Abstract
Polyprenol phosphate phosphoglycosyl transferases (PGTs) catalyze the first membrane-committed step in assembly of essential glycoconjugates. Currently there is no structure-function information to describe how monotopic PGTs coordinate the reaction between membrane-embedded and soluble substrates. We describe the structure and mode of membrane association of PglC, a PGT from Campylobacter concisus. The structure reveals a unique architecture, provides mechanistic insight, and identifies ligand-binding determinants for PglC and the monotopic PGT superfamily.
Phosphoglycosyl transferases (PGTs) catalyze the first membrane-committed step in biosynthetic pathways leading to a wide array of biologically important glycoconjugates, including glycoproteins, glycolipids, and peptidoglycan1. The polytopic and monotopic PGT superfamilies, which mediate phosphosugar transfer from a sugar nucleoside diphosphate to a membrane-resident polyprenol phosphate (Pren-P), are strikingly different1,2. Polytopic PGTs, exemplified by MraY and WecA, include 10–11 transmembrane helices (TMH) and an active site crafted from inter-TMH loops2. Of this superfamily, MraY is the only member to be structurally characterized3. In contrast, the monotopic PGT superfamily members, exemplified by the Campylobacter PglCs, are predicted from primary structure to adopt a dual-domain architecture with a globular soluble domain and single, small membrane-associated domain4. Due to the pivotal roles played by complex glycoconjugates, for example in bacterial survival and virulence5, both of the PGT superfamilies include members that are important pharmacological targets6. Despite the importance of PGTs as a functional class, there is no structural information to address the monotopic PGTs as a strategic point of pathway intervention. Monotopic membrane proteins are located on one face of the membrane encroaching into one or both leaflets of the lipid bilayer and are poorly represented in the PDB (< 0.05% of the non-redundant structures).
PglC from Campylobacter concisus is a representative member of the monotopic PGT superfamily because it encompasses the minimal functional core (ca. 200 residues)1 of the three known families (Supplementary Figure 1). Recent biochemical studies provide evidence that PglC catalysis involves a two-step ping-pong mechanism7. Specifically, a strictly conserved Asp–Glu dyad is essential for function, wherein the Asp serves as the nucleophile forming a covalent phosphosugar intermediate. This catalytic strategy fundamentally differs from the ternary complex mechanism known for WecA and MraY of the polytopic PGT superfamily8.
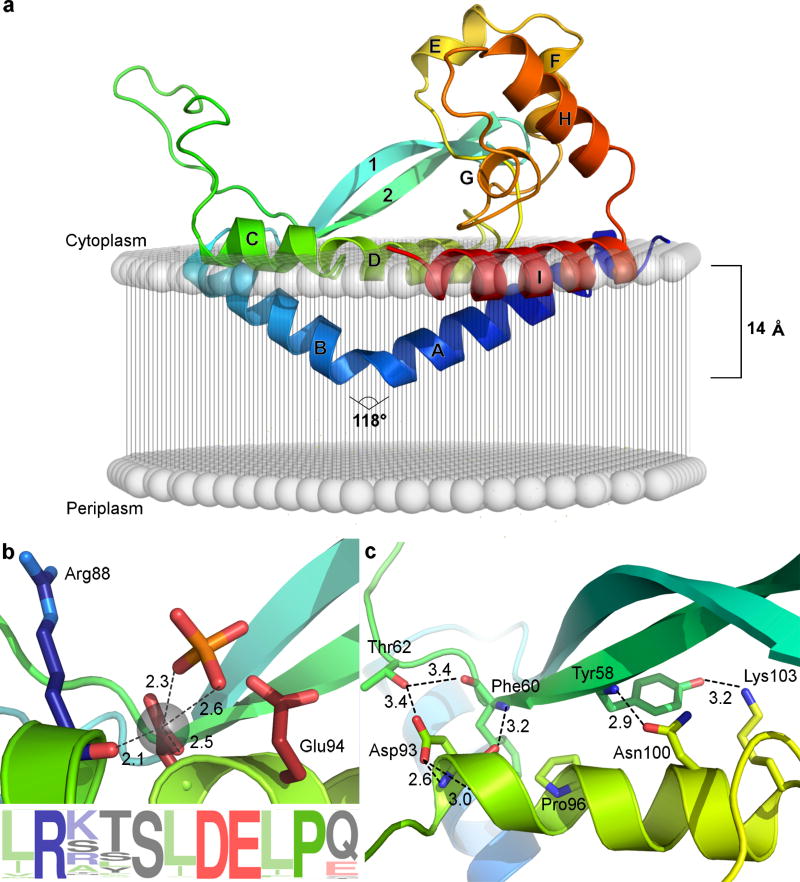
The structure of PglC reveals new paradigms for membrane association and membrane-dependent enzyme function. The full-length structure (Fig. 1a; Supplementary Fig. 2a,b) was determined via single-wavelength anomalous dispersion phasing using wild-type selenomethionine-substituted protein and I57M/I87M and I57M/Q175M variants (Online Methods; Supplementary Fig. 3). The catalytically active I57M/Q175M variant is designated as PglC throughout. The structure of PglC, comprising residues 1–183 (of 201 total, including a GSG linker at the N terminus), showed clear electron density (Supplementary Fig. 4a) and was refined to 2.74 Å resolution with excellent geometry (Supplementary Table 1). PglC crystallizes with two protomers in the asymmetric unit (Supplementary Fig. 4b); however, previous characterization in lipid bilayer nanodiscs supports a functional monomeric biological assembly9. Tracing from the N terminus, α-helices A and B form a helix-break-helix motif which extends into a long β-hairpin structure (strands 1 and 2, residues 42–60). An extended loop structure links the β-hairpin motif to co-planar helices C and D. The C-terminus of helix D supports the base of a globular double twisted loop domain (residues 105–140) formed by helices E, F, G, and H and the loops connecting them. Lastly, helix I, co-planar with helices C and D, is at the C-terminus of the observed structure. The structure also defines the locations of the conserved Asp–Glu catalytic dyad7, the essential Mg2+ cofactor, a phosphate-binding subsite (Fig. 1b), and the head group of phosphatidylethanolamine (PE).
