Abstract
Zika virus (ZIKV) is a mosquito-borne flavivirus that leads to devastating consequences for fetal development. However, accurate diagnosis of ZIKV is made difficult by the fact that most infected patients are asymptomatic or present with symptoms similar to those of other febrile illnesses. Thus, the development of a simple, accurate, highly sensitive, and reliable method for the biomedical analysis and diagnosis of ZIKV is needed. Herein, a novel ZIKV liquid biopsy system was constructed via a dendritic Ru(bpy)32+-polymer-amplified electro-chemiluminescence (ECL) strategy. This system accomplished amplification-free analysis of ZIKV using a drop of blood, and simultaneously achieved a high sensitivity of 500 copies and superior specificity. This strategy adopted the humoral biomarker as the diagnostic index, which greatly simplified the analysis process, and established a nondestructive detection mode. Furthermore, the performance index for biomedical analysis of clinical ZIKV samples was investigated, and the results indicated that the dendritic Ru(bpy)32+-polymer-amplified ECL strategy reliably responded to ZIKV from the body fluid (blood, saliva, and urine). Hence, this system suitably met the strict clinical requirements for ZIKV detection and thus has the potential to serve as a new paradigm for the biomedical analysis and diagnosis of ZIKV.
Short abstract
A dendritic Ru(bpy)32+-polymer-amplified ECL diagnosis strategy for the Zika virus using a drop of blood is presented.
Introduction
The Zika virus (ZIKV) is an Aedes mosquito-borne flavivirus that could produce devastating consequences for the process of fetal development.1−3 Furthermore, ZIKV has been declared a public health emergency of international concern by the World Health Organization (WHO) because of the large-scale outbreak of the virus in the Americas.4,5 ZIKV mainly spreads via infected mosquito bite, but can also be transmitted by mother-to-fetus transmission, sexual contact, or blood transfusion.6−10 Additionally, it has been indicated that the infection of ZIKV was the main cause of Guillain-Barré syndrome,11,12 congenital microcephaly,13,14 and neurological defects in newborns.15,16 Thus, the development of a simple, accurate, highly sensitive, and reliable method for the biomedical analysis and diagnosis of ZIKV would be of great significance for the prevention and control of ZIKV. However, biomedical analysis and accurate diagnosis of ZIKV are made difficult by the fact that most infected patients are asymptomatic or present symptoms similar to those of other febrile illnesses.17
The existing immunoassays for ZIKV detection, such as the enzyme-linked immunosorbent assay (ELISA),18,19 provide an inexpensive and instrumentless approach, but their poor sensitivity and specificity limited the application of these immunoassays applied to the clinical detection and diagnosis of ZIKV.20,21 In particular, the antibody used in the immunoassay for ZIKV detection would also respond to homologous flaviviruses, such as Dengue virus.22,23 Thus, the specificity of immunoassays cannot meet the requirements for the accurate detection and early diagnosis of ZIKV.24 Conversely, the enzymatic amplification-based detection assays, such as reverse-transcription polymerase chain reaction (RT-PCR)25,26 and nucleic acid sequence-based amplification (NASBA),27,28 are endowed with the properties of high sensitivity and desirable specificity. In particular, the PCR-based assays are considered the gold standard for ZIKV detection. However, the labor-intensive sample pretreatment steps, expensive equipment, centralized laboratory facilities, and trained personnel required by PCR greatly reduced the popularization rate of its clinical application.29,30 Therefore, it is of significance to develop a novel method for biomedical analysis and diagnosis of ZIKV.
In recent years, the rapid developments in biomedical analysis and analytical chemistry have led to the emergence of many new detection platforms.31−34 This advancement inspired us to construct a new methodology for biomedical analysis and diagnosis of ZIKV. Herein, a novel ZIKV liquid biopsy system was constructed by integrating a dendritic Ru(bpy)32+-polymer-amplified electro-chemiluminescence (ECL) strategy as an effective signal giving-out pattern. This system accomplished amplification-free analysis of ZIKV using a drop of blood, and simultaneously achieved a high sensitivity of 500 copies and desirable specificity. This strategy adopted the humoral biomarker as the diagnostic index, which greatly simplified the biomedical analysis process, and established a nondestructive detection mode. Furthermore, we investigated the performance index for the biomedical analysis of clinical ZIKV samples, and the results indicated that the Ru(bpy)32+-polymer-amplified ECL strategy steadily responded to ZIKV from the body fluid (blood, saliva, and urine). Hence, this system suitably met the strict clinical requirements for ZIKV detection and thus has the potential to serve as a new paradigm for the biomedical analysis and diagnosis of ZIKV.
Results and Discussion
Design of a Dendritic Ru(bpy)32+-Polymer-Amplified ECL Assay
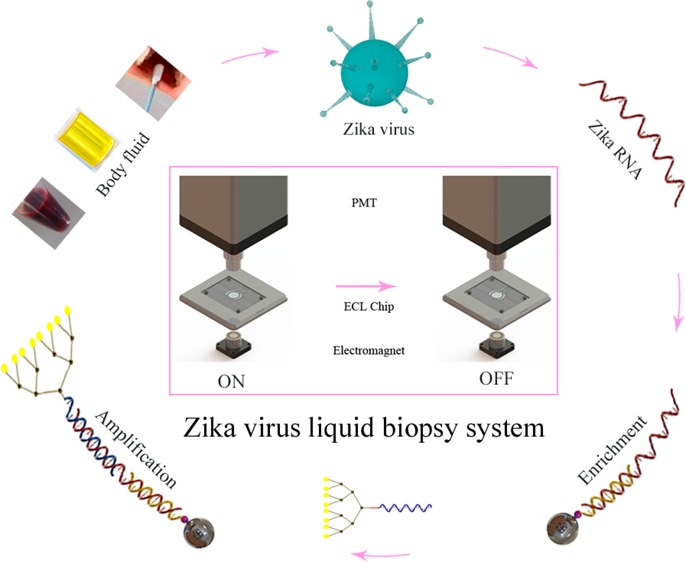
The constructed ZIKV liquid biopsy system was composed of the sample pretreatment, RNA enrichment, and ECL signal readout steps (Figure 1). First, ZIKV samples from body fluids (blood, saliva, and urine) were pretreated by dissociation and magnetic bead enrichment. Biomedical analysis and diagnosis of ZIKV via body fluid samples can provide a nondestructive detection mode that greatly simplifies the analysis process and alleviates the damage to the patients. Subsequently, The RNA of ZIKV was immobilized on magnetic beads by capture probe. The magnetic bead enrichment step can concentrate low-concentration ZIKV RNA from the ZIKV samples and increase the specificity via the recognition induced by the capture probe, which immobilized the ZIKV RNA on the magnetic beads. Finally, the RNA captured by the magnetic beads was recognized by the dendritic Ru(bpy)32+-polymer-amplified ECL probe after the washing process. The amplified probe provides a stable, intense response signal for ZIKV, and serves as the foundation of the high sensitivity of the method. The distinguishable ECL signals were then detected by an ECL instrument.
Figure 1.

