Abstract
Ethylene regulates numerous aspects of plant growth and development. Multiple external and internal factors coordinate ethylene production in plant tissues. Transcriptional and post-translational regulations of ACC synthases (ACSs), which are key enzymes mediating a rate-limiting step in ethylene biosynthesis have been well characterized. However, the regulation and physiological roles of ACC oxidases (ACOs) that catalyze the final step of ethylene biosynthesis are largely unknown in Arabidopsis. Here, we show that Arabidopsis ACO1 exhibits a tissue-specific expression pattern that is regulated by multiple signals, and plays roles in the lateral root development in Arabidopsis. Histochemical analysis of the ACO1 promoter indicated that ACO1 expression was largely modulated by light and plant hormones in a tissue-specific manner. We demonstrated that point mutations in two E-box motifs on the ACO1 promoter reduce the light-regulated expression patterns of ACO1. The aco1-1 mutant showed reduced ethylene production in root tips compared to wild-type. In addition, aco1-1 displayed altered lateral root formation. Our results suggest that Arabidopsis ACO1 integrates various signals into the ethylene biosynthesis that is required for ACO1’s intrinsic roles in root physiology.
Keywords: ACC oxidase 1, Arabidopsis thaliana, ethylene biosynthesis, lateral root, transcriptional regulation
INTRODUCTION
Ethylene is a gaseous plant hormone that participates in a variety of processes throughout the plant life cycle from seed germination to organ senescence and fruit ripening (Beaudoin et al., 2000; Ghassemian et al., 2000; Li et al., 2013; Maunders et al., 1987; Picton et al., 1993).
In plants, ethylene is biosynthesized from S-adenosyl-L-methionine (SAM) through the intermediate 1-aminocyclopropane-1-carboxylic acid (ACC). ACC synthases (ACSs) and ACC oxidases (ACOs) catalyze the conversion of SAM to ACC followed by the conversion of ACC to ethylene, respectively (Yang and Hoffman, 1984). In the ethylene biosynthesis pathway, the conversion of SAM to ACC, which is catalyzed by ACS, is considered as a rate-limiting step. For this reason, studies on the regulation of ethylene biosynthesis by multiple signals, such as MAP kinases and plant hormones, have focused on ACS genes and proteins (Barry et al., 2000; Chae et al., 2003; Liu and Zhang, 2004; Skottke et al., 2011; Thain et al., 2004; Ye et al., 2015; Yi et al., 1999; Yoon and Kieber, 2013). Arabidopsis ACS family members show diverse affinities for SAM, implying that these enzymes might be optimized to perform different functions in various tissues and cell types (Yamagani et al. 2003).
The conversion of ACC to ethylene catalyzed by ACO is the final regulatory step in ethylene biosynthesis (Alonso and Ecker, 2001; Kende and Zeevaart, 1997; Lasserre et al., 1996; Ruduś et al., 2013; Vriezen et al., 1999). ACO is a member of a large Fe(II)-requiring dioxygenase/oxidase superfamily. However, studies on the function and regulation of ACO have been hindered by the general perception of ACS as the key regulatory enzyme in ethylene biosynthesis, along with an incomplete gene family definition and limited biochemical and physiological studies (Booker and DeLong, 2015). A query for ACO in the TAIR (www.arabidopsis.org) yields 13 loci for Arabidopsis. Among these, ACO4 was the first enzyme predicted to have ACC oxidase catalytic activity (Gόmez-Lim et al., 1993), while ACO2 was recognized for its function of counteracting ABA-induced endosperm rupture inhibition during seed germination (Linkies et al., 2009). Although transcript levels for ACOs have been analyzed under various conditions (Gόmez-Lim et al., 1993; Linkies et al., 2009; Thain et al., 2004; Qin et al., 2007), biochemical, functional, and genetic studies on ACOs remain to be conducted.
Here, we investigated tissue-specific expression pattern of ACO1 using a promoter-GUS reporter system, as well as the biochemical and physiological analysis in the regions where ACO1 is expressed. We found that the ACO1 promoter displays a tissue-specific expression pattern that is distinctly modified by different light conditions and treatments with several plant hormones. We demonstrated that the ACO1 promoter contains two E-box motifs that are indispensable for transcriptional activity and regulation by light signal. Biochemical and physiological studies of the aco1-1 mutant showed that ACO1 is required for ethylene production and for the control of lateral root development. Our results suggest that ACO1 is an active ethylene biosynthetic enzyme required for normal root development in Arabidopsis and its expression integrates multiple signals into ethylene biosynthesis.
MATERIALS AND METHODS
Plant materials, growth conditions, and hormone treatments
Arabidopsis thaliana ecotype Col-0 was used as the wild-type in this study. The aco1-1 (SALK_127963) line was obtained from SALK. T-DNA insertion in aco1-1 was confirmed by genotyping PCR with ProACO1-GUS 1154 For (Supplementary Table S1) and T-DNA left border (5′-CTTTGACGTTGGAGTCCACGTTCTTTAATA-3′) primers. Homozygous aco1-1 was screened by genotyping PCR using ProACO1-GUS 1154 For and ProACO1-GUS 1 Rev primers (Supplementary Table S1). Seeds were surface-sterilized with ethanol-distilled water (70:30, v/v) for 5 min, washed with distilled water, and stratified at 4°C for 3 days. Surface-sterilized seeds were planted on 0.8% phytagel (Sigma) containing 0.5× Murashige and Skoog (MS)-salt medium and 1% (w/v) sucrose and grown in the light (120 μmol m−2 s−1) at 22 ± 1°C for 16 hr and in the dark at 20 ± 1°C for 8 h in an environmental growth chamber (Sanyo, Osaka). For etiolated conditions, the plates were wrapped with aluminum foil after exposure to white light for 1 day. Two-week-old seedlings were transferred from agar plates to soil to obtain adult plants. For hormone treatment, seedlings were grown on 1× MS medium containing 0.8% Phytagel for 7 days and then transferred to 1× MS liquid medium for 6 hr. For ethylene treatment, ethylene gas was injected into plates.
Root length and lateral root measurement
To measure root length and lateral root number, seedlings were incubated on vertical plates. Lateral root number was counted as the total number of emerging and emerged lateral roots under a dissecting microscope (Olympus SZ-PT). Seedlings were photographed to measure root length.
