Abstract
The nanopore-based single-molecule biosensor has been extensively investigated for various biomedical detections. It has demonstrated the potential in gene sequencing and diagnosis-oriented biomarker detection such as microRNAs. In real-time detection, however, samples extracted from bio-fluids contain various non-target nucleic acids components. These components can cause severely influence the target detection accuracy. We have discovered that a polycationic probe can solve this issue. The polycationic peptide domain of the probe can separate the target•probe complex from free nucleic acids, and only lead the complex into the pore, therefore realizing simultaneous enrichment and detection of target microRNAs. This study establishs a universal approach to detecting any short pathogenic nucleic acids fragment in complex samples
I. Introduction
Nanopores are a type of molecular scale pore structures fabricated in an insulating membrane. A transmembrane voltage applied can drive an ion current across the pore. Single molecules that interact with the nanopore can specifically block the ion pathway, resulting in characteristic change in the nanopore conductance, called signatures. The signature profile such as conductance level and duration is the identity of individual target molecules, while counting the number of signatures allows target quantification.
Nanopores have been developed as a powerful singlemolecule detector in life sciences exploration [1-14]. In genetic detection, the nanopore represents a potential new generation technology for gene sequencing[15-18], and in epigenetic detection, the nanopore has been shown to be able to analyze DNA methylation[19] and gene damage [20]. We have utilized the nanopore to investigate single aptamer folding/unfolding mechanism and its regulation[21,22]. In particular, the nanopore has been applied in quantification of microRNAs (miRNAs) [23,24], a group of extremely important small regulatory RNAs[25-28] that are promising cancer biomarkers[29-37]. Therefore in the future the nanopore sensor could offer a non-invasive approach to cancer diagnostics.
However, there are challenges in nanopore clinical applications. In microRNA detection, the clinical samples are total RNA extractions from tissue or plasma. RNA extractions are a complex mixture of various RNA species such as miRNAs, mRNAs, tRNA etc. All of these components have chances to bind the nanopore. Their interaction with the nanopore will cause contaminative nanopore signals that could severely influence the recognition and measurement of target microRNA signals. Therefore, a priority in real-time applications is eliminating these interferences.
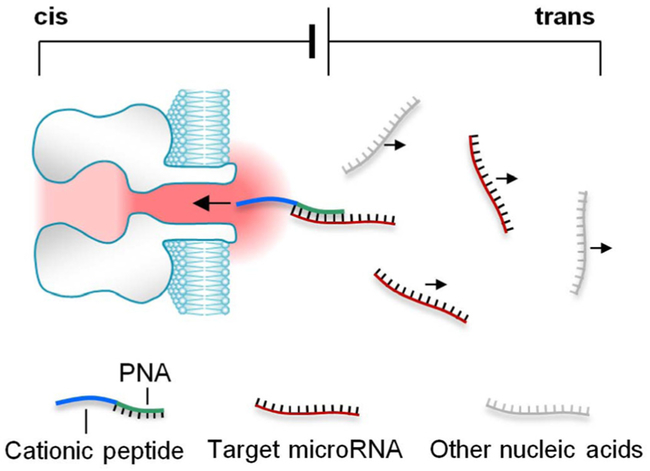
Here we uncovered that this issue can be universally solved by using a unique polycationic probe. This probe can simultaneously enrich and detect the target microRNA in the presence of non-target nucleic acids (Fig. 1). The probe contains a peptide nucleic acids (PNA) attached with a polycationic peptide lead. When the target microRNA is bound by PNA, the peptide lead can drives the complex moving toward the nanopore in the electrical field, whereas any free nucleic acids without probe binding carry negative charge, thus migrate against the pore. Therefore only the target microRNA is enriched around the nanopore opening, and simultaneously detected from the signatures generated by the microRNA•probe complex. There will be no any interference signal originating from free nucleic acids. This simple and universal approach, once validated, could be expanded to detect any short pathogenic nucleic acids in complex samples.
Figure 1.
Simultaneous enrichment and single-molecule detection of microRNAs in the nanopore by using a polycationic probe.
II. Results
A. Identifying microRNA with a polycationic probe
We selected microRNA Let-7b as the target to test this proof-of-concept. The PNA domain of the probe P7b is fully matched with the 3’ 10 bases of Let-7b. The cationic peptide domain of P7b contains 11 amino acids, including 6 arginines and 2 lysines. Both Let-7b and P7b was presented in trans side of the multant α-hemolysin protein pore K131D. Trans opening of this pore has been constructed with an anionic Asp 131 ring to attract cationic molecules. A positive voltage was applied from trans side with cis side grounded. In this configuration, only cationic molecules in trans solution can move toward the pore, while anionic molecules move oppositely against the pore.
