Highlights
-
•
Bacterial pathways have previously been elucidated for trimethylamine formation.
-
•
We explain how to identify human-associated bacteria that produce trimethylamine.
-
•
How to identify which gut bacteria are responsible for producing trimethylamine.
-
•
Are we missing anything – unknown bacterial trimethylamine formation pathways?
Abstract
The bacterial formation of trimethylamine (TMA) has been linked to cardiovascular disease. This review focuses on the methods employed to investigate the identity of the bacteria responsible for the formation of TMA from dietary choline and carnitine in the human gut. Recent studies have revealed the metabolic pathways responsible for bacterial TMA production, primarily the anaerobic glycyl radical-containing, choline-TMA lyase, CutC and the aerobic carnitine monooxygenase, CntA. Identification of these enzymes has enabled bioinformatics approaches to screen both human-associated bacterial isolate genomes and whole gut metagenomes to determine which bacteria are responsible for TMA formation in the human gut. We centre on several key methodological aspects for identifying the TMA-producing bacteria and report how these pathways can be identified in human gut microbiota through bioinformatics analysis of available bacterial genomes and gut metagenomes.
1. Introduction
The human gastro-intestinal tract is an ecosystem rich in microbial diversity. Gut microbiota encompasses diverse groups of bacteria, archaea, viruses, fungi and other microeukaryotes [1], [2], [3], [4], [5]. It has been estimated that, on average, 3.8 × 1013 bacteria inhabit the human gut, which encompasses over 1000 species [6], [7]. It is now evident that gut microbiota have an important role in human health and disease. In a healthy gut, these microbes form a stable community whereas during gut dysbiosis, opportunistic pathogens and parasites thrive [8]. Understanding the complex interactions and metabolic capacity of the gut microbiome will help us to examine the workings of this microbiome and better manage disease.
Recent work on the role of the human gut microbiome in disease has linked the bacterial metabolite trimethylamine (TMA) with atherosclerotic cardiovascular disease (ACVD). Previous studies on humans and experimental animals have indicated that choline and carnitine, both of which are conditional B-type vitamins, are the major dietary precursors of TMA in the gut [9], [10], [11], [12], [13], [14], [15], [16], [17]. TMA formation from choline and carnitine is linked to ACVD through hepatic formation of trimethylamine N-oxide (TMAO) although the underlying molecular and cellular mechanisms remain to be fully established [9], [15], [18].
The metabolic pathways responsible for bacterial transformation of choline and carnitine to TMA were unknown until very recently [10], [17]. It is interesting that choline and carnitine degradation to TMA involves some unique chemistry – the former requires a glycyl radical, encoded in the choline-TMA lyase, CutC protein for the breakage of the carbon-nitrogen bond, whereas the latter employs a mononuclear iron in the active centre in the CntA protein. CutC belongs to a larger family of enzymes sharing the same glycyl radical chemistry and shows significant sequence homology to other members of the family such as glycerol dehydratase and pyruvate formate lyase [10], [19]. Similarly, CntA is a new member of the Rieske-containing oxygenases and has significant sequence homology to several members of the Rieske protein family [17]. Although the enzymes responsible for TMA formation from these compounds have now been established, identifying of the microbiota species in the human gut that are responsible for TMA formation remains a challenging task. We focus on several key methodological aspects, which require particular attention when identifying the TMA-producing bacteria and report how to identify these TMA-production pathways in the human gut microbiome through analyses of published bacterial genomes and microbiomes.
2. Pathways of TMA production by gut microbiota
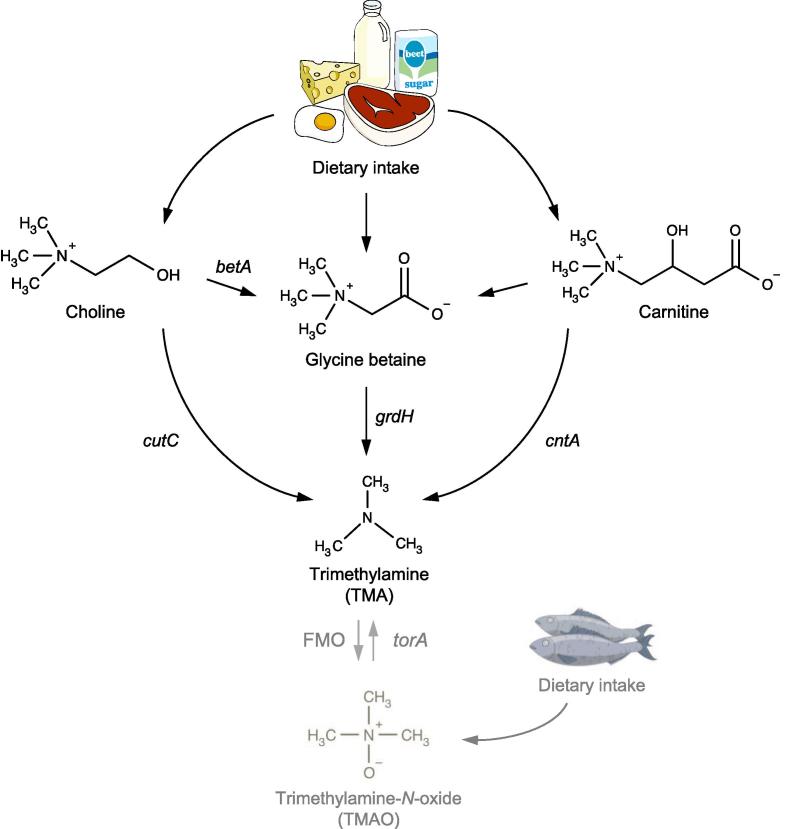
Several pathways for bacterial TMA formation are currently known (Fig. 1), involving a choline-TMA lyase, CutC [10], [19], a carnitine monooxygenase, CntAB [17], a glycine betaine reductase, GrdH [20], or additionally via the reduction of TMAO [21], [22], [23]. Here we focus on the production of TMA from choline and carnitine, since they have been shown to be relevant in cardiovascular disease [9], [24]. These two pathways have been determined using a combination of comparative omics, bioinformatics, molecular genetics and biochemistry based on several model laboratory bacterial strains [10], [17], [19].
