Abstract
Biological interpretation of GWAS data frequently involves assessing whether SNPs linked to a biological process, e.g., binding of a transcription factor (TF), show unsigned enrichment for disease signal. However, signed annotations quantifying whether each SNP allele promotes or hinders the biological process can enable stronger statements about disease mechanism. We introduce a method, signed LD profile regression, for detecting genome-wide directional effects of signed functional annotations on disease risk. We validate the method via simulations and application to molecular QTL in blood, recovering known transcriptional regulators. We apply the method to eQTL in 48 GTEx tissues, identifying 651 TF-tissue associations including 30 with robust evidence of tissue specificity. We apply the method to 46 diseases and complex traits (average N=290K), identifying 77 annotation-trait associations representing 12 independent TF-trait associations, and characterize the underlying transcriptional programs using gene-set enrichment analyses. Our results implicate new causal disease genes and new disease mechanisms.
Introduction
Mechanistic interpretation of GWAS data has become a central challenge for understanding the biological underpinnings of disease. One successful paradigm for such efforts has been GWAS enrichment, in which a genome annotation containing SNPs that affect some biological process is shown to be enriched for GWAS signal1–7. However, there are instances in which experimental data allow us not only to identify SNPs that affect a biological process, but also to predict which SNP alleles promote the process and which SNP alleles hinder it, enabling us to assess whether there is a systematic association between SNP alleles’ direction of effect on the process and their direction of effect on a trait. Transcription factor (TF) binding, which plays a major role in human disease1,8–12, represents an important case in which such signed functional annotations are available: because TFs have a tendency to bind to specific DNA sequences, it is possible to estimate whether the sequence change introduced by a SNP allele will increase or decrease binding of a TF1,13–19.
Detecting genome-wide directional effects of TF binding on disease would constitute a significant advance in terms of both evidence for causality and understanding of biological mechanism. Regarding causality, this is because directional effects are not confounded by simple co-localization in the genome (e.g., of TF binding sites with other regulatory elements), and thus provide stronger evidence for causality than is available using unsigned enrichment methods. Regarding biological mechanism, it is currently unknown whether disease-associated TFs regulate only a few key disease genes or whether broad transcriptional programs comprising many target genes are responsible for TF associations; a genome-wide directional effect implies the latter model (Discussion).
Here we introduce a new method, signed LD profile (SLDP) regression, for quantifying the genome-wide directional effect of a signed functional annotation on polygenic disease risk, and apply it with 382 annotations each reflecting predicted binding of a particular TF. Our method requires only GWAS summary statistics20, accounts for linkage disequilibrium and untyped causal SNPs, and is computationally efficient. We validate the method via extensive simulations and further validate it by applying it to molecular QTL in blood21, recovering known transcriptional regulators. We then apply the method to eQTL in 48 tissues from the GTEx consortium22 and to 46 diseases and complex traits, demonstrating genome-wide directional effects of TF binding in both settings. We further characterize the transcriptional programs underlying our complex trait associations via gene-set enrichment analyses using gene sets from the Molecular Signatures Database23,24 (MSigDB).
Results
Overview of methods
Our method for quantifying directional effects of signed functional annotations on disease risk, SLDP regression, relies on the fact that the signed marginal association of a SNP to disease includes signed contributions from all SNPs tagged by that SNP. Given a signed functional annotation with a directional effect on disease risk, the vector of marginal SNP effects on disease risk will therefore be proportional (in expectation) to a vector quantifying each SNP’s aggregate tagging of the signed annotation, which we call the signed LD profile of the annotation. Thus, our method detects directional effects by assessing whether the vector of marginal SNP effects and the signed LD profile are systematically correlated genome-wide.
More precisely, under a polygenic model25 in which true causal SNP effects are correlated with a signed functional annotation, we show that
| (1) |
where is the vector of marginal correlations between SNP alleles and a trait, v is the signed functional annotation (re-scaled to norm 1) reflecting, e.g., the signed effect of a SNP on TF binding, R is the LD matrix, is the SNP-heritability of the trait, and rf is the correlation between the vector v and the vector of true causal effects of each SNP, which we call the functional correlation. Equation (1), together with an estimate of , allows us to estimate rf by regressing on the signed LD profile Rv of v. To improve power, we use generalized least-squares regression to account for redundancy among linked SNPs. We assess statistical significance by randomly flipping the signs of entries of v, with consecutive SNPs being flipped together in large blocks (~300 blocks total), to obtain a null distribution and corresponding P-values and false discovery rates (FDRs). We perform a multiple regression that explicitly conditions on a “signed background model” corresponding to directional effects of minor alleles in five equally sized minor allele frequency (MAF) bins, which could reflect confounding due to genome-wide negative selection or population stratification. We note that SLDP regression requires signed effect size estimates and quantifies directional effects, in contrast to stratified LD score regression5, which analyzes unsigned χ2 statistics and quantifies unsigned heritability enrichment. Details of and intuition for the method are described in the Methods section and the Supplementary Note; we have released open-source software implementing the method (URLs).
We applied SLDP regression using a set of 382 signed annotations v, constructed using the Basset software19, each quantifying the predicted effects of SNP alleles on binding of a particular TF in a particular cell line. The resulting annotations were sparse, with only 0.2% of SNPs having nonzero entries on average. (Methods and Supplementary Table 1.)
Simulations
We performed simulations with real genotypes, simulated phenotypes, and our 382 signed TF binding annotations to assess null calibration, robustness to confounding, and power (Methods).
We first performed null simulations involving a heritable trait with no unsigned enrichment or directional association to any of our 382 annotations. The resulting P-values were well-calibrated (Figure 1a, Supplementary Table 2, and Supplementary Figure 1a).
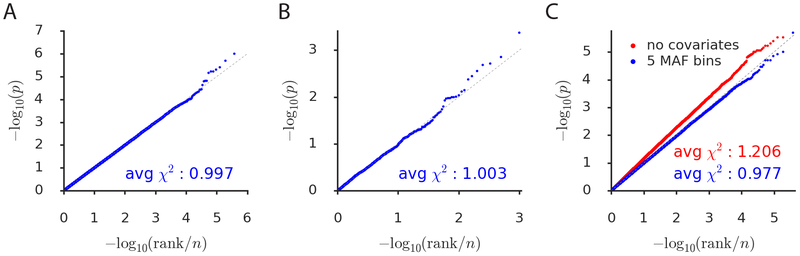
Figure 1: Simulations assessing null calibration.
We report null calibration (q-q plots of two-sided SLDP regression −log10(p) values) in simulations of (a) no enrichment, (b) unsigned enrichment, and (c) directional effects of minor alleles. The q-q plots are based on (a) 382 annotations × 1,000 simulations = 382,000, (b) 1,000, and (c) two sets of 382 × 1,000 = 382,000 P-values. A 5-MAF-bin signed background model is included in all cases except for the red points in part (c), which are computed with no covariates. We also report the average χ2-statistic corresponding to each set of P-values. Numerical results are reported in Supplementary Table 2.
We next performed null simulations involving a trait with unsigned enrichment but no directional effects; these simulations were designed to mimic unsigned genomic confounding as might arise from the co-localization of TF binding sites with other enriched regulatory elements5,13 (Methods). We again observed well-calibrated P-values (Figure 1b). It is notable that our method is well-calibrated even though it has no knowledge of the unsigned genomic confounder; this contrasts with unsigned enrichment approaches, in which unsigned genomic confounders must be carefully accounted for and modeled5.
We next performed null simulations to assess whether our method remains well-calibrated in the presence of confounding due to genome-wide directional effects of minor alleles on both disease risk and TF binding, which could arise due to genome-wide negative selection or population stratification (Methods). P-values were well-calibrated for the default version of the method, which conditions on the 5-MAF-bin signed background model, but were not well-calibrated without conditioning on this model (Figure 1c). The incorrect calibration that we observe when we do not include our signed background model could potentially be explained by genome-wide negative selection against decreased TF binding26 resulting in a bias in the sign of the entries of our annotations (Supplementary Figure 2). We condition on the signed background model in all analyses in this paper unless stated otherwise.
Finally, we performed causal simulations with true directional effects to assess the power and establish the unbiasedness of SLDP regression (Methods). The method is well-powered to detect directional effects corresponding to a functional correlation of 2–6% (Figure 2a, Supplementary Table 3, and Supplementary Figures 3–5), similar to values observed in analyses of real traits (see below). Notably, the power of the method is improved dramatically by its use of generalized least-squares to account for redundant information (Figure 2a) as well as by its modelling of untyped causal SNPs via the signed LD profile (Supplementary Figure 3). In all instances, our method produced either unbiased or nearly unbiased estimates of functional correlation and related quantities (Figure 2b and Supplementary Figure 6).
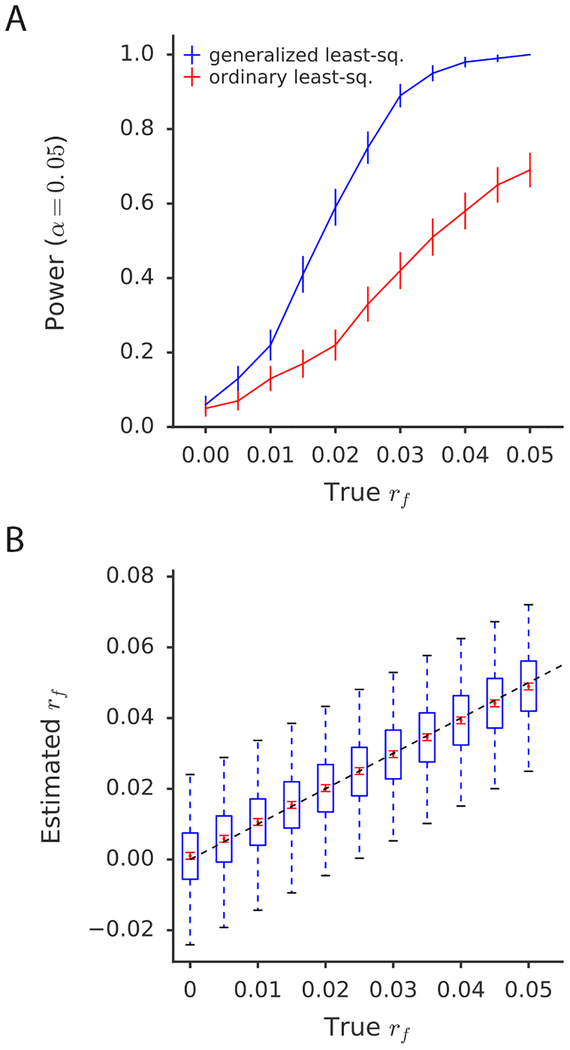
Figure 2: Simulations assessing power, bias, and variance.
