Introductory Paragraph
MCR-1-positve E. coli (MCRPEC) have been reported in human worldwide; however, thus far it’s prevalence is low and potential sources for human mcr-1 carriage have not yet been identified. Herein, we analysed a nationwide epidemiological data set on MCRPEC in humans throughout China and assessed factors associated with MCRPEC carriage using natural and national anthropogenic data. We identified 774 non-duplicate MCRPEC isolates from 774 stool samples collected from 5,159 healthy individuals in 30 provinces/municipalities in 2016, with a prevalence of MCRPEC ranging from 3.7% to 32.7% (average of 15.0%), substantially higher than previously reported. MCRPEC carriage was associated with provincial regions, production of sheep and freshwater aquaculture, annual consumption of total meat, pork and mutton, and daily intake of aquaculture products. MCRPEC was significantly more prevalent in provinces with higher aquaculture industries. WGS analysis revealed that the MCRPEC isolates were clustered into four distinct lineages, two of which were dominant and harbored most of the MCRPEC isolates. The high prevalence of MCRPEC in community poses a substantial risk for colistin usage in clinical practice and suggests the need for intestinal screening of mcr-1 carriers in intensive care units in Chinese hospitals. Furthermore, our data suggests that aquaculture is a significant reservoir of mcr-1.
Keywords: MCR-1, prevalence, factors, colistin resistance, Enterobacteriaceae, foodborne source, aquaculture, WGS analysis
Introduction
Since the discovery of the plasmid-mediated colistin resistance determinant, MCR-1, in Enterobacteriaceae in 2015, it has been reported globally; over five continents and 50 countries within two years1,2. So far, nine MCR-1 variants (mcr-1.2 to mcr-1.10)3–6 have been reported. Additionally, mcr has been identified in extensively drug resistant (XDR) Enterobacteriaceae isolates carrying plasmid-borne carbapenemase and ESBL genes7. Given that polymixins are a last resort treatment for XDR bacterial infections, the spread of mcr across pathogenic bacterial clones is likely to pose a serious public health crisis. In July 2016, the European Medicines Agency updated the risk level of colistin resistance transfer from low to high8. In November 2016, the Ministry of Agriculture of China (article No. 2428) withdrew colistin as a feed additive and growth promoter which was officially enforced in April of 20179. In Feb of 2017, the Department of Livestock Development in Thailand also banned the use of colistin as a feed additive10. Despite these regulatory edicts, colistin is still widely used in animal production and will continue to be used in farms either metaphylatically or for treating individual animals.
Unlike the high detection rates of MCR-positive Enterobacteriaceae (MCRPE) in animals7,11, in human populations MCRPE has been reported to be substantially lower. Among E. coli isolates associated with human infections, 1.3-1.7% were positive for mcr-12,12,13, while only 0.2%-0.7% of the clinical K. pneumoniae isolates harbored mcr-11,2,12,13. For E. coli isolates derived from human colonisation, the prevalence of mcr-1-positve E. coli (MCRPEC) varied between inpatients (0.4%-2.9%)2,14 and healthy individuals (0.7-6.2%)2,15,16. Most of these studies focused on MCRPE from clinical samples in inpatients, while the studies on MCRPE were from healthy individuals and were of a small sample size involving one or two hospitals2,16. Recently, we reported the widespread distribution of MCRPEC among Chinese farms and proposed the term “phantom resistome” - a phenomena where mcr-1 rates were considerably higher in DNA extracted from samples than isolates grown on media containing colistin17. A recent study examining the acquisition of MCRPE when traveling to India used an enrichment method and showed higher rates than non-enrichment methods which may, in part, explain the “phantom resistome”18.
Our previous MCRPEC data showed that carriage among humans had a weak but positive association with living distance from commercial animal (pig and poultry) farms, but not associated with diet preference (vegetarian vs. non-vegetarian)2. These findings suggested a possible link of MCRPEC colonisation/carriage between humans and animals, but this link remains unproven due to the limited sampling size and narrow geographic scope of previous studies2,14. Accordingly, we have determined the prevalence and molecular epidemiology of MCRPEC carriage in healthy individuals across 30 (out of overall 34) provinces/municipalities in China using an enrichment method to capture a larger sector of the “phantom resistome” and a truer prevalence of human carriage. Furthermore, given the potential spread of MCRPEC in Chinese communities we analysed the anthropogenic and environmental factors associated with human MCRPEC carriage. In addition, as mcr genes may have originated in aquatic environments19,20, the 30 Chinese provinces/municipalities were further divided into low, medium and high areas of aquaculture industrial intensity21, to access the impact of aquaculture-associated anthropogenic factors on MCRPEC carriage.