Preparation of transgenic plants expressing the GUS gene fused to truncated ACO1 promoters
Genomic DNA was extracted from Col-0 wild-type according to the method described previously (Park et al., 2010). ACO1 promoter fragments were cloned from genomic DNA by PCR amplification using primer pairs designed to introduce a HindIII restriction site into the 5′-end and a BamHI site into the 3′-end of each promoter fragment, respectively (Supplementary Table S1). For the ProACO1-GUS Δ312 construct, ProACO1-GUS 1843 For primer and ProACO1-GUS Δ312 Rev primer were used (Supplementary Table S1). Pro-ACO1-GUS 1843mE2mE4 was generated from ProACO1-GUS 1843 plasmids by site-directed mutagenesis (Stratagen). The primers used for mutagenesis are shown in Supplementary Table S1. Promoter fragments were cloned into pGEM T easy vector (Promega) and subcloned into pBI121 after confirmation by DNA sequencing. The vector constructs were transformed into Agrobacterium tumefaciens strain GV3101::pMP90 and transformed into Arabidopsis following the method described previously (Clough and Bent, 1998).
Total RNA isolation, and semi-quantitative and quantitative RT-PCR analysis
Total RNA was extracted from the wild-type, mutant, and chemical-treated seedlings using TRI reagent (Sigma). For semi-quantitative RT-PCR, first-strand cDNA was synthesized using 5 μg of total RNA and M-MLV reverse transcriptase (Promega). PCR was performed in a reaction containing 1 μl cDNA, 0.25 μl Real Taq (RBC, Taiwan), 2.5 μl 2.5 mM dNTP mixture, 2.5 μl 10× buffer (Takara, Japan), and 1 μl of each primer (10 pmol) in a 25 μl reaction. The gene-specific primers for ACOs are listed in Table S2. Ten μl samples of the 50 μl PCR products were electrophoresed on 0.8 or 1% agarose gels and stained with ethidium bromide (EtBr). Stained bands were scanned and densitometrically analyzed using the Quantity One program (Bio-Rad). For quantitative real-time PCR (qPCR), total RNA was extracted from 10 DAG long day-grown Col-0 and aco1-1 using the Spectrum Plant Total RNA kit (Sigma). RevertAid reverse transcriptase (Thermo Scientific) was used to synthesize complementary DNA from 2 ug total RNA. qPCR was carried out using LightCycler 480 (Roche) and SensiMix SYBR & Fluorescein Kit (Bioline). ACO1 primers (ACO1 q-Forward primer: 5′-GTGGGGATTCTTCATGGTTGATAATCATGG-3′ and ACO1 q-Reverse primer: 5′-CATCGTCTTGCTGAGTTCCTCTGAAATG-3′) were used for qPCR. PP2A (Forward primer: 5′-TATCGGATGACGATTCTTCGTGCAG-3′, Reverse primer: 5′-GCTTGGTCGACTATCGGAATGAGAG-3′) was used as an internal control.
Histochemical GUS staining
Histochemical GUS staining was performed as described previously (Weigel and Glazebrook, 2002). Briefly, samples were incubated in staining solution containing 2 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid (Duchefa) in 50 mM Na2HPO4 buffer, pH 7.2, 2 mM potassium ferrocyanide, 2 mM potassium ferricyanide, 0.2% Triton X-100 at 37°C overnight after infiltrating under vacuum on ice for 20 min. The samples were dehydrated using 20, 35 and 50% ethanol and fixed with fixative (50% ethanol, 10% acetic acid and 5% formaldehyde). The samples were cleared with 70% ethanol and observed with dissecting (Olympus SZ-PT) and light (Olympus CX21) microscopes.
GUS activity measurement
To measure GUS activity, plant tissues were ground in liquid nitrogen. Crude plant extracts were resolved in GUS extraction buffer (50 mM Na2HPO4 buffer, pH 7.0, 10 mM EDTA, pH 8.0, 0.1% SDS, 0.1% Triton X-100, 10 mM β-mercaptoethanol and 25 μg ml−1 PMSF). The crude extracts were mixed with a reaction buffer (1 mM 4-methylumbelliferyl-β-D-glucuronide [Duchefa] in GUS extraction buffer). After exactly 10 and 20 min, reactions were stopped by Na2CO3. Fluorescence was measured with a spectrofluorometer.
Ethylene production quantification
One hundred root segments (approximately 10 mm from root tip) or root tip segments (approximately 2 mm long) from 5 DAG seedlings were harvested in 25 mL silicon-capped vials containing 200 μL buffer (100 mM MES, pH 6.8 and 1.5 mM chloramphenicol). Vials containing root tissues were incubated at 27 ± 1°C in the dark with shaking (170 rpm). Ethylene measurement was performed as described previously (Yun et al., 2009). Air samples (1 ml) from these vials were withdrawn with a syringe and injected into a gas chromatograph equipped with a column containing alumina (HP5890 series II; Hewelett Packard, USA, 80/100 Porapak-Q column, oven temperature: 120°C, injector temperature: 150°C, detector temperature: 280°C).
RESULTS
Spatio-temporal expression patterns for ACO1 in Arabidopsis
Given that multiple ACO genes are encoded in the Arabidopsis genome, we speculated that each ACO member might have specific function(s) in various tissues, as is the case with ACS family members (Tsuchisaka el al., 2009). Among the 13 ACOs found in Arabidopsis, only ACO2 and ACO4 have been shown to be functional enzymes (Gόmez-Lim et al., 1993; Linkies et al., 2009). Thus, we focused on ACO1 (At2g19590), a close homolog of ACO2 and ACO4, which might be functional in ethylene production.
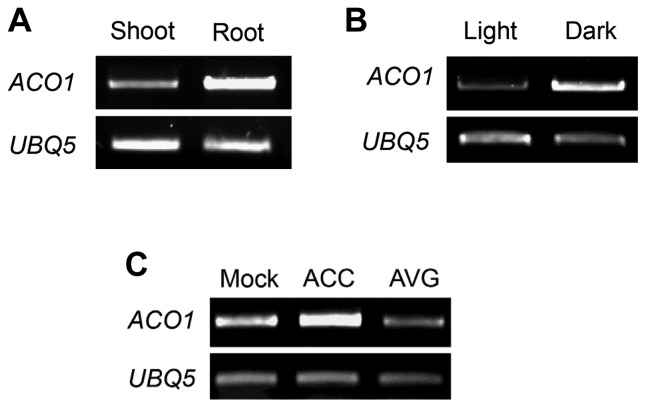
RT-PCR analysis indicated that ACO1 was more highly expressed in roots compared to shoots in Arabidopsis seedlings grown under light (Fig. 1A). In addition, comparison of ACO1 transcript levels between light- and dark-grown seedlings showed that ACO1 mRNA was more highly accumulated in dark-grown seedlings compared to light-grown seedlings (Fig. 1B). As ACOs catalyze ethylene production using ACC as a substrate, we examined whether the ACC level might affect ACO1 expression. RT-PCR analysis indicated that exogenous ACC application to 5 days after germination (DAG) seedlings promoted ACO1 expression whereas treatment with an ACS inhibitor, aminoethoxyvinylglycine (AVG), inhibited its expression (Fig. 1C). The ACC and AVG treatment results indicated that ACO1 expression was positively regulated by ACC level in plant cells. Taken together, these results suggest that ACO1 expression is tissue-specific, and is regulated by light as well as the ethylene precursor, ACC.