Principle of the Zika virus liquid biopsy system based on the dendritic Ru(bpy)32+-polymer-amplified ECL strategy. (A) Process of dissociation, recognition and capture of ZIKV RNA. SDS lysis buffer was used to smear the ZIKV and cells in the body fluids. DRP represents the dendritic Ru(bpy)32+-polymer. Magnetic bead enrichment can concentrate low-concentration ZIKV RNA from ZIKV samples and increase the specificity by the recognition induced by the capture probe. (B) ECL signal-producing step of the dendritic Ru(bpy)32+-polymer-amplified strategy. Excess TPA (10 μM) was added to the ECL chip to assist ECL generation by the dendritic Ru(bpy)32+-polymer.
The construction of the dendritic Ru(bpy)32+-polymer-amplified ECL probe for ZIKV comprises the synthesis of Ru(bpy)32+ and probe assembly. The synthetic route and characterization of activated Ru(bpy)32+ are shown in Figure 2A (the complete synthetic process is outlined in section 1.1 of the Supporting Information, Figures S1 and S2). The assembly of the dendritic Ru(bpy)32+-polymer-amplified ECL probe (Figure 2B) was initiated via the sulfhydryl and double-bond interaction between the dendritic polymer and the DNA recognition domain (the structural formula of the dendritic polymer and the reaction process are presented in Figures S3 and S4). In this structure, the amino groups of the dendritic polymer provided the conjugation site for carboxyl-activated Ru(bpy)32+ (the complete synthetic route for the dendritic Ru(bpy)32+-polymer-amplified ECL probe is outlined in section 1.2 of the Supporting Information). The differences in properties between the dendritic Ru(bpy)32+-polymer and Ru(bpy)32+ were compared, and are listed below.
Figure 2.
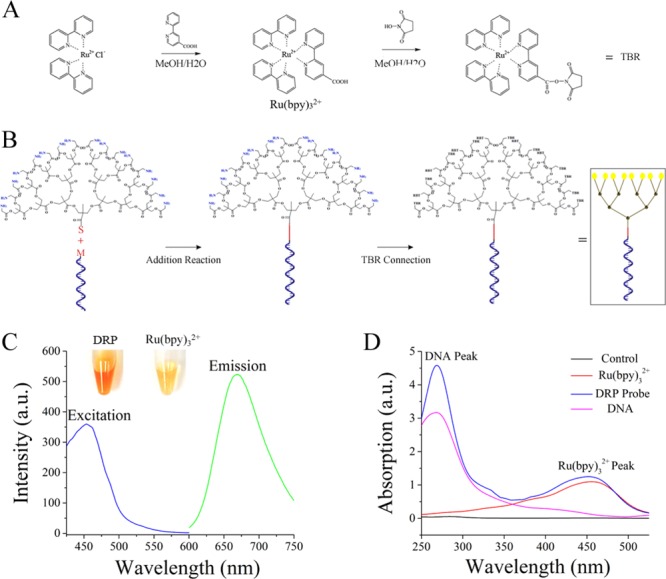
Synthetic routes for Ru(bpy)32+ and the dendritic Ru(bpy)32+-polymer. (A) Synthetic route and activation process of Ru(bpy)32+. The concentration of MeOH was 80%. The details of the crystallization, separation, and purification processes are provided in section 1.1 of the Supporting Information. (B) Synthetic route for the dendritic Ru(bpy)32+-polymer probe. The DNA recognition domain was labeled with maleimide, abbreviated as M. The dendritic polymer was labeled with amino and sulfhydryl group. Sulfhydryl, abbreviated as S, could bond with maleimide via an addition reaction. The amino group provided the conjugation site for Ru(bpy)32+. The details of the purification process and the complete structure of the dendritic Ru(bpy)32+-polymer are shown in section 1.1 of the Supporting Information. (C) Excitation and emission of the dendritic Ru(bpy)32+-polymer. The concentrations of the solutions were identical. DRP represents the dendritic Ru(bpy)32+-polymer. The excitation peak of the dendritic Ru(bpy)32+-polymer was at 450 nm, and the emission peak was at 660 nm. (D) Absorption spectrum of the dendritic Ru(bpy)32+-polymer-amplified ECL probe. Absorption peaks simultaneously appeared at 260 and 450 nm after the dendritic Ru(bpy)32+-polymer was linked with nucleic acids.
The photograph of the dendritic Ru(bpy)32+-polymer and Ru(bpy)32+ in Figure 2C indicated that the dendritic Ru(bpy)32+-polymer exhibited a light-brown color, and that Ru(bpy)32+ was yellow. This presents the obvious distinctions between the dendritic Ru(bpy)32+-polymer and Ru(bpy)32+ in character. The spectral characterization results (Figure 2C) reveal that the dendritic Ru(bpy)32+-polymer possesses excitation and emission wavelengths identical to those of Ru(bpy)32+ (Figures S5 and S6). The maximum absorption peak of the dendritic Ru(bpy)32+-polymer appeared at 450 nm, whereas the emission peak appeared at 660 nm. These results indicate that the synthetic route for the dendritic Ru(bpy)32+-polymer is feasible, and that Ru(bpy)32+ and the dendritic polymer can be connected. Meanwhile, we evaluated the junction between the dendritic Ru(bpy)32+-polymer and the specific recognition domains to confirm the feasibility of the assembly step for the dendritic Ru(bpy)32+-polymer-amplified ECL probe. Specifically, the dendritic Ru(bpy)32+-polymer-amplified ECL probe was characterized on the basis of its absorption spectrum. The results in Figure 2D indicate that absorption peaks simultaneously appeared at 260 nm (DNA peak) and 450 nm (Ru(bpy)32+ peak) after the dendritic Ru(bpy)32+-polymer was linked with nucleic acids. These results are consistent with the aforementioned predictions, thereby demonstrating the feasibility of bond formation between the dendritic Ru(bpy)32+-polymer and a specific recognition domain.
The labeling rate of the dendritic Ru(bpy)32+-polymer was evaluated by simultaneously recording the fluorescence and absorption spectra. Herein, the labeling rate represents the rate at which the amino group on the dendritic polymer connects with Ru(bpy)32+. This labeling rate was achieved with an excess of Ru(bpy)32+ and thus represents the saturated labeling amount of Ru(bpy)32+ on dendritic Ru(bpy)32+-polymer. Additionally, the absorption and fluorescence spectra of the dendritic Ru(bpy)32+-polymer were recorded to calculate the extent of labeling via comparison with the standard curves (Figures S7 and S8). Both the absorption and fluorescence spectra indicated that a labeling rate of approximately 50% Ru(bpy)32+ on the dendritic polymer was achieved. The particle size and ζ potential are listed in Figures S9 and S10. These results also preliminarily verified the feasibility of the synthetic route for the dendritic Ru(bpy)32+-polymer. In addition, we also investigated the stability of the ECL intensity of the dendritic Ru(bpy)32+-polymer probe. The ECL signal of the dendritic Ru(bpy)32+-polymer probe was detected at different time points (an excess of the reactant, terephthalic acid, was used). The results in Figure S11 show that the dendritic Ru(bpy)32+-polymer probe provided an intense ECL signal. Additionally, the ECL intensity of the dendritic Ru(bpy)32+-polymer probe remained stable for different times from 10 to 60 min, revealing the stability of the luminescence from the dendritic Ru(bpy)32+-polymer probe.
