Sir,
We read with great interest the article recently published in Brain by Nibbeling et al. (2017) reporting five novel spinocerebellar ataxia (SCA) genes—FAT2, PLD3, KIF26B, EP300 and FAT1—identified by exome sequencing and targeted resequencing of exome candidates in genetically undiagnosed families with autosomal dominant SCA. Of particular interest for us was the identification of a point mutation, presumably leading to loss of function, in PLD3 (coding for phospholipase D3). This new variant was assigned as SCA46 (OMIM #617770). We recently characterized PLD3 as a resident lysosomal protein that is synthesized as a transmembrane protein that undergoes proteolytic cleavage, thereby releasing a stable soluble lysosomal enzyme with 5′ exonuclease activity (Gavin et al., 2018; Gonzalez et al., 2018).
While there is little doubt about the genetic data, we have concerns regarding the cell biology follow up experiments, particularly the localization studies and functional assay for PLD3 performed by Nibbeling et al. Importantly, we generated PLD3 knockout (KO) mice that do not present signs of cerebellar degeneration or spinocerebellar ataxia, challenging the interpretation of the suggested loss-of-function mechanism or PLD3 as the SCA46-causative gene.
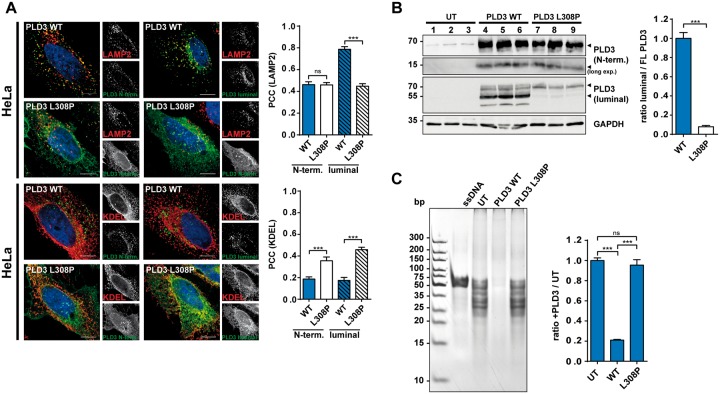
Nibbeling et al. used GFP-tagged PLD3 constructs to localize PLD3 in HeLa cells upon overexpression. They found both the wild-type and the L308P-mutant protein in the endoplasmic reticulum (ER). In contrast, in our experiments expressing untagged PLD3, followed by immunofluorescence staining using antibodies directed against the N-terminus or the luminal domain, PLD3-WT was found in LAMP2-positive endolysosomes, compatible with its function in lysosomes (Gavin et al., 2018; Gonzalez et al., 2018). Hardly any PLD3-WT was detected in the ER (Fig. 1A). However, detection of the luminal domain of PLD3-L308P revealed a significantly reduced lysosomal localization, but increased co-localization with the ER marker KDEL, implicating sorting or folding defects due to the patient's point mutation. Immunoblot analysis of HeLa cells transfected with PLD3-WT and detection with the PLD3-specific antibodies showed the typically-observed cleavage pattern of the ~70 kDa full-length protein, a ~50 kDa luminal fragment detected with the luminal antibody and a ~12 kDa N-terminal fragment (Gonzalez et al., 2018). In contrast, the PLD3-L308P mutation led to strikingly reduced proteolytic cleavage of full-length PLD3, and both the 50 kDa and 12 kDa fragment were almost completely absent (Fig. 1B). This data is in good agreement with our previous findings, showing cleavage of PLD3 in late endosomes and lysosomes and supports our localization data of the ER retention of PLD3-L308P. PLD3 acts as a lysosomal 5′ exonuclease (Gavin et al., 2018). Testing 5′ exonuclease activity against single stranded DNA after transfection of HeLa cells with wild-type and L308P-mutant PLD3 revealed efficient cleavage of the DNA-oligo-substrate in wildtype-transfected cells, but not in the L308P-mutant-transfected cells, demonstrating a loss of exonuclease activity of the L308P mutation (Fig. 1C).
Figure 1.
