Abstract
Negative selection against deleterious alleles produced by mutation influences withinpopulation variation as the most pervasive form of natural selection. However, it is not known whether deleterious alleles affect fitness independently, so that cumulative fitness loss depends exponentially on the number of deleterious alleles, or synergistically, so that each additional deleterious allele results in a larger decrease in relative fitness. Negative selection with synergistic epistasis should produce negative linkage disequilibrium between deleterious alleles and, therefore, an underdispersed distribution of the number of deleterious alleles in the genome. Indeed, we detected underdispersion of the number of rare loss-of-function alleles in eight independent data sets from human and fly populations. Thus, selection against rare protein-disrupting alleles is characterized by synergistic epistasis, which may explain how human and fly populations persist despite high genomic mutation rates.
Negative, or purifying, selection prevents the unlimited accumulation of deleterious mutations and establishes a mutation-selection equilibrium (1). The properties of negative selection are determined by the corresponding fitness landscape, the map that relates fitness to the “mutation burden” in an individual. Because of the difficulty of ascribing precise selection coefficients to different alleles, the mutation burden can be approximated by the total number of putatively deleterious mutations in an individual. Under the null hypothesis of no epistasis, selection acts on different mutations independently, so that each additional mutation causes the same decline in relative fitness and fitness declines exponentially with their number. By contrast, if synergistic, or narrowing (2), epistasis between deleterious alleles is present, each additional mutation causes a larger decrease in relative fitness. Synergistic epistasis reduces the mutation load under a given genomic rate of deleterious mutations (1, 3, 4) and canmake sex and recombination advantageous (5). However, because neither the mutation burden nor fitness can be easily measured, data on fitness landscapes of negative selection remain inconclusive (6). Theory suggests that narrowing epistasis may emerge as a result of pervasive pleiotropy and the modular organization of biological networks (7). Some genome-wide investigations have found epistasis but no consistent directionality of effect (6, 8, 9).
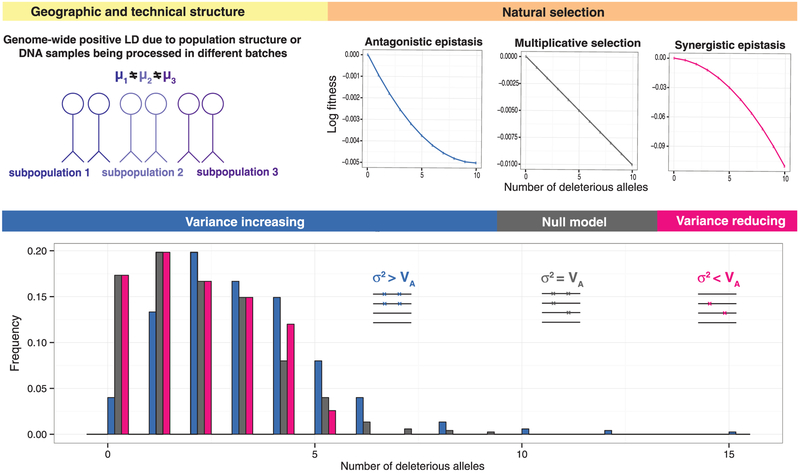
We examined the distribution of the mutation burden in human and Drosophila melanogaster populations. In the absence of epistasis, alleles should contribute to the mutation burden independently (3), such that the variance of the mutation burden is equal to the sum of the variances at all loci or the additive variance (VA) (10, 11) (Fig. 1). If mutant alleles are rare, the mutation burden follows a Poisson distribution with a variance (σ2) equal to its mean (μ) (fig. S1).
Fig. 1. Rare mutation burden under natural selection (orange, right) and population structure (yellow, left).
The mutation burden (bottom panel) is shown under the null model (gray, the absence of epistasis and population structure) and under variance-increasing (blue, antagonistic epistasis and population structure) and variance-reducing (pink, synergistic epistasis) models. μk is the mean of the mutation burden in subpopulation k within the population.