Target Sequence Design Strategy
ZIKV belongs to a series of flaviviruses that are homologous in genetic information. Thus, the existing methods based on immunoassays cannot effectively differentiate different flaviviruses, especially the dengue virus and ZIKV.35−37 In this work, we designed the conserved sequence from the ZIKV NS1 gene.38,39 The genomes of ZIKV strains used in the present study were downloaded from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/genome/viruses/variation/Zika/), and aligned using MEGA 7. Conserved ZIKV-specific sequences that were divergent from other flaviviruses were identified. The alignment results of some ZIKV strains and other related flavivirus strains are shown in Figure 3. For the ZIKV strains, the Asian lineage is in purple, while the African lineage is in red. Herein, we verified the specificity of the ZIKV liquid biopsy system for flaviviruses with the designed sequence. The results in Figure 4A indicated that the ZIKV liquid biopsy system could specifically respond to ZIKV, and that the other flavivirus group did not provide an ECL signal that could differentiate them from the control group. Therefore, the specificity of the ZIKV liquid biopsy system obtained a better proof. Additionally, the sequence was distinctive from those of the other flaviviruses. The results proved that this system could respond to different ZIKV strains (Figure 4B). Five ZIKV strains were simultaneously detected, and stable and differentiable ECL signals were acquired. Thus, the ZIKV liquid biopsy system provided outstanding specificity and desirable ZIKV recognition ability. These performance indexes indicate that this system can suitably meet the clinical requirements for a ZIKV liquid biopsy method.
Figure 3.

Analysis of sequence alignment between ZIKV and other flaviviruses. The genomes of ZIKV strains used in the present study were downloaded from the NCBI database, and aligned using MEGA 7. Conserved ZIKV-specific sequences that were divergent from other flaviviruses were identified. For the ZIKV strains, the African lineage is in red, while the others are of Asian lineage.
Figure 4.
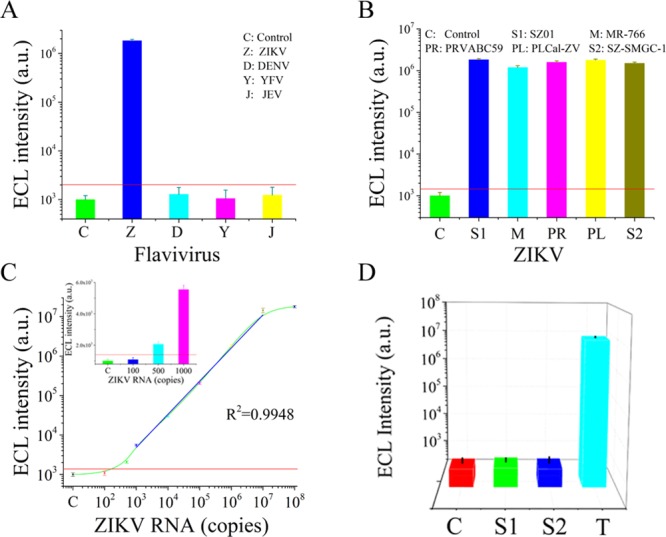
(A) Specificity of the ZIKV liquid biopsy system for flaviviruses. Zika virus is abbreviated as ZIKV, and DENV is dengue virus; YFV is yellow fever virus, and JEV is Japanese encephalitis virus. The concentration of all virus RNA was set as 106 copies. (B) Biomedical analysis of different strains of ZIKV. All the ZIKV strains were set at a concentration of 106 copies. MR-766 was the African strain, and the others (SZ01, PRVABC59, PLCal-ZV, SZ-SMGC-1) were Asian strains. (C) Sensitivity of the dendritic Ru(bpy)32+-polymer. The target ZIKV RNA concentration varied from 102 to 108 copies. A linear dynamic range was observed from 103 to 107 copies. (D) Specificity of the dendritic Ru(bpy)32+-polymer-amplified ECL method for target ZIKV RNA over random sequences. The sequences of the random sequences are listed in Table S2.
Performance Index of the Dendritic Ru(bpy)32+-Polymer-Amplified ECL Strategy
Performance indexes of the dendritic Ru(bpy)32+-polymer-amplified ECL strategy were evaluated in this section. First, we evaluated the sensitivity of the dendritic Ru(bpy)32+-polymer. The results in Figure 4C revealed that the dendritic Ru(bpy)32+-polymer reached a high sensitivity of 500 copies. Therefore, the dendritic Ru(bpy)32+-polymer provided an excellent luminescence signal over a wide response range, providing a foundation for the clinical detection and diagnosis of ZIKV. We also investigated the specificity of the dendritic Ru(bpy)32+-polymer-amplified ECL assay by comparing the ECL intensity produced in response to the specific sequence and random sequences (listed in Table S2). The results in Figure 4D indicated that only the specific ZIKV sequence resulted in intense ECL; the random sequences showed essentially the same ECL intensity as that of the control group. This result verifies the specificity of the polymer-amplified ECL assay for nucleic acids.
Verification of the Zika Virus Liquid Biopsy System for Infected Mice
In this section, we constructed a mouse mode of the ZIKV infection. Blood and urine samples were collected and detected for the preliminary verifications of the ZIKV liquid biopsy system in clinical sample detection. However, the detection of saliva samples was not executed because saliva samples were very difficult to collect. We first detected blood samples from the mice infected with ZIKV. The blood samples were treated with a 5-fold volume of sodium dodecyl sulfate (SDS) buffer. The results in Figure 5A revealed that the ZIKV liquid biopsy system achieved a high detection ratio of 90%. Then, the urine samples from the infected mice were collected by a simple device constructed by our group (Figure 5B). Subsequently, the solution of ZIKV was cracked by SDS buffer; the RNA of ZIKV was concentrated and purified by magnetic bead enrichment. Finally, the response signal was obtained by the dendritic Ru(bpy)32+-polymer probe. The results in Figure 5C indicated that the ZIKV liquid biopsy system could stably respond to all the ZIKV samples from the urine of infected mice. Stable signals were acquired from most of the urine samples. Thus, it can be proven that this ZIKV liquid biopsy system possesses the ability to detect ZIKV from the urine and blood of the mice that were artificially infected with ZIKV. The results in Figure 5D indicated that the ZIKV liquid biopsy system could specifically respond to ZIKV in blood and urine, and that the other flavivirus groups did not provide an ECL signal that could differentiate from the control group. Therefore, the specificity of the ZIKV liquid biopsy system for body fluid samples was proven.
Figure 5.
