Abstract
Background & Aims
Crohn’s disease is an inflammatory bowel disease that affects the ileum and is associated with increased cytokines. Although interleukin (IL)6, IL17, IL21, and IL22 are increased in Crohn’s disease and are associated with disrupted epithelial regeneration, little is known about their effects on the intestinal stem cells (ISCs) that mediate tissue repair. We hypothesized that ILs may target ISCs and reduce ISC-driven epithelial renewal.
Methods
A screen of IL6, IL17, IL21, or IL22 was performed on ileal mouse organoids. Computational modeling was used to predict microenvironment cytokine concentrations. Organoid size, survival, proliferation, and differentiation were characterized by morphometrics, quantitative reverse-transcription polymerase chain reaction, and immunostaining on whole organoids or isolated ISCs. ISC function was assayed using serial passaging to single cells followed by organoid quantification. Single-cell RNA sequencing was used to assess Il22ra1 expression patterns in ISCs and transit-amplifying (TA) progenitors. An IL22-transgenic mouse was used to confirm the impact of increased IL22 on proliferative cells in vivo.
Results
High IL22 levels caused decreased ileal organoid survival, however, resistant organoids grew larger and showed increased proliferation over controls. Il22ra1 was expressed on only a subset of ISCs and TA progenitors. IL22-treated ISCs did not show appreciable differentiation defects, but ISC biomarker expression and self-renewal–associated pathway activity was reduced and accompanied by an inhibition of ISC expansion. In vivo, chronically increased IL22 levels, similar to predicted microenvironment levels, showed increases in proliferative cells in the TA zone with no increase in ISCs.
Conclusions
Increased IL22 limits ISC expansion in favor of increased TA progenitor cell expansion.
Keywords: Intestinal Stem Cells, Interleukin-22, Inflammatory Bowel Disease
Abbreviations used in this paper: BSA, bovine serum albumin; cDNA, complementary DNA; EGFP, enhanced green fluorescent protein; FACS, fluorescence-activated cell sorter; IBD, inflammatory bowel disease; IL, interleukin; ILC, innate lymphoid cell; ILC3, IL22-secreting lymphocyte; IL22RA1, IL22 receptor A1; IL22TG, IL22 transgenic; ISC, intestinal stem cell; mRNA, messenger RNA; OFE, organoid forming efficiency; STAT3, signal transducer and activator of transcription 3; TA, transit-amplifying; TBS, Tris-buffered saline
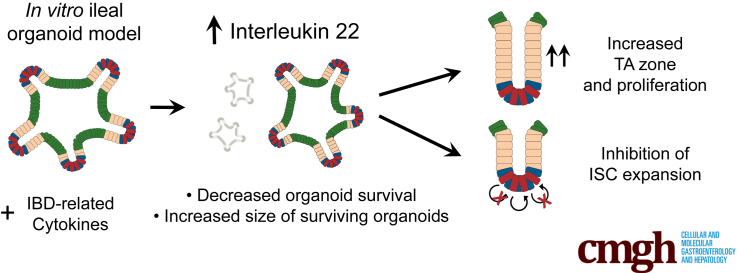
Graphical abstract
Summary.
In a screen of inflammatory bowel disease–related cytokines, interleukin 22 stimulated a concentration-dependent increase in ileal organoid size along with decreased survivability. Further characterization showed a novel role for interleukin 22 in controlling progenitor cell numbers and stem cell expansion.
Inflammatory bowel disease (IBD) features exaggerated immune responses that can affect the small intestine (Crohn’s disease) or colon (ulcerative colitis).1, 2 Early initiation of inflammation is complex and influenced by a number of contributing factors such as underlying genetics and composition of the gut microbiota. Ultimately, however, it is chronic inflammation-driven breaches in the epithelial barrier that fuel the continuous cycle of inflammation and impaired epithelial regeneration that culminates in clinical sequelae, such as ulcerated regions of submucosa, pain, and bleeding.1, 3 Intestinal epithelial stem cells (ISCs) typically regenerate the epithelial lining in a tightly coordinated balance between ISC self-renewal and differentiation into transit-amplifying (TA) progenitor cells and their postmitotic descendants. How the inflammatory microenvironment in the ISC zone impacts this stereotypical process is poorly understood.
Local immune cells in the gut secrete a wide array of cytokines that mediate initiation, progression, and resolution of inflammation in the small intestine and colon. During an inflammatory response, T cells and innate lymphoid cells (ILCs) secrete a subset of the cytokines called interleukins (ILs) into the ISC microenvironment.3 Dysregulated cytokine responses are associated with IBD and often are characterized by chronically increased serum levels of IL6, IL17, IL21, and IL22 and their associated signaling pathways.3 For instance, in healthy individuals serum IL22 levels are reported at approximately 2 pg/mL, whereas in Crohn’s disease patients those levels are approximately 12 times higher at approximately 24 pg/mL.4 Importantly, these values represent systemic IL22 levels, which may not accurately reflect actual concentrations and the phenotypic consequences close to the mucosal inflammation site. This concept is particularly highlighted in the case of IL23, which at low concentrations promotes proliferation of human lung cancer cells, but inhibits proliferation at high concentrations.5 In the case of IBD, it is logical that IL concentrations would be considerably higher in the mucosal microenvironment, where the cytokine is produced by ILCs, and systemic levels would be reduced effectively by systemic diffusion and first-pass removal by the liver. Although empiric measurements of IL levels in the microenvironment currently are beyond the limits of technology, prediction of microenvironment levels may guide more physiologically relevant dose-response experiments in vitro and provide more accurate interpretations related to the mechanisms of action of ILs in the microenvironment.
In this study, we used an ileal organoid model to screen the impact that a subset of IBD-related ILs have on ISCs. Similar to a recent study,6 our screen showed that IL22 has a profound influence on organoid size and survival in a concentration-dependent manner, and confirms that IL22 regulates epithelial proliferation and differentiation. Our study extends this prior work to model and investigate the role of IL22 in the ISC microenvironment, and, importantly, draws a distinction between systemic levels of IL22 reported in serum from IBD patients and predicted IL22 concentrations in the ISC microenvironment. Our study offers an alternative interpretation as to how increased levels of IL22 in the microenvironment affect proliferative epithelial populations in the crypt.
Results
Screening IBD-Related Cytokines Using an Ileal Organoid Model
Crohn’s disease, an IBD, typically is restricted to the ileum in the small intestine. We sought to test the impact of a number of IBD-related cytokines specifically on the ileal epithelium and observe the effects on ISCs in vitro. To do this, we used an ileal organoid model in which organoids were derived specifically from the terminal mouse ileum, a region that is involved in a majority of patients with Crohn’s disease. Intestinal organoids, also known as enteroids,7 are spherical, ISC-driven, epithelial structures that form in vitro when single ISCs or crypts are suspended in extracellular matrix and supplemented with defined growth factors that mimic the in vivo stem cell niche.8 Organoids are composed of ISCs that differentiate into all the epithelial absorptive and secretory lineages found in vivo and self-pattern with crypt buds containing ISCs that differentiate and migrate into intervening villus-like zones.8 Because of these physiologically relevant properties and functions, organoids represent an excellent model to study the dynamics of ISC-driven regeneration in vitro.
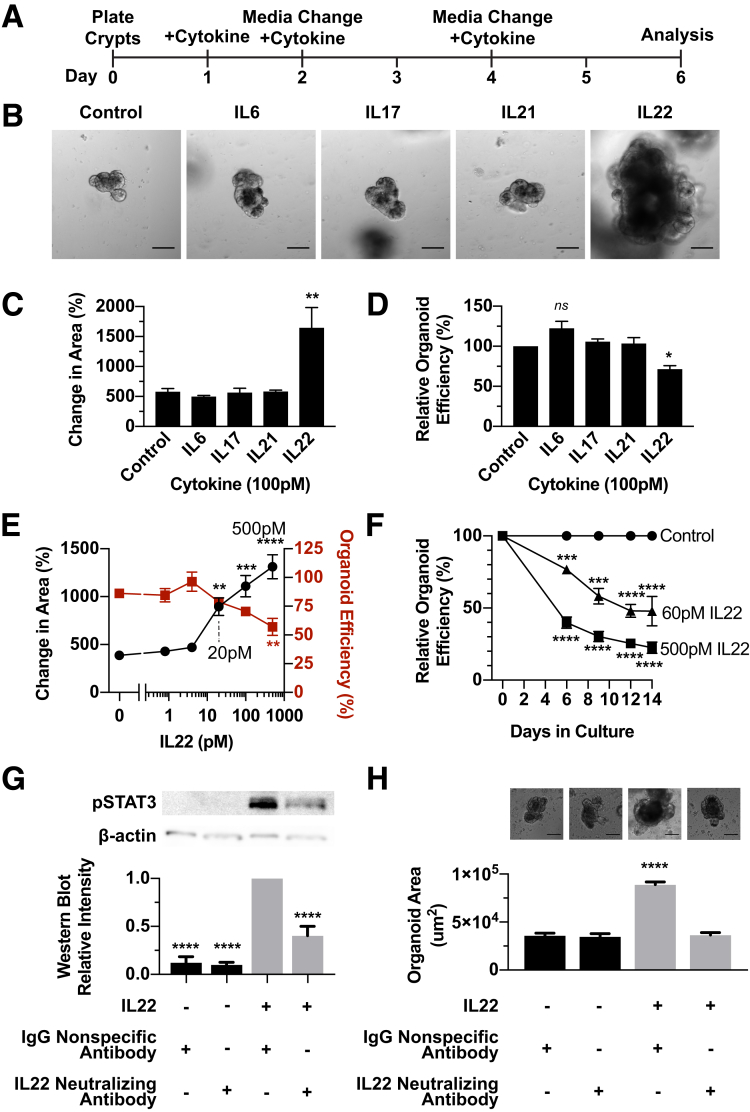
Organoids can be used as a rapid and easy screen to visually detect changes in properties that are regulated by ISCs, including growth and long-term self-renewal. We used this ileal organoid model system to test whether a subset of Crohn’s disease–related ILs would decrease ISC activity using organoid survival as a proxy for ISC survival and organoid size as a measure of ISC proliferation. IL16, IL1β, and tumor necrosis factor α levels in patients with IBD average approximately 100 pmol/L (1500 pg/mL), a concentration that was used to screen for IL effects on the ileal organoid epithelium.9 Ileal crypts were allowed to establish in vitro for 1 day, and then supplemented with 100 pmol/L IL6, IL17, IL21, or IL22 for 6 days (Figure 1A). Although there was no significant size increase caused by IL6, IL17, and IL21, organoids treated with 100 pmol/L IL22 grew approximately 3 times larger (Figure 1B and C). IL6, IL17, and IL21 had no significant effect on organoid survival, but IL22 caused an approximately 28% decrease in organoid survival compared with controls (Figure 1D). In this case, organoid survival was defined by organoid forming efficiency (OFE), which is the number of organoids that derive from a single ileal crypt. Although there may have been more subtle effects of IL6, IL17, and IL21 on ISCs, we focused on investigating how IL22 affected ISC self-renewal and differentiation based on the overt organoid growth and death phenotype observed in the screen.
