Abstract
Background
Acinetobacter baumannii is a healthcare-associated pathogen with high rates of carbapenem resistance. Colistin is now routinely used for treatment of infections by this pathogen. However, colistin use has been associated with development of resistance to this agent.
Objectives
To elucidate the phylogenomics of colistin-susceptible and -resistant A. baumannii strain pairs from a cohort of hospitalized patients at a tertiary medical centre in the USA.
Methods
WGS data from 21 pairs of colistin-susceptible and -resistant, XDR clinical strains were obtained and compared using phylogeny of aligned genome sequences, assessment of pairwise SNP differences and gene content.
Results
Fourteen patients had colistin-resistant strains that were highly genetically related to their own original susceptible strain with a median pairwise SNP distance of 5.5 (range 1–40 SNPs), while seven other strain pairs were divergent with ≥84 SNP differences. In addition, several strains from different patients formed distinct clusters on the phylogeny in keeping with closely linked transmission chains. The majority of colistin-resistant strains contained non-synonymous mutations within the pmrAB locus suggesting a central role for pmrAB mutations in colistin resistance. Excellent genotype–phenotype correlation was also observed for carbapenems, aminoglycosides and tetracyclines.
Conclusions
The findings suggest that colistin resistance in the clinical setting arises through both in vivo evolution from colistin-susceptible strains and reinfection by unrelated colistin-resistant strains, the latter of which may involve patient-to-patient transmission.
Introduction
Acinetobacter baumannii has emerged as an opportunistic pathogen associated with bacteraemia, respiratory, skin and soft tissue infection due to the high frequency with which it acquires antimicrobial resistance, and its ability to survive in the healthcare environment.1 It is one of the six pathogens that account for a majority of healthcare-associated infections in the United States.2 Of particular concern, the majority of A. baumannii clinical strains are now resistant to carbapenems.3 Colistin belongs to the polymyxin class and is one of the few agents that remain active against most carbapenem-resistant A. baumannii strains. However, colistin use has been associated with development of resistance to this agent, which has been linked to mutations in the two-component transcriptional regulator genes pmrAB in most instances.4
Several genomic studies have demonstrated that A. baumannii can rapidly evolve antimicrobial resistance by chromosomal mutations as well as by plasmid acquisition.5,6 For example, mutations are frequently identified in pmrAB involved in colistin resistance, adeRS involved in tigecycline resistance, as well as iron acquisition genes.5 In addition, there is rapid, extensive gain and loss of antibiotic resistance genes through acquisition and loss of plasmids, transposons and integrons.7
We previously described a cohort of hospitalized patients at a large tertiary medical centre who were initially infected by colistin-susceptible, carbapenem-resistant A. baumannii strains and subsequently developed infection or colonization with colistin-resistant A. baumannii strains after being treated with colistin.8 Conventional genotyping using PFGE showed that colistin-resistant strains were more similar to the colistin-susceptible strains from the same patients compared with colistin-resistant strains from other patients, supporting in vivo emergence of colistin resistance through evolution of persistently infecting or colonizing strains. However, a recent genomic study of within-patient evolution of A. baumannii reported 13 of 40 (32.5%) patients thought to have persistent infections in fact had reinfection by different strains.5 These results suggest that even though colistin resistance is often acquired through evolution of the original infecting strain, some patients may acquire colistin-resistant infection through reinfection by genetically distinct strains, which can only be resolved by genome sequencing. The aim of this work was to elucidate the genomic basis for the emergence of resistance following colistin therapy among a well-characterized set of patients at a tertiary care hospital.