Fig. 1.
Known pathways for the formation of trimethylamine (TMA) from dietary choline and carnitine. The key enzymes responsible for TMA production indicated in the figure are: CntAB, carnitine monooxygenase [17] which is analogous to YeaW; CutCD, choline-TMA lyase [10] and GrdH, glycine betaine reductase [20]. Choline to glycine betaine is mediated by the Bet pathway [48], a pathway from carnitine to glycine betaine has been proposed, but has yet to be elucidated [49], [50]. Additionally shown in dark grey the TorA, trimethylamine N-oxide reductase [21] and TMAO formation pathway FMO, flavin-containing monooxygenase are critical to TMA cycling, but are not the focus of this review [51], [52].
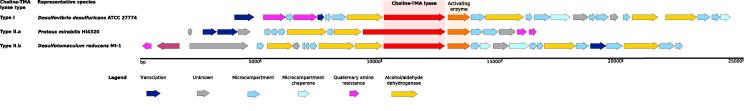
The enzyme that catalyses the degradation of choline to TMA under anaerobic conditions has been identified as the glycyl radical-containing enzyme CutC [10], [19]. Interestingly, the cut gene cluster also houses a set of genes encoding a microcompartment [10], [19]. Although the cut gene cluster was originally characterized from a sulfate-reducing deltaproteobacterium, Desulfovibrio desulfuricans [10], subsequent analyses have shown that cutC homologues and the shell proteins involved in microcompartment formation also occurred in gut Gammaproteobacteria, Actinobacteria and Firmicutes [12], [19], [25]. The formation of a functional microcompartment in choline metabolism has been experimentally demonstrated in Proteus mirabilis [19].
Further to the Cut pathway, it has been previously hypothesized that other pathways capable of degrading choline to TMA may also exist, as exemplified by Edwardsiella tarda ATCC 23685. It has been demonstrated that although this bacterium was capable of producing TMA from choline, it appeared lacking the characterized cut gene cluster [14]. A close examination of the recently published genomes of Edwardsiella tarda has revealed the presence of several glycyl radical family proteins (Edwardsiella tarda ATCC 23685 EDWATA_00359: E-value 9e-136; identity 33% to characterized CutC of P. mirabilis [19]), which are annotated as pyruvate-formate lyases (EC: 2.3.1.54) (Fig. 2). However the key choline-binding site residues showed very low consensus to confirmed CutC proteins (Fig. 2). It therefore remains to be tested whether these so-called pyruvated-formate lyases are functional choline-TMA lyases, or indeed a novel glycyl radical-independent pathway exists in Edwardsiella tarda for choline degradation to TMA.
Fig. 2.
A sequence alignment of characterized CutC proteins from Klebsiella pneumonia[34], Desulfovibrio alaskensis[35] and Proteus mirabilis[19] aligned with Clustal Omega [53] to blast matching genes from Edwardsiella tarda. Despite showing a global high matching sequence similarity, when we compare the key binding site residues (coloured), we observe very little concensus. Sequences are visualised in JalView [54].