(a) Power curves comparing SLDP regression using generalized least-squares (i.e., weighting) to an ordinary (i.e., unweighted) regression of the summary statistics on the signed LD profile. Error bars indicate standard errors of power estimates. (b) Assessment of bias and variance of the SLDP regression estimate of rf, across a range of values of the true rf. Blue box and whisker plots depict the sampling distribution of the statistic, while the red dots indicate the estimated sample mean and the red error bars indicate the standard error around this estimate. Both (a) and (b) are conducted at realistic sample size (47,360) and heritability (0.5). Numerical results are reported in Supplementary Table 3.
Analysis of molecular traits in blood
TF binding is known to affect gene expression and other molecular traits27, and regulatory relationships in blood are particularly well-characterized28. We therefore applied SLDP regression to 12 molecular traits in blood with an average sample size of N = 149, to further validate the method.
We first analyzed cis-eQTL data based on RNA-seq experiments in three blood cell types from the BLUEPRINT consortium21 (Supplementary Table 4). We tested each of our 382 TF binding annotations for a directional effect on aggregate expression in each of the three blood cell types (Methods). We detected 409 significant associations at per-trait FDR<5%, representing 107 distinct TF-blood cell type expression associations (Figure 3a and Supplementary Table 5a). All of the detected associations were positive, implying that greater binding of these TFs leads to greater expression (in aggregate across genes); 78% of the associations involved TFs annotated as activating but not repressing in UnitProt29 (Figure 3a and Methods). As expected, many of the detected associations recapitulate known aspects of transcriptional regulation, including the pro-transcriptional roles of RNA polymerase II and other members of the transcription pre-initiation complex (PIC) as well as roles of TFs unrelated to the PIC but known to have activating activity30–32. We obtained similar results in an independent set of whole-blood eQTL based on expression array experiments from the Netherlands Twin Registry33 (Figures 3b–c and Supplementary Table 5b–c).
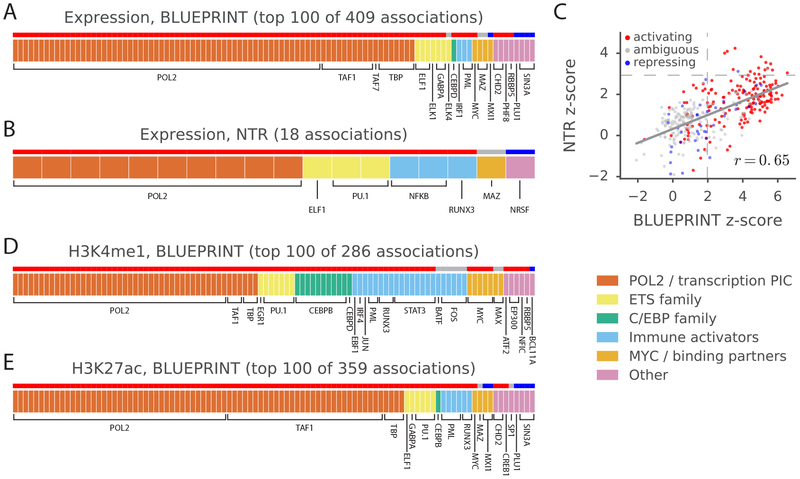
Figure 3: Analysis of blood molecular traits using signed LD profile regression.
Each segmented bar in (a,b,d,e) represents the set of significant annotations (or top 100 annotations) at a per-trait FDR<5% for the indicated traits, with each annotation corresponding to a particular TF profiled in a particular cell line. Results in (a,d,e) are aggregated across the 3 BLUEPRINT cell types. The stripe above each segmented bar is colored red for TFs with activating activity and no repressing activity in UniProt (see main text and Methods), blue for TFs with repressing activity and no activating activity, and gray for ambiguous TFs. (c) z-scores from the analyses of expression in the NTR data set and neutrophil expression in the BLUEPRINT data set, respectively, for each of the 382 annotations tested; red, blue, and gray again indicate UniProt (unambiguously) activating TFs, (unambiguously) repressing TFs, and ambiguous TFs, respectively. Dashed lines represent significance thresholds for 5% FDR. GWAS data are described in Supplementary Table 4, and the statistical method and multiple comparisons adjustments are described in the “Overview of methods” section and the Methods. Numerical results are reported in Supplementary Table 5.
We next conducted a similar analysis using histone QTL (H3K27me1 and H3K27ac) and methylation QTL for the three cell types in the BLUEPRINT data set. We detected 645 significant associations at per-trait FDR<5%, four of which were negative (Figure 3d,e and Supplementary Table 5d,e). Again, the majority of the positive associations (82%) involved unambiguously activating TFs29. The four negative associations involved MAFK and MAFF, both of which lack a transactivation domain34, and CTCF, which is known to act as an insulator35,36. Many of the detected associations recover known aspects of histone mark biology36–39,40,41,43,44 and match a prior analysis of allelic imbalance in ChIP-seq data45.
Analysis of gene expression across 48 GTEx tissues
We next applied SLDP regression to GTEx eQTL across 48 tissues22 (average N = 149). We first tested each of our 382 TF binding annotations for a directional effect on expression in each of the 48 tissues in turn, analogous to our previous analysis of molecular traits in blood (Supplementary Table 6). For each significant association, we then assessed for tissue specificity by checking whether the association remained at least as significant when conditioning on average eQTL effects across tissues (Methods).
Our analysis yielded 2,330 annotation-tissue expression associations at per-trait FDR<5%, representing 651 distinct TF-tissue expression associations of which 30 were robustly tissue-specific in our conditional analysis (Figure 4 and Supplementary Table 7). We detected both known and novel associations. For example, our results recapitulate known activating roles for FOXA1 and FOXA2 in pancreas and other gastrointestinal tissues37–39, early B-cell factor 1 (EBF-1) in lymphocytes40,41, hepatocyte nuclear factors 4γ and 4α (HNF4G and HNF4A) in liver42,43, PU.1 in spleen44, and FOS in fibroblasts45 and nerve tissue46–48. We also detected ubiquitous activating signatures for the transcription pre-initiation complex members POL2, TAF1, and TBP (90% of the 28 tissues with a sample size above 150). Our results were concordant with TF-tissue associations identified via a purely gene expression-based analysis (Methods and Supplementary Figure 7).
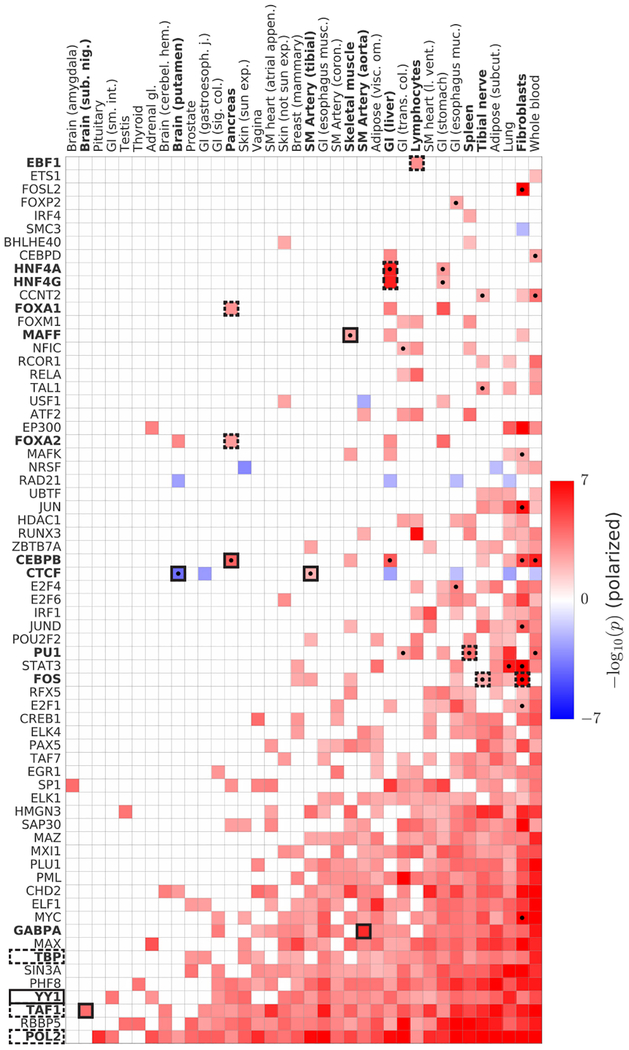
Figure 4: Analysis of GTEx eQTLs using signed LD profile regression.
We plot polarized −log10(p) values for all significant associations as a heatmap. Columns denote the 36 GTEx tissues (of 48 GTEx tissues tested) with significant associations. Rows denote the 67 TFs (of 75 TFs tested) with significant associations, collapsing all annotations corresponding to a single TF into one row and displaying in each cell the most significant result. Cells with dots indicate associations that show robust evidence for tissue-specificity in our conditional analysis (see main text and Methods). Cells indicated in outline correspond to associations described in the main text, with dashed outline indicating known associations and solid outline indicating previously unknown associations or associations supporting emerging theories. GWAS data are described in Supplementary Table 6, and the statistical method and multiple comparisons adjustments are described in the “Overview of methods” section and the Methods. Numerical results are reported in Supplementary Table 7.
