Abstract
Introduction
Bovine Norovirus (BoNeV) which has been confirmed in Asia, America, and Europe, seems to be distributed worldwide, even though only reported from a number of countries. Bovine noroviruses are predominantly detected in diarrhoeic animals rather than neboviruses. The study reveals the importance of noro- and neboviruses in early age diarrhoea of calves.
Material and Methods
A total of 127 stool samples were collected from three provinces located in the central region of Turkey. Samples were subjected to nucleic acid isolation and reverse transcription and polymerase chain reaction (PCR). Positive samples were sequenced and analysed.
Results
According to PCR, five samples (3.93%) were found to be positive for bovine norovirus while 32 (25.19%) samples were found to be positive for bovine nebovirus. Phylogenetic analysis indicated that the novel Turkish norovirus strains were found to be of genotype III.2 and all novel neboviruses were substituted under Nebraska-like strains.
Conclusion
Although predominantly bovine noroviruses are detected worldwide, the study indicated that bovine neboviruses were more prevalent in the studied area. We suggest that bovine neboviruses are more frequently responsible for calf diarrhoea than supposed by virologists. This is also the first report of neboviruses other than Kirklareli virus which is distantly related to neboviruses detected in Turkey.
Keywords: calf, diarrhoea, nebovirus, bovine norovirus, Turkey
Introduction
The Caliciviridae family includes five genera (Norovirus, Sapovirus, Lagovirus, Vesivirus, and Nebovirus) according to the International Committee on Taxonomy of Viruses (ICTV). In addition, two novel genera have recently been proposed: Recovirus which was formerly known as Tulanevirus, and St-Valérien-like porcine viruses known as Valovirus (5, 10). Caliciviruses have been detected in a number of organisms such as humans, cattle, swine, chickens, fish, and amphibians. These viruses consist of a non-enveloped, hexagonal/spherical capsid about 35–40 nm in diameter and a linear single-strand positive-sense RNA genome which is 7 to 8.5 kb in size (12).
The first bovine enteric caliciviruses were described in the late seventies in Europe. After the first description, Jena-like (GIII.1) and Newbury2-like (GIII.2) noroviruses have been increasingly detected in cattle with enteric or respiratory diseases worldwide (8, 17, 21, 22). After full length genome analysis of Bo/Newbury1/76/UK and Bo/Nebraska/80/US strains, they are designated under a novel genus as Nebovirus. Neboviruses are also bovine enteric caliciviruses associated with enteric diseases in calves (3), in the aetiology of which several reports claim noroviruses and neboviruses may play a role (18).
In Turkey, bovine noroviruses were detected in diarrhoeic calves for the first time in 2011. Among 70 stool samples collected from calves, 6/70 (8.5%) samples were found to be positive in the Marmara region of Turkey (23). In another survey study, 4/235 (1.7%) samples were found to be positive for bovine norovirus in 2016 (7). There is no report of neboviruses in Turkey other than one describing a newly detected virus, designated as Kirklareli virus which is distantly related to neboviruses and lagoviruses (1).
The aims of the study are to examine bovine norovirus (BoNoV) and nebovirus (BoNeV) in diarrhoeic calves in Turkey and to suggest a new RT-PCR assay for the detection of these viruses. We believe that this is also the first report of bovine nebovirus detected in Turkey.
Material and Methods
Samples and RNA isolation A total of 127 stool samples from diarrhoeic calves up to one month of age were collected by rectal swab during a one-year period (2014–2015) from three cities (Sivas, Malatya, and Elaziğ) located in inland Central Turkey (Fig. 1). Collected stool samples were transported to the laboratory just after the sampling and stored at −80ºC until being subjected to RNA isolation.
Fig. 1.
Sampling area
Faecal samples were diluted 1:10 with 1 M phosphate buffered saline and centrifuged for 5 min at 5,000 rpm to remove large cellular debris. After the centrifugation, supernatants were submitted to a nucleic acid extraction procedure using a GF-1 Viral Nucleic Acid Extraction Kit (Vivantis Technologies, Malaysia) according to the manufacturer’s instructions. Eluted nucleic acids were stored at −80ºC until use.