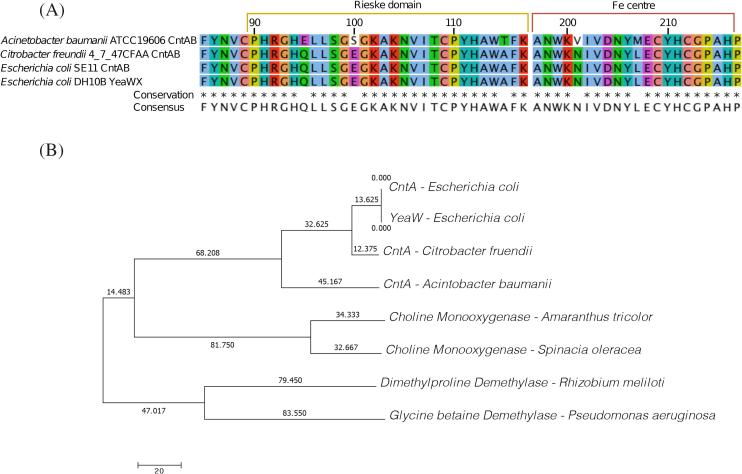
An aerobic pathway for the degradation of carnitine to TMA has been identified by our groups using Acinetobacter baumannii as a model. The enzyme, carnitine monooxygenase encoded by the cntA gene, represents a novel protein of the Rieske oxygenase family, which is best known for several enzymes involved in the oxidation of aromatic compounds [17]. In Escherichia coli and closely related bacteria, CntA is sometime also referred as YeaW in the literature [26]. YeaW is sometimes described as a novel enzyme targeting γ-butyrobetaine [27], however, we and others have shown that E. coli YeaW is homologous to the CntA of A. baumannii through phylogenetic analysis, PFAM domain analyses and substrate specificity tests (Fig. 3) [28], [29], [30]. The CntA enzyme has been identified in several key gut microbiota groups, including Proteobacteria, Actinobacteria and Firmicutes [17], [27], [31], [32].
Fig. 3.
A. Protein alignment of CntA and YeaW showing sequence conservation of the functional domains based on representative CntA sequences from functional confirmed Citrobacter freundii CntA, Acinetobater baumanii CntA, Escherichia coli CntA and Escherichia coli YeaW. The alignment shows a high level on sequence conservation between CntA sequences and the functional domain of YeaW is indistinguishable. B. Phylogenetic tree of Rieske oxygenases based on full length protein sequences with bootstrap values. A. and B. combined show that the CntA from Escherichia coli SE11 [17] and YeaW from Escherichia coli DH10B [26], [27] have an identical protein sequence. CntA of Citrobacter freundii and Acinetobater baumanii cluster with the other CntA proteins and are separate from other Rieske oxygenases used as outgroups.
3. Identification of TMA formation potential in bacterial isolates
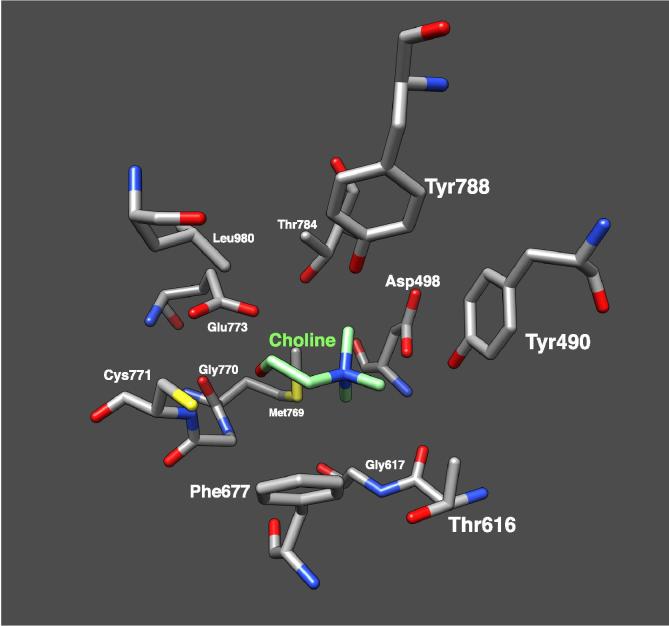
One of the most widely-used and simplest approaches to identify gut-associated microbes with the potential to metabolise choline and carnitine to TMA is in silico BLAST searches of the key signature genes against microbial genome databases such as IMG (https://img.jgi.doe.gov/cgi-bin/m/main.cgi) and NCBI [12], [29]. BLAST searches are carried out using single amino acid sequences of a key gene (with a proven function, Table 1) as the query and comparing this, by means of local alignments, to all of the sequences in a given database. The BLAST searches result in hits to similar gene sequences and provides measures of their statistical significance and can be carried out directly through databases sites such as IMG and NCBI. Using key genes from the choline (cutC) and carnitine (cntA) to TMA pathways as BLAST search queries, a number of hits, or gene homologues, can be identified in fully sequenced human-associated bacterial genomes. However, caution must be used when using this approach. It has been well-documented that the presence of a cutC homologue alone is insufficient to predict choline usage, due to a number of closely related genes with different substrate specificity, even within a single bacterial genome [12], [14], [19], [29], [32]. The gene synteny of the cut cluster provides a genetic context [12], the presence of the activator enzyme gene (cutD) and microcompartment genes, in addition to cutC, may be a better predictor of function in genome sequenced bacteria than cutC alone (Fig. 4). Again, this approach is not without pitfalls. It has been documented that several microbial species of the Pelobacter genus can metabolism choline to TMA [33], [34] and yet the genomes do not appear to contain microcompartment genes in the neighborhood of the cutC/D genes. Alternatively the protein structure is available for CutC (Fig. 5), which would help to determine the active site, enabling the validation of choline-binding CutC (Fig. 2) [35], [36].
Table 1.