Our analysis also uncovered many previously unknown associations. For example, our most significant association in aorta is a previously unreported activating role for GABPA, one of several TFs whose binding sites are enriched near aortic aneurysm-specific genes49. In addition, our top — and only — association in the brain tissue substantia nigra is TAF1. Neurodegeneration in the substantia nigra is a hallmark of Parkinson’s disease50 and TAF1 was shown earlier this year to be the causal gene in a rare form of Parkinsonism51. Our analysis links these two facts, potentially shedding light on the mechanism of TAF1’s role in Parkinsonism.
Our tissue-specific analysis (Methods) also suggests new master-regulatory relationships for further exploration (Figure 4). For example, we detected a robust tissue-specific activating role for CEBPB in pancreas, where it was our top result. Though CEBPB is not a classic pancreatic TF52, it is expressed in pancreatic beta cells specifically under metabolic stress52. We also identified a robust tissue-specific activating role in skeletal muscle for MAFF, a TF whose expression is increased by an order of magnitude in muscle tissue after exercise53 (Methods). MAFF is typically considered a repressor, and we identified it as such in our blood hQTL analysis; the positive association here suggests a tissue-specific function in muscle, perhaps via recruitment of an as-yet uncharacterized activator. Finally, we identified robust tissue-specific roles for CTCF as a repressor in tibial artery and an activator in the brain tissue putamen. While CTCF is known to be capable of both repression and activation35,36,54, these results suggest that its repressive/activating role varies meaningfully across tissues.
Our results also demonstrate how our method can offer insights into non-tissue-specific aspects of transcriptional regulation. For example, YY1, a pioneer TF that has recently attracted considerable interest55–58, has been theorized via detailed experimental work to mediate enhancer-promoter interaction59. However, YY1 knockdown experiments have shown a mix of up-regulation and down-regulation of many genes59, presumably due to downstream regulatory cascades. In contrast, our analysis, which due to its use of eQTLs is able to focus exclusively on cis-regulatory effects, shows a robust, predominantly activating role for YY1 across 25 tissues.
Analysis of 46 diseases and complex traits
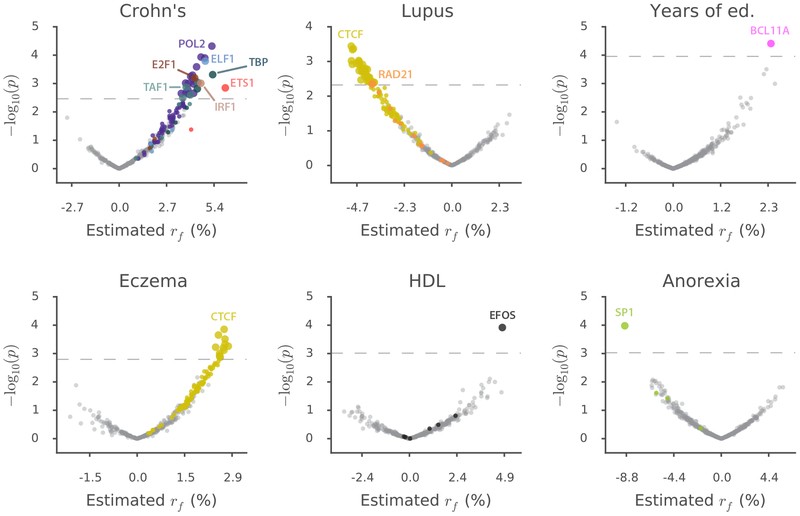
We applied SLDP regression to 46 diseases and complex traits with an average sample size of 289,617 (URLs and Supplementary Table 8). We first tested each of our 382 TF binding annotations against each of the 46 traits in turn (Table 1 and Supplementary Table 9). For each significant association, we then characterized the implicated transcriptional programs by evaluating 10,325 gene sets from MSigDB23,24 (URLs) for enrichment among the genomic regions driving the association (controlling for LD and co-localizing genes; Methods) (Table 1 and Supplementary Table 10).
Table 1. Independent TF-trait associations from analysis of diseases and complex traits using signed LD profile regression.
For each of 12 independent associations at per-trait FDR<5% after pruning correlated annotations (R2 ≥ 0.25), we report the associated trait; the TF of the most significant annotation and the number of correlated annotations with significant associations; the estimated functional correlation rf, P-value, q-value, and minimum number of TF binding sites contributing to the association; and the top two significant MSigDB gene-set enrichments among loci driving the association.
| Trait | Top TF (#) | rf | p | q | Min. # sites | Top 2 significant MSigDB enrichments |
|---|---|---|---|---|---|---|
| Years of ed. | BCL11A (1) | 2.4% | 3.9×10−5 | 1.5×10−2 | 104 |
|
| Crohn’s | POL2* (20) | 5.3% | 4.8×10−5 | 1.5×10−2 | 74 |
|
| Anorexia | SP1 (1) | −8.9% | 1.1×10−4 | 4.0×10−2 | 30 |
|
| HDL | FOS (1) | 4.8% | 1.2×10−4 | 4.6×10−2 | 19 |
|
| Eczema | CTCF (12) | 2.7% | 1.4×10−4 | 3.4×10−2 | 106 |
|
| Crohn’s | ELF1 (1) | 4.9% | 1.6×10−4 | 1.5×10−2 | 58 |
|
| Crohn’s | POL2 (1) | 4.4% | 2.6×10−4 | 1.5×10−2 | 50 |
|
| Lupus | CTCF** (36) | −5.0% | 3.6×10−4 | 4.4×10−2 | 100 |
|
| Crohn’s | TBP (1) | 5.4% | 4.9×10−4 | 1.5×10−2 | 54 |
|
| Crohn’s | E2F1 (1) | 4.3% | 6.4×10−4 | 2.7×10−2 | 90 |
|
| Crohn’s | IRF1 (1) | 4.7% | 9.8×10−4 | 1.5×10−2 | 90 |
|
| Crohn’s | ETS1 (1) | 6.1% | 1.4×10−3 | 1.5×10−2 | 114 |
|
Linked TFs producing significant associations:
TAF1, TBP;
RAD21. See Supplementary Table 10 for full gene set names and enrichment q-values (all <5×10−2). GWAS data are described in Supplementary Table 8, gene set data are described in the Methods, and the statistical method and multiple comparisons adjustments are described in the “Overview of methods” section and the Methods. LMPP: lymphoid-primed pluripotent progenitor; GMP: granulocyte-monocyte precursor.
Our analysis yielded 77 significant annotation-trait associations at per-trait FDR<5%, spanning six diseases and complex traits (Figure 5 and Supplementary Table 9a) and representing 12 independent TF-trait associations (after pruning correlated annotations; Table 1 and Supplementary Note). Our results were 4.3× enriched for auto-immune disease associations (Supplementary Note). We verified empirically that our results are not driven by directional effects of minor alleles (Supplementary Table 9b and Supplementary Note), and we computed a lower bound on the number of independent TF binding sites contributing to each association (74 on average; Table 1, Supplementary Figure 8, and Methods).
Figure 5: Analysis of diseases and complex traits using signed LD profile regression.
For each disease or complex trait with at least one significant result, we plot −log10(p) against estimated effect size for each of the 382 annotations analyzed. Points are colored by TF identity, with TFs with no significant associations for each trait colored in gray. Larger points denote significant results. The number of significant results for each trait is: Crohn’s, 26; Lupus, 36; Years of education, 1; Eczema, 12; HDL, 1; Anorexia, 1. GWAS data are described in Supplementary Table 8, and the statistical method and multiple comparisons adjustments are described in the “Overview of methods” section and the Methods. Numerical results are reported in Supplementary Table 9a.
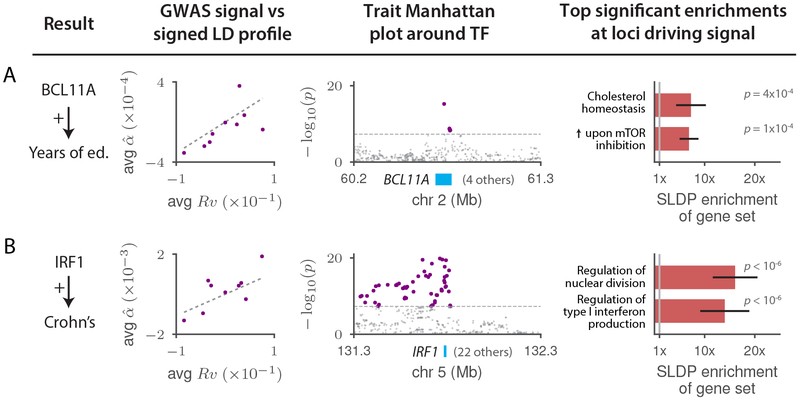
Of our 12 independent TF-trait associations, five refine emerging theories of disease while seven are previously unknown. Due to space restrictions, we highlight two relationships from each category (Figure 6 and Supplementary Table 11; Figure 7 and Supplementary Table 12), providing discussion of additional relationships in the Supplementary Note. We begin with the TF-trait associations that build on previous knowledge (Figure 6). First, we detected a positive association between genome-wide binding of BCL11A and years of education (Figure 6a) that aligns with existing evidence from educational attainment GWAS60, rare-variant studies of intellectual disability61–64, experimental knockout work in mice64, and fine-mapping of the BCL11A GWAS locus (Supplementary Table 13). Our result suggests that BCL11A causes intellectual disability not via regulation of a few key disease genes but rather via binding throughout the genome causing modulation (in cis) of genes comprising a broad transcriptional program relevant to brain function or development (Discussion). Furthermore, our MSigDB gene-set enrichment analysis allows us to characterize this putative transcriptional program as being significantly enriched for genes involved in mTOR signaling and in cholesterol metabolism (Figure 6a and Supplementary Table 10). MTOR is an intellectual disability gene65,66 with links to cholesterol67,68, defects in brain cholesterol metabolism have been linked to central nervous system disease69,70, and BCL11A has also been linked to lipid levels71–73. These observations raise the possibility that mTOR causes intellectual disability by interacting with BCL11A to regulate cholesterol metabolism in the developing brain (Supplementary Note).