Reverse transcription polymerase chain reaction (RT-PCR) The cDNA synthesis was carried out in a 25 μL final volume containing 4 μL of RNA extract, 10 mM of deoxynucleoside triphosphate (dNTP), 2.5 μL of 10× RT buffer (50 mM Tris-HCl (pH 8.3 at 25°C), 75 mM of KCl, 3 mM of MgCl2, and 10 mM of DTT), 50 ng of the random hexamer, 40 U of RNasin, and 200 U of M-MuLV Reverse Transcriptase RNase H- (Vivantis, Germany). The reverse transcription was performed at 37°C for 1 h. Obtained cDNA samples were amplified with two sets of primers which were Nebo2F/Nebo1R and BoNoV851-F/BoNoV1350-R, and their sequences are presented in Table 1
Table 1.
Oligonucleotide data used in the study
| Virus | Name | Sequence | Amplicon size (bp) | Location | Reference |
|---|---|---|---|---|---|
| Nebo2F | TYTAYGACGGGCGCTWTTATGG | ||||
| Nebovirus | Nebo1R | AGACAGTGCCGAAAAGGGTGTA | 282 | 4984–5265a | This study |
| BoNoV851-F | ATGTGGTVCAGGCNAACAGCTA | ||||
| BoNoV1350-R | ACTACCTTCCCACARHGACARA | 500 | 4538–5037b | This study | |
| Norovirus | IIIBovNo-F | CGCTCCATGTTYGCBTGG | |||
| IIIBovNo-R | ATCAGCACATGRGGRAACTG | 516 | 4972–5487c | (13) |
PCR was conducted in a 50 μl final volume using 5 μL of the RT reaction mixture as a template. The PCR mixture contained 5 μL of 10× PCR buffer, 10 mM of dNTP, 10 pmol/μL of each set of sense/antisense primers, and 5 U of Taq DNA polymerase (Vivantis, Germany). PCR was conducted under the following conditions: 1 cycle at 95°C for 2 min; 40 cycles at 94°C for 40 s, 5°C below primer melting temperature for 30 s, and 72°C for 40 s; and a final elongation step at 72°C for 10 min. PCR products were analysed by electrophoresis in 2% agarose gels stained with ethidium bromide.
Sequencing and phylogenetic analysis The PCR amplicons were purified with the Wizard SV Gel and PCR Clean-Up System (Promega, USA) and sequenced using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, USA) on an ABI 3100 automated sequencer (Applied Biosystems, USA). All sequenced products were used to obtain phylogenetic data. Partial sequences of polyprotein genes were compared with other Calicivirus sequence data which were provided by the National Center for Biotechnology Information (NCBI: https://www.ncbi.nlm.nih.gov/) Sequence alignment and phylogenetic analysis being based on both nucleotide sequences was achieved with the use of Unipro UGENE, version 1.21 (15).
Table 2.