Bacterial strains confirmed to degrade choline or carnitine to TMA and containing either the cutCD or cntAB pathways respectively.
| Organism | Reference | Choline | Carnitine | |
|---|---|---|---|---|
| Proteobacteria | Acinetobacter baumannii (ATCC 19606) | Zhu 2014 [17] | + | |
| Acinetobacter calcoaceticus (ATCC 39647) | Ditullio 1994 [56] | + | ||
| Acinetobacter calcoaceticus 69/V | Kleber 1977 [57] | + | ||
| Citrobacter freundii 4_7_47CFAA | + | |||
| Desulfovibrio alaskensis G20 | Weimer 1988 [58] | + | ||
| Desulfovibrio desulfuricans (ATCC 27774) | Craciun 2012 [10] | + | ||
| Desulfovibrio desulfuricans subsp. aestuarii (DSM 17919) | Rath 2017 [29] | + | ||
| Escherichia coli BL21-DE3 | Kalnins 2018 [28] | + | ||
| Escherichia coli DH10b | Koeth 2014 [24] | + | ||
| Escherichia coli K12 (DSM 10517) | Rath 2017 [29] | + | ||
| Escherichia coli MS 200-1 | Romano 2017 [59] | + | ||
| Escherichia coli MS 69-1 | Campo 2015 [12] | + | ||
| Escherichia coli SE11 | Zhu 2014 [17] | + | ||
| Escherichia fergusonii (ATCC 35469) | Romano 2015 [14] | + | ||
| Klebsiella pneumoniae (MSCL 535) | Kuka 2014 [16]/Kalnins 2018 [28] | + | ||
| Klebsiella pneumoniae (MSCL) | Kalnins 2015 [35] | + | ||
| Klebsiella sp. MS 92-3 | Campo 2015 [12] | + | ||
| Pelobacter acetylenicus | Schink 1985 [34] | + | ||
| Pelobacter carbinolicus | Aklujkar 2012 [33] | + | ||
| Proteus mirabilis (ATCC 29906) | Campo 2015 [12] | + | ||
| Proteus mirabilis (DSM 4479) | Jameson 2016 [19] | + | ||
| Proteus mirabilis BB2000 | Campo 2015 [12] | + | ||
| Proteus mirabilis HI4320 | Campo 2015 [12] | + | ||
| Proteus penneri (ATCC 35198) | Romano 2015 [14] | + | ||
| Proteus vulgaris | Seim 1982 [60] | + | ||
| Providencia rettgeri (DSM 1131) | Romano 2015 [14] | + | ||
| Providencia rettgeri (MSCL 730) | Kalnins 2018 [28] | + | ||
| Serratia marcescens (MSCL 1476) | Unemoto 1966 [61]/Kalnins 2018 [28] | + | ||
| Vibrio cholinicus | Hayward 1959 [62] | + | ||
| Firmicutes | Anaerococcus hydrogenalis (DSM 7454) | Romano 2015 [14] | + | |
| Clostridium asparagiforme (DSM 15981) | Romano 2015 [14] | + | ||
| Clostridium citroniae WAL-17108 | Campo 2015 [12] | + | ||
| Clostridium hathewayi (DSM 13479) | Rath 2017 [29] | + | ||
| Clostridium hathewayi (DSM 13749) | Romano 2015 [14] | + | ||
| Clostridium sporogenes (ATCC 15579) | Romano 2015 [14] | + | ||
| Streptococcus dysgalactiae (ATCC 12394) | Campo 2015 [12] | + | ||
| Actinobacteria | Olesnella uli (DSM 7084) | Campo 2015 [12] | + | |
Fig. 4.
Alignment of the cut gene clusters for bacterial isolates representative of the three choline-TMA lyase (cutC) types [19]. The clusters are aligned to the homologous regions of the glycyl radical enzyme (GRE), cutC and activating enzyme, cutD. Gene functions are denoted by colour and the legend shown at the bottom.
Fig. 5.
The active site of CutC choline-trimethylamine lyase from Klebsiella Pneumoniae (PDB 5A0U) showing the key residues involved in substrate recognition and enzymatic activity, visualised in Chimera [55].
It is important to note that such homology based phylogenetic analysis of cutC and cntA in bacterial isolates takes us one step away from the functionally verified genes and inevitably introduces margins for errors. The identified homologues may or may not encode a functional enzyme and additional experiments (e.g. by heterologous expression in a non-TMA producer) are required to validate the functionality of CutC/CntA proteins in TMA formation [10], [17], [19]. Wherever it is possible to obtain the bacterial isolate, the bacterium’s ability to produce TMA from choline should be experimentally tested [10], [17].
4. Identification of TMA formation potential in microbiome studies
Recent studies have begun to resolve which microbial species are responsible for TMA formation and degradation in the human gut through analysis of metagenomic datasets [12], [29], [31], [32], [37], [38]. This approach has involved analysing existing human gut metagenomes in search of key TMA metabolic genes [31], [32], screening fecal samples for the presence of these key genes [12], [29] or sequencing the faecal microbiomes of patients suffering with atherosclerotic cardio vascular disease [37].