In contrast, epistatic selection creates dependencies between deleterious alleles, so the total variance of the mutation burden is no longer equal to the additive variance (12). Selection with synergistic epistasis creates repulsion, or negative linkage disequilibrium (LD). As a result of this LD, the variance of the mutation burden is reduced by a factor of ρ (<1), which is determined by the strength of selection and the extent of epistasis, leading to an underdispersion (σ2 < VA) (12, 13) (fig. S2). Antagonistic (diminishing returns) epistasis, instead, creates positive LD between deleterious alleles and increases the variance of the mutation burden leading to its overdispersion (σ2 > VA). Also, the difference between σ2 and VA is a genome-wide estimate of the net LD in fitness (11, 14). Using fully sequenced individual genomes from a population, we tested for synergistic epistasis without needing to measure fitness directly.
The ideal population for our test would be single-ancestry, outbred, nonadmixed, and randomly mating. We analyzed the Genome of the Netherlands (GoNL) Project (15), the Alzheimer’s Disease Neuroimaging Initiative (ADNI), and Dutch controls from Project MinE, an amyotrophic lateral sclerosis study. For each of these, we obtained whole-genome sequences of unrelated individuals of European descent. We obtained similar data for Zambian flies from phase 3 of the Drosophila Population Genomics Project (DPGP3) (16). For each population, after applying stringent quality control filters (tables S8 to S12), we computed the mutation burden and corresponding σ2 and VA values, focusing on rare alleles for coding synonymous,missense, and loss-of-function (LoF) mutations (here defined as splice site disrupting or nonsense). For all of these data sets, the distribution of rare LoF alleles was underdispersed (Table 1).
Table 1. Negative linkage disequilibrium (LD) between rare LoF alleles in human and D. melanogaster genomes.
For humans, only singletons, and for flies, only alleles up to a minor allele count of 5, are included (see tables S2 and S3 for other frequency cut-offs). Net LD is normalized per pair of alleles and per pair of loci (11). A one-sided P value was obtained for σ2/VA by permutation, and a joint P value for all three human data sets shown (GoNL, ADNI, MinE) was computed by meta-analysis using Stouffer’s method (11) (coding synonymous P = 0.999, missense P = 5.155 × 10−4, LoF P = 0.002). The number of samples is given in parentheses for each data set.
| Variant type | Mean | σ2/VA | Net LD | |
|---|---|---|---|---|
| Per pair of derived alleles |
Per pair of loci |
|||
| Humans | ||||
| Genome of the Netherlands GoNL (495) | ||||
| Synonymous | 30.26 | 1.675 | 0.022 | 4.554 × 10−8 |
| Missense | 60.88 | 2.077 | 0.018 | 3.609 × 10−8 |
| Nonsense | 1.67 | 0.929 | −0.039 | –8.013 × 10−8 |
| Splice | 0.90 | 0.953 | −0.049 | –1.008 × 10−7 |
| LoF | 2.58 | 0.930 | −0.029 | –5.848 × 10−8 |
| European ancestry ADNI (714) | ||||
| Synonymous | 38.99 | 2.077 | 0.028 | 2.709 × 10−8 |
| Missense | 77.98 | 2.008 | 0.013 | 1.268 × 10−8 |
| Nonsense | 2.10 | 0.933 | −0.032 | –3.126 × 10−8 |
| Splice | 1.16 | 0.878 | −0.104 | −1.020 × 10−7 |
| LoF | 2.83 | 0.930 | −0.022 | −2.126 × 10−8 |
| Dutch MinE (601) | ||||
| Synonymous | 42.93 | 1.749 | 0.017 | 2.414 × 10−8 |
| Missense | 79.34 | 1.960 | 0.012 | 1.675 × 10−8 |
| Nonsense | 1.89 | 1.057 | 0.028 | 3.898 × 10−8 |
| Splice | 0.95 | 0.972 | −0.033 | −4.641 × 10−8 |
| LoF | 2.83 | 0.996 | −0.001 | −1.727 × 10−9 |
| D. melanogaster | ||||
| Zambian DPGP3 (191) | ||||
| Synonymous | 3577.06 | 57.473 | 0.016 | 1.658 × 10−6 |
| Missense | 2051.52 | 18.536 | 0.008 | 6.710 × 10−7 |
| Nonsense | 10.21 | 0.928 | −0.007 | −4.139 × 10−7 |
| Splice | 2.60 | 0.948 | −0.020 | −1.308 × 10−6 |
| LoF | 12.81 | 0.929 | −0.005 | −3.298 × 10−7 |
On average, rare LoF alleles displayed variance (σ2) reduced by a factor of ~0.95, compared to additive variance (VA). In contrast, rare synonymous and missense alleles were overdispersed. The GoNL project also provided a set of highquality short insertions and deletions (indels), and in this data set, we observed an underdispersed distribution for the combined set of LoF alleles and frameshift indels (table S19). Overlaying the mutation burden distributions with Poisson distributions having identical means shows that the underdispersion is due to a depletion of individuals with a high number of deleterious alleles (figs. S12 to S17).
