ABSTRACT
Early mammalian development is crucially dependent on the establishment of oxidative energy metabolism within the trophectoderm (TE) lineage. Unlike the inner cell mass, TE cells enhance ATP production via mitochondrial oxidative phosphorylation (OXPHOS) and this metabolic preference is essential for blastocyst maturation. However, molecular mechanisms that regulate establishment of oxidative energy metabolism in TE cells are incompletely understood. Here, we show that conserved transcription factor TEAD4, which is essential for pre-implantation mammalian development, regulates this process by promoting mitochondrial transcription. In developing mouse TE and TE-derived trophoblast stem cells (TSCs), TEAD4 localizes to mitochondria, binds to mitochondrial DNA (mtDNA) and facilitates its transcription by recruiting mitochondrial RNA polymerase (POLRMT). Loss of TEAD4 impairs recruitment of POLRMT, resulting in reduced expression of mtDNA-encoded electron transport chain components, thereby inhibiting oxidative energy metabolism. Our studies identify a novel TEAD4-dependent molecular mechanism that regulates energy metabolism in the TE lineage to ensure mammalian development.
KEY WORDS: Mammalian development, Mitochondrial transcription, POLRMT, TEAD4, Trophoblast stem cell, Electron Transport Chain
Highlighted Article: The transcription factor TEAD4 promotes mtDNA transcription and oxidative energy metabolism in the trophectoderm lineage to ensure early mammalian development.
INTRODUCTION
During early mammalian embryogenesis, totipotent blastomeres differentiate to inner cell mass (ICM) and trophectoderm (TE) lineages. The ICM develops into the embryo proper, whereas the TE is essential for blastocyst maturation and implantation (Frum and Ralston, 2015; Cockburn and Rossant, 2010; Zernicka-Goetz et al., 2009). Development of the TE and ICM lineages are regulated by restricted expression of cell type-specific transcription factors and spatiotemporal integration of cell signaling events (Frum and Ralston, 2015; Knott and Paul, 2014). Interestingly, along with differential gene expression programs, TE and ICM development is also associated with establishment of altered metabolic preferences. ICM cells largely use a glycolytic metabolic pathway for ATP production (Trimarchi et al., 2000; Kaneko and DePamphilis, 2013). In contrast, TE cells undergo a metabolic switch and use oxidative phosphorylation (OXPHOS) for enhanced ATP production. This metabolic switch in TE cells is necessary for sufficient ATP supply for distinct metabolic processes and for the activity of the Na+, K+-ATPase pump, both of which ensure blastocoel formation and blastocyst maturation (Houghton, 2006). Although the necessity of metabolic transition in the TE lineage during early embryogenesis is well characterized, much less is understood about the molecular mechanism that controls this glycolytic to OXPHOS metabolic transition.
OXPHOS is dependent on proper redox reactions at the electron transport chain (ETC) complexes, which are localized at the mitochondrial inner membrane. The components of ETC are encoded by both nuclear and mitochondrial DNA (mtDNA). Mammalian mtDNA, which is a ∼16.5 kbp closed circular molecule, encodes 13 proteins involved in the ETC (Gustafsson et al., 2016). Therefore, mtDNA transcription is a key regulatory step for OXPHOS. The mtDNA transcription is mediated by a mitochondria-specific RNA polymerase, POLRMT, that initiates transcription at both light- and heavy-strand mtDNA promoters (Gaspari et al., 2004).
POLRMT cannot initiate transcription on its own from a double-stranded mtDNA promoter and requires mitochondrial transcription factor A (TFAM) and mitochondrial transcription factor B2 (TFB2M) or B1 (TFB1M) to start promoter-specific transcription initiation (Yakubovskaya et al., 2014; Falkenberg et al., 2002; Metodiev et al., 2009; Larsson et al., 1998; Kuhl et al., 2016). In vitro reconstitution and structural analyses with TFAM, POLRMT and TFB2M indicates a multi-step transcription initiation process, which starts with binding of TFAM near the transcription start sites (TSS) on mtDNA (Hillen et al., 2017). POLRMT is then recruited to TFAM-bound mtDNA and positioned near the TSS. Subsequently, TFB2M is recruited at the initiation complex, which induces opening up the double-stranded DNA at the promoter region, leading to initial RNA synthesis by POLRMT. Thus, TFAM and TFB2M are implicated in initiating POLRMT-mediated transcription at the double-stranded promoter region (Hillen et al., 2017; Sologub et al., 2009). However, TFAM and TFB2M are not required for the transcription of a single-stranded mtDNA template or a template with a single-stranded DNA bubble covering the TSS (Wanrooij et al., 2008). It has been proposed that TFAM and TFB2M are also not required for the transition from initiation to transcriptional elongation. Rather, the transition involves the dissociation of the initiation factor TFB2M (Hillen et al., 2017; Sologub et al., 2009) and the formation of the elongation complex involving the elongation factor TEFM (Minczuk et al., 2011). TEFM increases the POLRMT affinity to an elongation-like DNA-RNA template and also increases POLRMT processivity to form longer mtDNA transcripts (Posse et al., 2015). Thus, along with POLRMT, TFAM, TFB2M and TEFM constitute the basic machinery for mtDNA transcription.
The importance of TFAM in mtDNA transcription, replication and packaging are well documented from multiple studies (Bogenhagen, 2012; Kukat and Larsson, 2013; Rebelo et al., 2011; Kukat et al., 2015), However, the function of TFAM in the context of mtDNA transcription remains incompletely understood. A study with a human recombinant mtDNA transcription system indicates that TFAM is dispensable for transcription initiation from the HSP1 and LSP promoters (Shutt et al., 2010), indicating that TFAM is not absolutely essential to initiate mtDNA transcription. Rather, it appears that TFAM acts as a regulator of efficiency of mtDNA transcription, in which varying concentration of TFAM dictate mtDNA transcript level. Gene knockout studies in mice indicate that TFAM is essential for embryonic development (Larsson et al., 1998). However, importance of TFAM in the regulation of mitochondrial function during pre-implantation mammalian development is yet to be determined, as TFAM mutant mouse embryos do not show a pre-implantation phenotype or an implantation defect. Rather, they die during post-implantation embryonic development (Larsson et al., 1998).
It has been established that several other nuclear transcription factors shuttle from nucleus to cytoplasm and then to the mitochondria, and could regulate mtDNA transcription (Rebelo et al., 2011; Garcia-Ospina et al., 2003; Meier and Larner, 2014). For example, NFATc1 localizes to mitochondria and binds to mtDNA only during osteogenic differentiation in human mesenchyme stem cells and negatively regulates mitochondrial transcription (Lambertini et al., 2015). Cyclic AMP response element-binding protein binds to the mtDNA and directly activates mtDNA-encoded transcripts (Lee et al., 2005). Mitochondrial MEF2D is required selectively for mitochondrial gene ND6 transcriptional activation in neuronal cells (She et al., 2011). In addition, the transcription factor signal transducer and activator of transcription 3 (STAT3), which localizes to mitochondria, has been shown to regulate mtDNA transcription in a cell type-specific manner. It has been shown that STAT3 negatively regulates mtDNA transcription in keratinocytes (Macias et al., 2014) but promotes mtDNA transcription during reprogramming and maintenance of naïve pluripotent stem cells (Carbognin et al., 2016). These observations indicate that transcription factors beyond the members of the core mtDNA transcriptional machinery could regulate mtDNA transcription in a cell type-specific manner.
In a recent study, Kaneko et al. (Kaneko and DePamphilis, 2013) elegantly showed that transcription factor TEAD4 is crucial for energy homeostasis during pre-implantation mouse development. TEAD4 is a member of a highly conserved family of transcription factors containing the TEA/ATTS DNA-binding domain and is a component of the Hippo signaling pathway (Bürglin, 1991). TEAD4 expression is conserved in the developing TE lineage across multiple mammalian species, including human. In mouse pre-implantation embryos, TEAD4 is expressed beginning at the four-cell stage and expression is maintained throughout TE lineage development. TEAD4 is highly expressed in mouse TSCs, and is also expressed in the ICM lineage and ICM-derived embryonic stem cells (ESCs). However, in comparison with TE and TSCs, TEAD4 expression is significantly reduced in the ICM and ESCs (Home et al., 2012). Gene knockout studies in mice showed that TEAD4 is essential for the development of the TE lineage during pre-implantation development. Mouse developing embryos lacking functional TEAD4 fail to form blastocoel cavity and fail to implant in the uterus (Kaneko and DePamphilis, 1998; Kaneko and DePamphilis, 2013; Yagi et al., 2007; Nishioka et al., 2008; Nishioka et al., 2009; Ito and Suda, 2014). These embryos also fail to maintain expression of Cdx2, Gata3 and other TE-specific genes, and multiple studies implicated that transcriptional activity of TEAD4 is crucial to establish a TE/trophoblast-specific gene expression program during pre-implantation development (Home et al., 2012; Ralston et al., 2010; Wu et al., 2010). Loss of TEAD4 in pre-implantation mouse embryos results in reduction of mitochondrial activity and excessive production of reactive oxygen species (ROS) (Kaneko and DePamphilis, 2013). However, a TEAD4-dependent molecular mechanism that promotes mitochondrial activity and oxidative energy metabolism in the developing TE/trophoblast lineage have yet to be identified.
To understand how TEAD4 regulates energy metabolism during early mammalian development, we studied mouse pre-implantation embryos and mouse TSCs. We show that higher mitochondrial activity and oxidative energy metabolism in the TE and TSCs are crucially dependent on TEAD4, which directly regulates expression of mitochondrial electron transport chain (ETC) components by promoting POLRMT recruitment at the mtDNA. Loss of TEAD4 inhibits POLRMT recruitment, thereby impairing expression of mtDNA-encoded ETC components. Our results identify TEAD4 as a positive regulator of the mitochondrial transcription in mammalian cells.
RESULTS
TE and TSCs contain matured mitochondria and, similar to TE cells, TSCs also rely on oxidative energy metabolism
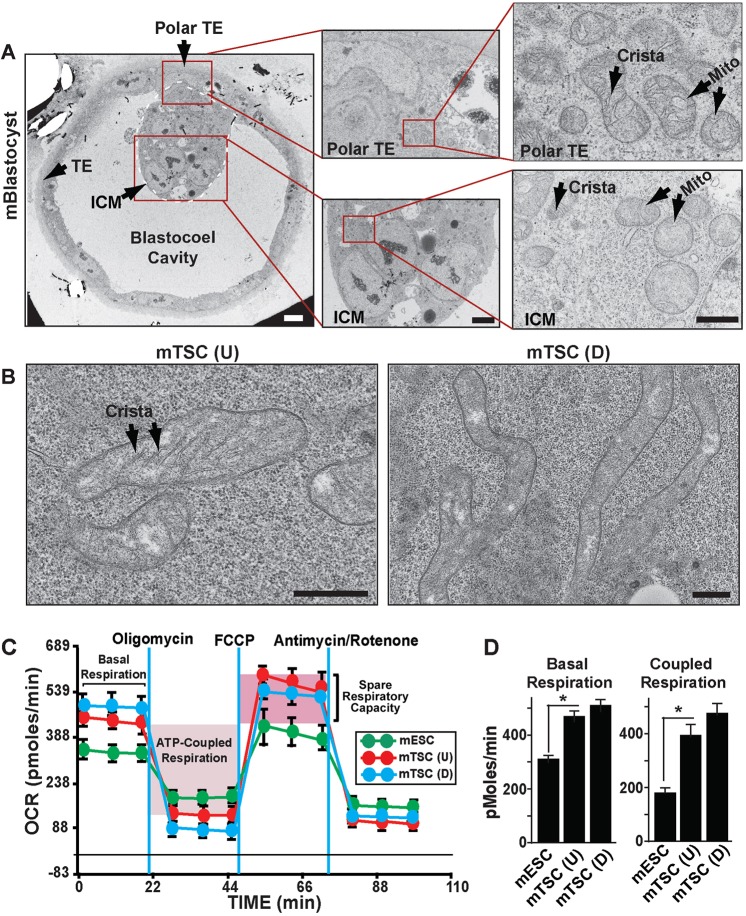
During pre-implantation development, ICM cells maintain a glycolytic metabolic pathway for ATP production. As the developing TE is characterized with enhanced OXPHOS, it has been proposed that the TE contains more matured mitochondria compared with the ICM (Houghton, 2006). However, no published study exists showing a comparative spatial analysis of mitochondrial structure in the TE versus ICM of a mouse blastocyst. Therefore, we performed electron microscopy and found that differences in mitochondrial morphology do indeed exist between the TE and the ICM (Fig. 1A). We confirmed that the mitochondria within the ICM cells are mostly globular in shape and lack proper cristae formation (Fig. 1A), whereas relatively elongated mitochondria with proper cristae formation are common within the TE cells, including the polar TE cells (Fig. 1A).