Results
Distribution of mcr-1
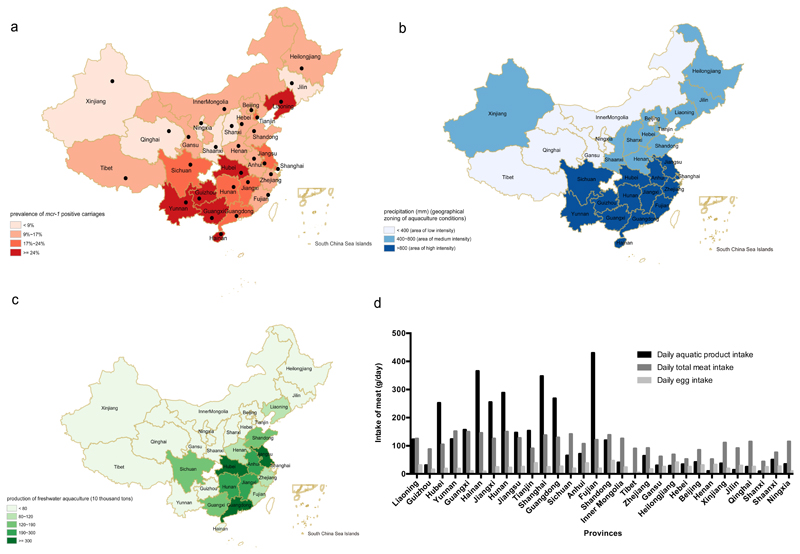
Our inclusion/exclusion criteria for subjects and criteria for processing of samples (whole genome sequencing and bioinformatics analysis) is given in Supp Fig 1. From 1st June 2016 to 30th September 2016, we collected 5159 rectal swabs from healthy individuals across 30 provinces/municipalities in China. Zhejiang and Beijing contributed the biggest number of samples (both > 470); Sichuan province and Shanghai contributed the smallest (both ≤ 50). The ratio of male to female in this study was 1.2 to 1. Age of subjects ranged from 12 to 89 years old (yo) and 83.8% were between 21 and 50 yo (Table 1). A total of 774 fecal samples were positive for the mcr-1 gene (15.0%), and MCRPEC (774 non-duplicate isolates) from each sample were further characterised. Liaoning province displayed the highest mcr-1 prevalence of 32.7% (95% CI: 20.7%-46.7%), while Ningxia province showed the lowest prevalence at 3.7% (95% CI: 1.5%-7.6%). The mean positive prevalence of mcr-1 carriages was 15.0% (95% CI: 14.0%-16.0%) among the 30 provinces/municipalities (Figure 1, Figure 2A and Supplementary Table 1).
Table 1. Descriptive and univariable analysis on the variables of interests.
| Variables | Categories | Percentage (n=5159) | Prevalence% (95%CI) | OR (95% CI) | P |
|---|---|---|---|---|---|
| Total samples in this study | 100% | 15.0 (14.0-16.0) | N/A¶ | N/A | |
| Gender | 0.899 | ||||
| Male | 53.9% | 15.1 (13.8-16.4) | 1.0 (0.9-1.2) | ||
| Female | 46.1% | 14.9 (13.5-16.4) | 1.0 | ||
| Age * | 0.048 | ||||
| 10~20 | 3.5% | 12.7 (8.2-18.5) | 1.0 | ||
| 21~30 | 26.2% | 13.8 (12.0-15.7) | 1.1 (0.7-1.7) | ||
| 31~40 | 36.4% | 15.3 (13.7-17.0) | 1.2 (0.8-2.0) | ||
| 41-50 | 21.2% | 15.1 (13.0-17.3) | 1.2 (0.8-1.9) | ||
| 51~60 | 7.6% | 15.7 (12.3-19.7) | 1.3 (0.8-2.1) | ||
| 61~70 | 3.6% | 18.1 (12.9-24.3) | 1.5 (0.9-2.7) | ||
| 70~80 | 1.2% | 19.7 (10.6-31.8) | 1.7 (0.8-3.6) | ||
| 81~90 | 0.2% | 36.4 (10.9-69.2) | 3.9 (1.1-14.5) | ||
| Geographical zoning of aquaculture conditions * | <0.001 | ||||
| low | 13.7% | 10.1 (8.0-12.6) | 1.0 | ||
| medium | 43.3% | 10.6 (9.3-11.9) | 1.1 (0.8-1.4) | ||
| high | 43.0% | 21.0 (19.4-22.8) | 2.4 (1.8-3.1) | ||
| Annual freshwater aquaculture production (10 thousand tons)D * | <0.001 | ||||
| ≤100 | 66.3% | 13.1 (12.0-14.3) | 1.0 | ||
| >100 | 33.7% | 18.8 (17.0-20.7) | 1.5 (1.3-1.8) | ||
| Annual aquatic product consumption (kg/person)A * | <0.001 | ||||
| ≤10 | 54.8% | 11.0 (9.9-12.1) | 1.0 | ||
| >10 | 45.2% | 19.9 (18.3-21.5) | 2.0 (1.7-2.3) | ||
| Daily aquatic product intake(g/day)E * | <0.001 | ||||
| ≤100 | 62.5% | 10.7 (9.7-11.8) | 1.0 | ||
| > 100 | 37.5% | 22.1 (20.3-24.0) | 2.4 (2.0-2.8) | ||
| GDP (100 million RMB)A | 0.985 | ||||
| Population number (10 thousands)B * | 0.040 | ||||
| Population density (10 thousands per km2)B | 0.580 | ||||
| Pollutants (10 thousand tons)A | smoke dust * | 0.002 | |||
| household garbage | 0.208 | ||||
| chemical oxygen demand | 0.213 | ||||
| ammonia nitrogen * | 0.018 | ||||
| total nitrogen | 0.908 | ||||
| total phosphorus | 0.275 | ||||
| Annual animal production (10 thousand tons)C | pig * | <0.001 | |||
| chicken * | <0.001 | ||||
| cattle | 0.906 | ||||
| sheep * | 0.001 | ||||
| Annual animal-derived food consumption(kg/person)A | meat * | <0.001 | |||
| pork * | <0.001 | ||||
| beef | 0.300 | ||||
| mutton * | <0.001 | ||||
| chicken meat * | <0.001 | ||||
| egg * | <0.001 | ||||
| Daily animal-derived food intake(g/day)E | total meat * | <0.001 | |||
| egg * | <0.001 | ||||
N/A= not applicable
Significant variable in univariable analysis (p≤0.05)
China Agriculture Yearbook, 2016
National Institute for Nutrition and Health, Chinese Center for Disease Control and Prevention (2010-2012
Figure 1.