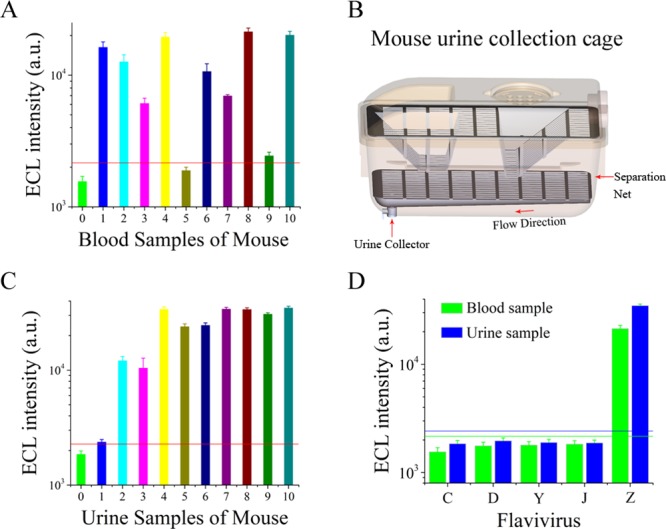
Verification of the ZIKV liquid biopsy system on infected mice. (A) Biomedical analysis of ZIKV from blood samples of mice. The blood samples were derived from mice infected with ZIKV. (B) Biomedical analysis of ZIKV from urine samples of mice. The urine samples from infected mice were collected by a simple collection device constructed by our group. (C) Urine collector for infected mice. (D) Specificity of the ZIKV liquid biopsy system for flaviviruses in blood and urine samples from infected mice.
Instance for Biomedical Analysis of ZIKV from Clinical Samples
Previous studies have established that ZIKV can be detected in body fluids (blood, saliva, and urine) of infected patients. In symptomatic patients, viremia ranges from 7.28 × 106 to 9.3 × 108 copies/mL, and in asymptomatic patients, it ranges from 2.5 × 103 to 8 × 106 copies/mL.13,40,41 These results inspired us to construct a novel detection mode (ZIKV liquid biopsy system) that adopted the humoral biomarker as the diagnostic index, which greatly simplified the biomedical analysis process, and established a nondestructive approach. For verification of whether this system can be applied to the clinical samples, the sensitivity of this ZIKV liquid biopsy system for blood samples was assessed, and the results are recorded in Figure 6A. The sensitivity of this system reached 103 copies, indicating that it could detect the viral load in most ZIKV patients.
Figure 6.
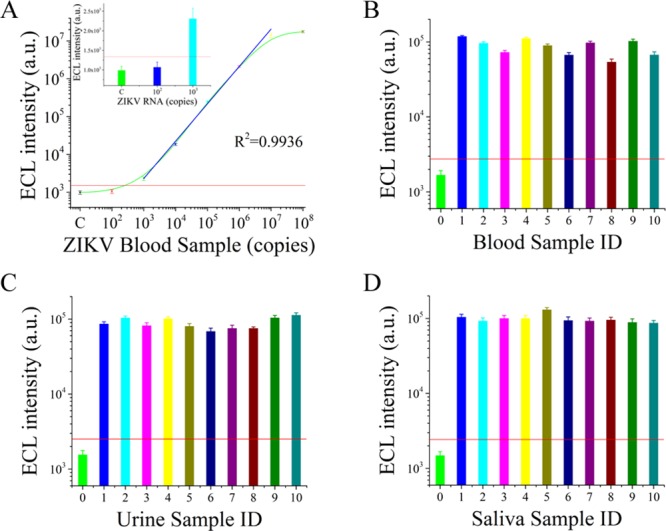
Biomedical analysis of ZIKV from clinical human body fluid samples and modeling samples. (A) Sensitivity of the ZIKV liquid biopsy system for blood samples. (B) Biomedical analysis of ZIKV in spiked blood samples. (C) Biomedical analysis of ZIKV in spiked urine samples. (D) Biomedical analysis of ZIKV in spiked saliva samples.
Moreover, we simulated spiked ZIKV samples by adding ZIKV to body fluids (blood, saliva, and urine). Specifically, 106 ZIKV copies were added to the 120 samples from healthy adults. We proceeded to carry out stability estimations with the spiked ZIKV samples (50 blood samples, 50 saliva samples, and 20 urine samples). The results in Figure 6B–D indicate that the ZIKV in body fluids (blood, saliva, and urine) can be steadily detected by the ZIKV liquid biopsy system. This finding confirmed the performance of the ZIKV liquid biopsy system on clinical samples. In Figure 6, only 30 samples are listed; the remaining 90 samples are shown in Figures S12–S14. The positive rates achieved in spiked blood, saliva, and urine samples were 94%, 96%, and 90%, respectively. Thus, these results indicated that this system could be applied to the liquid biopsy of ZIKV samples. Although we intended to use the detection process for assessing clinical samples from patients who were infected with ZIKV to further evaluate the properties of the ZIKV liquid biopsy system, patients infected with ZIKV in China are very rare. We sought to collect body fluid samples from patients infected with ZIKV, but did not find them. Thus, this strategy remains to be examined by body fluid samples from patients infected with ZIKV. However, the spiked samples in blood, saliva, and urine confirmed the feasibility for clinical samples to some extent.
Comparisons with Existing ZIKV Detection Methods
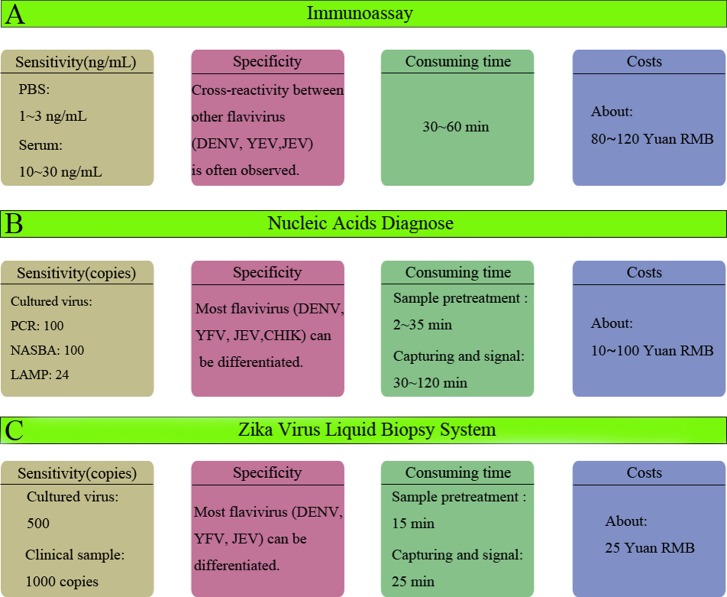
The existing ZIKV detection methodology focused on the biomarkers such as virus-specific antigens, immunoglobulins, antibodies, and viral nucleic acids. Herein, we systematically compared the performance of existing ZIKV detection methods and the ZIKV liquid biopsy system. As shown in Figure 7A, the ZIKV detection methods that hinged on quantitative information obtained from proteic biomarkers were mostly based on the immunoassay strategy42−45 (such as ELISA and immunochromatography) which could not differentiate ZIKV from the other homologous flaviviruses, such as the Dengue virus. The homology of flaviviruses greatly limits the application of ZIKV detection methods based on the immunoassay strategy to clinical analysis. In addition to this limitation, the main shortcomings of immunoassays for ZIKV are the low sensitivity and the high costs of antibodies. Therefore, ZIKV detection methods based on viral nucleic acids46−51 (Figure 7B) provided superior alternative to immunoassays that could differentiate the other flaviviruses via sequence analysis and could provide improved specificity. Furthermore, the sensitivity and the whole costs were considerably improved. However, these methods require time-consuming, complex sample preparation and nucleic acid extraction procedures. The commercial kits of sample preparation and nucleic acid extraction procedures require more than 40 min and skilled technicians, and the subsequent PCR usually requires approximately 40 min. Thus, a whole PCR process for ZIKV detection took more than 80 min.