Fig. 1.
Trophectoderm (TE) and TE-derived TSCs contain mature mitochondria for oxidative energy metabolism. (A) Electron microscopy (EM) showing ultrastructural differences in mitochondria (Mito) between the ICM and TE in a mouse blastocyst. Scale bars: 10 µm. (B) EM showing mitochondria in undifferentiated [mTSC(U)] and differentiated [mTSC(D)] mouse TSCs. Scale bars: 500 nm. (C) Undifferentiated and differentiated mouse TSCs and mouse ESCs were subjected to a mitochondrial stress test by adding oligomycin, FCCP and AntimycinA/Rotenone at different time intervals, and changes in oxygen consumption rates (OCRs) were measured. Basal respiration, mitochondrial ATP synthesis-coupled respiration (light-pink shade) and spare respiratory capacity (deep-pink shade) are indicated. (D) Undifferentiated mouse TSCs maintain significantly (*P<0.001, three independent experiments) higher oxidative respiration compared with undifferentiated ESCs, and oxidative respiration does not significantly alter upon TSC differentiation. Data are mean±s.e.m.
Although TSCs are used as a model system for understanding regulatory mechanisms of TE development and function, mitochondrial morphology, function and energy metabolism in them are poorly understood. The morphological differences between ICM and TE mitochondria prompted us to further investigate mitochondrial structure in mouse TSCs. Electron microscopy revealed that, similar to the TE cells, proliferating TSCs also contain morphologically matured, elongated mitochondria with increased cristae formation (Fig. 1B). Upon spontaneous differentiation (without FGF4 in culture), mouse TSCs predominantly lead to trophoblast giant cell (TGCs) formation (Ray et al., 2009). We also looked at mitochondrial morphology upon spontaneous TSC differentiation and found large elongated mitochondria (Fig. 1B, right panel) in differentiated TSCs. Thus, mouse TSC differentiation into TGCs is associated with extensive mitochondrial fusion, an observation supported by another earlier study, which showed loss of TGC formation in mitofusin 2 mutant mouse embryos (Chen et al., 2003).
Because mitochondrial OXPHOS-dependent oxygen consumption substantially increases between cleavage-stage embryos and blastocysts, and TE development is associated with induction of OXPHOS (Trimarchi et al., 2000; Houghton, 2006; Kaneko and DePamphilis, 2013), and both TE and TSCs contain morphologically matured mitochondria, we tested whether preferences for cellular energy metabolism differ in undifferentiated versus differentiated mouse TSCs versus mouse ESCs. To characterize the metabolic profiles in TSCs versus ESCs, we measured the oxygen consumption rate (OCR), which indicates cellular aerobic respiration. We found that, under standard culture conditions that maintain stemness of ESCs and TSCs, basal OCR is higher in TSCs compared with that in ESCs (Fig. 1C,D). To further confirm whether TSCs are associated with higher OXPHOS, OCR was monitored after cells were metabolically stressed by adding oligomycin, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) and antimycin A/rotenone in succession. In TSCs, treatment with oligomycin, an ATP synthase inhibitor, induced greater loss of OCR (Fig. 1C). The greater loss of OCR upon inhibition of mitochondrial ATP synthesis indicated that higher levels of mitochondrial respiration are coupled with ATP production in TSCs (Fig. 1D). To measure reserve respiratory capacity, which is an indicative parameter of maximal respiratory efficiency of mitochondria, we next added FCCP, an ionophore that induces high proton conductance into the mitochondrial membrane with rapid acceleration of the ETC. FCCP treatment resulted in significantly higher OCR increase in TSCs than that in ESCs (Fig. 1C,D), indicating higher maximal respiratory capacity of mitochondria in TSCs. Finally, treatment with antimycin A and Rotenone, inhibitors of ETC complex III and complex I, respectively, revealed that the rate of oxygen consumption due to non-mitochondrial sources was minimal in both TSCs and ESCs. We also found that the OCR in mouse TSCs does not change significantly upon induction of differentiation (Fig. 1C,D), indicating that both undifferentiated and differentiated TSCs rely upon oxidative energy metabolism. These findings indicated that, similar to TE, mouse TSCs maintain a mitochondrial respiration and a preference toward oxidative energy metabolism. The presence of matured mitochondria and a preferential oxidative energy metabolism also suggested that mouse TSCs could be used as a model system to understand molecular mechanisms that regulate mitochondrial function within the developing TE lineage.
TEAD4 is crucial for maintaining oxidative energy metabolism and mitochondrial ETC function in TSCs
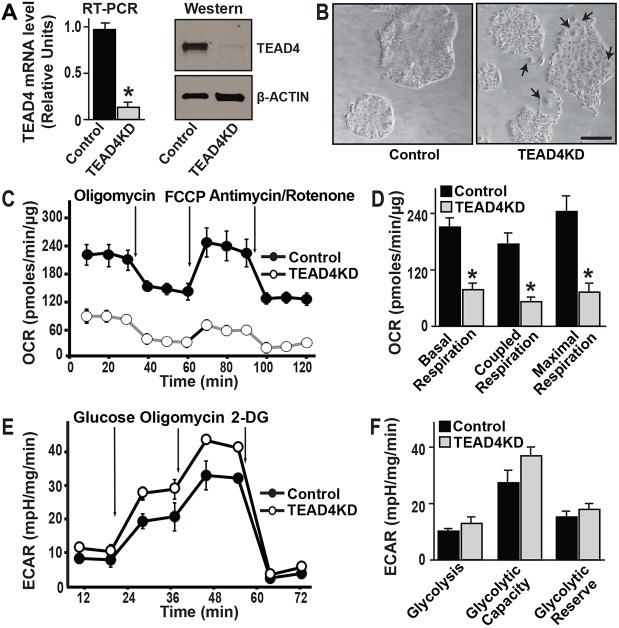
To understand the molecular mechanisms that are crucial for preferential oxidative energy metabolism in TSCs, we tested the importance of TEAD4 in energy homeostasis in the developing TE lineage. TEAD4 is highly expressed in TSCs (Fig. 2A) and regulates TSC-specific gene expression (Home et al., 2012; Yagi et al., 2007; Nishioka et al., 2008; Ralston et al., 2010; Nishioka et al., 2009). To test whether depletion of TEAD4 affects oxidative energy metabolism in TSCs, we stably depleted TEAD4 in mouse TSCs by RNA interference using short hairpin RNAs (shRNAs) (Fig. 2A). Depletion of TEAD4 induced a loss of stem-state colony morphology in TSCs after prolonged culturing in contrast to the TSCs stably transfected with scramble shRNA (Fig. 2B), indicating that TEAD4 is important to maintain TSC self-renewal. Gene expression analyses showed that depletion of TEAD4 resulted in loss of stem-state regulators such as Cdx2, Gata3 and Elf5 (Fig. S1). However, differentiation markers were not significantly induced (Fig. S1). Therefore, when TSCs are maintained in culture condition with FGF4, loss of TEAD4 affects the self-renewal ability of mouse TSCs but does not induce their differentiation. To characterize the metabolic profiles of TEAD4-depleted TSCs (TEAD4KD), we measured OCR and found that loss of TEAD4 strongly reduced OCR in TSCs (Fig. 2C). TEAD4KD TSCs showed a significantly reduced basal respiration compared with control TSCs (Fig. 2C,D). Treatment of oligomycin induced only minor loss of OCR (Fig. 2C) in TEAD4KD TSCs, indicating that very low levels of mitochondrial respiration are coupled with ATP production in those cells (Fig. 2C,D). Furthermore, analyses of ATP production confirmed that depletion of TEAD4 resulted in loss of ATP production in TSCs (Fig. 2D). We also compared the extracellular acidification rate (ECAR), an indicative parameter for glycolytic energy metabolism, in control versus TEAD4KD TSCs. ECAR was monitored after addition of glucose, oligomycin and 2-deoxy-D-glucose (2DG) in succession (Fig. 2E). We found that, unlike oxidative respiration, glycolytic capacity in TSCs is not significantly dependent on TEAD4 (Fig. 2E,F). Collectively, these results indicate that TEAD4 is important for maintaining oxidative energy metabolism in TSCs.
Fig. 2.
TEAD4 is important for oxidative respiration in mouse TSC. (A) Quantitative RT-PCR and western blot analyses showing depletion of TEAD4 mRNA and protein expression in TSCs upon shRNA-mediated RNAi (TEAD4KD). (B) Micrographs of control and TEAD4-depleted TSC colonies (passage 2 after RNAi) in TSC culture conditions. The TEAD4KD TSCs are characterized with more visible cellular boundaries in cell colonies and the presence of higher numbers of differentiated trophoblast giant cells (TGCs, arrows), indicating propensity toward differentiation. Scale bar: 100 µm. (C) A mitochondrial stress test was performed to measure oxygen consumption rates (OCRs) in control and TEAD4KD TSCs. (D) Quantitative analyses of OCR in control versus TEAD4KD TSCs. Plots show strong reduction in oxidative respiration in TEAD4KD TSCs. (E) Control and TEAD4KD TSCs were subjected to a glycolysis stress test by adding glucose, oligomycin and 2-deoxy glucose (2-DG) at different time intervals and changes in the extracellular acidification rate (ECAR) were measured. The graphs show representative ECAR profiles. (F) Plots show only modest changes in glycolysis rate and maximal glycolytic capacity in TSCs upon TEAD4 depletion. *P<0.001. Data are mean±s.e.m.
Loss of Tead4 induces mitochondrial ETC dysfunction in TSCs
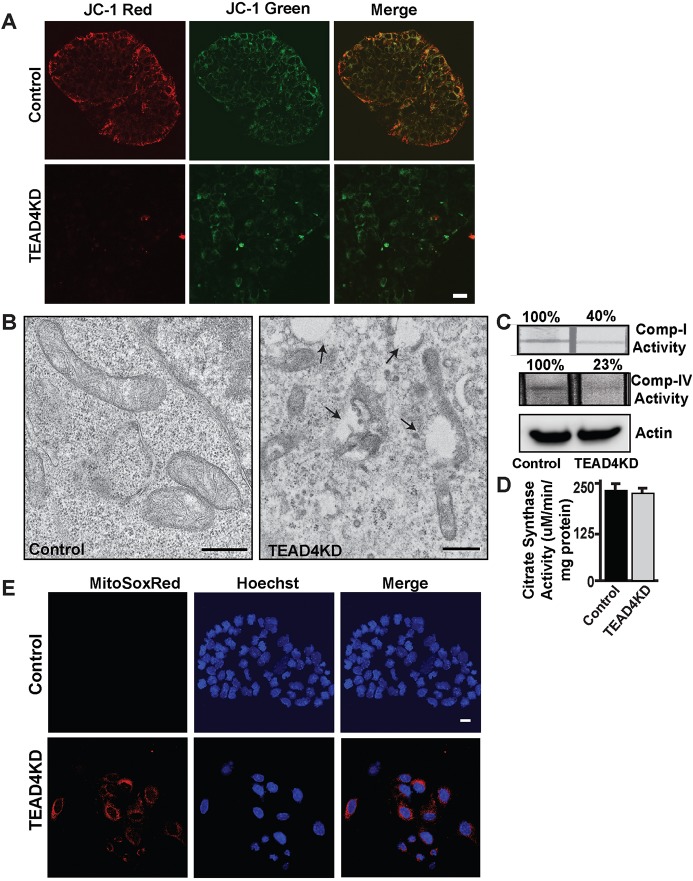
Oxidative ATP synthesis relies on a proton gradient across the inner mitochondrial membrane. As the TEAD4KD TSCs displayed reduction in OCR and ATP production, we tested whether loss of TEAD4 is associated with reduction of mitochondrial membrane potential in TSCs. We used a fluorescent vital dye JC-1, which accumulates within mitochondria in a membrane potential-dependent fashion, resulting in a fluorescence emission shift from green (∼529 nm) to red (∼590 nm) (St John et al., 2006). TSCs stably transfected with scramble shRNA treated with JC1 exhibited bright red-orange fluorescence, indicating the presence of healthy mitochondria (Fig. 3A). In contrast, TEAD4KD TSCs displayed mostly green fluorescence, representing hypo-polarized mitochondria (Fig. 3A).
Fig. 3.