To screen existing human gut metagenomes for TMA-production pathways and TMA-degradation pathways, metagenome data can be retrieved from public repositories, such as MG-RAST (http://metagenomics.anl.gov/) or specific metagenomes, such as the Karlsson data set (http://sra.dnanexus. com/studies/SRP016067) [29], [31], [32], [38]. Alternatively, Jie et al. [37] undertook targeted clinical sample collection, focusing on the fecal metagenome of ACVD patients to link the gut microbiome and TMA-production pathways with ACVD [39]. The primary methods utilised to identify TMA-production pathways from metagenome data are BLAST and Profile Hidden Markov Models (profile-HMM) [29], [31], [32]. The basis of BLAST (see Section 3) is a single representative amino acid sequence, while profile-HMMs, adapted originally from speech recognition algorithms, use multiple aligned amino acid sequences that are representative of a key enzyme or a specific protein family. The profile-HMMs rely on probabilistic models and take into account key amino acids and gaps, rather than identifying the percentage match of an entire sequence, as BLAST does. The two alternate methods have different advantages, BLAST only requires a single confirmed sequence, while profile-HMMs are based on the conserved motifs within a functional protein family, which reduces the bias innate to the BLAST approach [40]. BLAST and profile-HMM searches can be carried out locally using BLAST+ (NCBI) and HMMER (hmmer.org), respectively. After generating BLAST or profile-HMM hits it is crucial to reduce false positive hits, to homologues with unrelated functions, which can account for >90% of the total hits. This can be achieved through phylogenetic comparison of the hits to reference strains with known functions [31]. Likewise false negatives can further confuse results, e.g. assigning cutC homologues as pyruvate-formate lyase may account for results of Jie et al. [37], who report that ACVD patients had a significantly higher incidence of cntA than cutC (contrary to previous findings [29], [31], [32]), and a significant enrichment of pyruvate-formate lyase homologues. Wherever possible, one should 1) examine the key signature residuces of metagenome-derived CutC/CntA for substrate co-ordinnation; 2) confirm the function of metagenome-derived CutC/CntA by over-expression in a foreign hosts; and 3) obtain the (most closely related) bacterial isolate and test the bacterium’s ability to produce TMA [10], [17].
An alternative approach to using metagenome data is to utilise the abundant 16S rRNA gene sequencing data available and infer microbiome function, i.e. TMA degradation, based on taxonomy. There are various programs available, such as PICRUSt [41], Tax4Fun [42], Piphillin [43] and Vikodak [44] that aim to predict function from 16S rRNA gene sequences. These allow the user to get an idea of the potential metabolic function of a microbial community. The function prediction programs rely on how accurate and comprehensive the databases are, fortunately for studying the human gut, these programs have been developed using the human microbiome project and gut microbiome databases are sufficiently comprehensive, resulting in 75–85% average correlations to metagenome data, at predicting community level functions [3], [41], [42], [43], [44]. Another consideration for specific functions is how evenly distributed the functions of interest are across bacterial families or genera. Matching 16S rRNA gene taxonomy and metabolic function can be difficult to reconcile for some functions, due to gene loss between closely related strains and lateral gene transfer [45], [46]. The carnitine degradation genes cntAB have been detected in Proteobacteria, while the choline degradation cutCD genes have been detected in Proteobacteria, Firmicutes and Actinobacteria (Table 1), however these functional genes are unevenly distributed between bacterial phyla preventing accurate prediction of function through 16S rRNA genes alone [10], [12], [32]. PICRUSt has been applied to predict significant differences in genes involved in choline, carnitine and trimethylamine (TMA) metabolism in chronic kidney disease gut microbiomes [47], however these predictions should inform, rather than replace shotgun metagenome sequencing for accurate analysis of metabolic functions in a community.
5. Summary
In the rapidly developing and expanding field of gut microbiome-associated disease, microbial approaches are vital to understanding the causes of disease and the complex community interactions. The identification of metabolic pathways responsible for TMA production has informed molecular and clinical approaches that identify the species responsible for generating TMA in the human gut. A more in-depth understanding of these processes and an exhaustive identification of all bacterial TMA-production pathways will help to complete the picture of gut microbiome structure and function in terms of TMA-related disease pathogenesis. Direct links between diet, bacterial communities and human disease have been discovered, through a combination of molecular microbial approaches, these tools pave the way for future studies to expand the field of human microbiome caused disease.
Acknowledgements
We are grateful for a Leverhulme Trust research project grant (RPG-2016-307) and a BBSRC/EPSRC-funded career development fellowship (EJ – BB/M017982/1) for funding.