Methods
Strain collection
A total of 42 carbapenem-resistant A. baumannii strains, two each from 21 different patients, were collected during the course of routine clinical care at a tertiary hospital in Western Pennsylvania between 2007 and 2016, including the previously described patients.8 One of each strain pair was colistin susceptible (MIC ≤2 mg/L; ‘S’ strains) while the second strain was resistant (MIC >2 mg/L; ‘R’ strains). Eighteen patients (serial numbers 1–7 and 9–19) had a colistin-susceptible strain first, were treated with colistin, and then had a colistin-resistant strain. One patient was treated with colistin before the colistin-susceptible strain was isolated, then had a colistin-resistant strain later (patient 8); one patient had a colistin-resistant strain first then colistin-susceptible strain later, but did not receive colistin around or before this time (patient 20). One patient received colistin earlier, had a colistin-resistant strain, then a colistin-susceptible strain later (patient 21). Colistin resistance was assessed using broth microdilution MICs determined by the CLSI method and confirmed by detection of the characteristic mass peaks representing phosphoethanolamine modification of hepta-acetylated lipid A using MALDI-TOF/MS as previously described.8 MICs of other antibiotics were determined by broth microdilution using Sensititre GNX3F plates (TREK Diagnostic Systems, Oakwood Village, OH, USA). Results were interpreted according to the CLSI susceptibility breakpoints.9
Genome sequencing, assembly and annotation
The genomic DNA was extracted with the Qiagen Blood and Tissue Kit and sequenced using Illumina HiSeq 150 bp or MiSeq 250 bp paired-end sequencing. Genome sequences were assembled using SPAdes v3.510 and annotated using prokka v1.11.11 A high-quality reference genome sequence for S1, the colistin-susceptible strain of the first strain pair identified, was obtained using the Pacific Biosciences single molecule real time platform using P4-C2 sequencing chemistry and assembled using Hierarchical Genome Assembler v2.0 into a single contiguous chromosome and three plasmid sequences.12
A number of methods were used to identify genetic variation among study strains. Nullarbor (https://github.com/tseemann/nullarbor) provided high-quality, core SNPs shared among all strains. Breseq v0.25 provided additional details on pairwise SNP comparisons of strains from the same patient.13 Within-patient SNPs were defined as SNPs that were present in only one isolate within a patient pair while subclade-specific SNPs were shared by all strains within a particular subclade. A core SNP phylogenetic tree was reconstructed with RAxML using the general time-reversible model of evolution, Γ distributed rate variation among sites (GTRGAMMA) and 1000 rapid bootstrap replicates.14
Gene content was compared using BLASTn searches of predicted genes against assembled contigs using an 80% sequence identity cut-off and confirmed by mapping sequencing reads to the query gene using bwa-mem v0.7.15 Gene content matrices were then generated using custom python scripts. Pangenome analysis was conducted using the orthologue clustering program Roary v3.6 with the default 95% sequence identity cut-off.16 Antibiotic gene content was assessed using ResFinder and NCBI β-lactamase (www.ncbi.nlm.nih.gov/pathogens/beta-lactamase-data-resources/) databases based on 70% nucleotide sequence identity cut-off.17 The capsule gene locus (KL) was compared using assembled contigs and mapping of raw sequence reads to reference KL sequences as described by Kenyon et al.18 Sequence alignments were manually curated and compared on CLC genomics workbench v9.0 (www.clcbio.com).
