A new alga-bacterium model system sheds light on mortality factors affecting phytoplankton bloom demise and sulfur cycling.
Abstract
Emiliania huxleyi is a bloom-forming microalga that affects the global sulfur cycle by producing large amounts of dimethylsulfoniopropionate (DMSP) and its volatile metabolic product dimethyl sulfide. Top-down regulation of E. huxleyi blooms has been attributed to viruses and grazers; however, the possible involvement of algicidal bacteria in bloom demise has remained elusive. We demonstrate that a Roseobacter strain, Sulfitobacter D7, that we isolated from a North Atlantic E. huxleyi bloom, exhibited algicidal effects against E. huxleyi upon coculturing. Both the alga and the bacterium were found to co-occur during a natural E. huxleyi bloom, therefore establishing this host-pathogen system as an attractive, ecologically relevant model for studying algal-bacterial interactions in the oceans. During interaction, Sulfitobacter D7 consumed and metabolized algal DMSP to produce high amounts of methanethiol, an alternative product of DMSP catabolism. We revealed a unique strain-specific response, in which E. huxleyi strains that exuded higher amounts of DMSP were more susceptible to Sulfitobacter D7 infection. Intriguingly, exogenous application of DMSP enhanced bacterial virulence and induced susceptibility in an algal strain typically resistant to the bacterial pathogen. This enhanced virulence was highly specific to DMSP compared to addition of propionate and glycerol which had no effect on bacterial virulence. We propose a novel function for DMSP, in addition to its central role in mutualistic interactions among marine organisms, as a mediator of bacterial virulence that may regulate E. huxleyi blooms.
INTRODUCTION
Phytoplankton are unicellular, photosynthetic microorganisms that contribute to about half of the estimated global net primary production and therefore serve as the basis of the marine food web (1). Biotic interactions can control the fate of phytoplankton blooms in the ocean, namely, predation by zooplankton, viral infections, and potentially algicidal activity of bacteria (2–4). One bacterial group highly associated with phytoplankton blooms is the Roseobacter clade (α-proteobacteria) (5–7), which inhabits diverse marine environments and has a wide variety of metabolic capabilities (8–11). Moreover, Roseobacters have been found to have a range of direct interactions, from cooperative to pathogenic, with phytoplankton species (12–15). These interactions are thought to be mediated by secreted infochemicals (16). Infochemical signaling occurs within the phycosphere, the microenvironment that surrounds algal cells where molecules can accumulate to relatively high effective concentrations (17, 18). The organosulfur compound dimethylsulfoniopropionate (DMSP), as well as its metabolic products, plays a key role in trophic-level interactions (16) and was suggested to act as an infochemical within the phycosphere (19). It is produced by diverse phytoplankton species and is known to mediate algal-bacterial interactions by acting as a chemoattractant (20, 21) and as sulfur and carbon sources for bacterial growth (14, 22, 23).
Emiliania huxleyi is a cosmopolitan coccolithophore species that forms massive annual blooms and plays an important role in the global carbon cycle (24, 25). E. huxleyi produces and accumulates DMSP intracellularly (up to 250 mM) (26). It harbors the gene alma1 that encodes a DMSP-lyase responsible for high production of the volatile metabolic product dimethyl sulfide (DMS) (27). Therefore, E. huxleyi blooms contribute to DMS emission to the atmosphere and are thought to largely affect the global sulfur biogeochemical cycle (28). Once emitted to the atmosphere, DMS can undergo oxidation and induce subsequent formation of cloud condensation nuclei (29). The turnover of E. huxleyi blooms is often mediated by infection of E. huxleyi virus that leads to rapid lysis of host cells (3, 30, 31). During the demise of E. huxleyi blooms, an increase in bacterial abundance is observed (32, 33); however, bacterial regulation of the fate of phytoplankton blooms and the cellular mechanisms governing it are largely unknown (4, 6, 7, 34).
Activity of algicidal bacteria can be mediated by physical attachment (15, 34) or by secretion of toxins or hydrolytic exo-enzymes (12, 35) or by combining both strategies (36). For example, chemical cues from E. huxleyi trigger the production of roseobacticides by Phaeobacter inhibens, which leads to algal cell death (12, 37). Although co-occurrence of algicidal bacteria with their algal host was demonstrated in the environment (15, 34), there is still limited knowledge on how these algicidal interactions are manifested and what their impact is on phytoplankton blooms.
In the current work, we isolated a Sulfitobacter strain (D7) from a North Atlantic E. huxleyi bloom. We established a robust coculturing system in which Sulfitobacter D7 exhibited algicidal activity against E. huxleyi while consuming algal DMSP and producing high amounts of volatile organic sulfur compounds (VOSCs). We further examined how the level of DMSP exudation by a suite of E. huxleyi strains may affect their differential susceptibility to Sulfitobacter D7 infection. In a complementary approach, we show that addition of DMSP promoted bacterial pathogenicity against E. huxleyi in a dose-dependent manner and induced susceptibility in a resistant algal strain. Finally, we discuss the routes by which DMSP can promote bacterial virulence and the potential role of pathogenic bacteria in regulating algal bloom dynamics.
RESULTS
We obtained a bacterial consortium associated with copepods collected during an E. huxleyi bloom in the North Atlantic with the notion that grazers co-ingest microorganisms that interact with the algal prey (Fig. 1A) (38). Inoculation of this copepod-associated microbiome (CAM) into E. huxleyi 379 cultures led to algal cell death. Upon application of antibiotics, the effect of CAM on E. huxleyi was abolished (fig. S1A). This provided a first indication for the presence of pathogenic bacteria in CAM.
Fig. 1. Coculturing of E. huxleyi with Sulfitobacter D7 isolate exhibits distinct phases of pathogenicity.
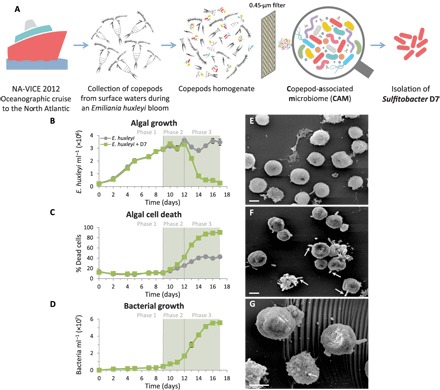
(A) A scheme describing the origin of the CAM bacterial consortium and isolation of Sulfitobacter D7 from an E. huxleyi bloom in the North Atlantic. (B to D) A detailed time course of E. huxleyi 379 monocultures (gray line) and during coculturing with Sulfitobacter D7 (green line). The following parameters were assessed: (B) algal growth, (C) algal cell death, and (D) bacterial growth. No bacterial growth was observed in control cultures. The green background represents the presence of a pungent scent in cocultures. Algae-bacteria coculturing had distinct dynamics characterized by defined phases (1 to 3) of pathogenicity. (E to G) Scanning electron microscopy (SEM) images of (E) uninfected E. huxleyi 379 and (F and G) Sulfitobacter D7–infected E. huxleyi cells at phase 2 (scale bars, 2 μm). Arrows in (F) point to Sulfitobacter D7 attachment to E. huxleyi cells. Arrow in (G) points to a membrane blebbing–like feature. Results depicted in (B) to (D) represent average ± SD (n = 3). Error bars smaller than the symbol size are not shown. Statistical differences in (B) to (D) were tested using repeated-measures analysis of variance (ANOVA). P < 0.001 for the differences between control and cocultures.
A new algicidal E. huxleyi-bacterium model system
To study the interaction of E. huxleyi with a specific pathogenic bacterium, we isolated from CAM a Sulfitobacter (termed Sulfitobacter D7) that belongs to the Roseobacter clade and sequenced its genome (GenBank accession numbers CP20694 to CP20699) (figs. S1B and S2). Sulfitobacter D7 showed algicidal effects against E. huxleyi cultures upon coculturing. Time-course experiments of E. huxleyi cultures incubated with 103 Sulfitobacter D7 ml−1 revealed a three-phase dynamics (Fig. 1, B to D). In phase 1, both control and cocultures grew exponentially, until day 9, followed by a stationary phase (namely, phase 2) (Fig. 1B). During phase 3 (12 to 15 days) of coculturing, algal abundance declined rapidly, and algal cell death occurred in ~90% of the population, while in control cultures, it reached only ~40% during stationary phase (Fig. 1C). Rapid bacterial growth coincided with algal cell death during coculturing, reaching 5.5 × 107 bacteria ml−1 by day 16 (overall growth of four orders of magnitude), while we observed no bacteria in control cultures (Fig. 1D). During phases 2 and 3 of coculturing, we reproducibly detected a distinct pungent scent of volatiles that emerged only from Sulfitobacter D7–treated cultures (Fig. 1, B to D, represented by the green background). E. huxleyi cultures incubated with CAM exhibited features similar to those of Sulfitobacter D7 cocultures (fig. S1, C to E, and text S1). Moreover, Sulfitobacter D7 abundance during coculturing with CAM increased steadily by three orders of magnitude, as quantified by quantitative polymerase chain reaction (qPCR) (fig. S1E, inset). This finding strengthens the possible role of Sulfitobacter D7 as a major pathogenic component within CAM.