Fig. 1. ACO1 gene expression under various conditions.
(A) Comparison of ACO1 expression in the shoot and root of 5 DAG seedlings. Seedlings were cut into the shoot (cotyledon and hypocotyl) and root by a razor blade. The shoot and root fragments were subjected to total RNA extraction. (B) ACO1 expression in seedlings grown under different light conditions. Seedlings were grown in the light or in the dark for 5 DAG. (C) Ethylene precursor effect on ACO1 expression. Five DAG seedlings were treated with 1 μM ACC or 1 μM AVG for 6 hr. Semi-quantitative RT-PCR was performed with total RNA. PCR products were electrophoresed on 1% agarose gels and detected by ethidium bromide (EtBr) staining. UBQ5 was used to normalize the expression level.
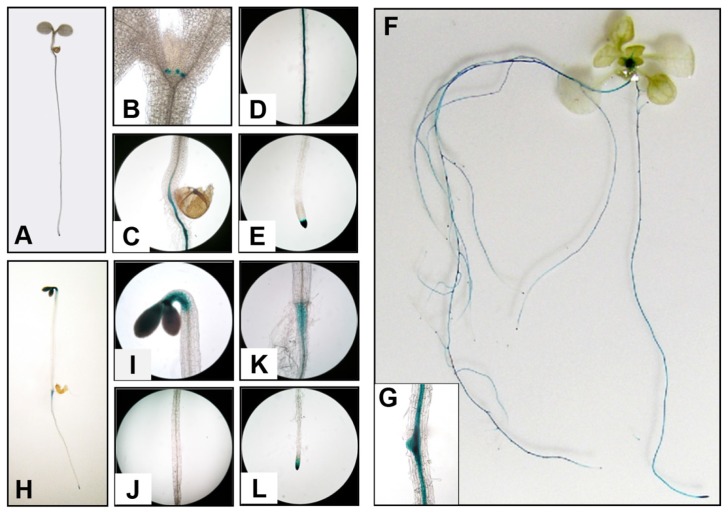
To further explore the physiological and functional relevance of ACO1 in Arabidopsis, we generated transgenic Arabidopsis plant expressing the β-glucuronidase (GUS) gene driven by the 1,843 bp ACO1 promoter. Using ProA-CO1-GUS 1843 transgenic plants, the spatio-temporal expression of ACO1 was further analyzed. Consistent with the RT-PCR results for ACO1, in 3 DAG seedlings grown under light, GUS activities were predominant in roots (Figs. 2A–2E). On the other hand, ACO1 expression in aerial parts was restricted to the stipule (Fig. 2B). In the root, ACO1 was expressed in vascular tissue from the hypocotyl-root junction to the maturation zone and root tip, but not in the elongation zone (Figs. 2C–2E). The expression pattern of ACO1 in the root was consistent with previously reported in silico gene expression analysis (Dugardeyn et al., 2008). In 12 DAG seedlings, GUS activity was detected only in the rosette leaf basal region but was ubiquitously observed in roots (Fig. 2F). Additionally, enhanced ACO1 expression was detected at the lateral root primordial formation sites (Fig. 2G). When ProACO1-GUS 1843 was grown in the dark, strong GUS activities were detected in the cotyledon and the concave side of the apical hook (Figs. 2H and 2I) but were not detected in other hypocotyl regions (Figs. 2I and 2J). Furthermore, ACO1 in etiolated roots was expressed in the hypocotyl-root junction and root tip, which was similar to the pattern observed in light-grown seedlings (Figs. 2K and 2L). Collectively, these results indicated that ACO1 is predominantly expressed in roots and its expression is strictly regulated by light. In adult plants, GUS expression was detected in apical primary and lateral inflorescence stems, the petiole, the basal pedicel, and the leaf vein in addition to the whole root (Supplementary Fig. S1). No ACO1 promoter activity was detected in flowers, silique, or rosette leaf (Supplementary Fig. S1).
Fig. 2. Spatio-temporal expression pattern for ACO1.
(A–G) ACO1 promoter tissue-specific activity under light conditions. Images show whole plant (A), shoot apex (B), hypocotyl-root junction (C), root maturation zone (D), and root tip (E) for 3 DAG ProACO1-GUS 1843 seedlings. (F) GUS expression pattern for 12 DAG ProACO1-GUS 1843 plants. (G) Insert, ACO1 expression in lateral root primordium from 5 DAG ProACO1-GUS 1843 seedlings. (H–L) Tissue-specific activity for the ACO1 promoter under dark conditions. Images show whole plant (H), cotyledon and apical hypocotyl (I), basal hypocotyl (J), hypocotyl-root junction (K) and root tip (L) from 4 DAG dark-grown ProACO1-GUS 1843 seedlings.
Tissue-specific regulation of ACO1 expression by external and internal stimuli in Arabidopsis
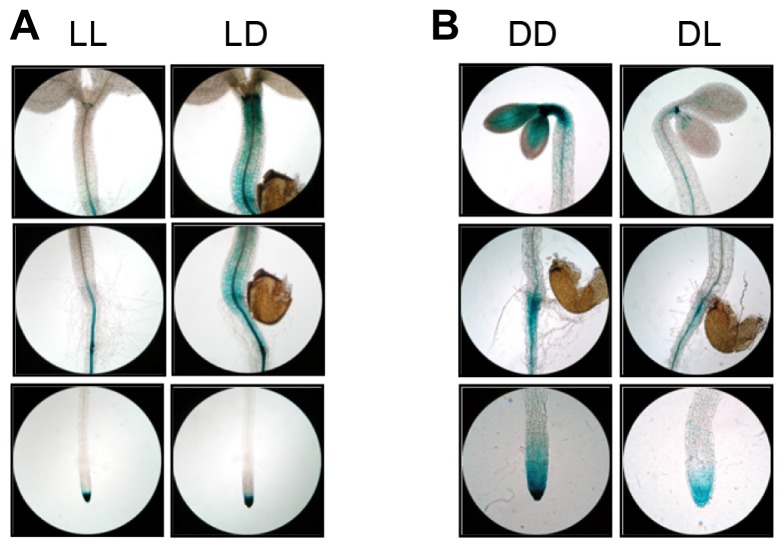
We further investigated the effect of external and internal stimuli on ACO1 promoter activity. For this purpose, we first examined whether light could regulate ACO1 expression. Consistent with prominent ACO1 expression in the dark, ACO1 expression in the hypocotyl and the hypocotyl-root junction of light-grown seedling was greatly increased by dark treatment for 6 hours, although it was not altered in roots and cotyledons (Fig. 3A). In contrast, the dark to light transition strongly reduced ACO1 expression (Fig. 3B), suggesting that ACO1 expression was strongly regulated by light signals.