Impaired lysosomal delivery, proteolytic processing and loss of function of the PLD3-L308P mutation. (A) Confocal microscopy of HeLa cells transfected with the PLD3-WT or PLD3-L308P mutation followed by staining with the indicated antibodies. Pearson correlation coefficient (PCC) for co-localization for the indicated antibodies is depicted; n = 12 cells. (B) Immunoblot of PLD3-WT or PLD3-L308P mutation with indicated PLD3-specific antibodies and GAPDH. Quantification of the fragments relative to full-length protein is shown. UT = untransfected. (C) Ethidium-bromide stained polyacrylamide gel analysis of untreated ssDNA substrate and treated with untransfected HeLa cell lysates or transfected with PLD3-WT or PLD3-L308P-mutation transfected HeLa cell lysates. Quantification represents n = 3. ***P < 0.001; ns = not significant.
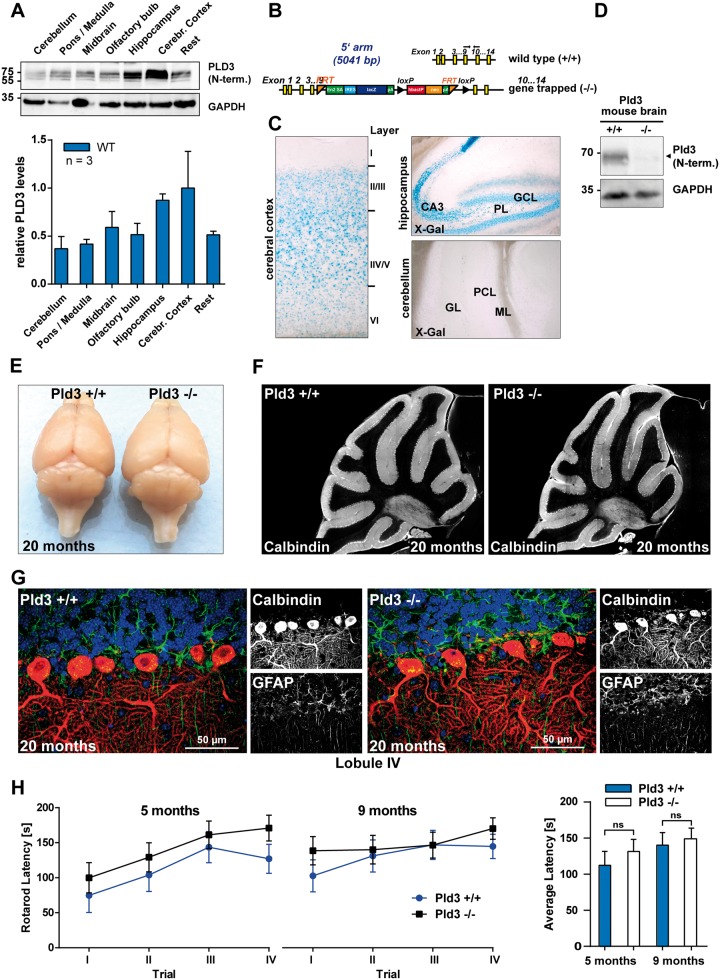
We were surprised at the suggested causative role of PLD3 for cerebellar ataxia, since our protein expression data generated by immunoblot with a PLD3-specific antibody showed lowest PLD3 expression levels in the adult cerebellum compared to all other brain regions (Fig. 2A). We generated PLD3 KO mice to analyse loss-of-function effects in vivo. Integration of a targeting vector containing LacZ coding for β-galactosidase in intron 9 and artificial splice-acceptor sites (Fig. 2B) led to a gene-trapped allele and complete loss of detectable PLD3 protein (Fig. 2C). The β-galactosidase reporter under the endogenous PLD3 promotor allowed us to follow the region-specific expression pattern. Again, hardly any PLD3 expression was observed in the cerebellum, while extensive β-galactosidase activity was detected in the cerebral cortex and the hippocampus (Fig. 2D). Our expression analyses are in good agreement with a previously published in situ hybridization analysis (Pedersen et al., 1998).
Figure 2.