FIGURE 1. PglC reveals a distinct architecture and topology for monotopic membrane proteins.
a, Predicted position of PglC with respect to the membrane, including the reentrant membrane helix (RMH) formed by the helix-break-helix motif of helices A and B (N to C termini colored blue to red). b, Depiction of the PglC active site showing the conserved Asp–Glu dyad with Mg2+ and phosphate ligands and sequence logo. c, The AHABh (alpha-helix-associated beta-hairpin)-motif that defines the superfamily fold is formed by a β-hairpin comprising β-strands 1 and 2 packing against helix D.
Overall, the structure of PglC reveals a new protein fold, which features a unique α-helix-associated β-hairpin (AHABh) motif composed of strands 1 and 2 and helix D (Fig. 1c). There are no relevant matches to the fold reported by DALI10 (Supplementary Table 2). The membrane interaction modality includes a reentrant helix-break-helix11 (helices A and B), which together with coplanar membrane-associated helices (C, D, and I) at the membrane interface act to stabilize the minimal functional unit (Fig. 1a; reentrant membrane helix discussed below). The structure is independent of large domains, relying instead on short-range features such as proline-kinks and hydrogen-bond networks (Supplementary Fig. 5a–d). Although the structure appears “open”, the full-length monomeric PglC has a surface-to-volume ratio (SVR) of 0.44 Å−1 similar to that of other proteins of the same size. Moreover, there are no cavities of substantial volume found within the double-twisted loop motif, showing that the domain is well packed (Supplementary Fig. 6). In addition, comparison of the structure to the published model based on covariance4 suggests that the loop between β strand 2 and helix C is likely to close onto the active site upon ligand binding.
The N-terminal helix of PglC had been predicted to adopt a canonical single-pass transmembrane helix geometry4,12; however, the structure shows this segment to be broken into two helices (A and B) by a Ser–Pro motif with an inter-helix angle of 118° (Fig. 1a and Supplementary Fig. 5a). The occurrence of proline at this position is almost universally conserved in the superfamily4,11,13,14 and proline has been ascribed a similar structural role in the unrelated protein caveolin11. This geometry allows the helix to penetrate 14 Å into the cytoplasmic face of the membrane and reemerge on the same face – thus it is termed a reentrant membrane helix (RMH). This topology has been proposed based on biochemical studies in a related superfamily member13 and herein is validated for PglC.
The distinctive fold of PglC contrasts with previously reported structures of monotopic proteins that utilize known soluble scaffolds (e.g. Rossmann-like, α-β-α sandwich-like), which have subsequently evolved to function at the membrane interface. As there are no known soluble homologs, evolution of the PglC scaffold may have been intimately dependent on membrane association. The structure of PglC will enable discovery of inhibitory agents and computation of high-quality homology models of diverse monotopic PGTs, as the scaffold encompasses the functional core of all superfamily members (Supplementary Fig. 1). Sequences with >65% identity to the functional core, comprising 724 non-redundant members of the superfamily, are listed in Supplementary Dataset 1.
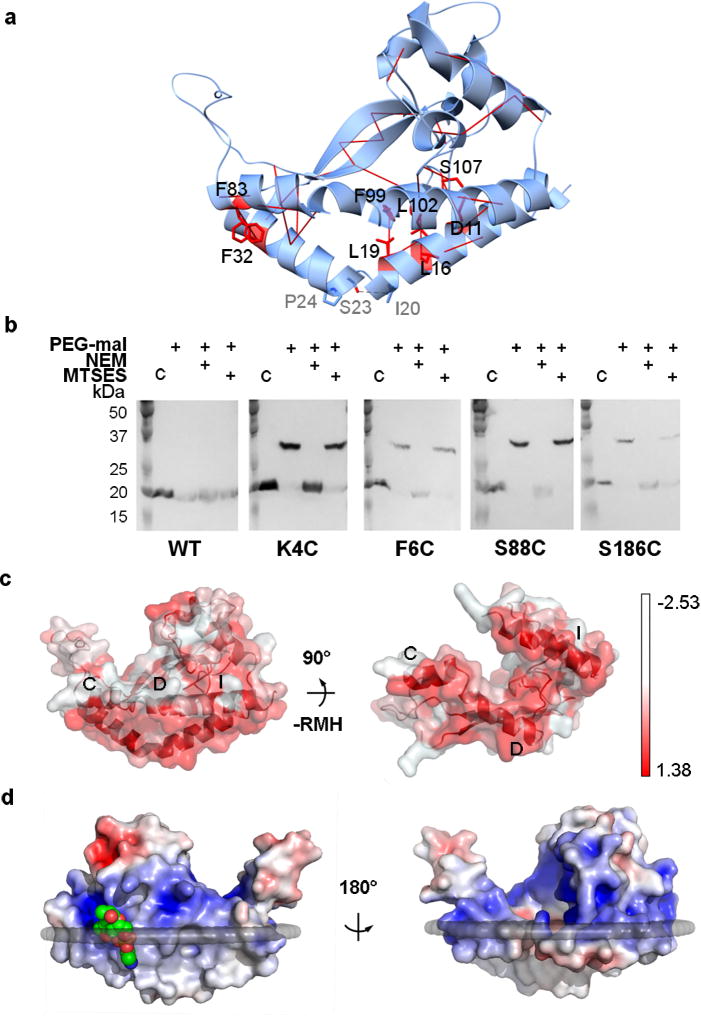
Several lines of evidence indicate that the structure is catalytically relevant. Covariance analyses establish contacts (at ≥99% probability), consistent with interactions observed in the structure between the RMH and co-planar helices C, D, and I (Fig. 2a). The topology of the N-terminal domain of PglC was independently assessed in vivo using the substituted-cysteine accessibility method (SCAM)15 on PglC constructs without SUMO-tags (Fig. 2b; Supplementary Fig. 7, Online Methods) as described previously13. SCAM supports a model of PglC in which the membrane-inserted domain forms a reentrant helix with N- and C-termini located in the cytoplasm. Additionally, highly conserved basic residues at the N terminus of PglC are consistent with the “positive-in” rule for a topology that faces the cytoplasm12. Hydrophobic surface analysis16 supports the interaction of helices C, D, I and the RMH with the membrane (Fig. 2c). Furthermore, helical wheel analysis17 positions helices C, D, and I relative to the membrane interface (Supplementary Fig. 8).