Prevalence of mcr-1-positive E. coli among 30 provinces/municipalities in China. Sample size is listed in parenthesis after name of each province. Error bars represent 95% confidence interval. Dashed line indicates average prevalence of mcr-1 positive carriages in China. Mean and median prevalence are 16.40% and 15.11%, respectively across China.
Figure 2.
The mcr-1-positive prevalence in E. coli isolates (A), the precipitation and zoning of distinct aquaculture conditions. (B), freshwater aquaculture (C), and daily dietary intake of animal products (D) among 30 provinces/ municipalities in China. Each region is marked with the province’s name. In figure 2(A), the black dots indicate locations of both capital cities of provinces and the sample collection sites. Mean intake of daily aquatic product, total meat and egg are 120.72, 107.32 and 24.44 g/day, respectively; median intake of daily aquatic product, total meat and egg are 64.68, 112.89 and 24.95 g/day, respectively across China.
Correlation of mcr-1 carriages with environmental and anthropogenic factors
Given that previous studies have suggested that MCRPEC started and spread within Chinese farming communities, we analyzed the potential association of mcr-1 carriage with behavioral and non-behavioral factors including environmental and anthropogenic (Figure 2B, 2C, 2D, and Supplementary Table 2). When analysed by univariable analysis, 18/27 associations were shown to be significant (p ≤ 0.05) (Table 1). No observable difference was seen between male and female (OR=1.0, 95%CI: 0.9-1.2). The odds of MCRPEC for ages 81 to 90 (OR=3.9, 95%CI: 1.1-14.5) was significantly higher than other age brackets but accounted for 0.2% of the samples. After removing highly correlated variables, 13 variables remained, seven of which were significantly (P<0.05) associated with mcr-1 positivity by multivariable logistic analysis (Table 2). In this model, geographical zones with low aquaculture industry had significantly lower odds (OR=0.5, 95%CI: 0.3-0.7) of mcr-1 positivity compared to those with higher aquaculture activity. Populations with lower levels (≤100 g/day) of aquatic food intake also had significantly lower odds (OR=0.6, 95%CI: 0.5-0.7) of mcr-1 positivity. Unsurprisingly, meat, especially pork and mutton consumption was significantly associated with higher mcr-1 prevalence (Table 2). Hosmer-Lemeshow “goodness of fit test” χ2 = 0.6 (df = 8, p = 1 > 0.05) indicates the model was a robust fit of the data. The estimated area under the Receiver Operating Characteristic curve was 0.65, indicating good predictability.
Table 2. Multivariable logistic regression analysis of factors associated with mcr-1 positivity (n=5159).
| Variables | Categories | OR (95% CI) | P |
|---|---|---|---|
| Geographical zoning of aquaculture conditions | low* | 0.5 (0.3-0.7) | 0.002 |
| medium* | 0.7 (0.5-0.9) | 0.016 | |
| high* | 1.0 | 0.005 | |
| Daily aquatic product intake(g/day)* | ≤100* | 0.6 (0.5-0.7) | <0.001 |
| >100* | 1.0 | <0.001 | |
| Total population* | 0.003 | ||
| Annual animal production (10 thousand tons) | sheep* | <0.001 | |
| Annual animal-derived food consumption (kg/person) | meat* | <0.001 | |
| pork* | 0.014 | ||
| mutton* | 0.003 | ||
significant variable in multivariable logistic regression analysis (p<0.05)
Minimum inhibitory concentration (MIC) profiles of MCRPEC
We tested antimicrobial susceptibility of all MCRPEC isolates from the 30 provinces/municipalities to nine clinical antibiotics (cefepime, amikacin, piperacillin/tazobactam, ceftazidime, ticarcillin/clavulanic acid, ciprofloxacin, imipenem, colistin and cefoperazone/sulbactam). In general, MCRPEC was more commonly resistant to ticarcillin/clavulanic acid (57.88%) and ciprofloxacin (38.89%) than other antimicrobial agents, and only a small number of mcr-1 positive isolates were resistant to amikacin (1.16%), imipenem (1.29%) and cefoperazone/sulbactam (2.32%, Supplementary Table 3). Significant difference of MIC profiles among three aquaculture conditional areas was only found in cefepime (P=0.002), ceftazidime (P=0.018) and ciprofloxacin (P=0.011) (Supplementary Table 4).