Fig. 3. Light effect on ACO1 promoter activity.
(A) GUS expression in light-grown ProACO1-GUS 1843 seedlings before (LL) and after (LD) transition to dark. (B) GUS expression in dark-grown ProACO1-GUS 1843 seedlings before (DD) and after (DL) transition to light. Transitions between dark and light were performed for 6 hr. Three or more seedlings were used for GUS staining and showed similar staining patterns.
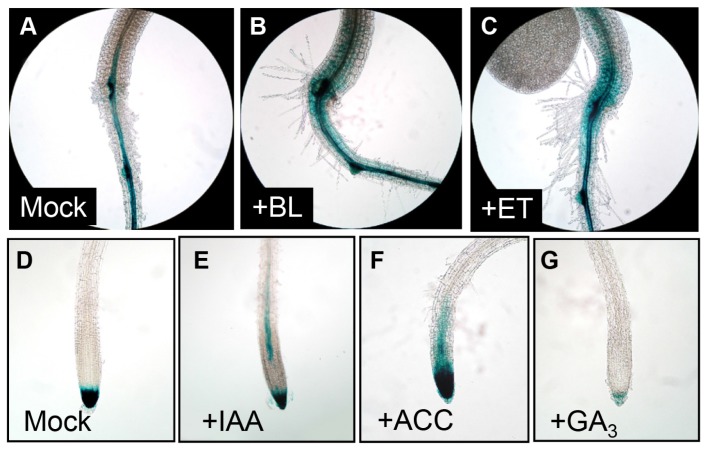
As multiple plant hormones regulate ethylene production (Chae et al., 2003; Yi et al., 1999; Yun et al, 2009), we examined whether plant hormones alter ACO1 promoter activity. Thus, we treated ProACO1-GUS 1843 seedlings with brassinosteroid (BR), ethylene, auxin, ACC, and gibberellin (GA). Brassinolide (BL), the most bioactive BR, and ethylene enhanced ACO1 promoter activity in roots and the hypocotyl compared to the controls (Figs. 4A–4C). Exogenous application of auxin specifically activated the promoter in the root elongation zone of vascular tissues (Fig. 4E). Consistent with RT-PCR result (Fig. 1C), ACC treatment enhanced promoter activity from the root tip to the elongation zone (Fig. 4F). In contrast, AVG treatment reduced GUS expression in root maturation zone but not in the root tip (Supplementary Fig. S2). GA repressed promoter activity in the root tip (Fig. 4G). These results indicated that plant hormone signals regulated ACO1 expression differently with distinct tissue specificity. Taken together, these findings imply that multiple signals can modulate ethylene production by altering ACO1 transcription in a tissue-specific manner.
Fig. 4. ACO1 promoter tissue-specific regulation by various plant hormones.
(A–C) Hypocotyl-root junction in ProACO1-GUS 1843 seedlings. (D–G) Roots from ProACO1-GUS 1843 seedlings. 5 DAG seedlings were left untreated (A, D) or treated with 100 nM BL (B), 1 ppm ethylene (C), 1 μM IAA (E), 10 μM ACC (F), and 50μM GA3 (G) for 6 h.
Determination of the E-box motif on the ACO1 promoter required for ACO1 expression
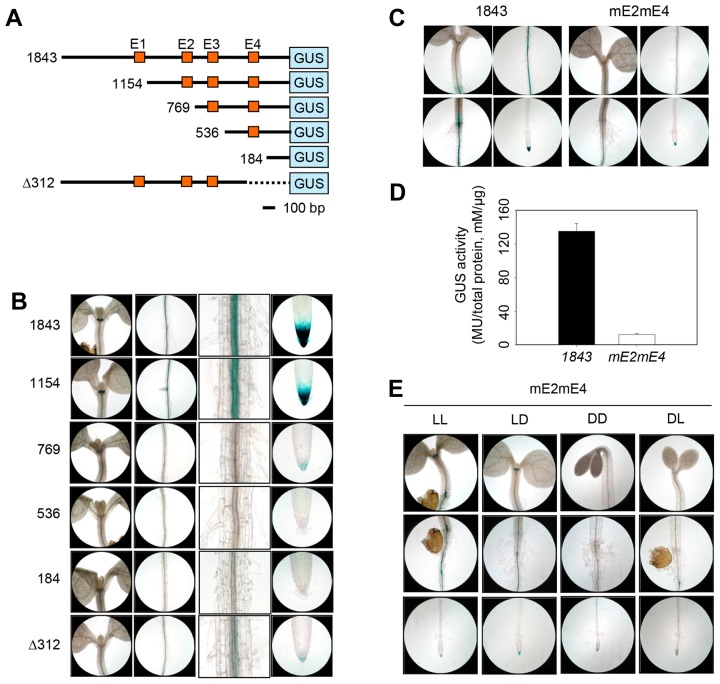
To define the regulatory region required for tissue-specific ACO1 expression, we analyzed cis-regulatory elements in the ACO1 promoter. This promoter contains four E-box motifs (CANNTG, E1~E4) known as BR/light-regulated cis-regulatory elements (Fig. 5A; Martínez-García et al., 2000; Sun et al., 2010). Thus, we generated transgenic plants expressing the GUS reporter gene driven by four different truncated ACO1 promoter lengths (ProACO1-GUS 1154, 769, 536, and 184) in which the cis-regulatory elements were deleted (Fig. 5A). In addition, transgenic plants possessing the ProACO1-GUS Δ312 construct were prepared. The ProACO1-GUS Δ312 plants contained the entire 5′-intergenic regions but 312-bp fragment upstream of the start codon was deleted (Fig. 5A). ProACO1-GUS 1154 seedlings showed indistinguishable GUS activity compared to the ProACO1-GUS 1843 (Fig. 5B). However, ProACO1-GUS 769 seedlings showed significantly reduced GUS activity compared to the ProACO1-GUS 1843 implying that the region between 1,154 and 769 bp from the start codon was required for ACO1 promoter activity. Consistently, plants with ProACO1-GUS 536 or ProACO1-GUS 184 showed no detectable GUS expression (Fig. 5B). ProACO1-GUS Δ312 showed only weak GUS activity in the maturation region and root tips (Fig. 5B), suggesting that the 312-bp fragment upstream of the start codon was also important for ACO1 promoter activity. These results suggest that at least two regulatory regions containing E2 and E4 motifs are essential for ACO1 expression. We mutated the E-box motifs (CANNTG to TCTGAA; mE2mE4) without creating novel motif(s) when the mutagenized sequences were combined with adjacent sequences. Histochemical and quantitative analysis further demonstrated that GUS expression driven by ProACO1-GUS 1843 was greatly abolished by point mutations in the two E-box motifs (Figs. 5C and 5D). Furthermore, ProACO1-GUS 1843 mE2mE4 GUS activity was not altered by light-dark transition, suggesting that E2 and E4 elements were also involved in light regulation of ACO1 expression (Fig. 5E).