Figure 1.
A focused screen of IBD-related cytokines shows that IL22 causes a dose-dependent decrease in organoid survival and an increase in organoid size. (A–D) Ileal organoid screen for cytokine effects on intestinal epithelium. (A) Schematic of experimental design in which ileal organoids were treated for 6 days with 100 pmol/L IL6, IL17, IL21, or IL22. (B) Representative images of treated organoids after 6 days. Scale bar: 100 μm. (C) Percentage change in area of organoids comparing day 0 with day 6. Technical replicate n = 10+ organoids; biological N = 3 mice; significance is relative to untreated control. (D) Organoid efficiency relative to control organoids. Technical replicate n = 3 wells; biological N = 3 mice; significance is relative to untreated control. (E) Organoid response to a range of concentrations of IL22 (0, 0.8, 4, 20, 100, and 500 pmol/L), measured by change in organoid area (left Y axis, black lines, technical replicate n = 10+ organoids) and organoid survival (right Y axis, red lines, technical replicate n = 3+ wells) after 6 days. Biological N = 3 mice/treatment; significance is in relation to 0 pmol/L IL22. (F) Organoid survival with 0, 60, or 500 pmol/L IL22 treatment. Technical replicate n = 3 wells; biological N = 3 mice. Asterisks denote significance between the treatment group and control at the designated time point. (G and H) Organoids were treated with 500 pmol/L IL22 in the presence of 206 ng/mL IL22 neutralizing antibody or 206 ng/mL IgG of the same species. (G) Top: Representative images of treated organoids. Bottom: Quantification of organoid area. Technical replicate n = 10+ organoids; biological N = 3 mice; significance is relative to IgG-only control. (H) Top: Representative Western blot images for pSTAT3 and β-actin. Scale bar: 100 μm. Bottom: Quantification of intensity of Western blot bands normalized within each blot to the band with the highest intensity. Technical replicate n = 3 blots, biological N = 3 mice. Significance was calculated by 1-way analysis of variance with a Bonferroni correction for multiple comparisons. *P < .05, **P < .01, ***P < .001, ****P < .0001. pSTAT3, phosphorylated signal transducer and activator of transcription 3.
IL22 Imparts Concentration-Dependent Effects on Ileal Organoids
IL22-dependent changes in organoid size and survival have been reported in organoids derived from a mixture of crypts isolated from full-length intestine.6 It remains to be determined whether IL22 affects ileal-specific epithelium in the same way. A dose-response experiment showed that 20 pmol/L of IL22 was the lowest dose that caused a significant increase in organoid size (Figure 1E). Organoid size continued to increase in a dose-dependent manner at each tested concentration up to 500 pmol/L (Figure 1E). Interestingly, although ileal organoid size increased as a function of IL22 concentrations, there was a decrease in OFE from crypts cultured in increasing concentrations of IL22 (up to 500 pmol/L) (Figure 1E).
Next, we tested whether a reduction in OFE persisted over time. OFE was quantified in ileal organoid cultures exposed to 2 different IL22 doses: 60 pmol/L, a lower dose reported to cause increased organoid size but no decrease in OFE,6 and 500 pmol/L, a higher dose shown to cause both increased organoid size and decreased OFE (Figure 1F). There was a clear decrease in the ability of crypts to generate organoids at both concentrations and this trend persisted over the 2-week time course (Figure 1F). To determine whether these effects were IL22-dependent, ileal organoids were treated with the higher IL22 concentration (500 pmol/L) in the presence or absence of an IL22-neutralizing antibody (Figure 1G and H). The neutralizing antibody blocked IL22-dependent phosphorylation of signal transducer and activator of transcription 3 (STAT3),10 a key downstream signaling mediator of IL22 (Figure 1G). Importantly, the IL22 neutralizing antibody blocked size increases in IL22-treated ileal organoids, showing the phenotype was IL22-dependent (Figure 1H). Although 1 study showed that lower levels of IL22 did not impart a negative effect on organoids derived from a mixture of crypts derived from full-length intestine,6 our results showed a significant decrease in ileal OFE at both concentrations. Interestingly, although IL22 impaired organoid formation from some ileal crypts, the crypts that survived grew bigger and developed faster than untreated control crypts.
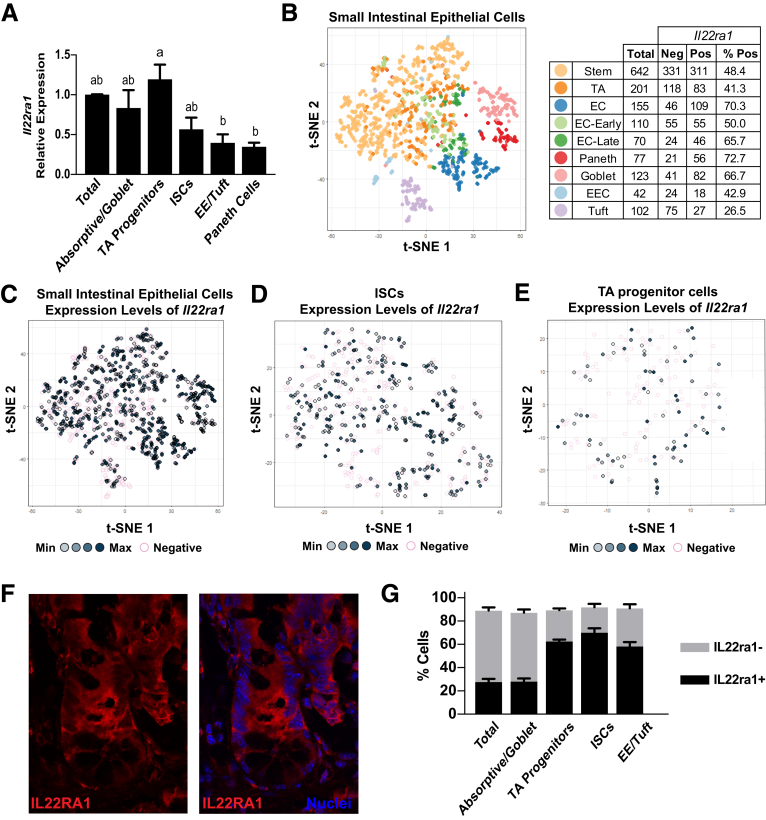
IL22 Receptor Is Expressed Only in Subsets of Intestinal Epithelial ISCs and TA Progenitors
The heterogeneous response of some crypts to increased IL22 levels suggested ISCs in the base of each crypt might have differential responses to increased IL22. IL22 signals through the receptor IL22RA1 and activates STAT3 signaling,10 so we reasoned that ISCs might have variable responses to IL22 based on differential expression of its cognate receptor. To test this, the ileal epithelium was dissociated to single cells and populations were differentially FACS-enriched for ISCs, TA progenitors, enteroendocrine/tuft cells, Paneth cells, and Goblet/absorptive cells based on established methods using a Sox9-EGFP mouse model.11 Across these populations, Il22ra1 messenger RNA (mRNA) was detected at the highest levels in the TA progenitor cells, but also was detected in each of the other populations, albeit at significantly lower levels (Figure 2A). We next investigated Il22ra1 mRNA expression at cellular resolution using single-cell RNA sequencing. A previously published data set that surveyed the full transcriptome of 1522 single mouse small intestinal cells was investigated to define the extent of Il22ra1 expression heterogeneity in different lineages (Figure 2B–E).12 Expression of Il22ra1 mRNA was quantified in a binary on/off manner for each ISC, progenitor, and differentiated cell population (Figure 2B). Expression of Il22ra1 was observed only in subsets in each population, and, moreover, in those cells that expressed Il22ra1, there was a broad range of expression levels (Figure 2C). A higher resolution view of just the ISC and TA progenitor cells further highlights the heterogeneous expression patterns of Il22ra1 in these populations (Figure 2D and E). We did not observe distinct clustering of Il22ra1-positive or Il22ra1-negative cells in the t-distributed stochastic neighbor embedding (t-SNE) clustering analysis, suggesting that although Il22ra1 expression is heterogeneous, it does not identify discrete subpopulations of ISCs or TA progenitors based on this type of analysis (Figure 2C–E). These data suggest that during homeostasis, only a subset of ISCs and TA progenitor cells are receptive to IL22 stimulation.
Figure 2.
Il22ra1 is expressed heterogeneously throughout the crypt. (A) The Il22ra1 gene expression profile characterized in FACS-isolated total epithelium (CD326+), absorptive/goblet differentiated cells (Sox9-EGFPneg), TA progenitor cells (Sox9-EGFPsublow), ISCs (Sox9-EGFPlow), enteroendocrine/tuft cells (Sox9-EGFPhigh), and Paneth cells (Sox9-EGFPhigh, CD24high). Technical replicate n = 3; biological N = 3 mice. Significance was calculated by 1-way analysis of variance with Tukey multiple comparisons; bars that are not connected by the same letter are statistically significant (P < .05). (B) Left: t-SNE analysis of single-cell RNA sequence analysis of mouse small intestinal epithelium. Each color represents a different population defined in the original analysis based on lineage-specific transcriptomic signatures. Right: Table depicts the number of cells in each lineage category expressing IL22ra1. (C) Cells from the t-SNE profiles in the graph in panel B are highlighted specifically for the expression of IL22ra1 levels in all epithelial cells. Darker shades of grey represent higher expression levels. Pink circles represent no expression. (D) The same analysis in panel C except only ISCs are shown. (E) The same analysis in panel C except only TA progenitors are shown. (F) Representative immunohistochemistry of IL22RA1 (red) and cell nuclei (blue) in a mouse ileal crypt. (G) FACS analysis of fixed cell populations described in panel A stained for IL22RA1. Technical replicate n = 3; biological N = 3 mice. Bars represent parts of whole. EC, enterocyte; EE, enteroendocrine; EEC, enteroendocrine; Max, maximum; Min, minimum.