GenBank accession numbers of noro- and nebovirus strains used in genetic analysis
| Genus | Strain | Accession number | Genotype |
|---|---|---|---|
| Mu/NoV/GV/MNV1/2002/USA | AY228235.2 | GV | |
| NLV/SaintCloud/624/1998/US | AF414427.1 | GIV. 1 | |
| NLV/Brattleboro/321/1995/US | AF414415.1 | GII.3 | |
| Toronto/ CAN | U02030 | GII.3 | |
| SOV/ UK | L07418 | GI.2 | |
| NV/USA | M87661 | GI.1 | |
| Bo/Thirsk10/00/UK | AY126468.2 | GIII. 1 | |
| Bo/Jena/DE | AJ011099 | GIII. 1 | |
| Bo/ DuzceN2/2010/TUR | KF218823.1 | GIII.2 | |
| Bo/SivasN6/2011/TUR | KF218824.1 | GIII.2 | |
| bovine/ DijonA077/06/FR | GU259577.1 | GIII.2 | |
| bovine/ DijonA436/08/FR | GU259579.1 | GIII.2 | |
| Bo/Newbury2/1976/UK | AF097917.5 | GIII.2 | |
| Bo/NoV/JN-MA156/04/Korea | DQ912792.1 | GIII.2 | |
| Bo/ Dumfries/94/UK | AY126474.2 | GIII.2 | |
| Bo/AdiyamanN6/2011/TUR | KF218825.1 | GIII.2 | |
| Bo/BalikesirN9/2009/TUR | KF218822.1 | GIII.2 | |
| bovine/GIII.2/471_0790/2005/NOR | FM242193.1 | GIII.2 | |
| bovine/GIII.2/718_0785/2006/NOR | FM242191.1 | GIII.2 | |
| bovine/GIII.2/312_0529/2006/NOR | FM242186.1 | GIII.2 | |
| bovine/GIII.2/240_0243/2005/NOR | FM242189.1 | GIII.2 | |
| Bo/Nov-33/USA/2010 | JN585051.1 | GIII.2 | |
| BoNoV/ Bolat7/2016/TR | MG022084 | GIII.2 | |
| BoNoV/Bolat85/2016/TR | MG022085 | GIII.2 | |
| Bo/Nov-45/USA/2010 | JN585060.1 | GIII.2 | |
| Bo/Nov-6/USA/2010 | JN585033.1 | GIII.2 | |
| Norovirus | Bo/Nov-16/USA/2010 | JN585041.1 | GIII.2 |
| Bo/Nov-10/USA/2010 | JN585036.1 | GIII.2 | |
| Bo/Nov-2/USA/2010 | JN585029.1 | GIII.2 | |
| Bo/Nov-1/USA/2010 | JN585028.1 | GIII.2 | |
| Bo/Nov-13/USA/2010 | JN585038.1 | GIII.2 | |
| Bo/NoV/JN-MA140/04/Korea | DQ912791.1 | GIII.2 | |
| Bo/NoV/JN-MA302/04/Korea | DQ912797.1 | GIII.2 | |
| Bo/NoV/JN-MA271/04/Korea | DQ912795.1 | GIII.2 | |
| Bo/NoV/JN-SA296/04/Korea | DQ912796.1 | GIII.2 | |
| bovine/ DijonA407/08/FR | GU259575.1 | GIII.2 | |
| Bo/MonastirB139/2009/TUN | JN418489.1 | GIII.2 | |
| bovine/Belgium/B52/2002/Be | AY686489.2 | GIII.2 | |
| bovine/XJSHZ148/2011/CN | JQ905268.1 | GIII.2 | |
| BEC200/IT | HM745911.1 | GIII.2 | |
| Bo/Penrith55/00/UK | AY126476.1 | GIII.2 | |
| BEC830/IT | HM745913.1 | GIII.2 | |
| Bovine/ Wasme/B200/2003/Be | AY686494.1 | GIII.2 | |
| Bovine/Wasme/B199/2003/Be | AY686493.1 | GIII.2 | |
| CV186-OH | AF542084.1 | GIII.2 | |
| CV95-OH | AF542083.1 | GIII.2 | |
| Bo/ 21Z90/GIII.2/2012/Iran | KM650007.1 | GIII.2 | |
| BEC483/IT | HM745909.1 | GIII.2 | |
| Bovine/ Mohiville/B 123/2002/Be | AY686490.2 | GIII.2 | |
| Bo/4Z90/GIII.2/2012/Iran | KM650013.1 | GIII.2 | |
| Bo/6P90/GIII.2/2011/Iran | KM650011.1 | GIII.2 | |
| Bo/ 58B89/GIII.2/2010/Iran | KM650009.1 | GIII.2 | |