Fig. 5. Histochemical GUS staining in truncated series from ProACO1-GUS transgenic seedlings.
(A) Schematic diagram from truncated ACO1 promoter-GUS constructs. Numbers represent the number of nucleotides from truncated ACO1 promoters fused to GUS gene (blue box), except for Δ312, which indicates promoter deletion size. Orange boxes indicate localization of E-box motif (E1~E4). The bar represents 100 nucleotides in length. (B) GUS expression in truncated ProACO1-GUS seedlings. Seedlings from ProACO1-GUS 1843 (1843), ProACO1-GUS 1154 (1154), ProACO1-GUS 769 (769), ProACO1-GUS 536 (536), ProACO1-GUS 184 (184), and ProACO1-GUS Δ312 ( 312) were grown on vertically oriented plates for 5 DAG then subjected to GUS staining. From left to right, each image shows hypocotyl, root maturation zone, magnified root maturation zone, and root tip, respectively. Three or more seedlings were used for GUS staining and showed similar staining patterns. (C) Histochemical analysis of GUS activity in ProACO1-GUS 1843 and ProACO1-GUS 1843 mE2mE4 (mE2mE4). GUS staining was repeated with three independent transgenic plants and the representative of similar results are shown. (D) GUS activity quantification in ProACO1-GUS 1843 and ProACO1-GUS 1843 mE2mE4 (mE2mE4). Columns indicate mean values from two independent experiments. Error bars show standard deviation. (E) GUS expression in ProACO1-GUS 1843 mE2mE4 (mE2mE4) seedlings under different light conditions; continuous light-grown (LL), light-grown seedlings treated with dark (LD), continuous dark-grown (DD) and dark-grown seedling treated with light (DL).
ACO1 is required for ethylene production and regulates lateral root development
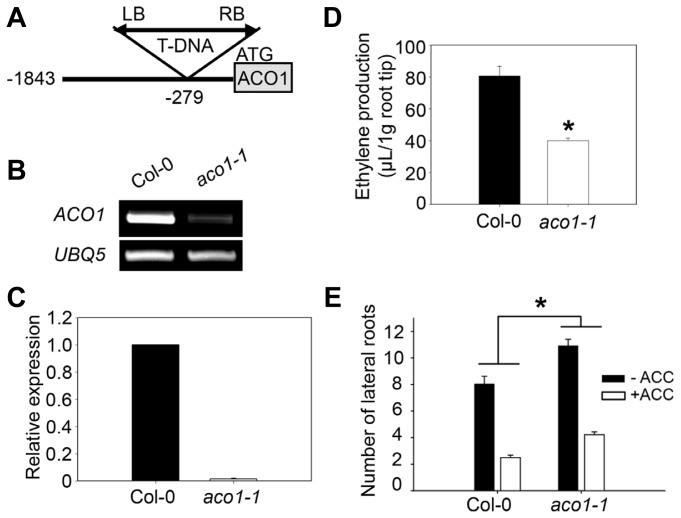
To address the biochemical and physiological functions of ACO1, an Arabidopsis mutant harboring a T-DNA insertion 279-bp upstream of the ACO1 start codon was obtained, and a homozygous line was isolated (referred to as aco1-1; Fig. 6A). aco1-1 was the only mutant that was publically available. The T-DNA insertion significantly reduced ACO1 transcript levels in the mutant (Fig. 6B). The reduced ACO1 transcript levels in the aco1-1 mutant are consistent with the histochemical evidence that deletion of the 312-bp fragment upstream of ACO1 start codon greatly reduced ACO1 promoter activity (Fig. 5B). Quantitative RT-PCR analysis showed that ACO1 transcript level in aco1-1 was decreased by more than 60-fold compared to that in the wild-type (Fig. 6C). When we examined the expression of other ACO-like genes in the aco1-1 mutant, no gene showed a significantly altered expression level (Supplementary Figs. S3 and S4).
Fig. 6. Biochemical and physiological phenotypes of aco1-1.
(A) T-DNA insertion map. T-DNA was inserted at 279 bp upstream of the start codon in the ACO1 gene. LB, left border; RB, right border. Numbers represent the number of nucleotides upstream (−) of the start codon. (B) Transcript level for ACO1 gene in aco1-1 seedlings. The wild-type and aco1-1 seedlings were grown for 5 DAG in the light. Semi-quantitative RT-PCR was performed with total RNA extracted from the wild-type and aco1-1 seedlings. PCR products were detected by EtBr staining. UBQ5 was used to normalize the expression level. (C) Quantitative real time-PCR was performed with total RNA extracted from 10 DAG seedlings grown in the light. ACO1 expression level in aco1-1 was normalized to that of PP2A and is shown relative to the expression levels in the wild-type. Two biological repeats along with three technical repeats were performed for quantification of ACO1 expression level. The error bars indicate the S.D. (n=2). (D) Ethylene production in aco1-1 root tips. Ethylene production from 2 mm root tip segments was analyzed after 6 hr incubation. Values are expressed as the mean of three biological replications ± S.E. Each biological replicate was performed with two technical repeats; each repeat used 100 root tips weighing approximately 0.001 g. The asterisk indicates significant differences from the wild-type at P<0.01 using t-test. (E) Effect of ACC treatment on aco1-1 lateral root formation. Lateral root numbers of 9 DAG seedlings grown on control (−ACC) or 1 μM ACC (+ACC)-containing media were counted under a dissecting microscope. Each column represents the average lateral root number. Error bars represent the S.E. (n ≥ 22). The asterisks indicate significant difference between the wild-type and aco1-1 at P < 0.001 using t-test. Three biological repeats were performed and showed similar results. The average value of the three repeats is shown.