Loss of TEAD4 results in impaired mitochondrial function in mouse TSCs. (A) Control and TEAD4KD TSCs were stained with a mitochondrial membrane potential probe, JC-1, and excited simultaneously for observing hypo-polarized membrane potential (monomeric form excited by 488 nm laser, green) and hyper-polarized membrane potential (J-aggregate form excited using the 568 nm argon-krypton laser, red). Scale bar: 20 µm. (B) TEM pictures showing mitochondrial ultrastructural differences in control and TEAD4KD TSCs. Micrographs show the presence of an increased number of vacuoles and mitochondrial structural abnormalities in TEAD4KD TSCs. Scale bars: 500 nm. (C,D) Activities of mitochondrial electron-transport chain (ETC) complex I, ETC complex IV, and actin and citrate synthase were measured in control and TEAD4KD TSCs. Data show reduced ETC complex I and IV activities in TEAD4KD TSCs without any significant change in citrate synthase activity. Data are mean±s.e.m. (E) Control and TEAD4KD TSCs were stained with the mitochondrial ROS indicator MitoSox Red (red) and Hoechst (blue) to monitor mitochondrial distribution of reactive oxygen species accumulation. Scale bar: 20 µm.
Next, we tested whether mitochondrial ultrastructure in TSCs is altered upon TEAD4 depletion. Transmission electron microscopy revealed that, unlike control TSCs, mitochondria in TEAD4KD TSCs are more electroluscent, without prominent cristae and often associated with cellular vacuoles (Fig. 3B). These changes are indicative of mitochondria with reduced folding of the mitochondrial inner membrane, which contains the five complexes of the mitochondrial OXPHOS system. The OXPHOS system includes four ETC complexes (complex I to complex IV) and the ATP-producing FoF1-ATPase complex, which is involved in ATP generation. Inhibition of mitochondrial respiration and alteration of mitochondrial morphology are often associated with ETC dysfunction. Therefore, we tested whether loss of TEAD4 results in functionally impaired ETC complexes in TSCs. We measured complex I activity with control and TEAD4KD TSCs and found that complex I activity was significantly reduced in TEAD4KD TSCs (Fig. 3C). We also observed drastic reduction of complex I activity upon TEAD4 depletion (Fig. 3C). However, we did not notice any significant change in the activity of citrate synthase, an enzyme associated with the Krebs cycle, which is localized in the mitochondrial matrix and an indicator of intact mitochondrial mass (Fig. 3D).
Impaired activities of ETC complexes, including complex I leads to excessive ROS production (Zhou et al., 2011). So we tested whether loss of TEAD4 is associated with induction of ROS production in TSCs. Using MitoSox, a probe that detects mitochondrial ROS production (Zhou et al., 2011), we observed induced mitochondrial ROS production in TEAD4KD TSCs when compared with that of TSCs stably transfected with scramble shRNA (Fig. 3E). This is in line with the findings from a study by Kaneko and DePamphilis (2013), which showed that loss of TEAD4 in pre-implantation mouse embryos induces ROS production. Collectively, these results indicate that TEAD4 is important to maintain proper ETC function in TSCs.
TEAD4 promotes mtDNA transcription by facilitating POLRMT recruitment at the mtDNA
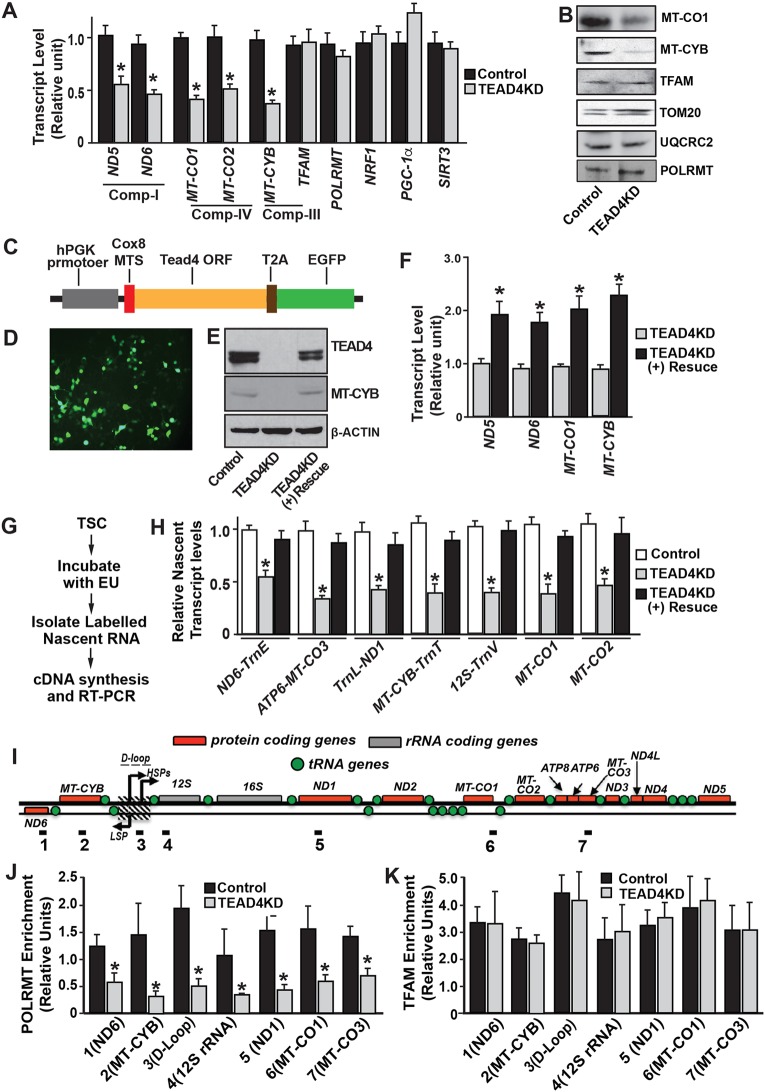
Impaired function of mitochondrial ETC complexes in TEAD4KD TSCs prompted us to investigate a TEAD4-dependent mechanism that regulates expression of mtDNA-encoded ETC components. Therefore, we asked whether mtDNA transcription is altered upon TEAD4 depletion in TSCs. Quantitative RT-PCR analyses revealed that loss of TEAD4 in TSCs inhibits expression of mtDNA-encoded genes (Fig. 4A). However, mRNA expressions of nuclear DNA encoded mitochondrial regulators, including transcription factor A mitochondrial (TFAM), nuclear respiratory factor 1 (NRF1), co-factor PGC1α, deacetylase sirtuin-3 (SIRT3) and POLRMT, were not altered upon TEAD4 depletion (Fig. 4A).
Fig. 4.
TEAD4 promotes mtDNA transcription and POLRMT recruitment. (A) RT-PCR analysis of mtDNA-encoded transcript in control and TEAD4KD TSCs (*P<0.01, three independent experiments). (B) Western blot analyses showing expressions of mtDNA-encoded ETC complex members, MT-CO1 and MT-CYB, and nuclear DNA-encoded mitochondrial proteins, TFAM, TOM20 and UQCRC2, in control and TEAD4KD TSCs. (C) Schematic representation of the ectopic TEAD4-expressing lentiviral construct. A mitochondrial transport signal was added to the N-terminal end of the Tead4-coding sequence to ensure efficient mitochondrial localization of the ectopically expressed TEAD4 protein. A reporter EGFP was attached to the C-terminal end of Tead4 along with a self-cleaving T2A peptide. (D) Protein expression from the ectopic TEAD4-expressing construct in TEAD4KD mTSCs was monitored via EGFP expression. (E) Western blot analyses showing rescue of TEAD4 and MTCYB expression in TEAD4KD TSCs after transduction with ectopic TEAD4-expressing lentiviral construct. (F) RT-PCR analysis of mtDNA-encoded transcripts in TEAD4KD TSCs without and with the rescue of TEAD4 expression (*P<0.01, three independent experiments). (G) Schematic of the method for measurement of nascent mtDNA transcripts in TSCs. (H) Plots show reduction of nascent mtDNA transcripts in TEAD4KD TSCs (*P<0.01, three independent experiments) and rescue of nascent transcripts upon expression of ectopic TEAD4. Primers were designed to amplify polycistronic cDNA [e.g. ND6-TrnE, a primer pair amplifying NADH dehydrogenase subunit 6 and the adjacent transfer RNA (Glu) (tRNA)]. (I) Schematic diagram showing mtDNA-encoded genes and localization of primer pairs (1-7), which are used for the quantitative ChIP assay. (J,K) Quantitative ChIP assays in control and TEAD4KD TSCs were performed to determine POLRMT and TFAM occupancy at different regions of the mtDNA. Plots show a significant reduction in POLRMT occupancy (J) but maintenance of TFAM occupancy at different regions of mitochondrial genome upon TEAD4 depletion in mTSCs (*P<0.01, three independent experiments). Data are mean±s.e.m.
To further test TEAD4-dependent regulation of mtDNA-encoded ETC complex members, we tested protein expressions of MT-CYB, a component of ETC complex III, and MT-CO1, a component of ETC complex IV. We observed strong downregulation of both MT-CYB and MT-CO1 protein expression in TEAD4KD TSCs (Fig. 4B). In contrast, protein expression of UQCRC2 (required for the assembly of ETC complex II), translocase of outer mitochondrial membrane 20 (TOM20), POLRMT and TFAM, which are expressed from nuclear DNA and localize to mitochondria for their function, were unchanged upon TEAD4 depletion (Fig. 4B). These results confirmed that TEAD4 is important to optimize expression of mtDNA-encoded ETC components in TSCs.
To further confirm the specificity of TEAD4-dependent regulation of mtDNA transcription in mouse TSCs, we wanted to rescue TEAD4KD mouse TSCs with a mutant TEAD4 lacking nuclear localization signal (NLS). However, analyses of mouse TEAD4 protein sequence revealed that the NLS in TEAD4 overlaps with the functional TEA domain (Fig. S2A), making the approach to selectively delete the NLS without affecting TEAD4 DNA-binding activity not feasible. Thus, we performed two following experiments. First, we depleted YAP1, a co-factor for TEAD4 to regulate nuclear DNA-encoded genes and tested mtDNA transcription. We found that depletion of the TEAD4 co-factor YAP1 reduced the mRNA expression of TSC-specific genes Cdx2 and Gata3, but did not reduce transcript levels of mtDNA-encoded ETC components (Fig. S2B-E). These results indicated that a TEAD4-YAP1 axis, which regulates nuclear genes in mouse TSCs, is not involved in the regulation of mtDNA-encoded ETC component genes. Second, we rescued TEAD4 function in TEAD4KD mTSCs by ectopically expressing a modified TEAD4 protein, in which a mitochondrial targeting sequence (MTS) from subunit VIII of human cytochrome c oxidase was attached (Fig. 4C). The modified TEAD4 was also attached to a reporter EGFP, separated by the self-cleaving 2A peptide from Thosea asigna virus (T2A peptide). Thus, we could monitor expression of ectopic TEAD4 from EGFP expression (Fig. 4D). We found that rescue of TEAD4 expression in TEAD4KD mouse TSCs rescued expression of mtDNA-encoded ETC components (Fig. 4E,F).
Transcription from mtDNA generates polycistronic transcripts, which are processed to generate matured transcripts. However, the processed transcripts of mtDNA-encoded genes often have a shorter half-life (Wolf and Mootha, 2014; Piechota et al., 2006; Chujo et al., 2012). Therefore, to better assess inhibition of mtDNA transcription in TEAD4KD TSCs, we analyzed nascent mtDNA transcripts using a nascent transcript capture kit. Analyses of nascent mtDNA transcripts further confirmed significant downregulation of mtDNA transcription in TEAD4KD TSCs and expression of ectopic TEAD4 rescued the process (Fig. 4G,H and Table S1). Collectively, these experiments indicated specific importance of a TEAD4-dependent mechanism that promotes mtDNA transcription in mouse TSCs.
Mitochondrial transcripts are generated by POLRMT from two heavy strand promoters (HSPs) and one light-strand promoter (LSP), which are localized at the D-loop region of the mtDNA (Taanman, 1999) (Fig. 4E). In addition, establishment of the transcription initiation complex requires additional transcription factors, such as TFAM (Falkenberg et al., 2002; Larsson et al., 1998). However, our findings above indicated that, in TSCs, TEAD4 promotes mtDNA transcription via a mechanism, which is independent of the regulation of TFAM and POLRMT expression (Fig. 4B), mtRNA splicing and mtRNA degradation (Fig. 4H). Therefore, we asked whether POLRMT recruitment at the mtDNA is dependent upon TEAD4 function. To test this, we isolated mitochondria from control and TEAD4KD TSCs, and performed chromatin immunoprecipitation (ChIP) analyses to assess POLRMT recruitment at different regions of the mtDNA (Fig. 4I). Intriguingly, ChIP analyses detected significant reduction in POLRMT recruitment at the mtDNA upon loss of TEAD4 (Fig. 4J). We also tested TFAM recruitment at the mtDNA in TEAD4KD TSCs. We found that, unlike POLRMT, TFAM binding at the mtDNA was not inhibited upon TEAD4 depletion (Fig. 4K). These findings strongly indicated a novel function of TEAD4, in which it promotes transcription of mtDNA-encoded ETC components by facilitating POLRMT recruitment to the mtDNA.