Figure 6: Highlighted TF binding-complex trait associations that either provide biological insight into established genetic associations or refine emerging theories.
For each of (a) BCL11A-Years of education and (b) IRF1-Crohn’s disease, we display plots of the marginal correlation of SNP to trait versus the signed LD profile Rv of the annotation in question, with SNPs averaged in bins of 4,000 SNPs of similar Rv values and a larger bin around Rv = 0; Manhattan plots of the trait GWAS signal near the associated TF; and the top two significant MSigDB gene-set enrichments among the loci driving the association, with error bars indicating standard errors. GWAS data are described in Supplementary Table 8, gene set data are described in the Methods, and the statistical method and multiple comparisons adjustments are described in the “Overview of methods” section and the Methods. Numerical results are reported in Supplementary Table 11.
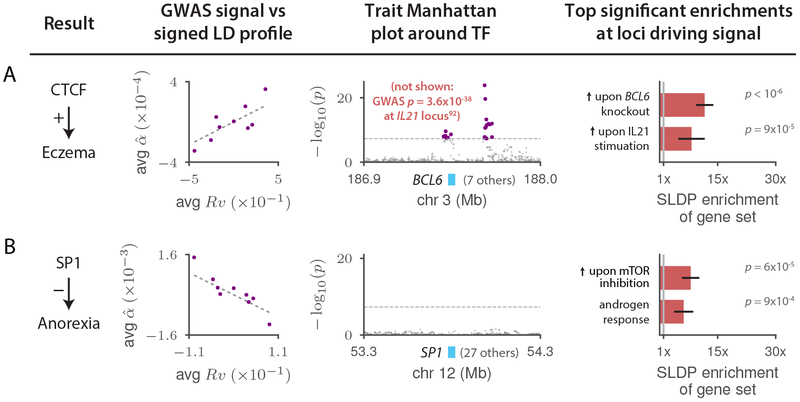
Figure 7: Highlighted previously unknown TF binding-complex trait associations.
For each of (a) CTCF-Eczema and (b) SP1-Anorexia, we display plots of the marginal correlation of SNP to trait versus the signed LD profile Rv of the annotation in question, with SNPs averaged in bins of 4,000 SNPs of similar Rv values and a larger bin around Rv = 0; Manhattan plots of the trait GWAS signal near the associated TF or, in the case of CTCF-Eczema, the BCL6 gene (see main text; there is no GWAS peak at CTCF); and the top two significant MSigDB gene-set enrichments among the loci driving the association, with error bars indicating standard errors. GWAS data are described in Supplementary Table 8, gene set data are described in the Methods, and the statistical method and multiple comparisons adjustments are described in the “Overview of methods” section and the Methods. Numerical results are reported in Supplementary Table 12.
Second, we detected a positive association between genome-wide binding of interferon regulatory factor 1 (IRF1) and Crohn’s disease (CD) (Figure 6b), a case in which existing GWAS evidence has been suggestive but not conclusive. Although IRF1 lies in a locus associated with CD in multiple GWAS74–76 (one of the earliest CD associations77), this locus remains mysterious. Strong LD makes it challenging to determine which variant(s) are causal, and high gene density (23 protein-coding genes within 500kb of IRF1) complicates the task of determining which gene is affected by any putative causal variant, resulting in several genes74,78,79 being previously nominated as potentially causal. For example, a recent large-scale fine-mapping study80 narrowed the causal signal to 8 SNPs including rs2188962, an eQTL for SLC22A5 in immune and gut epithelial cells22,80 but also for IRF1 in blood33. Transcriptome-wide association studies have also been inconclusive81–83. Our result provides genome-wide evidence for a causal link between IRF1 and CD that, unlike single-locus approaches, is not fundamentally limited by LD and pleiotropy near the IRF1 gene (Discussion). The top results in our MSigDB gene-set enrichment analysis strengthen our finding: the regions driving this association are most significantly enriched for genes involved in production of type I interferon and regulation of nuclear division (Figure 6b and Supplementary Table 10), matching well-known roles of IRF184,85. We note that several other TF-trait associations from our analysis implicate causal genes at established GWAS loci, including ELF1-CD and ETS1-CD, with gene-set enrichments suggesting connections to existing CD drugs and to the role of autophagy in CD pathogenesis, respectively (Table 1 and Supplementary Note).
We next discuss two selected TF-trait associations that were previously unknown (Figure 7). First, we detected a positive association between genome-wide binding of CTCF and eczema (Figure 7a). We do not observe a GWAS signal for eczema at the CTCF locus. This could be because the CTCF gene is under strong selective constraint (probability of loss-of-function intolerance86 = 1.00, greater than 99.9% of genes) and highlights the potential of our method to uncover causal roles for genes that harbor relatively little variation. The top two significant MSigDB gene-set enrichments for CTCF-Eczema are convergent: genes up-regulated in Treg cells upon knockout of the inflammatory regulator BCL6; and genes up-regulated in response to stimulation by the immune signaling molecule IL21, which is a known regulator of BCL6 activity87,88 (Figure 7a and Supplementary Table 10). These enrichments, because they pertain to genes putatively regulated in cis by CTCF to cause eczema, suggest a detailed cascade that we hypothesize to modulate eczema risk: IL21 signaling regulates BCL6, which in turn regulates CTCF to activate a broad transcriptional program that increases eczema risk. This hypothesis makes three predictions: it predicts that BCL6 modulates CTCF activity, and it predicts that IL21 and BCL6 each affect eczema risk. Indeed, we determined that BCL6 has many binding sites near the CTCF promoter in publicly available ChIP-seq data69,89–91 (Supplementary Table 14), and the IL21 and BCL6 genes each fall in eczema GWAS loci92–94 (in each case along with 7 other protein-coding genes within 500kb). Thus, the association between CTCF binding and eczema that we detected nominates causal genes at two different existing eczema GWAS loci and provides a parsimonious mechanism that explains their effect on eczema via a regulatory cascade that activates a CTCF-mediated transcriptional program.
Second, we detected a negative association between genome-wide binding of SP1 and anorexia (Figure 7b), a heritable trait for which no loci reach genome-wide significance in our GWAS data95. SP1 levels observationally correlate negatively with psychiatric conditions such as schizophrenia96,97 (which is positively genetically correlated with anorexia98), but this association has not been shown to be causal and has not previously been observed in GWAS of psychiatric traits. Our MSigDB gene-set enrichment results for this association yielded significant enrichments for an androgen response gene set and an mTOR signaling gene set (Figure 7b and Supplementary Table 10). (Years of education, for which an mTOR signaling gene-set was also among the top two MSigDB enrichments, is also significantly positively genetically correlated with anorexia98; the median rank of the top-scoring mTOR gene set across the 10 other independent TF-complex trait associations was 1,123, of 10,325.) The androgen response result is intriguing given the sex-imbalanced nature of this phenotype99. The mTOR signaling result is noteworthy given the well-established connections between mTOR, caloric restriction, and growth100; it also suggests that a link between SP1 and mTOR could explain prior observations tying SP1 to insulin101,102, appetite103,104, and energy metabolism105. mTOR has also been shown to play an important role in androgen signaling106, suggesting a potential unification of these two signals.
We provide additional discussion of other TF-trait associations in the Supplementary Note (Supplementary Figure 9 and Supplementary Tables 15 and 16).
Discussion
We have introduced a method, signed LD profile regression, for identifying genome-wide directional effects of signed functional annotations on diseases and complex traits. Our approach allows us to draw fine-grained biological conclusions that are not confounded by simple genomic co-localization of functional elements. The directional relationships we identify concretely implicate broad disease-relevant transcriptional programs. Our characterization of these programs via gene-set enrichment analyses yields detailed hypotheses about disease mechanisms that in several cases mechanistically link existing GWAS loci and disparate molecular evidence into a parsimonious mechanism mediated by the associated TF.
Our method differs from unsigned GWAS enrichment methods1–7 by assessing whether a systematic genome-wide correlation exists between a signed functional annotation and the (signed) true causal effects of SNPs on disease, rather than assessing whether a set of SNPs have large effects on a disease without regard to the directions of those effects. Our method also differs from single-locus GWAS methods11,12,81 in that a consistent genome-wide directional effect across a large set of TF binding sites (Table 1) is less susceptible to pleiotropy, LD, and allelic heterogeneity81,82. Finally, our method differs from genetic correlation and Mendelian randomization98 analyses, which can be confounded by reverse causality and pleiotropic effects107–109; in contrast, the sequence-based nature of our annotations makes them ideal instrumental variables for the effect of TF binding on the trait of interest. (Supplementary Note.)
The genome-wide nature of our method means that our results constitute instances in which TFs affect traits via coordinated regulation of gene expression throughout the genome110 (a “genome-wide” model) rather than via regulation of one or a small number of key disease genes111 (a “local” model). This distinction has potential implications for drug development as well as attempts to elucidate disease mechanisms (Supplementary Note). For example, as we have shown, the genome-wide nature of the putative transcriptional programs identified by our method allows us to characterize and interpret these programs by aligning them with existing gene sets, leading in some cases to detailed mechanistic hypotheses.
There exist many potentially effective methods for constructing signed TF binding annotations1,13–16,18,112,113 and many potential data sets on which to train them114–116. We present an initial exploration of alternative annotations generated using some of these, along with a discussion of potential signed annotations besides TF binding annotations, in the Supplementary Note (Supplementary Figures 10–14 and Supplementary Tables 17–20).
We note several limitations of signed LD profile regression. First, though our results are less susceptible to confounding due to their signed nature, they are not immune to it: in particular, our method cannot distinguish between two TFs that are close binding partners and thus share sequence motifs, and it likewise cannot distinguish between binding of the same TF in different cell types, as the resulting annotations could be highly correlated. Second, we used annotations constructed using data from cell lines, which is non-ideal because chromatin dynamics in cell lines do not necessarily match those in real tissue; we note, however, that though this reduces our power and the effect sizes we see, it does not introduce false positives into our results. Third, the interpretability of our MSigDB gene-set enrichment analysis is limited by the potential for distinct gene sets to have overlapping membership and for co-expressed genes to be included in the same gene sets; however, we believe this is somewhat ameliorated by the fact that we treat blocks of genes together in our empirical null (Methods). Due to space restrictions, additional limitations are discussed in the Supplementary Note.