We examined the specificity of algicidal activity in Sulfitobacter D7 by comparing the dynamics of coculturing with an additional bacterial strain, Marinobacter D6, which was also isolated from CAM (fig. S3). Although bacterial growth was prominent and reached similar concentrations to those of Sulfitobacter D7, the algal culture persisted in stationary growth and no increase in algal cell death was observed. Here, we used the term “bacterial infection” to describe the algicidal impact of Sulfitobacter D7 on E. huxleyi.
Scanning electron microscopy (SEM) analysis of E. huxleyi–Sulfitobacter D7 interaction revealed membrane blebbing–like features in infected E. huxleyi cells at phase 2 of the infection (Fig. 1G), likely corresponding to early stages of cell death (Fig. 1C). Furthermore, some E. huxleyi cells had bacteria attached to their surface in a polar manner (Fig. 1F).
With an attempt to shed light on the ecological significance of this interaction, we analyzed samples collected during the North Atlantic E. huxleyi bloom from which Sulfitobacter D7 was isolated [North Atlantic Virus Infection of Coccolithophore Expedition (NA-VICE) cruise, 2012] (31, 39). We detected E. huxleyi cells by microscopic observations, flow cytometry, molecular analyses, and satellite imagery of chlorophyll fluorescence and particulate inorganic carbon, representing the calcium carbonate exoskeleton of E. huxleyi (31). Using qPCR, we detected the coexistence of E. huxleyi and Sulfitobacter bacteria in the water column. E. huxleyi cells were prevalent in surface waters, peaking at 30 to 40 m, with cell concentrations typical for oceanic E. huxleyi blooms (up to ~103 cells ml−1) (Fig. 2). Sulfitobacter bacteria were abundant mainly at the surface, reaching a maximum level of 8.4 × 103 bacteria ml−1, and were also found in deeper waters (Fig. 2). This evidence of co-occurrence during bloom succession, along with the isolation of Sulfitobacter D7 from the same bloom patch, suggests the potential existence of this algicidal interaction during E. huxleyi blooms. Further exploration is needed to determine the extent and impact of this interaction in natural settings. Together, the reproducibility of laboratory cocultures and the natural coexistence of these organisms in the wild lay the foundation for establishing this E. huxleyi–Sulfitobacter D7 system as an attractive, ecologically relevant model for studying algicidal alga-bacterium interaction in the oceans.
Fig. 2. Co-occurrence of E. huxleyi and Sulfitobacter during a natural algal bloom.
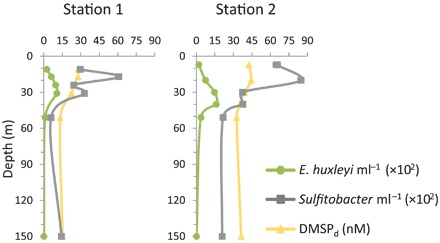
Depth profiles of E. huxleyi and Sulfitobacter abundances and DMSPd concentration at sampling stations in the North Atlantic during an E. huxleyi bloom, July 2012 (NA-VICE cruise). Station 1 (61.8172°N/33.4682°W) was sampled on 3 July, and station 2 (61.5413°N/34.1067°W) was sampled on 5 July. Both stations were within “Early Infection” station according to Laber et al. (39). Results of E. huxleyi and Sulfitobacter quantification represent an average of three technical repeats ± SD. Error bars smaller than the symbol size are not shown.
Characterization of the metabolic basis of E. huxleyi–Sulfitobacter D7 interaction
We sought to reveal the nature of the emitted volatiles during bacterial infection (Fig. 1, B to D, represented by the green background). We performed an untargeted headspace analysis using solid-phase microextraction (SPME) coupled to gas chromatography–mass spectrometry (GC-MS). We detected significant amounts of methanethiol (MeSH) and dimethyl disulfide (DMDS) in the headspace of Sulfitobacter D7– and CAM-infected E. huxleyi cultures, as well as small amounts of dimethyl trisulfide (DMTS) and methyl methylthiomethyl disulfide that did not appear in the headspace of control cultures (fig. S4). A targeted analysis of the major volatile organic sulfur compunds (VOSCs) dissolved in the media showed that DMS, MeSH, and DMDS were present in Sulfitobacter D7–infected E. huxleyi cultures, while only DMS was found in control cultures (Fig. 3A). The concentration of DMS in the media did not significantly differ between control and Sulfitobacter D7–infected cultures throughout the time course of infection (Fig. 3B). In contrast, MeSH and DMDS were detected only in media of infected cultures, as early as phase 1, followed by a sharp increase (>10-fold) during phases 2 and 3 (Fig. 3, C and D). MeSH is known to be readily oxidized to DMDS (fig. S5C) (40) and subsequently to DMTS and methyl methylthiomethyl disulfide during sample handling (41, 42). We therefore consider these volatiles to be part of the MeSH pool.
Fig. 3. A major shift in the composition of VOSCs during the pathogenic phase of Sulfitobacter D7 infection of E. huxleyi.
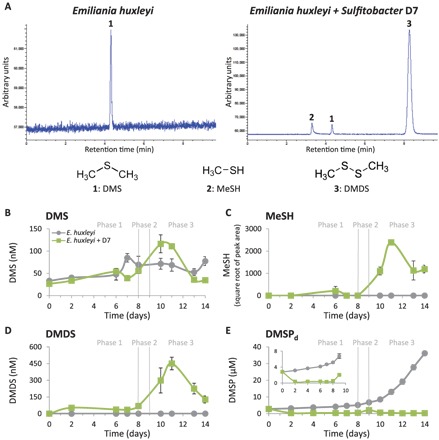
(A) Representative GC–flame photometric detector (GC-FPD) chromatograms of VOSCs detected in media of monocultures and Sulfitobacter D7–infected E. huxleyi 379 cultures at phase 3 (t = 11 days). Peaks are marked by numbers that represent different compounds, as indicated below. DMDS is presumably an oxidation product of MeSH (fig. S5C) and therefore considered as part of the MeSH pool. (B to E) Quantification of VOSCs; (B) DMS, (C) MeSH, (D) DMDS, and (E) dissolved DMSP (DMSPd) in media of control (gray line) and Sulfitobacter D7–infected (green line) E. huxleyi 379 cultures during defined phases (1 to 3), as described in Fig. 1. Inset in (E): zoomed-in view of DMSPd concentration during phases 1 and 2. Algal growth, algal cell death, and bacterial growth are presented in fig. S9. Results depicted in (B) to (E) represent average ± SD (control, n = 4; Sulfitobacter D7–infected, n = 2). Error bars smaller than the symbol size are not shown. Statistical differences in (B) to (E) were tested using repeated-measures ANOVA. P < 0.001 for the differences between control and cocultures, except for DMS.
MeSH and DMS are known products of competing catabolic pathways of DMSP (fig. S6) (43). The “DMSP demethylation” pathway involves enzymatic demethylation of DMSP [encoded by dmdA genes (44)] and subsequent production of MeSH, which can be incorporated into bacterial proteins (45). The “DMSP-cleavage” pathway is catalyzed by a DMSP-lyase enzyme [encoded by various bacterial ddd genes (43) and by E. huxleyi alma1 gene (27)] and involves cleavage of DMSP and release of DMS. Since both MeSH and DMS were produced during Sulfitobacter D7 infection, we measured the concentration of their common precursor, dissolved DMSP (DMSPd), in the media of E. huxleyi cultures. DMSPd accumulated from ~2 to ~36 μM in control E. huxleyi cultures as they aged (Fig. 3E). In contrast, upon Sulfitobacter D7 infection, DMSPd concentration was comparatively low, reaching a maximal level of ~2 μM (Fig. 3E). This implies that algae-derived DMSPd was consumed by Sulfitobacter D7 during coculturing.
To identify pathways involved in DMSP catabolism, we performed gene mining of Sulfitobacter D7 genome, which revealed all the putative genes of the DMSP demethylation pathway and none of the known genes in the DMSP-cleavage pathway (fig. S6). Accordingly, Sulfitobacter D7 grown in monocultures in the presence of DMSP or in algae-derived conditioned medium (CM) consumed DMSP and produced MeSH but not DMS (text S2 and table S1). We suggest that during infection, Sulfitobacter D7 consume E. huxleyi–derived DMSP and produce MeSH, which can be assimilated into bacterial biomass. DMS found in both control and infected E. huxleyi cultures was most likely a product of the activity of the DMSP-lyase, Alma1, encoded by E. huxleyi (27).
DMSPd was detected during the E. huxleyi bloom that we sampled in the North Atlantic Ocean (Fig. 2). The concentrations ranged between 13 and 45 nM, which were comparable with previous studies of E. huxleyi blooms (5). The presence of this metabolic currency along with E. huxleyi and Sulfitobacter suggests that this interaction, mediated by algal DMSP, may occur in the natural environment.