Endogenous TEAD4 localizes to mitochondria and physically occupy mtDNA
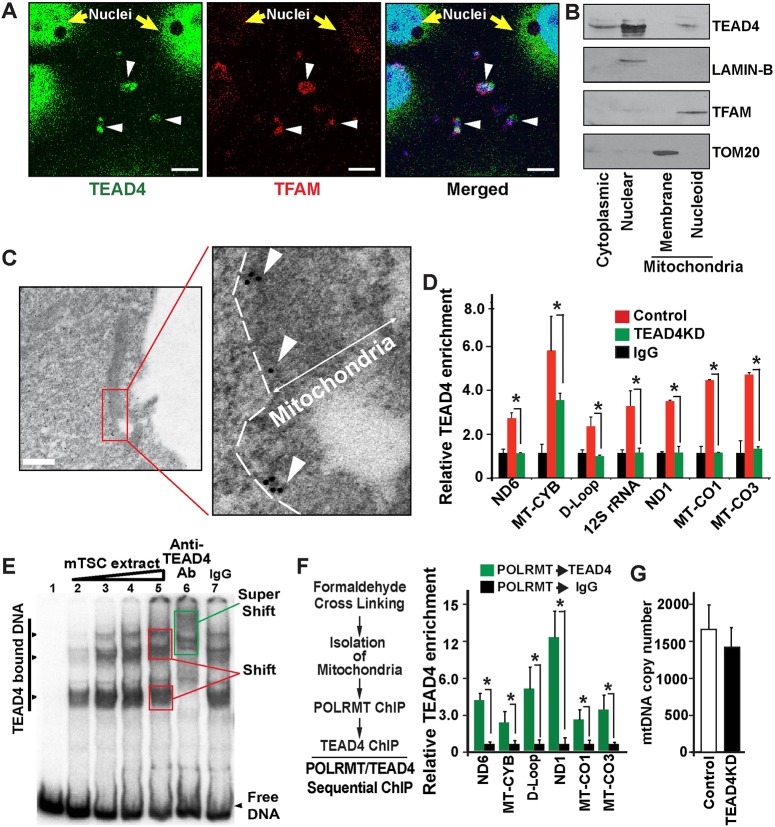
Transcription factors often shuttle from the nucleus to the cytoplasm. Although a majority of transcription factors mediate their function within the nucleus, several of them localize to the mitochondria and are implicated in regulation of mitochondrial genome transcription in different cellular contexts (Ralston et al., 2010; Wu et al., 2010; Mahato et al., 2014; Leppens et al., 1996; Leese and Barton, 1984). TEAD4 is a nuclear-cytoplasmic transcription factor (Home et al., 2012) and Kaneko and DePamphilis showed that ectopic TEAD4 colocalizes with mitochondria in the NIH3T3 cell line (Kaneko and DePamphilis, 2013). Therefore, we investigated whether endogenous TEAD4 localizes to mitochondria and regulates mtDNA transcription in TSCs. Co-immunostaining studies with an anti-TEAD4 antibody along with anti-TFAM antibody indicated that endogenous TEAD4 localizes to mitochondria in proliferating mouse TSCs (Fig. 5A and Fig. S3). To further confirm TEAD4 localization in mitochondria in the mouse TSCs, we performed additional experiments. First, we isolated purified mitochondria from mouse TSCs, sub-fractionated these purified mitochondria and confirmed the presence of TEAD4 within the mitochondrial nucleoid fraction via western blot analyses (Fig. 5B). Finally, we performed immunogold transmission electron microscopy (immuno-TEM) using an anti-TEAD4 antibody and found that TEAD4 is localized in the mitochondrial matrix surrounded by the inner membrane (Fig. 5C). These experiments confirmed mitochondrial localization of TEAD4 in mouse TSCs.
Fig. 5.
TEAD4 localizes to mitochondria and occupies mtDNA along with POLRMT. (A) Mouse TSCs were co-stained with TEAD4 (green) and TFAM (red). Confocal images show both TEAD4 and TFAM localization in mitochondria (arrowheads). Scale bars: 10 µm. TEAD4 also localizes at high levels in the nuclei, whereas TFAM localization in the nuclei is at a much reduced level. (B) Purified mitochondria from mTSCs were sub-fractionated and western blot analyses were performed to determine localizations of TEAD4 and other mitochondrial proteins. The cytoplasmic and nuclear fractions of mTSCs were used as controls. (C) Immuno-TEM showing TEAD4 within mitochondria (arrowheads). Dotted line shows the boundary of the mitochondrial membrane. Scale bar: 500 nm. (D) Quantitative ChIP analysis showing TEAD4 occupancy along mitochondrial genome (*P<0.001, three independent experiments). For simplicity, only one IgG is shown, although IgG was used for both control and TEAD4KD chromatin. (E) EMSA to test TEAD4 binding at TEA motifs of mtDNA. A ∼200 bp mtND1 fragment, containing TEA motifs, was incubated without (lane 1) or with (lanes 2-5) increasing amounts of mTSC extract. TEAD4-containing DNA-protein complexes were tested by monitoring mobility super shifts with anti-TEAD4 antibody (lane 6) or IgG (lane 7). (F) Sequential ChIP showing co-occupancy of TEAD4 and POLRMT in different mtDNA regions (*P<0.001, three independent experiments). (G) mtDNA copy numbers in mouse TSCs with or without TEAD4 depletion. Data are mean±s.e.m.
Multiple recent studies indicate that site-specific occupancy of nuclear transcription factors on the mitochondrial genome is crucial for the regulation of mtDNA transcription (Wu et al., 2010; Larsson et al., 1998; Taanman, 1999; Piechota et al., 2006; St John et al., 2006; Zhou et al., 2011; Chujo et al., 2012; Wolf and Mootha, 2014). As we detected TEAD4 localization in the mitochondria, and presence of several consensus TEAD motifs throughout the mtDNA (Fig. S4A), we investigated whether endogenous TEAD4 occupies the mtDNA in mouse TSCs. We performed TEAD4 ChIP with isolated mitochondria from mouse TSCs and screened the mtDNA for TEAD4 binding using multiple primer pairs. Our screening detected TEAD4 binding at different regions of the mtDNA, including the D-loop region where mtDNA transcription starts (Fig. 5D). The specificity of TEAD4 occupancy was further confirmed by monitoring loss of TEAD4 occupancy at the mtDNA in TEAD4KD TSCs.
Our analyses showed that the TEAD4-binding regions contained several sequences that resemble the putative consensus TEA motifs (Fig. S4B). Therefore, to further gain additional evidence that TEAD4 binds to the TEA target sequences within the mitochondrial genome, we performed electrophoretic mobility shift assay (Fig. 5E). We incubated mTSC extracts with a ∼200 bp mtND1 DNA fragment, containing a single high-affinity TEA (GGAATG) motif along with three other overlapping putative low-affinity TEA motifs (Fig. S4C). Formation of multiple DNA-protein complexes with increasing amounts of mTSC extract (Fig. 5E, lanes 2-5) were confirmed via mobility shift of the mtND1 fragment. To confirm that TEAD4 is responsible for the mobility shifts with mTSC extracts, we tested whether the shifted band could be supershifted by the addition of antibodies directed against the TEAD4 protein. As shown for a fragment the gel-shifted band seen in the mTSC extracts is supershifted by the addition of anti-TEAD4 factor antibody (Fig. 5E, lane 6, green box) but not IgG (Fig. 5E, lane 7).
As TEAD4 is an evolutionarily conserved transcription factor and is expressed in the TE layers within the developing blastocyst of multiple mammalian embryos (Home et al., 2012), we asked whether TEAD4 localization to the mitochondria is a conserved event in trophoblast progenitors of other mammalian species. To test this, we studied RCHO-1 trophoblast cells, which represent the rat trophoblast stem cell state (Sahgal et al., 2006). We also studied primary cytotrophoblasts (hCTB) cells of first-trimester human placenta, which are considered as human trophoblast progenitors (Genbacev et al., 2016; Zdravkovic et al., 2015; Knofler and Pollheimer, 2013) and depend on OXPHOS for ATP production (Maloyan et al., 2012). We observed mitochondrial localization of TEAD4 in both RCHO1 and human first-trimester CTB cells (Fig. S5A,B). Furthermore, ChIP analyses with isolated mitochondria from RCHO-1 confirmed TEAD4 binding to the mtDNA (Fig. S5C,D). Our results from mouse TSC, rat RCHO-1 and human CTB indicated that TEAD4 localization to mitochondria and binding to mtDNA are conserved events in mammalian trophoblast progenitors.
Two findings, that TEAD4 binds the mtDNA (Fig. 5D) and that loss of TEAD4 leads to loss of POLRMT recruitment and mtDNA transcription in mouse TSCs, indicated that TEAD4 could be a part of the mitochondrial transcriptional machinery in mouse TSCs and could directly regulate POLRMT binding to the mtDNA. Therefore, we next asked whether TEAD4 interacts with POLRMT on the mtDNA. We performed a sequential ChIP assay (SeqChIP) with isolated mitochondria from mouse TSCs and confirmed a TEAD4 and POLRMT interaction on mtDNA (Fig. 5F). Collectively, these results indicated that endogenous TEAD4 facilitates mtDNA transcription in mouse TSCs by directly facilitating POLRMT recruitment to the mtDNA.
We also tested whether loss of TEAD4 affects mtDNA replication in mouse TSCs. The mtDNA replication is not primed by dedicated primases. Rather, processed mtRNA transcripts, synthesized by POLRMT, serve as a primer for mtDNA replication. It has been shown that loss of different factors that control mtDNA transcription leads to varying outcomes on mtDNA replication. Inhibition of mtDNA transcription could be associated with either enhanced mtDNA replication or inhibition of mtDNA replication. For example, loss of TFAM is shown to reduce both mtDNA transcript levels and inhibits mtDNA replication in multiple cellular contexts (Wang et al., 1999; Li et al., 2000; Kang et al., 2007). In contrast, loss of the elongation factor TEFM in mammalian cells reduces promoter-distal mtDNA transcripts, but does not have a significant effect on mtDNA replication (Minczuk et al., 2011). So we tested whether loss of TEAD4 alters mtDNA copy numbers in mouse TSCs. However, under our experimental conditions, we did not notice any significant alteration of mtDNA copy number when TEAD4 was depleted in mouse TSCs (Fig. 5G), indicating that TEAD4 mediated regulation of mtDNA transcription might not be associated with the mtDNA replication process.
Loss of Tead4 impairs POLRMT recruitment and mtDNA transcription in developing pre-implantation embryo
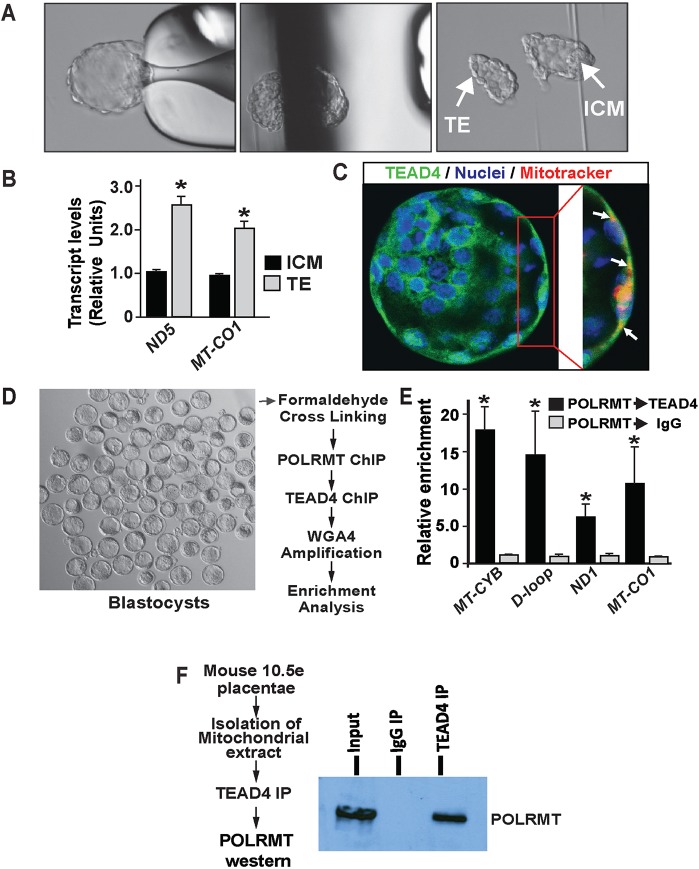
Establishment of mitochondrial OXPHOS in the TE lineage is essential for blastocyst maturation. Therefore, we asked whether development of the TE lineage is associated with enhanced mtDNA transcription. We performed blastocyst microsurgery of mouse blastocysts to isolate TE cells (Fig. 6A) and immunosurgery (Saha et al., 2013) to isolate ICM cells from mouse blastocysts, and confirmed higher RNA expressions of mtDNA-encoded genes in TE lineage cells (Fig. 6A,B). Like mTSCs and hCTB cells, TEAD4 also localized within mitochondria in the TE of the developing blastocyst (Fig. 6C).