Relationship of the MCRPEC isolates determined by whole genome sequencing
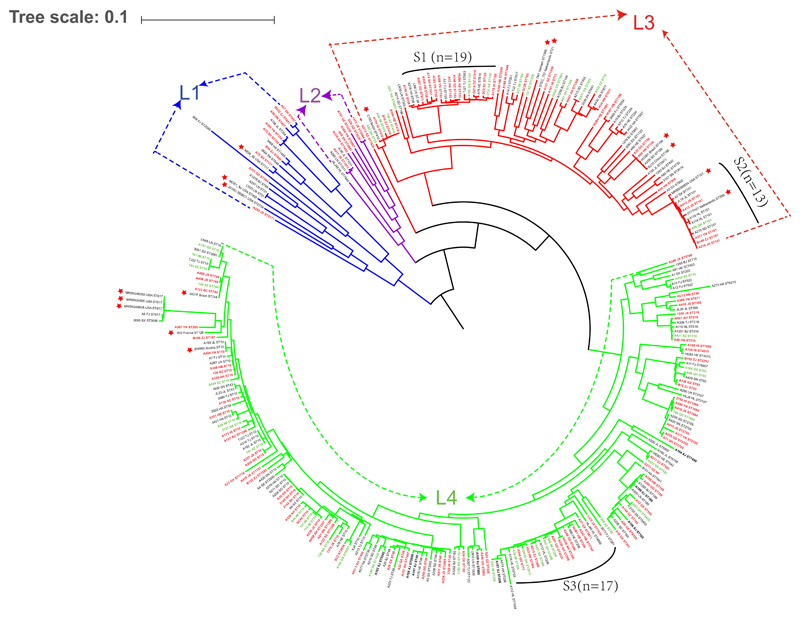
The core-genome based phylogenetic analysis together with Bayesian analysis of the population structure (BAPS) was employed to define lineages within the 287 sequenced MCRPEC isolates from 30 provinces/municipalities across China (Supplementary Table 1). Genomic data from an additional 16 clinical MCRPEC isolates from Asia, Europe, North and South America were added for comparison (Supplementary Table 5). The BAPS analysis revealed four distinct lineages among the 287 MCRPEC isolates (Figure 3). The main lineages, L3 (n=78) and L4 (n=183), comprised of MCRPEC isolates from 29 (except Xinjiang province) and 30 provinces, respectively. The additional 16 international MCRPEC isolates fell into lineages L1 (n=3), L3 (n=7) and L4 (n=6) and displayed high similarity to the isolates analysed in this study. There was no obvious correlation between lineages and provinces. Notably, the isolates from the low, medium and high aquaculture areas correlated within lineages L3 and L4, in which three highly homologous sublineages S1, S2 and S3 harbored 19, 13 and 17 MCRPEC isolates from 15, 9 and 10 provinces, respectively. Two isolates, each from USA and Netherlands, also fell into cluster S2 (Figure 3).
Figure 3.
Genetic relationship of the MCRPEC isolates. Phylogenic tree of 287 MCRPEC isolates from the 30 provinces in China and 16 human-originated isolates from other countries. Each isolate is labeled with its province or country and ST types. Isolates from the 14, 11 and 5 provinces/municipalities within the low-high aquaculture regions are marked with red, black and green colour, respectively. Branches color are indicated with blue, purple, red and green for four lineages, respectively. Red stars represent the sixteen isolates from other countries.
Distribution of antibiotic resistance genes and virulence associated genes among MCRPEC isolates
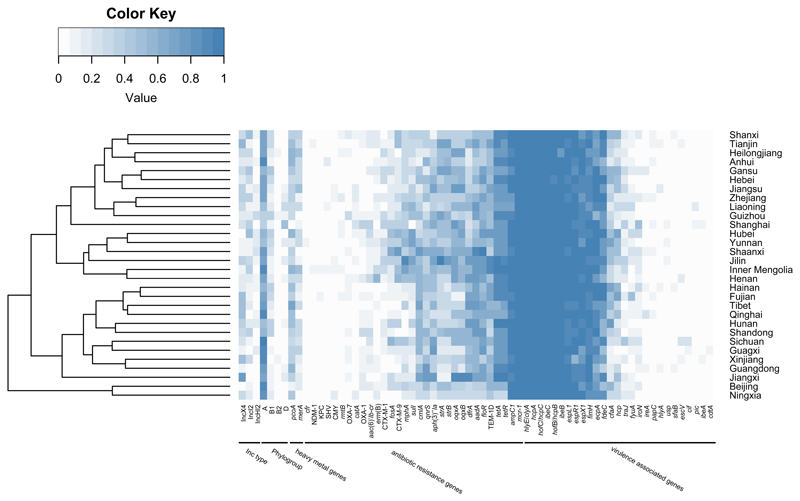
Antibiotic resistance genes (ARGs) and virulence associated genes (VAGs) in the 287 MCRPEC isolates were determined from the whole genome sequences. A heatmap showing the percentage of presences of both RGs and VAGs for each province/municipality was constructed (Figure 4). No obvious association of ARGs or VAGs was found between provinces. Noticeably, most isolates belong to phylogenetic group A and only a few isolates were of phylogroup B2 and D, but phylogroup B2 and D isolates harbored a larger number of VAGs than those from group A and B1. However, ARGs of the isolates from B2 and D group, were not strongly associated with VAGs of them (Figure 4 and Supplementary Table 6). For VAGs, the mcr-1 gene co-existed more with hcpA/B/C that encodes the hemorrhagic pilus of E. coli, ecpA (common pilus), hlyE/clyA (hemolysin/cytolysin A), espL/R/X (Type III secretion system), fimH (Type I fimbriae) and ibeB/C. Whereas VAGs of pathogenic E. coli such as ETEC, EAEC, EIEC, EPEC, EHEC, DAEC etc. were rarely found in MCRPEC isolates from our study (Supplementary Table 6). In addition, resistance (floR) to florfenicol, a veterinary drug, was commonly associated with mcr-1. The association of tetracycline RGs (tetA and tetR), plasmid-mediated quinolone resistance genes (oqxA/B), β-lactamase RG (blaTEM) and some aminoglycoside RGs (aadA, strA/B and aph(3’)-Ia) with mcr-1 were higher than other ARGs; namely, other β-lactamase RGs (blaCTX, blaOXA, blaCMY and blaSHV, etc.) and carbapenem RGs (blaKPC and blaNDM) (Figure 4 and Supplementary Table 6).