Despite these limitations, signed LD profile regression is a powerful new way to leverage functional genomics data to draw mechanistic conclusions from GWAS about both diseases and underlying cellular processes.
Online Methods
Signed LD profile regression
Intuition
Our method for quantifying directional effects of signed functional annotations on disease risk, signed LD profile regression, relies on the following intuition. Suppose there are M SNPs and we are given a signed functional annotation, specified by a length-M vector v, with a directional linear effect on disease risk. For example, v might be a vector whose m-th entry is the effect of SNP m on binding of some TF. If we knew the length-M vector β of the true causal effects of the same SNPs on a trait, we could simply regress β on v to evaluate whether there is a non-trivial signed association across SNPs m between vm and βm. In reality, we cannot do this because we do not observe β; instead we observe a vector, denoted , of GWAS summary statistics describing the marginal correlation of every SNP to our trait of interest. This vector differs from β because it includes both causal and tagging effects, plus statistical noise. Specifically, it can be shown mathematically that, in expectation, will equal the matrix-vector product Rβ where R is the M × M LD matrix. Therefore, just as β would be proportional to v in the presence of a signed effect, would likewise be proportional to Rv, which is a vector capturing each SNP’s aggregate tagging of the signed annotation. This means that instead of regressing β on v (which is impossible since we do not observe β), we can regress on Rv. We call the vector Rv the signed LD profile of v, and thus our method is called signed LD profile regression. The remainder of our technical material is oriented toward i) weighting this regression to achieve optimal power, ii) being able to efficiently perform the required computations, iii) determining the proper way to test the null hypothesis of no signed effect, and iv) controlling for potential confounding due to directional effects of minor alleles.
Model and estimands
Let M be the number of SNPs in the genome. We assume a linear model:
| (2) |
where and are the standardized genotype vector and phenotype, respectively, of a randomly chosen individual from some population, is a vector of true causal effects of each SNP on phenotype, and represents environmental noise. Given a signed functional annotation , we then model
| (3) |
where the scalar μ represents the genome-wide directional effect of v on β, σ2 represents other sources of heritability unrelated to v, and the notation [⋅,⋅] is used to specify the mean and covariance of the distribution without specifying any higher moments.
Though we can estimate μ, its value depends on the units of the annotation and the heritability of the trait. Because of this, we focus instead on the functional correlation rf, which re-scales μ to be dimensionless and is defined as
| (4) |
where is the SNP-heritability of the phenotype and is the (signed) population LD matrix of the genotypes. The quantity rf can be interpreted as a form of genetic correlation; the value of cannot exceed the proportion of SNP-heritability explained by SNPs with non-zero values of v. (Note that rf can also be defined as a correlation between β and v; this definition is approximately equivalent in expectation under our random effects model, provided vT Rv ≈ |v|2.) We additionally estimate , the total phenotypic variance explained by the signed contribution of v to β, as well as . For annotations with small support, these quantities are expected to be small in magnitude. To see this, notice that cannot exceed the total (unsigned) phenotypic variance explained by SNPs with non-zero values of v. It follows that cannot exceed the proportion of (unsigned) SNP-heritability explained by SNPs with non-zero values of v. For more detail on the model and estimands, see the Supplementary Note.
Main derivation
Let be the genotype matrix in a GWAS of N individuals, with standardized columns, and let be the phenotype vector. In the Supplementary Note, we show that under the above model the following identity approximately holds:
| (5) |
where is a vector whose m-th entry contains the marginal correlation of SNP m to the phenotype and is the population LD matrix. Equation (1) from the main text can be derived from Equation (5) by re-scaling v so that vT Rv = 1, then substituting for μ.
We call Rv the signed LD profile of v. Equation (5) means that we can estimate μ by regressing on the signed LD profile using generalized least-squares with Ω: = σ2R2 + R/N as the inverse weight matrix. (We assign a regression weight of zero to SNPs in the major histocompatibility complex region.) It can be shown that if a) all causal SNPs are typed, b) sample size is infinite, and c) R is invertible, this method is equivalent to estimating β via and then regressing this estimate on v to obtain μ, which is the optimal regression-based approach in that setting. Note that because we generate P-values for hypothesis testing empirically (see below), we are guaranteed that our generalized least-squares scheme will remain well-calibrated even if our estimate of the matrix Ω is inaccurate due to, e.g., mis-match between the reference panel and the study population. Once we have estimated μ, we re-scale this estimate to yield an estimate of rf and other estimands of interest. For more detail on derivations and computational considerations, see the Supplementary Note.
Null hypothesis testing
To test the null hypothesis H0:μ = 0 (or, equivalently, H0:rf = 0), we split the genome into approximately 300 blocks of approximately the same size with the block boundaries constrained to fall on estimated recombination hotspots117. We then define the null distribution of our statistic as the distribution arising from independently multiplying v by one independent random sign per block. We perform this empirical sign-flipping many times to obtain an approximation of the null distribution and corresponding P-values. Our use of sign-flipping ensures that any true positives found by our method are the result of genuine first-moment effects; if in contrast we estimated standard errors using least-squares theory or a re-sampling method such as the jackknife or bootstrap, our method might inappropriately reject the null hypothesis only because the variance of β is higher in parts of the genome where Rv is large in magnitude. This would make our method susceptible to confounding due to unsigned enrichments, as might arise from the co-localization of TF binding sites with enriched regulatory elements such as enhancer regions. Additionally, the fact that we flip the signs of SNPs in each block together ensures that our null distribution preserves any potential association of our annotation to the LD structure of the genome. See the Supplementary Note for more details.
Controlling for covariates and the signed background model
Given a signed covariate , we can perform inference on the signed effect of v conditional on u by first regressing Ru out of and out of Ru using the generalized least-squares method outlined above, and then proceeding as usual with the residuals of and Ru.
Unless stated otherwise, all analyses in this paper control in this fashion for a “signed background model” consisting of 5 annotations u1, …,u5, defined by
| (6) |
where MAFm is the minor allele frequency of SNP m and αs is a parameter describing the MAF-dependence of the signed effect of minor alleles on phenotype. Based on the literature on MAF-dependence of the unsigned effects var(βm), we set αs = −0.3118.
382 TF annotations
Briefly, we constructed the annotations by training a sequence-based neural network predictor of ChIP-seq peak calls, using the Basset software19, to predict the results of 382 TF binding ChIP-seq experiments from ENCODE119 and comparing the neural network’s predictions for the major and minor allele of each SNP in the ChIP-seq peaks.
Specifically, we downloaded every ChIP-seq and DNase I hypersensitivity experiment in ENCODE and trained the Basset model to jointly predict each downloaded track on a set of held-out genomic segments. (We included tracks other than TF binding tracks because training predictions using all tracks slightly improved prediction accuracy for the TF binding tracks.) After training the joint predictor, we retained the predictions for every TF binding track for which a) the number of SNPs in the set of ChIP-seq peaks with non-zero difference in Basset predictions between the major and minor allele was at least 5,000 in our 1000G reference panel, and b) Basset’s estimated area under the precision-recall curve (AUPRC) was at least 0.3. This yielded a set of 382 TF ChIP-seq experiments that spanned 75 distinct TFs and 84 distinct cell lines. For each experiment, we constructed an annotation via
| (7) |
where C is the set of SNPs in the ChIP-seq peaks arising from the experiment, is the Basset prediction for the 1,000 base-pair sequence around SNP m when the minor allele is placed at SNP m, and is the Basset prediction for the 1,000 base-pair sequence around SNP m when the major allele is placed at SNP m. (We always used the minor allele as the reference allele in both our TF binding annotations and our GWAS summary statistics.)
Simulations
All simulations were carried out using real genotypes of individuals with European ancestry from the GERA cohort120 (N = 47,360). The set of M = 2.7 million causal SNPs was defined as the set of genome-wide very well imputed SNPs (INFO ≥ 0.97) that had very low missingness (< 0.5%) and non-negligible MAF (MAF ≥ 0.1%) in the GERA data set, and were represented in our 1000G Phase 3 European reference panel107,121. We simulated traits using normally distributed causal effect sizes (with annotation-dependent mean and variance in some cases), with .
Null simulations
For the simulations in Figure 1a, we simulated 1,000 independent null phenotypes with the architecture with and . For each phenotype, we computed GWAS summary statistics using plink2122 (URLs), adjusting for 3 principal components as well as GERA chip type as covariates. For each of our 382 TF annotations, we then ran SLDP regression on each of these 1,000 phenotypes, yielding a set of 382,000 P-values. (Supplementary Figure 1a shows an analysis of the P-value distribution for each annotation individually, confirming correct calibration for these annotations.)
For the simulations in Figure 1b, we simulated 1,000 independent traits in which each trait had an unsigned enrichment for a randomly chosen annotation: after choosing an annotation v, we set where σ2 and τ2 were set to achieve and a 20× unsigned enrichment for the SNPs with non-zero values of v. We then computed summary statistics as above and ran SLDP regression to assess v for a genome-wide directional effect. This procedure yielded 1,000 P-values.
For the simulations in Figure 1c, we simulated 1,000 independent phenotypes with a directional effect of minor alleles: we set where is non-zero if SNP m is in the bottom quintile of the MAF spectrum of the GERA sample and 0 otherwise, as in the signed background model. We set μ such that 10% of heritability would be explained by this directional effect, and then set σ2 to achieve . We then computed summary statistics as above and ran SLDP regression to assess for a directional effect of each of our 382 annotations on each of the 1,000 phenotypes, yielding a set of 382,000 P-values. (We note that this represents a best-case scenario in which the background model exactly matches the confounding being simulated, up to differences in MAF between the reference panel and the GWAS sample, and we caution that our method may not be appropriate for annotations with much stronger correlations to minor alleles than the annotations that we analyze here; Supplementary Figure 1b.) Finally, we repeated the same computation but running SLDP regression without the 5-MAF-bin signed background model to obtain an additional set of 382,000 P-values.
