Stein DJ, Paul R, Neuroimaging markers of human immunodeficiency virus infection in south africa, Journal of neurovirology 18 (3) (2012) 151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Kallianpur KJ, Kirk GR, Sailasuta N, Valcour V, Shiramizu B, Nakamoto BK, Shikuma C, Regional cortical thinning associated with detectable levels of hiv dna, Cerebral Cortex 22 (9) (2011) 2065–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Wilson TW, Heinrichs-Graham E, Becker KM, Aloi J, Robertson KR, Sandkovsky U, White ML, O’neill J, Knott NL, Fox HS, et al. , Multimodal neuroimaging evidence of alterations in cortical structure and function in hiv-infected older adults, Human brain mapping 36 (3) (2015) 897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Corrêa DG, Zimmermann N, Tukamoto G, Doring T, Ventura N, Leite SC, Cabral RF, Fonseca RP, Bahia PR, Gasparetto EL, Longitudinal assessment of subcortical gray matter volume, cortical thickness, and white matter integrity in hiv-positive patients, Journal of Magnetic Resonance Imaging 44 (5) (2016) 1262–1269. [DOI] [PubMed] [Google Scholar]
- [81].du Plessis S, Vink M, Joska JA, Koutsilieri E, Bagadia A, Stein DJ, Emsley R, Prefrontal cortical thinning in hiv infection is associated with impaired striatal functioning, Journal of Neural Transmission 123 (6) (2016) 643–651. [DOI] [PubMed] [Google Scholar]
- [82].Castillo D, Ernst T, Cunningham E, Chang L, Altered associations between pain symptoms and brain morphometry in the pain matrix of hiv-seropositive individuals, Journal of Neuroimmune Pharmacology (2017) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Sanford R, Cruz ALF, Scott SC, Mayo NE, Fellows LK, Ances BM, Collins DL, Regionally specific brain volumetric and cortical thickness changes in hiv-infected patients in the haart era, JAIDS Journal of Acquired Immune Deficiency Syndromes 74 (5) (2017) 563–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Foley J, Ettenhofer M, Wright M, Hinkin CH, Emerging issues in the neuropsychology of hiv infection, Current HIV/AIDS Reports 5 (4) (2008) 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Filippi CG, Ulu AM, Ryan E, Ferrando SJ, van Gorp W, Diffusion tensor imaging of patients with hiv and normal-appearing white matter on mr images of the brain, American Journal of Neuroradiology 22 (2) (2001) 277–283. [PMC free article] [PubMed] [Google Scholar]
- [86].Pomara N, Crandall DT, Choi SJ, Johnson G, Lim KO, White matter abnormalities in hiv-1 infection: a diffusion tensor imaging study, Psychiatry Research: Neuroimaging 106 (1) (2001) 15–24. [DOI] [PubMed] [Google Scholar]
- [87].Chang L, Ernst T, Leonido-Yee M, Walot I, Singer E, Cerebral metabolite abnormalities correlate with clinical severity of hiv-1 cognitive motor complex, Neurology 52 (1) (1999) 100–100. [DOI] [PubMed] [Google Scholar]
- [88].Towgood KJ, Pitkanen M, Kulasegaram R, Fradera A, Kumar A, Soni S, Sibtain NA, Reed L, Bradbeer C, Barker GJ, et al. , Mapping the brain in younger and older asymptomatic hiv-1 men: frontal volume changes in the absence of other cortical or diffusion tensor abnormalities, Cortex 48 (2) (2012) 230–241. [DOI] [PubMed] [Google Scholar]
- [89].Thurnher MM, Castillo M, Stadler A, Rieger A, Schmid B, Sundgren PC, Diffusion-tensor mr imaging of the brain in human immunodeficiency virus–positive patients, American journal of neuroradiology 26 (9) (2005) 2275–2281. [PMC free article] [PubMed] [Google Scholar]
- [90].Zhu T, Zhong J, Hu R, Tivarus M, Ekholm S, Harezlak J, Ombao H, Navia B, Cohen R, Schifitto G, Patterns of white matter injury in hiv infection after partial immune reconstitution: a dti tract-based spatial statistics study, Journal of neurovirology 19 (1) (2013) 10–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Stebbins GT, Smith CA, Bartt RE, Kessler HA, Adeyemi OM, Martin E, Cox JL, Bammer R, Moseley ME, Hiv-associated alterations in normal-appearing white matter: a voxel-wise diffusion tensor imaging study, JAIDS Journal of Acquired Immune Deficiency Syndromes 46 (5) (2007) 564–573. [DOI] [PubMed] [Google Scholar]
