Abstract
Despite advances in diagnosis and treatment, the survival of non-small cell lung cancer (NSCLC) patients is poor. Further understanding of the disease mechanism and treatment strategies is required. Copines are a family of calcium-dependent phospholipid-binding proteins that are evolutionally conserved in various eukaryotic organisms and protists. Copine 1, encoded by CPNE1, is a soluble membrane-binding protein, which includes two tandem C2 domains at the N-terminus and an A domain at the C-terminus. A previous study reported that Copine 1 binds with various intracellular proteins via its A domain and C omain. However, the role of CPNE1 in lung cancer remains unclear. In the presented study, CPNE1 expression level was demonstrated to be positively associated with the stage (P=0.002) and significantly associated with lymph node status (P=0.011) and distant metastasis (P=0.042). Furthermore, the function of CPNE1 in regulation of cell growth, migration and invasion was investigated, and it was demonstrated that knockdown of CPNE1 inhibits the cell cycle in NSCLC cells. Collectively, these data suggest that CPNE1 is an oncogene in NSCLC and serves an important role in tumorigenesis of NSCLC progression.
Keywords: Copine 1, clinical, promotes cell growth, metastasis, lung adenocarcinoma
Introduction
Lung cancer is the leading cause of cancer-associated mortality worldwide and in China (1,2). Non-small cell lung cancer (NSCLC) accounts for 85% of lung cancer (3). Despite advances in cancer research and treatment, the prognosis of NSCLC remains poor, with the 5-year survival rate being only 15% (4). Although novel drugs including epithelial growth factor receptor have been demonstrated to be beneficial, particularly in patients with sensitive target mutations, the survival and outcome have not altered dramatically (5). Therefore, it is important to elucidate the pathogenesis of NSCLC and identify novel treatment targets.
Copines are a family of calcium-dependent phospholipid-binding proteins that are evolutionally conserved in various eukaryotic organisms and protists (6). Currently, nine family members have been identified (7,8). It has been reported that Copine 1, encoded by CPNE1, is a soluble membrane-binding protein, which includes two tandem C2 domains at the N-terminus and an A domain at the C-terminus (6). The C2 domains act as calcium-dependent phospholipid binding motifs and may be associated with cell signalling and/or membrane trafficking pathways (9). The A domain is named from von Willebrand Factor, a plasma and extracellular matrix protein, and have been studied in integrins and several extracellular matrix proteins and appear to function as protein-binding domains (10). It was previously reported that Copine 1 binds with various intracellular proteins via its A domain (7,8). To date, the role of CPNE1 in lung cancer remains unclear.
In the present study, the expression of CPNE1 in 128 lung adenocarcinoma patient tissues was initially investigated by immunohistochemical staining. Higher expression of CPNE1 was observed to be significantly associated with advanced tumor, node and metastasis (TNM) stage, lymph node metastasis and distant metastasis in lung adenocarcinoma. In addition, the results of the present study indicated that knockdown of CPNE1 inhibits cell cycle progression, cell growth and migration of NSCLC cells. In conclusion, it was hypothesized that CPNE1 serves an important role in tumorigenesis of human lung adenocarcinoma.
Materials and methods
Tissue samples
Following approval by the Ethics Committee of The First Affiliated Hospital of Soochow University (Suzhou, China), a group of 128 patients with lung adenocarcinoma were recruited consecutively from The First Affiliated Hospital of Soochow University, from March 2009 to December 2012. Those patients included 67 males and 61 females, and their ages ranged from 20-79 years. Patients had not received chemotherapy or radiotherapy prior to diagnosis and tissue sampling. Patients were diagnosed with NSCLC based on their histological and pathological characteristics according to the Revised International System for Staging Lung Cancer (11). Lung adenocarcinoma and adjacent normal tissues from the 128 patients were snap-frozen and stored at −80°C. All participants provided written informed consent for the whole study.