Figure 4.
Distribution of phylogenetic group, Inc type, antibiotic resistance genes and virulence associated genes among mcr-1-positive E. coli from 30 provinces/ municipalities in China. Each box represents positive percentage of corresponding item among sequenced isolates in corresponding province.
Diversity of MCRPEC isolates and plasmid Inc types
MLST groups of MCRPEC were assessed by minimum spanning trees for all 287 isolates from 30 provinces/municipalities, which were divided into three sectors according to distinct aquaculture activities (Supplementary Figure 2). The 287 MCRPEC strains were separated into 135 MLST clades showing marked diversity across each province (Supplementary Table 6). ST10 was the dominant clade containing 46 (16.0%) MCRPEC and other MLST clades, including ST48, ST206, ST93, ST101, ST1684 and ST216 were also widely distributed across low, medium and high regions of aquaculture industry. ST410 (n=5) and ST155 (n=9) were not identified in the medium-high regions of aquaculture activity, while ST189 (n=5) and ST58 (n=5) were not present in low areas of aquaculture (Supplementary Figure 2). Furthermore, mcr-1 was identified in plasmids containing a variety of different Inc types - IncX4-type (n=83, 28.9%), IncI2-type (n=60, 21.0%), IncHI2-type (n=23, 8.0%), IncP-type (n=8, 2.8%), IncY-type (n=8, 2.8%), IncF-type (n=1), and syncretic (n=9, 3.1%) plasmids (Supplementary Table 6).
Distinct prevalence of MCRPEC between human and chicken origin
Given the significant correlation between the MCRPEC carriage in humans and the animal food chain as shown above, we further tried to analyse the association between human MCRPEC isolates and other MCRPEC isolates of animal origins by whole genome phylogenetic analysis. But in NCBI database, we only found 88 whole genomes of MCRPEC from Chinese chickens from previous studies17,22 (All sequences are available in NCBI under BioProject accession numbers PRJNA349231 and PRJNA417344). Unfortunately, there are no MCRPEC whole genome sequences available for swine, sheep, cattle or fish from China. Although MLST diversity was also found in MCRPEC from chickens, minimum spanning tree analysis showed distinct clustering between human and chicken isolates (Supplementary Figure 3). ST10 was also the dominant clade; 46 and 5 MCRPEC from human and chicken, respectively. However, ST10 MCRPEC was more common in human carriage than in chickens (16.0% vs 5.7%, P=0.013). In contrast, ST156 was more prevalent in chicken carriage than from human intestines (23.9% vs 1.0%, P<0.0001). Interestingly, several ST clades each containing ≥ 5 isolates were only observed in human carriage, such as ST155, ST58, ST189, ST410, ST216, ST2705 and ST1684 (Supplementary Figure 3). Core-genome phylogenetic analysis supported the commonality of ST156 clades among disparate origins, provinces and even countries (Supplementary Figure 4). ST345 and ST101 also overlapped for MCRPEC isolates from humans and chickens.
Discussion
A key finding of this study is the high prevalence of MCRPEC carriage as human normal flora (15.0%), which is considerably higher than previously reported (0.7-6.2%)2,15,16. Our results suggest that the true prevalence of MCRPEC was previously underestimated probably as a consequence of directly screening for mcr-1 positive Enterobacteriaceae without an enrichment step. In contrast to previous studies, we first determined the presence of mcr-1 by PCR and then isolated MCRPEC with an enrichment step using selective media with colistin, which resulted in a significant increase in MCRPEC detection. The higher MCRPEC prevalence will have major clinical implications as nosocomial outbreaks of MCRPEC are likely to occur if appropriate screening in Chinese ICUs, HDUs and transparent/haematology units are not enforced. Although few isolates were from phylogenetic group B2, patients presenting with significant co-morbidities will be at risk of infection and there is the high possibility of mcr-1 transfer in-vivo into B2 pathogens. Given that polymyxin B has recently been approved for human medicine by the China Food and Drug Administration (January 5th, 2017),23 further studies are urgently needed to assess the impact of MCRPEC carriage prevalence on endogenous infections and treatment failure.
Using microbiological and anthropogenic data, we identified possible associations with human MCRPEC carriage. The positive correlation between human MCRPEC carriage and the consumption of total meat, pork and sheep is in accordance with the notion that the spread of mcr-1 probably occurred from farm animals to human beings1,11. However, the identification of aquatic-food as a correlated factor of MCRPEC is unexpected (Supplementary Figure 5). Although colistin is not officially used in Chinese aquaculture, the aquatic environment is likely to be contaminated by colistin through contamination from farms as the drug is very poorly absorbed after oral administration to pigs and poultry24, and can be excreted in high-levels with mcr-1/MCRPEC from the animal faeces25. Consequently, when manure is used either for irrigation in agriculture, feeding farmed fish, or directly discharged into the aquatic environment, it will pollute rivers and lakes with MCRPEC. The high stability of colistin in water26 will further exacerbate the persistence and dissemination of MCRPEC in the aquatic environment by providing a selective pressure. Aquatic food produce may also be contaminated by MCRPEC, which is supported by the daily intake of aquatic food products as a correlated factor for MCRPEC carriage.