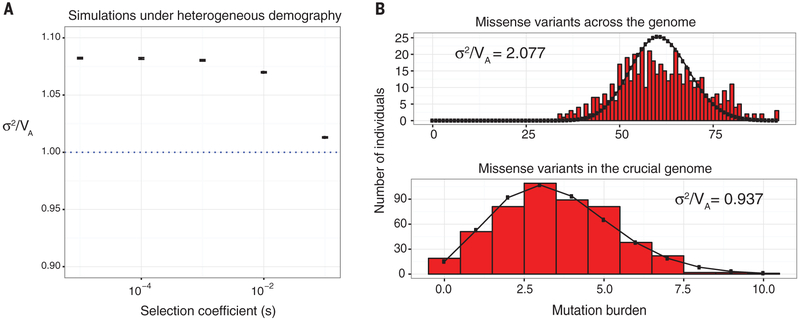
Even without epistasis, overdispersion in the mutation burden would be observed if genomewide positive LD is present owing to population structure (Fig. 1) (17). If the population has a cline in average rare mutation burden (μ) due to, for example, a south-to-north expansion (15) followed by assortative mating, this may translate into an excess of σ2 over VA (figs. S3 and S4). Overdispersion may also be caused by DNA samples being sequenced or processed in different batches. A large proportion of the overdispersion in rare mutation burden computed on synonymous or missense alleles in the detailed GoNL samples could be attributed to geographic origin and sequencing batch (fig. S5 and tables S4 and S15). In contrast, LoF alleles were not significantly overdispersed by confounders (table S16). This is consistent with the results obtained for populations simulated under heterogeneous demography, which show that overdispersion in mutation burden decreases with the strength of negative selection (Fig. 2A) (11).
Fig. 2. Simulated and empirical distributions of rare missense mutation burden.
(A) Simulations using SLiM 2.0 of unlinked sites under multiplicative selection in a finite population with heterogeneous demography (11). σ2/VA was calculated for the rare mutation burden computed on singletons at equilibrium, with the null expectation as shown (blue dotted line). Error bars show SEM (100 replicates). (B) Missense rare mutation burden (red) computed on singletons across the genome (σ2/VA = 2.077) and only in the crucial genome (σ2/VA = 0.937) in the GoNL data set, overlaid with Poisson distributions (black) having identical means. The crucial genome for humans was constructed by selecting only genes with an estimated selection coefficient against heterozygous protein-truncating variants exceeding 0.2 (11).
Given that overdispersion scales with selection strength, we constructed a “crucial” genome for humans, selecting only genes with an estimated selection coefficient against heterozygous protein-truncating variants exceeding 0.2 (11). An analogous essential genome was constructed for D. melanogaster using the Database of Essential Genes (11). When only their crucial or essential genomes were considered, both humans (Fig. 2B and fig. S8) and D. melanogaster (fig. S9) showed an underdispersion in their missense mutation burden. In contrast, synonymous alleles remained overdispersed. Accordingly, we also observed that σ2/ VA scales inversely with the strength of selection acting on a gene for missense but not for synonymous alleles in the fly data sets (fig. S18) (11).