Cell culture
The human lung adenocarcinoma A549, H1299, SPC-A1 and HCC827 cell lines and human lung squamous carcinoma cell line H226 and human immortalized normal epithelial cell line BEAS-2B and the 293T (293) cell line were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). H1299 and A549 cells were identified by short tandem repeat method (12). Both A549 and H1299 cells were grown in Dulbecco's modified Eagle's medium (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) containing 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). All cells were maintained in a humidified atmosphere with 5% CO2 at 37°C. Beijing Microread Genetics Co., Ltd. (Beijing, China) determined the genetic characteristics of the cells using a Goldeneye™ 20A kit (Peoplespot, Beijing, China) and an ABI 3100 genetic analyser (Thermo Fisher Scientific, Inc.). All cell lines were passaged for <3 months and tested in February 2017.
Immunohistochemical assay
Immunohistochemical analyses of 128 lung adenocarcinoma and normal tissues were conducted as described in the present authors' previous study (13). The sections were incubated with anti-Copine 1 antibodies (1:100; cat. no. ab155675; Abcam, Cambridge, UK) overnight at 4°C. Then incubated with pre-diluted biotinylated secondary antibodies for 40 min at 37°C (SV0002; Wuhan Boster Biological Technology, Ltd., Wuhan, China) according to the manufacturer's protocol. The reactions were developed using the DAB Substrate Kit (cat. no. 550880; BD Biosciences, San Jose, CA, USA), and the sections were counterstained for 2 min at room temperature with hematoxylin.
The scoring of immunostaining was evaluated on the basis of staining intensity and percentages of three positively stained areas at random by two pathologists in a double-blinded manner. Briefly, the proportion of positive cells in each specimen was quantitatively evaluated and scored as follows: 0, Staining in 0% of the cells examined; 1, staining in 0.01-10% of the cells examined; 2, staining in 10.01-50% of the cells examined; 3, staining in 50.01-75% of the cells examined; and 4, staining in >75% of the cells examined. The staining intensity was graded as follows: 0, No signal; 1, weak; 2, moderate; and 3, strong. The histological score for each section was computed using the following formula: Histological score = proportion score x intensity score. A total score with a possible range of 0-12 was calculated and graded as follows: Negative (-; score, 0), weak (+; score, 1-4), moderate (++; score, 5-8) or strong (+++; score, 9-12). Scores of - and + were considered to indicate low expression levels, whereas scores of ++ and +++ were considered to indicate high expression levels.
Establishment of CPNE1-silenced stable cell lines
The human CPNE1 (GenBank accession no. NM_003915) specific small interfering RNA (siRNA) sequence, which was designed with BLOCK-iT RNAi Designer (Thermo Fisher Scientific, Inc.) was 5′-CACACAACTGGTCTCATACTT-3′. The non-silencing sequence generated by Shanghai GeneChem Co., Ltd. (Shanghai, China; 5′-TTCTCCGAACGTGTCACGT-3′) was used as a scrambled (scr) control. Pairs of complementary oligonucleotides were synthesized, annealed, and ligated into linear pGCSIL-green fluorescent protein (GFP) lentiviral plasmids generated by Shanghai GeneChem Co., Ltd. and digested by enzyme AgeI and EcoRI. Subsequently, lentiviral vectors that expressed the CPNE1-specific siRNA or negative control siRNA (Shanghai GeneChem Co., Ltd.) were generated in 293T packaging cells by co-transfection of the recombinant pGCSIL-GFP vector (20 μg), pHelper 1.0 (15 μg; Shanghai GeneChem Co., Ltd.) and pHelper 2.0 plasmids (10 μg; Shanghai GeneChem Co., Ltd.) using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Viral particles from the media were harvested at 48 h following transfection and purified by ultracentrifugation, centrifugation for 2 h at 21,000 x g and 4°C. The viral titer was determined by counting the numbers of infected green cells under fluorescence microscopy following transfection of 293T cells with serial dilutions of concentrated lentivirus.