FIGURE 2. PglC crystallizes in a native conformation.
a, Co-evolution and covariance analyses across the monotopic PGT superfamily predict 10 interactions with ≥ 99% probability of contact (red solid lines). Phe32/Phe83 exhibit π-π-stacking; Leu19/Phe99, Leu16/Leu102, Thr29/Leu92, Tyr34/Val41, and Ala30/Val41 make hydrophobic interactions between the RMH and the coplanar helices; and Asp11/Ser107 form a hydrogen bond. b, In vivo SCAM analysis indicates that the N and C termini of PglC are localized on the cytoplasmic face (*, native PglC; **, PglC labeled with PEG-mal; C, control, no PEG-mal labeling). PEG-mal, PEG-maleimide; NEM, N-ethylmaleimide; MTSES, 2-sulfonatoethyl methanethiosulfonate. c, The hydrophobic surfaces (red) of amphipathic helices labeled C, D, and I establish a planar hydrophobic surface (left); also shown rotated 90° with RMH removed (right). d, A phosphatidylethanolamine (shown as space-filling, green) head group binding site at Arg8 locates this position to the membrane interface. The surface is colored by electrostatic potential from +5 kT/e (blue) to −5 kT/e (red).
Thin-layer chromatography analysis of the purified protein prior to crystallization shows that PglC co-purifies with endogenous PE and a lesser amount of endogenous phosphatidylglycerol (PG), but the electron density and binding interactions are more consistent with PE (Supplementary Fig. 9). The location of the PE head group corroborates the localization of helix D and the N-terminus of helix A at the membrane interface (Fig. 2d). Taken together, the structural features of PglC define a new modality for the interaction between a monotopic membrane protein and the lipid bilayer. The structure has evolved to simultaneously allow effective interactions with both soluble and membrane-embedded substrates at the membrane interface.
Numerous basic residues establish favorable electrostatics (Fig. 2d) for binding and orienting the negatively charged, phosphate-rich substrates in the active site. The presence of phosphate in the structure provides a putative location for the phosphate-binding subsite of the UDP-diNAcBac or phosphosugar intermediate (Fig. 1b). Phosphate was not added in the protein preparation or crystallization conditions but resulted from hydrolysis of exogenously-added 5-iodo-UDP during crystallization (Supplementary Fig. 10). The strictly conserved PRP motif (111–113) orients Arg112 towards the phosphate binding subsite, potentially positioning the Arg112 side chain for interaction with the uracil nucleobase of the UDP-sugar18. Notably, in one of the two chains in the asymmetric unit, electron density is observed consistent with a polyethylene glycol (PEG) molecule or, alternatively, the alkyl chain of DDM (see Online Methods). In either case, this indicates a narrow, hydrophobic volume that would be consistent with the Pren-P binding site. This binding site would position Pren-P proximal to the catalytic dyad and Arg112 and would allow the embedded Pren-P access to the phosphosugar intermediate at the membrane interface (Supplementary Fig. 11a,b).
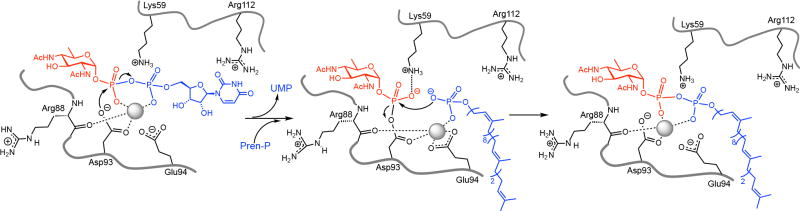
The catalytic Asp–Glu dyad in PglC is reminiscent of the conserved Asp–Asp dyad of polytopic MraY and WecA19,20. Structural analysis of Mg2+-bound MraY3 shows the dyad within a typical α-helix, wherein adjacent residue side chains are not co-facial. In contrast, in PglC, residues with low helical propensity (Ser91, Asp93, Glu94)21 distort the first turn of helix D into a 310-helix geometry ending in Pro96 (Fig. 1c, Supplementary Table 3) and orient the two acidic residue side chains into a co-facial configuration poised for catalysis (Fig. 1b). In addition, the Asp side chain is resident at the N-cap position of the 310-helix, stabilizing the observed side-chain rotamer. The Asp position is enforced by formation of a coordinate bond with the Mg2+ cofactor7. Together, these interactions contribute to the nucleophilic reactivity of the non-coordinating oxygen of Asp93, which is an important feature of covalent catalysis at phosphoryl groups22. The difference in geometries enforced by the polytopic and monotopic PGT scaffolds may underlie the mechanistic divergence between the two superfamilies. Overall, the locations of the catalytic dyad, Mg2+ cofactor and substrate binding residues afford a mechanistic scheme for PglC catalysis consistent with formation of a covalent sugar-phosphate intermediate7 (Fig. 3).
FIGURE 3. The active site structure supports the proposed mechanism of PglC.
Binding subsites defined through structural analysis are consistent with the ping-pong (covalent intermediate) mechanism7, which predicts that the binding subsite of the β-phosphate of the UDP-sugar and the phosphate of Pren-P would be coincident. Grey sphere, Mg2+.
The monotopic PGTs address the lipophilic nature of polyprenol-linked substrate and product by positioning the reaction components at the membrane interface. This strategy is energetically advantageous as it obviates the need for a membrane extraction step and generates the membrane-bound product ready for processing by the next enzyme in the pathway. In contrast, the polyprenol phosphate glycosyltransferase GtrB, which mediates biosynthesis of a polyprenol monophosphosugar, includes an extensive oligomeric intramembrane structure and a soluble globular glycosyltransferase domain, which collaborate to translocate the polyprenol phosphate from the membrane to the active site (15 Å away from the membrane interface)23. The monotopic PGTs masterfully accomplish a similar reaction through tactical placement of the active site at the membrane interface, avoiding the need for substrate translocation in the catalytic cycle.
ONLINE METHODS
Expression and purification of PglC
Expression and purification of both the wild type PglC from C. concisus and the double mutants (I57M/Q175M and I57M/I87M) PglCs were carried out in a modification of previously published protocols4,7 (Supplementary Table 4). The final yields of all three proteins were similar.