To determine if the heterogeneous Il22ra1 expression extended to the protein level, we immunostained ileal tissue sections to assess IL22RA1 localization, and quantified the number of IL22RA1-expressing cells by flow cytometry (Figure 2F and G). Immunostaining showed broad expression of IL22RA1 in crypt-based epithelial cells with higher levels apparent in the TA progenitor zone6 (Figure 2F). Flow cytometry confirmed at the protein level that only subpopulations of epithelial cells express the IL22 receptor (Figure 2G). We attempted to test functional differences between IL22RA1+ and IL22RA1- by FACS isolation and OFE assays on single ISCs; however, all commercially available antibodies detect cytoplasmic epitopes and were not suitable for isolation of live cells. Although technical limitations preclude testing functional differences in IL22RA1+ and IL22RA1- ISCs, the heterogeneous expression of IL22RA1 in ISC and TA progenitors suggest that there are mechanistic differences that could impact ISC behavior during an inflammatory response in vivo.
Computational Modeling Predicts Higher Levels in the ISC Microenvironment Than in Serum
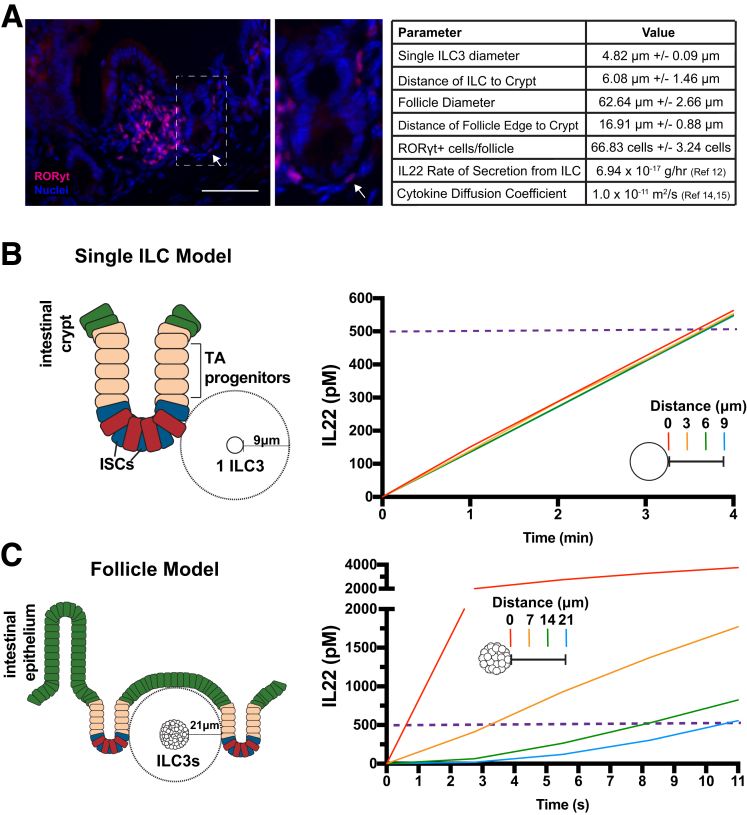
A fundamental question in studies investigating the role of ILs in IBD pathogenesis is: what are the physiologically relevant IL concentrations that contribute to the disease? IL22 levels reported for IBD patients are based on peripheral blood serum levels,4 which likely do not accurately reflect the IL22 levels at the site of secretion in the intestinal or colonic mucosa. Currently, there is no accurate method to measure absolute levels of IL22 in the ISC microenvironment, which we define as the distance of ILC3s from the crypt base (∼6–17 μm) (Figure 3A). Therefore, we used COMSOL Multiphysics to computationally model and predict IL22 concentrations that ISCs might experience in their microenvironment during an inflammatory episode.
Figure 3.
COMSOL Multiphysics simulation of IL22 diffusion. (A) Representative in vivo image of mouse ILC3 follicle with RORγt+ ILC3 immune cells (pink) and cell nuclei (blue). Parameters in table were determined empirically, n = 10+ measured values per mouse, N = 3 mice. Scale bar: 100 μm. (B) Left: Single ILC3 IL22 secretion model in which values of IL22 concentration were calculated by a computational model along a line segment with a length of 9 μm surrounding 1 ILC3 cell. Right: Concentration of IL22 over time at 4 points at 3-μm increments. (C) Left: Follicle model in which values of IL22 concentration were calculated along a line segment with a length of 21 μm. Right: Concentration of IL22 from a lymphoid follicle over time at 4 points at 7-μm increments.
Empirically derived values for factors that could influence IL22 concentrations, such as IL22 secretion rates from ILC3s, distances of ILC3s to ISC, number of ILC3s in a lymphoid follicle, distance of the lymphoid follicle to ISCs, and the radius of ILC3s were applied to the model (Figure 3A). Previous work established that isolated and induced ILCs can secrete IL22 in culture,13 and, from this study, we calculated that a single ILC can secrete IL22 at an average rate of 4 fg/h. An estimated diffusion coefficient for IL22 was based on its molecular weight.14, 15, 16 IL22-secreting ILC3s express Retinoic acid-related orphan receptor gamma-t (RORgt),17 which was used as a marker to measure the distances of ILC3s to the ISCs in the base of the crypt. ILC3s are as close as 6 μmol/L from the base of the crypt (average distance, ∼6.0 μm) (Figure 3A). ILC3s also exist in the mucosal microenvironment localized to innate lymphoid follicles,18 which were measured to be as close as 17 μm from the crypt base (average distance, ∼17 μm) (Figure 3A). These data show that cells expressing IL22 can be in very close proximity (on the single-cell diameter scale) to the ISC zone.
By using these parameters, we first asked how long it would take for a single ILC3 cell to secrete enough IL22 to achieve a concentration of 500 pmol/L at the ISC zone (Figure 3B). The model predicted that IL22 can accumulate to 500 pmol/L within a distance of 0–9 μm within 4 minutes (Figure 3B). In the setting of Crohn’s disease and chronic inflammation, IL22 is up-regulated and an influx of IL22-secreting ILC3s was observed19; therefore, it is expected that the single-ILC3 model was a conservative estimate and likely under-represents concentrations of IL22 in the inflammatory microenvironment. Because ILC3s also exist in innate lymphoid follicles,18 we next modeled IL22 microenvironment concentrations up to 21 μm, which included the average distance of a lymphoid follicle from the ISC zone (Figure 3C). We also used secretion values for 67 ILCs, which is the average number of ILC3s in a cross-section of a lymphoid follicle (Figure 3A). Although the model only takes the IL22 secretion from the number of ILC3s in a 2-dimensional space into consideration, the model effectively approximated the microenvironment concentrations in 3-dimensional space. By using the follicle model prediction, ISCs in close proximity to the follicle (0–21 μm) would detect 500 pmol/L IL22 within 11 seconds of initiation of secretion (Figure 3C). Although the IL22 concentrations were modeled in a closed system and the rates of IL22 removal and degradation are unknown, the computational models suggested the 500 pmol/L IL22 concentration used in the ileal organoid model was not unreasonable to achieve in the ISC/ILC3 inflammatory microenvironment, therefore this IL22 concentration was used throughout the remainder of the study.
IL22 Does Not Cause Appreciable Lineage Bias of ISCs
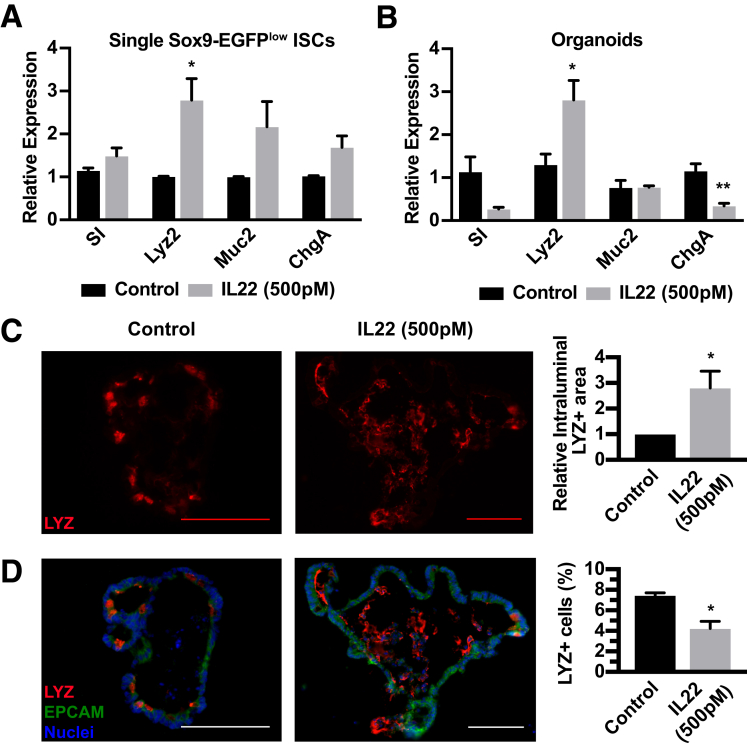
Although 500 pmol/L of IL22 has a clear impact on ileal organoid survival and size, these metrics do not reflect IL22-dependent influences directly on ISC differentiation. Aberrant differentiation of ISCs caused by increased levels of IL22 could explain a loss of ISC self-renewal and regenerative capacity of the gut epithelium in inflammatory conditions. We performed a study using isolated ISCs to test whether increased IL22 influenced early differentiation programs directly on ISCs. ISCs were FACS-isolated and incubated with 500 pmol/L IL22 for 6 hours followed by gene expression analysis for lineage-restricted biomarkers: absorptive enterocyte (sucrose isomaltase: SI), Paneth (lysozyme 2: Lyz2), goblet (mucin 2: Muc2), and enteroendocrine (chromogranin A: ChgA) (Figure 4A). In these studies, only lysozyme mRNA, which is associated with Paneth cell lineage, was increased significantly. To determine the impact of increased IL22 on the tissue level, we tested ileal organoids treated with 500 pmol/L IL22 for expression of all key genes that mark the main differentiated cell types. In this case, increased IL22 concentrations led to reduced or unchanged levels of all genes except for the Paneth cell biomarker Lyz2 (Figure 4B). To determine whether this increase in lysozyme mRNA was associated with increased Paneth cells, IL22-treated ileal organoids were immunostained for lysozyme and Paneth cell numbers were quantified. Although there was an approximately 2.5-fold increase in intraluminal lysozyme staining, there was a slight reduction in the relative number of Paneth cells positioned in the ileal organoid epithelial monolayer (Figure 4C and D). Together, these data suggest that IL22 does not cause appreciable differentiation defects of ISCs, but promotes the production and secretion of lysozyme from existing Paneth cells.