References
- 1.Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M., Gill S.R., Nelson K.E., Relman D.A. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gill S.R., Pop M., DeBoy R.T., Eckburg P.B., Turnbaugh P.J., Samuel B.S., Gordon J.I., Relman D.A., Fraser-Liggett C.M., Nelson K.E. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grice E.A., Segre J.A. The human microbiome: our second genome. Annu. Rev. Genomics Human Genet. 2012;13:151–170. doi: 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marchesi J.R. Chapter 2 – Prokaryotic and Eukaryotic Diversity of the Human Gut. In: Laskin A.I., Sariaslani S., Gadd G.M., editors. Advances in Applied Microbiology. Academic Press; 2010. pp. 43–62. [DOI] [PubMed] [Google Scholar]
- 5.Breitbart M., Hewson I., Felts B., Mahaffy J.M., Nulton J., Salamon P., Rohwer F. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 2003;185(20):6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooper L.V., Macpherson A.J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 2010;10(3):159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 7.Sender R., Fuchs S., Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14(8):e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Org E., Mehrabian M., Lusis A.J. Unraveling the environmental and genetic interactions in atherosclerosis: central role of the gut microbiota. Atherosclerosis. 2015;241(2):387–399. doi: 10.1016/j.atherosclerosis.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z., Klipfell E., Bennett B.J., Koeth R., Levison B.S., DuGar B., Feldstein A.E., Britt E.B., Fu X., Chung Y.-M. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craciun S., Balskus E.P. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. 2012;109(52):21307–21312. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang W.W., Wang Z., Levison B.S., Koeth R.A., Britt E.B., Fu X., Wu Y., Hazen S.L. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 2013;368(17):1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martínez-del Campo A., Bodea S., Hamer H.A., Marks J.A., Haiser H.J., Turnbaugh P.J., Balskus E.P. Characterization and detection of a widely distributed gene cluster that predicts anaerobic choline utilization by human gut bacteria. mBio. 2015;6(2) doi: 10.1128/mBio.00042-15. e00042–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ierardi E., Sorrentino C., Principi M., Giorgio F., Losurdo G., Di Leo A. Intestinal microbial metabolism of phosphatidylcholine: a novel insight in the cardiovascular risk scenario. Hepatobiliary Surg. Nutr. 2015;4(4):289. doi: 10.3978/j.issn.2304-3881.2015.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romano K.A., Vivas E.I., Amador-Noguez D., Rey F.E. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the Proatherogenic metabolite Trimethylamine-N-Oxide. mBio. 2015;6(2) doi: 10.1128/mBio.02481-14. e02481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koeth R.A., Wang Z., Levison B.S., Buffa J.A., Sheehy B.T., Britt E.B., Fu X., Wu Y., Li L., Smith J.D. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013;19(5):576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuka J., Liepinsh E., Makrecka-Kuka M., Liepins J., Cirule H., Gustina D., Loza E., Zharkova-Malkova O., Grinberga S., Pugovics O. Suppression of intestinal microbiota-dependent production of pro-atherogenic trimethylamine N-oxide by shifting L-carnitine microbial degradation. Life Sci. 2014;117(2):84–92. doi: 10.1016/j.lfs.2014.09.028. [DOI] [PubMed] [Google Scholar]
- 17.Zhu Y., Jameson E., Crosatti M., Schäfer H., Rajakumar K., Bugg T.D., Chen Y. Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc. Natl. Acad. Sci. 2014;111(11):4268–4273. doi: 10.1073/pnas.1316569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu W., Gregory J.C., Org E., Buffa J.A., Gupta N., Wang Z., Li L., Fu X., Wu Y., Mehrabian M. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. 2016;165(1):111–124. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jameson E., Fu T., Brown I.R., Paszkiewicz K., Purdy K.J., Frank S., Chen Y. Anaerobic choline metabolism in microcompartments promotes growth and swarming of Proteus mirabilis. Environ. Microbiol. 2016 doi: 10.1111/1462-2920.13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreesen J.R. Glycine metabolism in anaerobes. Antonie Van Leeuwenhoek. 1994;66(1–3):223–237. doi: 10.1007/BF00871641. [DOI] [PubMed] [Google Scholar]
- 21.Méjean V., Lobbi-Nivol C., Lepelletier M., Giordano G., Chippaux M., Pascal M.C. TMAO anaerobic respiration in Escherichia coli: involvement of the tor operon. Mol. Microbiol. 1994;11(6):1169–1179. doi: 10.1111/j.1365-2958.1994.tb00393.x. [DOI] [PubMed] [Google Scholar]
- 22.McCrindle S.L., Kappler U., McEwan A.G. Microbial dimethylsulfoxide and trimethylamine-N-oxide respiration. Adv. Microb. Physiol. 2005;50:147–201e. doi: 10.1016/S0065-2911(05)50004-3. [DOI] [PubMed] [Google Scholar]