Amino acid sequences of PglC constructs for crystallography (linker between SUMO domain and PglC in bold)
Wild-type SUMO-SGSG-PglC (C. concisus)
MGHHHHHHGSLQDSEVNQEAKPEVKPEVKPETHINLKVSDGSSEIFFKIK KTTPLRRLMEAFAKRQGKEMDSLRFLYDGIRIQADQAPEDLDMEDNDIIE AHREQIGGSGSGMYRNFLKRVIDILGALFLLILTSPIIIATAIFIYFKVS RDVIFTQARPGLNEKIFKIYKFKTMSDERDANGELLPDDQRLGKFGKLIR SLSLDELPQLFNVLKGDMSFIGPRPLLVEYLPIYNETQKHRHDVRPGITG LAQVNGRNAISWEKKFEYDVYYAKNLSFMLDVKIALQTIEKVLKRSGVSK EGQATTEKFNGKN
I57M/I87M SUMO-SGSG-PglC (C. concisus)
MGHHHHHHGSLQDSEVNQEAKPEVKPEVKPETHINLKVSDGSSEIFFKIK KTTPLRRLMEAFAKRQGKEMDSLRFLYDGIRIQADQAPEDLDMEDNDIIE AHREQIGGSGSGMYRNFLKRVIDILGALFLLILTSPIIIATAIFIYFKVS RDVIFTQARPGLNEKIFKMYKFKTMSDERDANGELLPDDQRLGKFGKLMR SLSLDELPQLFNVLKGDMSFIGPRPLLVEYLPIYNETQKHRHDVRPGITG LAQVNGRNAISWEKKFEYDVYYAKNLSFMLDVKIALQTIEKVLKRSGVSK EGQATTEKFNGKN
I57M/Q175M SUMO-SGSG-PglC (C. concisus)
MGHHHHHHGSLQDSEVNQEAKPEVKPEVKPETHINLKVSDGSSEIFFKIK KTTPLRRLMEAFAKRQGKEMDSLRFLYDGIRIQADQAPEDLDMEDNDIIE AHREQIGGSGSGMYRNFLKRVIDILGALFLLILTSPIIIATAIFIYFKVS
RDVIFTQARPGLNEKIFKMYKFKTMSDERDANGELLPDDQRLGKFGKLIR
SLSLDELPQLFNVLKGDMSFIGPRPLLVEYLPIYNETQKHRHDVRPGITG
LAQVNGRNAISWEKKFEYDVYYAKNLSFMLDVKIALMTIEKVLKRSGVSK
EGQATTEKFNGKN
PglC was cloned into pET-His6-SUMO vector4. A codon sequence (TCTGGCTCTGGG) encoding a SGSG linker was incorporated between the SUMO-tag and PglC sequence to allow efficient cleavage of PglC from the SUMO-tag using SUMO protease in later stages of protein purification. BL21-CodonPlus (DE3)-RIL cells (Agilent) were transformed with pET-His6-SUMO-PglC plasmid construct. The Studier auto-induction method24 was used for expression of the protein. Freshly transformed cells were grown overnight in 3 mL MDG media (0.5% (w/v) glucose, 0.25 (w/v) % aspartate, 2 mM MgSO4, 25 mM Na2HPO4, 25 mM KH2PO4, 50 mM NH4Cl, 5 mM Na2SO4 and 0.2× trace metal mix (from 1000× stock, Teknova, cat. # T1001) at 37 °C using kanamycin and chloramphenicol (30 µg/mL each). The overnight culture was transferred into 500 mL auto-induction media (1% (w/v) tryptone, 0.5% (w/v) yeast extract, 0.5% (v/v) glycerol, 0.05% (w/v) glucose, 0.2% (w/v) α-D-lactose, 2 mM MgSO4, 25 mM Na2HPO4, 25 mM KH2PO4, 50 mM NH4Cl, and 5 mM Na2SO4, 0.2× trace metal mix) containing kanamycin (90 µg/mL) and chloramphenicol (30 µg/mL). Cells were grown in a baffled Fernbach culture flask (2800 mL) at 200 rpm at 37 °C for 3h, after which time the temperature was reduced to 16 °C. The culture was allowed to grow for another 20 h and the cells were harvested at 3,700 × g for 30 min. The resulting cell pellet (~20 g/L of culture) was washed with a buffer containing 50 mM HEPES, pH 7.5, 150 mM NaCl and used for protein purification.
Protein purification was carried out at 4 °C. A 20 g batch of cells was re-suspended in 100 mL buffer A (50 mM HEPES, pH 7.5, 150 mM NaCl) containing 50 mg lysozyme (RPI, cat. # L38100), 100 µL EDTA-free protease inhibitor cocktail (EMD cat. # 539134) and 50 µL DNase I (NEB, cat. # M0303S). Cells were placed on a rotating mixer to tumble for 15 min at 4 °C followed by sonication (Sonics Vibra-Cell; 50% amplitude, 1 sec ON – 2 sec OFF, 2 × 1.5 min) for effective cell lysis. Cells were always kept on ice during sonication and rested for 5 min in between the two sonication cycles. The resultant suspension was tumbled for 15 min at 4 °C followed by centrifugation at 9,000 × g for 45 min at 4 °C using a Ti45 rotor. The resulting supernatant was further centrifuged at 140,000 × g for 65 min at 4 °C. The membrane pellet, also known as cell envelope fraction (CEF), was resuspended in 2 mL of buffer A. The total volume of the solution was ~5 mL. To this resuspended CEF, 23 mL of buffer containing 50 mM HEPES, pH 7.5, 100 mM NaCl, 1% DDM (Anatrace, cat. # D310A) and 28 µL protease inhibitor cocktail solution was added. The suspension was tumbled overnight at 4 °C. The solution was centrifuged at 150,000 × g for 65 min at 4 °C using a Ti70 rotor. The supernatant was incubated with 1 mL Ni-NTA resin that was pre-equilibrated with an equilibration buffer containing 50 mM HEPES, pH 7.5, 100 mM NaCl, 20 mM imidazole and 5% glycerol. After tumbling the protein solution with the resin for 1 h, the flow-through was separated. The column was washed with 20 mL of wash-1 buffer (equilibration buffer + 0.03% DDM), followed by 20 mL of wash-2 buffer (equilibration buffer containing 45 mM imidazole + 0.03% DDM). The protein was eluted from the column using elution buffer (equilibration buffer containing 500 mM imidazole + 0.03% DDM). Elution fractions (2 × 1 mL) were combined and immediately desalted using a 5 ml HiTrap desalting column (GE Healthcare, cat. # 17-1408-01) that was pre-equilibrated with a desalting buffer containing 50 mM HEPES, pH 7.5, 100 mM NaCl, 0.03% DDM and 5% glycerol. The purified SUMO-tagged PglCs were analyzed by SDS-PAGE and the purity of the proteins was judged to be > 90% (Supplementary Figure 3). The protein yields were ~14 mg/L of culture.