Causal simulations
For the simulations in Figure 2, we fixed a representative annotation v (binding of IRF4 in GM12878), and simulated traits using , with μ set to achieve rf = {0,0.005,0.01, …,0.05} and σ2 set to achieve in each case. For each value of rf, we simulated 100 independent traits, computed summary statistics using plink2, and then ran each of the methods under consideration using the annotation v. In addition to the findings stated in the main text, our simulations also show that the power of our method increases with sample size and SNP-heritability (Supplementary Figure 4), and is only minimally affected by within-Europe reference panel mismatch (Supplementary Figure 5).
Analysis of molecular traits in blood
We downloaded BLUEPRINT consortium QTL data for gene expression, H3K4me1, H3K27ac, and methylation in three different blood cell types with sample sizes of N = 158, 165, and 125 for monocytes, neutrophils, and T cells, respectively21 (Supplementary Table 4 and URLs). For each of the 3 gene expression traits, we constructed one summary statistics vector by meta-analyzing, for each SNP, the marginal effect sizes of that SNP for the expression of all nearby genes. Specifically, we set
| (8) |
where Gm is the set of all genes within 500kb of SNP m, and is the marginal correlation of SNP m to the expression of gene k. Assuming independence of expression across genes this is analogous to a fixed-effects meta-analysis across genes at every SNP to determine that SNP’s effect on aggregate expression, though our results do not rely on this theoretical characterization because of the empirical, signed nature of our null hypothesis testing procedure. Since in practice gene expression is not independent across genes, the scale of the resulting vector is arbitrary. Therefore, we placed all such vectors on the same scale by scaling them so that they have an estimated SNP-heritability of 0.5. (This only affects the regression weights used by SLDP regression.) Applying the same procedure to the two histone marks and to methylation in addition to gene expression yielded 12 sets of summary statistics (Supplementary Table 4).
We ran SLDP regression using each of our 382 TF annotations for each of these 12 traits. We obtained results at FDR< 5% using the Benjamini-Hochberg procedure123 within each of the 12 traits and reported the union of significant results across cell types for each trait. We determined the top 100 associations to display in Figure 3a by choosing the significant associations with the highest estimated values of rf.
For our replication analysis, we used expression array-based whole blood eQTL data from the NTR33, which we obtained by downloading the set of TWAS weights81 computed for that data set (Supplementary Table 4 and URLs). We then proceeded as above. 196 of the 409 BLUEPRINT gene expression associations replicated (same direction of effect with nominal P< 0.05). We note, however, that because TWAS weights were only available for genes with a significantly heritable cis-expression in NTR, we only had data for 2,454 genes compared with 15,023 – 17,081 genes for the BLUEPRINT traits, thereby lowering our power in this analysis.
Comparison to UniProt annotations
For each TF represented in our annotations, we queried the UniProt database29 to establish whether the TF was: (unambiguously) “activating”, defined as all TFs annotated as having activating activity but not repressing activity in UniProt; (unambiguously) repressing, defined as all TFs with repressing activity but not activating activity; or “ambiguous”, defined as all TFs with both or neither activities. 78% and 82% of our positive associations in the BLUEPRINT eQTL and chromatin QTL, respectively, were unambiguous activators. The set of significant positive SLDP associations for eQTL/chromatin QTL were enriched for (unambiguously) “activating” TFs compared to the set of annotations as a whole (P = 7.9 × 10−43 for eQTL results and P = 1.9 × 10−9 for chromatin QTL results). See Supplementary Note.
Analysis of gene expression across 48 GTEx tissues
We downloaded GTEx v7 eQTLs for all 48 tissues for which data were available and processed them using the same procedure described for the blood molecular traits, resulting in one vector of summary statistics per GTEx tissue (Supplementary Table 6 and URLs). We ran SLDP regression using each of our 382 TF annotations for each of these tissues. We obtained results at FDR<5% using the Benjamini-Hochberg procedure123 within each of the 48 tissues.
Conditional analysis for tissue-specific effects
We obtained a set of eQTL summary statistics for a fixed-effect meta-analysis across the GTEx tissues from ref.124 and processed these via the procedure described above into a single vector . For each tissue t, we then residualized out of the vector of eQTL data for tissue t to obtain a residualized vector . This amounts to subtracting a scalar multiple of from , with the scalar determined to remove as much signal as possible from . For each significant association between an annotation v and a vector from our main GTEx analysis, we then compared the p-value of that association to the p-value obtained for the association between v and the residualized vector , declaring as tissue-specific any association for which the latter was at least as significant as the former. See Supplementary Note.
This criterion for tissue-specificity is conservative and stands in contrast to, e.g., reporting associations that remain significant at a specified threshold after conditioning. The latter approach is susceptible to the fact that conditioning on a noisily measured confounder can produce false positives125; associations meeting the former criterion are likely to be robustly tissue-specific.
Assessment for concordance with absolute expression levels in GTEx tissues
Briefly, we assessed whether the proportion of significant TF associations in which the TF was expressed above a minimum threshold in the associated GTEx tissue was greater than the corresponding proportion for non-significant TFs. This held in 32 out of the 34 tissues for which we could perform the comparison (p = 2.1 × 10−15 for trend across tissues; see Supplementary Figure 7 for breakdown by tissue.) See Supplementary Note.
Analysis of 46 diseases and complex traits
We applied SLDP regression to 46 diseases and complex traits with an average sample size of 289,617, including 16 traits with publicly available summary statistics and 30 UK Biobank traits for which we have publicly released summary statistics computed using BOLT-LMM v2.392 (Supplementary Table 8 and URLs). We ran SLDP regression using each of our 382 TF annotations for each of these traits. We obtained results at per-trait FDR<5% using the Benjamini-Hochberg procedure123. We report as significant results at a per-trait FDR<5%, following standard practice. However, when many traits are analyzed, per-trait FDR control does not imply global FDR control, and we estimate the global FDR of our results to be 9.4% (Supplementary Note).
MSigDB gene-set enrichment analysis of results on diseases and complex traits
We downloaded all 10,235 MSigDB gene sets, which are organized into eight distinct tranches based on their origin, from the MSigDB online portal (URLs). We also downloaded a set of LD blocks in Europeans derived from estimated recombination hotspots117 and converted each gene set into a length-1693 vector s with one entry per LD block whose i-th entry equaled the number of genes from the set that are present in the i-th LD block. We then converted each significant SLDP regression association between an annotation v and a trait summary statistics vector into a length-1693 vector q whose i-th entry equaled the covariance between and the SLDP Rv within the i-th LD block. To assess the SLDP result for enrichment of a gene-set vector s, we computed a weighted mean of the qi whose weights were given by s. That is, we computed . The idea is that if the LD blocks in which s is large correspond to the LD blocks in which the SLDP regression signal is the strongest, the weighted mean a should be large in magnitude and have the same sign as the overall SLDP regression association. We assess this via an empirical null distribution constructed by permuting the LD blocks to obtain “shuffled” versions of s and q. This enrichment method is more conservative than ordinary gene-set enrichment methods for two reasons. First, by permuting only LD blocks and not genes, it accounts for correlations induced by LD as well as co-regulation of nearby genes and gene overlap in the genome. Second, because a significant SLDP regression association cannot arise as a result of a strong signal in only one genomic location, this method is more robust to outliers and cannot, e.g., produce a rejection simply because of a very strong signal at just one gene. In comparison to gene-set enrichment methods for GWAS data, this method also has the advantage that it will not cause gene sets containing large genes to produce signals of enrichment. Separately from null hypothesis testing, we computed heuristic standard errors for use in Figures 6 and 7 by computing the closed-form standard deviation of assuming that the si are fixed and the qi are i.i.d.
To quantify effect size, we computed a fold-enrichment by dividing by the average value of q at LD blocks containing no genes. That is the enrichment is defined as This quantity e is the number reported in Figures 6 and 7.
We conducted our hypothesis test for gene-set enrichment for each of our 77 significant TF-complex trait associations against each of the 10,325 MSigDB gene sets. For every TF-complex trait association and every tranche of gene-sets from MSigDB, we assessed significance at FDR< 5% using the Benjamini-Hochberg procedure123. This detected 6,379 significant enrichments in total (0.8% of all 795,025 tests conducted). We ranked these enrichments by q-value, except for the 15 enrichments whose p-values were less than the resolution of our empirical null hypothesis testing procedure, which we ranked by fold-enrichment.
Auto-immune enrichment among complex trait associations
Of the 12 independent TF-complex trait associations, 9 involve an auto-immune disease, representing a 4.3× enrichment (P = 1.9 × 10−5 using one-sided binomial test) and providing additional evidence for the relevance of TF binding to these phenotypes in particular126.
Estimation of lower bound on number of independent TF binding sites contributing to each association
For intuition, we first examined, for each annotation, the estimated covariance between the GWAS summary statistics and the signed LD profile in each of 300 independent genomic blocks, finding agreement with the genome-wide direction of association in 59% of the blocks on average across our 12 independent associations, and in 85% of the blocks with estimated covariances of large magnitude (Supplementary Figure 8).
For further quantification, we then converted each of the 12 independent TF-trait associations reported in Table 1 into a vector q of length ~300 whose i-th entry equaled the covariance between the GWAS in question and the signed LD profile in question within the i-th of the ~300 independent genomic blocks used for our null hypothesis testing. For every threshold , we then computed the number Kt of the entries of q with magnitude at least t, as well as the number St of those entries whose sign agreed with that of the genome-wide trend. Our estimated lower bound on the number of independent TF binding sites contributing to the association was then given by
| (9) |
The intuition is that the distribution of the signs of the entries of q can be modeled as a mixture of a uniform distribution (for genomic chunks with no signal) and a distribution with all of its mass on the sign of the genome-wide trend (for genomic chunks with signal). The number of entries drawn from the latter distribution gives the number of independent genomic blocks contributing to the association, which is a lower bound on the number of independent TF binding sites contributing to the association. Estimating this number naively without thresholding yields the expression 2S0 – K0. However, this is an under-estimate in the presence of noise in q. We therefore repeat this argument considering only the subset of entries of q with magnitude at least t for a small number of thresholds t and retain the largest estimate.