Role of DMSP in algicidal E. huxleyi–Sulfitobacter D7 interaction
We further aimed to assess the interplay between accumulation of algae-derived DMSPd and the dynamics of Sulfitobacter D7 growth and pathogenicity. We used a suite of axenic E. huxleyi strains that differentially accumulated DMSPd in media of monocultures (Fig. 4A). This difference was most prominent in stationary phase (11 days) when media of E. huxleyi strain 379 had the highest DMSPd concentration, followed by strains 1216, 373, and 2090 (72, 27, 13, and 5.5 μM, on average, respectively). Inoculation of Sulfitobacter D7 into CM derived from all E. huxleyi strains in stationary phase (11 days) revealed that Sulfitobacter D7 consumed alga-derived DMSP (Table 1), and bacterial growth was highly correlated with initial DMSPd concentration (Fig. 4B). CM derived from E. huxleyi 379 had the highest bacterial yield after 24 hours of growth followed by CM from E. huxleyi 1216, 373, and lastly 2090 (1.1 × 108, 7.5 × 107, 2 × 107, and 1.7 × 107 bacteria ml−1, on average, respectively) (Fig. 4B). Intriguingly, we detected substantial variability in infection dynamics among E. huxleyi strains (Fig. 4, C to F). All strains were infected at various degrees, presenting all three phases of pathogenicity (Fig. 4, C to E), except for E. huxleyi 2090 that was unaffected by the presence of bacteria (Fig. 4F and table S2). Pronounced differences were observed for the dynamics of phase 3 in which E. huxleyi 379 cultures declined most rapidly (within 5 days), followed by strains 1216 (within 7 days) and 373 (within 10 days). In all cases, the decline in algal abundance correlated with the growth of bacteria that reached ~108 bacteria ml−1, corresponding to phase 3, except in strain 2090 where bacterial abundance was 10-fold lower. Moreover, the duration of phase 3 had an inverse correlation with the concentration of DMSPd in the media of uninfected E. huxleyi strains (table S2). Namely, strains that accumulated more DMSPd in the medium during algal growth in monocultures were more susceptible to Sulfitobacter D7 infection during coculturing. This raised the hypothesis that DMSP not only is an important carbon and reduced sulfur source for bacterial growth but also may promote Sulfitobacter D7 pathogenicity against E. huxleyi.
Fig. 4. E. huxleyi strain-specific DMSP exudation and susceptibility to Sulfitobacter D7.
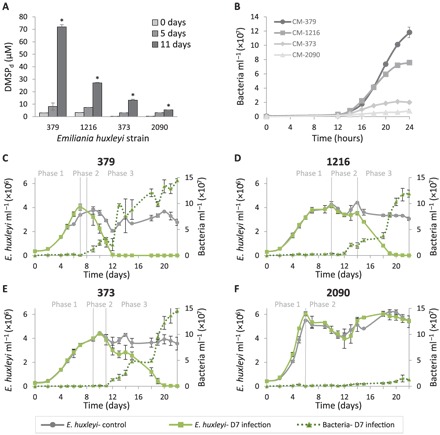
(A) Concentration of DMSPd in media of monocultures of four axenic E. huxleyi strains (379, 1216, 373, and 2090) at different stages of growth. (B) Growth curves of Sulfitobacter D7 in CM obtained from E. huxleyi cultures from (A) at 11 days of growth. (C to F) Differential dynamics of cocultures of Sulfitobacter D7 with a suite of E. huxleyi strains. Time course of algal and bacterial growth (left and right axes, solid and dotted lines, respectively) in monocultures (gray) and Sulfitobacter D7–infected (green) cultures of E. huxleyi strains (C) 379, (D) 1216, (E) 373, and (F) 2090. No bacterial growth was observed in control cultures. Defined phases (1 to 3) of pathogenicity are denoted. Results represent average ± SD (n = 3). Error bars smaller than the symbol size are not shown. Statistical differences between strains in (A) were tested using one-way ANOVA for each time point, followed by a Tukey post hoc test. *P < 0.001 for the differences between all strains on day 11. P values in (B) to (F) were calculated using repeated-measures ANOVA, followed by a Tukey post hoc test. (B) P < 0.001 for the differences between all CM. (C to E) P < 0.001 for the differences between control and cocultures. (F) P < 0.001 only for the differences in bacterial growth between control and cocultures.
Table 1. Concentration of DMSPd and its consumption by Sulfitobacter D7 grown for 24 hours in CM derived from various E. huxleyi strains at 11 days of growth (Fig. 4A).
| DMSPd (μM) |
DMSP consumed (μM)* |
||
| t = 0 hours† | t = 24 hours‡ | ||
| CM-379 | 71.3 | 44.4 ± 1.6 | 26.9 |
| CM-1216 | 26.5 | <0.15§ | 26.3|| |
| CM-373 | 13 | <0.15§ | 12.8|| |
| CM-2090 | 5 | 3.4 ± 0.01 | 1.6 |
*Estimated by subtraction of the concentration at 24 hours from 0 hours.
†Results represent average ± SD (n = 1).
‡Results represent average ± SD (n = 3).
§Not detected. Detection limit was 150 nM.
||Underestimation. Because of detection limits, we assumed a concentration of 150 nM at t = 24 hours.
Intriguingly, the addition of DMSP to E. huxleyi 379 cultures inoculated with Sulfitobacter D7 expedited the dynamics of infection in a dose-dependent manner (fig. S7). Cocultures supplemented with 500 and 100 μM DMSP collapsed after 5 and 7 days, respectively, while cocultures in which DMSP was not added declined only after day 11. Algal monocultures were not affected by the addition of DMSP. To test the specificity of DMSP in promoting bacterial virulence, we supplemented algal monocultures and cocultures with additional 3-carbon substrates, glycerol and propionate (Fig. 5). Once again, the addition of DMSP promoted Sulfitobacter D7 infection dynamics, while glycerol and propionate had a minor effect (Fig. 5A). The DMSP-supplemented cocultures reached phase 2 after only 4 days and completely collapsed at day 8. The cocultures supplemented with glycerol and propionate had similar dynamics to cocultures with no substrate addition; all entered phase 2 at day 5 and fully collapsed at day 12. Bacterial growth in all cocultures was similar until day 5, with slightly more bacteria in the glycerol-treated cocultures at day 3 (Fig. 5B). Although bacterial density was similar between all the substrate-supplemented cultures, the early virulence of Sulfitobacter D7 was invoked only in the presence of DMSP. These results provide a direct link between DMSP and algicidal activity of Sulfitobacter D7 against E. huxleyi.
Fig. 5. DMSP promotes Sulfitobacter D7 virulence toward E. huxleyi.
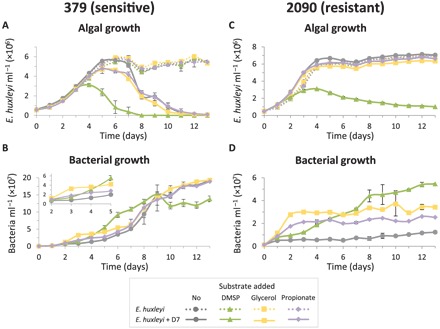
Time course of algal and bacterial growth in cultures of (A and B) the sensitive E. huxleyi strain 379 and (C and D) the resistant E. huxleyi strain 2090, monocultures (dotted lines) and during coculturing with Sulfitobacter D7 (solid lines). Cultures were supplemented at day 0 with 100 μM of the following substrates: DMSP (green, triangle), glycerol (yellow, square), propionate (purple, diamond), or none (gray, circle). Inset in (B): zoomed-in view of bacterial growth at days 2 to 5. No bacterial growth was observed in control cultures. Results represent average ± SD (n = 3). Error bars smaller than the symbol size are not shown. Statistical differences were tested using two-way repeated-measures ANOVA, accounting for infection and the different substrates. (A) P < 0.001 for the differences between control and cocultures and for the differences between the DMSP treatment and the other treatments in cocultures. (B) P < 0.05 only for the differences between the glycerol treatment and the rest of the treatments in cocultures. (C) P < 0.001 for the differences between the DMSP treatment in cocultures and the other treatments. (D) P < 0.01 for the differences between all treatments in cocultures.
We further hypothesized that the observed resistance of E. huxleyi 2090 to Sulfitobacter D7 infection may be explained by the low level of DMSPd in the media of 2090 cultures (Fig. 4A). Therefore, we added exogenous DMSP to E. huxleyi 2090 and examined its susceptibility to bacterial infection. Intriguingly, algal growth arrest was induced at day 4 of E. huxleyi 2090–Sulfitobacter D7 cocultures supplemented with 100 μM DMSP (Fig. 5C). Glycerol and propionate did not affect the dynamics of cocultures at all, although bacterial growth was more prominent compared to the nonsupplemented cocultures (Fig. 5D). High inoculum of Sulfitobacter D7 did not affect E. huxleyi 2090 growth, unless DMSP was present (Fig. 5C). This strengthens the pivotal role of DMSP in mediating Sulfitobacter D7 virulence toward E. huxleyi.
DISCUSSION
A new role for DMSP in algicidal interactions
In this study, we aimed to shed light on the possible role of bacteria as mortality agents during E. huxleyi bloom succession. We established a robust model system for studying the algicidal interactions between Sulfitobacter D7 and E. huxleyi and demonstrated that DMSP produced by the alga is a key metabolite for this interaction. DMSP has many suggested cellular functions, including osmoregulation and antioxidant activity (46, 47), and is often considered as a metabolic currency during mutualistic interactions (12, 14, 21, 22, 48). Our results place DMSP as a mediator of bacterial virulence via several suggested cellular pathways (Fig. 6). First, DMSP promotes growth of Sulfitobacter D7 (fig. S8), and it is consumed and metabolized to MeSH by the bacterial demethylation pathway (Fig. 3). DMSP-degrading bacteria can produce more cellular energy from demethylation rather than cleavage of DMSP (49). In both pathways, there is an increase of reduced carbon, but the assimilation of reduced sulfur into amino acids (methionine and cysteine) can only occur through the demethylation pathway (45). As demonstrated in P. inhibens, these amino acids can subsequently be incorporated into bacterial algicides, roseobacticides, that kill E. huxleyi cells (12, 37). Therefore, DMSP and its metabolic products can promote bacterial virulence by acting as precursors for the synthesis of bacterial algicides.