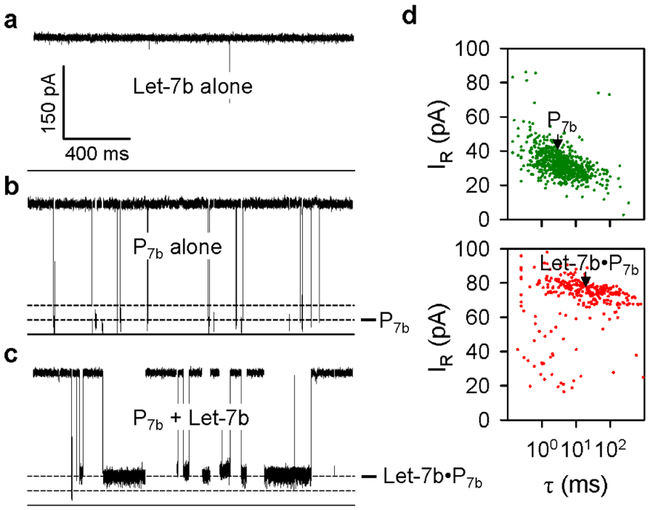
The current trace in Fig. 2a shows that Let-7b alone in never blocked the pore under a positive voltage, suggesting that anionic microRNA was prevented from interacting with the pore. In contrast, the current trace in Fig. 2b shows that the robe P7b alone in trans solution produced a large number of Level 1 blocks, due to the negative charge on the peptide that drives the polymer toward the nanopore. These blocks with duration of 4.8 ms reduced the pore conductance to 8.2% (Fig. 2d upper). However, when the Let-7b / P7b mixture was added in trans solution, as Fig. 2c, Level 1 blocks were replaced with a large number of distinct Level 2 blocks. By comparison, the Level 2 block duration was prolonged by 4 times to 21 ms, and featured a higher residual conductance to 26% (Fig. 2d lower). Because the Level 2 blocks were only observed in the Let-7b/P7b mixture, they should be generated by the Let-7b•P7b complex that binds with the pore from trans entrance. Therefore Level 2 blocks can serve as the signature for Let-7b identification.
Figure 2.
Detection of microRNAs with a polycationic probe, a-c. Current traces for the K131D pore in the presence of microRNA Let-7b alone (a), probe P7b alone (b), and the mixture of Let-7b and P7b (c), recorded at + 180 mV in 1 M KCl solutions (pH7.2). Concentrations of Let-7b and P7b were 300 nM and 100 nM respectively, d. Scattering plots showing the duration (τ) and residual currents (IR) for P7b and the complex Let-7b•P7b.
B. Configuration for microRNA•probe complex in nanopore
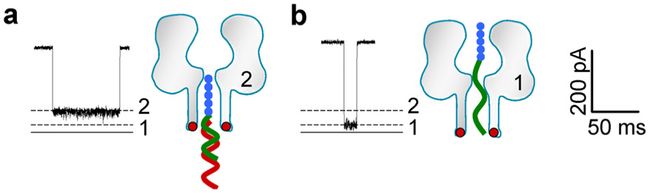
The molecular configuration for the Let-7b•P7b complex in the pore is proposed in Fig. 3a. When the Let-7b•P7b, complex interacts with the pore, the peptide domain (~1.7 nm wide) of the complex will enter the pore from trans entrance (~2 nm), while the wider Let-7b•PNA duplex domain (~2.3 nm) is anchored at the entrance without entering the pore. This configuration suggests that the Level 2 signature is generated by the peptide domain occupying the pore’s β-barrel. Because the duration of Level 2 blocks was consistently extended as the voltage increased (data not shown), it is expected that the trapped Let-7b•P7b, complex is released back to trans solution at the end of Level 2, rather than translocates through the pore. Distinguished from Level 2, the Level 1 block conductance is contributed by trapping the entire P7b in the pore (Fig. 3b). PNA itself can be rarely trapped in the pore, so the trapping of PNA should be led by the peptide domain.
Figure 3.
Molecular configurations for the complex Let-7b•P7b and the probe P7b alone in the nanopore. a. Signature and configuration model for Let-7b•P7b, which generate Level 2 block. b. Signature and configuration model for P7b translocation through the pore, which yield Level 1 block.
