Abstract
Tissue engineering approaches for the repair of osteochondral defects using biomaterial scaffolds and stem cells have remained challenging due to the inherent complexities of inducing cartilage-like matrix and bone-like matrix within the same local environment. Members of the transforming growth factor β (TGFβ) family have been extensively utilized in the engineering of skeletal tissues, but have distinct effects on chondrogenic and osteogenic differentiation of progenitor cells. The goal of this study was to develop a method to direct human bone marrow-derived mesenchymal stem cells (MSCs) to deposit either mineralized matrix or a cartilaginous matrix rich in glycosaminoglycan and type II collagen within the same biochemical environment. This differential induction was performed by culturing cells on engineered three-dimensionally woven poly(ɛ-caprolactone) (PCL) scaffolds in a chondrogenic environment for cartilage-like matrix production while inhibiting TGFβ3 signaling through Mothers against DPP homolog 3 (SMAD3) knockdown, in combination with overexpressing RUNX2, to achieve mineralization. The highest levels of mineral deposition and alkaline phosphatase activity were observed on scaffolds with genetically engineered MSCs and exhibited a synergistic effect in response to SMAD3 knockdown and RUNX2 expression. Meanwhile, unmodified MSCs on PCL scaffolds exhibited accumulation of an extracellular matrix rich in glycosaminoglycan and type II collagen in the same biochemical environment. This ability to derive differential matrix deposition in a single culture condition opens new avenues for developing complex tissue replacements for chondral or osteochondral defects.
Keywords: : cartilage, cartilage defects, cartilage tissue engineering, regenerative medicine
Introduction
Articular cartilage is the connective tissue that lines the ends of long bones in diarthrodial joints, providing a low-friction, load-bearing surface to help distribute loads between opposing bones. While cartilage provides protection against load-bearing and impact upon motion between the articular surfaces, it is an aneural, avascular tissue that has little intrinsic capacity for repair.1 Thus, focal chondral or osteochondral injuries result in significant pain and disability, and may lead to osteoarthritis (OA), a painful and disabling joint disease characterized by progressive degenerative changes in joint tissues.2 The definitive treatment for end-stage OA is total joint arthroplasty that seeks to replace the diarthrodial joint with a metal and plastic prosthesis.3–7 Furthermore, there are few treatments currently available for focal osteochondral defects within the joint, and current therapies rely on techniques such as microfracture to enhance intrinsic repair.8 Despite significant progress toward developing cartilage substitutes in recent years, there remains a need for regenerative medicine approaches that can enhance the repair of large cartilage or osteochondral defects using biomimetic implants with improved fixation properties and similar mechanical and biochemical properties to those of native articular cartilage. The development of an osteochondral construct is attractive in this aspect since the osseous and mineralized phase could provide an anchoring layer to affix the cartilaginous phase within the joint.9
In the field of tissue engineering, there has been extensive interest in creating cartilage, bone, or combined osteochondral constructs that can thus provide enhanced fixation of engineered tissues into defect sites. Several previous approaches have demonstrated the ability to develop osteochondral constructs by combining multiple cells types,10–14 multilayered scaffolds,15–18 or multistep differentiation protocols.11,19–25 However, it is still a major challenge to differentially direct cell fate determination into distinct lineages (i.e., cartilage and bone) from a single cell source, in a single culture system, while utilizing only one scaffold material. If proven efficacious, a single stage approach could streamline the engineering of multiphase tissues by circumventing the need for multiple cell types or multiple differentiation culture conditions.
Mesenchymal stem cells (MSCs) are an attractive cell source for cartilage and bone tissue engineering mainly because of their easy access, high capability of in vitro expansion, and multipotent ability to express various cellular phenotypes, including osteogenic and chondrogenic lineages.26,27 For example, previous work from our group has demonstrated the capability of growing engineered cartilage or bone on three-dimensionally (3D) woven poly(ɛ-caprolactone) (PCL) scaffolds seeded with human MSCs under separate, prescribed chondrogenic or osteogenic conditions.28–32 However, MSCs require high concentrations of exogenous growth factors to enter distinct lineage programs in vitro.33–35 For example, transforming growth factor β 3 (TGFβ3) is commonly used to induce MSC chondrogenesis,36–39 while bone morphogenic protein 2 (BMP2) is often used to induce osteogenesis.40–42 These growth factors and environmental cues that define chondrogenic differentiation may interfere with osteogenic differentiation,43–46 and vice-versa. Thus, it remains challenging to direct chondrogenesis and osteogenesis simultaneously and in a site-specific manner within one in vitro culture environment due to inhibitory effects of chondrogenic-inducing TGFβ3 and osteogenic-inducing BMP2 on one another. For example, upon TGFβ3 signaling, Mothers against DPP homolog 3 (Smad3) is phosphorylated and translocates into the nucleus to repress Runt-related transcription factor (Runx2) and Runx2-induced transcriptional activation of osteoblast differentiation genes.46 The inhibition is achieved by recruiting Class IIa histone deacetylases (HDAC) 4 and 5 to the Smad3/Runx2 complex at Runx2-binding DNA sequences, repressing the expression of Runx2 and its downstream targets.45,47
It had been suggested that medium supplemented with TGFβ3 can induce chondrogenesis in MSCs through the canonical SMAD2/3 pathway.37,39,48 In addition, RUNX2 is a transcription factor with multiple binding sites in the promoter regions of bone matrix markers,49 and its osteogenic potential has been suggested in several studies in vivo or in vitro with defined osteogenic conditions.50–55 However, it remains unclear whether RUNX2 would exert its osteogenic effect in a chondrogenic environment with TGFβ3 stimulation. Here, we developed a method to concomitantly induce osteogenic and chondrogenic differentiation from human bone marrow-derived MSCs on two separate 3D woven PCL scaffolds in one single culture system. We employed the TGFβ3/SMAD3 axis to produce a cartilaginous matrix on one scaffold. Meanwhile, we engineered MSCs to potentiate mineralized matrix by overexpressing RUNX2 with SMAD3 modulation on the other scaffold. We hypothesized that a combination of SMAD3 knockdown and RUNX2 overexpression would significantly enhance mineral deposition even under the influence of TGFβ3 stimulation. The novelty of this research lies within our capability to, under the same biochemical cues, produce two scaffolds with distinct extracellular matrix (ECM) compositions by modulating intracellular signaling pathway of TGFβ3.
Materials and Methods
MSC culture and differentiation
Bone marrow was obtained from discarded and de-identified waste tissue from adult bone marrow transplant donors in accordance with the Institutional Review Board of Duke University Medical Center. Adherent cells were expanded and maintained in expansion medium: DMEM-low glucose (Gibco), 1% penicillin/streptomycin (Gibco), 10% fetal bovine serum (FBS) (ThermoFisher), and 1 ng/mL basic fibroblast growth factor (Roche).56 MSCs from three donors were expanded until end of passage 2, and then an equal number of MSCs from each donor was combined to make a superlot. This approach has been shown to increase experimental throughput and utilize assay resources more efficiently, while effectively representing the average differentiation behavior of each of their contributing cell populations.57 Experiments with individual MSC donors showed similar responses to SMAD3 knockdown and RUNX2 overexpression with respect to glycosaminoglycan (GAG) or mineral deposition (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/tea). MSCs were used at passage 5 for all experiments, unless otherwise noted.
Cells were then induced to differentiate in a defined medium consisting of DMEM-high glucose (Gibco), 1% ITS+ (Corning), 1% penicillin/streptomycin (Gibco), 100 nM dexamethasone (Sigma), 50 μg/mL ascorbic acid (Sigma), 40 μg/mL L-proline (Sigma), and 100 nM β-glycerophosphate (Chem-Impex International).37,58,59 rhTGF-β3 (R&D Systems) was supplemented at 5 ng/mL.
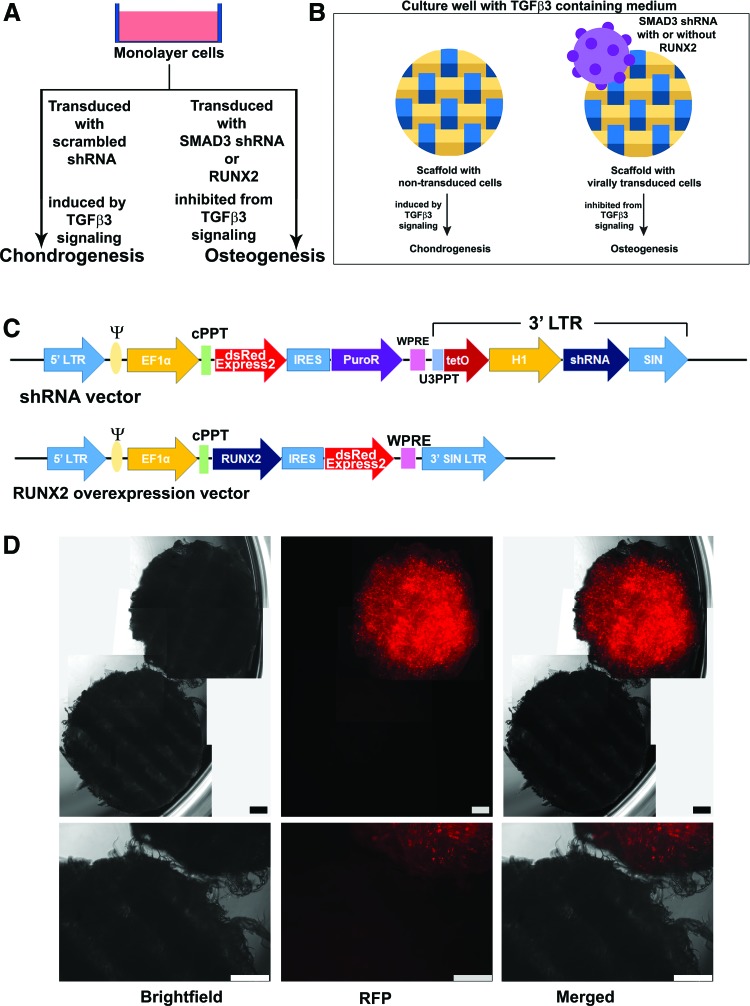
We examined the effect of TGFβ3 signaling manipulation in both monolayer and scaffold culturing system, as outlined in Figure 1A, B. All media were exchanged every 3 days. Monolayer cells were harvested at 1 week postinduction for quantitative real-time polymerase chain reaction (qRT-PCR) analysis, and 3 weeks postinduction for alcian blue staining/quantification, and alizarin red staining/quantification. Scaffolds were harvested 5 weeks postinduction for histological and biochemical analysis.