Nucleotide sequence accession numbers
Raw sequencing reads from this study are deposited in NCBI sequence reads archive under accession numbers SRR6256389–SRR6256430; accession numbers for complete sequence assemblies of the reference genome S1 chromosome and three plasmids are CP026943–CP026946 while 41 draft genome assemblies are available under accession numbers PUCB00000000–PUDP00000000 (see Table S1 available as Supplementary data at JAC Online). GenBank accession numbers for two new β-lactamase gene sequences are MG452937 (blaADC-161) and MG452939 (blaOXA-162).
Results
Complete genome of A. baumannii strain S1
A complete genome sequence was generated for reference strain S1 using Hierarchical Genome Assembler into a single chromosome and three separate plasmid sequences pAbS1_01–pAbS1_03.19 Plasmid pAbS1_01 was a novel 108 kb plasmid containing predominantly phage-related genes; a second 110 kb plasmid pAbS1_02 was very similar to the pABTJ2 found in the reference strain MDR-TJ and contained no known resistance elements. A third plasmid pAbS1_03 was a 72 kb plasmid highly similar (95%) to pAba3207b found in an A. baumannii ST422 strain identified in Mexico,20 except that it contained Tn2008A carrying the blaOXA-23 carbapenemase gene.21
Sequence typing of colistin-susceptible and -resistant A. baumannii strains
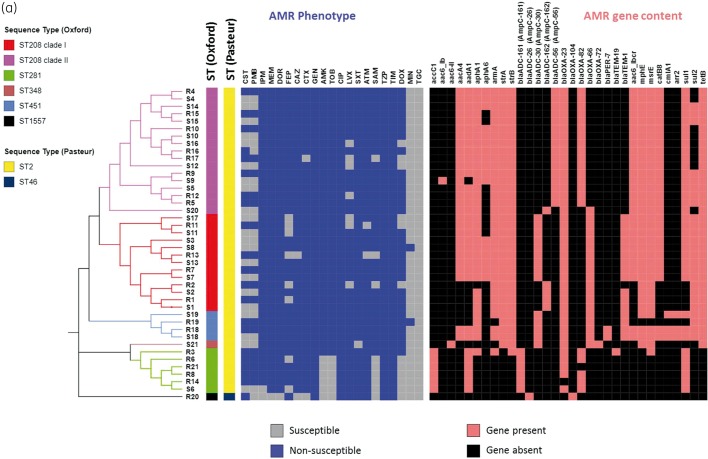
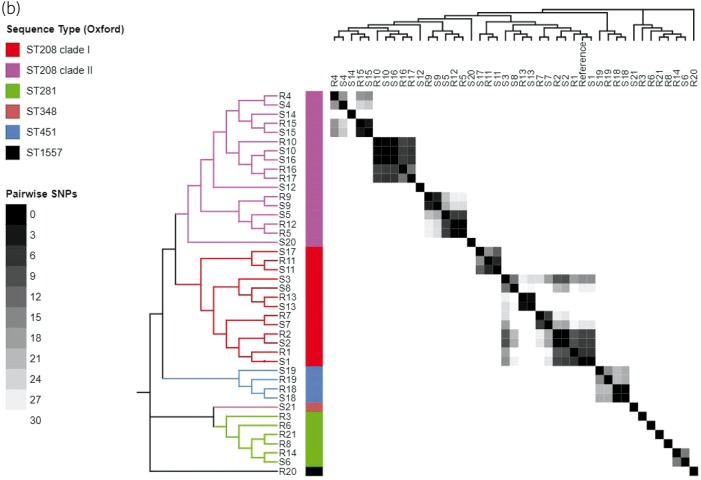
All except one strain (41 of 42) were ST2 belonging to previously described global clone 2 (GC2) as defined by the Pasteur Institute MLST scheme,22 with only one strain (R20) belonging to ST46, which differs from ST2 in six of seven loci (Figure 1 and Table 1). On the other hand, the Oxford MLST scheme captured substantially more diversity among strains,23 clustering the 42 strains into five distinct STs: the majority of strains (30 of 42, 71.4%) belonged to ST208 while ST281 and ST451 made up 11.8% (n = 4) and 5.9% (n = 2), and one isolate (R20) belonged to a novel ST, ST1557. ST208 is now understood as identical to ST92, which is defined by the Oxford MSLT scheme when Sanger sequencing is used instead of high-throughput sequencing,24 and includes strains that were previously reported as ST92 based on Sanger sequencing.8 In fact, ST208, ST281 and ST451 all belonged to clonal group 92 (CG92) based on this scheme. Phylogenomic analyses based on high-quality shared SNPs and aligned core gene sequences (Figure 1) supported the distinction of the A. baumannii genomes into five major clades, corresponding to ST208 clade I, ST208 clade II, ST281/348, ST451 and ST1557. The Oxford MLST scheme therefore captures the population structure of our study isolates with better granularity and will be used henceforth.
Figure 1.
(a) Phylogeny, AMR and AMR gene content of 42 A. baumannii genomes showing core SNP maximum likelihood tree (left), STs (coloured bars), AMR susceptibility profile and presence/absence of AMR genes. AMR, antimicrobial resistance; CST, colistin; PMB, polymyxin B; IPM, imipenem; MEM, meropenem; DOR, doripenem; FEP, cefepime; CAZ, ceftazidime; CTX, cefotaxime; GEN, gentamicin; AMK, amikacin; TOB, tobramycin; CIP, ciprofloxacin; LVX levofloxacin; SXT trimethoprim/sulfamethoxazole; ATM, aztreonam; SAM, ampicillin/sulbactam; TZP, piperacillin/tazobactam; TIM, ticarcillin/clavulanic acid; DOX, doxycycline; MIN, minocycline; TGC, tigecycline. (b) Phylogeny of 42 A. baumannii genomes (left and top panels) juxtaposed with heatmaps of STs and pairwise SNP differences between strains.
Table 1.