Figure 4.
Cell lineage analysis in single ISCs and ileal organoids treated with IL22. (A) Gene expression analysis of single Sox9-EGFPlow single ISCs after 6 hours with or without 500 pmol/L IL22 examining expression of differentiated cell genes. Technical replicate n = 3, biological N = 3 mice. (B) Gene expression analysis of organoids after 6 days with or without 500 pmol/L IL22 examining expression of differentiated cell genes including SI (enterocytes), Lyz2 (Paneth cells), Muc2 (goblet cells), and ChgA (enteroendocrine cells). (C and D) Immunohistochemistry staining and quantification of organoids treated for 6 days with IL22 then stained for lysozyme (LYZ, red), (EPCAM) (green), and nuclei (blue). Technical replicate n = 10+ organoids; biological N = 3 mice. (C) Left: Representative intraluminal LYZ staining (red). Right: Quantification of intraluminal stain. (D) Left: Representative organoid staining. Right: Quantification of total number of LYZ+ cells relative to total nuclei per organoid. Significance was calculated using an unpaired t test relative to the untreated control. Scale bar: 100 μm. *P < .05, **P < .01, ***P < .001, and ****P < .0001.
IL22 Causes a Decrease in ISC Biomarkers and Pathways That Maintain ISC Self-Renewal
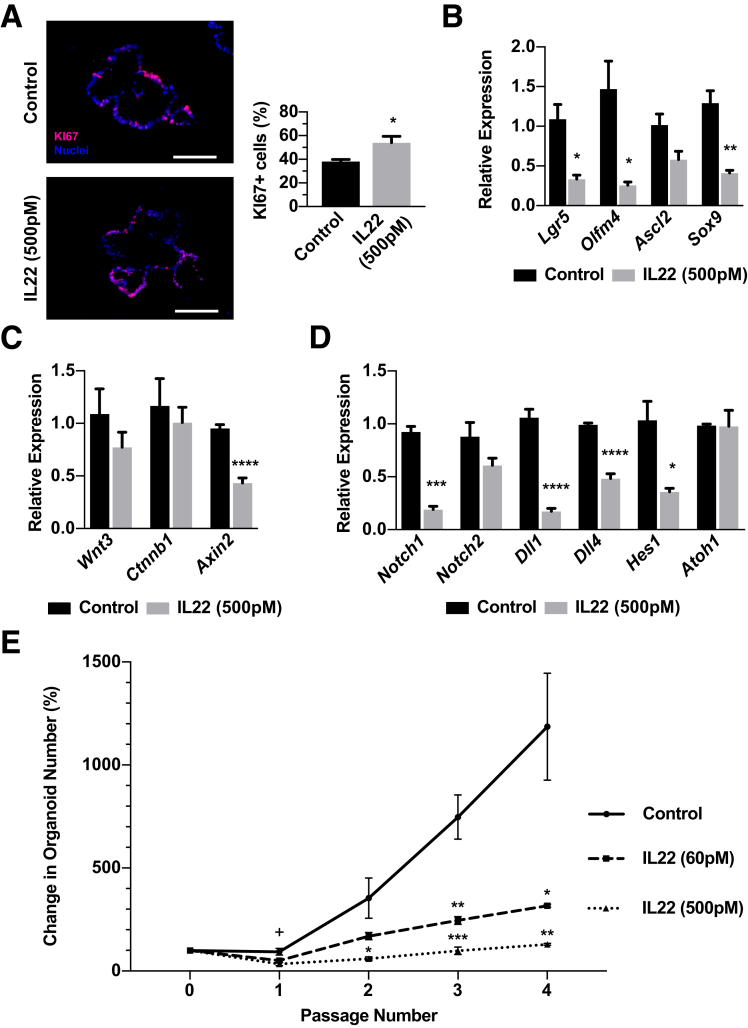
We next questioned whether IL22 increased the proliferation and self-renewal properties of ISCs because ileal organoids showed significantly increased size when treated with increased IL22. Ileal organoids treated with 500 pmol/L of IL22 showed a significantly higher number of KI67+ cells in the epithelial monolayer (Figure 5A). We evaluated the mRNA levels for ISC biomarkers Lgr5 and Olfm4, and, contrary to predictions, found these ISC biomarkers to be down-regulated significantly, suggesting there was a decrease in ISC numbers (Figure 5B). Expression of key Wnt and Notch pathway genes, which support ISC survival, proliferation, and self-renewal,20, 21, 22 were down-regulated significantly in response to IL22 (Figure 5C and D). Although Wnt3 and β-catenin (Ctnnb1), which tightly control ISC function and proliferative capacity, were not changed significantly in response to IL22, the Wnt responsive target Axin2 was down-regulated 2-fold in response to IL22, suggesting a reduction of ISC self-renewal pathway inputs (Figure 5C). The Notch1 receptor ligands Dll1 and Dll4 and downstream target Hes1 were down-regulated after exposure to IL22 (Figure 5D); however, there was no significant change in Atoh1 (Figure 5D), which is directly inhibited by Notch and required for secretory cell differentiation.23 This result is consistent with no substantial changes in secretory lineage allocation in organoids and single ISCs exposed to IL22. Overall, down-regulation of ISC biomarkers and signaling pathways that regulate and maintain functional ISCs suggest there are lower ISC numbers or impaired ISC self-renewal properties in ileal organoids exposed to increased IL22.
Figure 5.
IL22 limits ISC expansion. (A) Representative immunohistochemistry for the proliferation marker KI67 (red) and nuclei (blue) with quantification of the proportion of KI67+ nuclei. Technical n = 10+ organoids; biological N = 3 mice. Scale bar: 100 μm. (B–D) Gene expression analysis of organoids after 6 days with or without 500 pmol/L IL22 for (B) ISC-associated genes including Lgr5, Olfm4, Ascl2, and Sox9; (C) Wnt signaling pathway-associated genes including Wnt3, Ctnnb1, and Axin2; and (D) Notch signaling pathway-associated genes including Notch1, Notch2, Dll1, Dll4, Hes1, and Atoh1. Technical replicate n = 3, biological N = 3 mice. Significance was calculated using an unpaired t test relative to the untreated control. (E) Percentage organoid increase in response to 0, 60, or 500 pmol/L IL22 at each passage compared with the number of organoids at initial plating at p0. Technical replicate n = 3, biological N = 3 mice. Significance was calculated using 1-way analysis of variance with Bonferroni correction at each time point in comparison with control. +P < .05 for 500 pmol/L IL22 compared with control at passage 1. *P < .05, **P < .01, ***P < .001, and ****P < .0001.
IL22 Limits ISC Expansion
Because gene expression studies suggested IL22 caused a reduction in ISCs, we sought to test ISC functional properties when ISCs were exposed to increased levels of IL22. Serial passaging is used extensively in the hematopoietic stem cell field to assay stem cell function,24 and here we used this strategy to assay ISC function in the presence of increased IL22. In vivo studies strongly suggest that ISCs primarily divide symmetrically to generate 2 ISCs, and thus are able to expand their numbers to maintain the proper balance of ISCs in the crypt base.25 With this mechanism in mind, every passage of organoids to single cells should produce a clonal organoid derived from the number of ISCs found in the original organoid. During organoid ontogeny from a single ISC, ISCs undergo symmetric expansion to produce more ISCs per organoid, and, conceptually, upon each passage the extent of the symmetric ISC expansion is measured by the increase in organoid numbers. Ileal organoids were allowed to establish in culture for 1 day, then were exposed to IL22 for 6 days, followed by dissociation to single cells and replating (defined as passaging). Organoids were passaged 4 times over approximately 4 weeks. In untreated controls, organoid numbers increased with each passage, indicating symmetric division and expansion of ISCs (Figure 5E). By contrast, treatment with IL22 suppressed the ability for organoid numbers to increase with passage (Figure 5E). Although IL22 limited ISC expansion, it did not completely ablate ISCs because a similar number of organoids were observed throughout all 4 passages in response to IL22, suggesting that IL22 is not explicitly toxic to all ISCs.
Increased IL22 In Vivo Causes an Increase in Proliferative Cells in the TA Progenitor Zone
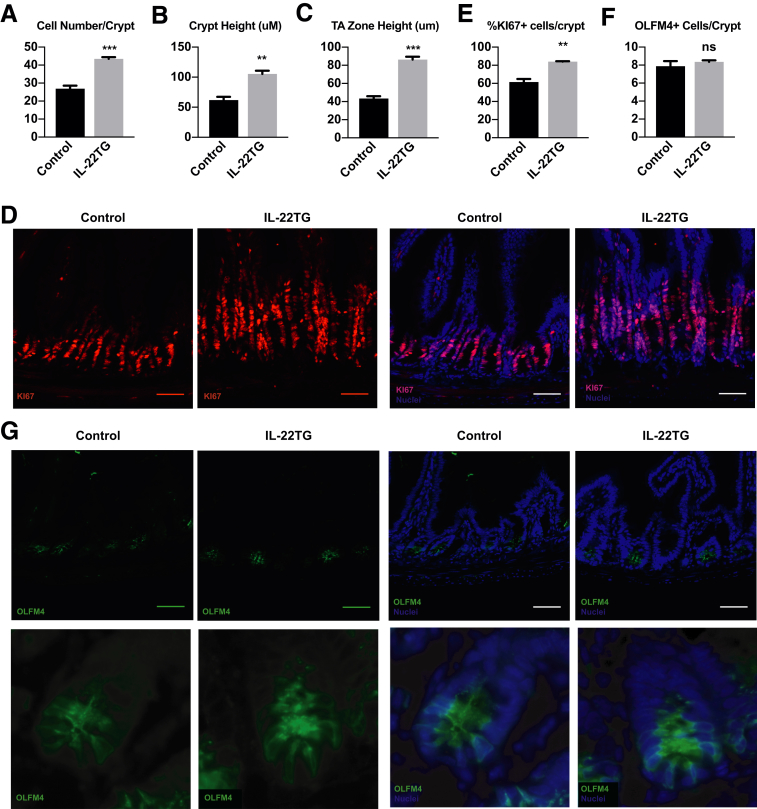
Although there was a significant increase in proliferating cells in ileal organoids treated with increased levels of IL22, there was no increase in functional ISCs based on organoid passaging experiments. Based on this, we hypothesized that the increase in proliferating cells observed in IL22-treated organoids was owing to an increase in TA progenitors, not ISCs. We identified an IL22 transgenic (IL22TG) mouse model that was used to study the role of IL22 in liver disease.26 These mice express high levels of IL22 from an albumin promoter, and IL22 serum levels reached approximately 4–7 ng/mL, which is in the range of approximately 8 ng/mL IL22 used in our ileal organoid studies.26 Intestines from these mice were obtained and the number of proliferating cells, their location in the crypt, and the number of ISCs was quantified (Figure 6).