FIG. 1.
Overview of experimental approach. (A) MSCs were cultured in monolayer. The use of SMAD3 shRNA and RUNX2 overexpression was hypothesized to induce MSCs toward osteogenesis, despite the presence of TGFβ3. (B) MSCs were pretransduced with SMAD3 shRNA or SMAD3 shRNA with RUNX2 before being seeded on the woven scaffolds. Scrambled shRNA and scrambled shRNA with RUNX2 were used as control groups. Square box indicates two scaffolds cultured in the same well. Yellow and blue bars represent warp and weft fibers. (C) Schema of viral backbones used. A detailed description of the backbone components is provided in Supplementary Table S2. We expect transduced cells to emit a strong signal under the red fluorescent protein (RFP) channel (due to dsRedExpress2 in our transfer vector construct). (D) Distinct differences between NT and virally transduced scaffolds cultured in the same well. Representative microscopic images of scaffolds in culture (week 5). Only the scaffold with virally transduced cells (scrambled control in this image) emitted RFP signal, while the NT scaffold did not. Upper panel: stitched image from multiple views to capture the entire scaffold pair. Lower panel: single images of scaffold pairs. Scale bar = 500 μm. TGFβ3, transforming growth factor β; MSCs, mesenchymal stem cells; SMAD3, Mothers against DPP homolog 3; shRNA, short hairpin RNA; NT, nontransduced. Color images available online at www.liebertpub.com/tea
PCL scaffold processing
PCL scaffold fabric was produced as previously described,30,60 and cylindrical samples (4 mm Ø) were obtained, aseptically processed, and coated with 0.002% poly-L lysine (Sigma).28
SMAD3 knockdown with short hairpin RNA
Sequences for short hairpin RNA (shRNA) targeting the human SMAD3 transcript were designed using the RNAi consortium (TRC) GPP Web Portal (Broad Institute).61 shRNA specific to the SMAD3 transcript (SMAD3 shRNA) was selected for its superior efficiency compared to two other sequences (data not shown), and it had the following target sequence: 5′-TGAGCAGAACAGGTAGTATTA-3′. A vector delivering a scrambled sequence was used as control. The scrambled shRNA had the following sequence: 5′-TTCTCCGAACGTGTCACGTT-3′. Both the target and the scrambled sequences have been confirmed to have no homology that is longer than 15 base pairs with any other human transcripts (NCBI Blast, Homo sapiens annotation release 108). shRNA sequences were cloned into a modified lentiviral vector (Fig. 1C) (Addgene #12247)62 as described elsewhere63 using MluI and ClaI restriction sites. This modified lentiviral transfer vector also contains a dsRedExpress2 expression cassette, which enabled fluorescence microscopy imaging. Although there was a puromycin resistant cassette in this vector, we did not perform antibiotic selection of transduced MSCs, as we have found that puromycin supplementation in the culture can inhibit the ability of MSCs to proliferate and differentiate postselection (data not shown).
RUNX2 overexpression
The coding sequence of RUNX2 (NM_001024630.3) was synthesized and cloned into a modified lentiviral transfer vector (Addgene #12250)62 using MluI and XmaI restriction sites. An overview of this vector is outlined in Figure 1C.
Lentivirus production
To produce vesicular stomatitis virus glycoprotein pseudotyped lentivirus, HEK293T cells were plated at 3.8 × 106 cells per 10 cm dish in DMEM-high glucose (Gibco) supplemented with 10% heat inactivated FBS (Gibco). The following day, cells were co-transfected with the appropriate transfer vector (20 μg), the second-generation packaging plasmid psPAX2 (Addgene #12260, 15 μg), and the envelope plasmid pMD2.G (Addgene #12259, 6 μg) by calcium phosphate precipitation.64 After 14–16 h of incubation, 12 mL of fresh medium was exchanged. Twenty-four hours later, supernatant was harvested (Harvest 1) and stored at 4°C, and another 12 mL of fresh medium was again exchanged. After another 24 h, supernatant was harvested a second time (Harvest 2). Harvest 1 and harvest 2 were pooled together, filtered through 0.45 μm cellulose acetate filters (Corning) to clear out producer cells, aliquoted, and stored at −80°C until future use.
Lentivirus transduction of MSC
One day before transduction, MSCs were plated at 4,500 cells/cm2. The next day, MSCs were transduced in the presence of 4 μg/mL polybrene (Sigma). Four groups of lentivirus were used: scrambled shRNA, SMAD3 shRNA, scrambled shRNA with RUNX2, and SMAD3 shRNA with RUNX2, and these transductions were carried out separately in individual culture vessels. Twenty-four hours post transduction, MSCs were washed once with phosphate-buffered saline (PBS), and fresh expansion medium was exchanged. Nontransduced (NT) and transduced MSCs were expanded separately, each in expansion medium for 1 week. Subsequently, MSCs were either cultured in induction medium in monolayer experiments, or digested with 0.05% Trypsin-EDTA (Gibco) and seeded on 4-mm Ø poly-L-lysine-coated scaffolds at 250,000 cells per scaffold. Scaffolds seeded with NT or virally transduced cells were combined at this step and cultured in expansion medium for 1 week, and then in induction medium for 5 weeks. Due to the dsRedExpress2 expression cassette in our shRNA transfer vector (Fig. 1C), scaffolds seeded with NT and virally transduced MSCs could be easily distinguished by fluorescence microscopy (Fig. 1D).
Quantitative real-time polymerase chain reaction
Monolayer cells were harvested for qRT-PCR 1 week postinduction. RNA isolation was carried out following manufacturer's protocol (Norgen). Reverse transcription was performed immediately after RNA was obtained, using Superscript VILO cDNA master mix (Invitrogen). qRT-PCR was performed using Fast SyBR Green master mix (Applied Biosystems) following manufacturer's protocol. Primer pairs (Supplementary Table S1) were synthesized by Integrated DNA Technologies, Inc. (IDT). Results are reported as log10 of fold change in expression of the gene of interest, normalized to ribosomal 18S expression by the ΔΔCt method.
Alcian blue and alizarin red staining and quantification
Three weeks postinduction, monolayer cells were washed once with PBS and fixed in 4% paraformaldehyde for 30 min at room temperature.
Wells were then washed with DI H2O, and submerged in 1 mL of alcian blue (0.1% w/v in 0.1 N hydrochloric acid) (Acros) for 4 h or in 1 mL of alizarin red (40 mM, pH 4.1–4.3) (EMD Millipore) for 20 min at room temperature with gentle shaking. After staining, nonspecific dye was rinsed off by washing with 1 mL of DI H2O three times.
For quantification of the amount of alcian blue in stained wells, wells were incubated in 500 μL of 6 M guanidine hydrochloride solution (Sigma) overnight at room temperature. The next morning, supernatant was collected into clear bottom 96-well plates. Known concentrations of alcian blue were used as standards, and absorbance was recorded at 595 nm with the Cytation 5 microplate reader (BioTek).
For quantification of the amount of alizarin red in stained wells, dye was released as previously described.65 Known concentrations of alizarin red were used as standards, and absorbance was recorded at 405 nm with the Cytation 5 reader (BioTek).
Biochemical analyses for DNA and GAG content of engineered scaffolds
On the day of harvest, fluorescence microscopy was used to distinguish between virally transduced (red fluorescent signal) and NT (no fluorescent signal) scaffolds for harvest. Scaffolds were subsequently transferred to individual wells, washed once with PBS, and stored at −20°C until processing. Scaffolds were lyophilized overnight, then digested in 125 μg/mL papain at 58°C for 20 h for biochemical analyses. DNA content was measured with the PicoGreen assay (ThermoFisher), and GAG was measured using the 1,9-dimethylmethylene blue assay66 at 525 nm wavelength.
Histological processing of scaffold samples
Virally transduced and NT scaffolds were separated as described above. Scaffolds were washed once with PBS, fixed in 4% paraformaldehyde for 48 h, paraffin embedded, and sectioned at 10 μm thickness. Slides were stained for Safranin O and Fast Green using a standard protocol,67 and for von Kossa following manufacturer's protocol (Abcam #ab150687). For immunohistochemistry, the following primary monoclonal antibodies were used: type I collagen (Abcam #ab90395), type II collagen (Developmental Studies Hybridoma bank #II-II6B3), and type X collagen (Sigma #C7974). All immunohistochemistry staining procedures were carried out with biotinylated goat anti-mouse secondary antibodies (Abcam #ab97021) as described previously.28 Human osteochondral sections were used as positive controls, and sample sections were incubated without primary antibody for negative controls.
Alkaline phosphatase assay
Virally transduced and NT scaffolds were separated as described above. Scaffolds were washed once with PBS, snap frozen in liquid nitrogen, and stored at −80°C until processing. Scaffolds were homogenized with a biopulverizer (Biospec Products) chilled in liquid nitrogen. Subsequent steps were carried out following manufacturer's protocol (Abcam #ab83371).