Considering that ACO1 was predominantly expressed in roots, we speculated that ACO1 might be required for ethylene biosynthesis to regulate root developmental and physiological responses. Thus, we measured ethylene production in roots from the wild-type and aco1-1 mutant plants. Ethylene production in root segments from 5 DAG aco1-1 was not significantly different from that from the wild-type plants (Supplementary Fig. S5A). However, when we analyzed ethylene production in root tips where the expression level was high (Fig. 2E), the aco1-1 mutant showed greatly reduced ethylene production compared to the wild-type plants (Fig. 6D), suggesting that ACO1 is required for ethylene biosynthesis in Arabidopsis seedling root tips.
To test whether ACO1 expression in roots contributed to ethylene-mediated root growth inhibition, we measured primary root lengths of aco1-1 and the wild-type plants. However, we could not detect statistically significant differences in root length (Supplementary Fig. S5B), indicating that ACO1 may not be a major enzyme involved in ethylene-regulated root growth.
Based on ACO1 tissue-specific expression in root vascular tissue and increased promoter activity at the lateral root primordia formation sites (Figs. 1D–1F), we measured the lateral root number of aco1-1 and the wild-type. Although the phenotypic differences between the wild-type and aco1-1 mutant primary root lengths were subtle, the number of lateral roots was significantly higher in aco1-1 compared to that in the wild-type (Fig. 6E). It is well known that exogenous treatment of ACC negatively impacts on lateral root formation (Negi et al., 2008). As ACOs catalyze conversion of ACC to ethylene, we tested whether aco1-1 reduces the ACC-induced inhibition of lateral root development. The wild-type displayed 68.9% (5.5/8.0) reduction in lateral root formation upon ACC treatment, whereas the aco1-1 mutant showed 61.5% (6.7/10.9) reduction (Fig. 6E). aco1-1 grown on ACC-treated media showed increased lateral root number compared to the wild-type on the same media (Fig. 6E), suggesting that ACO1 expression is partially required for lateral root development control by ACC.
DISCUSSION
Previous studies suggest that conversion of ACC to ethylene, mediated by ACO, is a critical regulatory step in ethylene biosynthesis (Alonso and Ecker, 2001; Kende and Zeevaart, 1997; Lasserre et al., 1996; Van de Poel et al., 2014; Vriezen et al., 1999). ACC is considered a long-distance transported substance but ethylene has limited transport capacity (Bradford and Yang, 1980). Thus, tissue-specific functions of ethylene should be investigated with respect to corresponding ACO activity in each tissue, rather than ACS-catalyzed ACC biosynthesis. Thus, it has been suggested that determination of ACO transcript level in each tissue may serve as an indicator of ethylene content (Ruduś et al., 2013).
It is known that ACO genes display distinct expression patterns in various plant organs and at different developmental stages associated with ethylene activity in rice, tomato, and garden cress (Barry et al., 1996; Iwamoto et al., 2010; Linkies et al., 2009). However, tissue-specific expression patterns and regulation of ACO genes in Arabidopsis are poorly understood, although ACO gene transcription is known to be differentially regulated by various biotic and abiotic stresses, light signaling, and multiple hormones (García et al., 2010; Gόmez-Lim et al., 1993; Mazzella et al., 2005; Qin et al., 2007; Thain et al., 2004).
In this study, we demonstrated the tissue-specific expression of ACO1 and its physiological role in Arabidopsis. Using transcriptional fusion of the ACO1 promoter and GUS reporter gene, we showed that Arabidopsis ACO1 exhibits a specific spatio-temporal expression pattern (Fig. 2 and Supplementary Fig. S1). Its transcription appeared to be under the control of an intrinsic signaling network involved in responses to light and hormones (Figs. 3 and 5). Light-to-dark transition strongly increased ACO1 expression whereas dark-to-light transition reduced ACO1 promoter activity. Furthermore, we identified two E-box motifs that were essential for ACO1 promoter activity and its regulation by light signal (Fig. 5). Both BR-regulated BZR1 and light-regulated PIF transcription factors are known to interact with each other and bind E-box motifs in their target genes (Oh et al., 2012). In addition, a recent study demonstrated that auxin-regulated ARF6 transcription factor shares its target genes with BZR1, and their transcriptional activity is interdependent as well (Oh et al., 2014). In this study, we showed that ACO1 promoter activity was regulated by all three signals in a tissue-specific manner (Figs. 3 and 4). Mutations in two E-box motifs (E2 and E4) in the ACO1 promoter compromised its regulatory activity by light, suggesting that E2 and E4 are essential cis-elements required for ACO1 expression via binding with PIF and BZR1 (Figs. 5C–5E). Interestingly, the ACO1 promoter also contains core AuxREs that are the binding sites for ARFs (Supplementary Fig. S6; Ulmasov et al., 1999) and the ACO1 expression pattern was altered by IAA treatment (Fig. 4E). Therefore, two core AuxREs in the transcriptional regulatory region (1,154 and 769 bp from the ACO1 start codon) might also regulate the promoter activity. Taken together, these results imply that tissue-specific ACO1 expression might be mediated by combinatorial activation of these transcription factors that coordinate multiple signals.
We isolated the aco1-1 mutant that had a T-DNA insertion at 279 bp upstream from the ATG codon. ACO1 expression in aco1-1 was greatly reduced by the T-DNA mutation that displaced regulatory cis-elements on the ACO1 promoter (Figs. 6A and 6B; Supplementary Fig. S6). The reduced ACO1 expression in aco1-1 further supports the importance of cis-elements for ACO1 promoter activity.
Although ACO1 has a conserved Fe2+-binding motif that is required for binding to the substrate ACC (Linkies et al., 2009), no evidence has supported that ACO1 is a functional enzyme producing ethylene in Arabidopsis. We showed that ethylene production by the aco1-1 mutant was greatly reduced in root tips (Fig. 6D). This finding provides an evidence that ACO1 functions as an active ACC oxidase in Arabidopsis roots.