Fig. 6.
Endogenous TEAD4 localizes to mitochondria and occupies mtDNA within the TE lineage of a pre-implantation mammalian embryo. (A) Micrographs showing isolation of TE via microsurgery from a mouse blastocyst. (B) Plots show induction of mtDNA-encoded transcripts (ND5 and MT-CO1) in TE lineage cells during blastocyst development (*P<0.05, three independent experiments). (C) Mouse blastocyst stained with TEAD4 (green), mitotracker (red) and DAPI (blue). Higher magnification of the TE cell shows TEAD4 colocalization with mitotracker (arrows). (D-F) Quantitative assessment method for TEAD4 and POLRMT co-occupancy at mtDNA-encoded genes within mouse blastocysts. (D) Method for sequential ChIP-WGA with mouse blastocysts to test TEAD4-POLRMT co-occupancy at mtDNA. (E) Induction of mtDNA-encoded transcripts (ND5 and MT-CO1) in TE lineage cells during blastocyst (*P<0.01, three independent experiments). (F) Western blot showing co-IP between TEAD4 and POLRMT in trophoblast progenitor/stem cells isolated from mouse placenta (E10.5). Data are mean±s.e.m.
As we found that TEAD4 colocalizes with POLRMT on mtDNA and is required for optimum POLRMT recruitment at the mtDNA-encoded ETC genes in mTSCs, we tested whether a similar event is associated with mtDNA transcription during mouse blastocyst development. We performed sequential ChIP (SeqChIP) with blastocysts and confirmed a TEAD4 and POLRMT interaction on the same mtDNA regions in mouse blastocyst (Fig. 6D,E). Second, we performed co-immunoprecipitation analyses with isolated mitochondrial protein from trophoblast progenitor/stem cells of developing mouse placentas. We chose to perform this experiment in trophoblast progenitor/stem cells, as we found that it is technically challenging to isolate mitochondria and perform co-immunoprecipitation analyses with limited amount of cells from the TE of mouse blastocysts. Co-immunoprecipitation experiments confirmed that endogenous TEAD4 physically interacts with endogenous POLRMT in mitochondria of mouse trophoblast progenitors (Fig. 6F). We also observed a similar interaction in mouse TSCs (data not shown). Collectively, our results indicate that, similar to the observations in mouse TSCs, a TEAD4-dependent mechanism could regulate POLRMT binding to the mtDNA and regulate mitochondrial transcription in TE cells during blastocyst maturation. Therefore, we used a Tead4 conditional knockout system (Tead4F/F/UBC-CreERT2 mice) to delete Tead4 in mouse embryos (Yagi et al., 2007) and tested its importance in mtDNA transcription and expression of mtDNA-encoded genes during pre-implantation development.
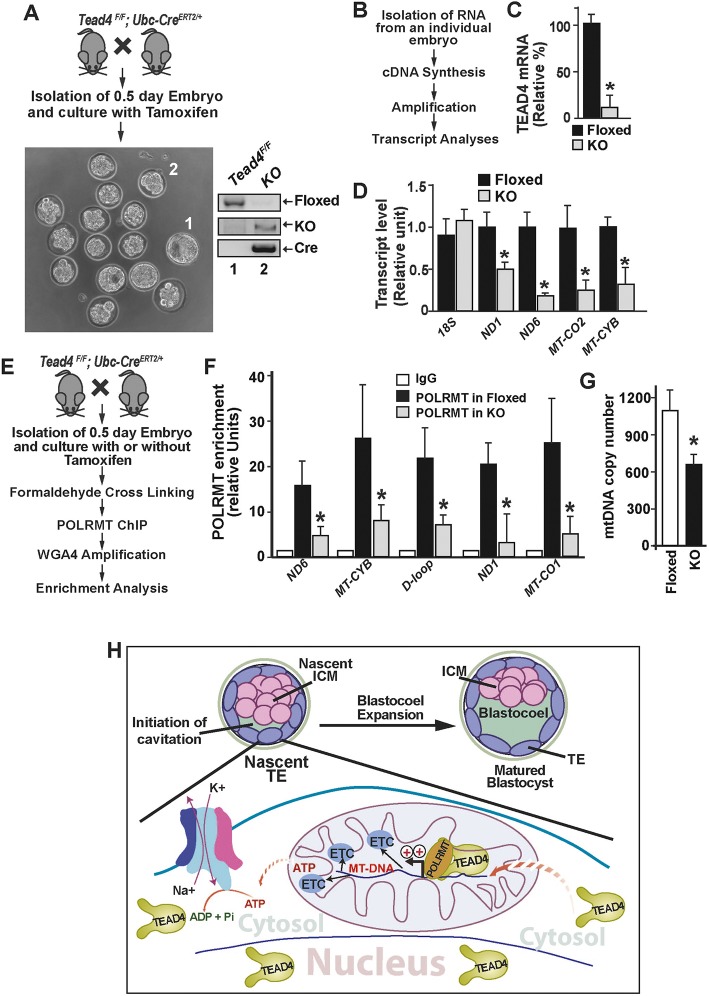
We isolated pre-implantation embryos from Tead4F/F/UBC-CreERT2 mice, treated with 4(OH) tamoxifen (MHT) to delete Tead4 floxed alleles, and cultured them ex vivo (Fig. 7A). Interestingly, we noticed conditional deletion of Tead4 floxed alleles resulted in a mixed phenotype (Fig. S6A) when compared with that of wild-type embryos cultured under similar ex vivo conditions (Fig. 6D) or with Tead4F/F embryos (Fig. 7A, embryo 1; Fig. S6, embryo 1). Several Tead4-deleted embryos failed to mature to the blastocyst stage even when they were cultured at 5% O2 levels, and several embryos that matured to the blastocyst stage showed the presence of a very small blastocoel cavity (Fig. S6, embryos 2 and 3). In addition, several embryos that matured to the blastocyst stage showed partial presence of the Tead4-floxed alleles even after MHT treatment, reflecting inefficient cre-mediated recombination efficiency of Tead4-floxed alleles (Fig. S6B) in those embryos. These results indicated that incomplete deletion of the TEAD4 floxed allele could lead to mixed phenotype in Tead4-conditional knockout system. To avoid these ambiguities, we used higher concentration (1.5 µM instead of 1 µM) of MHT and cultured embryos in a condition that mimics the in vivo condition proposed by Kaneko and DePamphilis (2013). We noticed that these experimental modifications ensured efficient cre-mediated excision of Tead4 floxed alleles and resulted in impaired blastocyst maturation (Fig. 7A). However, these conditions did not affect the maturation of embryos that lacked Cre expression.
Fig. 7.
TEAD4 directly regulates POLRMT recruitment to promote mtDNA transcription during pre-implantation mammalian development. (A) Conditional deletion of Tead4 in pre-implantation mouse embryos. Loss of Tead4 expression abrogated blastocyst maturation in ex vivo culture conditions. Deletion of Tead4 alleles was confirmed by genotyping. (B) Strategy for quantitative RT-PCR analyses in Tead4-deleted embryos. (C) RT-PCR analyses showing loss of Tead4 mRNA expression upon Cre-mediated excision of Tead4F/F alleles (*P<0.001, ten individual embryos). (D) Loss of mtDNA-encoded transcripts in Tead4-deleted pre-implantation embryos (*P<0.01, ten individual embryos). (E) Schematic of POLRMT recruitment analysis in pre-implantation embryos. (F) Significant reduction in POLRMT recruitment at mtDNA-encoded genes in Tead4-deleted embryos (*P<0.001, three individual experiments with up to 25 embryos in each experiment). Data are mean±s.e.m. (G) Reduced mtDNA copy numbers in Tead4-deleted pre-implantation embryos (*P<0.01, 12 individual embryos). (H) Model illustrating the significance of mitochondrial TEAD4. In nascent TE cells of an early blastula, TEAD4 promotes expression of mtDNA-encoded ETC components by facilitating POLRMT recruitment. This promotes oxidative phosphorylation and ATP synthesis. The induced ATP production facilitates cellular processes, including the activity of the Na+, K+-ATPase pump, thereby ensuring blastocyst maturation.
We tested RNA expression of mtDNA-encoded ETC components in Tead4-null (knockout) embryos, which failed to mature to the blastocyst stage. Our analyses confirmed loss of Tead4 mRNA expression as well as inhibition of mtDNA transcription upon deletion of Tead4 floxed alleles (Fig. 7B-D). Furthermore, ChIP analyses confirmed a strong loss of POLRMT binding to the mtDNA in Tead4-null embryos (Fig. 7E-F). Interestingly, we also noticed a significant reduction of mtDNA copy number in Tead4 null embryos (Fig. 7G), a result that differs from the observation inTEAD4-depleted mouse TSCs, in which loss of TEAD4 does not alter mtDNA copy number. However, during pre-implantation development, the first mtDNA replication event is initiated at the blastocyst stage (Thundathil et al., 2005) due to upregulated expression of the catalytic subunit of mitochondria DNA polymerase γ (POLGA) in the TE lineage (Thundathil et al., 2005; Kelly et al., 2012). As TE development and blastocyst maturation is impaired in Tead4-null embryos, it is possible that the reduced mtDNA copy number in Tead4-null embryos is due to impaired activation of the replication machinery.
Nevertheless, collectively our results indicated that TEAD4 facilitates POLRMT recruitment and enhances mtDNA transcription during pre-implantation development. We propose that this mitochondrion-associated function of TEAD4 ensures optimal expression of mtDNA-encoded ETC components in TE cells (as presented in Fig. 7H) and promotes mitochondrial energy metabolism, which is essential for blastocyst maturation.
DISCUSSION
Over the years, numerous studies have implicated mitochondrial regulation in successful embryonic development. These studies noted dynamic changes in cellular metabolism as well as mitochondrial structure, rearrangement and function during the course of pre-implantation development (Wilding et al., 2009; Houghton, 2006). However, almost nothing is known about the molecular control of mtDNA transcription in the context of extra-embryonic TE/trophoblast lineage development. In this study, we show that a TEAD4-dependent molecular mechanism is crucial to optimize mtDNA transcription in the TE and TE-derived TSCs. Our findings are consistent with previous reports that TEAD4 localizes to both cytoplasm and nuclei in the developing TE lineage and is essential for blastocyst maturation (Kaneko and DePamphilis, 2013; Home et al., 2012).
TEAD4 and mitochondrial energy metabolism
Previous work identified the essential role for TEAD4 in early mouse embryogenesis (Hirate et al., 2012; Home et al., 2012; Kaneko and DePamphilis, 2013; Nishioka et al., 2008; Ralston et al., 2010; Yagi et al., 2007; Mihajlović et al., 2015), and we and others showed that TEAD4 directly regulates transcription of key TE-specific nuclear encoded genes in the TE and TSCs (Ralston et al., 2010; Home et al., 2012). These findings implicated the importance of TEAD4 in establishment of the TE-specific transcriptional program. TEAD4 is a nucleo-cytoplasmic transcription factor and ectopic overexpression studies by Kaneko and DePamphilis, showed that, among all TEAD family members, TEAD4 has the unique ability to localize to the mitochondria (Kaneko and DePamphilis, 2013). In addition, conditional gene deletion studies by Kaneko and DePamphilis showed that TEAD4-null pre-implantation embryos have defective mitochondrial function. Therefore, it was proposed that the establishment of energy homeostasis rather than the establishment of TE-specific gene expression could be the main function of TEAD4 during early mammalian development. However, discrepancies exist in different studies with different experimental approaches. All studies that used the RNAi to deplete TEAD4 (Home et al., 2012; Mihajlović et al., 2015; Wu et al., 2010; Alarcon, 2010) in pre-implantation embryos resulted in impairment of blastocyst maturation and loss of TE-specific factors upon TEAD4 depletion, whereas variable results are obtained with Tead4 conditional knockout systems. Some studies reported impaired blastocyst development and strong loss of TE-specific genes upon Tead4 deletion (Nishioka et al., 2008; Yagi et al., 2007), whereas Kaneko et al. showed that the blastocyst maturation in Tead4-deleted embryos is dependent upon exposure to the oxygen level when cultured ex vivo. The study also showed heterogeneity in TE-specific gene expression in Tead4-null embryos. However, our findings of the variation of the cre-mediated recombination efficiency in TEAD4-conditional knockout system indicates that further studies are needed to draw definitive conclusions about the importance of TEAD4 in establishing the TE-specific transcriptional program. Nevertheless, the fact that endogenous TEAD4 migrates to mitochondria in the TE and TSCs, and regulates mtDNA transcription, further support the hypothesis that TEAD4 is a crucial regulator of mitochondrial function in early mammalian embryos. Most probably during the initiation of blastocoel cavity formation, TEAD4 localization within mitochondria in nascent TE cells and facilitates POLRMT recruitment at the mtDNA, This results in induced expression of mtDNA-encoded ETC complex components, which are required to establish OXPHOS to meet the excessive requirement of ATP in TE cells (Fig. 7G).