PLD3 expression in the cerebellum is low and PLD3 KO mice do not show any signs of cerebellar degeneration. (A) Immunoblot analysis of PLD3 expression in different brain regions of adult wild-type mice with an antibody against the N-terminus of PLD3; n = 3. (B) Targeting vector for PLD3 LacZ reporter and PLD3 KO mice. (C) β-Galactosidase activity in different brain regions of an adult PLD3-reporter mouse with X-Gal as colorimetric substrate. (D) Immunoblot analysis of brain lysates with an antibody against the N-terminus of PLD3. (E) Photograph of the brain of wild-type and PLD3 KO mice, 20 months old. (F) Calbindin-immunofluorescence of the cerebellum of a wild-type and a PLD3 KO mouse, 20 months old. (G) Immunofluorescence staining for calbindin (red), GFAP (green) of the molecular layer in lobule IV of 20-month-old wild-type and PLD3 KO mice. Nuclei are stained with DAPI (blue). (H) Rotarod latency in 5- and 9-month-old animals (n = 9–11).
Nibbeling et al. suggest a dominant loss-of-function mechanism due to the PLD3-L308P mutation, suggesting that the PLD3 knockout reflects the situation observed in SCA patients. We did not observe any alterations in the size or morphology of the cerebellum in aged (>20 months) PLD3-deficient animals (Fig. 2E). Immunohistology for calbindin did not reveal loss of Purkinje cells in the cerebellum (Fig. 2F). Similarly, no signs of Bergmann astrogliosis or demyelination (Fig. 2G and Supplementary Fig. 1) of the cerebellum were observed. Behavioural testing for rotarod performance in adult mice (5 months and 9 months) moreover failed to reveal signs of cerebellar dysfunction (Fig. 2H). These data do not reveal a major degenerative phenotype of the cerebellum in PLD3 KO mice.
Nibbeling et al. show a reduced phospholipase D (PLD) activity for PLD3-L308P. However, we failed to show PLD activity even for PLD3-WT (unpublished). To investigate possible changes on the lipid profile of the brain due to impaired catabolism of possible PLD3 substrates, we performed shotgun lipidomics on brain lysates covering 285 lipids (Supplementary Table 1). We did not observe any major changes for lipid species associated with PLD activity, neither on substrates nor products site including phosphatidic acid species or phosphatidylcholine. This result rather shows that PLD3 does not perform a typical PLD activity.
In summary our data support the finding by Nibbeling et al. that the PLD3-L308P mutation leads to a loss of function. We show that PLD3 is not an ER-resident protein and is localized and active in lysosomes acting as a 5′ exonuclease. The PLD3 mutation leads to ER retention and lack of nuclease function. Importantly, the loss of PLD3 does not interfere with lipid catabolism. Our PLD3 KO mouse analysis challenges the interpretation of PLD3 mutations as the causative SCA46 gene and further characterization of additional patients will be required to clarify a role of PLD3 in spinocerebellar ataxia.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary material.
Supplementary Material
Acknowledgement
We acknowledge Inken Beck (Czech Centre for Phenogenomics) for revitalization of the PLD3 KO mouse strain.
Funding
Funding was provided by the Alzheimer Forschung Initiative e.V. (M.D.) and the Deutsche Forschungsgemeinschaft (GRK 1459) (P.S.). AC.G. is supported by the Hans & Ilse Breuer foundation. D.M. is supported by National Institutes of Health (NIH) grant R21AI126011.
Competing interests
The authors report no competing interests
References
- Gavin AL,, Huang D,, Huber C,, Martensson A,, Tardif V,, Skog PD, et al. PLD3 and PLD4 are single-stranded acid exonucleases that regulate endosomal nucleic-acid sensing. Nat Immunol 2018; 19: 942–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez AC,, Schweizer M,, Jagdmann S,, Bernreuther C,, Reinheckel T,, Saftig P, et al. Unconventional trafficking of mammalian phospholipase D3 to lysosomes. Cell Rep 2018; 22: 1040–53. [DOI] [PubMed] [Google Scholar]
- Nibbeling EAR,, Duarri A,, Verschuuren-Bemelmans CC,, Fokkens MR,, Karjalainen JM,, Smeets C, et al. Exome sequencing and network analysis identifies shared mechanisms underlying spinocerebellar ataxia. Brain 2017; 140: 2860–78. [DOI] [PubMed] [Google Scholar]
- Pedersen KM,, Finsen B,, Celis JE,, Jensen NA. Expression of a novel murine phospholipase D homolog coincides with late neuronal development in the forebrain. J Biol Chem 1998; 273: 31494–504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary material.