- [92].Corrêa DG, Zimmermann N, Doring TM, Wilner NV, Leite SC, Cabral RF, Fonseca RP, Bahia PR, Gasparetto EL, Diffusion tensor mr imaging of white matter integrity in hiv-positive patients with planning deficit, Neuroradiology 57 (5) (2015) 475–482. [DOI] [PubMed] [Google Scholar]
- [93].Leite SC, Corrêa DG, Doring TM, Kubo TT, Netto TM, Ferracini R, Ventura N, Bahia PR, Gasparetto EL, Diffusion tensor mri evaluation of the corona radiata, cingulate gyri, and corpus callosum in hiv patients, Journal of Magnetic Resonance Imaging 38 (6) (2013) 1488–1493. [DOI] [PubMed] [Google Scholar]
- [94].Wade BS, Valcour V, Busovaca E, Esmaeili-Firidouni P, Joshi SH, Wang Y, Thompson PM, Subcortical shape and volume abnormalities in an elderly hiv+ cohort, in: Proceedings of SPIE, Vol. 9417, NIH Public Access, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Li S, Wu Y, Keating SM, Du H, Sammet CL, Zadikoff C, Mahadevia R, Epstein LG, Ragin AB, Matrix metalloproteinase levels in early hiv infection and relation to in vivo brain status, Journal of neurovirology 19 (5) (2013) 452–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Dewey J, Hana G, Russell T, Price J, McCaffrey D, Harezlak J, Sem E, Anyanwu JC, Guttmann CR, Navia B, et al. , Reliability and validity of mri-based automated volumetry software relative to auto-assisted manual measurement of subcortical structures in hiv-infected patients from a multisite study, Neuroimage 51 (4) (2010) 1334–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Becker JT, Maruca V, Kingsley LA, Sanders JM, Alger JR, Barker PB, Goodkin K, Martin E, Miller EN, Ragin A, et al. , Factors affecting brain structure in men with hiv disease in the post-haart era, Neuroradiology 54 (2) (2012) 113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Sullivan EV, Rosenbloom MJ, Rohlfing T, Kemper CA, Deresinski S, Pfefferbaum A, Pontocerebellar contribution to postural instability and psychomotor slowing in hiv infection without dementia, Brain imaging and behavior 5(1) (2011) 12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Andrews S, Tsochantaridis I, Hofmann T, Support vector machines for multiple-instance learning, in: Neural Information Processing Systems, 2003. [Google Scholar]
- [100].Berlinet A, Thomas-Agnan C, Reproducing kernel Hilbert spaces in probability and statistics, Springer Science & Business Media, 2011. [Google Scholar]
- [101].Hofmann T, Scholkopf B, Smola AJ, Kernel methods in machine learning, The annals of statistics (2008) 1171–1220. [Google Scholar]
- [102].Maji S, Berg AC, Malik J, Classification using intersection kernel support vector machines is efficient, in: CVPR, 2008. [Google Scholar]
- [103].Scholkopf B, Sung K-K, Burges CJ, Girosi F, Niyogi P, Poggio T, Vapnik V, Comparing support vector machines with gaussian kernels to radial basis function classifiers, IEEE transactions on Signal Processing 45 (11) (1997) 2758–2765. [Google Scholar]
- [104].Xu Y, Yin W, A block coordinate descent method for regularized multiconvex optimization with applications to nonnegative tensor factorization and completion, SIAM Journal on imaging sciences 6 (3) (2013) 1758–1789. [Google Scholar]
- [105].Chang C-C, Lin C-J, Libsvm: a library for support vector machines, ACM transactions on intelligent systems and technology (TIST) 2 (3) (2011) 27. [Google Scholar]
- [106].Calamai PH, Moré JJ, Projected gradient methods for linearly constrained problems, Mathematical programming 39 (1) (1987) 93–116. [Google Scholar]