List of mutations associated with colistin resistance in this study. Mutations were detected by pairwise SNP comparisons between one pair of colistin-susceptible and -resistant strains from each patient and sequence alignment of pmrAB genes
| Patient | ST (Oxford) | Core SNP differencesa | pmr mutationsb | Studies reporting identical pmrAB mutations (first author and reference number) | SNPs outside pmr |
|---|---|---|---|---|---|
| 1 | 208/208 | 6 | lptF, cyaA, yjaC, topA and deaD | ||
| 2 | 208/208 | 3 | L20F (pmrA) | cyaA, adeI | |
| 3 | 281/208 | 5720 | |||
| 4 | 208/208 | 17 | M12I (pmrA) | Arroyo28 | pgaC |
| 5 | 208/208 | 7 | R134S, T235I | Arroyo28 | cyaA, mmgC, mexA and itrA2 |
| 6 | 281/281 | 40 | A226T | pilY and zinc binding alcohol dehydrogenase | |
| 7 | 208/208 | 7 | – | znuA, yaiI, smvA, cyaA, ubiH, yecA and edd | |
| 8 | 281/208 | 7273 | F105L (pmrA) | ||
| 9 | 208/208 | 4 | T232A, F267L, L271F | gtr5, mfs | |
| 10 | 208/208 | 2 | Q277K | ||
| 11 | 208/208 | 5 | znuA, lptC, mfs and nlhH | ||
| 12 | 208/208 | 84 | P119L, R134S, T235I | Arroyo28 | |
| 13 | 208/208 | 3 | L208R, R263C | Arroyo28 | itrA2 |
| 14 | 281/208 | 7642 | pilY | ||
| 15 | 208/208 | 3 | P233T, L292H | Adams29 | |
| 16 | 208/208 | 7 | clpA, tolR, sel1 and mfs | ||
| 17 | 208/208 | 313 | A14V (pmrA) | ||
| 18 | 451/451 | 1 | P233S | Adams29; Beceiro30; Arroyo28; Kim31 | gna |
| 19 | 451/451 | 16 | G315V | tetR, wzc, gna, asnC and pgap1 | |
| 20 | 1557/208 | 44 830 | V444A | ||
| 21 | 281/348 | 7488 | – |
Core SNP counts include only genome sequences shared by all 42 isolates. Upstream part of pmrB was truncated in reference genome S1 and was excluded from core SNP analysis.
Mutations refer to pmrB except when pmrA is indicated inside parenthesis.
Pairwise SNP analyses
A total of 93 359 shared core SNP positions were identified among the 42 strains. When pairwise SNP differences were compared, 14 pairs of strains from the same patients were very highly related with a median of 5.5 (range 1–40) SNP differences from each other (Table 1). These paired strains belonged to highly related branches of the phylogenetic tree (Figure 1). As such, these monophyletic strain pairs were considered to represent within-patient emergence of colistin resistance through the persistence and evolution of a single infecting strain (Figure 1 and Table 1). The vast majority (98 of 110) of these 1–40 SNPs from within patient pairs were unique to individual patients, while 11 SNPs were shared by resistant isolates from two patients. All 11 SNPs shared by more than one resistant isolate were localized to two intergenic and hypothetical gene sequences clustered within two chromosomal regions containing phage-like sequences at positions 1898 and 3339 kb on the S1 reference genome. No SNPs were shared by more than two resistant isolates. The other seven isolate pairs, including those from two patients whose resistant strains were identified before the susceptible strains, had much higher genetic distances indicated by 7273 median pairwise SNP distances (range 84–44 830) and belonged to genetically distinct subclades on the phylogeny (Table 1 and Figure 1). These strains most likely represented reinfection of the patients with new strains of A. baumannii.
Pairwise genomic comparison of same-patient strains
Assessment of pairwise SNP differences and gene content provided insight into the genomic basis of colistin resistance among strains. A number of studies have linked non-synonymous mutations within pmrAB with colistin resistance.4 Therefore, we aligned pmrAB sequences comparing gene sequences within our data set and with the ACICU reference strain, to identify whether colistin-resistant strains contained mutations within pmrAB that may be linked to colistin resistance. In the pmr locus, strain S1 had a 2.3 kb deletion involving 94% of pmrC (phosphoethanolamine transferase), all of pmrA and the first 44 amino acids of pmrB, all replaced by a single copy of ISAba22. Colistin-resistant strain R1 had intact pmrAB genes. In addition, strain R1 had acquired mutations in the lptF gene involved in lipooligosaccharide transport, and cyaA, associated with reduced susceptibility to fosfomycin, quinolones and cefoxitin.25 Two mutations present in S1 (located in topoisomerase gene topA, and deaD gene involved in Type III secretion system) were not present in R1.26 Additional mutations were found in yjaC likely involved in quorum sensing.27 In addition, two of three plasmids found in S1 (pAbS1_01 and pAbS1_02) were also contained in R1. The overall picture suggests a scenario where both S1 and R1 evolved from a shared ancestor.
The majority of colistin-resistant strains (15 of 21, 71.4%) contained non-synonymous mutations within the pmrAB locus suggesting a central role for pmrAB mutations in the emergence of colistin resistance as reported by previous studies (Table 1).4,28 For example, M12I mutation within pmrA and P233S, P233T, T235I and R263C pmrB mutations found among colistin-resistant strains in our study were previously linked to colistin resistance by at least one study.28–31
In addition, several colistin-resistant strains had additional mutations outside pmrAB. These mutations more commonly affected genes involved in the Type III secretion system (cyaA), major facilitator family of transport proteins (mfs), surface structures (pilY, wzc, itrA2, gtr5, gna), zinc uptake (znuA) and efflux pumps (adeI, mexA).32,33 The wzc gene is part of the capsule polysaccharides synthesis K locus where mutations have been shown to affect virulence.34–37 Several strain pairs showed significant differences in gene content despite very low SNP differences in the core genome, in keeping with rapid loss and gain of genetic elements. For example, strain R6 contained a 12 kb genomic island not found in S6. This genomic island is similar to TnAbaR23 previously linked to unpredictable changes in drug resistance.8,38 Within S6 there is a 14 kb prophage-like chromosomal sequence that was not found in R6. An alternative explanation is that S6 and R6 may represent independent acquisition of distinct, yet highly similar strains.