- 23.Hoyles L., Jiménez-Pranteda M.L., Chilloux J., Brial F., Myridakis A., Aranias T., Magnan C., Gibson G.R., Sanderson J.D., Nicholson J.K. Metabolic retroconversion of trimethylamine N-oxide and the gut microbiota. bioRxiv. 2017:225581. doi: 10.1186/s40168-018-0461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koeth R.A., Levison B.S., Culley M.K., Buffa J.A., Wang Z., Gregory J.C., Org E., Wu Y., Li L., Smith J.D., Tang W.H., DiDonato J.A., Lusis A.J., Hazen S.L. gamma-Butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of L-carnitine to TMAO. Cell Metab. 2014;20(5):799–812. doi: 10.1016/j.cmet.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Axen S.D., Erbilgin O., Kerfeld C.A. A Taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput. Biol. 2014;10(10):e1003898. doi: 10.1371/journal.pcbi.1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boxhammer S., Glaser S., Kühl A., Wagner A., Schmidt C.L. Characterization of the recombinant Rieske [2Fe–2S] proteins HcaC and YeaW from E. coli. Biometals. 2008;21(4):459–467. doi: 10.1007/s10534-008-9134-y. [DOI] [PubMed] [Google Scholar]
- 27.Koeth R.A., Levison B.S., Culley M.K., Buffa J.A., Wang Z., Gregory J.C., Org E., Wu Y., Li L., Smith J.D. γ-Butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of L-carnitine to TMAO. Cell Metab. 2014;20(5):799–812. doi: 10.1016/j.cmet.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalnins G., Sevostjanovs E., Hartmane D., Grinberga S., Tars K. CntA oxygenase substrate profile comparison and oxygen dependency of TMA production in Providencia rettgeri. J. Basic Microbiol. 2018;58(1):52–59. doi: 10.1002/jobm.201700428. [DOI] [PubMed] [Google Scholar]
- 29.Rath S., Heidrich B., Pieper D.H., Vital M. Uncovering the trimethylamine-producing bacteria of the human gut microbiota. Microbiome. 2017;5(1):54. doi: 10.1186/s40168-017-0271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z., Roberts A.B., Buffa J.A., Levison B.S., Zhu W., Org E., Gu X., Huang Y., Zamanian-Daryoush M., Culley M.K. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell. 2015;163(7):1585–1595. doi: 10.1016/j.cell.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jameson E., Doxey A.C., Airs R., Purdy K.J., Murrell J.C., Chen Y. Metagenomic data-mining reveals contrasting microbial populations responsible for trimethylamine formation in human gut and marine ecosystems. Microb. Genomics. 2016 doi: 10.1099/mgen.0.000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falony G., Vieira-Silva S., Raes J. Microbiology Meets Big Data-The Case of Gut Microbiota-Derived TMA. Annu. Rev. Microbiol. 2015;69(1) doi: 10.1146/annurev-micro-091014-104422. [DOI] [PubMed] [Google Scholar]
- 33.Aklujkar M., Haveman S.A., DiDonato R., Chertkov O., Han C.S., Land M.L., Brown P., Lovley D.R. The genome of Pelobacter carbinolicus reveals surprising metabolic capabilities and physiological features. BMC Genomics. 2012;13(1):690. doi: 10.1186/1471-2164-13-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schink B. Fermentation of acetylene by an obligate anaerobe, Pelobacter acetylenicus sp. nov. Arch. Microbiol. 1985;142(3):295–301. [Google Scholar]
- 35.Kalnins G., Kuka J., Grinberga S., Makrecka-Kuka M., Liepinsh E., Dambrova M., Tars K. Structure and function of CutC choline lyase from human microbiota bacterium Klebsiella pneumoniae. J. Biol. Chem. 2015 doi: 10.1074/jbc.M115.670471. JBC. M115. 670471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bodea S., Funk M.A., Balskus E.P., Drennan C.L. Molecular basis of C-N bond cleavage by the glycyl radical enzyme choline trimethylamine-lyase. Cell Chem. Biol. 2016;23(10):1206–1216. doi: 10.1016/j.chembiol.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jie Z., Xia H., Zhong S.L., Feng Q., Li S., Liang S., Zhong H., Liu Z., Gao Y., Zhao H., Zhang D., Su Z., Fang Z., Lan Z., Li J., Xiao L., Li J., Li R., Li X., Li F., Ren H., Huang Y., Peng Y., Li G., Wen B., Dong B., Chen J.Y., Geng Q.S., Zhang Z.W., Yang H., Wang J., Wang J., Zhang X., Madsen L., Brix S., Ning G., Xu X., Liu X., Hou Y., Jia H., He K., Kristiansen K. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017;8(1):845. doi: 10.1038/s41467-017-00900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borrel G., McCann A., Deane J., Neto M.C., Lynch D.B., Brugère J.-F., O'Toole P.W. Genomics and metagenomics of trimethylamine-utilizing Archaea in the human gut microbiome. ISME J. 2017;11(9):2059. doi: 10.1038/ismej.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J., Jia H. Metagenome-wide association studies: fine-mining the microbiome. Nat. Rev. Microbiol. 2016;14(8):508–522. doi: 10.1038/nrmicro.2016.83. [DOI] [PubMed] [Google Scholar]