Fig. 6. Conceptual model of the possible routes in which algal DMSP promotes bacterial virulence in E. huxleyi phycosphere.
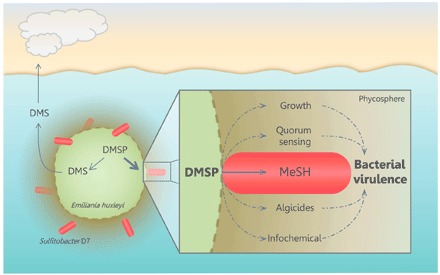
During interaction, Sulfitobacter D7 consumes E. huxleyi–derived DMSP and transforms it into MeSH, which facilitates bacterial growth. DMSP and its metabolic products can promote production of QS molecules (19) and bacterial algicides (37), which were proposed to be involved in bacterial virulence. Furthermore, DMSP may facilitate bacterial chemoattraction to algal cells (20, 21). The algicidal effect of Sulfitobacter and other members of the Roseobacter clade [e.g., P. inhibens (15)] may have a broader-scale impact on the dynamics of E. huxleyi blooms. These blooms are an important source for DMSP and its cleavage product DMS, which is emitted to the atmosphere. By consuming large amounts of DMSP, bacteria may reduce DMS production by the algal DMSP-lyase (Alma1). Accordingly, we propose that the balance between competing DMSP catabolic pathways, driven by microbial interactions, may regulate oceanic sulfur cycling and feedback to the atmosphere.
Roseobacticide biosynthesis by P. inhibens is regulated by quorum sensing (QS) (37), as is virulence of many other pathogenic bacteria (50). Moreover, the production of QS molecules in Roseobacters can be stimulated by DMSP (19). Thus, the involvement of QS may also be applicable in the E. huxleyi–Sulfitobacter D7 system described here. Genomes of Sulfitobacter spp., including Sulfitobacter D7, encode genes involved in N-acyl-l-homoserine lactone (AHL)–based QS (51). It was also shown that a precursor for QS molecules produced by the bacterium Pseudoalteromonas piscicida induced mortality of E. huxleyi in cultures (52). Therefore, biosynthesis of QS molecules can regulate expression of virulence-related genes and may also contribute to pathogenicity by producing intermediate compounds that function as algicides themselves. Further investigation is needed to assess the involvement of QS and algicides in the pathogenicity of Sulfitobacter D7.
DMSP can also mediate Sulfitobacter D7 virulence by acting as a chemotaxis cue toward E. huxleyi phycosphere. Marine bacteria can sense DMSP and use it as a signal for chemotaxis (20) in pathogenesis (53) and symbiosis (21, 48). Sulfitobacter D7 is a motile bacterium, and its genome encodes flagella biosynthesis genes. Therefore, DMSP released from E. huxleyi cells can serve as a cue in which Sulfitobacter D7 can locate algal cells and subsequently attach and consume DMSP (Figs. 1, F and G, and 3E). Previous studies suggested that physical attachment of Roseobacters to phytoplankton mediated algal cell death (15, 34). We speculate that Sulfitobacter D7 attachment to E. huxleyi cells may promote its algicidal activity; however, further research on the role of physical attachment in Sulfitobacter D7 virulence is required. The presence of genes encoding type IV secretion system in Sulfitobacter D7 and other Roseobacter genomes (9) may facilitate interactions with eukaryotic microorganisms and may regulate bacterial virulence.
Since DMSP is not a specific metabolite for E. huxleyi and can be produced by diverse algal species, it is likely that other infochemicals also mediate the specificity of this interaction. Such infochemical could convey information regarding the physiological state of the algal cell. For example, p-coumaric acid, a molecule released from senescing E. huxleyi cells, was shown to trigger the production of roseobacticides by P. inhibens (12, 37). Specificity in DMSP signaling can also be achieved by differential exudation rates among E. huxleyi strains (Fig. 4A). We found correlation between patterns of DMSP exudation and the response of E. huxleyi strains to Sulfitobacter D7 infection. Strains exhibiting higher exudation were more susceptible and died faster upon Sulfitobacter D7 infection (Fig. 4). Therefore, the extent of metabolite exudation by algal strains would shape an “individual phycosphere,” which can potentially determine the susceptibility to bacterial infection. This algicidal microscale interaction may shape the population of E. huxleyi strains during algal bloom dynamics.
Ecological impact of algicidal bacteria on E. huxleyi blooms
E. huxleyi bloom demise is thought to be mediated by viral infection (3, 30, 31). However, if viruses were the only mortality agent regulating bloom demise, E. huxleyi strains resistant to viral infection should have taken over the bloom under viral pressure. Our study reveals an important algicidal control by bacteria that possibly constrain the outgrowth of virus-resistant E. huxleyi strains. Strains of E. huxleyi resistant to viral infection (373 and 379) (54) were highly susceptible to Sulfitobacter D7. Conversely, E. huxleyi 2090, which is highly susceptible to viral infection (54), was resistant to Sulfitobacter D7. We propose that a trade-off between susceptibility to viral infection and bacterial pathogenicity, mediated by DMSP, may affect the fate of E. huxleyi cells during bloom dynamics. Moreover, lysis of E. huxleyi cells by viral infection leads to the release of dissolved organic matter, including DMSP (55), which, in turn, can boost bacterial growth and virulence of pathogens, such as Sulfitobacter D7 (5). Therefore, algae-bacteria interaction may have an underappreciated active role in phytoplankton bloom demise. Further research is required to assess the impact of algicidal bacteria on phytoplankton bloom dynamics. Determination of algal bloom demise dominated by viruses or bacteria would encompass many challenges. Mechanistic understanding of these microbial interactions is essential to assess their relative metabolic and biogeochemical imprint.
E. huxleyi blooms are an important source of DMS emission (56). The balance between competing DMSP catabolic pathways, driven by microbial interactions (bacterium-bacterium, alga-bacterium, and alga-virus), may regulate oceanic sulfur cycling (Fig. 6) (57). Interactions of algae with pathogenic bacteria may shunt DMSP catabolism toward high amounts of MeSH, at the expense of DMS, and can boost bacterial growth by incorporation of this reduced sulfur and carbon source. This metabolic switch may constitute a profound biogeochemical signature during algal blooms by affecting the cycling of sulfur and feedback to the atmosphere.
MATERIALS AND METHODS
Oceanographic cruise sampling and isolation of CAM bacterial consortium
Waters were collected from 61.5° to 61.87°N/33.5° to 34.1°W in June to July 2012, during the NA-VICE (KN207-03), aboard the R/V Knorr (www.bco-dmo.org/project/2136). Samples were obtained from five to six depths using a Sea-Bird SBE 911plus CTD carrying 10-liter Niskin bottles. Biomass from 1 to 2 liters of seawater was prefiltered through a 200-μm mesh, collected on 0.8-μm polycarbonate filters (Millipore), flash frozen in liquid nitrogen, and stored at −80°C until further processing. Copepods were collected from surface waters (0 to 5 m) using 100-μm mesh nets on 29 June (57.7°N/32.2°W) and 11 July (61.9°N/33.7°W), as described by Frada et al. (38). Single copepods were thoroughly washed with clean artificial seawater (ASW) and kept at 4°C. Between 2 weeks and 1 month later, single copepod individuals were homogenized with a sterile pestle and inoculated into 2 ml of various E. huxleyi strains growing exponentially (38). Lysis of E. huxleyi strain NCMA379 was observed within 1 week. The supernatant of the culture lysate was passed through a 0.45-μm filter and reinoculated into E. huxleyi 379, resulting in the collapse of the culture. The addition of penicillin and streptomycin (20 U ml−1 and 20 μg ml−1, respectively) abolished culture lysis, indicating the presence of bacterial pathogens (fig. S1A). A suspension (<0.45 μm) of the culture lysate (CAM) was kept at 4°C for further analyses.
Isolation of Sulfitobacter D7 and Marinobacter D6
Sulfitobacter D7 and Marinobacter D6 were isolated from a coculture of E. huxleyi 379 with CAM at 7 days of growth. Bacterial populations in cocultures were stained with the live nucleic acid fluorescent marker SYTO 13 (Molecular Probes). Two distinct subpopulations were observed in CAM-treated cultures (fig. S1B) and were sorted at room temperature on the basis of green fluorescence intensity (530/30 nm) in purity mode using a BD FACSAria II cell sorter equipped with a 488-nm laser. Sorted populations were independently plated on marine agar 2216 plates (Difco) and incubated in the dark at 18°C. Sulfitobacter D7 and Marinobacter D6 were each isolated from a single colony and streaked three times from a single colony to ensure isolation of a single bacterial strain. For identification, DNA was extracted from a single colony of Sulfitobacter D7 and Marinobacter D6 using REDExtract-N-Amp Plant PCR kit (Sigma-Aldrich) according to the manufacturer’s instructions and was used as a template for PCR with general primers for bacterial 16S ribosomal RNA (rRNA): 5′-agtttgatcctggctcag-3′ (forward) and 5′-taccttgttacgacttcacccca-3′ (reverse) (58). Amplicons were paired-end sequenced using the ABI 3730 DNA Analyzer and manually assembled. Sulfitobacter D7 and Marinobacter D6 were grown in marine broth 2216 (Difco) and stored in 15% glycerol at −80°C.