Figure 7.
Performance comparison of existing ZIKV detection methods. (A) Performance of immunoassays for ZIKV detection. (B) Performance of nucleic-acid-based diagnostics for ZIKV detection. (C) Performance of the ZIKV liquid biopsy system.
Compared with these methods, the ZIKV liquid biopsy system (Figure 7C) achieved a sensitivity that was slightly lower than the PCR and NASBA methods. However, the sensitivity is sufficient to meet the clinical needs for ZIKV liquid biopsy. Meanwhile, the process was substantially simplified because no amplification step is required by the ZIKV liquid biopsy system; only 40 min was required to analyze each sample. Furthermore, the ZIKV liquid biopsy system can differentiate ZIKV from other flaviviruses, and desirable specificity was obtained. The cost of the ZIKV liquid biopsy system was 25 Yuan RMB for one test (the details are listed in Table S3). This cost is much lower than those of an immunoassay and most of the nucleic acid-based diagnoses. In addition, direct comparison with existing techniques (immunoassay, PCR, NASBA) using commercial kits was performed under the same experimental conditions. The results and descriptions are detailed in the Supporting Information. In conclusion, this ZIKV liquid biopsy system suitably met the strict clinical requirements for ZIKV detection and thus has the potential to serve as a new paradigm for the biomedical analysis and diagnosis of ZIKV.
Conclusions
A novel ZIKV liquid biopsy system was constructed by integrating a Ru(bpy)32+-polymer-amplified ECL strategy as an effective signal giving-out pattern. The amplification-free analysis of ZIKV was performed using a drop of blood, and a high sensitivity of 500 copies and desirable specificity were simultaneously achieved. This strategy adopted the humoral biomarker as the diagnostic index, which greatly simplified the biomedical analysis process, and established a nondestructive detection mode. Furthermore, we investigated the performance index for the biomedical analysis of clinical ZIKV samples, and the results indicated that the Ru(bpy)32+-polymer-amplified ECL strategy reliably responded to ZIKV in body fluids (blood, saliva, and urine). Hence, this system suitably met the strict clinical requirements for ZIKV liquid biopsy and thus has the potential to serve as a new paradigm for the biomedical analysis and diagnosis of ZIKV.
Experimental Section
Reagents
All of the oligonucleotides and probes used in this paper were synthesized and purified by Invitrogen. cis-Bis(2,2′-bipyridine)dichlororuthenium(II), 2,2′-bipyridine-4, 4′-dicarboxylic acid, N,N′-dicyclohexylcarbodiimide (DCC), sodium borate, sodium hexafluorophosphate, N-(3-(dimethylamino)propyl)-N′-ethylcarbodiimide hydrochloride (EDC), and N-hydroxysuccinimide (NHS) were obtained from Alfa Aesar Co., Ltd. Streptavidin magnetic beads were synthesized by New England BioLabs. Diethylpyrocarbonate-treated (DEPC-treated) water and RNAase inhibitor were obtained from Takara Biotechnology (Dalian) Co., Ltd.
Synthetic Routes for Ru(bpy)32+ and Dendritic Ru(bpy)32+-Polymer
The Ru(bpy)32+-NHS ester was synthesized by the synthetic procedure for Ru(bpy)2(dcbpy)(PF6)2 and activation of the carboxyl group on Ru(bpy)32+. Ru(bpy)2(dcbpy)(PF6)2 was synthesized by refluxing cis-dichlorobis(2,2′-bipyridine)ruthenium(II) with an excess of the corresponding 2,2′-bipyridine-4,4′-dicarboxylic acid (1:1.2) in an ethanol/water (80%) mixture and incubating the mixture at 80 °C for one night. The solution was subsequently cooled and acidified (pH = 4.4) to crystallize out the unreacted 2,2′-bipyridine-4,4′-dicarboxylic acid. The sediment was washed and removed with a qualitative filter-paper-based suction filter. A precooled solution of sodium hexafluorophosphate (NaPF6) was added to the resultant filtrate isolates of the ruthenium complexes as PF6 salts. The mixture was cooled with an ice–water mixture. Then, the crystal products were treated with the freeze-drying process to eliminate the influence of volatile solvents. At this point, the synthetic procedure for Ru(bpy)2(dcbpy)(PF6)2 was terminated.
The core of the dendritic polymer was obtained from Sigma and then modified with amino acids via lipid reactions. The molecular ratio between maleimide-labeled DNA and the amino-labeled dendritic polymer was set as 10:1. A mixture of maleimide-labeled DNA and the amino-labeled dendritic polymer was incubated at 37 °C for 12 h. Then, the dissociated DNA was removed with an ultrafiltration device. The obtained products were mixed with carboxyl-activated Ru(bpy)32+ at a molecular ratio of 1:1000 and incubated at 37 °C for 12 h. The products were purified by sedimentation with precooled alcohol solution and centrifugation.
Dissociation Process of ZIKV and Magnetic Beads Enrichment
The ZIKV samples were separated with a commercial virus-lysis kit, the key constituent of which was SDS. To achieve a better separation effect, the solution could be heated to 60 °C for 5 min. A 10 μL portion of magnetic beads (1 mg/mL) labeled with capture probe was added to each sample for 10 min. Meanwhile, the mixture was processed by magnetic separation. The captured products were collected for later ECL steps.
ECL Process
The magnetic beads used in this assay were labeled with streptavidin and could recognize and bind biotin from the capture probe. The signal probe, composed of the dendritic Ru(bpy)32+-polymer and recognition domain, was employed as the ECL-generating group. When the target is present, the capture probe and signal probe form the classic sandwich structure, which can be adapted to nucleic acid detection. The gradient cooling process was more effective. Eventually, the streptavidin magnetic beads captured the sandwich structure. After the washing steps, the ECL signal was detected in the presence of the coreactant tripropylamine (TPA).
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31470877, 81261160323), the National Key Research and Development Program of China (2016YFC1200105), the National Science and Technology Key Projects for Major Infectious Diseases (2017ZX10302301 and 2013ZX10003001), the Science and Technology Planning Project of Guangzhou (201704020226, 2016A020250001, and 201604020006), and the Guangdong Natural Science Foundation (2015A030311009).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.8b00471.
Characterization of dendritic Ru(bpy)32+-polymer, biomedical analysis of ZIKV from clinical samples, and performance of the existing method for ZIKV detection (PDF)
The authors declare no competing financial interest.
Notes
Safety statement: no unexpected or unusually high safety hazards were encountered.