The high diversity of ST clades of MCRPEC across China further confirms the wide distribution of mcr-1-carrying E. coli and that colistin resistance is mediated more by plasmids than dominant bacterial clones. The unique main ST clusters in human (ST10) and chicken (ST156) implied the different colonized MCRPEC and transmission route from each origin, but this finding might be restricted by the limited animal sampling size and locations in the two previous studies17,22. However, all MCRPEC were restricted to a limited number of distinct E. coli lineages; the examined 375 (287 from human and 88 from chicken) isolates can be placed into 4 broad lineages (Figure 3). This limited diversity contrasts with the transmission of the blaNDM gene, which was found in eight distinct E. coli lineages from a single poultry production chain in one province (Shandong)17. These data suggest that the host spectrum of mcr-1-carrying plasmids is considerably narrower than blaNDM-carrying plasmids, which would also suggest why mcr-1 is mainly found in E. coli and has yet to be reported outside Enterobacteriaceae. The predominant association of mcr-1 with narrow-host type of plasmids, e.g. IncI2, IncHI2, and IncX47, is also likely to contribute to the limited number of clusters of MCRPEC isolates in China and worldwide.
Although this is the largest and most comprehensive analysis of its type, we acknowledge several limitations in this study. First, for the sampling we only utilized one hospital per province, which may not be fully congruent with the social-economic parameters representative of each province. Second, total meat consumption consisted of all meat types not just pork; however, pork accounted for 62.4% of the annual consumption of total meat in China27. Third, the possible transmission of MCRPEC to humans through aquatic food was only identified by correlation analysis, and has not been proven by detailed epidemiological data. However, it has been hypothesized that mcr-1 originated from an aquacultural environment19,20, and several recent studies have reported the widespread distribution of waterborne mcr-1 gene in Asia, Europe and South America7,28,29. Fourth, all human MCRPEC analyzed in this study were collected in 2016; however, most of the environmental and anthropogenic data were from 2015, although we would anticipate little variation. Lastly, we collected 774 MCRPEC from 30 provinces/municipalities but provided complete genomic data on 37.1% of the isolates (n=287).
Despite these limitations, our study provides new evidence on a China-wide scale that foodborne transmission of MCRPEC is a major correlated factor for human carriage. We would also suggest that the methods applied in this study be fully adopted to understand the true prevalence of MCRPEC; not just in China but globally. It is hoped that the newly released polices to withdrawal colistin as feed addictive, in combination with the prudent colistin usage as disease treatments in farm animals, might reduce the risk of MCRPEC in humans. We have shown that aquatic food production, a previously unknown mcr-1 source, has a significantly associated with human MCRPEC carriage highlighting the need for a broader one-health approach to AMR studies. Hitherto, the primary focus of mcr surveillance programs has been pig and poultry farms, and thus in China, Southeast Asia and South America, regions of freshwater and marine food produce, should be incorporated into future surveillance programs 28. The clinical impact of the high MCRPEC carriage in healthy people remains unknown and should be examined further not least to inform current Chinese hospital infection control programs and screening programs supporting high-risk Chinese patients.
Methods
Subject Enrollment
We undertook a cross-sectional study to investigate the prevalence of MCRPEC from humans in China, from 1st June, 2016 to 30th September, 2016. Thirty hospitals from 30 provincial capital cities were recruited in this study, which covered all Chinese provinces/municipalities except for Chongqing, Hong Kong, Macao and Taiwan (Figure 2A and Supplementary Table 1). Hospitals were asked to enroll subjects over a five-day period. A schematic diagram summarising our sampling strategy and our inclusion/exclusion criteria is shown in Supplementary Figure 1. We included healthy individuals attending the routine physical examination, during which stool samples were collected. The inclusion criteria were subjects that were considered generally healthy and consented to the study. Exclusion criteria included neonates, pregnancy, gastroenteritis and subjects presenting with chronic illnesses e.g. cancer. Vegetarians were also excluded in keeping with data from previous studies2. Ethical permission for this study was agreed on 22nd February 2016 by the Zhejiang University ethics committee under the project DETER-XDR-China. A brief survey of the selected individuals was conducted regarding meat consumption and antibiotics used in the past three months. Individual consent forms were translated into Mandarin and consent was obtained for all healthy individuals. Due to the uneven economical and social development in different provinces in China, the number of individuals attending physical examinations during a five-day period varied significantly in different provinces. Therefore, the number of samples from each province were varied.
Sample collection
All colistin resistant isolates were recovered from rectal swabs by three steps. First, stool samples were cultured in LB broth (Luqiao, Beijing, China) and the DNA was extracted from the enriched broth by the boiling method. Second, the broth suspension of a mcr-1 positive sample was enriched by the Enterobacteriaceae Enrichment broth (Luqiao, Beijing, China), and then the enrichment was inoculated onto MacConkey agar (Luqiao, Beijing, China) plate containing 2 mg/L colistin. Thirdly, colonies on selected agar were confirmed for mcr-1 gene by PCR and the species identified by MALDI-TOF mass spectrometry (BrukerDaltonik GmbH, Bremen, Germany)30.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing of all non-duplicated MCRPEC to nine clinical antimicrobial agents (cefepime, amikacin, piperacillin/tazobactam, ceftazidime, ticarcillin/clavulanic acid, ciprofloxacin, imipenem, colistin and cefoperazone/sulbactam), all of which are often prescribed to patients as part of the Chinese national formulary, was undertaken by the broth microdilution method using ATCC25922 as a quality control strain. Interpretation of the results was according to the European Committee on Antimicrobial Susceptibility Testing clinical breakpoints (version 6.0) and the Clinical and Laboratory Standards Institute document M100-S2531.