In this study, we demonstrated that ACO1 expression is required for lateral root development. aco1-1 displayed enhanced lateral root development (Fig. 6E). Auxin is a key hormone that promotes lateral root formation through direct LBD gene activation in the lateral root primordia (Okushima et al., 2007). It is known that ethylene inhibits lateral root formation through promoting PIN3 and PIN7 expression, leading to the disruption of local auxin maximum which is required for lateral root development (Lewis et al., 2011). In addition, ACC treatment increases these protein levels in root tips (Lewis et al., 2011). In our study, we showed that ACO1 was highly expressed in root tips and its expression was enhanced by ACC treatment (Figs. 2E and 4F). Consistently, ethylene production in aco1-1 root tips was reduced compared to that of the wild-type (Fig. 6D). Thus, our results suggest that ACO1 expression in root tips might be responsible for the negative regulation of lateral root development by ethylene through alteration of auxin transport.
aco1-1 was slightly less sensitive to ACC in lateral root formation rather than complete insensitivity (Fig. 6E). In addition, we could not detect significant changes in root ethylene production and in root length (Supplementary Fig. S5). These faint phenotypes might be due to incomplete disruption of the gene expression in the mutant. Notably, ACO1 is a close homolog of ACO2-4 (Supplementary Fig. S7). In silico analysis showed that these genes have the same expression pattern in the root (Dugardeyn et al., 2008). Thus, the weak phenotypes of aco1-1 may also be explained by genetic redundancy that compensates the lack of ACO1 activity. We tried to generate a transgenic plant that overexpressed ACO1 under control of the Cauliflower mosaic virus (CaMV) 35S promoter. Although over 50 antibiotic-resistant transgenic plants were selected from three independent transformations, we failed to isolate a transgenic plant over-expressing ACO1. Therefore, in future study, multiple knockout mutants of this family genes might be helpful to understand their intrinsic functions.
Although we observed specific ACO1 expression in the aerial stipule, we could not determine its physiological role. Interestingly, expression of other known ethylene biosynthetic genes was also observed in the stipule (Yun et al., 2009), indicating that ethylene plays a physiological role in this organ. Stipules have been identified to be the first major site for high free auxin production during the very early stages of leaf development (Aloni et al., 2003). It is possible that the physiological interaction between auxin and ethylene in the stipule is crucial for regulation of leaf development.
High expression of ACO1 in dark-grown seedlings might correlate with the triple responses of ethylene in the dark (Fig. 3B). However, the physiological role of ACO1 expression increased by light-to-dark transition in hypocotyls is unclear (Fig. 3A). Smale et al., reported that ethylene treatment promoted hypocotyl elongation when seedlings were grown under long day conditions (Smale. et al., 1997) In addition, recent study demonstrated that ethylene-induced hypocotyl elongation also occurred in the dark period (Das et al., 2016). Thus, it will be interesting to study whether ACO1 expression induced by the light-to-dark transition correlates with ethylene-stimulated hypocotyl elongation in the long day-grown seedling.
Supplementary data
ACKNOWLEDGMENTS
This work was supported by grants from the Next-Generation BioGreen 21 Programs (PJ0132082018 to S.K.K), Rural Development Administration. This research was also supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (NRF-2017R1A2B4004274 to T.W.K.).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Aloni R., Schwalm K., Langhans M., Ullrich C. Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta. 2003;216:841–853. doi: 10.1007/s00425-002-0937-8. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., Ecker J.R. The ethylene pathway: A paradigm for plant hormone signaling and interaction. Sci STKE. 2001;2001:RE1. doi: 10.1126/stke.2001.70.re1. [DOI] [PubMed] [Google Scholar]
- Barry C.S., Blume B., Bouzayen M., Cooper W., Hamilton A.J., Grierson D. Differential expression of the 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. Plant J. 1996;9:525–535. doi: 10.1046/j.1365-313x.1996.09040525.x. [DOI] [PubMed] [Google Scholar]
- Barry C.S., Llop-Tous M.I., Grierson D. The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 2000;123:979–986. doi: 10.1104/pp.123.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin N., Serizet C., Gosti F., Giraudat J. Interactions between abscisic acid and ethylene signaling cascades. Plant Cell. 2000;12:1103–1115. doi: 10.1105/tpc.12.7.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker M.A., DeLong A. Producing the ethylene signal: regulation and diversification of ethylene biosynthetic enzymes. Plant Physiol. 2015;169:42–50. doi: 10.1104/pp.15.00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford K.J., Yang S.F. Xylem transport of 1-aminocyclopropane-1-carboxylic acid, an ethylene precursor, in waterlogged tomato plants. Plant Physiol. 1980;65:322–326. doi: 10.1104/pp.65.2.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer C.S., Sukumar P., Muday G.K. Ethylene modulates flavonoid accumulation and gravitropic responses in roots of Arabidopsis. Plant Physiol. 2006;140:1384–1396. doi: 10.1104/pp.105.075671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae H.S., Faure F., Kieber J.J. The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell. 2003;15:545–559. doi: 10.1105/tpc.006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.C., Kim Y.-S., Lee J.Y., Kaufman P.B., Kirakosyan A., Yun H.S., Kim T.-W., Kim S.Y., Cho M.H., Lee J.S., Kim S.-K. Brassinolide interacts with auxin and ethylene in the root gravitropic response of maize (Zea mays) Physiol Plantarum. 2004;121:666–673. [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dugardeyn J., Vandenbussche F., Van Der Straeten D. To grow or not to grow: what can we learn on ethylene–gibberellin cross-talk by in silico gene expression analysis? J Exp Bot. 2008;59:1–16. doi: 10.1093/jxb/erm349. [DOI] [PubMed] [Google Scholar]
- Edelmann H.G., Sabovljevic A., Njio G., Roth U. The role of auxin and ethylene for gravitropic differential growth of coleoptiles and roots of rye- and maize seedlings. Adv Space Res. 2005;36:1167–1174. [Google Scholar]
- García M.J., Lucena C., Romera F.J., Alcántara E., Pérez-Vicente R. Ethylene and nitric oxide involvement in the up-regulation of key genes related to iron acquisition and homeostasis in Arabidopsis. J Exp Bot. 2010;61:3885–3899. doi: 10.1093/jxb/erq203. [DOI] [PubMed] [Google Scholar]
- Ghassemian M., Nambara E., Cutler S., Kawaide H., Kamiya Y., McCourt P. Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell. 2000;12:1117–1126. doi: 10.1105/tpc.12.7.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gόmez-Lim M.A., Valdés-López V., Cruz-Hernandez A., Saucedo-Arias L.J. Isolation and characterization of a gene involved in ethylene biosynthesis from Arabidopsis thaliana. Gene. 1993;134:217–221. doi: 10.1016/0378-1119(93)90096-l. [DOI] [PubMed] [Google Scholar]