| Bo/27T90/GIII.2/2011/Iran | KM650012.1 | GIII.2 | |
| Bo/BEC/PenrithC39/2000/UK | DQ228162 | ||
| Bo/M3641/2000/HUN | JX018212 | ||
| bovine/ DijonA290-2/07/FR | GU259551 | ||
| bovine/DijonA386/08/FR | GU259561 | ||
| bovine/ DijonA364/08/FR | GU259556 | ||
| Bo/Nebraska/80/US | AY082891 | ||
| bovine/DijonA058/05/FR | GU259542 | ||
| Bo/BEC/Penrith140/2000/UK | DQ228159 | ||
| Bo/ BEC/Starcross93/2000/UK | DQ228165 | ||
| Nebovirus | Bo/CV548-OH/2002/US | AY549172 | |
| Bo/ CV531-OH/2002/US | AY549171 | ||
| Bo/ CV526-OH/2002/US | AY549170 | ||
| Bo/ CV504-OH/2002/US | AY549168 | ||
| Bo/ CV562-OH/2002/US | AY549173 | ||
| Bo/BEC/Starcross117/2000/UK | DQ228164 | ||
| Bo/BEC/Penrith142/2000/UK | DQ228160 | ||
| Bo/BEC/Penrith143/2000/UK | DQ228161 | ||
| Bo/ BEC/Penrith 150/2000/UK | DQ228157 | ||
| Bo/Newbury1/76/UK | NC_007916 | ||
| Continued on next page | |||
| Bo/ DijonA216/06/FR | FJ687386 | ||
| BoNeV/ Sivas100/2016/TR | MG022086 | ||
| BoNeV/ Sivas120/2016/TR | MG022087 | ||
| BoNeV/ Sivas54/2016/TR | MG022088 | ||
| BoNeV/ Sivas60/2016/TR | MG022089 | ||
| BoNeV/ Sivas63/2016/TR | MG022090 | ||
| BoNeV/ Sivas64/2016/TR | MG022091 | ||
| BoNeV/ Sivas75/2016/TR | MG022092 |
Results
Prevalence of BoNoV and BoNeV among diarrhoeic calves in Turkey From tested 127 samples, using both the CDC’s recommendation and our generic primers, five of them were found to be positive for BoNoV (3.93%) and 32 (25.19%) were positive for BoNeV. The molecular survey was based on two primers designed anteriorly, BoNoV851-F/1350R amplifying a 500 bp fragment and Nebo2-F/1-R amplifying a 282 bp fragment, these fragments being in the genomes of noroviruses and neboviruses, respectively.
Assesment of genetic diversity Genetic diversities between obtained amplicons and similarity to other reported sequences were investigated for two caliciviruses. These two partial sequences of 274 bp and 422 bp represented a polyprotein gene of BoNeV and both RdRP and major capsid protein genes of BoNoV, respectively.
Partial sequences indicated that NeV/Sivas60/ 2017/TR, NeV/Sivas63/2017/TR, and NeV/Sivas64/ 2017/TR strains clustered together with 99.82% identity, while four other strains were clustered in another branch. The NeSV/Sivas75/2017/TR strain showed the highest identity with NeV/Sivas100/2017/TR (99.82%), and next highest identity with NeV/ Sivas120/2017/TR strain (99.45%). NeV/Sivas54/2017/TR strain was located outside of the other six NeV strains (Fig. 2). The NeV/Sivas60/2017/TR, NeV/Sivas63/2017/TR, and NeV/Sivas64/2017/TR triplet were also closely related to a partial polyprotein gene of a Nebraska-like strain, PenrithC39/2000/UK (97.99%).
Fig. 2.