Interestingly, TEAD4 is also expressed in the ICM and ICM-derived ESCs, and is present in the cytoplasm of those cells (Home et al., 2012). However, TEAD4 function in those cells is yet to be defined. Although ICM and ESCs use non-oxidative energy, they have metabolically matured mitochondria (Mahato et al., 2014; Leese and Barton, 1984; Mognetti et al., 1996). Furthermore, studies in mouse ESCs revealed that those mitochondria could mediate OXPHOS in ESCs, resulting in a mixed energy metabolism system (Carbognin et al., 2016). Compared with the TE and TSCs, TEAD4 expressions in ICM and ESCs are significantly low. Thus, reduced expression level might lead to less TEAD4 localization in the mitochondria of those cells, leading to inefficient mtDNA transcription, mitochondrial maturation and establishment of OXPHOS. Alternatively, the inefficiency of cellular processes that regulate TEAD4 localization to mitochondria could lead to defective mtDNA transcription in those cells. Nevertheless, it will be interesting to test whether a TEAD4 function is also important to promote mitochondrial maturation and OXPHOS in those contexts.
Our findings also raise at least two questions: (1) what mechanisms regulate TEAD4 localization to the mitochondria; and (ii) does TEAD4 regulate mtDNA transcription in other cell types: It is well known that cellular signaling mechanisms differ in TE and ICM lineage cells. In addition, TEAD4 is expressed in multiple other cell types, including endothelial cells and several cancer cells. Thus, defining signaling mechanisms that regulate TEAD4 localization to mitochondria and testing TEAD4-mediated regulation of mtDNA transcription in other cellular contexts are important areas of further study.
TE/trophoblast lineage and mitochondrial regulation:
Mammalian reproduction is crucially dependent on trophoblast cells, which assure embryo implantation and placentation. Specification of the TE in a pre-implantation embryo initiates trophoblast lineage development. After implantation of the blastocyst, the polar TE cells form the extra-embryonic ectoderm and its derivatives: the ectoplacental cone (EPC) and the chorionic ectoderm (chorion). The EPC and chorion contain trophoblast stem and progenitor cells (TSPCs), which generate diverse trophoblast cell types that are essential for establishing successful pregnancy. Improper development in TE/trophoblast leads to either pregnancy failure or pregnancy-associated complications such as intrauterine growth retardation and pre-eclampsia. Despite of this importance of the trophoblast cell lineage, our understanding of mitochondrial regulation and energy metabolism in trophoblast cells is extremely poor. For example, molecular mechanisms that regulate TE and trophoblast development largely came from studies with TSCs. However, prior to this study, metabolic preferences in TSCs were never tested. Our finding that TSCs rely on oxidative energy metabolism makes them different from ESCs and epiblast stem cells (Choi et al., 2015), which rely on glycolytic mechanisms for ATP production. During early stages of placenta development, TSPCs undergo extensive proliferation and differentiation in an environment that is associated with dynamic changes in oxygen tension. Thus, it is predictive that TSPC proliferation versus differentiation is associated with fine-tuning of mitochondrial energy metabolism. In addition, multiple studies indicated that mitochondrial dysfunction in trophoblast cells is associated with pregnancy-associated pathological conditions (Shi et al., 2013; Wang and Walsh, 1998; Widschwendter et al., 1998; Xie et al., 2014). However, at this moment, it is unknown to us whether or not primary TSPCs within post-implantation mammalian embryos also rely on mitochondrial energy metabolism. Given the conserved nature of TEAD4 expression in the developing trophoblast lineage, it will be interesting to study the importance of TEAD4 in mitochondrial energy metabolism in TSPCs of a post-implantation mammalian embryo.
MATERIALS AND METHODS
Cell culture and reagents
Mouse TSCs were derived following previously described procedures. Mouse TSCs were cultured on irradiated MEFs in TSC culture medium supplemented with 20% fetal bovine serum, 100 μM β-mercaptoethanol, 1 mM sodium pyruvate, 2 mM L-glutamine and primocin 1 μg/ml in the presence of 25 ng/ml fibroblast growth factor 4 and 1 μg/ml heparin, as described previously (Home et al., 2012). Mouse ESCs were cultured in ESC stem state culture conditions as previously described (Mahato et al., 2014). For experimental purposes, mouse TSCs were expanded in the proliferative state without MEF feeders by culturing in the presence of MEF-conditioned medium. RCHO-1 cells were cultured similarly to TSCs without CM, FGF4 and heparin (Sahgal et al., 2006). Human primary CTB cells were isolated from first trimester placenta and cultured in a hypoxia chamber as described previously (Douglas and King, 1989; Kliman et al., 1986; Knofler and Pollheimer, 2013). For this purpose, discarded and de-identified first trimester human placentae were used following approval from Institutional Review Board of the University of Kansas Medical Center.
Mouse embryos
Tead4f/f; UBC-cre/ERT2 female mice were super ovulated and mated with the same genotype males to collect one-cell embryos, as described previously (Saha et al., 2013). The embryos were cultured in KSOM (Millipore) with and without MHT (Sigma Cat. H7904) at 37°C in a 5% CO2 incubator at 5% oxygen. Embryos were collected for RNA and chromatin preparation at the blastocyst stage. RNA prepared from individual blastocyst was used to make cDNA. cDNA was amplified using WGA4 kit (Sigma Cat. WGA4-10RXN) and subsequently used for genotyping and transcript analysis. Experiments were approved by the Institutional Animal Care and Use committee of the University of Kansas Medical Center.
Isolation of ICMs and TEs from blastocysts
ICMs were isolated by embryo immunosurgery and TE samples were isolated after micro-dissection according to a method described previously (Saha et al., 2013; Home et al., 2009). Purified ICM cells and TE samples were further processed for generating samples for either ChIP-WGA or extraction of RNA.
Complex I and complex IV assay
Complex I and IV activity were determined using the Complex I Enzyme Activity Dipstick Assay Kit (Abcam, ab109720) and Complex IV Rodent Enzyme Activity Dipstick Assay Kit (Abcam, ab109878), respectively, according to the manufacturer's instructions. Band intensity was measured using ImageJ.
Oxygen consumption rate and extracellular acidification rate (ECAR) measurements
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were determined by XF cell mito-stress test kit (#101706-100, Seahorse Biosciences) and XF Glycolysis stress test kit (#102194-100, Seahorse Biosciences) as described previously (Zhou et al., 2012; Mahato et al., 2014). ECAR and OCR values were normalized against total protein per well.
Immunohistochemistry and microscopy
Cells were grown on coverslips and processed for immunofluorescence following a previously published protocol (Home et al., 2012; Mahato et al., 2014). For live cell microscopy, cells and embryos at blastocyst stage were stained with MitoSox red (Molecular Probes, M-36008), JC-1 (Molecular Probes, M34152) or MitoTracker Green (Molecular Probes, M-7514) along with Hoechst dye for nuclear staining. Confocal microscopy was performed on cells using a Zeiss LSM PASCAL. All antibodies are listed in Table S2.
TEM protocol for routine morphology and Immunogold:
Samples were fixed in 2% glutaraldehyde following a previously published protocol (Mahato et al., 2014). For immunogold labeling, samples were fixed with 4% paraformaldehyde and examined in a J.E.O.L. JEM 1400 transmission electron microscope using a previously published protocol (She et al., 2011).
RNA interference
For RNA interference, shRNAs against Tead4 and Yap1 were cloned in pLKO.1 lentiviral vectors following previously described protocols (Home et al., 2012). The Tead4 target sequence was 5′-GCTGAAACACTTACCCGAGAA-3′ and the Yap1 target sequence was 5′-GCAGACAGATTCCTTTGTTAACT-3′ were used. We used scrambled shRNA as a negative control (Addgene plasmid #1864). After culturing, samples were prepared for mRNA and protein analysis. For rescue of TEAD4 function in TEAD4KD TSCs, a modified TEAD4 protein with a mitochondrial targeting sequence (MTS) from subunit VIII of human cytochrome c oxidase was ectopically expressed. The MTS sequence was cloned upstream of mouse Tead4 cDNA in the vector pLKO.Tead4-T2A-GFP by Gibson assembly (Jo et al., 2015; Home et al., 2012). The targeting sequence is as follows: 5′-ATGTCCGTCCTGACGCCGCTGCTGCTGCGGGGCTTGACAGGCTCGGCCCGGCGGCTCCCAGTGCCGCGCGCCAAGATCCATTCGTTG-3′. Ectopic TEAD4 expression was confirmed via western blot.
Electrophoretic mobility shift assay
The mtND1 200 bp PCR fragment was purified after PCR using 32P dATP. For gel shift, radiolabeled DNA was incubated with mTSC extract for 30 min in 25 mm HEPES (pH 7.6), 100 mm NaCl, 1 mm DTT, 0.1 mm PMSF, 10% glycerol, 10 μg tRNA and 0.5 μg poly(dI.dC) at room temperature following a previously described protocol (Kumar et al., 2013). For super-shift, equal amounts of TEAD4 antibody or IgG were included in the incubation mix. The mixture was then separated on 4% PAGE (79:1 of acrylamide:bisacrylamide) containing 2.5% glycerol. The gel was autoradiographed after drying.
Mitochondria purification, sub-fractionation and immunoprecipitation
In brief, 20×106 cells were used to isolate mitochondria using mitochondria Isolation Kit for cultured cells (Thermo Scientific, 89874). The homogenate was centrifuged twice at 900 g for 5 min to remove nuclei and unbroken cells (cell lysate) and then the supernatant was centrifuged 3000 g for 15 min. The resultant pellet was used for the purer mitochondrial fraction for western analysis. Mitochondrial sub-fractionation was performed following published protocols after mitochondria purification (She et al., 2011). For immunoprecipitation, a mitochondrial fraction was prepared from E10.5 placentae or mTSC by simply incubating cell lysis buffer from a previously published protocol (Home et al., 2012). The cell lysate was centrifuged twice at 900 g for 5 min to remove nuclei and unbroken cells, and then the supernatant was centrifuged 3000 g for 15 min. The resultant pellet was resuspended in buffer similar to that used in ChIP. The different immunoprecipitated samples were directly boiled in protein sample preparation buffer and used for the western analysis.
Chromatin immunoprecipitation and sequential ChIP
Quantitative ChIP analysis was performed following published protocols (Home et al., 2012; Ray et al., 2009). In brief, 20×106 cells were used to isolate mitochondria using mitochondria isolation kit for cultured cells. Protein-DNA complexes were crosslinked by incubating with 1% formaldehyde (Sigma) for 2 h at 4°C temperature with gentle rotation (Kucej et al., 2008). Chromatin crosslinking was stopped by adding glycine (125 mM) to the reaction mix. These samples were sonicated. Crosslinked chromatin fragments were immunoprecipitated with different antibodies. Quantification of the precipitated DNA was performed using quantitative PCR (qPCR) amplification. A list of the primers used for ChIP analyses is provided in Table S3. For sequential ChIP, crosslinked chromatin fragment prepared from isolated mitochondria was immunoprecipitated with antibody rabbit anti-mtRNA polymerase. Eluted chromatin was re-incubated with mouse anti-Tead4 and mouse IgG1 for overnight at 4°C in the presence of 50 μg/ml yeast tRNA (Dutta et al., 2010). ChIP and SeqChIP on pre-implantation mouse blastocyst embryo were performed previously described (Saha et al., 2013). Immunoprecipitated chromatins were purified using a QIAquick PCR Purification Kit (Qiagen) and amplified using a WGA4 kit (Sigma Aldrich). Amplified DNA was used for target validation analysis. All the samples were normalized to their respective IgG controls for each cell types.