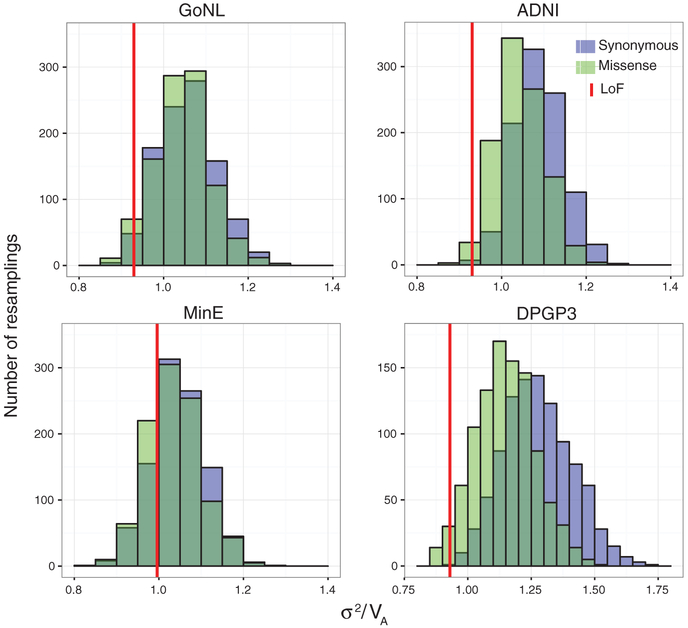
To investigate the significance of the underdispersion in rare LoF alleles, we generated an empirical null distribution for σ2/VA for each data set by resampling synonymous alleles at matched allele frequency as our test set of LoF alleles (Fig. 3) (11). We meta-analyzed the human data with three suitable (low inbreeding and admixture) non-European populations from phase I of the 1000 Genomes Project (18) (tables S1 and S2), and the fruit fly data with an American population from the D. melanogaster Genetic Reference Panel (DGRP) (19) (table S3). Meta-analysis across all data sets using Stouffer’s method indicates that rare LoF alleles were significantly underdispersed in humans (P = 0.0003) and flies (P = 9.43 × 10−6) (11). Permuting functional consequences across variants, we confirmed the significance of our underdispersion signal in rare protein-altering mutations in humans (missense P = 2.670 × 10−4, LoF P = 0.002) and D. melanogaster (missense P = 9.43 × 10−6, LoF P = 0.0001) (11). Furthermore, through regression analysis, resampling experiments, and simulations, we showed that the underdispersion signal persists after correcting for potential confounders and is not driven by outliers (tables S5, S17, and S18 and fig. S11) (11).
Fig. 3. Resampling distributions of σ2/VA for rare LoF mutation burden in humans and D. melanogaster.
Synonymous (purple) and missense (green) alleles were resampled at the same allele frequency as LoF alleles to obtain empirical null distributions for σ2/VA in each data set. For humans, only singletons, and for flies, only alleles up to a minor allele count of 5, are included. A one-sided P value for σ2/VA of the rare LoF mutation burden (red) was obtained, and a joint P value for all three human data sets shown (GoNL, ADNI, MinE) was computed by meta-analysis using Stouffer’s method (11) (P = 0.0003).
We also sought to determine the source of the observed negative LD and what it says about the shape of the fitness landscape. Directional selection with synergistic epistasis was proposed as a solution to the mutation load paradox (3, 4) and as a deterministic mechanism for the evolution of sex (5). However, as long as mutations are not unconditionally deleterious, they may be subject to stabilizing selection instead of directional selection, and this may also result in negative LD (20). Furthermore, in small populations, genetic drift in the presence of multiplicative selection may act as a random force to create negative LD, because mutations that arise as unique events at different sites will be in repulsion (21, 22).
Although stabilizing selection is always narrowing and can thus be regarded as simply another way of generating synergy, a far lower mutational load is generated under stabilizing selection compared with purely directional selection (20). However, LoF alleles are likely to be unconditionally deleterious. With regard to the role of genetic drift, we validated with simulations of finite populations with realistic human demography that negative LD between unlinked sites is quantitatively negligible under a model of multiplicative selection (fig. S10). We also demonstrated that most of our signal in rare LoF alleles comes from net negative LD between completely unlinked alleles on different chromosomes (table S6) and very distant alleles on the same chromosome (figs. S6 and S7). If the source of negative LD is narrowing selection, then sexual reproduction has an evolutionary advantage for purely deterministic reasons. Our analysis cannot preclude the role of random chance or genetic drift in aiding this advantage by creating negative LD, as our signal, in part, comes from linked sites in the genome, although the majority does not.