Phylogenetic analysis
A multiple sequence alignment was generated using MUSCLE (59) with the default parameters. A maximum likelihood phylogeny was inferred using RAxML (60) under the GTRCAT model. Nodal support was estimated from a rapid bootstrap analysis with 1000 replicates.
Sulfitobacter D7 whole-genome sequencing and assembly
Sulfitobacter D7 genomic DNA was extracted using a DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s instructions. Genomic DNA was prepared for sequencing using the Nextera XT kit (Illumina, San Diego, CA) according to the manufacturer’s instructions. After processing, libraries were assessed for size using an Agilent TapeStation 2000 automated electrophoresis device (Agilent Technologies, Santa Clara, CA) and for concentration by a Qubit fluorometer (Thermo Fisher Scientific Inc., Waltham, MA). Libraries were pooled in equimolar ratio and sequenced using an Illumina NextSeq 500 sequencer, with paired-end 2 × 150 base reads. Library preparation and sequencing were performed at the DNA Services Facility, University of Illinois at Chicago. Standard Pacific Biosciences large insert library preparation was performed. DNA was fragmented to approximately 20 kb using Covaris g-TUBEs. Fragmented DNA was enzymatically repaired and ligated to a PacBio adapter to form the SMRTbell template. Templates larger than 10 kb were BluePippin (Sage Science) size selected, depending on library yield and size. Templates were annealed to sequencing primer, bound to polymerase, and then bound to PacBio MagBeads and SMRTcell sequenced. Sequencing was performed at the Great Lakes Genomics Center at the University of Wisconsin–Milwaukee. De novo assembly was performed using the SPAdes assembler (61) on both raw Illumina and PacBio reads, with multiple k-mers specified as “-k 31,51,71,91”. Coverage levels were assessed by mapping raw Illumina reads back to the contigs with Bowtie2 (62) and computing the coverage as the number of reads aligning per contig times the length of each read divided by the length of the contig. We assessed the relationship between coverage and cumulative assembly length over coverage-sorted contigs and took 33% of the coverage level at half the total assembly length as a coverage threshold. Contigs with coverage less than this value or with a length shorter than 500 base pairs were removed. The sequence of Sulfitobacter D7 has been deposited in GenBank (accession numbers CP20694 to CP20699, BioProject PRJNA378866).
Culture maintenance, axenization, and bacterial infection
E. huxleyi strains were purchased from the National Center for Marine Algae (NCMA) and the Roscoff Culture Collection (RCC) and maintained in filtered seawater (FSW). NCMA379, RCC1216, and NCMA373 were cultured in f/2 medium (-Si) (63), and NCMA2090 was cultured in k/2 medium (-tris, -Si) (64). Cultures were incubated at 18°C with a 16-hour light/8-hour dark illumination cycle. A light intensity of 100 μmol photons m−2 s−1 was provided by cool white light-emitting diode lights. Cultures were made axenic by the following treatment: Cells were gently washed with autoclaved FSW on sterile 1.2-μm nitrocellulose membrane filters (Millipore). Cells were transferred to algal growth media containing the following antibiotic mix: chloramphenicol (20 μg ml−1), polymyxin B (120 U ml−1), penicillin (40 U ml−1), and streptomycin (40 μg ml−1). After 7 days, the cultures were diluted into fresh algal growth media, and the antibiotics mix was replenished. After another 7 days, the cultures were diluted again into fresh algal growth media without antibiotics. For strains 1216, 373, and 2090, cultures were treated again with the following antibiotics mix: ampicillin (50 μg ml−1), streptomycin (25 μg ml−1), and chloramphenicol (5 μg ml−1). Cultures were transferred one to two times a week. After 2 weeks, the cultures recovered and no bacteria could be detected by flow cytometry (see full description in the following section) or by plating on marine agar 2216 plates. Cultures were maintained with antibiotics and were transferred every 7 to 10 days. Before infection, E. huxleyi cultures were transferred three to four times to antibiotic-free algal growth media. For all experiments, E. huxleyi cultures were infected at early exponential growth phase (2 × 105 to 4 × 105 cells ml−1). For CAM infection, algal cultures were inoculated with 104 bacteria ml−1. For Sulfitobacter D7 infection, bacteria were inoculated from a glycerol stock (kept at −80°C) into 1/2 YTSS [2 g of yeast extract, 1.25 g of tryptone, and 20 g of sea salts (Sigma-Aldrich) dissolved in 1 liter of double distilled water (DDW)] and grown overnight at 28°C at 150 rpm. Bacteria were washed three times in FSW by centrifugation (10,000g, 1 min). Algal cultures were inoculated at t = 0 days with 103 bacteria ml−1. In the experiment presented in Fig. 5, E. huxleyi 2090 cultures were inoculated with 106 bacteria ml−1. When noted, DMSP, glycerol, or propionate were added at t = 0 days. DMSP was synthesized according to Steinke et al. (26).
Enumeration of algae and bacteria abundances and algal cell death by flow cytometry
Flow cytometry analyses were performed using an Eclipse iCyt flow cytometer (Sony Biotechnology Inc., Champaign, IL, USA) equipped with 405- and 488-nm solid-state air-cooled lasers and with a standard optic filter setup. E. huxleyi cells were identified by plotting the chlorophyll fluorescence (663 to 737 nm) against side scatter and were quantified by counting the high-chlorophyll events. For bacterial counts, samples were fixed with a final concentration of 0.5% glutaraldehyde for at least 30 min at 4°C, then plunged into liquid nitrogen, and stored at −80°C until analysis. After thawing, samples were stained with SYBR Gold (Invitrogen) that was diluted 1:10,000 in tris-EDTA buffer, incubated for 20 min at 80°C, and cooled to room temperature. Samples were analyzed by flow cytometry (excitation, 488 nm; emission, 500 to 550 nm). For algal cell death analysis, samples were stained with a final concentration of 1 μM SYTOX Green (Invitrogen), incubated in the dark for 30 min at room temperature, and analyzed by flow cytometry (excitation, 488 nm; emission, 500 to 550 nm). An unstained sample was used as control to eliminate the background signal.
Scanning electron microscopy
Samples of 0.5 ml were mixed with 0.5 ml of fixation medium (3% paraformaldehyde, 2% glutaraldehyde, and 400 mM NaCl, final) and stored at 4°C. Samples were adhered to silicon chips coated with poly-l-lysine (0.01%; Sigma-Aldrich). After three washes in 0.1 M cacodylate buffer, samples were postfixed with 1% OsO4 for 1 hour, followed by three washes in 0.1 M cacodylate buffer and three washes in Milli-Q water. Samples were dehydrated by a series of increasing concentration of ethanol (30 to 100%). Ethanol was replaced by liquid CO2 and critical point dried in BAL-TEC Critical Point Dryer 030. Last, samples were coated with gold/palladium (Edwards, S150) and imaged using the high-tension mode of XL30 ESEM.
Enumeration of E. huxleyi and Sulfitobacter D7 by qPCR
For environmental samples, genomic DNA was extracted using an adapted phenol-chloroform method previously described by Schroeder et al. (54). Filters were cut into small, easily dissolved pieces and placed in a 2-ml Eppendorf tube. Following the addition of 800 μl of GTE buffer [50 mM glucose, 25 mM tris-HCl (pH 8.0), and 10 mM EDTA], proteinase K (10 μg ml−1), and 100 μl of 0.5 M filter-sterilized EDTA, samples were incubated at 65°C for 1 to 2 hours. Following incubation, 200 μl of a 10% (v/v) stock solution of SDS was added, and DNA was then purified by phenol extraction and ethanol precipitation. For laboratory samples, DNA was extracted using REDExtract-N-Amp Plant PCR kit (Sigma-Aldrich) according to the manufacturer’s instructions. E. huxleyi abundance was determined by qPCR for the cytochrome c oxidase subunit 3 (cox3) gene: 5′-agctagaagccctttgaggtt-3′ (Cox3F1) and 5′-tccgaaatgatgacgagttgt-3′ (Cox3R1). Sulfitobacter D7 abundance was determined by qPCR for the 16S rRNA gene using primers designed in this study: 5′-cttcggtggcgcagtgac-3′ (16S-D7bF) and 5′-tcatccacaccttcctcccg-3′ (16S-D7bR). The specificity of 16S-D7b primers was evaluated using TestPrime (www.arb-silva.de/search/testprime/) against the Silva SSU Ref database (65). The primers matched only few Sulfitobacter sp. other than Sulfitobacter D7. All reactions were carried out in technical triplicates. For all reactions, Platinum SYBR Green qPCR SuperMix-UDG with ROX (Invitrogen) was used as described by the manufacturer. Reactions were performed on the StepOnePlus Real-Time PCR System (Applied Biosystems) as follows: 50°C for 2 min, 95°C for 2 min, 40 cycles of 95°C for 15 s, 60°C for 30 s, followed by a melting curve analysis. Results were calibrated against serial dilutions of E. huxleyi (NCMA374 or NCMA2090) and Sulfitobacter D7 DNA at known concentrations, enabling exact enumeration of cell abundance. Samples showing multiple peaks in melting curve analysis or peaks that were not corresponding to the standard curves were discarded.