Quantitative RT-PCR
Total RNA from cells was prepared using a RNeasy Kit (Qiagen) after DNAseI digestion. Purified RNA was used to prepare cDNA following a previously published protocol (Home et al., 2009). For analysis of expression in blastocyst embryos, total RNA was isolated using PicoPure RNA isolation kit (MDS Analytical Technology) and processed as described previously (Home et al., 2009). All these samples were analyzed by qRT-PCR using ABI7500. The oligonucleotides used for qRT-PCR are provided in Table S3. All the samples were normalized to their respective 18S RNA controls for each cell types.
Nascent transcript assay
Total RNA from cells was prepared as described above. Before preparing cDNA, the nascent RNA was biotin labeled and purified from total RNA (Molecular Probes, Click-iT Nascent RNA Capture Kit C-10365). After purification of nascent RNA, the residual RNA was precipitated using glycogen and ethanol. Purified RNA was used to prepare cDNA by following a previously published protocol (Home et al., 2009). All these samples were analyzed by qRT-PCR. The oligonucleotides used for qRT-PCR are provided in Table S3.
Quantitation of mtDNA copy number
The number of mtDNA copies per cell was determined using real-time PCR absolute quantification. For absolute quantitation, fragments of the mouse mtND2 and β2 microglobulin genes were cloned in pCR 2.1 plasmid vector and the number of mtND2 and β2 microglobulin genes in a sample was measured against a standard curve of known amount of plasmid vectors. The number of mtDNA copies per cell was calculated using the following formula: mtDNA copy number/cell=2× (copies of mtND2 gene/copies of β2 microglobulin gene).
Genotyping
Genomic DNA was prepared using tail tissue from mouse using a REDExtract-N-Amp Tissue PCR kit (Sigma-Aldrich). Genotyping was carried out using REDExtract-N-Amp PCR ReadyMix (Sigma-Aldrich) and respective primers (see Table S3). Genomic DNA from individual blastocysts was prepared using the following technique with a REDExtract-N-Amp Tissue PCR kit (Sigma-Aldrich). Each blastocyst was collected into separate PCR tubes and was lysed with 16 µl of extraction buffer and 4 µl of tissue prep buffer. Briefly, they were incubated at 42°C for 10 min followed by heat inactivation at 98°C for 3 min and neutralization with 16 µl of neutralization buffer. 4 µl of this genomic DNA was used for a 20 µl PCR reaction.
Statistical significance
Statistical significance for experimental data was analyzed using Student's paired t-test. Although in few figures, studies from multiple groups are presented, the statistical significance was tested by comparing data of two groups and significantly altered values (P≤0.05) between control and TEAD4-delpeted conditions are highlighted in the figures.
Supplementary Material
Acknowledgements
We thank Jackson laboratory for providing mice; Michael J Soares from KUMC for providing rat RCHO-1 cells; Sumedha Gunewardena from KUMC for TEAD4 motif analysis; the KUMC Electron Microscopy Research Lab (EMRL) facility for assistance with the electron microscopy (the JEOL JEM-1400 TEM used in the study was purchased with funds from the National Institutes of Health); Melissa Larson from KUMC transgenic core facility for assistance with the mouse blastocyst isolation, microsurgery and in vitro culture; Drs Inge Kühl and Nils-Göran Larsson from Max-Planck-Institute for Biology of Ageing for providing anti mouse POLRMT antibody and valuable comments.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: R.P.K., S.P.; Methodology: R.P.K., S.R., P.H., B.B., H.M.W., S.P.; Software: S.P.; Validation: R.P.K., S.P.; Formal analysis: R.P.K., S.P.; Investigation: R.P.K., S.P.; Resources: R.P.K., S.R., P.H., B.S., B.B., H.M.W., H.C., A.G., J.M.-F., P.K., R.H.S., A.P., S.P.; Data curation: R.P.K., S.P.; Writing - original draft: R.P.K.; Writing - review & editing: R.P.K., S.P.; Visualization: R.P.K., S.P.; Supervision: R.P.K., S.P.; Project administration: R.P.K., S.P.; Funding acquisition: S.P.
Funding
This work was supported by National Institutes of Health research grants (HD062546 and HD079363) and a National Institutes of Health Centers of Biomedical Research Excellence grant (GM104936). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.162644.supplemental
References
- Alarcon V. B. (2010). Cell polarity regulator PARD6B is essential for trophectoderm formation in the preimplantation mouse embryo. Biol. Reprod. 83, 347-358. 10.1095/biolreprod.110.084400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D. F. (2012). Mitochondrial DNA nucleoid structure. Biochim. Biophys. Acta 1819, 914-920. 10.1016/j.bbagrm.2011.11.005 [DOI] [PubMed] [Google Scholar]
- Bürglin T. R. (1991). The TEA domain: a novel, highly conserved DNA-binding motif. Cell 66, 11-12. 10.1016/0092-8674(91)90132-I [DOI] [PubMed] [Google Scholar]
- Carbognin E., Betto R. M., Soriano M. E., Smith A. G. and Martello G. (2016). Stat3 promotes mitochondrial transcription and oxidative respiration during maintenance and induction of naive pluripotency. EMBO J. 35, 618-634. 10.15252/embj.201592629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Detmer S. A., Ewald A. J., Griffin E. E., Fraser S. E. and Chan D. C. (2003). Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160, 189-200. 10.1083/jcb.200211046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H. W., Kim J. H., Chung M. K., Hong Y. J., Jang H. S., Seo B. J., Jung T. H., Kim J. S., Chung H. M., Byun S. J. et al. (2015). Mitochondrial and metabolic remodeling during reprogramming and differentiation of the reprogrammed cells. Stem Cells Dev. 24, 1366-1373. 10.1089/scd.2014.0561 [DOI] [PubMed] [Google Scholar]
- Chujo T., Ohira T., Sakaguchi Y., Goshima N., Nomura N., Nagao A. and Suzuki T. (2012). LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res. 40, 8033-8047. 10.1093/nar/gks506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn K. and Rossant J. (2010). Making the blastocyst: lessons from the mouse. J. Clin. Invest. 120, 995-1003. 10.1172/JCI41229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas G. C. and King B. F. (1989). Isolation of pure villous cytotrophoblast from term human placenta using immunomagnetic microspheres. J. Immunol. Methods 119, 259-268. 10.1016/0022-1759(89)90405-5 [DOI] [PubMed] [Google Scholar]
- Dutta D., Ray S., Home P., Saha B., Wang S., Sheibani N., Tawfik O., Cheng N. and Paul S. (2010). Regulation of angiogenesis by histone chaperone HIRA-mediated incorporation of lysine 56-acetylated histone H3.3 at chromatin domains of endothelial genes. J. Biol. Chem. 285, 41567-41577. 10.1074/jbc.M110.190025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg M., Gaspari M., Rantanen A., Trifunovic A., Larsson N.-G. and Gustafsson C. M. (2002). Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 31, 289-294. 10.1038/ng909 [DOI] [PubMed] [Google Scholar]
- Frum T. and Ralston A. (2015). Cell signaling and transcription factors regulating cell fate during formation of the mouse blastocyst. Trends Genet. 31, 402-410. 10.1016/j.tig.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ospina G. P., Jimenez-Del Rio M., Lopera F. and Velez-Pardo C. (2003). Neuronal DNA damage correlates with a positive detection of c-Jun, nuclear factor kB, p53 and Par-4 transcription factors in Alzheimer's disease. Rev. Neurol. 36, 1004-1010. [PubMed] [Google Scholar]
- Gaspari M., Falkenberg M., Larsson N.-G. and Gustafsson C. M. (2004). The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J. 23, 4606-4614. 10.1038/sj.emboj.7600465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genbacev O., Larocque N., Ona K., Prakobphol A., Garrido-Gomez T., Kapidzic M., Bárcena A., Gormley M. and Fisher S. J. (2016). Integrin alpha4-positive human trophoblast progenitors: functional characterization and transcriptional regulation. Hum. Reprod. 31, 1300-1314. 10.1093/humrep/dew077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson C. M., Falkenberg M. and Larsson N.-G. (2016). Maintenance and Expression of Mammalian Mitochondrial DNA. Annu. Rev. Biochem. 85, 133-160. 10.1146/annurev-biochem-060815-014402 [DOI] [PubMed] [Google Scholar]
- Hillen H. S., Morozov Y. I., Sarfallah A., Temiakov D. and Cramer P. (2017). Structural basis of mitochondrial transcription initiation. Cell 171, 1072-1081.e10. 10.1016/j.cell.2017.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirate Y., Cockburn K., Rossant J. and Sasaki H. (2012). Tead4 is constitutively nuclear, while nuclear vs. cytoplasmic Yap distribution is regulated in preimplantation mouse embryos. Proc. Natl. Acad. Sci. USA 109, E3389-E3390; author reply E3391-2 10.1073/pnas.1211810109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Home P., Ray S., Dutta D., Bronshteyn I., Larson M. and Paul S. (2009). GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J. Biol. Chem. 284, 28729-28737. 10.1074/jbc.M109.016840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Home P., Saha B., Ray S., Dutta D., Gunewardena S., Yoo B., Pal A., Vivian J. L., Larson M., Petroff M. et al. (2012). Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc. Natl. Acad. Sci. USA 109, 7362-7367. 10.1073/pnas.1201595109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton F. D. (2006). Energy metabolism of the inner cell mass and trophectoderm of the mouse blastocyst. Differentiation 74, 11-18. 10.1111/j.1432-0436.2006.00052.x [DOI] [PubMed] [Google Scholar]
- Ito K. and Suda T. (2014). Metabolic requirements for the maintenance of self-renewing stem cells. Nat. Rev. Mol. Cell Biol. 15, 243-256. 10.1038/nrm3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo A., Ham S., Lee G. H., Lee Y. I., Kim S., Lee Y. S., Shin J. H. and Lee Y. (2015). Efficient mitochondrial genome editing by CRISPR/Cas9. Biomed. Res. Int. 2015, 305716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko K. J. and Depamphilis M. L. (1998). Regulation of gene expression at the beginning of mammalian development and the TEAD family of transcription factors. Dev. Genet. 22, 43-55. [DOI] [PubMed] [Google Scholar]
- Kaneko K. J. and Depamphilis M. L. (2013). TEAD4 establishes the energy homeostasis essential for blastocoel formation. Development 140, 3680-3690. 10.1242/dev.093799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D., Kim S. H. and Hamasaki N. (2007). Mitochondrial transcription factor A (TFAM): roles in maintenance of mtDNA and cellular functions. Mitochondrion 7, 39-44. 10.1016/j.mito.2006.11.017 [DOI] [PubMed] [Google Scholar]
- Kelly R. D. W., Mahmud A., Mckenzie M., Trounce I. A. and St John J. C (2012). Mitochondrial DNA copy number is regulated in a tissue specific manner by DNA methylation of the nuclear-encoded DNA polymerase gamma A. Nucleic Acids Res. 40, 10124-10138. 10.1093/nar/gks770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman H. J., Nestler J. E., Sermasi E., Sanger J. M. and Strauss J. F. III (1986). Purification, characterization, and in vitro differentiation of cytotrophoblasts from human term placentae. Endocrinology 118, 1567-1582. [DOI] [PubMed] [Google Scholar]
- Knofler M. and Pollheimer J. (2013). Human placental trophoblast invasion and differentiation: a particular focus on Wnt signaling. Front Genet. 4, 190 10.3389/fgene.2013.00190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott J. G. and Paul S. (2014). Transcriptional regulators of the trophoblast lineage in mammals with hemochorial placentation. Reproduction 148, R121-R136. 10.1530/REP-14-0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucej M., Kucejova B., Subramanian R., Chen X. J. and Butow R. A. (2008). Mitochondrial nucleoids undergo remodeling in response to metabolic cues. J. Cell Sci. 121, 1861-1868. 10.1242/jcs.028605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl I., Miranda M., Posse V., Milenkovic D., Mourier A., Siira S. J., Bonekamp N. A., Neumann U., Filipovska A. Polosa P. L. et al. (2016). POLRMT regulates the switch between replication primer formation and gene expression of mammalian mtDNA. Sci. Adv. 2, e1600963 10.1126/sciadv.1600963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukat C. and Larsson N.-G. (2013). mtDNA makes a U-turn for the mitochondrial nucleoid. Trends Cell Biol. 23, 457-463. 10.1016/j.tcb.2013.04.009 [DOI] [PubMed] [Google Scholar]