- 40.Eddy S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011;7(10):e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langille M.G., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A., Clemente J.C., Burkepile D.E., Thurber R.L.V., Knight R. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31(9):814. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aßhauer K.P., Wemheuer B., Daniel R., Meinicke P. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics. 2015;31(17):2882–2884. doi: 10.1093/bioinformatics/btv287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwai S., Weinmaier T., Schmidt B.L., Albertson D.G., Poloso N.J., Dabbagh K., DeSantis T.Z. Piphillin: Improved prediction of metagenomic content by direct inference from human microbiomes. PloS One. 2016;11(11):e0166104. doi: 10.1371/journal.pone.0166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagpal S., Haque M.M., Mande S.S. Vikodak-A modular framework for inferring functional potential of microbial communities from 16S metagenomic datasets. PloS One. 2016;11(2):e0148347. doi: 10.1371/journal.pone.0148347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meehan C.J., Beiko R.G. Lateral gene transfer of an ABC transporter complex between major constituents of the human gut microbiome. BMC Microbiol. 2012;12(1):248. doi: 10.1186/1471-2180-12-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huttenhower C., Gevers D., Knight R., Abubucker S., Badger J.H., Chinwalla A.T., Creasy H.H., Earl A.M., FitzGerald M.G., Fulton R.S. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu K.-Y., Xia G.-H., Lu J.-Q., Chen M.-X., Zhen X., Wang S., You C., Nie J., Zhou H.-W., Yin J. Impaired renal function and dysbiosis of gut microbiota contribute to increased trimethylamine-N-oxide in chronic kidney disease patients. Sci. Rep. 2017;7(1):1445. doi: 10.1038/s41598-017-01387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andresen P.A., Kaasen I., Styrvold O.B., Boulnois G. Molecular cloning, physical mapping and expression of the bet genes governing the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Microbiology. 1988;134(6):1737–1746. doi: 10.1099/00221287-134-6-1737. [DOI] [PubMed] [Google Scholar]
- 49.Lindstedt G., Lindstedt S. On the biosynthesis and degradation of carnitine. Biochem. Biophys. Res. Commun. 1961;6(5):319–323. doi: 10.1016/0006-291x(61)90137-1. [DOI] [PubMed] [Google Scholar]
- 50.Uanschou C., Frieht R., Pittner F. What to learn from a comparative genomic sequence analysis of L-carnitine dehydrogenase. Monatshefte für Chemie/Chemical Monthly. 2005;136(8):1365–1381. [Google Scholar]
- 51.Chen Y., Patel N.A., Crombie A., Scrivens J.H., Murrell J.C. Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc. Natl. Acad. Sci. 2011;108(43):17791–17796. doi: 10.1073/pnas.1112928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoyles L., Jiménez-Pranteda M.L., Chilloux J., Brial F., Myridakis A., Aranias T., Magnan C., Gibson G.R., Sanderson J.D., Nicholson J.K., Gauguier D., McCartney A.L., Dumas M.-E. Metabolic retroconversion of trimethylamine N-oxide and the gut microbiota. Microbiome. 2018;6(1):73. doi: 10.1186/s40168-018-0461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McWilliam H., Li W., Uludag M., Squizzato S., Park Y.M., Buso N., Cowley A.P., Lopez R. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013;41(W1):W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 56.Ditullio D., Anderson D., Chen C.-S., Sih C.J. L-carnitine via enzyme-catalyzed oxidative kinetic resolution. Bioorg. Med. Chem. 1994;2(6):415–420. doi: 10.1016/0968-0896(94)80009-x. [DOI] [PubMed] [Google Scholar]
- 57.Kleber H., Seim H., Aurich H., Strack E. Utilization of trimethylammonium-compounds by Acinetobacter calcoaceticus (author's transl) Arch. Microbiol. 1977;112(2):201–206. doi: 10.1007/BF00429336. [DOI] [PubMed] [Google Scholar]
- 58.Weimer P.J., Van Kavelaar M.J., Michel C.B., Ng T.K. Effect of phosphate on the corrosion of carbon steel and on the composition of corrosion products in two-stage continuous cultures of Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 1988;54(2):386–396. doi: 10.1128/aem.54.2.386-396.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Romano K.A., Martinez-del Campo A., Kasahara K., Chittim C.L., Vivas E.I., Amador-Noguez D., Balskus E.P., Rey F.E. Metabolic, epigenetic, and transgenerational effects of gut bacterial choline consumption. Cell Host Microbe. 2017;22(3) doi: 10.1016/j.chom.2017.07.021. 279-290. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seim H., Löster H., Claus R., Kleber H.P., Strack E. Formation of γ-butyrobetaine and trimethylamine from quaternary ammonium compounds structure-related to l-carnitine and choline by Proteus vulgaris. FEMS Microbiol. Lett. 1982;13(2):201–205. [Google Scholar]
- 61.Unemoto T., Hayashi M., Miyaki K., Hayashi M. Formation of trimethylamine from DL-carnitine by Serratia marcescens. Biochimica et Biophysica Acta (BBA)-General Subjects. 1966;121(1):220–222. doi: 10.1016/0304-4165(66)90382-5. [DOI] [PubMed] [Google Scholar]
- 62.Hayward H.R., Stadtman T.C. Anaerobic degradation of choline I. Fermentation of choline by an anaerobic, cytochrome-producing bacterium Vibrio cholinicus N. sp. J. Bacteriol. 1959;78(4):557. doi: 10.1128/jb.78.4.557-561.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]