Figure 6.
Increased IL22 causes an increase in TA progenitors. Quantification of (A) total number of cells per crypt, (B) total crypt height, and (C) height of TA zone in control and IL22TG mice. (D) Representative immunohistochemistry for the proliferation marker KI67 (red) and nuclei (blue). Quantification of (E) the percentage of positive KI67 cells per crypt and (F) the total number of OLFM4+ cells per crypt. (G) Representative immunohistochemistry for the ISC marker OLFM4 (green) and nuclei (blue). All quantification: Technical replicate n = 10+ crypts/mouse, N = 3 mice/treatment. Scale bar: 100 μm. **P < .01, ***P < .001. Significance was calculated using an unpaired t test relative to the untreated control.
In the IL22TG mice, the number of cells per crypt and crypt height increased (Figure 6A and B). The TA progenitor cell zone increased 2-fold (Figure 6C), and the number of KI67+ proliferating cells increased 1.4-fold (Figure 6D and E). To determine if this increase in proliferating cells was owing to an increase in ISC numbers, intestinal sections were stained with the ISC marker OLFM4. No difference was observed in the number of OLFM4+ cells per crypt between control and IL22TG mice (Figure 6F and G), suggesting that IL22 does not expand ISC numbers, but rather acts to expand the progenitor cell population.
Discussion
Defining how inflammation of the gastrointestinal tract impairs ISC-driven epithelial renewal could have profound impacts on understanding the mechanisms regulating initiation, progression, and resolution of IBD. Because inflammation-induced cytokines that mediate the immune response are found in close proximity to the ISC niche, we used an ileal organoid model to screen a subset of IBD-related ILs for their impact on ISCs. The screen showed that IL22 in particular decreased OFE, however, the organoids that survived were substantially larger and had more proliferating cells compared with controls. These observations served as a foundation to test the hypothesis that IL22 was regulating ISC survival and proliferation.
The IL22-dependent increase in growth was observed at levels approximately 14 times those found in the serum of patients with IBD, but, interestingly, the reduced OFE was observed at levels only approximately 340 times those measured in serum.4 Although the serum levels of cytokines are commonly used as a guide for physiologic levels, we questioned this assumption and reasoned that IL22 levels might be substantially higher in the ISC microenvironment. Supporting this concept, Crohn’s disease patients have an influx of IL22-secreting ILC3 lymphocytes in inflamed lesions,19 and in colitis ILC3s increase IL22 production in response to inflammatory stimuli.27 These studies suggest there could be much higher IL22 concentrations in the ISC niche compared with serum IL22 levels, which likely would be effectively reduced owing to systemic dilution and first-pass liver effects. Direct measurement of IL22 in the ISC microenvironment is currently not technically possible; however, computational modeling of IL22 concentrations in the ISC niche indicated that IL22 could achieve the highest levels used in this study within seconds to minutes after an inflammatory stimulus. Together, these findings indicated that pathophysiologic IL22 concentrations could be much higher at the site of secretion compared with those measured in the peripheral circulation. These results challenge conventional assumptions regarding the definition of physiologically relevant levels based on serum cytokine concentrations and provide a method to predict microenvironment concentrations, which can guide experimental design for testing IL doses in organoid culture systems.
A recent study concluded that lower levels of IL22 promoted ISC-mediated epithelial regeneration in a mouse model of graft-versus-host disease.6 Our study showed that IL22 concentrations approximately 1.7 times higher inhibited ISC expansion in ileal organoids. Down-regulation of ISC-associated biomarkers and a decrease in ISC self-renewal pathway gene expression also supported the interpretation of an IL22-dependent loss of ISC expansion, which was functionally confirmed in organoid passaging experiments. However, surviving ileal organoids showed a seemingly contradictory, but clear, increase in organoid size and number of proliferating cells. In an attempt to reconcile the results, we explored whether high IL22 levels had a similar effect in vivo. Consistent with interpretations from ileal organoid passaging experiments, high levels of IL22 in an IL22TG mouse showed no ISC expansion based on OLFM4 ISC biomarker expression. However, the mice showed a substantial and significant increase in proliferating cells in the TA progenitor zone. Together, our results suggest that high IL22 levels might selectively increase expansion of proliferative TA progenitor cells at the expense of ISC expansion.
Serial organoid passaging assays, in which new organoids are clonally derived after passaging organoids to single cells, indicated there was no ISC expansion in the presence of high IL22 levels. These assays also showed that the number of organoids remained constant over time and multiple passages, suggesting that ISC numbers were not reduced but remained constant. In vivo studies have indicated that ISCs expand primarily by symmetric self-renewal.25 In this case, ISC commitment is driven not by asymmetric division dynamics, but rather by migration of ISCs out of a self-renewal niche supporting high Wnt and Notch signaling.28 A potential explanation for an IL22-dependent increase in proliferating cells with no increase in ISCs is that IL22 shifts the balance of symmetric self-renewal to asymmetric division where ISC division produces 1 ISC and 1 TA progenitor. In this case, a cellular mechanism would produce equivalent numbers of ISCs and TA progenitor cells after division. Our in vitro and in vivo results indicated that increased IL22 levels primarily expand TA progenitor cells, which is a reasonable conclusion considering that the highest Il22ra1 expression was found in the TA progenitor cell population. Increasing the pool of cells that will soon become differentiated epithelium may be an IL22-dependent cellular mechanism for producing the epithelial tissue bulk necessary to maintain barrier function in the face of chronic inflammation.
Interestingly, inactivation of STAT3 in cochlear hair cell differentiation results in a shift from asymmetric to symmetric divisions of immature cells that give rise to hair cells, thus activation of STAT3 signaling preserves the asymmetric division mode in this particular context.29 In the intestinal epithelium, IL22 signals through its cognate receptor, IL22ra1, and activates STAT3 signaling.10 It is possible that in the context of ISCs, IL22 signals through STAT3 to promote asymmetric division of ISCs resulting in the loss of ISC expansion properties. We show that IL22ra1 is expressed in the majority of ISCs, however, there was a small subset of IL22RA1-negative ISCs (23%). This could produce a scenario in which IL22ra1-negative ISCs regenerate the epithelium through symmetric self-renewal, whereas the IL22-receptor–positive ISCs would be influenced to undergo asymmetric division to increase the TA progenitor pool, and, ultimately, the number of differentiated epithelial cells. This mechanism is consistent with altered stem cell division observed in psoriasis, in which IL22 levels are increased,30 the percentage of stem cells decreases, and the TA progenitor cell compartment becomes enlarged.31 This also would explain the increased TA progenitor cell zone in IL22TG mice and mice injected with IL22 to enhance epithelial repair during graft-versus-host disease.6
Although IL22-induced epithelial repair was enhanced in the mouse model of graft-versus-host disease,6 studies have shown that increased levels of IL22 mRNA in inflamed lesions of patients with Crohn’s disease correlate with impaired regeneration.32, 33 These reports indicate that IL22 alone is insufficient to resolve impaired epithelial renewal in chronic inflammatory conditions and may point to a more complex and deleterious role for high IL22 levels in IBD. High IL22 levels failed to support organoid development in approximately 43% of ileal crypts put into culture, possibly indicating that IL22-responsive ISCs in the crypt base become incompetent to drive ISC self-renewal, and thus organoid development. We attempted to test this hypothesis by FACS isolation of IL22ra1-positive ISCs and single-cell organoid assays, however, there are no commercially available antibodies that facilitate FACS isolation of IL22RA1-expressing cells. Because increased IL22 impairs ISC expansion in ileal organoids, chronically increased IL22 actually may inhibit ISC-driven epithelial repair because of reduced expansion capabilities of ISCs in a high IL22 environment. This potential mechanism has important implications for establishing a role of IL22 in acute vs chronic inflammation, in which IL22 in acute inflammation enhances epithelial repair and rapidly replaces lost tissue through expansion and differentiation of the TA progenitors, whereas in chronic inflammation epithelial repair is inhibited by the loss of ISC expansion and perhaps even loss of crypts owing to IL22-induced commitment to TA progenitors over time. Because IL22 therapies have been proposed as a therapy to enhance epithelial repair of the intestinal epithelium,34 it is important to take the potentially negative IL22-dependent effects on ISC expansion under consideration.
Materials and Methods
Mice
All organoid experiments were conducted on C57Bl6 mice obtained from Jackson Laboratory (stock number: 000664, Bar Harbor, ME). Sox9–enhanced green fluorescent protein (EGFP) mice, which originally were generated by the Gene Expression Nervous System Atlas Brain Atlas Project35 and have been characterized previously,11, 36 were maintained on an outbred C57Bl/6 background. All mice used in these studies were 8–12 weeks old. All animal use was reviewed and approved by the Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill.