Supplementary Material
References
- Pardi N.; Hogan M. J.; Pelc R. S.; Muramatsu H.; Andersen H.; DeMaso C. R.; Dowd K. A.; Sutherland L. L.; Scearce R. M.; Parks R.; Wagner W.; Granados A.; Greenhouse J.; Walker M.; Willis E.; Yu J. S.; McGee C. E.; Sempowski G. D.; Mui B. L.; Tam Y. K.; Huang Y.-J.; Vanlandingham D.; Holmes V. M.; Balachandran H.; Sahu S.; Lifton M.; Higgs S.; Hensley S. E.; Madden T. D.; Hope M. J.; Karikó K.; Santra S.; Graham B. S.; Lewis M. G.; Pierson T. C.; Haynes B. F.; Weissman D. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature 2017, 543, 248–251. 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraki R.; Jurado K. A.; Hwang J.; Szigeti-Buck K.; Horvath T. L.; Iwasaki A.; Fikrig E. Fetal Growth Restriction Caused by Sexual Transmission of Zika Virus in Mice. J. Infect. Dis. 2017, 215, 1720–1724. 10.1093/infdis/jix204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauci A. S.; Morens D. M. Zika virus in the Americas-yet another arbovirus threat. N. Engl. J. Med. 2016, 374, 601–604. 10.1056/NEJMp1600297. [DOI] [PubMed] [Google Scholar]
- Stephen P.; Baz M.; Boivin G.; Lin S. X. Structural Insight into NS5 of Zika Virus Leading to the Discovery of MTase Inhibitors. J. Am. Chem. Soc. 2016, 138, 16212–16215. 10.1021/jacs.6b10399. [DOI] [PubMed] [Google Scholar]
- Ferguson N. M.; Cucunubá Z. M.; Dorigatti I.; Nedjati-Gilani G. L.; Donnelly C. A.; Basáñez M. G.; Nouvellet P.; Lessler J. Countering the Zika epidemic in Latin America. Science 2016, 353, 353–354. 10.1126/science.aag0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richner J. M.; Himansu S.; Dowd K. A.; Butler S. L.; Salazar V.; Fox J. M.; Julander J. G.; Tang W. W.; Shresta S.; Pierson T. C.; Ciaramella G.; Diamond M. S. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell 2017, 168, 1114–1125. 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapparapu G.; Fernandez E.; Kose N.; Cao B.; Fox J. M.; Bombardi R. G.; Zhao H.; Nelson C. A.; Bryan A. L.; Barnes T.; Davidson E.; Mysorekar I. U.; Fremont D. H.; Doranz B. J.; Diamond M. S.; Crowe J. E. Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature 2016, 540, 443–447. 10.1038/nature20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richner J. M.; Jagger B. W.; Shan C.; Fontes C. R.; Dowd K. A.; Cao B.; Himansu S.; Caine E. A.; Nunes B. T. D.; Medeiros D. B. A.; Muruato A. E.; Foreman B. M.; Luo H.; Wang T.; Barrett A. D.; Weaver S. C.; Vasconcelos P. F. C.; Rossi S. L.; Ciaramella G.; Mysorekar I. U.; Pierson T. C.; Shi P. Y.; Diamond M. S. Vaccine Mediated Protection Against Zika Virus-Induced Congenital Disease. Cell 2017, 170, 273–283. 10.1016/j.cell.2017.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X.; Ha Nam N.; Song M. M.; Hadiono C.; Ogden S. C.; Hammack C.; Yao B.; Hamersky G. R.; Jacob F.; Zhong C.; Yoon K. J.; Jeang W.; Lin L.; Li Y.; Thakor J.; Berg D. A.; Zhang C.; Kang E.; Chickering M.; Nauen D.; Ho C.-Y.; Wen Z.; Christian K. M.; Shi P.-Y.; Maher B. J.; Wu H.; Jin P.; Tang H.; Song H.; Ming G. L. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 2016, 165, 1238–1254. 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud D.; Gubler D. J.; Schaub B.; Lanteri M. C.; Musso D. An update on Zika virus infection. Lancet 2017, 390, 2099–2109. 10.1016/S0140-6736(17)31450-2. [DOI] [PubMed] [Google Scholar]
- Parra B.; Lizarazo J.; Jimenez-Arango J. A.; Zea-Vera A. F.; Gonzalez-Manrique G.; Vargas J.; Angarita J. A.; Zuniga G.; Lopez-Gonzalez R.; Beltran C. L.; Rizcala K. H.; Morales M. T.; Pacheco O.; Ospina M. L.; Kumar A.; Cornblath D. R.; Munoz L. S.; Osorio L.; Barreras P.; Pardo C. A. Guillain-Barre Syndrome Associated with Zika Virus Infection in Colombia. N. Engl. J. Med. 2016, 375, 1513–1523. 10.1056/NEJMoa1605564. [DOI] [PubMed] [Google Scholar]
- Pardee K.; Green A. A.; Takahashi M. K.; Braff D.; Lambert G.; Lee J. W.; Ferrante T.; Ma D.; Donghia N.; Fan M.; Daringer N. M.; Bosch I.; Dudley D. M.; O’Connor D. H.; Gehrke L.; Collins J. J. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266. 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- Musso D.; Gubler D. J. Zika virus. Clin. Microbiol. Rev. 2016, 29, 487–524. 10.1128/CMR.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlakar J.; Korva M.; Tul N.; Popovic M.; Poljsak-Prijatelj M.; Mraz J.; Kolenc M.; Rus K. R.; Vipotnik T. V.; Vodusek V. F.; Vizjak A.; Pizem J.; Petrovec M.; Zupanc T. A. Zika Virus Associated with Microcephaly. N. Engl. J. Med. 2016, 374 (10), 951–958. 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- Rasmussen S. A.; Jamieson D. J.; Honein M. A.; Petersen L. R. Zika virus and birth defects—reviewing the evidence for causality. N. Engl. J. Med. 2016, 374, 1981–1987. 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- Cugola F. R.; Fernandes I. R.; Russo F. B.; Freitas B. C.; Dias J. L.; Guimarães K. P.; Benazzato C.; Almeida N.; Pignatari G. C.; Romero S. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 2016, 534, 267–271. 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocca R. A.; Abbink P.; Peron J. P. S.; Zanotto P. M. d. A.; Iampietro M. J.; Badamchi-Zadeh A.; Boyd M.; Ng’ang’a D.; Kirilova M.; Nityanandam R.; Mercado N. B.; Li Z.; Moseley E. T.; Bricault C. A.; Borducchi E. N.; Giglio P. B.; Jetton D.; Neubauer G.; Nkolola J. P.; Maxfield L. F.; De La Barrera R. A.; Jarman R. G.; Eckels K. H.; Michael N. L.; Thomas S. J.; Barouch D. H. Vaccine protection against Zika virus from Brazil. Nature 2016, 536, 474–478. 10.1038/nature18952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huzly D.; Hanselmann I.; Schmidt-Chanasit J.; Panning M. High specificity of a novel Zika virus ELISA in European patients after exposure to different flaviviruses. Euro Surveill 2016, 21, 30203. 10.2807/1560-7917.ES.2016.21.16.30203. [DOI] [PubMed] [Google Scholar]