Statistical analysis
Statistical analysis was performed using Graphpad Prism version 7.03 (GraphPad Software). Alizarin red (n = 4 per group) and alcian blue (n = 5 per group) samples were analyzed using two-way ANOVA with Tukey post hoc test (α = 0.05) using samples from two independent experiments. For biochemical and alkaline phosphatase assays, two-tailed paired t-tests were performed on scaffold pairs of NT and virally transduced samples of each experimental group. These two experiments were independent from each other. Three scaffold pairs were collected for biochemical assays per experimental group. Additional scaffold pairs (n = 4–5) were collected for alkaline phosphatase assay per experimental group. Results were reported with mean values ± standard error of the means. For qRT-PCR analysis, calculated fold change values were log-transformed before statistical analysis with Student's t test, and standard errors were calculated using the standard propagation of error method.68
Results
SMAD3 knockdown or RUNX2 overexpression inhibits TGFβ3-induced cartilaginous matrix deposition
We examined the effect of SMAD3 knockdown and RUNX2 overexpression on MSCs in monolayer (Fig. 1A). We utilized this as a rapid and simple screening system before moving to 3D scaffold culture (Fig. 1B).
Human MSCs transduced with SMAD3 shRNA exhibited a significant reduction in SMAD3 level compared to those transduced with scrambled shRNA (p = 0.002) (Supplementary Fig. S2A). Bright field and fluorescence microscopy suggested that transduction efficiency was high (Supplementary Fig. 2C, D). We also investigated the efficiency of the RUNX2 overexpression, with the result showing a ∼6-fold increase in the level of RUNX2 compared to NT cells (p < 0.0001) (Supplementary Fig. S2B).
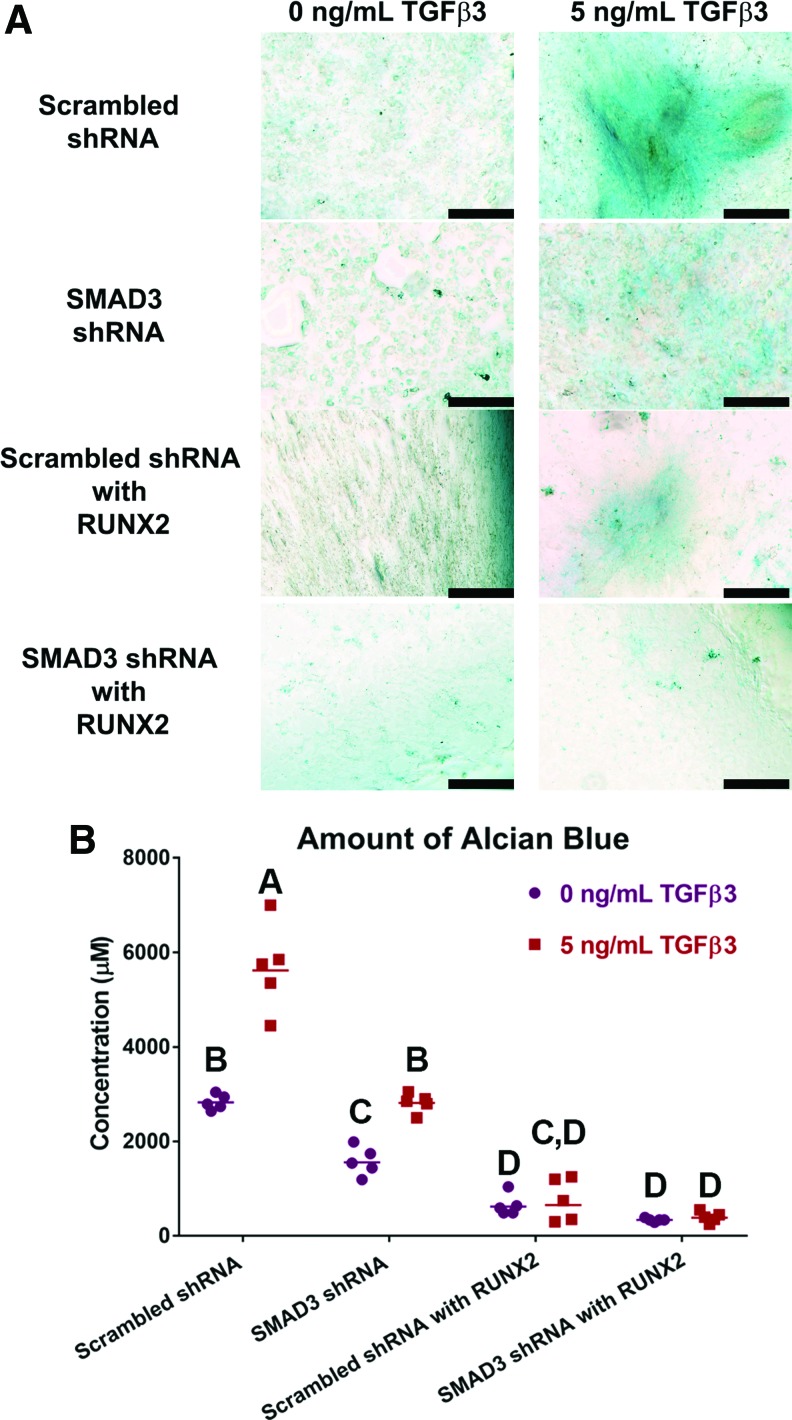
In the absence of TGFβ3, there was minimal GAG deposition, as indicated by low level of alcian blue staining and quantification (Fig. 2). When TGFβ3 was supplemented in the medium, GAG-rich matrix was observed in scrambled shRNA transduced wells as expected (5680 ± 412 μM) (Fig. 2A, C). On the other hand, GAG production was inhibited by knocking down SMAD3 (SMAD3 shRNA) (2820 ± 90 μM), overexpressing RUNX2 (scrambled shRNA with RUNX2) (770 ± 202 μM), or both (SMAD3 shRNA with RUNX2) (400 ± 50 μM) (p < 0.0001). Our results suggested that TGFβ3-induced GAG production could be inhibited. This raised the question whether these MSCs, inhibited from propagating TGFβ3 signaling, would start producing mineralized matrix instead.
FIG. 2.
Reduction in GAG deposition with SMAD3 knockdown or RUNX2 overexpression. (A) Alcian blue staining of monolayer cells cultured without (left) and with (right) TGFβ3. Distinct GAG rich matrix was observed in the scrambled shRNA transduced wells when stimulated with 5 ng/mL TGFβ3. All other wells exhibited very low levels of alcian blue staining. Scale bar = 500 μm. (B) Quantification of alcian blue dye in wells with 0 ng/mL (purple dots) or 5 ng/mL (red squares) of TGFβ3, n = 5. Points represent independent specimen. Scale bars represent geometric means for each group. Two-way ANOVA with Tukey post hoc test. Effect of TGFβ3 dose: p < 0.0001. Effect of virus type: p < 0.0001. Interaction between TGFβ3 dose and virus type: p < 0.0001. Groups of different letters are statistically different from one another. RUNX2, runt-related transcription factor; GAG, glycosaminoglycan. Color images available online at www.liebertpub.com/tea
SMAD3 knockdown and RUNX2 overexpression act to enhance mineral deposition in a chondrogenic environment
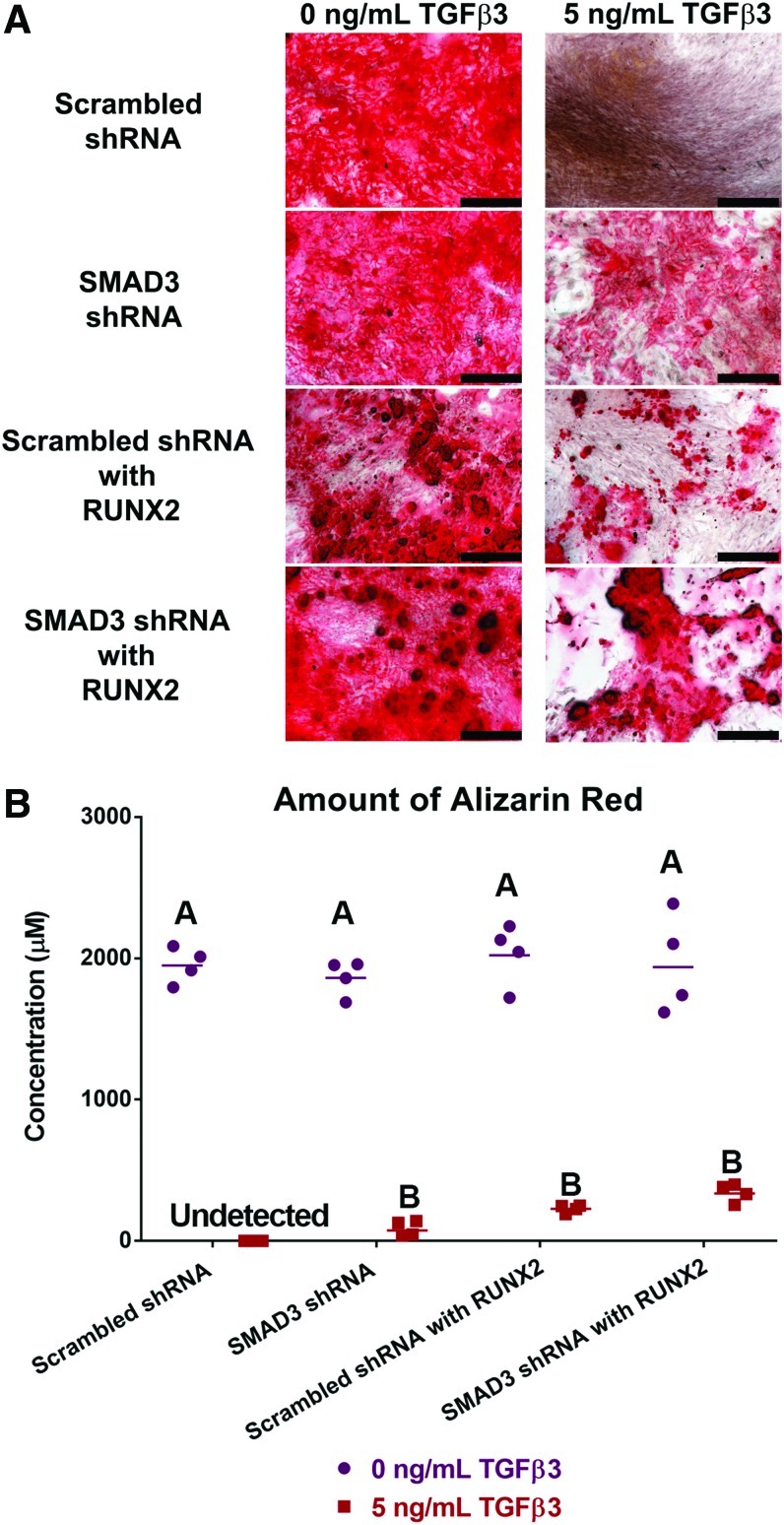
In monolayer, when MSCs were induced in the absence of TGFβ3, mineral deposition was observed in all samples by alizarin red staining (Fig. 3A). Quantitative analysis confirmed that there was no statistical difference among the groups in the amount of mineral produced when TGFβ3 was absent (Fig. 3B). However, in the presence of 5 ng/mL of TGFβ3, no staining was observed for the scrambled shRNA transduced wells. This was expected since TGFβ3 would induce MSCs toward a chondrogenic fate, and thus abolish mineral production. When SMAD3 level was reduced in SMAD3 shRNA transduced wells, TGFβ3 signaling was dampened, and thus mineral deposition was detected to some extent, as observed with both alizarin red staining and quantification (87.83 ± 27.10 μM) (Fig. 3A, C). When RUNX2 level was elevated in the scrambled shRNA with RUNX2-transduced group, mineral deposition was also detected (228.25 ± 13.80 μM). When SMAD3 shRNA was used in conjunction with RUNX2 overexpression (SMAD3 shRNA with RUNX2), mineral deposition was achieved at the highest level (342.00 ± 32.17 μM) within the groups under 5 ng/mL of TGFβ3, although the detected difference was not significant. Altogether, these data suggest that engineered MSCs could undergo matrix mineralization in a chondrogenic environment.