Crypt Isolation, Organoid Culture, and Quantification
The distal 8 cm of mouse small intestine (ileum) was isolated, filleted open, and rinsed in ice-cold Dulbecco's phosphate-buffered saline (DPBS). The intestine was placed in 3 mmol/L EDTA in DPBS for 15 minutes at 4°C with gentle agitation, then villi were gently scraped off using a pipette tip. The intestine was cut into 2- to 3-cm pieces and placed into 3 mmol/L fresh EDTA in DPBS with 10 mmol/L Y27632 (S1049; Selleck Chemicals, Houston, TX) for 30 minutes at 4°C with gentle agitation. Intestinal pieces were transferred to fresh DPBS with 10 μmol/L Y27632 and shaken by hand for 3 minutes at 2 shakes per second to release crypts. Isolated crypts were filtered through 100- and 70-μm filters to remove villus fragments and rinsed once with DPBS. Approximately 100–200 crypts were plated per 10 uL of Growth Factor Reduced Corning Matrigel Matrix (354230; Corning, Corning, NY), which was polymerized for 30 minutes at 37°C in a tissue culture incubator before addition of overlay media. Overlay media consisted of the following: advanced Dulbecco's modified Eagle medium (12634-028; Gibco, Gaithersburg, MD), 1× N2 (17502-048; Gibco), 1× B27 without vitamin A (16704-044; Gibco), 1 mmol/L HEPES (15630-056; Gibco), 1× GlutaMax (35050-079; Gibco), 100 U/mL penicillin/streptomycin (15140-122; Gibco), 500 mmol/L N-acetyl-cysteine (A9165; Sigma, St. Louis, MO), 50 ng/mL recombinant murine epidermal growth factor receptor (PMGG8043; Invitrogen, Carlsbad, CA), 100 ng/mL recombinant murine Noggin (250-38; Peprotech, Rocky Hill, NJ), 250 μg/mL recombinant mouse R-spondin1 (4645RS025/CF; R&D Systems, Minneapolis, MN), and 10 μmol/L Y-27632. Y-27632 was present only in the first 24–48 hours of cell culture and was not added to subsequent media changes. Organoids were allowed to establish in culture for 1 day before addition of IL6, IL17, IL21, or IL22 (406-ML, 421-ML, 594-ML, and 582-ML; R&D Systems) to cultures. Media then was changed every 2 days after plating and cytokines were added to overlay media after each media change (Figure 1A). To determine organoid efficiency, the number of living organoids were counted manually on days 0 (before cytokine addition), and 6, 9, 12, and 14 after plating. To determine organoid area, 10+ organoids were chosen and imaged before cytokine addition on day 1 and stage positions were saved to image the same organoid on day 6. Organoids were chosen randomly throughout the culture simply based on organoid morphology (spherical shape with clearly defined borders) to determine that the organoid was alive and its proximity to other organoids to be able to determine day 6 individual organoid size. The organoid area was measured from images using ImageJ software37 (National Institutes of Health, Bethesda, MD) and day 6 measurements were compared with day 0 to determine the percentage area increase.
For serial passaging, organoids were allowed to establish in culture for 1 day before addition of 60 or 500 pmol/L IL22. Media then was changed every 2 days after plating and cytokines were added to overlay media after each media change. The number of living organoids was recorded before passage and each well of organoids was passaged every 6 days for 4 passages. To passage, organoids and Matrigel were scraped up in 250 μL TrypLE Express (12605-036; Gibco) with 10 μmol/L Y-27632 and triturated with a p1000 pipette tip 75 times to dissociate Matrigel and organoids. Organoids then were transferred to a 1.7-mL microcentrifuge tube containing 250 uL TrypLE Express with 10 μmol/L Y-27632 and incubated at 37°C for 2.5 minutes. Cells then were triturated using a p1000 20 times to further dissociate Matrigel and organoids, then incubated at 37°C for another 2.5 minutes. Cells then were pelleted, resuspended in the appropriate amount of Matrigel for calculated passage ratio, and plated. Cells were replated at a similar density to previous passage. Matrigel was allowed to polymerize for 30 minutes at 37°C in a tissue culture incubator before overlay media and IL22 was added.
Western Blot
Organoids were rinsed 3 times with sterile DPBS. Cell Recovery Solution (354253; Corning) supplemented with 10 μmol/L Y-27632 then was added to each well and organoids and Matrigel were gently scraped from the bottom of the tissue culture plate. Organoids were incubated with end-over-end rotation for 45 minutes at 4°C, then pelleted and resuspended in 2× RIPA buffer with 1% protease inhibitor cocktail (P8340; Sigma), 1% phosphatase inhibitor cocktail (P2850; Sigma), and 1 μmol/L phenylmethylsulfonyl fluoride (P7626; Sigma). Samples were homogenized by passing through a 21-gauge needle 10 times. The protein concentration was determined using the Bradford Protein Assay Kit (SK3031; BioBasic, Amherst, NY). Proteins were separated using electrophoresis in a 10% acrylamide gel and transferred to GE Healthcare Amersham Hybond P 0.45-μm polyvinylidene difluoride membrane (10600023; GE Healthcare, Chicago, IL). The membrane was blocked with 5% bovine serum albumin (BSA) in 1× Tris-buffered saline (TBS) with 0.1% Tween-20 for 1 hour at room temperature, then incubated overnight with rotation at 4°C with phosphorylated signal transducer and activator of transcription 3 primary antibody (1:500, 9145; Cell Signaling Technology, Danvers, MA) in 5% BSA in 1× TBS with 0.1% Tween-20. The membrane was rinsed with 1× TBS with 0.1% Tween-20, then incubated with the secondary antibody anti-rabbit horseradish peroxidase (1:1000, 111-035-003; Jackson ImmunoResearch, West Grove, PA) for 2 hours at room temperature. Clarity Western ECL Substrate (BioRad, Hercules, CA) was used to visualize protein bands. Western blots were imaged using the FluorChem E system (Protein Simple, San Jose, CA). After phosphorylated signal transducer and activator of transcription 3 bands were imaged, the membrane was reblotted with β-actin primary antibody (1:1000, ab8225; Abcam, Cambridge, MA) and anti-rabbit horseradish peroxidase secondary antibody. Western blot images were quantified using ImageJ software.37
Computational Modeling
IL22 cytokine diffuses from IL22-secreting lymphocyte (ILC3) cells to influence the intestinal crypt. The estimated IL22 concentration used in this model was derived from literature values describing IL22 secretion from human ILC cells.14 Briefly, enzyme-linked immunoassay was used to measure secreted IL22 in supernatant derived from co-cultures of lipopolysaccharide-stimulated macrophages and a population of ILCs composed of 22%–35% ILC3 cells, as determined by positive staining for IL22. From enzyme-linked immunoassay measurements obtained with a detection limit of 15.6 pg/mL, a single ILC3 cell was determined to secrete IL22 at an average rate of 4 fg/h. By using Fick’s first law at steady state, diffusion flux from a single ILC3 was modeled. Flux was calculated using the surface area occupied by a single ILC3 cell (19.635 μm2), the diffusion of a single molecule of IL22 (1 × 10-11 m2/s), and the average secretion rate of IL22 (6.94 × 10-17 g/h), assuming uniform secretion across the entire surface area and constant secretion over time. The flux of IL22 produced by a single ILC3 cell represented in the first model as a circle with a radius of 2.5 μm was calculated as 5.78 × 10-14 mol/m2/s. The flux of IL22 produced by 67 cells in a lymphoid follicle represented in the second model as a circle with a radius of 20 μm was calculated as 3.87 × 10-12 mol/m2/s.
Finite-element analysis software COMSOL Multiphysics (Burlington, MA) was used to simulate the secretion rate and concentration of IL22 generated from a single ILC3 cell near the intestinal crypt and a lymphoid follicle containing 67 ILC3 cells. With previously reported values of human IL22 levels generated from patient-derived ILC3 cells,13 the Transport of Dilute Species Interface was used to generate a model of IL22 diffusion. The model includes 2 simulations: the first is a single ILC3 cell with a radius of 2.5 μm secreting IL22, and the second is a circular lymphoid follicle with a radius of 20 μm containing 67 tightly packed ILC3 cells secreting IL22 from each cell. Both simulations model cell secretion as isotropic diffusion without convective mixing. The diffusion coefficient of IL22 (1 × 10-11 m2/s) in liquid solution was conservatively estimated based on its molecular weight (17 kilodaltons) from a range of diffusion rates (from 1 × 10-10 m2/s to 1 × 10-11 m2/s) of signaling molecules in the 10–100 kilodalton range. IL22 has a small molecular weight of 17 kilodaltons in this range, and therefore its diffusion coefficient is expected to be toward the lower end of the numeric range reported.14, 15
In a previous computational model used to predict the spatial distribution of IL4, it was found that IL4-secreting cells communicate with possible target cells within a range of approximately 100 μm.38 The authors included local molecular processes of diffusion, degradation, internalization, and dissociation to determine that communication distance from a cytokine-producing cell to a target cell depends on several parameters but only a few variables. Parameters of low ligand diffusion, long half-life of the ligand, and high affinity of the target cell’s receptors can contribute to an increased effective communication distance, but these are biophysical properties that the target cell cannot regulate. Variables of the source cell, such as rate of internalization and dissociation, have little effect on the local cytokine distribution even when set to extremely fast levels. The major variable responsible for effective communication distance is the rate of secretion. These previous findings support the model presented here to simulate diffusion-dominant secretion of IL22 with parameter exclusions. The model presented does have its limitations, including a simplification of a closed system, despite a lymphoid follicle existing in an open system with continual inflow and outflow of cells. The flow of liquid through the modeling space was not considered, however, because liquid flow likely would reduce the effective concentration of the cytokine, we present a conservative model of IL22 diffusion and concentration dynamics.
Immunohistochemistry
Organoids in Matrigel were rinsed once with DPBS and fixed with room temperature 4% paraformaldehyde for 20 minutes at room temperature. Organoids then were rinsed 3 times with 30% sucrose and incubated overnight at 4°C in 30% sucrose. Organoids were embedded into cryomolds with OCT Compound (4583; Tissue-Tek, Sakura Finetek USA Inc, Torrance, CA), 8-μm sections were cut using an OTF5000 Cryostat Microtome (Bright Instruments, Luton, United Kingdom), and placed onto positively charged microscope slides and stored at -80°C. For immunostaining, slides were rinsed with PBS to remove the OCT Compound. Antigen retrieval and incubation in a pressure cooker for 30 seconds at 120°C and 10 seconds at 95°C, was performed only on slides being stained for KI67 and IL22 receptor A1 (IL22RA1). Slides were blocked with protein block (X090930-2; Dako, Agilent, Santa Clara, CA; or 15019S; Cell Signaling Technology) for 1 hour at room temperature. Primary antibodies were diluted in antibody diluent (S080981-2; Dako; or 15019S; Cell Signaling Technology) and incubated overnight at 4°C. Primary antibodies and dilutions used were as follows: KI67 (rabbit, 1:100, M7249; Dako), LYZ (goat, 1:500, sc-12091; Santa Cruz, Dallas, TX), CD326 epithelial cell adhesion molecule (EPCAM)/CD326 (rat, 1:500, H8201, clone G8.8; Biolegend, San Diego, CA), IL22RA1 (rat, 1:50, FAB42941P; R&D Systems), OLFM4 (rabbit, 1:250, 39141; Cell Signaling Technology). Secondary antibodies were diluted in antibody diluent and incubated on slides for 2 hours at room temperature. Secondary antibodies and dilutions used were as follows: anti-rabbit Cy3 (sheep, 1:1000, C2306; Sigma), anti-goat Cy3 (donkey, 1:1000, 705-165-003; Jackson ImmunoResearch), anti-rat Alexa Flour 488 (donkey, 1:1000, 712-546-153; Jackson ImmunoResearch), and anti-rat Cy3 (goat, 1:1000, 112-165-003; Jackson ImmunoResearch). Nuclei were stained with bis-benzamide (1:1000) diluted in PBS. Slides were mounted using Hydromount (HS-106; National Diagnostics, Atlanta, GA). Images were collected using an Olympus (Waltham, MA) IX81 or Zeiss (Oberkochen, Germany) LSM 700 confocal microscope. Images were analyzed in Metamorph Basic (Molecular Devices, San Jose, CA) and ImageJ software.37
Intraluminal lysozyme content was quantified using the open-source image analysis platform CellProfiler (Cambridge, MA).39 Images of sectioned organoids were taken marking the epithelial monolayer (EPCAM), the epithelial nuclei (4′,6-diamidino-2-phenylindole), and lysozyme protein after immunohistochemical staining as previously described. Images of each organoid were loaded into CellProfiler in sequence and the pixel intensity of each image was rescaled. Binary images of each marker were formed by applying an intensity threshold using a 3-class Otsu’s40 method with the midlevel class considered background. Thresholded 4′,6-diamidino-2-phenylindole and EPCAM images were combined to create binary images, with the foreground depicting the entire epithelial signal. Gaps in the foreground of the combined images were closed using a 10-pixel top-hat transformation resulting in a binary image with the epithelial element represented as a smoothed foreground.41 The lysozyme binary image was masked by the epithelial element to remove the intracellular lysozyme signal. The foreground area of the masked image was quantified and categorized by organoid treatment.