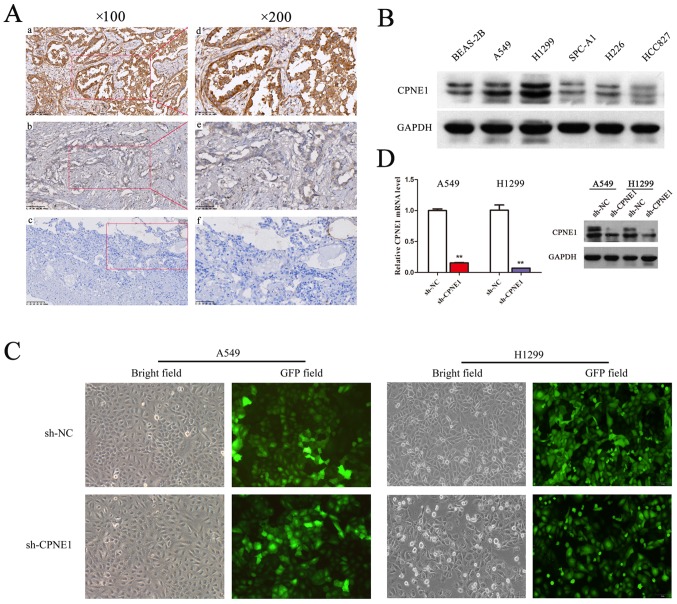
For lentivirus transduction, H1299 and A549 cells were subcultured into 6-well culture plates at 70-80% confluence and infected with CPNE1-specific siRNA lentivirus or control lentivirus at a multiplicity of infection of 20. The infection efficiency was detected using fluorescence microscopy to observe cells expressing GFP after 5 days. Each cell line was divided into the following two groups: The sh-NC group and the sh-CPNE1 group.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
RNA isolation, cDNA synthesis and RT-qPCR analysis were performed as previously described (13). The primer sequences used for CPNE1 mRNA detection were 5′-ACCCACTCTGCGTC CTT-3′ (forward) and 5′-TGGCGTCTTGTTGTCTATG-3′ (reverse), Cq values for CPNE1 mRNA were equilibrated to ACTB mRNA, for which the following primer sequences were used: 5′-CACAGAGCCTCGCCTTTGCC-3′ (forward) and 5′-ACCCATGCCCACCATCACG-3′ (reverse), which were used as internal controls. The 2−ΔΔCq method (14) was applied to calculate the relative expression of these gene.
Western blotting assay
Western blot analysis was performed as previously described (13). The antibodies used in the analysis were anti-CPNE1 (1:500; Z6; Santa Cruz Biotechnology, Inc., Dallas, TX, USA); anti-phosphorylated (p)-protein kinase B (AKT; Ser473; 1:1,000 D9E), anti-AKT (1:1,000; 9272s; Cell Signaling Technology, Inc., Danvers, MA, USA), anti-extracellular signal-regulated kinase (ERK; 1:1,000; 137F5), anti-p-ERK (Thr202/Tyr202; 1:1,000; D13.14.4E), anti-matrix metalloproteinase MMP2 (1:1,000; D8N9Y), anti-MMP9 (1:1,000; 603H), anti-Snail (1:1,000; C15D3; Cell Signaling Technology, Inc.); anti-cyclin A1 (1:300; ab53699; Abcam), anti-cyclin B1 (1:300; Ab-147; Abcam), anti-cyclin E1 (1:300; ab33911, Abcam) and anti-GAPDH (1:5,000; D4C6R; Cell Signaling Technology, Inc.) primary antibodies, and peroxidase conjugated anti-mouse or anti-rabbit secondary antibodies (1:5,000; 14709 and 14708; Cell Signaling Technology, Inc.).
Cell proliferation analysis
Cell proliferation was examined using a Cell Counting Kit-8 (CCK-8; Wuhan Boster Biological Technology, Ltd.), and the A549 sh-CPNE1 cells or the corresponding negative control cells were seeded in 96-well plates at a density of 2×103 cells per well and further grown under normal culture conditions for 24, 48 and 72 h. Cell viability was determined according to the manufacturer's protocol and the experiments were performed in triplicate. Cell proliferation was also detected using a clonogenic assay. Briefly, stable cells were diluted in complete culture medium and 2000 cells were reseeded in a 60 mm plate. Following incubation for 7-14 days, depending on cell growth rate, foci formed by at least 50 cells were stained with Giemsa at room temperature for 2 h and counted via light microscopy (CKX41; Olympus Corporation, Tokyo, Japan) at ×200. Cell viability was measured according to the manufacturer's protocol at several time points (24, 48 and 72 h). Each experiment was performed in triplicate.