C. Single-base discrimination between different microRNAs
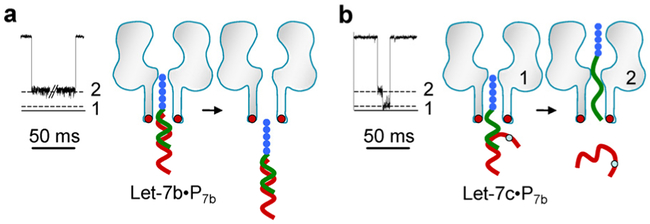
Disease diagnosis demands the nanopore sensor to be able to accurately determine microRNAs with similar sequences. We further uncovered that the PNA-peptide probe designed as above can build this capability to the nanopore sensor. In this study, the probe P7b was used to target microRNAs Let-7b and Let-7c. Both microRNAs in Let-7 family are different in one base at position 16. Thus the Let-7c•P7b, hybrid contains a single mismatched base pair (Table 1). Current traces in Fig. 4 show the duration of Level 2 signatures for fully-matched Let-7b•P7b was 380 ms (+130 mV in 3 M/0.5 M cis/trans KCl), whereas the duration was shortened over 30 times to 12 ms for one-mismatched Let-7c•P7b, complex. Also the Let-7c•P7b signatures featured two block levels, distinct from Let-7b•P7b’s single-level block. This suggests a different configuration for Let-7c•P7b in the pore: Driven by the voltage, Let-7c•P7b is de-hybridized in the pore. After de-hybridization, the free Let-7c returns to trans solution, but P7b passes through the pore, stepwise changing the conductance. Overall, the fully-matched miRNA•PNA duplex is sufficiently stable to resist dehybridization, while a single mismatch introduced in duplex significantly destabilizes the hybridization. This generates the 30 folds difference in block duration and distinct block profiles between two microRNAs, enabling us to accurately discriminate single Let-7b and Let-7c molecules by eyes.
Table 1.
Sequences of microRNAs and probe in this study
| P7b: | N′–YGRKKRRQRRR–AACCACACAA–C′ |
| Let–7b: | 3′–uugguguguuggaugauggagu–5′ |
| Let–7c: | 3′–uugguauguuggaugauggagu–5′ |
Figure 4.
Discrimination of microRNAs with single-base difference. The probe P7b was used to target Let-7b and Let-7c. The sequences of the two microRNAs contain a single-base difference (Table 1). The nanopore current was recorded at +130 mV in 3 M/0.5 M cis/trans KCl. a-b. Level 2 signatures produced by the fully-matched complex Let-7b•P7b (a), and Let-7c•P7b (b).
D. Optimized pore and probe for enhanced sensitivity
We finally investigated the sensitivity of the nanopore sensor. It was found that both probe design and nanopore engineering enables high sensitivity in microRNA detection. The anionic Asp 131 ring constructed at the trans entrance of the K131D pore can effectively attract cationic peptide. As a result, the target capture rate can be greatly enhanced. The capture rate for P7b (peptide-PNA) was sharply increased by 90-fold from 3.2 μM−1·s−1 in the wild-type pore without Asp 131 to 280 μM−1·s−1 in the K131D pore. The Let-7b•P7b complex can rarely be trapped in the wild-type pore, but it can be trapped in the K131D pore at a capture rate of 80±9 μM−1·s−1. This capture rate is 50-fold enhanced compared microRNA detection with a DNA probe in our earlier study[23]. Therefore the combined design polycationic probe and engineered nanopore can greatly enhance the microRNA capture rate.
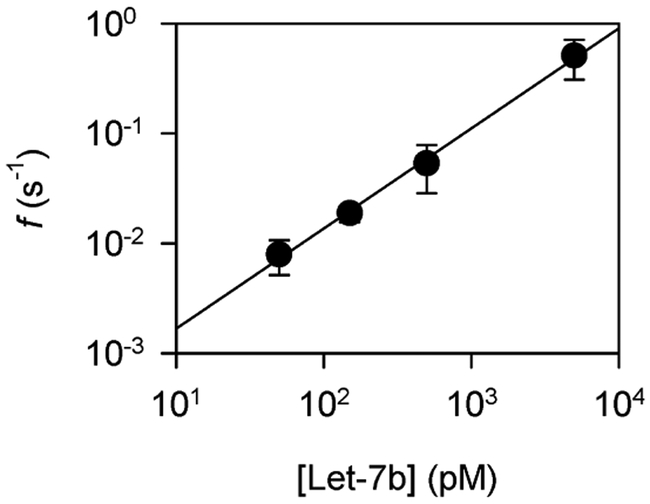
Moreover, the frequency of Let-7b•P7b signatures consistently increases along with the Let-7b concentration in a broad range from 50 pM to 5 nM (+180 mV, Fig. 5), and the data can be fitted to a straight line in the log-log scale. This result suggests the concentration-dependent frequency plot can be used for sensor calibration in microRNA detection.