- Kumar A.; Hou S.; Airo A. M.; Limonta D.; Mancinelli V.; Branton W.; Power C.; Hobman T. C. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep. 2016, 17, 1766–1775. 10.15252/embr.201642627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.; Kang N.; An K.; Kim D.; Koo J.; Kim M. S. MRPrimerV: a database of PCR primers for RNA virus detection. Nucleic Acids Res. 2017, 45, D475–D481. 10.1093/nar/gkw1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joguet G.; Mansuy J.-M.; Matusali G.; Hamdi S.; Walschaerts M.; Pavili L.; Guyomard S.; Prisant N.; Lamarre P.; Dejucq-Rainsford N.; Pasquier C.; Bujan L. Effect of acute Zika virus infection on sperm and virus clearance in body fluids: a prospective observational study. Lancet Infect. Dis. 2017, 17, 1200–1208. 10.1016/S1473-3099(17)30444-9. [DOI] [PubMed] [Google Scholar]
- Wong S. J.; Furuya A.; Zou J.; Xie X.; Dupuis A. P. II; Kramer L. D.; Shi P. Y. A multiplex microsphere immunoassay for Zika virus diagnosis. EBioMedicine 2017, 16, 136–140. 10.1016/j.ebiom.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug C. J.; Kieny M. P.; Murgue B. The Zika Challenge. N. Engl. J. Med. 2016, 374, 1801–1803. 10.1056/NEJMp1603734. [DOI] [PubMed] [Google Scholar]
- Faria N. R.; Quick J.; Claro I. M.; Theze J.; de Jesus J. G.; Giovanetti M.; Kraemer M. U. G.; Hill S. C.; Black A.; da Costa A. C.; Franco L. C.; Silva S. P.; Wu C. H.; Raghwani J.; Cauchemez S.; du Plessis L.; Verotti M. P.; de Oliveira W. K.; Carmo E. H.; Coelho G. E.; Santelli A. C. F. S.; Vinhal L. C.; Henriques C. M.; Simpson J. T.; Loose M.; Andersen K. G.; Grubaugh N. D.; Somasekar S.; Chiu C. Y.; Munoz-Medina J. E.; Gonzalez-Bonilla C. R.; Arias C. F.; Lewis-Ximenez L. L.; Baylis S. A.; Chieppe A. O.; Aguiar S. F.; Fernandes C. A.; Lemos P. S.; Nascimento B. L. S.; Monteiro H. A. O.; Siqueira I. C.; de Queiroz M. G.; de Souza T. R.; Bezerra J. F.; Lemos M. R.; Pereira G. F.; Loudal D.; Moura L. C.; Dhalia R.; Franca R. F.; Magalhaes T.; Marques E. T. Jr.; Jaenisch T.; Wallau G. L.; de Lima M. C.; Nascimento V.; de Cerqueira E. M.; de Lima M. M.; Mascarenhas D. L.; Moura Neto J. P.; Levin A. S.; Tozetto-Mendoza T. R.; Fonseca S. N.; Mendes-Correa M. C.; Milagres F. P.; Segurado A.; Holmes E. C.; Rambaut A.; Bedford T.; Nunes M. R. T.; Sabino E. C.; Alcantara L. C. J.; Loman N. J.; Pybus O. G. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature 2017, 546, 406–410. 10.1038/nature22401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppes C.; Rac M.; Dunn J.; Versalovic J.; Murray K. O.; Suter M. A.; Cortes M. S.; Espinoza J.; Seferovic M. D.; Lee W.; Hotez P.; Mastrobattista J.; Clark S. L.; Belfort M. A.; Aagaard K. M. Testing for Zika virus infection in pregnancy: key concepts to deal with an emerging epidemic. Am. J. Obstet. Gynecol. 2017, 216, 209–225. 10.1016/j.ajog.2017.01.020. [DOI] [PubMed] [Google Scholar]
- Duggal N. K.; Ritter J. M.; Pestorius S. E.; Zaki S. R.; Davis B. S.; Chang G.-J. J.; Bowen R. A.; Brault A. C. Frequent Zika Virus Sexual Transmission and Prolonged Viral RNA Shedding in an Immunodeficient Mouse Model. Cell Rep. 2017, 18, 1751–1760. 10.1016/j.celrep.2017.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos T.; Rodriguez A.; Almiron M.; Sanhueza A.; Ramon P.; de Oliveira W. K.; Coelho G. E.; Badaro R.; Cortez J.; Ospina M.; Pimentel R.; Masis R.; Hernandez F.; Lara B.; Montoya R.; Jubithana B.; Melchor A.; Alvarez A.; Aldighieri S.; Dye C.; Espinal M. A. Zika Virus and the Guillain-Barre Syndrome - Case Series from Seven Countries. N. Engl. J. Med. 2016, 375, 1598–1601. 10.1056/NEJMc1609015. [DOI] [PubMed] [Google Scholar]
- Pardee K.; Green A. A.; Takahashi M. K.; Braff D.; Lambert G.; Lee J. W.; Ferrante T.; Ma D.; Donghia N.; Fan M. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 2016, 165, 1255–1266. 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- Cordeiro M. T.; Pena L. J.; Brito C. A.; Gil L. H.; Marques E. T. Positive IgM for Zika virus in the cerebrospinal fluid of 30 neonates with microcephaly in Brazil. Lancet 2016, 387, 1811–1812. 10.1016/S0140-6736(16)30253-7. [DOI] [PubMed] [Google Scholar]
- Cordeiro M. T.; Brito C. A. A.; Pena L. J.; Castanha P. M. S.; Gil L. H. V. G.; Lopes K. G. S.; Dhalia R.; Meneses J. A.; Ishigami A. C.; Mello L. M.; Alencar L. X. E.; Guarines K. M.; Rodrigues L. C.; Marques E. T. A. Results of a Zika Virus (ZIKV) Immunoglobulin M-Specific Diagnostic Assay Are Highly Correlated With Detection of Neutralizing Anti-ZIKV Antibodies in Neonates With Congenital Disease. J. Infect. Dis. 2016, 214, 1897–1904. 10.1093/infdis/jiw477. [DOI] [PubMed] [Google Scholar]
- Lin S.; Gao W.; Tian Z.; Yang C.; Lu L.; Mergny J. L.; Leung C. H.; Ma D. L. Luminescence switch-on detection of protein tyrosine kinase-7 using a G-quadruplex-selective probe. Chem. Sci. 2015, 6, 4284–4290. 10.1039/C5SC01320H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori Y.; Otomura N.; Nishida A.; Nishiura M.; Umeno M.; Suetake I.; Kikuchi K. Synthetic-molecule/protein hybrid probe with fluorogenic switch for live-cell imaging of DNA methylation. J. Am. Chem. Soc. 2018, 140, 1686–1690. 10.1021/jacs.7b09713. [DOI] [PubMed] [Google Scholar]
- Collot M.; Fam T. K.; Ashokkumar P.; Faklaris O.; Galli T.; Danglot L.; Klymchenko A. S. Ultrabright and Fluorogenic Probes for Multicolor Imaging and Tracking of Lipid Droplets in Cells and Tissues. J. Am. Chem. Soc. 2018, 140, 5401–5411. 10.1021/jacs.7b12817. [DOI] [PubMed] [Google Scholar]