Finally, some strains from different patients were genetically highly similar and formed very closely related monophyletic clades suggesting transmission either directly between patients or through a common focal exposure. The median pairwise distance within these clusters was 20 SNPs, which substantially overlaps the SNP distance range of 1–40 SNPs seen among the same-patient monophyletic strain pairs, in contrast to the median SNP distance of 878 for the remaining strain pairs from different patients.
These genetic clusters were highly suggestive of between-patient transmission networks. Six of these phylogenetic clusters had a range of 0–35 pairwise SNP distances between strains within the same cluster, which was comparable to 3–40 SNPs observed between same-patient strain pairs. Five of six transmission clusters contained two or more colistin-resistant strains that were genetically very closely related. Only two strains (R5 and R12) shared common pmr mutations (R134S, T235I) suggesting possible direct transmission of a resistant strain between patients 5 and 12. Strains R5 and R12 were identified 425 days apart but from the same ICU, raising the possibility of an occult environmental reservoir given the ability of this organism to survive in the healthcare environment.1 None the less, the overall findings support the hypothesis that colistin resistance commonly arises in vivo in the course of colistin therapy.
Antimicrobial resistance genes and phenotype correlation
All A. baumannii strains in this study were MDR (resistant to three or more classes of antimicrobial agents) with all 42 strains also being XDR (resistant to all except up to two classes of antimicrobial agents).39 A large variety of β-lactamase genes was found among genomes in this study (Figure 1). All 42 genomes contained one or more OXA-type carbapenemase gene. Forty-one genomes contained the most commonly reported acquired carbapenemase gene blaOXA-23. All genomes contained an intrinsic blaOXA-51 group carbapenemase gene, where blaOXA-66 was tightly associated with ST208 clade I, ST348 and ST451; blaOXA-82 was associated with ST208 clade II and ST281, while one isolate (R20) contained blaOXA-104 (Figure 1). ISAba1 was found upstream of blaOXA-82 in all blaOXA-82-containing strains except S5 while blaOXA-66 and blaOXA-104 had no upstream IS. IS elements are known to provide strong promoter sequences for the expression of blaOXA-51-like. All genomes contained a variant of the chromosomal ADC group cephalosporinase gene.40,41 Genomes within the ST208 clade I and ST451 contained the blaADC-30 (corresponds to AmpC peptide, allele 30) or blaADC-162 (AmpC-162) gene that differed from blaADC-30 by A245E substitution, while all genomes belonging to ST208 clade II contained blaADC-56. Six ST281 strains contained blaADC-161 (AmpC-161) gene while the blaADC-26 (AmpC-26) gene was found in one genome (R20). blaADC had upstream ISAba1 sequences in 42 of 43 genomes in this study; the exception is blaADC-26 (R20), which had no upstream IS. Likewise, an allelic variant of the blaTEM-2 β-lactamase gene that encoded K37Q substitution was found predominantly among ST208 clade II genomes. Two genomes from the same patient (S18 and R18) contained blaPER-7, an ESBL gene. All but one genome (R20) contained two or more aminoglycoside resistance genes including aac(3)-Ia, aacA4, aadA1, aph(3′)-VIa, aph(3′)-Ic, armA, strA and strB.