Headspace analysis using SPME coupled to GC-MS
Headspaces of control, CAM-, and Sulfitobacter D7–infected E. huxleyi 379 cultures after 10 days of growth were sampled for 15 min using an SPME Divinylbenzene/Carboxen/Polydimethylsiloxane fiber (Supelco, Bellefonte, PA, USA). Samples were manually stirred before absorption. For desorption, the fiber was kept in the injection port for 5 min at 260°C. Agilent 7090A gas chromatograph combined with a time-of-fight (TOF) Pegasus IV mass spectrometer (Leco, USA) was used for GC-MS analysis. Carrier gas (helium) was set at a constant flow rate of 1.2 ml min−1. Chromatography was performed on an Rtx-5Sil MS column [30 m, 0.25 mm inner diameter (ID), 0.25 μm] (Restek, Bellefonte, PA, USA). The GC oven temperature program was 45°C for 0.5 min, followed by a 25°C min−1 ramp to a final temperature of 270°C with a 3-min hold time. The temperatures of the transfer line and source were 250° and 220°C, respectively. After a delay of 10 s, mass spectra were acquired at 20 scans s−1, with a mass range from 45 to 450 m/z. Peak detection and mass spectrum deconvolution were performed with ChromaTOF software (Leco). Identification was performed according to the National Institute of Standards and Technology Library. Identification of DMDS was proofed by injection of commercial standard (Sigma-Aldrich).
Evaluation of VOSCs
Samples were collected by small-volume gravity drip filtration (SVDF) (see full description in the following section) (66) and quickly diluted (1:10 in DDW) in a gas-tight vial. DMS, MeSH, and DMDS levels were determined using an Eclipse 4660 Purge-and-Trap Sample Concentrator system equipped with an Autosampler (OI Analytical). Separation and detection were done using GC-FPD (HP 5890) equipped with an Rt-XL sulfur column (Restek). The GC oven temperature program was 100°C for 1 min, followed by a 70°C min−1 ramp to a final temperature of 240°C with a 7-min hold time. All measurements were compared to standards (Sigma-Aldrich; fig. S5). For calibration curves, DMS and DMDS were diluted in DDW to known concentrations. For MeSH standard, we used MeS−Na+ dissolved in DDW and added HCl in 1:1 ratio by injection through the septa of the vials. We could not quantify MeSH since part of it was oxidized to DMDS during the procedure (fig. S5C). Therefore, MeSH abundance is presented as the square root of the area of the peak corresponding to MeSH. No VOSCs were present in blank (DDW) samples.
Determination of DMSP concentration
Laboratory experiments
Samples for DMSPd were obtained by SVDF (66). E. huxleyi cultures were filtered through Whatman GF/F filters by gravity using filtration towers. Filtrates (~3 ml) were acidified to 1.5% HCl for DMSPd preservation and stored at 4°C for >24 hours. Samples were diluted (typically 1:100) in DDW, and DMSPd was hydrolyzed to DMS by adding NaOH in a final concentration of 0.45 M and incubated for 1 hour at room temperature in the dark. Glycine buffer (pH 3) was added to a final concentration of 0.8 M for neutralization (pH 8 to 9, final). Samples were measured for DMS.
Field samples
Collection of water samples is described in the first section of Materials and Methods. To determine DMSPd, ≤20 ml was collected by SVDF and the filtrate was acidified with 50% sulfuric acid (10 μl per 1 ml of sample). Sample preparation was conducted at room temperature. All DMSP samples were stored at 4°C until analysis. Upon analysis, the samples were base hydrolyzed in strong alkali (sodium hydroxide; final concentration, 2 M) and analyzed for DMS. Instrumental determination of DMSP (as DMS) was carried out using the membrane inlet mass spectrometry (MIMS) (67) system that is composed of a Pfeiffer Vacuum quadrupole mass spectrometer equipped with a HiCube 80 pumping station, a QMA 200 analyzer, and a flow-through silicone capillary membrane inlet (Bay Instruments, Easton, MD). The inlet consisted of a glass vacuum line incorporating a U-tube and support for the 0.51-mm-ID Silastic tubing membrane and 0.5-mm-ID stainless steel capillary supply lines. The sample was pumped through the inlet system at 1.5 ml min-1 using a Gilson Minipuls 3 peristaltic pump. Before entering the membrane, the sample passed through a 75-cm length of capillary tubing immersed in a thermostated water bath (VWR, Suwanee, GA) and held at 30°C to ensure constant temperature (and membrane permeability) as the sample passed through the membrane. The U-tube section of the vacuum line (located between the membrane inlet and mass spectrometer) was immersed in an isopropanol bath (held at <-45°C) to remove water vapor from the gas stream before introduction of the stream into the mass spectrometer. In this configuration, the system maintained an operating vacuum pressure of 2.0 (±0.2) × 10−5 mbar. The sample liquid was pumped from the bottom of the sample test tube and through the membrane until the mass spectrometer signal stabilized (typically a minimum of 6 min). DMS was monitored semicontinuously by scanning at m/z 62 for 5 s every 15 s using a secondary electron multiplier detector. Calibration of the MIMS instrument was carried out with freshly prepared base-hydrolyzed DMSP standards made using ESAW (enriched seawater, artificial water) and commercially available DMSP powder (Research Plus, Bayonne, NJ). The detection limit for the system was 0.2 nM.
Sulfitobacter D7 growth in CM and MM supplemented with DMSP
Sulfitobacter D7 were grown overnight in 1/2 YTSS at 28°C. Bacteria were washed three times in ASW (68) by centrifugation (10,000g, 1 min). Media were inoculated with 104 bacteria ml−1. CM were obtained from monocultures of E. huxleyi strains by SVDF (66). This method was chosen to prevent lysis of algal cells during the procedure and release of intracellular components. Following SVDF, media were filtered through 0.22-μm syringe filters. In the experiment presented in Fig. 4B, bacterial growth was followed for 24 hours. Minimal medium (MM) was based on ASW supplemented with basal medium (-tris) (containing essential nutrients) (69) and vitamin mix (70). In the experiment presented in table S1, the MM was supplemented with glycerol (1 g liter−1) and 70 μM DMSP [synthesized according to Steinke et al. (26)]. Bacterial growth, DMSPd, and VOSC levels were measured at t = 0 hours and t = 24 hours. In the experiment presented in fig. S8, the MM was supplemented with glycerol (0.01 g liter−1), 0.5 mM NaNO3, metal mix of k/2 medium (64), and different concentrations of DMSP. Bacterial growth was measured at t = 16 hours.
Statistical analyses
For all time-course experiments, significant differences in the various parameters were determined using a one-way/two-way repeated-measures ANOVA. In other experiments, differences were tested by a one-way ANOVA. Tukey post hoc tests were used when more than two levels of a factor were compared.
Supplementary Material
Acknowledgments
We thank the chief scientist of the NA-VICE cruise, K. D. Bidle (Rutgers University), the captain and crew of the R/V Knorr, and the Marine Facilities and Operations at the Woods Hole Oceanographic Institution for assistance and cooperation at sea. We thank U. Alcolombri (ETH, Zurich) and A. Amrani (The Hebrew University of Jerusalem, Israel) for assistance in GC analyses. We thank S. Ben-Dor for contributing to the phylogenetic analysis. We thank G. Schleyer for assistance and constructive feedback on the manuscript. We thank A. R. Gavish for fruitful discussions and assistance in graphics. We thank the Rieger Foundation for granting a JNF Fellowship in Environmental Studies (to N.B.-G.). All SEM studies were conducted at the Moskowitz Center for Bio-Nano Imaging at the Weizmann Institute of Science. Funding: This research was supported predominantly by the European Research Council (ERC) StG (INFOTROPHIC grant no. 280991), CoG (VIROCELLSPHERE grant no. 681715) (to A.V.), and by the National Science Foundation (NSF) grants OCE-1061883 (to A.V.), OCE-1061876 (to G.R.D.), and OCE-1436458 and OCE-1428915 (to P.A.L.). Author contributions: N.B.-G., M.J.F., and A.V. conceptualized the project. N.B.-G. and A.V. designed the experiments and wrote the paper. N.B.-G. performed all experiments. M.J.F. isolated bacteria from cruise samples. C.K. performed genome and phylogenetic analyses. P.A.L. and G.R.D. measured DMSP in cruise samples. S.M. and A.A. performed the headspace analysis. S.J.G. sequenced the genome and assisted in analysis. R.R. performed statistical analyses. E.K. took part in SEM analysis. M.J.F., U.S., and D.S. assisted in analyzing cruise data. A.V. supervised the project. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/10/eaau5716/DC1
Text S1. Coculturing of E. huxleyi with the CAM exhibits similar phases of pathogenicity to that of Sulfitobacter D7.
Text S2. Sulfitobacter D7 consumes DMSP and produces MeSH but not DMS.
Fig. S1. Algicidal effect of the CAM on E. huxleyi.
Fig. S2. Phylogenetic analysis of Sulfitobacter D7 within the Roseobacter group.
Fig. S3. Marinobacter D6 isolated from CAM has no algicidal effect when cocultured with E. huxleyi.
Fig. S4. Headspace analysis of volatiles produced during algae-bacteria interactions using SPME coupled to GC-MS.
Fig. S5. Representative chromatograms of VOSC standards in GC-FPD analysis.