- Barragán F.; López-Senín P.; Salassa L.; Betanzos-Lara S.; Habtemariam A.; Moreno V.; Sadler P. J.; Marchán V. Photocontrolled DNA binding of a receptor-targeted organometallic ruthenium (II) complex. J. Am. Chem. Soc. 2011, 133, 14098–14108. 10.1021/ja205235m. [DOI] [PubMed] [Google Scholar]
- Cauchemez S.; Besnard M.; Bompard P.; Dub T.; Guillemette-Artur P.; Eyrolle-Guignot D.; Salje H.; Van Kerkhove M. D.; Abadie V.; Garel C.; Fontanet A.; Mallet H. P. Association between Zika virus and microcephaly in French Polynesia, 2013–15: a retrospective study. Lancet 2016, 387, 2125–2132. 10.1016/S0140-6736(16)00651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch I.; de Puig H.; Hiley M.; Carre-Camps M.; Perdomo-Celis F.; Narvaez C. F.; Salgado D. M.; Senthoor D.; O’Grady M.; Phillips E.; Durbin A.; Fandos D.; Miyazaki H.; Yen C.-W.; Gelvez-Ramirez M.; Warke R. V.; Ribeiro L. S.; Teixeira M. M.; Almeida R. P.; Munoz-Medina J. E.; Ludert J. E.; Nogueira M. L.; Colombo T. E.; Terzian A. C. B.; Bozza P. T.; Calheiros A. S.; Vieira Y. R.; Barbosa-Lima G.; Vizzoni A.; Cerbino-Neto J.; Bozza F. A.; Souza T. M. L.; Trugilho M. R. O.; de Filippis A. M. B.; de Sequeira P. C.; Marques E. T. A.; Magalhaes T.; Diaz F. J.; Restrepo B. N.; Marin K.; Mattar S.; Olson D.; Asturias E. J.; Lucera M.; Singla M.; Medigeshi G. R.; de Bosch N.; Tam J.; Gomez-Marquez J.; Clavet C.; Villar L.; Hamad-Schifferli K.; Gehrke L. Rapid antigen tests for dengue virus serotypes and Zika virus in patient serum. Sci. Transl. Med. 2017, 9, eaan1589. 10.1126/scitranslmed.aan1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J.; Tang W. W.; Sheets N.; Ellison J.; Sette A.; Kim K.; Shresta S. Identification of Zika virus epitopes reveals immunodominant and protective roles for dengue virus cross-reactive CD8+ T cells. Nat. Microbiol. 2017, 2, 17036. 10.1038/nmicrobiol.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista L. E.; Sethi A. K. Association between Guillain-Barre syndrome and Zika virus infection. Lancet 2016, 387, 2599–2600. 10.1016/S0140-6736(16)30844-3. [DOI] [PubMed] [Google Scholar]
- Baptista T.; Quaghebeur G.; Alarcon A. Neuroimaging findings of babies with microcephaly and presumed congenital Zika virus infection. Br. Med. J. 2016, 353, i2194. 10.1136/bmj.i2194. [DOI] [PubMed] [Google Scholar]
- Besnard M.; Lastere S.; Teissier A.; Cao-Lormeau V.; Musso D. Evidence of perinatal transmission of Zika virus, French Polynesia, December 2013 and February 2014. Euro Surveill 2014, 19, 20751. 10.2807/1560-7917.ES2014.19.13.20751. [DOI] [PubMed] [Google Scholar]
- Wæhre T.; Maagard A.; Tappe D.; Cadar D.; Schmidt-Chanasit J. Zika virus infection after travel to Tahiti, December 2013. Emerging Infect. Dis. 2014, 20, 1412–1414. 10.3201/eid2008.140302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. H.; Zeng H. Aptamer-Based ELISA Assay for Highly Specific and Sensitive Detection of Zika NS1 Protein. Anal. Chem. 2017, 89, 12743–12748. 10.1021/acs.analchem.7b02862. [DOI] [PubMed] [Google Scholar]
- Hage D. S. Development of Immunochromatographic Assays for the Selective Detection of Zika Virus or Dengue Virus Serotypes in Serum. Clin. Chem. 2018, 64, 991–993. 10.1373/clinchem.2017.282699. [DOI] [PubMed] [Google Scholar]
- Cecchetto J.; Fernandes F. C. B.; Lopes R.; Bueno P. R. The capacitive sensing of NS1 Flavivirus biomarker. Biosens. Bioelectron. 2017, 87, 949–956. 10.1016/j.bios.2016.08.097. [DOI] [PubMed] [Google Scholar]
- Bedin F.; Boulet L.; Voilin E.; Theillet G.; Rubens A.; Rozand C. Paper-based point-of-care testing for cost-effective diagnosis of acute flavivirus infections. J. Med. Virol. 2017, 89, 1520–1527. 10.1002/jmv.24806. [DOI] [PubMed] [Google Scholar]
- Eboigbodin K. E.; Brummer M.; Ojalehto T.; Hoser M. Rapid molecular diagnostic test for Zika virus with low demands on sample preparation and instrumentation. Diagn. Microbiol. Infect. Dis. 2016, 86, 369–371. 10.1016/j.diagmicrobio.2016.08.027. [DOI] [PubMed] [Google Scholar]
- Tian B.; Qiu Z.; Ma J.; Zardán Gómez de la Torre T.; Johansson C.; Svedlindh P.; Strömberg M. Attomolar Zika virus oligonucleotide detection based on loop-mediated isothermal amplification and AC susceptometry. Biosens. Bioelectron. 2016, 86, 420–425. 10.1016/j.bios.2016.06.085. [DOI] [PubMed] [Google Scholar]
- Song J.; Mauk M. G.; Hackett B. A.; Cherry S.; Bau H. H.; Liu C. Instrument-Free Point-of-Care Molecular Detection of Zika Virus. Anal. Chem. 2016, 88, 7289–7294. 10.1021/acs.analchem.6b01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotiwan N.; Brewster C. D.; Magalhaes T.; Weger-Lucarelli J.; Duggal N. K.; Rückert C.; Nguyen C.; Garcia Luna S. M.; Fauver J. R.; Andre B.; Gray M.; Black W. C.; Kading R. C.; Ebel G. D.; Kuan G.; Balmaseda A.; Jaenisch T.; Marques E. T. A.; Brault A. C.; Harris E.; Foy B. D.; Quackenbush S. L.; Perera R.; Rovnak J. Rapid and specific detection of Asian- and African-lineage Zika viruses. Sci. Transl. Med. 2017, 9, eaag0538. 10.1126/scitranslmed.aag0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee K.; Green A. A.; Takahashi M. K.; Braff D.; Lambert G.; Lee J. W.; Ferrante T.; Ma D.; Donghia N.; Fan M.; Daringer N. M.; Bosch I.; Dudley D. M.; O’Connor D. H.; Gehrke L.; Collins J. J. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266. 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- Lustig Y.; Mendelson E.; Paran N.; Melamed S.; Schwartz E. Detection of Zika virus RNA in whole blood of imported Zika virus disease cases up to 2 months after symptom onset, Israel, December 2015 to April 2016. Euro Surveill 2016, 21, 30269. 10.2807/1560-7917.ES.2016.21.26.30269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.