Molecular analysis
We selected 287 MCRPEC from all 30 provinces/municipalities (sequence all MCRPEC if 10 or less, or sequence 10 isolates from each province if number of MCRPEC is > 10, Supplementary Figure 1 and Supplementary Table 1). Genomic DNA of the selected isolates was extracted using the Wizard Genomic DNA Purification Kit (Promega, Beijing, China), following the manufacturer’s manuals. Indexed DNA libraries were constructed using the KAPA Hyper Prep Kit Illumina® platforms (Roche, Basle, Switzerland) with standard protocol and were sequenced on the Illumina Hiseq X ten platform with 150-bp paired-end strategy (Annoroad, Beijing, China). The draft genomes were assembled using SPAdes version 3.9.032 and MLST typing, antibiotic resistance genes and virulence associated genes were done using the SRST2 toolkit version 0.2.033. Minimum spanning tree of all sequence types was generated by BioNumerics version 7.0 (Applied Maths, Belgium) using BURST algorithm for related STs between different backgrounds. All assembled genomes were used for core-genome alignments to produce a phylogenetic tree by Harvest package version 1.1.234, and the corresponding features of each isolate were visualized by the online tool iTOL 335. The lineages of phylogenetic tree were defined by BAPS groups version 6.036. The mcr-1-carrying contigs generated by Illumina sequencing were examined for Inc types by PlasmidFinder version 1.337. Heatmap was generated using R3.3.2 (R Foundation for Statistical Computing) with the gplots 3.0.1 package. All WGS data have been deposited in NCBI database, Bioproject: PRJNA400107.
Data collection and statistical analysis
Natural (annual precipitation in capital city of each province) and anthropogenic data for the 30 provinces/municipalities in China were collected from National Bureau of Statistics of China, China Agriculture Yearbook, and National Institute for Nutrition and Health, Chinese Center for Disease Control and Prevention (Supplementary Table 2). The six anthropogenic factors included gross domestic product (GDP), population (number and density), pollutants (smoke dust, household garbage and industry discharge), animal production (farm animals and freshwater aquaculture, mainly including fish and shrimp), annual animal-derived food consumption, and daily animal-derived food intake. Most of the data were 2015 numbers, but the latest population data was from the 6th population census of China in 2010 and the latest data of daily animal-derived food intake was collected from 2010-2012 (Supplementary Table 2). Data were organised in Excel 2016 ©Microsoft. Descriptive analysis on the percentage and the prevalence (together with 95%CI) were calculated via functions provided by Excel 2016 ©Microsoft. Univariable analysis conducted by SPSS Version 22 ©IBM was adopted to select variables with a p≤0.05. Significant variables (p≤0.05) were subsequently assessed for collinearity via Cramer's coefficient phi (Φ). If a pair of variables were highly correlated (Φ>0.8), the more biologically plausible variable was kept for the multivariable logistic analysis. Significant variables on the univariable analyses were kept for the multivariable analysis. Multivariable logistic regression model using a forward stepwise process were then adopted for controlling confounders. Variables with a p < 0.05 were kept in the final model. The goodness of fit to the logistic regression model was tested via Hosmer-Lemeshow goodness-of-fit test. Subsequently, a receiver operating characteristic (ROC) curve was used to test the predictive ability of the mode.
Supplementary Material
Supplementary information is available online.
Acknowledgements
This work was supported in part by the grants from National Key Research and Development Program of China (2018YFD0500300), National Natural Science Foundation of China (81661138002 and 81772250), and Medical Research Council grant DETER-XDRE-CHINA (MR/P007295/1).
Footnotes
Data availability
WGS data that support the findings of this study have been deposited in NCBI database under Bioproject accession no. PRJNA400107. Other main data supporting the findings of this study are available within this Article and in the Supplementary Information files. Extra data supporting the findings of this study are available from the corresponding author upon reasonable request.
Additional information
Reprints and permissions information is available online at www.nature.com/reprints.
Authors’ contributions
YW, RZ and JZS designed the study. YS, HZ, JX, YH, LY, QS, YO, YLW and BS collected the data. YS, YW, YQW, HZ, RZ, QZ, CW, BS, ZS, ZW, SW, XX, YNW, CC, JL, TRW and JZS analysed and interpreted the data. YW, RZ, YS and TRW wrote the manuscript. All authors reviewed, revised, and approved the final report.
Competing interests
All authors declare that they have no conflicts of interest.
References
- 1.Liu YY, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, et al. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis. 2017;17:390–399. doi: 10.1016/S1473-3099(16)30527-8. [DOI] [PubMed] [Google Scholar]
- 3.Di Pilato V, et al. mcr-1.2, a new mcr cariant carried on a transferable plasmid from a colistin-resistant KPC carbapenemase-producing Klebsiella pneumoniae strain of sequence type 512. Antimicrob Agents Chemother. 2016;60:5612–5615. doi: 10.1128/AAC.01075-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai L, et al. A novel disrupted mcr-1 gene and a lysogenized phage P1-like sequence detected from a large conjugative plasmid, cultured from a human atypical enteropathogenic Escherichia coli (aEPEC) recovered in China. J Antimicrob Chemother. 2017;72:1531–1533. doi: 10.1093/jac/dkw564. [DOI] [PubMed] [Google Scholar]
- 5.Tijet N, et al. Molecular characteristics of mcr-1-carrying plasmids and new mcr-1 variant recovered from polyclonal clinical Escherichia coli from Argentina and Canada. PLoS One. 2017;12:e0180347. doi: 10.1371/journal.pone.0180347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu X, et al. MCR-1.6, a New MCR Variant Carried by an IncP Plasmid in a Colistin-Resistant Salmonella enterica Serovar Typhimurium Isolate from a Healthy Individual. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.02632-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel L, Jayol A, Nordmann P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin Microbiol Rev. 2017;30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2016/07/WC500211080.pdf.