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data availability
We have released all genome annotations we analyzed, as well as regression weight matrices for our 1000 genomes reference panel, at http://data.broadinstitute.org/alkesgroup/SLDP/.
Code availability
Open-source software implementing our approach is available at http://www.github.com/yakirr/sldp.
Code used to make all figures is available at http://www.github.com/yakirr/sldp-display.
Supplementary Material
Acknowledgements
We thank C de Boer, L Dicker, J Engreitz, T Finucane, N Friedman, R Gumpert, M Kanai, S Kim, X Liu, M Mitzenmacher, J Perry, S Reilly, D Reshef, S Raychaudhuri, A Schoech, P Sabeti, R Tewhey, O Troyanskaya, P Turley, O Weissbrod, J Zhou, and the CGTA discussion group for helpful discussions. We also thank the anonymous referees for their helpful comments. This research was conducted using the UK Biobank Resource under Application #16549 and was supported by US National Institutes of Health grants U01 HG009379, R01 MH101244, R01 MH109978, and R01 MH107649. Y.A.R. was supported by award number T32GM007753 from the National Institute of General Medical Sciences, the National Defense Science and Engineering Graduate Fellowship, and the Paul and Daisy Soros Foundation. H.K.F. was supported by the Fannie and John Hertz Foundation and by Eric and Wendy Schmidt. F.H. is supported by National Institute of Health award T32 DK110919. L.P. is supported by National Institutes of Health award R00HG008399. R.P.A. is supported by NSF IIS-1421780. Computational analyses were performed on the Orchestra High Performance Compute Cluster at Harvard Medical School, which is partially supported by grant NCRR 1S10RR028832–01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
URLs
Signed LD profile regression: open-source software is available at http://www.github.com/yakirr/sldp
Plink2: https://www.cog-genomics.org/plink2/
BLUEPRINT consortium data: ftp://ftp.ebi.ac.uk/pub/databases/blueprint/blueprint_Epivar/qtl_as/QTL_RESULTS/
TWAS weights for NTR data: https://data.broadinstitute.org/alkesgroup/FUSION/WGT/NTR.BLOOD.RNAARR.tar.bz2
GTEx eQTL data: https://www.gtexportal.org/home/datasets
MSigDB data: http://software.broadinstitute.org/gsea/msigdb
GTRD data: http://gtrd.biouml.org/
HOCOMOCO motif data: http://hocomoco11.autosome.ru/
Footnotes
Accession Codes
There are no accession codes.
Competing interests
D.R.K. is employed by the Calico Life Sciences LLC. The rest of the authors declare no competing interests.
References
- 1.Cowper-Sal lari R et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat. Genet 44, 1191–1198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maurano MT et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science 337, 1190–1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trynka G et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet 45, 124–130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickrell JK Joint Analysis of Functional Genomic Data and Genome-wide Association Studies of 18 Human Traits. Am. J. Hum. Genet 94, 559–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle EA, Li YI & Pritchard JK An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 169, 1177–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu X & Stephens M A large-scale genome-wide enrichment analysis identifies new trait-associated genes, pathways and tissues across 31 human phenotypes. bioRxiv 160770 (2017). doi: 10.1101/160770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karczewski KJ et al. Systematic functional regulatory assessment of disease-associated variants. Proc. Natl. Acad. Sci 110, 9607–9612 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathelier A, Shi W & Wasserman WW Identification of altered cis-regulatory elements in human disease. Trends Genet. TIG 31, 67–76 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Price AL, Spencer CCA & Donnelly P Progress and promise in understanding the genetic basis of common diseases. Proc R Soc B 282, 20151684 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitington T et al. Gene regulatory mechanisms underpinning prostate cancer susceptibility. Nat. Genet 48, 387–397 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Liu Y et al. Identification of breast cancer associated variants that modulate transcription factor binding. PLoS Genet 13, e1006761 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pique-Regi R et al. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res 21, 447–455 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee D et al. A method to predict the impact of regulatory variants from DNA sequence. Nat. Genet 47, 955–961 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J & Troyanskaya OG Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods 12, 931–934 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alipanahi B, Delong A, Weirauch MT & Frey BJ Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol 33, 831–838 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng H, Hashimoto T, Kang DD & Gifford DK GERV: a statistical method for generative evaluation of regulatory variants for transcription factor binding. Bioinformatics 32, 490–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelley DR, Snoek J & Rinn J Basset: Learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res gr.200535.115 (2016). doi: 10.1101/gr.200535.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasaniuc B & Price AL Dissecting the genetics of complex traits using summary association statistics. Nat. Rev. Genet 18, 117–127 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L et al. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell 167, 1398–1414.e24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.GTEx Consortium et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liberzon A et al. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang W et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet 6, e1000841 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arbiza L et al. Genome-wide inference of natural selection on human transcription factor binding sites. Nat. Genet 45, 723–729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst J et al. Genome-scale high-resolution mapping of activating and repressive nucleotides in regulatory regions. Nat. Biotechnol 34, 1180–1190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bodine DM Introduction to a review series on Transcription Factors in Hematopoiesis and Hematologic Disease. Blood blood-2017-02-766840 (2017). doi: 10.1182/blood-2017-02-766840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res 45, D158–D169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharrocks AD, Brown AL, Ling Y & Yates PR The ETS-domain transcription factor family. Int. J. Biochem. Cell Biol 29, 1371–1387 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Kimura T et al. Involvement of the IRF-1 Transcription Factor in Antiviral Responses to Interferons. Science 264, 1921–1924 (1994). [DOI] [PubMed] [Google Scholar]
- 32.Kakizuka A et al. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RARα with a novel putative transcription factor, PML. Cell 66, 663–674 (1991). [DOI] [PubMed] [Google Scholar]
- 33.Wright FA et al. Heritability and genomics of gene expression in peripheral blood. Nat. Genet 46, 430–437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedman JS et al. The minimal transactivation domain of the basic motif-leucine zipper transcription factor NRL interacts with TATA-binding protein. J. Biol. Chem 279, 47233–47241 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Bell AC, West AG & Felsenfeld G The Protein CTCF Is Required for the Enhancer Blocking Activity of Vertebrate Insulators. Cell 98, 387–396 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Xie X et al. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl. Acad. Sci 104, 7145–7150 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao N et al. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev 22, 3435–3448 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song Y, Washington MK & Crawford HC Loss of FOXA1/2 is Essential for the Epithelial-to-Mesenchymal Transition in Pancreatic Cancer. Cancer Res 70, 2115–2125 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao N et al. Foxa1 and Foxa2 Maintain the Metabolic and Secretory Features of the Mature β-Cell. Mol. Endocrinol 24, 1594–1604 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hagman J, Ramírez J & Lukin K B lymphocyte lineage specification, commitment and epigenetic control of transcription by early B cell factor 1. Curr. Top. Microbiol. Immunol 356, 17–38 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Somasundaram R, Prasad MAJ, Ungerbäck J & Sigvardsson M Transcription factor networks in B-cell differentiation link development to acute lymphoid leukemia. Blood 126, 144–152 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Odom DT et al. Control of Pancreas and Liver Gene Expression by HNF Transcription Factors. Science 303, 1378–1381 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonzo JA, Ferry CH, Matsubara T, Kim J-H & Gonzalez FJ Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4α in adult mice. J. Biol. Chem 287, 7345–7356 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolff L & Ruscetti S The spleen focus-forming virus (SFFV) envelope gene, when introduced into mice in the absence of other SFFV genes, induces acute erythroleukemia. J. Virol 62, 2158–2163 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Angel PE & Herrlich P The FOS and JUN Families of Transcription Factors (CRC Press, 1994). [Google Scholar]
- 46.Bullitt E Expression of c-fos-like protein as a marker for neuronal activity following noxious stimulation in the rat. J. Comp. Neurol 296, 517–530 (1990). [DOI] [PubMed] [Google Scholar]
- 47.Velazquez FN et al. Brain development is impaired in c-fos −/− mice. Oncotarget 6, 16883–16901 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J et al. c-fos regulates neuronal excitability and survival. Nat. Genet 30, 416–420 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Nischan J et al. Binding Sites for ETS Family of Transcription Factors Dominate the Promoter Regions of Differentially Expressed Genes in Abdominal Aortic AneurysmsCLINICAL PERSPECTIVE. Circ. Genomic Precis. Med 2, 565–572 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Triarhou LC Dopamine and Parkinson’s Disease (Landes Bioscience, 2013). [Google Scholar]