Expression and purification of selenomethionine (Se-Met)-labeled PglC
PglC was expressed as the His6-SUMO-PglC construct in BL21-CodonPlus (DE3)-RIL using Overnight Express™ Autoinduction System 2 (EMD Millipore, cat. # 71366-3). Media was prepared according to the manufacturer’s protocol and supplemented with 125 mg/L of selenomethionine (Sigma, cat. # S3132) and 100 nM vitamin B12 (Sigma, cat. # V2876). Freshly transformed cells were used for overnight growth at 37 °C in 1.5 mL MDG media containing kanamycin and chloramphenicol (30 µg/mL each). The overnight culture was centrifuged and cells were collected and washed with 4 × 1 mL sterile buffer containing 50 mM HEPES, pH 7.5, 150 mM NaCl. Cells were resuspended in 1 mL of the final auto-induction media and transferred into 250 ml of the same media containing kanamycin (90 µg/mL) and chloramphenicol (30 µg/mL) followed by a 26 h growth in a baffled Fernbach culture flask (2800 mL) at 250 rpm at 37 °C. Cells were harvested (~9.5 g of cells/1L culture) and washed with a buffer containing 50 mM HEPES, pH 7.5, 150 mM NaCl. Purification of the protein was carried out following the same protocol as described above for wild-type PglC. After purification, 6.4 mg Se-Met labeled His6-SUMO-PglC was obtained per liter of culture. The purity of the protein was judged to be > 90% by SDS-PAGE analysis (Supplementary Figure 3). The His6-SUMO-tag was removed from the protein using SUMO protease as described below. The purity of the protein was judged to be > 95% by SDS-PAGE analysis (Supplementary Figure 3). The final yield of the Se-Met labeled PglC was 1.6 mg/L of culture. The protein was concentrated to ~6.5 mg/mL for crystallographic studies and selenium incorporation was confirmed to be > 90% via mass spectrometry analysis.
SUMO Cleavage of His6-SUMO-PglCs
Purified His6-SUMO-PglC variants were incubated with 0.14 equivalents of SUMO protease (S. cerevisiae) at 16 °C with gentle shaking at 80 rpm for 6 h. The SUMO protease was expressed and purified following a previously published protocol25. The resulting solution was incubated with 250 µL Ni-NTA resin that was pre-equilibrated with a buffer containing 50 mM HEPES, pH 7.5, 100 mM NaCl, 20 mM imidazole, 0.03% DDM and 5% glycerol. After 45 min incubation, the flow-through was collected, the column was washed with two column volumes of the desalting buffer and the wash fractions were combined with the flow-through. The final yields of purified cleaved proteins were ~7.5 mg/L of culture. The purity of the proteins was judged to be >95% by SDS-PAGE analysis (Supplementary Figure 3). The proteins were concentrated to 6.5–7.0 mg/mL for crystallographic studies.
Crystallization of PglC
Initial crystallization screens of the wild type PglC (6.5 mg/mL) with 1 mM MgCl2 and/or 1 mM UDP were performed at the Hauptmann-Woodward Institute (www.hwi.buffalo.edu/crystallization/services.html) using the membrane protein screen, developed by the Malkowski lab26. From these screens, 0.1 M HEPES pH 7.5, 0.2 M MgCl2, and 25% PEG 3350 was selected as the preliminary crystallization condition for optimization via hanging-drop vapor-diffusion to yield final crystallization conditions of 0.1 M Bis-Tris pH 6.0, 0.4 M MgCl2, and 23% PEG 3350 for a protein concentration of 260–276 µM for the three variants of PglC. Additional detergent was not added during crystallization. Experiments were set up at 17 °C with temperature equilibrated solutions purchased from Hampton Research. I57M/I87M PglC at a concentration 260 µM was co-crystallized with 260 µM undecaprenol phosphate (UndP) (using a 10 mM stock solution of UndP in DMSO) following a 30-minute incubation step on ice. I57M/I87M PglC crystals used for data collection appeared within 7 days. I57M/Q175M PglC crystals were grown under similar conditions (0.1 M Bis-Tris pH 6.0, 0.4 M MgCl2, 23% PEG 3350) at 17 °C with temperature equilibrated solutions. Co-crystallization experiments with 5-iodo-UDP were carried following incubation of 1 mM 5-iodo-UDP in 260 µM protein on ice for 30 minutes. I57M/Q175M PglC crystals used for data collection appeared within 7 days. WT Se-Met PglC crystals were grown at 17 °C with temperature equilibrated crystallization conditions of 0.1 M Bis-Tris pH 6.0, 0.3 M MgCl2, 27% PEG3350, and 1 mM TCEP. 276 µM Se-Met PglC was co-crystallized with 1 mM UDP after incubation on ice for 30 minutes. WT Se-Met PglC crystals used for data collection appeared within 3 days and were fully grown after 14 days. All crystals were flash-cooled in liquid nitrogen for transport to the beamlines without additional cryoprotection.