FIG. 3.
Mineral deposition was achieved in the presence of TGFβ3. (A) Alizarin red staining of monolayer cells cultured without (left) and with (right) TGFβ3. The presence of TGFβ3 abolished mineral deposition. This effect was reversed in SMAD3 shRNA, scrambled shRNA with RUNX2, or SMAD3 shRNA with RUNX2. Scale bar = 500 μm. (B) Quantification of alizarin red dye in wells with 0 ng/mL (purple dots) or 5 ng/mL (red squares) of TGFβ3, n = 4. Points represent independent specimen. Scale bars represent geometric means for each group. Two-way ANOVA with Tukey post hoc test. Effect of TGFβ3 dose: p < 0.0001. Effect of virus type: NS. Interaction between TGFβ3 dose and virus type: NS Groups of different letters are statistically different from one another. NS, not significant. Color images available online at www.liebertpub.com/tea
Differential GAG deposition was achieved in the dual-scaffold culture system
To test whether these engineered MSCs could be applied toward a tissue engineering context, we explored the effect of SMAD3 knockdown and RUNX2 overexpression in a system where two 3D PCL scaffolds of NT and transduced cells were cultured adjacently in the same well (Fig. 1B).
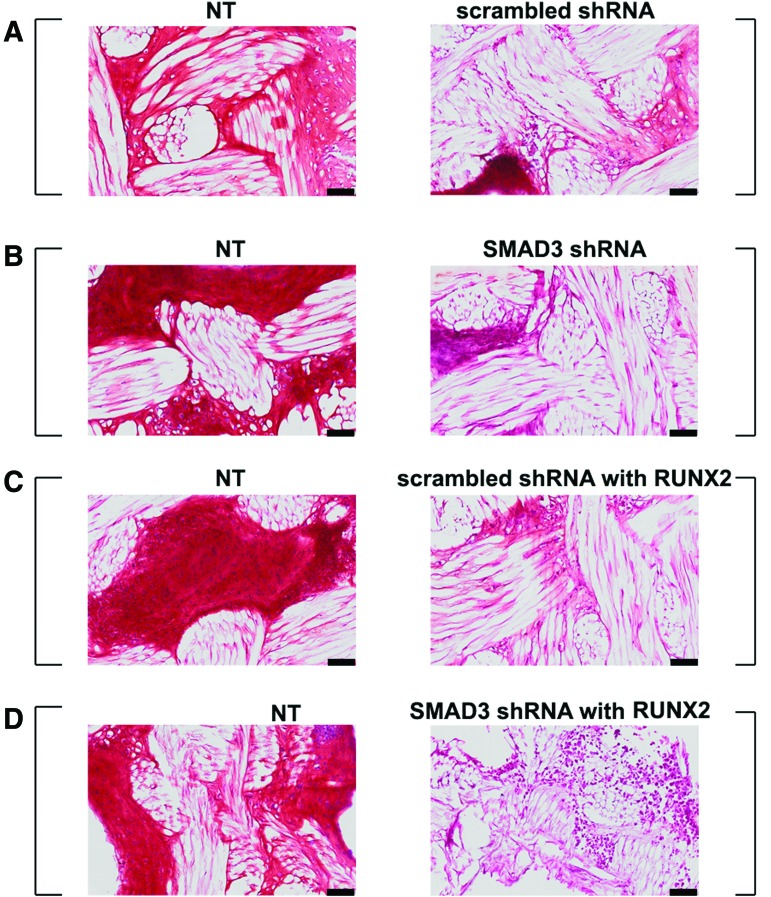
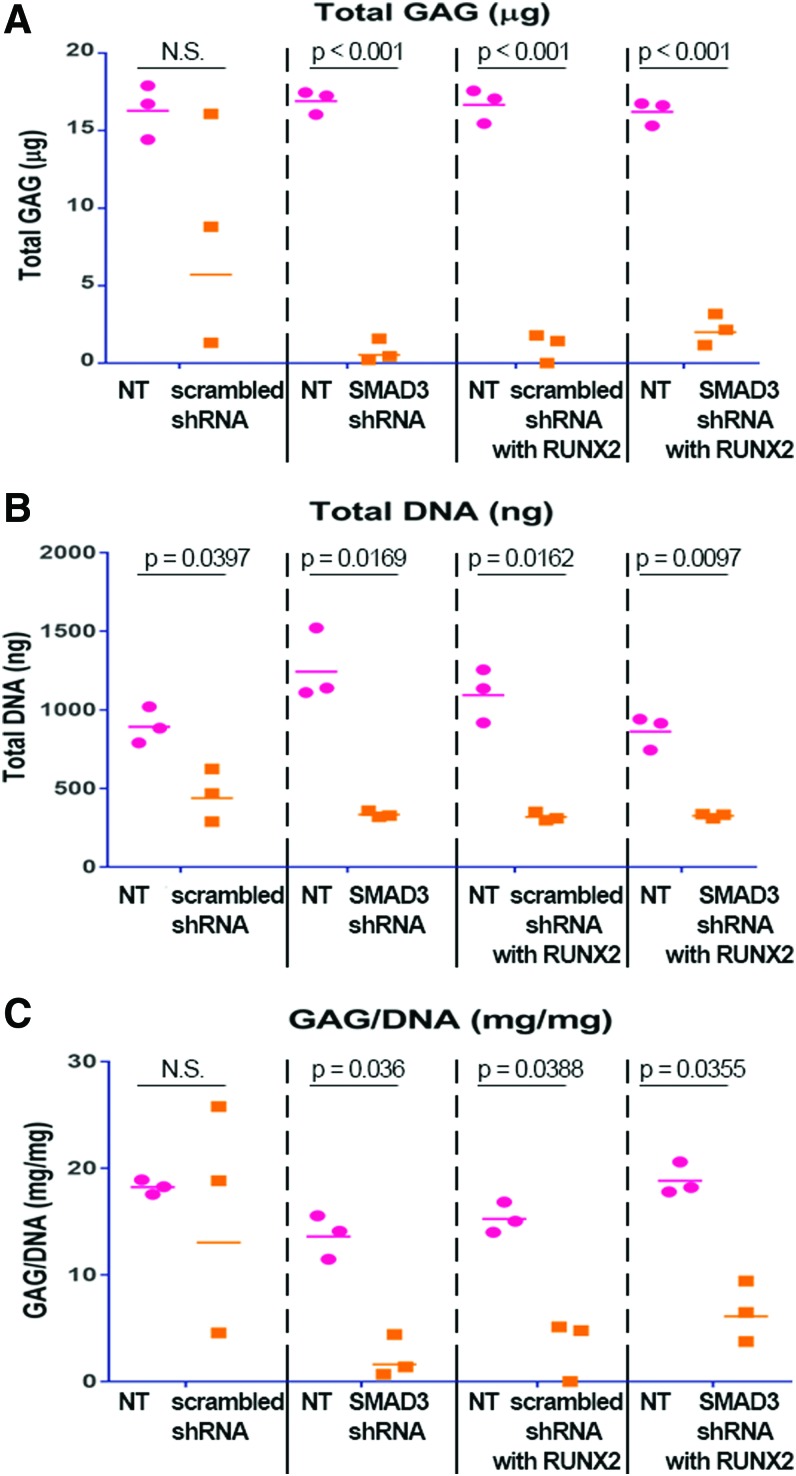
After 5 weeks in culture, NT scaffolds from all groups stained positive for Safranin-O (Fig. 4, left panel). There was a slight reduction in GAG deposition in scaffolds transduced with scrambled shRNA (Fig. 4A, right panel). However, GAG production was substantially reduced in scaffolds transduced with SMAD3 shRNA, RUNX2, or both (Fig. 4B, D, right panel). The finding was confirmed quantitatively by DMMB assay (Fig. 5A) (p < 0.001). In addition, there was a difference in the amount of total DNA between NT and virally transduced samples, although there was no difference among the virally transduced groups (Fig. 5B) (p < 0.05). When total GAG is normalized to the total amount of DNA produced, there was no statistical difference between the NT and the scrambled shRNA transduced scaffolds. On the contrary, a decrease in GAG production was apparent in the virally transduced scaffolds, when the SMAD3 shRNA with RUNX2 samples exhibited statistically significant difference from their NT counterparts (6.55 ± 1.64 mg/mg vs. 18.86 ± 0.87 mg/mg, p = 0.0355) (Fig. 5C). This suggested that NT MSCs could be induced to make GAG-rich matrix in the dual-scaffold culture condition. Moreover, SMAD3 knockdown and RUNX2 overexpression would inhibit the chondrogenic effect of TGFβ3 when used separately or together.