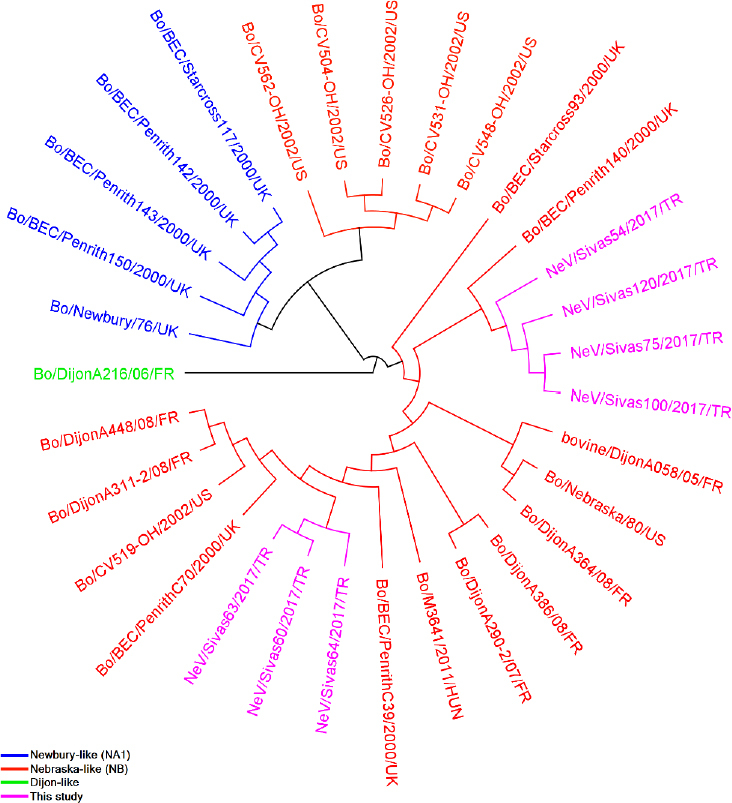
cladogram representing the phylogenic tree of bovine neboviruses based on 274 bp sequences. Each coloured branch indicates a different genotypes. Tree construction was built with use of Unipro UGENE v.1.21 software (14)
Two novel norovirus strains, Bo/NoV/Bolat7/ 2016/TR and Bo/NoV/Bolat85/2016/TR, were clustered together with 96.94% identity and merged with the Bo/Nov-6/USA/2010 and Bo/Nov-45/USA/2010 strains previously reported in the USA. Other related strains, Bo/Nov-1, -2, -10, -13 and -16, also intersected on the same root. On the other hand, comparison with reference strains implicated that these novel strains were in the GIII.2 genogroup (Fig. 3). Other norovirus members which were previously reported in Turkey were also matched in phylogenetic analysis. Bo/Balikesir/N9/ 2009/TUR showed the highest identity with the novel strains (90.80%) while Bo/SivasN6/2011/TUR showed 88.65%, Bo/DuzceN2/2010/TUR – 87.42%, and Bo/AdiyamanN6/2011/TUR – 86.81% identities.
Fig. 3.
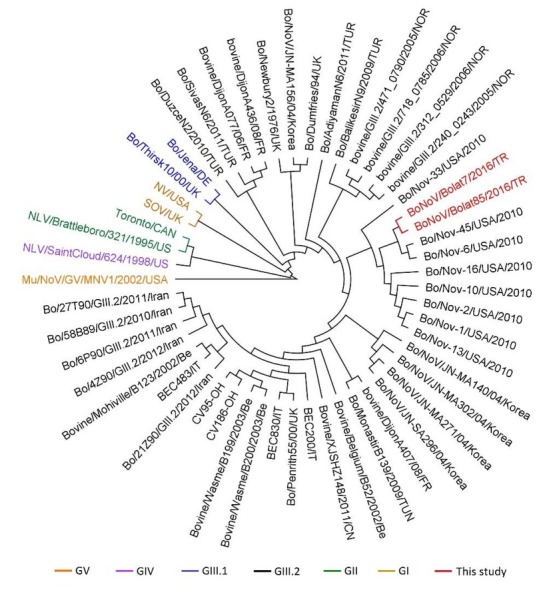
Norovirus phylogenic tree constructed on 422 bp sequences. Alignment and classification of all genome data were performed by the UPGMA method with Unipro UGENE v. 1.21 software (14). Strains were coloured according to their genogroups. Novel norovirus sequences are shown as ruby red colour
Discussion
In this study, the currency of two emerging pathogens related to bovine enteric caliciviruses has been evaluated. The pathogens emerged in the three Turkish provinces of Sivas, Malatya, and Elaziğ. We designed pairs of primers for each virus and used RT-PCR to survey the existing prevalence of these two viruses in diarrhoeic calves. As it is regarded as the “gold standard” for diagnosis, the RT-PCR-based molecular detection was preferred for faecal samples (19).