Phasing, model building and refinement of PglC
The datasets collected for both methionine variants indexed in the space group P 32 2 1 with unit-cell dimensions of a = b = 70.802, c = 188.442 for I57M/Q175M, and a = b = 71.61, c = 189.442 for I57M/I87M. These unit-cell dimensions will be referred to as the small unit cell. Matthews coefficient analyses for these data sets suggested two copies in the asymmetric unit (ASU). Conversely, derivatization of WT with SeMet resulted in the doubling of the unit cell axes a and b to give a unit cell with dimensions a = b = 143.375, c = 194.004 and a change in space group to P 31 2 1. These unit cell dimensions will be referred to as the large unit cell. As a result of the doubling of the a and b unit cell axes, Matthews coefficient analysis suggested that the ASU composition increased from 2 to 8 copies of PglC. The change in unit cell dimensions and space group are a result of subtle rearrangements that do not change the crystal contacts, but only change the contents of the asymmetric unit. A WT Se-Met PglC dataset collected from a crystal diffracting to 3.11 Å at beamline 24-ID-C (λ = 0.9791 Å/ 100 K) at the Advanced Photon Source (Chicago, IL) at the Se X-ray absorption energy peak (12665 eV) allowed initial phases to be solved by SAD using the Phenix suite27. Matthews coefficient analyses for the dataset suggested 8 copies in the ASU. Se-Met data were scaled and integrated using XDS. SHELXD28 was run for 5000 trials with a resolution cut-off of 4.5 Å to identify 16 Se sites. Fewer trials and higher-resolution cut-offs did not yield viable heavy-atom substructure solutions. Phenix.SOLVE was used to find an additional 6 Se sites and calculate subsequent Se substructure phases for 22 out of the expected 32 Se atoms in the ASU. Phenix.RESOLVE29 was used to perform initial solvent flattening and phase-extension. At this point, α-helical density apparent in the solvent-flattened map allowed for initial building of poly-Ala helical fragments manually. Using these α-helical fragments as a starting model, Phenix.AutoBuild was able to locate and assign sequence to two copies of the RMH in the ASU. Phenix.Find_NCS was used to find the two-fold non-crystallographic symmetry (NCS) operators relating the two helices, and the two-fold used for NCS map-averaging in Phenix.NCS_average. Phenix.Find_NCS was used to find all 8 NCS related positions from the electron density in the 2-fold averaged map. The two-fold averaged map was further averaged over all 8 NCS operators by Phenix.NCS_average. Additional model was built manually and sequence assigned, using as a guide computationally-derived models of PglC from EV-fold (Campylobacter jejuni)4 and RaptorX30, as well as the model of E. coli WcaJ (pfam accession P71241) computed by covariance using Rosetta31. A manually-extended model containing a dimer of 94 residues (AAs 3–60, 74–98, 165–175) in each chain was used as a search model for molecular replacement via Phenix.PhaserMR32 into a native I57M/I87M 2.59 Å dataset collected at beamline 24-ID-C (λ = 0.9792 Å/ 100 K) at the Advanced Photon Source (Chicago, IL) containing 2 copies in the ASU. Phenix.AutoBuild was used to complete building of 86% of the model and an additional 14 residues were built manually into the electron density using COOT33. This model containing two subunits with 185 residues in each chain was used to phase a more complete, higher I/σ(I) dataset of I57M/Q175M PglC at 2.74 Å resolution collected at the Advanced Photon Source (Chicago, IL) beamline 24-ID-C (λ = 1.5498 Å/ 100 K). The over-all fold of the WT and variant structures are identical despite crystallizing in enantiomeric space groups. Refinement against the electron density map was performed with Phenix.Refine34 to refine XYZ coordinates, real-space, rigid body, and group B-factors. Subsequent rounds of refinement included refinement of translation-libration-screw (TLS) parameters, manually placed waters, and simulated annealing of Cartesian coordinates and torsion angles.
The final model with two protomers in the asymmetric unit was refined to Rwork/Rfree of 0.2587/0.2815 with no significant outliers using Phenix.Refine. Both chains of the model include185 out of 205 amino acids. Chain A contains amino acids (−)3 to 182 and chain B contains amino acids (−)2 to 183. Chains A and B are highly similar with an RMSD of 0.31 Å. The extended loop structure encompassing residues 62–81 is well ordered in chain B owing to its participation in crystal contacts, however only weak density for this loop was observed in chain A. Residues 148–153 were not well resolved in either chain and were placed into 2Fo-Fc density contoured at 1 RMSD in COOT33. The final model of PglC was refined with two protein chains, four molecules of phosphatidylethanolamine (PE), 2 Mg2+ ions, one inorganic phosphate ion, and one PEG (tetraethylene glycol). The inorganic phosphate and the PEG were observed in only one of the two chains in the asymmetric unit (Chain A). The chemical structures of endogenous, native PE were built into the model using electron density maps calculated with the coefficients 2Fo-Fc and Fo-Fc resulting in four molecules with differing partial acyl chain lengths (two PE per monomer of PglC). PE molecules 303 (Chain A) and 302 (Chain B) lie within the predicted membrane interaction surface for each PglC monomer. Whereas PE molecules 304 (Chain A) and 303 (Chain B) are associated with hydrophobic patches distal to the predicted membrane plane, and so do not appear to be in physiologically relevant positions. Exclusion of the PE molecules with non-physiological positions increased both Rwork and Rfree statistics during refinement. Additionally, diffuse scattering from disordered detergent and additional unobserved lipid molecules could have contributed to slightly elevated Rwork/Rfree values. Examination of the space group, special projections and Matthews coefficient analyses determined that the apparent asymmetric unit contents were appropriate to the space group (the International Tables for Crystallography give the symmetry for three special projections for each space group in the standard orientation). The analysis showed that the observed intensities were consistent with high symmetry from the space group, and not from twinning. An inorganic phosphate (PO43−) ion and a Mg2+ ion were modeled in Chain A, and a Mg2+ ion and ordered water molecule were modelled in Chain B into positive Fo-Fc density contoured to 4 RMSD in COOT. The PEG molecule was modeled into positive Fo-Fc density in Chain A contoured to 2.5 RMSD in COOT. Refinement with the DDM detergent alkyl group (dodecane- Ligand ID:D12) yields results approximately equivalent to refinement with PEG. However, the absence of any observed density for the maltose disaccharide of the DDM detergent led us to continue the final refinement with the PEG moiety. Notably, the observed density cannot be the undecaprenol moiety of undecaprenol phosphate (Und-P) as the density was observed in all datasets including those collected from crystals which were not co-crystallized with added Und-P. The dihedral angles of residues in the final refined model are all in the favored (97.54%) or allowed (2.19%) regions of the Ramachandran plot.