- Kukat C., Davies K. M., Wurm C. A., Spåhr H., Bonekamp N. A., Kühl I., Joos F., Polosa P. L., Park C. B., Posse V. et al. (2015). Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc. Natl. Acad. Sci. USA 112, 11288-11293. 10.1073/pnas.1512131112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R. P., Krishnan J., Pratap Singh N., Singh L. and Mishra R. K. (2013). GATA simple sequence repeats function as enhancer blocker boundaries. Nat. Commun. 4, 1844 10.1038/ncomms2872 [DOI] [PubMed] [Google Scholar]
- Lambertini E., Penolazzi L., Morganti C., Lisignoli G., Zini N., Angelozzi M., Bonora M., Ferroni L., Pinton P., Zavan B. et al. (2015). Osteogenic differentiation of human MSCs: specific occupancy of the mitochondrial DNA by NFATc1 transcription factor. Int. J. Biochem. Cell Biol. 64, 212-219. 10.1016/j.biocel.2015.04.011 [DOI] [PubMed] [Google Scholar]
- Larsson N.-G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G. S. and Clayton D. A. (1998). Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 18, 231-236. 10.1038/ng0398-231 [DOI] [PubMed] [Google Scholar]
- Lee J., Kim C.-H., Simon D. K., Aminova L. R., Andreyev A. Y., Kushnareva Y. E., Murphy A. N., Lonze B. E., Kim K. S., Ginty D. D. et al. (2005). Mitochondrial cyclic AMP response element-binding protein (CREB) mediates mitochondrial gene expression and neuronal survival. J. Biol. Chem. 280, 40398-40401. 10.1074/jbc.C500140200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leese H. J. and Barton A. M (1984). Pyruvate and glucose uptake by mouse ova and preimplantation embryos. J. Reprod. Fertil. 72, 9-13. 10.1530/jrf.0.0720009 [DOI] [PubMed] [Google Scholar]
- Leppens G., Gardner D. K. and Sakkas D. (1996). Co-culture of 1-cell outbred mouse embryos on bovine kidney epithelial cells: effect on development, glycolytic activity, inner cell mass:trophectoderm ratios and viability. Hum. Reprod. 11, 598-603. 10.1093/HUMREP/11.3.598 [DOI] [PubMed] [Google Scholar]
- Li H., Wang J., Wilhelmsson H., Hansson A., Thoren P., Duffy J., Rustin P. and Larsson N. G. (2000). Genetic modification of survival in tissue-specific knockout mice with mitochondrial cardiomyopathy. Proc. Natl. Acad. Sci. USA 97, 3467-3472. 10.1073/pnas.97.7.3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias E., Rao D., Carbajal S., Kiguchi K. and Digiovanni J. (2014). Stat3 binds to mtDNA and regulates mitochondrial gene expression in keratinocytes. J. Invest. Dermatol. 134, 1971-1980. 10.1038/jid.2014.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahato B., Home P., Rajendran G., Paul A., Saha B., Ganguly A., Ray S., Roy N., Swerdlow R. H. and Paul S. (2014). Regulation of mitochondrial function and cellular energy metabolism by protein kinase C-lambda/iota: a novel mode of balancing pluripotency. Stem Cells 32, 2880-2892. 10.1002/stem.1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloyan A., Mele J., Muralimanohara B. and Myatt L. (2012). Measurement of mitochondrial respiration in trophoblast culture. Placenta 33, 456-458. 10.1016/j.placenta.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier J. A. and Larner A. C. (2014). Toward a new STATe: the role of STATs in mitochondrial function. Semin. Immunol. 26, 20-28. 10.1016/j.smim.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metodiev M. D., Lesko N., Park C. B., Camara Y., Shi Y., Wibom R., Hultenby K., Gustafsson C. M. and Larsson N. G. (2009). Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 9, 386-397. 10.1016/j.cmet.2009.03.001 [DOI] [PubMed] [Google Scholar]
- Mihajlović A. I., Thamodaran V. and Bruce A. W. (2015). The first two cell-fate decisions of preimplantation mouse embryo development are not functionally independent. Sci. Rep. 5, 15034 10.1038/srep15034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minczuk M., He J., Duch A. M., Ettema T. J., Chlebowski A., Dzionek K., Nijtmans L. G. J., Huynen M. A. and Holt I. J. (2011). TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res. 39, 4284-4299. 10.1093/nar/gkq1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mognetti B., Leppens G. and Sakkas D. (1996). The development of preimplantation mouse parthenogenones in vitro in absence of glucose: influence of the maternally inherited components. Mol. Reprod. Dev. 43, 421-427. [DOI] [PubMed] [Google Scholar]
- Nishioka N., Yamamoto S., Kiyonari H., Sato H., Sawada A., Ota M., Nakao K. and Sasaki H. (2008). Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech. Dev. 125, 270-283. 10.1016/j.mod.2007.11.002 [DOI] [PubMed] [Google Scholar]
- Nishioka N., Inoue K., Adachi K., Kiyonari H., Ota M., Ralston A., Yabuta N., Hirahara S., Stephenson R. O., Ogonuki N. et al. (2009). The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398-410. 10.1016/j.devcel.2009.02.003 [DOI] [PubMed] [Google Scholar]
- Piechota J., Tomecki R., Gewartowski K., Szczesny R., Dmochowska A., Kudla M., Dybczynska L., Stepien P. P. and Bartnik E. (2006). Differential stability of mitochondrial mRNA in HeLa cells. Acta Biochim. Pol. 53, 157-168. [PubMed] [Google Scholar]
- Posse V., Shahzad S., Falkenberg M., Hällberg B. M. and Gustafsson C. M. (2015). TEFM is a potent stimulator of mitochondrial transcription elongation in vitro. Nucleic Acids Res. 43, 2615-2624. 10.1093/nar/gkv105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston A., Cox B. J., Nishioka N., Sasaki H., Chea E., Rugg-Gunn P., Guo G., Robson P., Draper J. S. and Rossant J. (2010). Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137, 395-403. 10.1242/dev.038828 [DOI] [PubMed] [Google Scholar]
- Ray S., Dutta D., Rumi M. A. K., Kent L. N., Soares M. J. and Paul S (2009). Context-dependent function of regulatory elements and a switch in chromatin occupancy between GATA3 and GATA2 regulate Gata2 transcription during trophoblast differentiation. J. Biol. Chem. 284, 4978-4988. 10.1074/jbc.M807329200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo A. P., Dillon L. M. and Moraes C. T. (2011). Mitochondrial DNA transcription regulation and nucleoid organization. J. Inherit. Metab. Dis. 34, 941-951. 10.1007/s10545-011-9330-8 [DOI] [PubMed] [Google Scholar]
- Saha B., Home P., Ray S., Larson M., Paul A., Rajendran G., Behr B. and Paul S. (2013). EED and KDM6B coordinate the first mammalian cell lineage commitment to ensure embryo implantation. Mol. Cell. Biol. 33, 2691-2705. 10.1128/MCB.00069-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahgal N., Canham L. N., Canham B. and Soares M. J. (2006). Rcho-1 trophoblast stem cells: a model system for studying trophoblast cell differentiation. Methods Mol. Med. 121, 159-178. [PubMed] [Google Scholar]
- She H., Yang Q., Shepherd K., Smith Y., Miller G., Testa C. and Mao Z. (2011). Direct regulation of complex I by mitochondrial MEF2D is disrupted in a mouse model of Parkinson disease and in human patients. J. Clin. Invest. 121, 930-940. 10.1172/JCI43871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z., Long W., Zhao C., Guo X., Shen R. and Ding H. (2013). Comparative proteomics analysis suggests that placental mitochondria are involved in the development of pre-eclampsia. PLoS One 8, e64351 10.1371/journal.pone.0064351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutt T. E., Lodeiro M. F., Cotney J., Cameron C. E. and Shadel G. S. (2010). Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc. Natl. Acad. Sci. USA 107, 12133-12138. 10.1073/pnas.0910581107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sologub M., Litonin D., Anikin M., Mustaev A. and Temiakov D. (2009). TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell 139, 934-944. 10.1016/j.cell.2009.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John J. C., Amaral A., Bowles E., Oliveira J. F., Lloyd R., Freitas M., Gray H. L., Navara C. S., Oliveira G., Schatten G. P. et al. (2006). The analysis of mitochondria and mitochondrial DNA in human embryonic stem cells. Methods Mol. Biol. 331, 347-374. [DOI] [PubMed] [Google Scholar]
- Taanman J.-W. (1999). The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta 1410, 103-123. 10.1016/S0005-2728(98)00161-3 [DOI] [PubMed] [Google Scholar]
- Thundathil J., Filion F. and Smith L. C. (2005). Molecular control of mitochondrial function in preimplantation mouse embryos. Mol. Reprod. Dev. 71, 405-413. 10.1002/mrd.20260 [DOI] [PubMed] [Google Scholar]
- Trimarchi J. R., Liu L., Porterfield D. M., Smith P. J. S. and Keefe D. L. (2000). Oxidative phosphorylation-dependent and -independent oxygen consumption by individual preimplantation mouse embryos. Biol. Reprod. 62, 1866-1874. 10.1095/biolreprod62.6.1866 [DOI] [PubMed] [Google Scholar]
- Wang Y. and Walsh S. W. (1998). Placental mitochondria as a source of oxidative stress in pre-eclampsia. Placenta 19, 581-586. 10.1016/S0143-4004(98)90018-2 [DOI] [PubMed] [Google Scholar]
- Wang J., Wilhelmsson H., Graff C., Li H., Oldfors A., Rustin P., Brüning J. C., Kahn C. R., Clayton D. A., Barsh G. S. et al. (1999). Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat. Genet. 21, 133-137. 10.1038/5089 [DOI] [PubMed] [Google Scholar]
- Wanrooij S., Fuste J. M., Farge G., Shi Y., Gustafsson C. M. and Falkenberg M. (2008). Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc. Natl. Acad. Sci. USA 105, 11122-11127. 10.1073/pnas.0805399105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M., Schrocksnadel H. and Mortl M. G. (1998). Pre-eclampsia: a disorder of placental mitochondria? Mol. Med. Today 4, 286-291. [DOI] [PubMed] [Google Scholar]
- Wilding M., Coppola G., Dale B. and Di Matteo L. (2009). Mitochondria and human preimplantation embryo development. Reproduction 137, 619-624. 10.1530/REP-08-0444 [DOI] [PubMed] [Google Scholar]
- Wolf A. R. and Mootha V. K. (2014). Functional genomic analysis of human mitochondrial RNA processing. Cell Rep. 7, 918-931. 10.1016/j.celrep.2014.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Gentile L., Fuchikami T., Sutter J., Psathaki K., Esteves T. C., Arauzo-Bravo M. J., Ortmeier C., Verberk G., Abe K. et al. (2010). Initiation of trophectoderm lineage specification in mouse embryos is independent of Cdx2. Development 137, 4159-4169. 10.1242/dev.056630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Zhou S., Jiang Z., Dai J., Puscheck E. E., Lee I., Parker G., Hüttemann M. and Rappolee D. A. (2014). Hypoxic stress induces, but cannot sustain trophoblast stem cell differentiation to labyrinthine placenta due to mitochondrial insufficiency. Stem Cell Res. 13, 478-491. 10.1016/j.scr.2014.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R., Kohn M. J., Karavanova I., Kaneko K. J., Vullhorst D., Depamphilis M. L. and Buonanno A. (2007). Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 134, 3827-3836. 10.1242/dev.010223 [DOI] [PubMed] [Google Scholar]
- Yakubovskaya E., Guja K. E., Eng E. T., Choi W. S., Mejia E., Beglov D., Lukin M., Kozakov D. and Garcia-Diaz M. (2014). Organization of the human mitochondrial transcription initiation complex. Nucleic Acids Res. 42, 4100-4112. 10.1093/nar/gkt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdravkovic T., Nazor K. L., Larocque N., Gormley M., Donne M., Hunkapillar N., Giritharan G., Bernstein H. S., Wei G., Hebrok M. et al. (2015). Human stem cells from single blastomeres reveal pathways of embryonic or trophoblast fate specification. Development 142, 4010-4025. 10.1242/dev.122846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernicka-Goetz M., Morris S. A. and Bruce A. W. (2009). Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat. Rev. Genet. 10, 467-477. 10.1038/nrg2564 [DOI] [PubMed] [Google Scholar]
- Zhou R., Yazdi A. S., Menu P. and Tschopp J. (2011). A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221-225. 10.1038/nature09663 [DOI] [PubMed] [Google Scholar]
- Zhou W., Choi M., Margineantu D., Margaretha L., Hesson J., Cavanaugh C., Blau C. A., Horwitz M. S., Hockenbery D., Ware C. et al. (2012). HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 31, 2103-2116. 10.1038/emboj.2012.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.