FIG. 4.
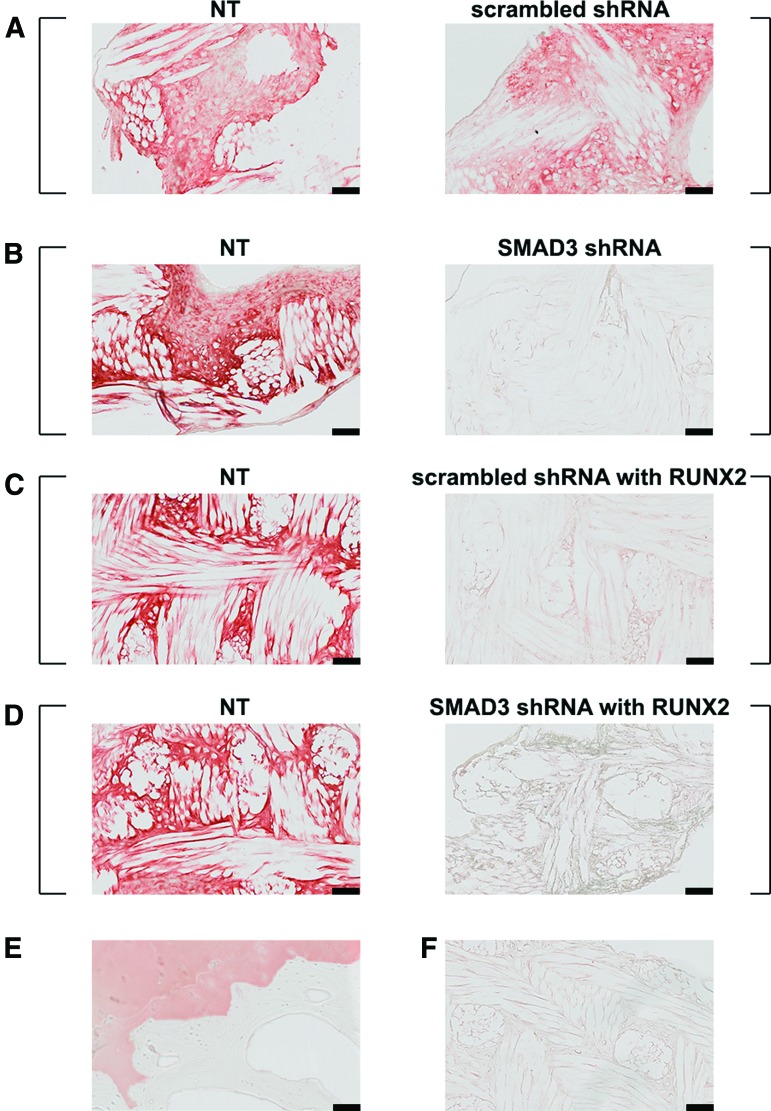
SMAD3 knockdown resulted in reduced GAG deposition. SafraninO-Fast Green staining of scaffold pairs cultured in 5 ng/mL TGFβ3. (A) NT and scrambled shRNA; (B) NT and SMAD3 shRNA; (C) NT and scrambled shRNA with RUNX2; (D) NT and SMAD3 shRNA with RUNX2. Square brackets indicate scaffold pairs cultured in the same well. GAG-rich matrix was observed in NT samples across all groups, while little GAG was observed in shRNA SMAD3 or RUNX2 transduced scaffolds. Scale bar = 100 μm. Color images available online at www.liebertpub.com/tea
FIG. 5.
SMAD3 knockdown resulted in reduced GAG deposition, quantified by biochemical assays. Dot plots representing the following: (A) Total GAG production as quantified by DMMB assay, (B) Double-stranded DNA production as quantified by PicoGreen assay, (C) GAG normalized to DNA amount. Points represent independent specimen; bars represent geometric means for each group. Differential GAG deposition was observed in culture of scaffold pairs. SMAD3 knockdown with RUNX2 overexpression resulted in reduced GAG accumulation in the extracellular matrix of virally transduced scaffolds (p < 0.001), n = 3. Points represent independent specimen. Bars represent geometric means for each group. Red dots represent scaffolds seeded with NT cells; orange squares represent scaffolds seeded with virally transduced cells. Two-tailed paired t tests for scaffold pairs in the same group (α = 0.05). Color images available online at www.liebertpub.com/tea
Differential type II collagen production was achieved in the dual-scaffold culture system
Type II collagen (COLII), another major component of articular cartilage, was observed in both the NT and the scrambled shRNA transduced scaffolds. This indicated that scrambled shRNA did not interfere with collagen production (Fig. 6A). On the contrary, COLII production was significantly reduced in scaffolds transduced with SMAD3 shRNA, RUNX2, or both (Fig. 6B, D). This recapitulated the differential GAG productions in NT versus virally transduced scaffolds, suggesting that TGFβ3 in the medium can induce nascent MSCs to make COLII-rich matrix in the dual-scaffold culture system. Moreover, either SMAD3 shRNA or RUNX2 may inhibit the chondrogenic effect of TGFβ3. It is important to note that this negative effect of SMAD3 knockdown and RUNX2 overexpression on the ability of MSCs to produce GAG and type II collagen rich matrix was expected, since these MSCs were specifically engineered for the purpose of producing mineralization instead.
FIG. 6.
Differential COLII expression. Immunohistochemistry staining of scaffold pairs cultured in 5 ng/mL TGFβ3. (A) NT and scrambled shRNA; (B) NT and SMAD3 shRNA; (C) NT and scrambled shRNA with RUNX2; (D) NT and SMAD3 shRNA with RUNX2. (E) Human osteochondral control. (F) Representative negative control with no primary antibody. Square brackets indicate scaffold pairs cultured in the same well. Type II collagen-rich matrix was observed in both NT and scrambled shRNA-transduced scaffolds, while differential type II collagen staining was observed for all the other groups. Scale bar = 100 μm. Color images available online at www.liebertpub.com/tea
We also investigated the expression of type I collagen (characteristic of bone) (Supplementary Fig. S3) and type X collagen (characteristic of hypertrophic cartilage) (Supplementary Fig. S4). Scaffolds cultured in the defined induction medium condition exhibited low levels of both collagens, and there was no observable difference in type I collagen staining between the NT and virally transduced scaffolds in all four groups. However, there was a slight increase in type X collagen staining in the group of SMAD3 shRNA with RUNX2 compared to their NT counterparts (Supplementary Fig. S4).