Structural Analyses of PglC
The webserver GREMLIN (http://gremlin.bakerlab.org/) was used to create a statistical model of the conservation and covariance in the PglC family alignment previously reported4. Contact pairs with greater than or equal to 99% probability of co-evolution were plotted as pseudo-bonds with UCSF Chimera onto the PglC structure for analysis. Hydrophobicity of residues of PglC was analyzed in PyMol according to the Eisenberg normalized consensus hydrophobicity scale16. Amphipathic α-helices identified from hydrophobic coloring in PyMol were analyzed via helical wheel projections created using the Helixator webserver (http://www.tcdb.org/progs/helical_wheel.php). The following helix sequences were used in construction of the projections: helix C – KFGKLMRSL (Residues 86–94), helix D – LDELPQLFNVLK (Residues 96–107), helix I – FMLDVKIALQTIEKVLK (Residues 170–186) (Supplementary Figure 8b). The free energy of transfer of PglC into the membrane was calculated as ΔGtransfer = −39.5 kcal/mol with the PPM (Positioning of Proteins in Membrane) server (http://opm.phar.umich.edu/server.php). Comparison of the membrane plane calculated by the PPM server and hydrophobicity analyses of the RMH and AHs suggests that PglC may be positioned approximately 5 Å deeper in the membrane than determined by the PPM server (Supplementary Figure 8c). Electrostatic surface analyses for PglC were performed using the Adaptive Poisson-Boltzmann Solver APBS35 plug-in for PyMol (Fig. 2d). APBS was run in Linearized Poisson-Boltzmann Equation mode with surface calculation by cubic B-splines with harmonic average smoothing. Electrostatic surface was visualized with contours at ± 5 kT/e.
Substituted-cysteine accessibility method (SCAM)
PglC from C. jejuni strain 11168 was used for SCAM analysis15. Unique cysteines were introduced either N-terminally (K4C, F6C) or C-terminally (S88C, S186C) to the membrane-associated domain at non-conserved, surface-exposed sites (residues correspond to N4, L6, S89, and S187 in PglC from C. concisus) (Supplementary Table 5). Wild-type PglC and the four cysteine variants were overexpressed in E. coli. Whole cells expressing each unique variant were treated with one of two thiol-blocking reagents, either N-ethylmaleimide (NEM), which is cell-permeant, or 2-sulfonatoethyl methanethiosulfonate (MTSES), which is only able to cross the outer cell membrane. Following cell lysis, any remaining free cysteines were reacted with PEG-maleimide (PEG-mal). Labeling of the target protein with PEG-mal was observed by Western blot as a band shift to higher molecular weight. Cysteines in the periplasm are thus distinguished by their ability to be blocked from PEGylation by both NEM and MTSES, while cytoplasmic cysteines are PEGylated following treatment with MTSES but not following treatment with NEM. Wild-type PglC has no native cysteines, and thus was not labeled with PEG-mal under any thiol-blocking conditions. All four cysteine variants were blocked from PEG-mal labeling by incubation with NEM but not by incubation with MTSES, indicating that all four are located in the cytoplasm (Fig. 2b). A unique cysteine variant of a periplasmic protein, PEB3 A204C, served as a positive control for thiol-blocking of cysteines in the periplasm.
Protein labeling and analysis for SCAM
For expression of the proteins, BL21(DE3)-RIL E. coli cells (Agilent) were transformed with the plasmid constructs. Labeling was performed using a protocol modified from that of Furlong et al.13. Cultures were grown overnight in 3 mL LB media with selection antibiotics (30 µg/mL kanamycin, 30 µg/mL chloramphenicol). Overnight cultures were diluted into 5 mL fresh LB (with the same selection antibiotic) to a final OD600 of 0.2. Cultures were grown for 1 h at 37 °C, then moved to 16 °C and induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 3–5 h. Following expression, cells were washed once with PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4), adjusted to a final OD600 of 1 in 200 µL PBS containing 1 mM EDTA, and divided into four 50 µL aliquots. Aliquots were treated with 5 µL of 55 mM N-ethylmaleimide (NEM, Alfa-Aesar, cat. # 40526) or sodium(2-sulfonatoethyl)methanethiosulfonate (MTSES, Cayman Chemical, cat. # 16529) for a final concentration of 5 mM, or water, and were incubated at room temperature in the dark for 1 h. Cells were harvested by centrifugation at 16,000 × g for 3 min and washed with 100 µL of PBS. Pellets were resuspended in 22.5 µL lysis buffer (50 mM HEPES, 5% SDS, pH 7.5) and 7.5 µL of 25 mM PEG-mal (Sigma, cat. # 63187) in DMSO. Samples not treated with PEG-mal were treated with 7.5 µL DMSO. Samples were mixed and incubated at room temperature in the dark for 1–1.5 h. Reactions were quenched by the addition of 30 µL of 2× loading buffer (100 mM Tris pH 6.8, 10% glycerol, 3% SDS, 6 M urea, 1% β-mercaptoethanol, 0.1% bromophenol blue) and frozen at −20 °C until needed.
Proteins were separated by SDS-PAGE on 12% acrylamide gels and transferred to nitrocellulose membranes for Western blot analysis against the C-terminal His6-tag (Fig. 2b). Blots were blocked with 3% bovine serum albumin in TBST (10 mM Tris, 150 mM NaCl, 0.05% Tween-20, pH 7.5) for 1 h. The primary antibody, mouse anti-His (LifeTein, cat. # LT0426) was applied in a 1:3,000 dilution for 1 h. The secondary antibody, goat anti-mouse AP (Invitrogen, cat. # 31328) was applied in a 1:10,000 dilution for 1 h. Immunoblots were developed using 1-Step NBT/BCIP Substrate Solution (ThermoFisher Scientific, cat. # 34042) and imaged using a Molecular Imager Gel Doc XR+ System (BioRad).
Statistics for phosphohydrolase activity assay
Phosphate release assays were carried out in triplicate. Error bars represent mean ± standard deviation.
Data availability
Data and coordinates for I57M/Q175M C. concisus PglC have been deposited in the Protein Data Bank with the accession code 5W7L.
Supplementary Material
Acknowledgments
We thank Dr. K. Rajashankar for assistance with phasing and the staff at NECAT (APS) for facilitating X-ray data collection. Financial support for this work was provided by NIH (R01-GM039334 to BI, the Predoctoral Training Program in the Biological Sciences (T32-GM007287) to SE and the Biomolecular Pharmacology Program Grant (T32-GM008541) to LCR. This work is also based upon research conducted at the Northeastern Collaborative Access Team beamlines 24-ID-E and 24-ID-C, which is funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403).
Footnotes
Author contributions: L.C.R crystallized, collected data, determined the structure, refined and analyzed the model of PglC and performed phosphate release kinetics; D.D. optimized expression, designed and made constructs, expressed and purified PglC, carried out lipid analysis, and analyzed the structure. S.E. designed and performed SCAM analyses. V.L. designed and purified original constructs for crystallization and A.J.L. obtained the original crystallization conditions. L.C.R., D.D., and S.E. wrote the manuscript. B.I. and K.N.A. conceived the project, designed experiments, assisted with data analysis and interpretation, and critically edited the manuscript.