Antimicrobial susceptibility phenotypes were compared with the corresponding gene content among genomes. All 41 blaOXA-23-containing strains were non-susceptible to all tested carbapenems (imipenem, meropenem and doripenem). The only strain without an acquired carbapenemase gene (R20) was carbapenem-susceptible but colistin-resistant, consistent with the aforementioned genotypes. All strains were non-susceptible to one or more cephalosporin, but two strains each (2 of 42, 4.8%) were susceptible to cefotaxime and ceftazidime while eight strains (19%) were susceptible to cefepime. All 16 blaADC-56-containing genomes were non-susceptible to cefepime compared with 69.2% (18 of 26) of strains containing other blaADC variants, consistent with the unique spectrum of blaADC-56 that includes cefepime.42
All 42 strains were non-susceptible to gentamicin and the 36 armA-containing strains were non-susceptible to amikacin and tobramycin as well, which is a phenotype consistent with production of the 16S ribosomal RNA methyltransferase, which protects the 30S ribosome from aminoglycoside binding. All 42 strains contained substitutions within the QRDR, namely S83L within gyrA (42 strains), S80L within parC (41 strains)43 and a previously unreported S80F parC mutation in one strain (R20). As expected, all strains were non-susceptible to ciprofloxacin while 83.3% were non-susceptible to levofloxacin. Non-susceptibility to doxycycline was high (71.4%), but only a small minority of strains (5.7%) were non-susceptible to minocycline. Genomes containing tetB had a significantly higher frequency of doxycycline non-susceptibility (82.8% versus 14.3%, P < 0.001) and the only two minocycline non-susceptible strains also contained tetB, reflecting the role of the TetB efflux pump in the non-susceptibility to these tetracyclines.44 All of the strains in this study were susceptible to tigecycline.
Capsule biosynthesis locus (KL) and correlation to ST
Gene content and arrangement within KL was strongly linked to Oxford STs. All ST208 genomes contained an identical KL corresponding to KL6a.18 ST281, ST348 and ST451 genomes shared a similar gene arrangement associated with a possible deletion of a 13 kb sequence within KL. This smaller KL locus differed from previously described KL types and was composed of regions A (capsule export genes, wza, wzb and wzc), B (pgm, gne1, gpi, ugd and galU), gna and itrA2 genes only. The ST1557 genome had a unique KL locus that shared 82% similarity to KL6a.
Discussion
Colistin resistance is a growing problem within the healthcare setting as colistin is considered an agent of last resort. It is only employed in the treatment of Gram-negative bacterial infection when other safer and more effective agents such as carbapenems cannot be used due to extensive drug resistance. Colistin resistance is in part driven by a global increase in carbapenem-resistant A. baumannii infections.45 A study from Greece reported an increase in colistin resistance from 1% in 2012 to >20% in 2014.46 Likewise, a survey of nosocomial infections in Italy found more than half of 26 carbapenem-resistant A. baumannii strains to be colistin resistant.47 In this study, we elucidated the genomic basis for the evolution of colistin resistance among patients with carbapenem-resistant A. baumannii infections. The majority of strains belonged to ST208, which is part of the International Clone 2, and possessed KL6a. Following exposure to colistin therapy, two-thirds of patients acquired colistin resistance through microevolution, primarily through mutation within the pmrAB genes of a previously susceptible infecting strain. In addition to pmrAB mutations, several strains contain mutations within cyaA, a gene previously implicated in resistance to fosfomycin and some β-lactams in Escherichia coli.25 In addition, an accumulation of mutations was observed in genes involved in surface structures including pilus, capsule and O antigen biosynthesis and transport in the colistin-resistant strains, which may reflect ongoing adaptation to the host defence systems.36
On the other hand, in some instances colistin resistance was linked to the presence of a strain that was genetically distinct from the corresponding susceptible strain suggesting that some patients acquired a new strain that was already colistin-resistant, or a susceptible strain that was acquired yet undetected, which subsequently evolved colistin resistance. Alternatively, these patients may initially have been coinfected by two strains, one colistin-susceptible and the other colistin-resistant, with colistin therapy later favouring dominance of the resistant strain.
In addition to being resistant to colistin, most of the A. baumannii strains in this study were XDR, highlighted by very high frequencies (>80%) of non-susceptibility to carbapenems, aminoglycosides, fluoroquinolones and tetracyclines. Of all tested antibiotics, strains in this study were only susceptible to minocycline (92% susceptible) and tigecycline (100% susceptible). These findings buttress the urgent need to develop new classes of agents against highly resistant organisms. The presence of tight genetic clusters containing strains from different patients also sheds additional light on the transmission of XDR A. baumannii infections in the hospital environment. The presence of diverse, co-evolving clusters is in keeping with multiple transmission events of carbapenem-non-susceptible A. baumannii. These clusters may also be associated with environmental reservoirs given the propensity of A. baumannii to survive in the hospital environment.48 None the less, there is limited overlap in specific mutations associated with resistant strains even within transmission clusters. This finding supports the hypothesis that colistin resistance largely develops independently within each patient as opposed to broad dissemination of a single resistant strain. A notable exception is the transmission of a resistant isolate between patients 5 and 12 with 0 SNP differences.