Differential mineral deposition was achieved in the dual-scaffold culture system
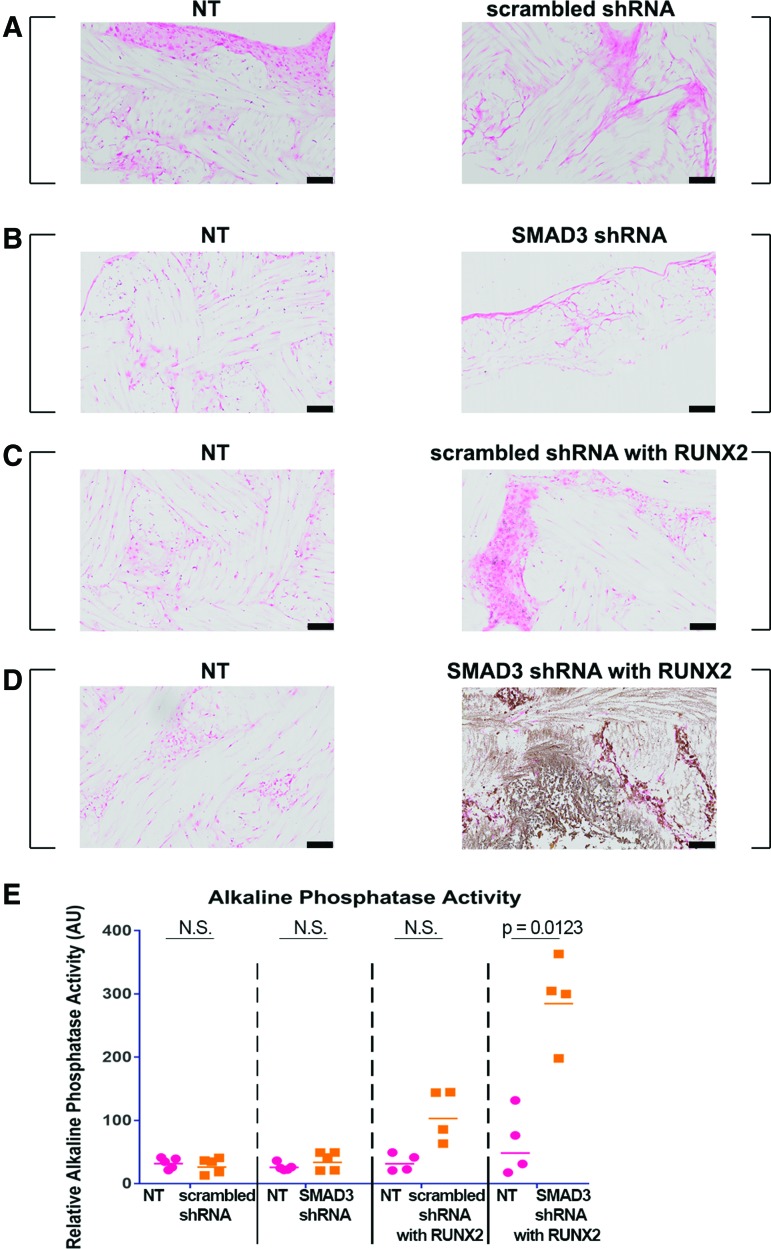
Mineral deposition in tissue-engineered constructs was first assessed by von Kossa staining. NT scaffolds from all groups did not exhibit positive staining (Fig. 7A–D, left panel), indicating that there was no mineral deposition when MSCs were cultured in the presence of TGFβ3. The strongest staining was observed in scaffolds containing cells transduced with SMAD3 shRNA and RUNX2 overexpression (Fig. 7D, right panel), suggesting that there was a synergistic effect of these two treatments on mineralized matrix in the culture condition. This phenomenon was confirmed quantitatively with alkaline phosphatase assay (p = 0.0123) (Fig. 7E). The amount of alkaline phosphatase was highest and most distinctive in the virally transduced scaffolds with both SMAD3 shRNA and RUNX2. Together, these data indicate that we could achieve matrix mineralization by knocking down SMAD3 in conjunction with overexpressing RUNX2 in our defined biochemical culture condition.
FIG. 7.
Synergistic effect of SMAD3 knockdown and RUNX2 overexpression for osteogenesis. (A–D) Von Kossa staining with nuclear red counterstain of scaffold pairs cultured in 5 ng/mL TGFβ3. (A) NT and scrambled shRNA; (B) NT and SMAD3 shRNA; (C) NT and scrambled shRNA with RUNX2; (D) NT and SMAD3 shRNA with RUNX2. Square brackets indicate scaffold pairs cultured in the same well. No mineral staining was observed except for the SMAD3 shRNA with RUNX2-transduced groups (brown). Scale bar = 100 μm. (E) Alkaline phosphatase activity quantification, n = 4–5. Points represent independent specimen. Bars represent geometric means for each group. Red dots represent scaffolds seeded with NT cells; orange squares represent scaffolds seeded with virally transduced cells. Two-tailed paired t tests for scaffold pairs in the same group (α = 0.05). Color images available online at www.liebertpub.com/tea
Discussion
A persistent challenge in the field of regenerative medicine has been the ability to engineer complex tissues comprised of multiple cell types and organized into regions of distinct ECM constituents, such as the osteochondral interface between cartilage and bone. Our study demonstrated that cartilage-like and bone-like matrix deposition can be achieved simultaneously by manipulation of the intracellular TGFβ3 signaling pathway and facilitation of RUNX2 expression. Here, we created a dual-scaffold single culture system by (1) supplementing exogenous TGFβ3 to induce chondrogenesis in one subset of nongenetically engineered MSCs (NT scaffolds) and (2) introducing shRNA to suppress SMAD3 level in conjunction with overexpressing RUNX2 to drive matrix mineralization in the other subset of MSCs (transduced scaffolds). Our histological and biochemical data showed distinct matrix phenotypes between the two scaffolds that were cultured in the same well. While the production of GAG and type II collagen was enriched in the NT scaffolds, transduced scaffolds with suppressed SMAD3 and enhanced RUNX2 expression did not display evidence of chondrogenesis. On the other hand, these genetically engineered scaffolds showed signs of mineralization, as demonstrated by von Kossa staining and alkaline phosphatase activity quantification. Our study presents novel findings that blocking a specific node in the TGFβ3 signaling pathway in conjunction with enhancing RUNX2 can lead to mineralization even under TGFβ3 stimulation. This method eliminates the need for prolonged culture in various differentiation conditions and multi-stage processes to generate two distinct tissue matrices. More importantly, it opens new avenues for functional interfacial tissue engineering for the treatment of osteochondral defects.
While our base medium condition was selected to allow MSCs to differentiate toward both chondrogenic and osteogenic lineages, the addition of TGFβ3 abolished the matrix mineralization potential. However, this inhibitory effect of TGFβ3 on mineral deposition was overcome by knocking down SMAD3. The effect was observed in monolayer and subsequently recapitulated in our dual-scaffold, single culture system. Our findings extend previous work on the intricate network of the TGFβ superfamily and their control of MSC differentiation. In addition to its stimulatory effects in chondrogenesis, TGFβ has also been suggested to play an inhibitory role in osteoblastic maturation through Smad3.47,49 Indeed, blocking of TGFβ type I receptor (TGFβRI) kinase through pharmacology has been demonstrated to enhance osteoblastic differentiation of mouse C2C12 cells.69 Pharmacologic inhibition of TGFβRI also increased bone mass and bone formation in postnatal mice.70 When we inhibited TGFβ signaling, although through the intracellular SMAD3 axis, we were able to detect comparable trends in MSC matrix mineralization and alkaline phosphatase activity.
A key finding of this study was the successful production of mineralized matrix under TGFβ3 stimulation, while preserving the potency to induce GAG and type II collagen-rich matrix in another subset of cells. We showed that medium supplemented with TGFβ3 can induce robust chondrogenesis in NT scaffolds, consistent with previous reports on cartilage tissue engineering using MSCs.37,39,48 In this same environment, we demonstrated the ability to produce mineralization by inducing RUNX2 overexpression with simultaneous SMAD3 modulation. To our knowledge, this is the first experiment implementing RUNX2 overexpression in human MSCs with successful matrix mineralization and high alkaline phosphatase activity.
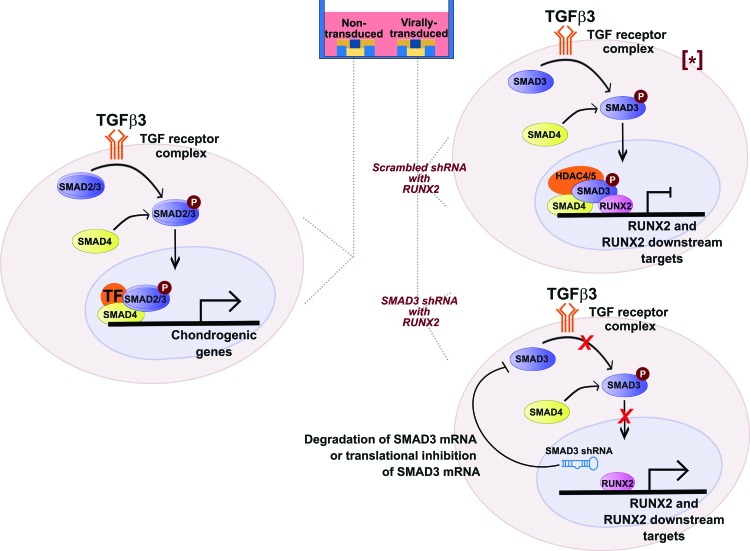
At the cellular level, TGFβ activates chondrogenic genes via the SMAD2/3-dependent pathway (Fig. 8, left). When TGFβ3 binds to the receptor, SMAD2/3 is phosphorylated, binds to SMAD4, and translocates into the nucleus, leading to transcriptional activation of chondrogenic genes. As a result, a cartilaginous matrix is produced. Simultaneously, TGFβ3 inhibits osteoblast differentiation genes via phosphorylated SMAD3 (Fig. 8, top right). After translocation into the nucleus, SMAD3 recruits HDAC4/5 to the promoters of RUNX2 and RUNX2 downstream targets, inhibiting their transcriptions.45,47 In cells genetically engineered to express SMAD3 shRNA (Fig. 8, bottom right), SMAD3 mRNA is either degraded or translationally inhibited, leading to de-repression of RUNX2 and its downstream targets. Removal of HDAC4/5 at these genomic regions makes the promoters accessible to exogenous RUNX2 and thus synergistically enhances transcriptions of bone matrix markers, resulting in a mineralized matrix.
FIG. 8.
Modulation of TGF signaling for simultaneous cartilaginous and mineralized matrices in one culture well. Left: On scaffold with NT cells, TGFβ3 binds to the receptor complex and phosphorylates SMAD2 and SMAD3. Phosphorylated SMAD2/3 binds to SMAD4, translocates into the nucleus and forms a complex with respective transcription factors to turn on chondrogenic genes. Top right: On scaffold with scrambled shRNA with RUNX2-transduced cells, TGF signaling phosphorylates SMAD3, which translocates into the nucleus and recruit HDAC4/5 to the promoters of RUNX2 and RUNX2 downstream targets, repressing their transcriptions. Bottom right: On scaffold with SMAD3 shRNA with RUNX2-transduced cells, SMAD3 shRNA is expressed, and thus degrades SMAD3 mRNA or inhibits its translation. This leads to de-repression at the promoters of RUNX2 and RUNX2 downstream targets, activating their expression. [*] Adapted from.47 TGF, transforming growth factor. Color images available online at www.liebertpub.com/tea
While scaffolds seeded cells transduced with SMAD3 shRNA and RUNX2 displayed matrix mineralization, they exhibited low levels of type I collagen. On the contrary, there was a slight increase in type X collagen staining in these scaffolds compared to the NT counterparts. While type I collagen is the most abundant protein of bone matrix,71 type X collagen is more characteristic of hypertrophic cartilage in the deep zone.72 This suggests that engineered MSCs may not have undergone full osteoblastic differentiation, but instead matured into hypertrophic chondrocytes that eventually progressed toward matrix mineralization. Future work focusing on dissecting the intracellular network responsible for endochondral or intramembranous ossification may shed light on the pathway underlying mineral deposition in these engineered MSCs, but are difficult to perform in vitro73 and thus will likely require in vivo studies.