- 51.Aneichyk T et al. Dissecting the Causal Mechanism of X-Linked Dystonia-Parkinsonism by Integrating Genome and Transcriptome Assembly. Cell 172, 897–909.e21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davis FP & Eddy SR Transcription Factors That Convert Adult Cell Identity Are Differentially Polycomb Repressed. PLOS ONE 8, e63407 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Popov Daniil V, Lysenko Evgeny A, Makhnovskii Pavel A, Kurochkina Nadia S & Vinogradova Olga L Regulation of PPARGC1A gene expression in trained and untrained human skeletal muscle. Physiol. Rep 5, e13543 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim S, Yu N-K & Kaang B-K CTCF as a multifunctional protein in genome regulation and gene expression. Exp. Mol. Med 47, e166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kleiman E, Jia H, Loguercio S, Su AI & Feeney AJ YY1 plays an essential role at all stages of B-cell differentiation. Proc. Natl. Acad. Sci. U. S. A 113, E3911–3920 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hwang SS et al. YY1 inhibits differentiation and function of regulatory T cells by blocking Foxp3 expression and activity. Nat. Commun 7, 10789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kwon H-K, Chen H-M, Mathis D & Benoist C Different molecular complexes that mediate transcriptional induction and repression by FoxP3. Nat. Immunol 18, 1238–1248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gabriele M et al. YY1 Haploinsufficiency Causes an Intellectual Disability Syndrome Featuring Transcriptional and Chromatin Dysfunction. Am. J. Hum. Genet 100, 907–925 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weintraub AS et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 171, 1573–1588.e28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okbay A et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Basak A et al. BCL11A deletions result in fetal hemoglobin persistence and neurodevelopmental alterations. J. Clin. Invest 125, 2363–2368 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Funnell APW et al. 2p15-p16.1 microdeletions encompassing and proximal to BCL11A are associated with elevated HbF in addition to neurologic impairment. Blood 126, 89–93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature 519, 223–228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dias C et al. BCL11A Haploinsufficiency Causes an Intellectual Disability Syndrome and Dysregulates Transcription. Am. J. Hum. Genet 99, 253–274 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lipton JO & Sahin M The Neurology of mTOR. Neuron 84, 275–291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reijnders MRF et al. Variation in a range of mTOR-related genes associates with intracranial volume and intellectual disability. Nat. Commun 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laplante M & Sabatini DM An emerging role of mTOR in lipid biosynthesis. Curr. Biol. CB 19, R1046–1052 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mathews ES & Appel B Cholesterol Biosynthesis Supports Myelin Gene Expression and Axon Ensheathment through Modulation of P13K/Akt/mTor Signaling. J. Neurosci 36, 7628–7639 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koudinov AR & Koudinova NV Cholesterol homeostasis failure as a unifying cause of synaptic degeneration. J. Neurol. Sci 229, 233–240 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Zhang J & Liu Q Cholesterol metabolism and homeostasis in the brain. Protein Cell 6, 254–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Macari ER, Schaeffer EK, West RJ & Lowrey CH Simvastatin and t-butylhydroquinone suppress KLF1 and BCL11A gene expression and additively increase fetal hemoglobin in primary human erythroid cells. Blood 121, 830–839 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.TANG L et al. BCL11A gene DNA methylation contributes to the risk of type 2 diabetes in males. Exp. Ther. Med 8, 459–463 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li S et al. Transcription Factor CTIP1/ BCL11A Regulates Epidermal Differentiation and Lipid Metabolism During Skin Development. Sci. Rep 7, 13427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Franke A et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet 42, 1118–1125 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jostins L et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Lange KM et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet 49, 256–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rioux JD et al. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat. Genet 29, 223–228 (2001). [DOI] [PubMed] [Google Scholar]
- 78.Silverberg MS OCTNs: Will the real IBD5 gene please stand up? World J. Gastroenterol. WJG 12, 3678–3681 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brant SR IBD5: the second Crohn’s disease gene? Inflamm. Bowel Dis 8, 371–372 (2002). [DOI] [PubMed] [Google Scholar]
- 80.Huang H et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547, 173–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gusev A et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet 48, 245–252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wainberg M et al. Vulnerabilities of transcriptome-wide association studies. bioRxiv 206961 (2017). doi: 10.1101/206961 [DOI] [Google Scholar]
- 83.Mancuso N et al. Integrating Gene Expression with Summary Association Statistics to Identify Genes Associated with 30 Complex Traits. Am. J. Hum. Genet 100, 473–487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Romeo G et al. IRF-1 as a negative regulator of cell proliferation. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res 22, 39–47 (2002). [DOI] [PubMed] [Google Scholar]
- 85.Honda K, Takaoka A & Taniguchi T Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25, 349–360 (2006). [DOI] [PubMed] [Google Scholar]
- 86.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Linterman MA et al. IL-21 acts directly on B cells to regulate Bcl-6 expression and germinal center responses. J. Exp. Med 207, 353–363 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chevrier S, Kratina T, Emslie D, Tarlinton DM & Corcoran LM IL4 and IL21 cooperate to induce the high Bcl6 protein level required for germinal center formation. Immunol. Cell Biol 95, 925–932 (2017). [DOI] [PubMed] [Google Scholar]
- 89.Hurtz C et al. BCL6-mediated repression of p53 is critical for leukemia stem cell survival in chronic myeloid leukemia. J. Exp. Med 208, 2163–2174 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hatzi K et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep 4, 578–588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang C, Hatzi K & Melnick A Lineage-specific functions of Bcl-6 in immunity and inflammation are mediated by distinct biochemical mechanisms. Nat. Immunol 14, 380–388 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Loh P-R, Kichaev G, Gazal S, Schoech AP & Price AL Mixed-model association for biobank-scale datasets. Nat. Genet 1 (2018). doi: 10.1038/s41588-018-0144-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ek WE, Rask-Andersen M, Karlsson T & Johansson A Genome-wide association analysis identifies 26 novel loci for asthma, hay fever and eczema. bioRxiv 195933 (2017). doi: 10.1101/195933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Portelli MA, Hodge E & Sayers I Genetic risk factors for the development of allergic disease identified by genome-wide association. Clin. Exp. Allergy 45, 21–31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Boraska V et al. A genome-wide association study of anorexia nervosa. Mol. Psychiatry 19, 1085–1094 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ben-Shachar D & Karry R Sp1 Expression Is Disrupted in Schizophrenia; A Possible Mechanism for the Abnormal Expression of Mitochondrial Complex I Genes, NDUFV1 and NDUFV2. PLoS ONE 2, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fusté M et al. Reduced expression of SP1 and SP4 transcription factors in peripheral blood mononuclear cells in first-episode psychosis. J. Psychiatr. Res 47, 1608–1614 (2013). [DOI] [PubMed] [Google Scholar]
- 98.Bulik-Sullivan B et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet 47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Striegel-Moore RH et al. Gender Difference in the Prevalence of Eating Disorder Symptoms. Int. J. Eat. Disord 42, 471–474 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Colman RJ et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science 325, 201–204 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pan X, Solomon SS, Borromeo DM, Martinez-Hernandez A & Raghow R Insulin deprivation leads to deficiency of Sp1 transcription factor in H-411E hepatoma cells and in streptozotocin-induced diabetic ketoacidosis in the rat. Endocrinology 142, 1635–1642 (2001). [DOI] [PubMed] [Google Scholar]
- 102.Yasui D, Peedicayil J & Grayson DR Neuropsychiatric Disorders and Epigenetics (Academic Press, 2016). [Google Scholar]
- 103.Zhang X et al. Hypermethylation of Sp1 binding site suppresses hypothalamic POMC in neonates and may contribute to metabolic disorders in adults: impact of maternal dietary CLAs. Diabetes 63, 1475–1487 (2014). [DOI] [PubMed] [Google Scholar]
- 104.Yang G et al. FoxO1 inhibits leptin regulation of pro-opiomelanocortin promoter activity by blocking STAT3 interaction with specificity protein 1. J. Biol. Chem 284, 3719–3727 (2009). [DOI] [PubMed] [Google Scholar]
- 105.Moreno-Aliaga MJ et al. Sp1-mediated transcription is involved in the induction of leptin by insulin-stimulated glucose metabolism. J. Mol. Endocrinol 38, 537–546 (2007). [DOI] [PubMed] [Google Scholar]
- 106.Audet-Walsh É et al. Nuclear mTOR acts as a transcriptional integrator of the androgen signaling pathway in prostate cancer. Genes Dev (2017). doi: 10.1101/gad.299958.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Davey Smith G & Hemani G Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet 23, R89–R98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Verbanck M, Chen C-Y, Neale B & Do R Widespread pleiotropy confounds causal relationships between complex traits and diseases inferred from Mendelian randomization. bioRxiv 157552 (2017). doi: 10.1101/157552 [DOI] [Google Scholar]
- 110.Michelson AM Deciphering genetic regulatory codes: A challenge for functional genomics. Proc. Natl. Acad. Sci 99, 546–548 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Deplancke B, Alpern D & Gardeux V The Genetics of Transcription Factor DNA Binding Variation. Cell 166, 538–554 (2016). [DOI] [PubMed] [Google Scholar]
- 112.Zeng H, Edwards MD, Liu G & Gifford DK Convolutional neural network architectures for predicting DNA-protein binding. Bioinforma. Oxf. Engl 32, i121–i127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kumar S, Ambrosini G & Bucher P SNP2TFBS – a database of regulatory SNPs affecting predicted transcription factor binding site affinity. Nucleic Acids Res 45, D139–D144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yevshin I, Sharipov R, Valeev T, Kel A & Kolpakov F GTRD: a database of transcription factor binding sites identified by ChIP-seq experiments. Nucleic Acids Res 45, D61–D67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kulakovskiy IV et al. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res 46, D252–D259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Venkataraman A et al. A toolbox of immunoprecipitation-grade monoclonal antibodies to human transcription factors. Nat. Methods (2018). doi: 10.1038/nmeth.4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 117.Berisa T & Pickrell JK Approximately independent linkage disequilibrium blocks in human populations. Bioinformatics 32, 283–285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schoech A et al. Quantification of frequency-dependent genetic architectures and action of negative selection in 25 UK Biobank traits. bioRxiv 188086 (2017). doi: 10.1101/188086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Banda Y et al. Characterizing Race/Ethnicity and Genetic Ancestry for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics 200, 1285–1295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Benjamini Y & Hochberg Y Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol 57, 289–300 (1995). [Google Scholar]
- 124.Hormozdiari F et al. Leveraging molecular QTL to understand the genetic architecture of diseases and complex traits. BioRxiv Press Nat Genet 203380 (2017). doi: 10.1101/203380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Carroll RJ Measurement Error in Epidemiologic Studies in Wiley StatsRef: Statistics Reference Online (American Cancer Society, 2014). doi: 10.1002/9781118445112.stat05178 [DOI] [Google Scholar]
- 126.Lambert SA et al. The Human Transcription Factors. Cell 172, 650–665 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have released all genome annotations we analyzed, as well as regression weight matrices for our 1000 genomes reference panel, at http://data.broadinstitute.org/alkesgroup/SLDP/.