Our empirical observations on properties of the fitness landscape for protein-disrupting variants have broader evolutionary implications, especially if the results extend to the broader class of mildly deleterious alleles. The question of how our species accommodates high deleterious mutation rates has long been pondered. Indeed, a newborn is estimated to have ~70 de novo mutations (23). The consensus for estimates for the fraction of the genome that is “functional” is that about 10% of the human genome sequence is selectively constrained (24). Thus, the average human should carry at least seven de novo deleterious mutations. If natural selection acts on each mutation independently, the resulting mutation load and loss in average fitness are inconsistent with the existence of the human population (1 – e−7 > 0.99). To resolve this paradox, it is sufficient to assume that the fitness landscape is flat only outside the zone where all the genotypes actually present are contained, so that selection within the population proceeds as if epistasis were absent (20, 25). However, our findings suggest that synergistic epistasis affects even the part of the fitness landscape that corresponds to genotypes that are actually present in the population.
Currently, although selection due to prereproductive mortality in humans is deeply relaxed, there is still a substantial opportunity for selection (26, 27). Thus, our results suggest that even humans are experiencing ongoing narrowing negative selection.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to L. Mirny, G. McVean, and I. Adzhubey for scientific discussions; all members of the Sunyaev lab and two anonymous reviewers for comments that improved the manuscript; D. Jordan for providing SLiM 2.0 simulation runs; C. Cassa, D. Jordan, D. Weghorn, and D. Balick for providing genic selection estimates for humans; J. Fan for helping with analyses as a summer student; and J. Lack for help with D. melanogaster inversion data. This project was supported by NIH grants R01GM078598, R01GM105857, R01MH101244, and U01HG009088. Analysis of fruit fly data was performed at IITP RAS and supported by the Russian Science Foundation (grant no. 14-50-00150). Data sets used in this study can be accessed as follows: GoNL: www.nlgenome.nl/; ADNI: http://adni.loni.usc.edu/; Project MinE: www.projectmine.com; The 1000 Genomes Phase I Project: ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/; DGRP and DPGP3: www.johnpool.net/genomes.html.
Footnotes
A complete listing of Alzheimer’s Disease Neuroimaging Initiative (ADNI) investigators can be found at http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
The members of the Genome of the Netherlands Consortium are listed in the supplementary materials.
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Muller HJ, Am. J. Hum. Genet 2, 111–176 (1950). [PMC free article] [PubMed] [Google Scholar]
- 2.Shnol EE, Kondrashov AS, Genetics 134, 995–996 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kimura M, Maruyama T, Genetics 54, 1337–1351 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crow JF, Kimura M, Proc. Natl. Acad. Sci. U.S.A 76, 396–399 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kondrashov AS, Nature 336, 435–440 (1988). [DOI] [PubMed] [Google Scholar]
- 6.de Visser JAGM, Elena SF, Nat. Rev. Genet 8, 139–149 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Chiu H-C, Marx CJ, Segrè D, Proc. Biol. Sci 279, 4156–4164 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bank C, Hietpas RT, Jensen JD, Bolon DNA, Mol. Biol. Evol 32, 229–238 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puchta O et al. , Science 352, 840–844 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bulmer M, The Mathematical Theory of Quantitative Genetics (Clarendon, 1980). [Google Scholar]
- 11.Materials and methods are available as supporting materials.
- 12.Kondrashov AS, Genet. Res 65, 113–121 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Charlesworth B, Genet. Res 55, 199–221 (1990). [DOI] [PubMed] [Google Scholar]
- 14.Bulmer MG, Genet. Res 28, 101–117 (1976). [DOI] [PubMed] [Google Scholar]
- 15.Francioli LC et al. , Nat. Genet 46, 818–825 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Lack JB et al. , Genetics 199, 1229–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang RC, Genetics 155, 1449–1458 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abecasis GR et al. , Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mackay TFC et al. , Nature 482, 173–178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charlesworth B, Evolution. 67, 3354–3361 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Felsenstein J, Genetics 78, 737–756 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barton NH, Otto SP, Genetics 169, 2353–2370 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Besenbacher S et al. , Nat. Commun 6, 5969 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rands CM, Meader S, Ponting CP, Lunter G, PLOS Genet 10, e1004525 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesecque Y, Keightley PD, Eyre-Walker A, Genetics 191, 1321–1330 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stearns SC, Byars SG, Govindaraju DR, Ewbank D, Nat. Rev. Genet 11, 611–622 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Larsen EC, Christiansen OB, Kolte AM, Macklon N, BMC Med 11, 154 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.