Previous work from our group and others have demonstrated the feasibility of biomaterial-mediated gene delivery using viral vectors. This technique allows for spatially defined and directed cellular differentiation and tissue development by utilizing poly-L-lysine to immobilize gene delivery vectors.28,53,74 An important next step in our dual-scaffold single-culture system would be to apply this scaffold-mediated lentiviral transduction method for derivation of an osteochondral construct from one cell source in a single culture system. This approach could be used to develop a bilayered scaffold with differential coating of lentivirus to induce a layer of cartilage (noncoated) on top of a layer of bone (coated with SMAD3 shRNA and RUNX2). Toward future clinical application, delivery of such cellularized or acellular bilayered scaffolds may be used to induce differential matrix deposition by endogenous homing MSCs in situ. Our findings provide proof-of-concept for the ability to simultaneously induce cartilaginous and mineralized matrices in spatially defined, bilayered scaffolds under such environment.
Conclusions
This study demonstrates the ability to genetically engineer MSCs for differential cartilaginous and mineralized matrix depositions on one type of scaffold in one culture system. A defined chondrogenic environment with medium-supplemented TGFβ3 facilitated the production of GAG and type II collagen on one scaffold. Simultaneously, engineered MSCs with enhanced RUNX2 levels in modulation of SMAD3 generated mineralized matrix on scaffolds cultured in the same conditions. Controlling differential cell fate determination by this approach would allow for construction of complex multiphasic tissues without the need for prolonged culture in various medium conditions or elaborative scaffold structures. In the future, the efficacy of this method could be applied to fabricate an osteochondral construct from one single cell source for enhanced graft osseo-integration.
Supplementary Material
Acknowledgments
The authors thank Sara Oswald for providing technical writing support for the article. This work was supported by the Arthritis Foundation, NIH grants no. AR061042, AR50245, AR48852, AG15768, AR48182, AR067467, AR065956, OD008586, the Nancy Taylor Foundation for Chronic Diseases, NSF CAREER Award CBET-1151035, and the Collaborative Research Center of the AO Foundation, Davos, Switzerland.
Authors' Contributions
N.P.T.H., J.M.B, C.A.G., and F.G. developed the concept and designed experiments. N.P.T.H., J.M.B., C.C.G., and F.T.M. performed experiments. N.P.T.H. and C.C.G. analyzed data. N.P.T.H. wrote the article. All authors edited the article.
Disclosure Statement
F.T.M. and F.G. are paid employees of Cytex Therapeutics, Inc. For the remaining authors, no competing financial interests exist.
References
- 1.Sophia Fox A.J., Bedi A., and Rodeo S.A. The basic science of articular cartilage: structure, composition, and function. Sports Health 1, 461, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loeser R.F., Goldring S.R., Scanzello C.R., and Goldring M.B. Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum 64, 1697, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hootman J.M., and Helmick C.G. Projections of US prevalence of arthritis and associated activity limitations. Arthritis Rheum 54, 226, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Lawrence R.C., Felson D.T., Helmick C.G., et al. . ; National Arthritis Data, Workgroup. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part II. Arthritis Rheum 58, 26, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Math K.R., Zaidi S.F., Petchprapa C., and Harwin S.F. Imaging of total knee arthroplasty. Semin Musculoskelet Radiol 10, 47, 2006 [DOI] [PubMed] [Google Scholar]
- 6.Mulcahy H., and Chew F.S. Current concepts in knee replacement: features and imaging assessment. Am J Roentgenol 201, W828, 2013 [DOI] [PubMed] [Google Scholar]
- 7.Mulcahy H., and Chew F.S. Current concepts in knee replacement: complications. Am J Roentgenol 202, W76, 2014 [DOI] [PubMed] [Google Scholar]
- 8.Anderson J.A., Little D., Toth A.P., et al. . Stem cell therapies for knee cartilage repair: the current status of preclinical and clinical studies. Am J Sports Med 42, 2253, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mikos A.G., Herring S.W., Ochareon P., et al. . Engineering complex tissues. Tissue Eng 12, 3307, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lam J., Lu S., Meretoja V.V., Tabata Y., Mikos A.G., and Kasper F.K. Generation of osteochondral tissue constructs with chondrogenically and osteogenically predifferentiated mesenchymal stem cells encapsulated in bilayered hydrogels. Acta Biomater 10, 1112, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaefer D., Martin I., Shastri P., et al. . In vitro generation of osteochondral composites. Biomaterials 21, 2599, 2000 [DOI] [PubMed] [Google Scholar]
- 12.Schaefer D., Martin I., Jundt G., et al. . Tissue-engineered composites for the repair of large osteochondral defects. Arthritis Rheum 46, 2524, 2002 [DOI] [PubMed] [Google Scholar]
- 13.Gao J., Dennis J.E., Solchaga L.A., Awadallah A.S., Goldberg V.M., and Caplan A.I. Tissue-engineered fabrication of an osteochondral composite graft using rat bone marrow-derived mesenchymal stem cells. Tissue Eng 7, 363, 2001 [DOI] [PubMed] [Google Scholar]
- 14.Cao T., Ho K.H., and Teoh S.H. Scaffold design and in vitro study of osteochondral coculture in a three-dimensional porous polycaprolactone scaffold fabricated by fused deposition modeling. Tissue Eng 9(Suppl. 1), S103, 2003 [DOI] [PubMed] [Google Scholar]
- 15.Moutos F.T., and Guilak F. Composite scaffolds for cartilage tissue engineering. Biorheology 45, 501, 2008 [PMC free article] [PubMed] [Google Scholar]
- 16.O'Shea T.M., and Miao X. Bilayered scaffolds for osteochondral tissue engineering. Tissue Eng B Rev 14, 447, 2008 [DOI] [PubMed] [Google Scholar]
- 17.Khanarian N.T., Haney N.M., Burga R.A., and Lu H.H. A functional agarose-hydroxyapatite scaffold for osteochondral interface regeneration. Biomaterials 33, 5247, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khanarian N.T., Jiang J., Wan L.Q., Mow V.C., and Lu H.H. A hydrogel-mineral composite scaffold for osteochondral interface tissue engineering. Tissue Eng A 18, 533, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo X., Park H., Liu G., et al. . In vitro generation of an osteochondral construct using injectable hydrogel composites encapsulating rabbit marrow mesenchymal stem cells. Biomaterials 30, 2741, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo X., Liao J., Park H., et al. . Effects of TGF-beta3 and preculture period of osteogenic cells on the chondrogenic differentiation of rabbit marrow mesenchymal stem cells encapsulated in a bilayered hydrogel composite. Acta Biomater 6, 2920, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng H.W., Luk K.D., Cheung K.M., and Chan B.P. In vitro generation of an osteochondral interface from mesenchymal stem cell-collagen microspheres. Biomaterials 32, 1526, 2011 [DOI] [PubMed] [Google Scholar]
- 22.Grayson W.L., Bhumiratana S., Grace Chao P.H., Hung C.T., and Vunjak-Novakovic G. Spatial regulation of human mesenchymal stem cell differentiation in engineered osteochondral constructs: effects of pre-differentiation, soluble factors and medium perfusion. Osteoarthritis Cartilage 18, 714, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Re'em T., Witte F., Willbold E., Ruvinov E., and Cohen S. Simultaneous regeneration of articular cartilage and subchondral bone induced by spatially presented TGF-beta and BMP-4 in a bilayer affinity binding system. Acta Biomater 8, 3283, 2012 [DOI] [PubMed] [Google Scholar]
- 24.Chen J., Chen H., Li P., et al. . Simultaneous regeneration of articular cartilage and subchondral bone in vivo using MSCs induced by a spatially controlled gene delivery system in bilayered integrated scaffolds. Biomaterials 32, 4793, 2011 [DOI] [PubMed] [Google Scholar]
- 25.Tuli R., Nandi S., Li W.J., et al. . Human mesenchymal progenitor cell-based tissue engineering of a single-unit osteochondral construct. Tissue Eng 10, 1169, 2004 [DOI] [PubMed] [Google Scholar]
- 26.Bhatia R., and Hare J.M. Mesenchymal stem cells: future source for reparative medicine. Congest Heart Fail 11, 87, 2005 [DOI] [PubMed] [Google Scholar]
- 27.Pittenger M.F., Mackay A.M., Beck S.C., et al. . Multilineage potential of adult human mesenchymal stem cells. Science 284, 143, 1999 [DOI] [PubMed] [Google Scholar]
- 28.Brunger J.M., Huynh N.P., Guenther C.M., et al. . Scaffold-mediated lentiviral transduction for functional tissue engineering of cartilage. Proc Natl Acad Sci U S A 111, E798, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glass K.A., Link J.M., Brunger J.M., Moutos F.T., Gersbach C.A., and Guilak F. Tissue-engineered cartilage with inducible and tunable immunomodulatory properties. Biomaterials 35, 5921, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moutos F.T., Freed L.E., and Guilak F. A biomimetic three-dimensional woven composite scaffold for functional tissue engineering of cartilage. Nat Mater 6, 162, 2007 [DOI] [PubMed] [Google Scholar]