Bovine Norovirus GIII.2 genogroup is the most widespread genogroup globally (4). Several countries from Europe have reported the prevalence of BoNoV for both diarrhoeic and clinically healthy calves. For instance, in the Netherlands 31.69% (77/243) of randomly selected 1–52-week-old veal calves were found to be positive. On the other hand, 300 stool samples from diarrhoeic calves collected in Belgium indicated 9.33% positivity while 11% of 398 samples were positive in the UK (11, 13). In Ohio state in the USA, 72% positivity was found in randomly selected animals (21). A similar study was conducted in a neighbouring state, Michigan, and 80% prevalence was reported from diarrhoeic samples (22). Several countries also reported that BoNoV could significantly affect calves. In South Korea 9.3% prevalence was found in 645 diarrhoeic faecal samples while in Argentina the prevalence was 3.3% in 90 samples, in Tunisia the prevalence was 16.6% in 169 samples, and in Egypt it was 24% in 25 samples (6, 8, 14, 17).
In Turkey, the first detection of BoNoV was reported in 2011. In this study faecal samples were collected from diarrhoeic calves in the Marmara region of Turkey and prevalence was detected at the rate of 8.57% (24). Later, a retrospective survey was conducted on 235 samples collected between 2009 and 2011 in Turkey and the prevelance rate was 1.57% (7). In this study the prevalence was determined as 3.93% in three provinces located in the Central-Eastern Anatolia, Turkey. This rate appears to be in agreement with previous reports in Turkey and other countries such as Argentina. In contrast, it is lower than in several countries, for instance the USA, the UK, South Korea, and Belgium. Distribution of noroviruses appears to be variable. Also animal age, sampling, and diagnostic methods may affect findings (19).
Nebovirus genus forms a different clade and includes three known genotypes of strains: Nebraska (NB), Newbury (NA1), and the recently suggested DijonA216 (9, 16). Phylogenetic studies indicate that these genotypes have 98% similarity based on their capsid protein sequences (16). We reported seven novel strains being clustered with the Nebraska-like genotypes. This is the first detection of neboviruses in Turkey.
The prevalence of nebovirus has been reported in several countries. For example, in France the prevalence rate was reported at 7% in diarrhoeic cattle (9) while the statistic was 8.4% in the UK (22), 9.2% in South Korea (17), 4.8% in Brazil (2), and 3.3% in Tunisia (8). On the other hand, Smiley et al (21) reported that the prevalence of nebovirus reached 28% in randomly selected animals (21). Surprisingly, we determined the ratio of prevalence at 25.19% from diarrhoeic samples, which was higher than expected.
Non-haemorrhagic enteritis and mild diarrhoea are clinically observable signs in calves infected with BoNoV and BoNeVs. NoV Newbury Agent 2 shows less virulence than Newbury Agent 1 which is known as Nebovirus (19). It was reported that gnotobiotic calves that were experimentally infected with nebovirus developed diarrhoea and intestinal lesions (20).
In summary, bovine norovirus (3.93%) and nebovirus (25.19%) were detected in diarrhoeic calves from three cities of Central Anatolia, Turkey. Phylogenic analysis of noroviruses showed that all of the novel sequences classified into the Norovirus GIII.2 genogroup. All novel neboviruses were grouped together with Nebraska-like strains as well. Nebovirus was also detected with relatively high incidence. The distribution of BoNeV has only been reported in a number of countries. On the other hand, in the literature bovine noroviruses were predominantly detected as compared to neboviruses. Interestingly, bovine neboviruses found to be more prevalent in the studied area. We suggest that bovine neboviruses are more often the cause of calf diarrhoea than is supposed by virologists.