- Iwamoto M., Baba-Kasai A., Kiyota S., Hara N., Takano M. ACO1, a gene for aminocyclopropane-1-carboxylate oxidase: effects on internode elongation at the heading stage in rice. Plant Cell Environ. 2010;33:805–815. doi: 10.1111/j.1365-3040.2009.02106.x. [DOI] [PubMed] [Google Scholar]
- Kende H., Zeevaart J.A.D. The five “classical” plant hormones. Plant Cell. 1997;9:1197–1210. doi: 10.1105/tpc.9.7.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasserre E., Bouquin T., Hernandez J., Pech J., Balagué C., Bull J. Structure and expression of three genes encoding ACC oxidase homologs from melon (Cucumis melo L.) Mol Gen Genet. 1996;251:81–90. doi: 10.1007/BF02174348. [DOI] [PubMed] [Google Scholar]
- Lewis D.R., Miller N.D., Splitt B.L., Wu G., Spalding E.P. Separating the roles of acropetal and basipetal auxin transport on gravitropism with mutations in two Arabidopsis multidrug resistance-like ABC transporter genes. Plant Cell. 2007;19:1838–1850. doi: 10.1105/tpc.107.051599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis D.R., Negi S., Sukumar P., Muday G.K. Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Development. 2011;138:3485–3495. doi: 10.1242/dev.065102. [DOI] [PubMed] [Google Scholar]
- Li Z., Peng J., Wen X., Guo H. ETHYLENE-INSENSITIVE3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell. 2013;25:3311–3328. doi: 10.1105/tpc.113.113340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkies A., Muller K., Morris K., Tureckova V., Wenk M., Cadman C.S.C., Corbineau F., Strnad M., Lynn J.R., Finch-Savage W.E., Leubner-Metzger G. Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using lepidium sativum and Arabidopsis thaliana. Plant Cell. 2009;21:3803–3822. doi: 10.1105/tpc.109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell. 2004;16:3386–3399. doi: 10.1105/tpc.104.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García J.F., Huq E., Quail P.H. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Maunders M.J., Holdsworth M.J., Slater A., Knapp J.E., Bird C.R., Schuch W., Grierson D. Ethylene stimulates the accumulation of ripening-related mRNAs in tomatoes. Plant Cell Environ. 1987;10:177–184. [Google Scholar]
- Mazzella M.A., Arana M.V., Staneloni R.J., Perelman S., Rodriguez Batiller M.J., Muschietti J., Cerdán P.D., Chen K., Sánchez R.A., Zhu T., Chory J., Casal J.J. Phytochrome control of the Arabidopsis transcriptome anticipates seedling exposure to light. Plant Cell. 2005;17:2507–2516. doi: 10.1105/tpc.105.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.-Y., Wang Z.-Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.-Y., Bai M.-Y., Arenhart R.A., Sun Y., Wang Z.-Y. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife. 2014;3:e03031. doi: 10.7554/eLife.03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.H., Kim T.-W., Son S.-H., Hwang J.-Y., Lee S.C., Chang S.C., Kim S.-H., Kim S.W., Kim S.-K. Brassinosteroids control AtEXPA5 gene expression in Arabidopsis thaliana. Phytochemistry. 2010;71:380–387. doi: 10.1016/j.phytochem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Picton S., Barton S.L., Bouzayen M., Hamilton A.J., Grierson D. Altered fruit ripening and leaf senescence in tomatoes expressing an antisense ethylene-forming enzyme transgene. Plant J. 1993;3:469–481. [Google Scholar]
- Qin Y.-M., Hu C.-Y., Pang Y., Kastaniotis A.J., Hiltunen J.K., Zhu Y.-X. Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell. 2007;19:3692–3704. doi: 10.1105/tpc.107.054437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruduś I., Sasiak M., Kępczyński J. Regulation of ethylene biosynthesis at the level of 11-aminocyclopropane-1-carboxylate oxidase (ACO) gene. Acta Physiol Plant. 2013;35:295–307. [Google Scholar]
- Skottke K.R., Yoon G.M., Kieber J.J., DeLong A. Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet. 2011;7:e1001370. doi: 10.1371/journal.pgen.1001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumar P., Edwards K.S., Rahman A., DeLong A., Muday G.K. PINOID kinase regulates root gravitropism through modulation of PIN2-dependent basipetal auxin transport in Arabidopsis. Plant Physiol. 2009;150:722–735. doi: 10.1104/pp.108.131607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Fan X.-Y., Cao D.-M., Tang W., He K., Zhu J.-Y., He J.-X., Bai M.-Y., Zhu S., Oh E., et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Developmental Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thain S.C., Vandenbussche F., Laarhoven L.J.J., Dowson-Day M.J., Wang Z.-Y., Tobin E.M., Harren F.J.M., Millar A.J., Van Der Straeten D. Circadian rhythms of ethylene emission in Arabidopsis. Plant Physiol. 2004;136:3751–3761. doi: 10.1104/pp.104.042523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A., Yu G., Jin H., Alonso J.M., Ecker J.R., Zhang X., Gao S., Theologis A. A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics. 2009;183:979–1003. doi: 10.1534/genetics.109.107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T.J. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- Van de Poel B., Van Der Straeten D. 1-aminocyclopropane-1-carboxylic acid (ACC) in plants: more than just the precursor of ethylene! Front. Plant Sci. 2014;5:640. doi: 10.3389/fpls.2014.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriezen W.H., Hulzink R., Mariani C., Voesenek L.A. 1-aminocyclopropane-1-carboxylate oxidase activity limits ethylene biosynthesis in Rumex palustris during submergence. Plant Physiol. 1999;121:189–196. doi: 10.1104/pp.121.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Glazebrook J. Arabidopsis: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- Yamagami T., Tsuchisaka A., Yamada K., Haddon W.F., Harden L.A., Theologis A. Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J Biol Chem. 2003;278:49102–49112. doi: 10.1074/jbc.M308297200. [DOI] [PubMed] [Google Scholar]
- Yang S.F., Hoffman N.E. Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol. 1984;35:155–189. [Google Scholar]
- Ye L., Li L., Wang L., Wang S., Li S., Du J., Zhang S., Shou H. MPK3/MPK6 are involved in iron deficiency-induced ethylene production in Arabidopsis. Front Plant Sci. 2015;6:953. doi: 10.3389/fpls.2015.00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H.C., Joo S., Nam K.H., Lee J.S., Kang B.G., Kim W.T. Auxin and brassinosteroid differentially regulate the expression of three members of the 1-aminocyclopropane-1-carboxylate synthase gene family in mung bean (Vigna radiata L.) Plant Mol Biol. 1999;41:443–454. doi: 10.1023/a:1006372612574. [DOI] [PubMed] [Google Scholar]
- Yoon G.M., Kieber J.J. 14-3-3 Regulates 1-Aminocyclopropane-1-Carboxylate synthase protein turnover in Arabidopsis. Plant Cell. 2013;25:1016–1028. doi: 10.1105/tpc.113.110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun H.R., Joo S.-H., Park C.H., Kim S.-K., Chang S.C., Kim S.Y. Effects of brassinolide and IAA on ethylene production and elongation in maize primary roots. J Plant Biol. 2009;52:268–274. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.