Gene Expression
Wells with organoids were rinsed once with DPBS and 200 μL RNA lysis buffer (from the RNAqueous-Micro Total RNA Isolation Kit, AM1931; ThermoFisher, Waltham, MA) was added to each well containing 50–200 organoid or 10,000 single ISCs. RNA was extracted from samples using the RNAqueous-Micro Total RNA Isolation Kit (AM1931; ThermoFisher) according to the manufacturer’s protocols and stored at -80°C. Complementary DNA (cDNA) was created using iScript Reverse Transcription Supermix for reverse-transcription quantitative polymerase chain reaction (170-8891; BioRad) according to the manufacturer’s protocols. cDNA was diluted 1:20 and 1 μL of diluted cDNA was used for Real-Time PCR using TaqMan probes (Applied Biosystems, Waltham, MA) (Table 1) and SsoAdvanced Universal Probes Supermix (1725281; BioRad) according to the manufacturer’s protocols.
Table 1.
List of TaqMan Probes for Quantitative Reverse-Transcription Polymerase Chain Reaction
| Gene symbol | Gene name | Life Technologies assay ID |
|---|---|---|
| 18S | Eukaryotic 18S ribosomal RNA | Hs99999901_s1 |
| SI | Sucrase isomaltase (α-glucosidase) | Mm01210305_m1 |
| ChgA | Chromogranin A | Mm00514341_m1 |
| Muc2 | Mucin 2 | Mm00458299_m1 |
| Lyz2 | Lysozyme 2 | Mm00727183_s1 |
| Lgr5 | Leucine-rich repeat containing G-protein–coupled receptor 5 | Mm00438890_m1 |
| Olfm4 | Olfactomedin 4 | Mm01320260_m1 |
| Ascl2 | Achaete-scute family bHLH transcription factor 2 | Mm01268891_g1 |
| Sox9 | SRY-box 9 | Mm00448840_m1 |
| Wnt3 | Wnt family member 3 | Mm00437336_m1 |
| Ctnnb1 | Catenin β 1 | Mm00483039_m1 |
| Axin2 | Axin 2 | Mm00443610_m1 |
| Notch1 | Notch 1 | Mm00435245_m1 |
| Notch2 | Notch 2 | Mm00803077_m1 |
| Dll1 | Delta-like canonical Notch ligand 1 | Mm01279269_m1 |
| Dll4 | Delta-like canonical Notch ligand 4 | Mm00444619_m1 |
| Hes1 | Hes family basic helix-loop-helix transcription factor 1 | Mm00468601_m1 |
| Atoh1 | Atonal basic helix-loop-helix transcription factor 1 | Mm00476035_s1 |
Single-Cell Isolation and Fluorescence-Activated Cell Sorter
For Sox9-EGFP cell isolation, crypts were isolated from whole small intestine from Sox9-EGFP+/- mice and single cells were isolated by fluorescence-activated cell sorter (FACS) by established methods.11, 42, 43 For wild-type cell isolation for IL22RA1 expression, crypts were isolated from the distal half of the small intestine from C57Bl6 mice. After crypt isolation, crypts were pelleted and resuspended in 9 mL calcium and magnesium-free Hank's balanced salt solution with 0.6 U/mL dispase (354235; Corning), 120 U/mL DNase (DN25; Sigma), and 10 μmol/L Y-27632. Cells then were shaken vigorously for 30 seconds every 2 minutes for 10–15 minutes until a majority of the cells were single, filtered through a 40-μm filter into ice-cold DPBS, and washed twice with DPBS. Cells were resuspended in ISC basal media (advanced Dulbecco's modified Eagle medium/F12, 1× N2, 1× B27 without vitamin A, 1 mmol/L HEPES, 1× GlutaMAX, 100 U/mL penicillin/streptomycin) and stained with FACS antibodies on ice for 1 hour protected from light.
For intracellular staining, cells were resuspended in 4% paraformaldehyde at room temperature, mixed thoroughly, and incubated for 15 minutes at room temperature. Cells were washed with 1% BSA in 1× PBS and intracellular antibodies were added to cells in 1× saponin permeabilization buffer in 1% BSA in 1× PBS, mixed and incubated for 30 minutes at room temperature protected from light. Cells then were rinsed with permeabilization buffer and resuspended in 1% BSA in 1× PBS for FACS analysis. Extracellular antibody staining was performed for allophycocyanin (APC)-conjugated anti-CD326 (1:250, 118218, clone G8.8; Biolegend). Intracellular antibody staining was performed for phycoerythrin (PE)-conjugated anti-IL22RA1 (1:100, FAB4291P; R&D Systems). Immediately before FACS analysis of live cells, AnnexinV Pac-Blue (1:100, 640918; Biolegend) was used for live/dead discrimination. All FACS and flow cytometry experiments were performed using a SH800Z Cell Sorter (Sony Biotechnology, San Jose, CA). For RNA isolation, 50,000 cells were sorted directly into 250 μL RNA lysis buffer (from the RNAqueous-Micro Total RNA Isolation Kit).
Statistical Analysis
All data are means and SEM for the various groups. Statistics are based on n = 3 biological replicates. For comparison of 1 group with a reference value (Western blot), a 1-sample t test was performed. For the comparison of 2 groups, an unpaired t test was performed. For the comparison of multiple groups, a 1-way analysis of variance followed by the Tukey multiple comparisons test or a Bonferroni correction was performed. All analyses of statistical significance were calculated and shown in reference with the control group unless otherwise stated. All graphs were made and statistics were performed using GraphPad Prism version 7.0b for Mac (GraphPad, San Diego, CA).
Acknowledgments
The authors would like to acknowledge all members of the Magness Laboratory, Drs Brian Diekman and Susan Henning for useful discussions, Dr John Serody for use of IL22 neutralizing antibody, Dr Bin Gao for providing IL22TG mouse intestines, and Dr Kathleen Caron for use of antibodies.
Footnotes
Author contributions Bailey Zwarycz was responsible for the study concept and design, acquisition of data, analysis and interpretation of data, drafting of the manuscript, and statistical analysis; Adam D. Gracz was responsible for the study concept and design, analysis and interpretation of data, and critical revision of the manuscript for important intellectual content; Kristina R. Rivera was responsible for the acquisition of data and the analysis and interpretation of data; Ian A. Williamson was responsible for the analysis and interpretation of data; Leigh A. Samsa was responsible for the study concept and design and critical revision of the manuscript for important intellectual content; Josh Starmer was responsible for the analysis and interpretation of data; Michael A. Daniele was responsible for the COMSOL modeling; Luisa Salter-Sid was responsible for the study concept and design; Qihong Zhao was responsible for the study concept and design and study supervision; and Scott T. Magness was responsible for the study concept and design, acquisition of data, analysis and interpretation of data, critical revision of the manuscript for important intellectual content, obtained funding, and study supervision.
Conflicts of interest These authors disclose the following: Josh Starmer and Scott T. Magness have a financial interest in SeqQuest, LLC; and Luisa Salter-Sid and Qihong Zhao are employed by Bristol Myers Squibb, who provided key financial support for this study. The remaining authors disclose no conflicts.
Funding This work was funded by National Institutes of Health grant R01 DK091427 (S.T.M.); Bristol Myers Squibb grants 5058894 (S.T.M.) and F31 DK107137 (B.Z.); and the University of North Carolina Center for Gastrointestinal Biology and Disease grant P30 DK34987.