Footnotes
Conflict of Interests Statement The authors declare that there is no conflict of interests regarding the publication of this article.
Financial Disclosure Statement This research was supported by the Cumhuriyet University Scientific Research Project Foundation (CÜBAP) project no: VET-029.
Animal Rights Statement None required.
References
- 1.Alkan F., Karayel I., Catella C., Bodnar L., Lanave G., B ányaiK., Di Martino B., Decaro N., Buonavoglia C., Martella V.. Identification of a bovine enteric calicivirus, Kírklareli virus, distantly related to neboviruses, in calves with enteritis in Turkey. J Clin Microbiol. 2015;53:3614. doi: 10.1128/JCM.01736-15. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Candido M., Alencar A.L.F., Almeida-Querioz S.R., Buzinaro M.G., Munin F.S., Godoy S.H., Livonesi M.C., Fernandes A.M., Sousa R.L.. First detection and molecular characterization of Nebovirus in Brazil. Epidemiol Infect. 2016;144:1876. doi: 10.1017/S0950268816000029. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carstens E.B.. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2009) Arch Virol. 2010;155:133. doi: 10.1007/s00705-009-0547-x. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Felice E., Mauroy A., Pozzo F.D., Thiry D., Ceci C., Di Martino B., Marsilio F., Thiry E.. Bovine noroviruses: A missing component of calf diarrhoea diagnosis. Vet J. 2016;207:53. doi: 10.1016/j.tvjl.2015.10.026. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farkas T., Sestak K., Wei C., Jiang X.. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. J Virol. 2008;82:5408. doi: 10.1128/JVI.00070-08. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferragut F., Vega C.G., Mauroy A., Conceição-Neto N., Zeller M., Heylen E., Uriarte E.L., Bilbao G., Bok M., Matthijnssens J., Thiry E., Badaracco A., Parreño V.. Molecular detection of bovine Noroviruses in Argentinean dairy calves: Circulation of a tentative new genotype. Infect Genet Evol. 2016;40:144. doi: 10.1016/j.meegid.2016.02.034. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gülaçtı İ Sözdutmaz I., Işıdan H.. Molecular characterization of the bovine noroviruses from diarrheic calves in Turkey. Turkish J Vet Anim Sci. 2016;40:428. –. [Google Scholar]
- 8.Hassine-Zaafrane M., Kaplon J., Sdiri-Loulizi K., Aouni Z., Pothier P., Aouni M., Ambert-Balay K.. Molecular prevalence of bovine noroviruses and neboviruses detected in central-eastern Tunisia. Arch Virol. 2012;157:1599. doi: 10.1007/s00705-012-1344-5. –. [DOI] [PubMed] [Google Scholar]
- 9.Kaplon J., Guenau E., Asdrubal P., Pothier P., Ambert-Balay K.. Possible novel Nebovirus genotype in cattle, France. Emerg Infect Dis. 2011;17:1120. doi: 10.3201/eid1706.100038. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.L’Homme Y., Sansregret R., Plante-Fortier É Lamontagne A.M., Ouardani M., Lacroix G., Simard C.. Genomic characterization of swine caliciviruses representing a new genus of Caliciviridae. Virus Genes. 2007;39:66. doi: 10.1007/s11262-009-0360-3. –. [DOI] [PubMed] [Google Scholar]
- 11.Mauroy A., Scipioni A., Mathijs E., Thys C., Thiry E.. Molecular detection of kobuviruses and recombinant noroviruses in cattle in continental Europe. Arch Virol. 2009;154:1841. doi: 10.1007/s00705-009-0518-2. –. [DOI] [PubMed] [Google Scholar]