- 9.Walsh TR, Wu YN. China bans colistin as a feed additive for animals. Lancet Infect Dis. 2016;16:1102–1103. doi: 10.1016/S1473-3099(16)30341-3. [DOI] [PubMed] [Google Scholar]
- 10. http://www.nationmultimedia.com/news/national/30305408.
- 11.Schwarz S, Johnson AP. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother. 2016;71:2066–2070. doi: 10.1093/jac/dkw274. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, et al. Molecular epidemiology of colistin-resistant Enterobacteriaceae in inpatients and avian from China: high prevalence of mcr-negative Klebsiella pneumoniae. Int J Antimicrob Agents. 2017;50:536–541. doi: 10.1016/j.ijantimicag.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Quan J, et al. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017;17:400–410. doi: 10.1016/S1473-3099(16)30528-X. [DOI] [PubMed] [Google Scholar]
- 14.Terveer EM, et al. Prevalence of colistin resistance gene (mcr-1) containing Enterobacteriaceae in feces of patients attending a tertiary care hospital and detection of a mcr-1 containing, colistin susceptible E. coli. PloS One. 2017;12:e0178598. doi: 10.1371/journal.pone.0178598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bi Z, et al. Prevalence of the mcr-1 colistin resistance gene in extended-spectrum beta-lactamase-producing Escherichia coli from human faecal samples collected in 2012 in rural villages in Shandong Province, China. Int J Antimicrob Agents. 2017;49:493–497. doi: 10.1016/j.ijantimicag.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 16.Zhong LL, et al. High rates of human fecal carriage of mcr-1-positive multi-drug resistant Enterobacteriaceae isolates emerge in China in association with successful plasmid families. Clin Infect Dis. 2017 doi: 10.1093/cid/cix885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol. 2017;2:16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 18.Bernasconi OJ, et al. Travelers Can Import Colistin-Resistant Enterobacteriaceae, Including Those Possessing the Plasmid-Mediated mcr-1 Gene. Antimicrob Agents Chemother. 2016;60:5080–5084. doi: 10.1128/AAC.00731-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabello FC, Godfrey HP, Buschmann AH, Dolz HJ. Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect Dis. 2016;16:e127–33. doi: 10.1016/S1473-3099(16)00100-6. [DOI] [PubMed] [Google Scholar]
- 20.Cabello FC, Tomova A, Ivanova L, Godfrey HP. Aquaculture and mcr Colistin Resistance Determinants. MBio. 2017;8 doi: 10.1128/mBio.01229-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao CS, et al. Geographical agglomeration characteristic and spatial evolution mechanism of aquaculture industry in China. Econ Geogr. 2016;36(9):118–127. [Article in Chinese] [Google Scholar]
- 22.Wu C, et al. Rapid rise of the ESBL and mcr-1 genes in Escherichia coli of chicken origin in China, 2008-2014. Emerg Microbes Infect. 2018;7:30. doi: 10.1038/s41426-018-0033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. http://app2.sfda.gov.cn/datasearchp/index1.do?tableId=25&tableName=TABLE25&tableView=%E5%9B%BD%E4%BA%A7%E8%8D%AF%E5%93%81&Id=214182.
- 24.Sato H, Ouchi M, Koumi J. Distribution of colistin sulfate in the body. Distribution and metabolism of orally administered colistin sulfate in chickens and pigs. Jpn J Antibiot. 1972;25:239–245. [Ariticle in Japenase] [PubMed] [Google Scholar]
- 25.Rhouma M, et al. Gastric stability and oral bioavailability of colistin sulfate in pigs challenged or not with Escherichia coli O149: F4 (K88) Res Vet Sci. 2015;102:173–181. doi: 10.1016/j.rvsc.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Milne RW, Nation RL, Turnidge JD, Coulthard K. Stability of colistin and colistin methanesulfonate in aqueous media and plasma as determined by high-performance liquid chromatography. Antimicrob Agents Chemother. 2003;47:1364–1370. doi: 10.1128/AAC.47.4.1364-1370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Qiong, Wang Ji-min. Current Situation and Future Trends of Meat Consumption in China. Food Nutr China. 2013;19:43–47. [Ariticle in Chinese] [Google Scholar]
- 28.Zhou HW, Zhang T, Ma JH, et al. Occurrence of Plasmid- and Chromosome-encoded mcr-1 in Water-borne Enterobacteriaceae in China. Antimicrob Agents Chemother. 2017 doi: 10.1128/AAC.00017-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandes MR, et al. Colistin-Resistant mcr-1-Positive Escherichia coli in Public Beaches, an Infectious Threat Emerging in Recreational Waters. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.00234-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu YY, et al. Colistin-resistance gene mcr-1 in children's gut flora. Intl J Antimicrob Agents. 2017 doi: 10.1016/j.ijantimicag.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 31.Wayne P, editor. Clinical and Laboratory Standards Institute. M100-S25. Performance standards for antimicrobial susceptibility testing. 25th Informational Supplement. USA: Clinical and Laboratory Standards Institute; 2015. [Google Scholar]
- 32.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inouye M, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/PREACCEPT-2573980311437212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng L, Connor TR, Siren J, Aanensen DM, Corander J. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol. 2013;30:1224–1228. doi: 10.1093/molbev/mst028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carattoli A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.