References
- 1.Fakhoury M., Negrulj R., Mooranian A., Al-Salami H. Inflammatory bowel disease: clinical aspects and treatments. J Inflamm Res. 2014;7:113–120. doi: 10.2147/JIR.S65979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Sloot K.W.J., Amini M., Peters V., Dijkstra G., Alizadeh B.Z. Inflammatory bowel diseases: review of known environmental protective and risk factors involved. Inflamm Bowel Dis. 2017;23:1499–1509. doi: 10.1097/MIB.0000000000001217. [DOI] [PubMed] [Google Scholar]
- 3.Neurath M.F. Cytokines in inflammatory bowel disease. Nat Rev Immunol. 2014;14:329–342. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- 4.Wolk K., Witte E., Hoffmann U., Wolf-Dietrich D., Endesfelder S., Asadullah K., Wolfram S., Volk H., Wittig B.M., Sabat R. IL-22 induces lipopolysaccharide-binding protein in hepatocytes: a potential systemic role of IL-22 in Crohn’s disease. J Immunol. 2007;178:5973–5981. doi: 10.4049/jimmunol.178.9.5973. [DOI] [PubMed] [Google Scholar]
- 5.Li J., Zhang L., Zhang J., Wei Y., Li K., Huang L., Zhang S., Gao B., Wang X., Lin P. Interleukin 23 regulates proliferation of lung cancer cells in a concentration-dependent way in association with the interleukin-23 receptor. Carcinogenesis. 2013;34:658–666. doi: 10.1093/carcin/bgs384. [DOI] [PubMed] [Google Scholar]
- 6.Lindemans C.A., Calafiore M., Mertelsmann A.M., O'Connor M., Dudakov J., Jenq R., Velardi E., Young L.F., Smith O.M., Lawrence G., Ivanov J.A., Fu Y., Takashima S., Hua G., Martin M.L., O'Rourke K.P., Lo Y., Mokry M., Romera-Hernandez M., Cupedo T., Dow L.E., Nieuwenhuis E.E., Shroyer N.F., Liu C., Kolesnick R., van den Brink M.R.M., Hanash A.M. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528:560–564. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stelzner M., Helmrath M., Dunn J.C.Y., Henning S.J., Houchen C.W., Kuo C., Lynch J., Li L., Magness S.T., Martin M.G., Wong M.H., Yu J., NIH Intestinal Stem Cell Consortium A nomenclature for intestinal in vitro cultures. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1359–G1363. doi: 10.1152/ajpgi.00493.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato T., Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 9.Singh U.P., Singh N.P., Murphy E.A., Price R.L., Fayad R., Nagarkatti M., Nagarkatti P.S. Chemokine and cytokine levels in inflammatory bowel disease patients. Cytokine. 2016;77:44–49. doi: 10.1016/j.cyto.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lejeune D., Dumoutier L., Constantinescu S., Kruijer W., Schuringa J.J., Renauld J.-C. Interleukin-22 (IL-22) activates the JAK/STAT, ERK, JNK, and p38 MAP kinase pathways in a rat hepatoma cell line. J Biol Chem. 2002;277:33676–33682. doi: 10.1074/jbc.M204204200. [DOI] [PubMed] [Google Scholar]
- 11.Formeister E.J., Sionas A.L., Lorance D.K., Barkley C.L., Lee G.H., Magness S.T. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1108–G1118. doi: 10.1152/ajpgi.00004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haber A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katx Y., Tirosh I., Beyaz S., Dionne D., Zhang M., Raychowdhury R., Garrett W.S., Rozenblatt-Rosen O., Shi H.N., Yilmaz O., Xavier R.J., Regev A. A single-cell survey of the small intestinal epithelium. Nature. 2017;16:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizuno S., Mikami Y., Kamada N., Handa T., Hayashi A., Sato T., Matsuoka K., Matano M., Ohta Y., Sugita A., Koganei K., Sahara R., Takazoe M., Hisamatsu T., Kanai T. Cross-talk between RORγt+ innate lymphoid cells and intestinal macrophages induces mucosal IL-22 production in Crohn's disease. Inflamm Bowel Dis. 2014;20:1426–1434. doi: 10.1097/MIB.0000000000000105. [DOI] [PubMed] [Google Scholar]
- 14.Young M.E., Carroad P.A., Bell R.L. Estimation of diffusion coefficients of proteins. Biotechnol Bioeng. 1980;22:947–955. [Google Scholar]
- 15.Francis K., Palsson B.O. Effective intercellular communication distances are determined by the relative time constants for cyto/chemokine secretion and diffusion. Proc Natl Acad Sci U S A. 1997;94:12258–12262. doi: 10.1073/pnas.94.23.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandrin D., Wagner D., Sitta C.E., Thoma R., Felekyan S., Hermes H.E., Janiak C., de Sousa Amadeu N., Kuhnemuth R., Lowen H., Egelhaaf S.U., Seidel C.A.M. Diffusion of macromolecules in a polymer hydrogel: from microscopic to macroscopic scales. Phys Chem Chem Phys. 2016;18:12860–12876. doi: 10.1039/c5cp07781h. [DOI] [PubMed] [Google Scholar]
- 17.Cortez V.S., Robinette M.L., Colonna M. Innate lymphoid cells: new insights into function and development. Curr Opin Immunol. 2015;32:71–77. doi: 10.1016/j.coi.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mowat A.M., Agace W.W. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 19.Geremia A., Arancibia-Cárcamo C.V., Fleming M.P., Rust N., Singh B., Mortensen N.J., Travis S.P., Powrie F. IL-23–responsive innate lymphoid cells are increased in inflammatory bowel disease. J Exp Med May. 2011;208:1127–1133. doi: 10.1084/jem.20101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.VanDussen K.L., Carulli A.J., Keeley T.M., Patel S.R., Puthoff B.J., Magness S.T., Tran I.T., Maillard I., Siebel C., Kolterud A., Grosse A.S., Gumucio D.L., Ernst S.A., Tsai Y., Dempsey P.J., Samuelson L.C. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carulli A.J., Keeley T.M., Demitrack E.S., Chung J., Maillard I., Samuelson L.C. Notch receptor regulation of intestinal stem cell homeostasis and crypt regeneration. Dev Biol. 2015;402:98–108. doi: 10.1016/j.ydbio.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mah A.T., Yan K.S., Kuo C.J. Wnt pathway regulation of intestinal stem cells. J Physiol. 2016;594:4837–4847. doi: 10.1113/JP271754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noah T.K., Shroyer N.F. Notch in the intestine: regulation of homeostasis and pathogenesis. Annu Rev Physiol. 2013;75:263–288. doi: 10.1146/annurev-physiol-030212-183741. [DOI] [PubMed] [Google Scholar]
- 24.Iscove N.N., Nawa K. Hematopoietic stem cells expand during serial transplantation in vivo without apparent exhaustion. Curr Biol. 1997;7:805–808. doi: 10.1016/s0960-9822(06)00341-1. [DOI] [PubMed] [Google Scholar]
- 25.Snippert H.J., van der Flier L.G., Sato T., van Es J.H., van den Born M., Kroon-Veenboer C., Barker N., Klein A.M., van Rheenen J., Simons B.D., Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 26.Park O., Wang H., Weng H., Feigenbaum L., Li H., Yin S., Ki S.H., Yoo S.H., Dooley S., Wang F.S., Young H.A., Gao B. In vivo consequences of liver-specific interleukin-22 expression in mice: implications for human liver disease progression. Hepatology. 2011;54:252–261. doi: 10.1002/hep.24339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Longman R.S., Diehl G.E., Victorio D.A., Huh J.R., Galan C., Miraldi E.R., Swaminath A., Bonneau R., Scherl E.J., Littman D.R. CX3CR1+ mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J Exp Med. 2014;211:1571–1583. doi: 10.1084/jem.20140678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 29.Chen Q., Quan Y., Wang N., Xei C., Ji Z., He H., Chai R., Li H., Yin S., Chin Y.E., Wei X., Gao W.-Q. Inactivation of STAT3 signaling impairs hair cell differentiation in the developing mouse cochlea. Stem Cell Rep. 2017;9:231–246. doi: 10.1016/j.stemcr.2017.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao J.-Q. Targeting interleukin-22 in psoriasis. Inflammation. 2014;37:94–99. doi: 10.1007/s10753-013-9715-y. [DOI] [PubMed] [Google Scholar]
- 31.Jia H.-Y., Shi Y., Luo L.-F., Jiang G., Zhou Q., Xu S.-Z., Lei T.-C. Asymmetric stem-cell division ensures sustained keratinocyte hyperproliferation in psoriatic skin lesions. Int J Mol Med. 2016;37:359–368. doi: 10.3892/ijmm.2015.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brand S., Beigel F., Olszak T., Zitzmann K., Eichhorst S.T., Otte J.-M., Diepolder H., Marquardt A., Jagla W., Popp A., Leclair S., Herrmann K., Seiderer J., Ochsenkuhn T., Goke B., Auerhammer C.J., Dambacher J. IL-22 is increased in active Crohn's disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am J Physiol Gastrointest Liver Physiol. 2006;290:G827–G838. doi: 10.1152/ajpgi.00513.2005. [DOI] [PubMed] [Google Scholar]
- 33.Martin J.C.J., Bériou G., Heslan M., Chauvin C., Utriainen L., Aumeunier A., Scott C.L., Mowat A., Cerovic V., Houston S.A., Leboeuf, Hubert F.X., Hémont C., Merad M., Milling S., Josein R. Interleukin-22 binding protein (IL-22BP) is constitutively expressed by a subset of conventional dendritic cells and is strongly induced by retinoic acid. Mucosal Immunol. 2014;7:101–113. doi: 10.1038/mi.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizoguchi A., Yano A., Himuro H., Ezaki Y., Sadanaga T., Mizoguchi E. Clinical importance of IL-22 cascade in IBD. J Gastroenterol. 2018;53:465–474. doi: 10.1007/s00535-017-1401-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong S., Zheng C., Doughty M.L., Losos K., Didkovsky N., Schambra U.B., Nowak N.J., Joyner A., Leblanc G., Hatten M.E., Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 36.Van Landeghem L., Santoro M.A., Krebs A.E., Mah A.T., Dehmer J.J., Gracz A.D., Scull B.P., McNaughton K., Magness S.T., Lund K.P. Activation of two distinct Sox9-EGFP-expressing intestinal stem cell populations during crypt regeneration after irradiation. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1111–G1132. doi: 10.1152/ajpgi.00519.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jansson A., Harlen M., Karlsson S., Nilsson P., Cooley M. 3D computation modelling of the influence of cytokine secretion on Th-cell development suggests that negative selection (inhibition of Th1 cells) is more effective than positive selection by IL-4 for Th2 cell dominance. Immunol Cell Biol. 2007;85:189–196. doi: 10.1038/sj.icb.7100023. [DOI] [PubMed] [Google Scholar]
- 39.Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., Golland P., Sabatini D.M. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern. 1979;9:62–66. [Google Scholar]
- 41.Berry M.V. Image analysis and mathematics morphology. Phys Bull. 1983;34:252. [Google Scholar]
- 42.Gracz A.D., Williamson I.A., Roche K.C., Johnston M.J., Wang F., Wang Y., Attayek P.J., Balowski J., Liu X.F., Laurenza R.J., Gaynor L.T., Sims C.H., Galanko J.A., Li L., Allbritton N.L., Magness S.T. A high-throughput platform for stem cell niche co-cultures and downstream gene expression analysis. Nat Cell Biol. 2015;17:340–349. doi: 10.1038/ncb3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gracz A.D., Ramalingam S., Magness S.T. Sox9 expression marks a subset of CD24-expressing small intestine epithelial stem cells that form organoids in vitro. Am J Physiol Gastrointest Liver Physiol. 2010;298:G590–G600. doi: 10.1152/ajpgi.00470.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]