Fig. S6. Sulfitobacter D7 genome encodes a DMSP catabolic pathway.
Fig. S7. DMSP promotes Sulfitobacter D7 virulence toward E. huxleyi in a dose-dependent manner.
Fig. S8. DMSP promotes growth of Sulfitobacter D7.
Fig. S9. E. huxleyi and Sulfitobacter D7 coculturing dynamics.
Table S1. Evaluation of DMSPd, MeSH, DMDS, DMS, and bacterial abundances after 24-hour incubation of Sulfitobacter D7 in CM obtained from uninfected E. huxleyi 379 cultures (E. huxleyi–CM) or MM supplemented with DMSP.
Table S2. Comparison of parameters related to Sulfitobacter D7 infection dynamics in various E. huxleyi strains.
Reference (71)
REFERENCES AND NOTES
- 1.Field C. B., Behrenfeld M. J., Randerson J. T., Falkowski P., Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 281, 237–240 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Calbet A., Landry M. R., Phytoplankton growth, microzooplankton grazing, and carbon cycling in marine systems. Limnol. Oceanogr. 49, 51–57 (2004). [Google Scholar]
- 3.Vardi A., Haramaty L., Van Mooy B. A., Fredricks H. F., Kimmance S. A., Larsen A., Bidle K. D., Host-virus dynamics and subcellular controls of cell fate in a natural coccolithophore population. Proc. Natl. Acad. Sci. U.S.A. 109, 19327–19332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayali X., Azam F., Algicidal bacteria in the sea and their impact on algal blooms. J. Eukaryot. Microbiol. 51, 139–144 (2004). [DOI] [PubMed] [Google Scholar]
- 5.González J. M., Simó R., Massana R., Covert J. S., Casamayor E. O., Pedrós-Alió C., Moran M. A., Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66, 4237–4246 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teeling H., Fuchs B. M., Becher D., Klockow C., Gardebrecht A., Bennke C. M., Kassabgy M., Huang S., Mann A. J., Waldmann J., Weber M., Klindworth A., Otto A., Lange J., Bernhardt J., Reinsch C., Hecker M., Peplies J., Bockelmann F. D., Callies U., Gerdts G., Wichels A., Wiltshire K. H., Glöckner F. O., Schweder T., Amann R., Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Buchan A., LeCleir G. R., Gulvik C. A., González J. M., Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Wagner-Döbler I., Biebl H., Environmental biology of the marine Roseobacter lineage. Annu. Rev. Microbiol. 60, 255–280 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Newton R. J., Griffin L. E., Bowles K. M., Meile C., Gifford S., Givens C. E., Howard E. C., King E., Oakley C. A., Reisch C. R., Rinta-Kanto J. M., Sharma S., Sun S., Varaljay V., Vila-Costa M., Westrich J. R., Moran M. A., Genome characteristics of a generalist marine bacterial lineage. ISME J. 4, 784–798 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Hahnke S., Brock N. L., Zell C., Simon M., Dickschat J. S., Brinkhoff T., Physiological diversity of Roseobacter clade bacteria co-occurring during a phytoplankton bloom in the North Sea. Syst. Appl. Microbiol. 36, 39–48 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Simon M., Scheuner C., Meier-Kolthoff J. P., Brinkhoff T., Wagner-Döbler I., Ulbrich M., Klenk H. P., Schomburg D., Petersen J., Göker M., Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 11, 1483–1499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seyedsayamdost M. R., Case R. J., Kolter R., Clardy J., The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 3, 331–335 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H., Tomasch J., Michael V., Bhuju S., Jarek M., Petersen J., Wagner-Döbler I., Identification of genetic modules mediating the Jekyll and Hyde interaction of Dinoroseobacter shibae with the dinoflagellate Prorocentrum minimum. Front. Microbiol. 6, 1262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amin S. A., Hmelo L. R., van Tol H. M., Durham B. P., Carlson L. T., Heal K. R., Morales R. L., Berthiaume C. T., Parker M. S., Djunaedi B., Ingalls A. E., Parsek M. R., Moran M. A., Armbrust E. V., Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522, 98–101 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Segev E., Wyche T. P., Kim K. H., Petersen J., Ellebrandt C., Vlamakis H., Barteneva N., Paulson J. N., Chai L., Clardy J., Kolter R., Dynamic metabolic exchange governs a marine algal-bacterial interaction. eLife 5, e17473 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pohnert G., Steinke M., Tollrian R., Chemical cues, defence metabolites and the shaping of pelagic interspecific interactions. Trends Ecol. Evol. 22, 198–204 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Bell W., Mitchell R., Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol. Bull. 143, 265–277 (1972). [Google Scholar]
- 18.Seymour J. R., Amin S. A., Raina J.-B., Stocker R., Zooming in on the phycosphere: The ecological interface for phytoplankton–bacteria relationships. Nat. Microbiol. 2, 17065 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Johnson W. M., Soule M. C. K., Kujawinski E. B., Evidence for quorum sensing and differential metabolite production by a marine bacterium in response to DMSP. ISME J. 10, 2304–2316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seymour J. R., Simó R., Ahmed T., Stocker R., Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science 329, 342–345 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Miller T. R., Hnilicka K., Dziedzic A., Desplats P., Belas R., Chemotaxis of Silicibacter sp. strain TM1040 toward dinoflagellate products. Appl. Environ. Microbiol. 70, 4692–4701 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller T. R., Belas R., Dimethylsulfoniopropionate metabolism by Pfiesteria-associated Roseobacter spp. Appl. Environ. Microbiol. 70, 3383–3391 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tripp H. J., Kitner J. B., Schwalbach M. S., Dacey J. W., Wilhelm L. J., Giovannoni S. J., SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452, 741–744 (2008). [DOI] [PubMed] [Google Scholar]
- 24.T. Tyrrell, A. Merico, Emiliania huxleyi: Bloom observations and the conditions that induce them, in Coccolithophores: From Molecular Processes to Global Impact, H. R. Thierstein, J. R. Young, Eds. (Springer Berlin Heidelberg, 2004), pp. 75–97. [Google Scholar]
- 25.B. Rost, U. Riebesell, Coccolithophore calcification and the biological pump: Response to environmental changes, in Coccolithophores: From Molecular Processes to Global Impact, H. R. Thierstein, J. R. Young, Eds. (Springer Berlin Heidelberg, 2004), pp. 99–125. [Google Scholar]
- 26.Steinke M., Wolfe G. V., Kirst G. O., Partial characterisation of dimethylsulfoniopropionate (DMSP) lyase isozymes in 6 strains of Emiliania huxleyi. Mar. Ecol. Prog. Ser. 175, 215–225 (1998). [Google Scholar]
- 27.Alcolombri U., Ben-Dor S., Feldmesser E., Levin Y., Tawfik D. S., Vardi A., Identification of the algal dimethyl sulfide–releasing enzyme: A missing link in the marine sulfur cycle. Science 348, 1466–1469 (2015). [DOI] [PubMed] [Google Scholar]
- 28.G. Malin, M. Steinke, Dimethyl sulfide production: What is the contribution of the coccolithophores? in Coccolithophores: From Molecular Processes to Global Impact, H. R. Thierstein, J. R. Young, Eds. (Springer Berlin Heidelberg, 2004), pp. 127–164. [Google Scholar]
- 29.Charlson R. J., Lovelock J. E., Andreae M. O., Warren S. G., Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature 326, 655–661 (1987). [Google Scholar]
- 30.Bratbak G., Egge J. K., Heldal M., Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar. Ecol. Prog. Ser. 93, 39–48 (1993). [Google Scholar]
- 31.Lehahn Y., Koren I., Schatz D., Frada M., Sheyn U., Boss E., Efrati S., Rudich Y., Trainic M., Sharoni S., Laber C., DiTullio G. R., Coolen M. J., Martins A. M., Van Mooy B. A., Bidle K. D., Vardi A., Decoupling physical from biological processes to assess the impact of viruses on a mesoscale algal bloom. Curr. Biol. 24, 2041–2046 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Castberg T., Larsen A., Sandaa R. A., Brussaard C. P. D., Egge J. K., Heldal M., Thyrhaug R., van Hannen E. J., Bratbak G., Microbial population dynamics and diversity during a bloom of the marine coccolithophorid Emiliania huxleyi (Haptophyta). Mar. Ecol. Prog. Ser. 221, 39–46 (2001). [Google Scholar]
- 33.Jacquet S., Heldal M., Iglesias-Rodriguez D., Larsen A., Wilson W., Bratbak G., Flow cytometric analysis of an Emiliania huxleyi bloom terminated by viral infection. Aquat. Microb. Ecol. 27, 111–124 (2002). [Google Scholar]
- 34.Mayali X., Franks P. J. S., Azam F., Cultivation and ecosystem role of a marine Roseobacter clade-affiliated cluster bacterium. Appl. Environ. Microbiol. 74, 2595–2603 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paul C., Pohnert G., Interactions of the algicidal bacterium Kordia algicida with diatoms: Regulated protease excretion for specific algal lysis. PLOS ONE 6, e21032 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X., Wang X., Li Z., Su J., Tian Y., Ning X., Hong H., Zheng T., Lysis of a red-tide causing alga, Alexandrium tamarense, caused by bacteria from its phycosphere. Biol. Control. 52, 123–130 (2010). [Google Scholar]