Limitations of this study include a relatively small sample size and that gene expression levels were not taken into account, which may explain some of the observed phenotypic diversity. In addition, some patients may have been coinfected or colonized by multiple strains in addition to the two included in the study. In addition, a limitation of short-read sequencing is that the full length of IS found upstream of several antibiotic resistance genes is often not completely resolved. Despite these limitations, this work provides insights into the genomic basis of colistin resistance in A. baumannii in clinical settings and adds to the body of evidence supporting the utility of routine genome-based surveillance for major hospital-associated infections. Genomic evidence for transmission events over a long period suggests the need for sustained environmental disinfection in addition to routine infection prevention practices to curtail the spread of colistin-resistant A. baumannii in the hospital setting. Furthermore, patients receiving colistin need to be closely monitored for the development of resistance.
Supplementary Material
Acknowledgements
We thank Jonas Korlach, Matthew Boitano and Tyson Clarke for their assistance in providing the internal reference genome sequences.
Transparency declarations
Y. D. has served on advisory boards for Meiji, The Medicines Company, Roche and Tetraphase, and has received research funding from Accelerate Diagnostics. All other authors have none to declare.
Funding
This study was supported by a research grant from the National Institutes of Health grant number R01AI104895. The effort of Y. D. was also supported by National Institutes of Health grant number R21AI123747.
Supplementary data
Table S1 appears as Supplementary data at JAC Online.
References
- 1. Peleg AY, Seifert H, Paterson DL.. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 2008; 21: 538–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boucher HW, Talbot GH, Bradley JS. et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 2009; 48: 1–12. [DOI] [PubMed] [Google Scholar]
- 3. Queenan AM, Pillar CM, Deane J. et al. Multidrug resistance among Acinetobacter spp. in the USA and activity profile of key agents: results from CAPITAL Surveillance 2010. Diagn Microbiol Infect Dis 2012; 73: 267–70. [DOI] [PubMed] [Google Scholar]
- 4. Olaitan AO, Morand S, Rolain JM.. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 2014; 5: 643.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wright MS, Iovleva A, Jacobs MR. et al. Genome dynamics of multidrug-resistant Acinetobacter baumannii during infection and treatment. Genome Med 2016; 8: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wen H, Wang K, Liu Y. et al. Population dynamics of an Acinetobacter baumannii clonal complex during colonization of patients. J Clin Microbiol 2014; 52: 3200–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wright MS, Haft DH, Harkins DM. et al. New insights into dissemination and variation of the health care-associated pathogen Acinetobacter baumannii from genomic analysis. MBio 2014; 5: e00963-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qureshi ZA, Hittle LE, O'Hara JA. et al. Colistin-resistant Acinetobacter baumannii: beyond carbapenem resistance. Clin Infect Dis 2015; 60: 1295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-seventh Edition M100-S27. CLSI, Wayne, PA, USA, 2017. [Google Scholar]
- 10. Bankevich A, Nurk S, Antipov D. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19: 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014; 30: 2068–9. [DOI] [PubMed] [Google Scholar]
- 12. Chin CS, Alexander DH, Marks P. et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 2013; 10: 563–9. [DOI] [PubMed] [Google Scholar]
- 13. Deatherage DE, Barrick JE.. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 2014; 1151: 165–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30: 1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013; https://arxiv.org/abs/1303.3997v2. [Google Scholar]
- 16. Page AJ, Cummins CA, Hunt M. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015; 31: 3691–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zankari E, Hasman H, Cosentino S. et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67: 2640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kenyon JJ, Nigro SJ, Hall RM.. Variation in the OC locus of Acinetobacter baumannii genomes predicts extensive structural diversity in the lipooligosaccharide. PLoS One 2014; 9: e107833.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liao YC, Lin SH, Lin HH.. Completing bacterial genome assemblies: strategy and performance comparisons. Sci Rep 2015; 5: 8747.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Castro-Jaimes S, Salgado-Camargo AD, Graña-Miraglia L. et al. Complete genome sequence of a multidrug-resistant Acinetobacter baumannii isolate obtained from a Mexican hospital (sequence type 422). Genome Announc 2016; 4: pii: e00583-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mugnier PD, Poirel L, Naas T. et al. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis 2010; 16: 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Diancourt L, Passet V, Nemec A. et al. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 2010; 5: e10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bartual SG, Seifert H, Hippler C. et al. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol 2005; 43: 4382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hamidian M, Nigro SJ, Hall RM.. Problems with the Oxford multilocus sequence typing scheme for Acinetobacter baumannii: do sequence type 92 (ST92) and ST109 exist? J Clin Microbiol 2017; 55: 2287–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ruiz C, Levy SB.. Use of functional interactions with MarA to discover chromosomal genes affecting antibiotic susceptibility in Escherichia coli. Int J Antimicrob Agents 2011; 37: 177–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. La MV, Jureen R, Lin RT. et al. Unusual detection of an Acinetobacter class D carbapenemase gene, blaOXA-23, in a clinical Escherichia coli isolate. J Clin Microbiol 2014; 52: 3822–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nouwen N, Driessen AJ.. SecDFyajC forms a heterotetrameric complex with YidC. Mol Microbiol 2002; 44: 1397–405. [DOI] [PubMed] [Google Scholar]
- 28. Arroyo LA, Herrera CM, Fernandez L. et al. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob Agents Chemother 2011; 55: 3743–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adams MD, Nickel GC, Bajaksouzian S. et al. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother 2009; 53: 3628–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beceiro A, Moreno A, Fernández N. et al. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob Agents Chemother 2014; 58: 518–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim Y, Bae IK, Lee H. et al. In vivo emergence of colistin resistance in Acinetobacter baumannii clinical isolates of sequence type 357 during colistin treatment. Diagn Microbiol Infect Dis 2014; 79: 362–6. [DOI] [PubMed] [Google Scholar]
- 32. Orans J, Johnson MD, Coggan KA. et al. Crystal structure analysis reveals Pseudomonas PilY1 as an essential calcium-dependent regulator of bacterial surface motility. Proc Natl Acad Sci U S A 2010; 107: 1065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park YK, Choi JY, Shin D. et al. Correlation between overexpression and amino acid substitution of the PmrAB locus and colistin resistance in Acinetobacter baumannii. Int J Antimicrob Agents 2011; 37: 525–30. [DOI] [PubMed] [Google Scholar]
- 34. Geisinger E, Isberg RR.. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog 2015; 11: e1004691.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kröger C, Kary SC, Schauer K. et al. Genetic regulation of virulence and antibiotic resistance in Acinetobacter baumannii. Genes (Basel) 2016; 8: pii: E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kenyon JJ, Hall RM.. Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS One 2013; 8: e62160.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vila J, Martí S, Sánchez-Céspedes J.. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J Antimicrob Chemother 2007; 59: 1210–5. [DOI] [PubMed] [Google Scholar]
- 38. Kochar M, Crosatti M, Harrison EM. et al. Deletion of TnAbaR23 results in both expected and unexpected antibiogram changes in a multidrug-resistant Acinetobacter baumannii strain. Antimicrob Agents Chemother 2012; 56: 1845–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Magiorakos AP, Srinivasan A, Carey RB. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18: 268–81. [DOI] [PubMed] [Google Scholar]
- 40. Zander E, Chmielarczyk A, Heczko P. et al. Conversion of OXA-66 into OXA-82 in clinical Acinetobacter baumannii isolates and association with altered carbapenem susceptibility. J Antimicrob Chemother 2013; 68: 308–11. [DOI] [PubMed] [Google Scholar]
- 41. Zong Z, Lü X, Valenzuela JK. et al. An outbreak of carbapenem-resistant Acinetobacter baumannii producing OXA-23 carbapenemase in western China. Int J Antimicrob Agents 2008; 31: 50–4. [DOI] [PubMed] [Google Scholar]
- 42. Tian GB, Adams-Haduch JM, Taracila M. et al. Extended-spectrum AmpC cephalosporinase in Acinetobacter baumannii: aDC-56 confers resistance to cefepime. Antimicrob Agents Chemother 2011; 55: 4922–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Adams-Haduch JM, Paterson DL, Sidjabat HE. et al. Genetic basis of multidrug resistance in Acinetobacter baumannii clinical isolates at a tertiary medical center in Pennsylvania. Antimicrob Agents Chemother 2008; 52: 3837–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang P, McElheny CL, Mettus RT. et al. Contribution of the TetB efflux pump to minocycline susceptibility among carbapenem-resistant Acinetobacter baumannii strains. Antimicrob Agents Chemother 2017; 61: pii: e01176-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cai Y, Chai D, Wang R. et al. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J Antimicrob Chemother 2012; 67: 1607–15. [DOI] [PubMed] [Google Scholar]
- 46. Oikonomou O, Sarrou S, Papagiannitsis CC. et al. Rapid dissemination of colistin and carbapenem resistant Acinetobacter baumannii in Central Greece: mechanisms of resistance, molecular identification and epidemiological data. BMC Infect Dis 2015; 15: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Agodi A, Voulgari E, Barchitta M. et al. Spread of a carbapenem- and colistin-resistant Acinetobacter baumannii ST2 clonal strain causing outbreaks in two Sicilian hospitals. J Hosp Infect 2014; 86: 260–6. [DOI] [PubMed] [Google Scholar]
- 48. Munoz-Price LS, Namias N, Cleary T. et al. Acinetobacter baumannii: association between environmental contamination of patient rooms and occupant status. Infect Control Hosp Epidemiol 2013; 34: 517–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.