- 31.Moutos F.T., Glass K.A., Compton S.A., et al. . Anatomically shaped tissue-engineered cartilage with tunable and inducible anticytokine delivery for biological joint resurfacing. Proc Natl Acad Sci U S A 113, E4513, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abrahamsson C.K., Yang F., Park H., et al. . Chondrogenesis and mineralization during in vitro culture of human mesenchymal stem cells on three-dimensional woven scaffolds. Tissue Eng A 16, 3709, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Somoza R.A., Welter J.F., Correa D., and Caplan A.I. Chondrogenic differentiation of mesenchymal stem cells: challenges and unfulfilled expectations. Tissue Eng B Rev 20, 596, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hillel A.T., Taube J.M., Cornish T.C., et al. . Characterization of human mesenchymal stem cell-engineered cartilage: analysis of its ultrastructure, cell density and chondrocyte phenotype compared to native adult and fetal cartilage. Cells Tissues Organs 191, 12, 2010 [DOI] [PubMed] [Google Scholar]
- 35.Zhai L.J., Zhao K.Q., Wang Z.Q., Feng Y., and Xing S.C. Mesenchymal stem cells display different gene expression profiles compared to hyaline and elastic chondrocytes. Int J Clin Exp Med 4, 81, 2011 [PMC free article] [PubMed] [Google Scholar]
- 36.Mackay A.M., Beck S.C., Murphy J.M., Barry F.P., Chichester C.O., and Pittenger M.F. Chondrogenic differentiation of cultured human mesenchymal stem cells from marrow. Tissue Eng 4, 415, 1998 [DOI] [PubMed] [Google Scholar]
- 37.Johnstone B., Hering T.M., Caplan A.I., Goldberg V.M., and Yoo J.U. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res 238, 265, 1998 [DOI] [PubMed] [Google Scholar]
- 38.Kruger J.P., Machens I., Lahner M., Endres M., and Kaps C. Initial boost release of transforming growth factor-beta3 and chondrogenesis by freeze-dried bioactive polymer scaffolds. Ann Biomed Eng 42, 2562, 2014 [DOI] [PubMed] [Google Scholar]
- 39.Madry H., Rey-Rico A., Venkatesan J.K., Johnstone B., and Cucchiarini M. Transforming growth factor Beta-releasing scaffolds for cartilage tissue engineering. Tissue Eng B Rev 20, 106, 2014 [DOI] [PubMed] [Google Scholar]
- 40.Lian J.B., Stein G.S., Javed A., et al. . Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endocr Metab Disord 7, 1, 2006 [DOI] [PubMed] [Google Scholar]
- 41.Kempen D.H., Lu L., Hefferan T.E., et al. . Retention of in vitro and in vivo BMP-2 bioactivities in sustained delivery vehicles for bone tissue engineering. Biomaterials 29, 3245, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Musgrave D.S., Bosch P., Ghivizzani S., Robbins P.D., Evans C.H., and Huard J. Adenovirus-mediated direct gene therapy with bone morphogenetic protein-2 produces bone. Bone 24, 541, 1999 [DOI] [PubMed] [Google Scholar]
- 43.Song B., Estrada K.D., and Lyons K.M. Smad signaling in skeletal development and regeneration. Cytokine Growth Factor Rev 20, 379, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim D.W., and Lassar A.B. Smad-dependent recruitment of a histone deacetylase/Sin3A complex modulates the bone morphogenetic protein-dependent transcriptional repressor activity of Nkx3.2. Mol Cell Biol 23, 8704, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang J.S., Alliston T., Delston R., and Derynck R. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO J 24, 2543, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alliston T., Choy L., Ducy P., Karsenty G., and Derynck R. TGF-beta-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J 20, 2254, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang S.Y., and Alliston T. Regulation of postnatal bone homeostasis by TGFbeta. BoneKEy Rep 2, 255, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bian L., Zhai D.Y., Tous E., Rai R., Mauck R.L., and Burdick J.A. Enhanced MSC chondrogenesis following delivery of TGF-beta3 from alginate microspheres within hyaluronic acid hydrogels in vitro and in vivo. Biomaterials 32, 6425, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stein G.S., Lian J.B., van Wijnen A.J., et al. . Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene 23, 4315, 2004 [DOI] [PubMed] [Google Scholar]
- 50.Byers B.A., Guldberg R.E., and Garcia A.J. Synergy between genetic and tissue engineering: runx2 overexpression and in vitro construct development enhance in vivo mineralization. Tissue Eng 10, 1757, 2004 [DOI] [PubMed] [Google Scholar]
- 51.Byers B.A., Pavlath G.K., Murphy T.J., Karsenty G., and Garcia A.J. Cell-type-dependent up-regulation of in vitro mineralization after overexpression of the osteoblast-specific transcription factor Runx2/Cbfal. J Bone Miner Res 17, 1931, 2002 [DOI] [PubMed] [Google Scholar]
- 52.Gersbach C.A., Byers B.A., Pavlath G.K., and Garcia A.J. Runx2/Cbfa1 stimulates transdifferentiation of primary skeletal myoblasts into a mineralizing osteoblastic phenotype. Exp Cell Res 300, 406, 2004 [DOI] [PubMed] [Google Scholar]
- 53.Gersbach C.A., Coyer S.R., Le Doux J.M., and Garcia A.J. Biomaterial-mediated retroviral gene transfer using self-assembled monolayers. Biomaterials 28, 5121, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gersbach C.A., Guldberg R.E., and Garcia A.J. In vitro and in vivo osteoblastic differentiation of BMP-2- and Runx2-engineered skeletal myoblasts. J Cell Biochem 100, 1324, 2007 [DOI] [PubMed] [Google Scholar]
- 55.Zheng H., Guo Z., Ma Q., Jia H., and Dang G. Cbfa1/osf2 transduced bone marrow stromal cells facilitate bone formation in vitro and in vivo. Calcif Tissue Int 74, 194, 2004 [DOI] [PubMed] [Google Scholar]
- 56.Hagmann S., Moradi B., Frank S., et al. . FGF-2 addition during expansion of human bone marrow-derived stromal cells alters MSC surface marker distribution and chondrogenic differentiation potential. Cell Prolif 46, 396, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bodle J.C., Teeter S.D., Hluck B.H., Hardin J.W., Bernacki S.H., and Loboa E.G. Age-related effects on the potency of human adipose-derived stem cells: creation and evaluation of superlots and implications for musculoskeletal tissue engineering applications. Tissue Eng C Methods 20, 972, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boskey A.L., Guidon P., Doty S.B., Stiner D., Leboy P., and Binderman I. The mechanism of beta-glycerophosphate action in mineralizing chick limb-bud mesenchymal cell cultures. J Bone Miner Res 11, 1694, 1996 [DOI] [PubMed] [Google Scholar]
- 59.Coe M.R., Summers T.A., Parsons S.J., Boskey A.L., and Balian G. Matrix mineralization in hypertrophic chondrocyte cultures. Beta glycerophosphate increases type X collagen messenger RNA and the specific activity of pp60c-src kinase. Bone Miner 18, 91, 1992 [DOI] [PubMed] [Google Scholar]
- 60.Moutos F.T., and Guilak F. Functional properties of cell-seeded three-dimensionally woven poly(epsilon-caprolactone) scaffolds for cartilage tissue engineering. Tissue Eng A 16, 1291, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moffat J., Grueneberg D.A., Yang X., et al. . A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283, 2006 [DOI] [PubMed] [Google Scholar]
- 62.Wiznerowicz M., and Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol 77, 8957, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Diekman B.O., Thakore P.I., O'Connor S.K., et al. . Knockdown of the cell cycle inhibitor p21 enhances cartilage formation by induced pluripotent stem cells. Tissue Eng A 21, 1261, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salmon P., and Trono D. Production and titration of lentiviral vectors. Curr Protoc Neurosci Chapter, Unit 4.21, 2006. Chapter 4. [DOI] [PubMed] [Google Scholar]
- 65.Diekman B.O., Wu C.L., Louer C.R., et al. . Intra-articular delivery of purified mesenchymal stem cells from C57BL/6 or MRL/MpJ superhealer mice prevents posttraumatic arthritis. Cell Transplant 22, 1395, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Farndale R.W., Buttle D.J., and Barrett A.J. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta 883, 173, 1986 [DOI] [PubMed] [Google Scholar]
- 67.Estes B.T., Diekman B.O., Gimble J.M., and Guilak F. Isolation of adipose-derived stem cells and their induction to a chondrogenic phenotype. Nat Protoc 5, 1294, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Livak K.J., and Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402, 2001 [DOI] [PubMed] [Google Scholar]
- 69.Maeda S., Hayashi M., Komiya S., Imamura T., and Miyazono K. Endogenous TGF-beta signaling suppresses maturation of osteoblastic mesenchymal cells. EMBO J 23, 552, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mohammad K.S., Chen C.G., Balooch G., et al. . Pharmacologic inhibition of the TGF-beta type I receptor kinase has anabolic and anti-catabolic effects on bone. PLoS One 4, e5275, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boskey A.L. Bone composition: relationship to bone fragility and antiosteoporotic drug effects. BoneKEy Rep 2, 447, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmid T.M., Bonen D.K., Luchene L., and Linsenmayer T.F. Late events in chondrocyte differentiation: hypertrophy, type X collagen synthesis and matrix calcification. In Vivo 5, 533, 1991 [PubMed] [Google Scholar]
- 73.Gimble J.M., Guilak F., Nuttall M.E., Sathishkumar S., Vidal M., and Bunnell B.A. In vitro differentiation potential of mesenchymal stem cells. Transfus Med Hemother 35, 228, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Phillips J.E., Burns K.L., Le Doux J.M., Guldberg R.E., and Garcia A.J. Engineering graded tissue interfaces. Proc Natl Acad Sci U S A 105, 12170, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.