- 37.Wang R., Gallant É., Seyedsayamdost M. R., Investigation of the genetics and biochemistry of roseobacticide production in the Roseobacter clade bacterium Phaeobacter inhibens. MBio 7, e02118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frada M. J., Schatz D., Farstey V., Ossolinski J. E., Sabanay H., Ben-Dor S., Koren I., Vardi A., Zooplankton may serve as transmission vectors for viruses infecting algal blooms in the ocean. Curr. Biol. 24, 2592–2597 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Laber C. P., Hunter J. E., Carvalho F., Collins J. R., Hunter E. J., Schieler B. M., Boss E., More K., Frada M., Thamatrakoln K., Brown C. M., Haramaty L., Ossolinski J., Fredricks H., Nissimov J. I., Vandzura R., Sheyn U., Lehahn Y., Chant R. J., Martins A. M., Coolen M. J. L., Vardi A., DiTullio G. R., Van Mooy B. A. S., Bidle K. D., Coccolithovirus facilitation of carbon export in the North Atlantic. Nat. Microbiol. 3, 537–547 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Lestremau F., Andersson F. A. T., Desauziers V., Investigation of artefact formation during analysis of volatile sulphur compounds using solid phase microextraction (SPME). Chromatographia 59, 607–613 (2004). [Google Scholar]
- 41.Trabue S., Scoggin K., Mitloehner F. M., Li H., Burns R. T., Field sampling method for quantifying volatile sulfur compounds from animal feeding operations. Atmos. Environ. 42, 3332–3341 (2008). [Google Scholar]
- 42.Jin Y., Wang M., Rosen R. T., Ho C., Thermal degradation of sulforaphane in aqueous solution. J. Agric. Food Chem. 47, 3121–3123 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Reisch C. R., Moran M. A., Whitman W. B., Bacterial catabolism of dimethylsulfoniopropionate (DMSP). Front. Microbiol. 2, 172 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howard E. C., Henriksen J. R., Buchan A., Reisch C. R., Bürgmann H., Welsh R., Ye W., González J. M., Mace K., Joye S. B., Kiene R. P., Whitman W. B., Moran M. A., Bacterial taxa that limit sulfur flux from the ocean. Science 314, 649–652 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Kiene R. P., Linn L. J., González J., Moran M. A., Bruton J. A., Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl. Environ. Microbiol. 65, 4549–4558 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirst G. O., Salinity tolerance of eukaryotic marine algae. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 40, 21–53 (1989). [Google Scholar]
- 47.Sunda W., Kleber D. J., Kiene R. P., Huntsman S., An antioxidant function for DMSP and DMS in marine algae. Nature 418, 317–320 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Kessler R. W., Weiss A., Kuegler S., Hermes C., Wichard T., Macroalgal-bacterial interactions: Role of dimethylsulfoniopropionate in microbial gardening by Ulva (Chlorophyta). Mol. Ecol. 27, 1–12 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Kiene R. P., Linn L. J., Bruton J. A., New and important roles for DMSP in marine microbial communities. J. Sea Res. 43, 209–224 (2000). [Google Scholar]
- 50.Antunes L. C. M., Ferreira R. B. R., Buckner M. M. C., Finlay B. B., Quorum sensing in bacterial virulence. Microbiology 156, 2271–2282 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Cude W. N., Buchan A., Acyl-homoserine lactone-based quorum sensing in the Roseobacter clade: Complex cell-to-cell communication controls multiple physiologies. Front. Microbiol. 4, 336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harvey E. L., Deering R. W., Rowley D. C., El Gamal A., Schorn M., Moore B. S., Johnson M. D., Mincer T. J., Whalen K. E., A bacterial quorum-sensing precursor induces mortality in the marine coccolithophore, Emiliania huxleyi. Front. Microbiol. 7, 59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garren M., Son K., Raina J. B., Rusconi R., Menolascina F., Shapiro O. H., Tout J., Bourne D. G., Seymour J. R., Stocker R., A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J. 8, 999–1007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schroeder D. C., Oke J., Malin G., Wilson W. H., Coccolithovirus (Phycodnaviridae): Characterisation of a new large dsDNA algal virus that infects Emiliania huxleyi. Arch. Virol. 147, 1685–1698 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Hill R. W., White B. A., Cottrell M. T., Dacey J. W. H., Virus-mediated total release of dimethylsulfoniopropionate from marine phytoplankton: A potential climate process. Aquat. Microb. Ecol. 14, 1–6 (1998). [Google Scholar]
- 56.Holligan P. M., Fernández E., Aiken J., Balch W. M., Boyd P., Burkill P. H., Finch M., Groom S. B., Malin G., Muller K., Purdie D. A., Robinson C., Trees C. C., Turner S. M., van der Wal P., A biogeochemical study of the coccolithophore, Emiliania huxleyi, in the North Atlantic. Global Biogeochem. Cycles 7, 879–900 (1993). [Google Scholar]
- 57.Simó R., Production of atmospheric sulfur by oceanic plankton: Biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. 16, 287–294 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Eyssen H. J., De Pauw G., Van Eldere J., Formation of hyodeoxycholic acid from muricholic acid and hyocholic acid by an unidentified gram-positive rod termed HDCA-1 isolated from rat intestinal microflora. Appl. Environ. Microbiol. 65, 3158–3163 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., Lesin V. M., Nikolenko S. I., Pham S., Prjibelski A. D., Pyshkin A. V., Sirotkin A. V., Vyahhi N., Tesler G., Alekseyev M. A., Pevzner P. A., SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guillard R. R. L., Ryther J. H., Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can. J. Microbiol. 8, 229–239 (1962). [DOI] [PubMed] [Google Scholar]
- 64.Keller M. D., Seluin R. C., Claus W., Guillard R. R. L., Media for the culture of oceanic ultraphytoplankton. J. Phycol. 23, 633–638 (1987). [Google Scholar]
- 65.Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., Glöckner F. O., Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kiene R. P., Slezak D., Low dissolved DMSP concentrations in seawater revealed by small-volume gravity filtration and dialysis sampling. Limnol. Oceanogr. Methods 4, 80–95 (2006). [Google Scholar]
- 67.Kana T. M., Darkangelo C., Hunt M. D., Oldham J. B., Bennett G. E., Cornwell J. C., Membrane inlet mass spectrometer for rapid high-precision determination of N2, O2, and Ar in environmental water samples. Anal. Chem. 66, 4166–4170 (1994). [Google Scholar]
- 68.Goyet C., Poisson A., New determination of carbonic acid dissociation constants in seawater as a function of temperature and salinity. Deep Sea Res. Part A Oceanogr. Res. Pap. 36, 1635–1654 (1989). [Google Scholar]
- 69.P. Baumann, L. Baumann, The marine gram-negative eubacteria: Genera Photobacterium, Beneckea, Alteromonas, Pseudomas and Alcaligene, in The Prokaryotes: A Handbook on Habitats, Isolation and Identification of the Bacteria, M. P. Starr, H. Stolp, H. G. Trüper, A. Balows, H. G. Schlegel, Eds. (Springer-Verlag, 1981), pp. 1302–1331. [Google Scholar]
- 70.González J. M., Mayer F., Moran M. A., Hodson R. E., Whitman W. B., Microbulbifer hydrolyticus gen. nov., sp. nov., and Marinobacterium georgiense gen. nov., sp. nov., two marine bacteria from a lignin-rich pulp mill waste enrichment community. Int. J. Syst. Bacteriol. 47, 369–376 (1997). [DOI] [PubMed] [Google Scholar]
- 71.Curson A. R. J., Rogers R., Todd J. D., Brearley C. A., Johnston A. W. B., Molecular genetic analysis of a dimethylsulfoniopropionate lyase that liberates the climate-changing gas dimethylsulfide in several marine α-proteobacteria and Rhodobacter sphaeroides. Environ. Microbiol. 10, 757–767 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/10/eaau5716/DC1
Text S1. Coculturing of E. huxleyi with the CAM exhibits similar phases of pathogenicity to that of Sulfitobacter D7.
Text S2. Sulfitobacter D7 consumes DMSP and produces MeSH but not DMS.
Fig. S1. Algicidal effect of the CAM on E. huxleyi.
Fig. S2. Phylogenetic analysis of Sulfitobacter D7 within the Roseobacter group.
Fig. S3. Marinobacter D6 isolated from CAM has no algicidal effect when cocultured with E. huxleyi.
Fig. S4. Headspace analysis of volatiles produced during algae-bacteria interactions using SPME coupled to GC-MS.
Fig. S5. Representative chromatograms of VOSC standards in GC-FPD analysis.
Fig. S6. Sulfitobacter D7 genome encodes a DMSP catabolic pathway.
Fig. S7. DMSP promotes Sulfitobacter D7 virulence toward E. huxleyi in a dose-dependent manner.
Fig. S8. DMSP promotes growth of Sulfitobacter D7.
Fig. S9. E. huxleyi and Sulfitobacter D7 coculturing dynamics.
Table S1. Evaluation of DMSPd, MeSH, DMDS, DMS, and bacterial abundances after 24-hour incubation of Sulfitobacter D7 in CM obtained from uninfected E. huxleyi 379 cultures (E. huxleyi–CM) or MM supplemented with DMSP.
Table S2. Comparison of parameters related to Sulfitobacter D7 infection dynamics in various E. huxleyi strains.
Reference (71)