- 12.Mikalsen A.B., Nilsen P., Frøystad-Saugen M., Lindmo K., Eliassen T.M., Rode M., Evensen O. Characterization of a novel calicivirus causing systemic infection in Atlantic salmon Salmo salar L. Proposal for a new genus of caliciviridae. PLoS One. 2014. 9, 9. doi:10.1371/journal.pone.0107132. [DOI] [PMC free article] [PubMed]
- 13.Milnes A.S., Binns S.H., Oliver S.L., Bridger J.C.. Retrospective study of noroviruses in samples of diarrhoea from cattle, using the Veterinary Laboratories Agency’s Farmfile database. Vet Rec. 2007;160:326. doi: 10.1136/vr.160.10.326. –. [DOI] [PubMed] [Google Scholar]
- 14.Mohamed F.F., Mansour S.M.G., El-Araby I.E., Mor S.K., Goyal S.M.. Molecular detection of enteric viruses from diarrheic calves in Egypt. Arch Virol. 2017;162:129. doi: 10.1007/s00705-016-3088-0. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okonechnikov K., Golosova O., Fursov M., UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics. 2012;28:1166. doi: 10.1093/bioinformatics/bts091. –. [DOI] [PubMed] [Google Scholar]
- 16.Oliver S.L., Asobayire E., Dastjerdi A.M., Bridger J.C.. Genomic characterization of the unclassified bovine enteric virus Newbury agent-1 (Newbury1) endorses a new genus in the family Caliciviridae. Virology. 2006;350:240. doi: 10.1016/j.virol.2006.02.027. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park S.I., Jeong C., Kim H.H., Park S.H., Park S.J., Hyun B.H., Yang D.K., Kim S.K., Kang M.I., Cho K.O.. Molecular epidemiology of bovine noroviruses in South Korea. Vet Microbiol. 2007;124:125. doi: 10.1016/j.vetmic.2007.03.010. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rabenau H.F., Stürmer M., Buxbaum S., Walczok A., Preiser W., Doerr H.W.. Laboratory diagnosis of norovirus: Which method is the best? Intervirology. 2003;46:232. doi: 10.1159/000072433. –. [DOI] [PubMed] [Google Scholar]
- 19.Scipioni A., Mauroy A., Vinjé J., Thiry E.. Animal noroviruses. Vet J. 2008;178:32. doi: 10.1016/j.tvjl.2007.11.012. –. [DOI] [PubMed] [Google Scholar]
- 20.Smiley J.R., Chang K.O., Hayes J., Vinjé J., Saif L.J.. Characterization of an enteropathogenic bovine calicivirus representing a potentially new calicivirus genus. J Virol. 2002;76:10089. doi: 10.1128/JVI.76.20.10089-10098.2002. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smiley J.R., Hoet A.E., Tråvén M., Tsunemitsu H., Saif L.J.. Reverse transcription-PCR assays for detection of bovine enteric caliciviruses (BEC) and analysis of the genetic relationships among BEC and human caliciviruses. J Clin Microbiol. 2003;41:3089. doi: 10.1128/JCM.41.7.3089-3099.2003. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wise A.G., Monroe S.S., Hanson L.E., Grooms D.L., Sockett D., Maes R.K.. Molecular characterization of noroviruses detected in diarrheic stools of Michigan and Wisconsin dairy calves: circulation of two distinct subgroups. Virus Res. 2004;100:165. doi: 10.1016/j.virusres.2003.11.014. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yilmaz A., Bostan K., Altan E., Muratoglu K., Turan N., Tan D., Helps C., Yilmaz H.. Investigations on the frequency of norovirus contamination of ready-to-eat food items in Istanbul, Turkey, by using real-time reverse transcription PCR. J Food Prot. 2011;74:840. doi: 10.4315/0362-028X.JFP-10-475. –. [DOI] [PubMed] [Google Scholar]
- 24.Yilmaz H., Turan N., Altan E., Bostan K., Yilmaz A., Helps C.R., Cho K.O.. First report on the phylogeny of bovine norovirus in Turkey. Arch Virol. 2011;156:143. doi: 10.1007/s00705-010-0833-7. –. [DOI] [PubMed] [Google Scholar]