The authors declare no competing interests.
References
- 1.Lukose V, Walvoort MTC, Imperiali B. Bacterial phosphoglycosyl transferases: Initiators of glycan biosynthesis at the membrane interface. Glycobiology. 2017;27:820–833. doi: 10.1093/glycob/cwx064. doi:doi.org/10.1093/glycob/cwx064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Price NP, Momany FA. Modeling bacterial UDP-HexNAc: polyprenol-P HexNAc-1-P transferases. Glycobiology. 2005;15:29R–42R. doi: 10.1093/glycob/cwi065. [DOI] [PubMed] [Google Scholar]
- 3.Chung BC, et al. Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science. 2013;341:1012–1016. doi: 10.1126/science.1236501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lukose V, et al. Conservation and Covariance in Small Bacterial Phosphoglycosyltransferases Identify the Functional Catalytic Core. Biochem. 2015;54:7326–7334. doi: 10.1021/acs.biochem.5b01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tytgat HL, Lebeer S. The sweet tooth of bacteria: common themes in bacterial glycoconjugates. Microbiol. Mol. Biol. Rev. 2014;78:372–417. doi: 10.1128/MMBR.00007-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bugg TD, Rodolis MT, Mihalyi A, Jamshidi S. Inhibition of phospho-MurNAc-pentapeptide translocase (MraY) by nucleoside natural product antibiotics, bacteriophage varphiX174 lysis protein E, cationic antibacterial peptides. Bioorg. Med. Chem. 2016;24:6340–6347. doi: 10.1016/j.bmc.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 7.Das D, Kuzmic P, Imperiali B. Analysis of a dual domain phosphoglycosyl transferase reveals a ping-pong mechanism with a covalent enzyme intermediate. Proc. Natl. Acad. Sci. U. S. A. 2017;114:7019–7024. doi: 10.1073/pnas.1703397114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Dabbagh B, et al. Catalytic mechanism of MraY and WecA, two paralogues of the polyprenyl-phosphate N-acetylhexosamine 1-phosphate transferase superfamily. Biochimie. 2016;127:249–257. doi: 10.1016/j.biochi.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Hartley MD, Schneggenburger PE, Imperiali B. Lipid bilayer nanodisc platform for investigating polyprenol-dependent enzyme interactions and activities. Proc. Natl. Acad. Sci. U. S. A. 2013;110:20863–20870. doi: 10.1073/pnas.1320852110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoki S, Thomas A, Decaffmeyer M, Brasseur R, Epand RM. The role of proline in the membrane re-entrant helix of caveolin-1. J. Biol. Chem. 2010;285:33371–33380. doi: 10.1074/jbc.M110.153569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heijne Gv. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. The EMBO Journal. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furlong SE, Ford A, Albarnez-Rodriguez L, Valvano MA. Topological analysis of the Escherichia coli WcaJ protein reveals a new conserved configuration for the polyisoprenyl-phosphate hexose-1-phosphate transferase family. Scientific reports. 2015;5:9178. doi: 10.1038/srep09178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel KB, Ciepichal E, Swiezewska E, Valvano MA. The C-terminal domain of the Salmonella enterica WbaP (UDP-galactose:Und-P galactose-1-phosphate transferase) is sufficient for catalytic activity and specificity for undecaprenyl monophosphate. Glycobiology. 2012;22:116–122. doi: 10.1093/glycob/cwr114. [DOI] [PubMed] [Google Scholar]
- 15.Nasie I, Steiner-Mordoch S, Schuldiner S. Topology determination of untagged membrane proteins. Methods Mol. Biol. 2013;1033:121–130. doi: 10.1007/978-1-62703-487-6_8. [DOI] [PubMed] [Google Scholar]
- 16.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 17.Zidovetzki R, Rost B, Armstrong DL, Pecht I. Transmembrane domains in the functions of Fc receptors. Biophys. Chem. 2003;100:555–575. doi: 10.1016/S0301-4622(02)00306-X. [DOI] [PubMed] [Google Scholar]
- 18.Jones S, Daley DT, Luscombe NM, Berman HM, Thornton JM. Protein-RNA interactions: a structural analysis. Nucleic acids Res. 2001;29:943–954. doi: 10.1093/nar/29.4.943. doi:doi.org/10.1093/nar/29.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lloyd AJ, Brandish PE, Gilbey AM, Bugg TDH. Phospho-N-Acetyl-Muramyl-Pentapeptide Translocase from Escherichia coli: Catalytic Role of Conserved Aspartic Acid Residues. J. Bacteriol. 2004;186:1747–1757. doi: 10.1128/jb.186.6.1747-1757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amer AO, Valvano MA. Conserved aspartic acids are essential for the enzymic activity of the WecA protein initiating the biosynthesis of O-specific lipopolysaccharide and enterobacterial common antigen in Escherichia coli. Microbiology (Reading, England) 2002;148:571–582. doi: 10.1099/00221287-148-2-571. [DOI] [PubMed] [Google Scholar]
- 21.Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. doi:doi.org/10.1016/S0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen KN, Dunaway-Mariano D. Catalytic scaffolds for phosphoryl group transfer. Curr Opin Struct Biol. 2016;41:172–179. doi: 10.1016/j.sbi.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ardiccioni C, et al. Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis. Nat. Commun. 2016;7:10175. doi: 10.1038/ncomms10175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 25.Weeks SD, Drinker M, Loll PJ. Ligation independent cloning vectors for expression of SUMO fusions. Protein Expr. Purif. 2007;53:40–50. doi: 10.1016/j.pep.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koszelak-Rosenblum M, et al. Determination and application of empirically derived detergent phase boundaries to effectively crystallize membrane proteins. Protein Sci. 2009;18:1828–1839. doi: 10.1002/pro.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1772–1779. doi: 10.1107/S0907444902011678. [DOI] [PubMed] [Google Scholar]
- 29.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. doi:doi.org/10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S, Sun S, Li Z, Zhang R, Xu J. Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model. PLoS Comp. Biol. 2017;13:e1005324. doi: 10.1371/journal.pcbi.1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ovchinnikov S, et al. Protein structure determination using metagenome sequence data. Science. 2017;355:294–298. doi: 10.1126/science.aah4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/s0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/s0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/s0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and coordinates for I57M/Q175M C. concisus PglC have been deposited in the Protein Data Bank with the accession code 5W7L.