Figure 5.
Concentration-dependent frequency of Let-7b•P7b signatures in the K131D pore.
III. Conclusions and Perspectives
In summary, we have discovered that a polycationic probe can realize simultaneous enrichment and detection of microRNAs in the nanopore. The positively charged domain of the probe can separate the microRNA•probe complex from free nucleic acids, and only lead the complex into the pore. PNA in the probe has been broadly applied in molecular biology, diagnostic assays and antisense therapies due to the guaranteed binding strength and specificity[38-41]. The use of PNA offers greater specificity in detecting miRNAs with single-nucleotide difference. The polycationic peptide domain of the probe realizes 1) separating the probe-bound microRNA from free nucleic acids for enrichment, 2) trapping the microRNA•probe complex into the nanopore, and 3) enhancing the sensitivity by promoting the capture rate. The peptide’s sequence and structure are programmable by tuning the polymer length and in particular the number of charged amino acids and their positions in the sequence. There are many chemical methods available to modify the peptide property, in order to generate varying signatures for multiplex microRNA detection. This simple nanopore method is universal to detecting any short pathogenic DNA or RNA fragments for disease prediction, dianostics and prognostics.
*.
Research is partially supported by NSF0546165 and NIH GM079613.
Contributor Information
Kai Tian, Department of Biological Engineering and Dalton Cardiovascular Research Center, University of Missouri, Columbia, MO 65211, USA..
Li-Qun Gu, Department of Biological Engineering and Dalton Cardiovascular Research Center, University of Missouri, Columbia, MO 65211, USA.
References
- [1].Bayley H et al. , "Functional engineered channels and pores -(Review)," Molecular Membrane Biology, Vol. 21, pp. 209–220, Jul. 2004 [DOI] [PubMed] [Google Scholar]
- [2].Bayley H et al. , "Droplet interface bilayers," Mol. Biosyst, Vol. 4, pp. 1191–1208, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gu LQ et al. , "Single molecule sensing by nanopores and nanopore devices," Analyst, Vol. 135, pp. 441–451, Mar. 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hall AR et al. , "Hybrid pore formation by directed insertion of alpha-haemolysin into solid-state nanopores," Nat Nanotechnol, Vol. 5, pp. 874–877, December 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hornblower B et al. , "Single-molecule analysis of DNA-protein complexes using nanopores," Nat Methods, Vol. 4, pp. 315–317, Apr. 2007 [DOI] [PubMed] [Google Scholar]
- [6].Howorka S et al. , "Nanopore analytics: Sensing of single molecules," Chemical Society Reviews, Vol. 38, pp. 2360–2384, 2009 [DOI] [PubMed] [Google Scholar]
- [7].Ma L et al. , "Biological nanopores for single-molecule biophysics," Chembiochem, Vol. 11, pp. 25–34, Jan. 2010 [DOI] [PubMed] [Google Scholar]
- [8].Majd S et al. , "Applications of biological pores in nanomedicine, sensing, and nanoelectronics," Current Opinion in Biotechnology, Vol. 21, pp. 439–476, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Movileanu L, "Interrogating single proteins through nanopores: challenges and opportunities," Trends Biotechnol, Vol. 27, pp. 333–341, 2009 [DOI] [PubMed] [Google Scholar]
- [10].Olasagasti F et al. , "Replication of individual DNA molecules under electronic control using a protein nanopore," Nat. Nanotechnol, Vol. 5, pp. 798–806, Nov. 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Venkatesan BM et al. , "Nanopore sensors for nucleic acid analysis," Nat Nanotechnol, Vol. 6, pp. 615–624, Oct. 2011 [DOI] [PubMed] [Google Scholar]
- [12].Wanunu M et al. , "Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient," Nat. Nanotechnol, Vol. 5, pp. 160–165, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wendell D et al. , "Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores," Nat Nanotechnol, Vol. 4, pp. 765–772, Nov. 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Langecker M et al. , "Synthetic lipid membrane channels formed by designed DNA nanostructures," Science, Vol. 338, pp. 932–936, Nov. 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Branton D et al. , "The potential and challenges of nanopore sequencing," Nature Biotechnology, Vol. 26, pp. 1146–1153, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cherf GM et al. , "Automated forward and reverse ratcheting of DNA in a nanopore at 5-A precision," Nat Biotechnol, Vol. 30, pp. 344–348, Apr. 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kasianowicz JJ et al. , "Characterization of individual polynucleotide molecules using a membrane channel," Proc. Natl. Acad. Sci. U. S. A, Vol. 93, pp. 13770–13773, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Manrao EA et al. , "Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase," Nat Biotechnol, Vol. 30, pp. 349–353, Apr. 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wallace EV et al. , "Identification of epigenetic DNA modifications with a protein nanopore," Chem. Commun. (Camb.), Vol. 46, pp. 8195–8197, Nov. 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].An N et al. , "Crown ether-electrolyte interactions permit nanopore detection of individual DNA abasic sites in single molecules," Proc Natl Acad Sci USA, Vol. 109, pp. 11504–11509, Jul. 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shim JW et al. , "Encapsulating a single G-quadruplex aptamer in a protein nanocavity," J Phys Chem B, Vol. 112, pp. 8354–8360, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Shim JW et al. , "Single-molecule detection of folding and unfolding of a single G-quadruplex aptamer in a nanopore nanocavity," Nucleic Acids Research, Vol. 37, pp. 972–982, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang Y et al. , "Nanopore-based detection of circulating microRNAs in lung cancer patients," Nat. Nanotechnol, Vol. 6, pp. 668–674, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wanunu M et al. , "Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors," Nat. Nanotechnol, Vol. 5, pp. 807–814, 2010 [DOI] [PubMed] [Google Scholar]
- [25].Carthew RW et al. , "Origins and Mechanisms of miRNAs and siRNAs," Cell, Vol. 136, pp. 642–655, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lee RC et al. , "The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14," Cell, Vol. 75, pp. 843–854, 1993 [DOI] [PubMed] [Google Scholar]
- [27].Kim VN et al. , "Biogenesis of small RNAs in animals," Nat. Rev. Mol. Cell Biol, Vol. 10, pp. 126–139, 2009 [DOI] [PubMed] [Google Scholar]
- [28].Wightman B et al. , "Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans," Cell, Vol. 75, pp. 855–862, 1993 [DOI] [PubMed] [Google Scholar]
- [29].Boeri M et al. , "MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer," Proc. Natl. Acad. Sci. U. S. A, Vol. 108, pp. 3713–3718, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hu Z et al. , "Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer," J Clin. Oncol, Vol. 28, pp. 1721–1726, Apr. 2010 [DOI] [PubMed] [Google Scholar]
- [31].Hunt EA et al. , "Direct detection and quantification of microRNAs," Anal. Biochem, Vol. 387, pp. 1–12, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Iorio MV et al. , "MicroRNAs in cancer: Small molecules with a huge impact," J. Clin. Oncol, Vol. 27, pp. 5848–5856, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Landi MT et al. , "MicroRNA expression differentiates histology and predicts survival of lung cancer," Clin. Cancer Res, Vol. 16, pp. 430–441, Jan. 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mitchell PS et al. , "Circulating microRNAs as stable blood-based markers for cancer detection," Proc. Natl. Acad. Sci. U. S. A, Vol. 105, pp. 10513–10518, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shen J et al. , "Plasma microRNAs as potential biomarkers for non-small-cell lung cancer," Lab Invest, Vol. 91, pp. 579–587, Apr. 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sozzi G et al. , "Plasma DNA quantification in lung cancer computed tomography screening: five-year results of a prospective study," Am. J Respir. Crit Care Med, Vol. 179, pp. 69–74, Jan. 2009 [DOI] [PubMed] [Google Scholar]
- [37].Zheng D et al. , "Plasma micrornas as novel biomarkers for early detection of lung cancer," Int. J. Clin. Exp. Pathol, Vol. 4, pp. 575–586, 2011 [PMC free article] [PubMed] [Google Scholar]
- [38].Nielsen PE et al. , "An introduction to peptide nucleic acid," Curr. Issues Mol. Biol, Vol. 1, pp. 89–104, 1999 [PubMed] [Google Scholar]
- [39].Nielsen PE et al. , "Peptide nucleic acid (PNA) cell penetrating peptide (CPP) conjugates as carriers for cellular delivery of antisense oligomers," Artif. DNA PNA. XNA, Vol. 2, pp. 90–99, Jul. 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Singer A et al. , "Nanopore based sequence specific detection of duplex DNA for genomic profiling," Nano Lett, Vol. 10, pp. 738–742, Feb. 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Natsume T et al. , "Hybridization energies of double strands composed of DNA, RNA, PNA and LNA," Chem Phys Lett, Vol. 434, pp. 133–138, Jan. 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]