Migration and invasion assays
The motility of cells analysis was evaluated as previously described (11). According to the manufacturer's protocol (BD Biosciences), 5×104 cells were seeded with medium containing 1% FBS into the upper chamber of a Transwell insert, and 800 μl medium containing 10% FBS was added to the lower chamber and then incubated at 37°C for 24 h. Cells that migrated to the lower chamber were stained with 1% crystal violet overnight at room temperature then washed with 1X PBS two times. For the invasion assay, the inserts were coated with Matrigel matrix (BD Biosciences) diluted in serum-free medium and incubated at 37°C for 2 h. The cells from three microscopic fields were imaged and counted via microscopy (CKX41; Olympus Corporation, Tokyo, Japan). The results were determined from three repeated experiments.
Cell cycle analysis
According to the protocol of the Cell Cycle and Apoptosis Analysis kit (Beyotime Institute of Biotechnology, Haimen, China), cells were cultured in 6-well plates for 72 h. The cells were subsequently collected, washed with cold PBS, fixed in 70% ethanol at 4°C for 24 h, washed with cold PBS again and stained in a propidium iodide/RNase A mixture. Following incubation in the dark at 37°C for 30 min, the cells were analyzed using a FACSCalibur flow cytometer (Beckman Coulter, Inc., Brea, CA, USA) and with MultiCycle 6-16-03-F32 (Beckman Coulter, Inc.).
Animal experiments and immunocytochemistry staining
A total of 12 female BALB/c athymic nude mice (age, 4-6 weeks old; weight, 16-20 g) were purchased from the Experimental Animal Center of Soochow University and bred under pathogen-free conditions. Mice were maintained in exhaust ventilated closed system cages in a specific pathogen-free environment, with 55±5% humidity, at 23±2°C, ad libitum access to food and water, and a 14/10 h light/dark cycle. To establish the lung carcinoma xenograft model, control A549 cells or CPNE1-silenced A549 cells were suspended at a concentration of 2×106 cells/100 μl PBS and inoculated subcutaneously into the flanks of nude mice. Mice were randomly divided into control group and sh-CPNE1 croup (n=6/group). Following 20 days, the tumor volume (V) was determined by measuring the length (L) and width (W) with a vernier caliper and applying the following formula: V = (LxW2)×0.5. All experiments were performed in accordance with protocols approved by the Animal Ethical and Welfare Committee of the First Affiliated Hospital of Soochow University.
Statistical analysis
The differences in CPNE1 expression between NSCLC tissues and normal tissues were analyzed using a paired Student's t-test (two-tailed). For cell lines, differences between two groups were assessed using an unpaired Student's t-test (two-tailed). Expression levels of mRNA and the clinicopathologic characteristics in the NSCLC samples were compared using non-parametric tests (Mann-Whitney U test for comparison between two groups, and the Kruskall-Wallis test for comparison between three or more groups). A two-way analysis of variance was used to determine the difference in cell growth between two groups. In all cases, P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were performed using GraphPad Prism 5.02 (GraphPad Software, Inc., La Jolla, CA, USA) and SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA).
Results
CPNE1 is frequently overexpressed in lung adenocarcinoma tissues and cell lines
It has been previously reported that Copines I, II and III are expressed ubiquitously in humans using cDNA probes (15), although the actual content of Copine proteins in human tissues has not been systematically investigated. A recent study demonstrated that the expression level of CPNE1 is increased in prostate cancer compared with normal prostate tissue (16); however, the expression of CPNE1 in NSCLC is unknown. Lung tumor tissue samples were surgically resected from 128 lung adenocarcinoma patients, and then were detected via immunohistochemistry (Fig. 1A). In the present study, higher expression of CPNE1 was observed to be significantly associated with advanced TNM stage, lymph node metastasis and distant metastasis in lung adenocarcinoma (17). CPNE1 expression level was demonstrated to be positively associated with stage (P=0.002) and significantly associated with lymph node status (P= 0.011) and distant metastasis (P=0.042; Table I). In addition, the results demonstrated that expression of CPNE1 in advanced patients was more frequently increased (III+IV vs. I+II; P<0.001). However, no significant association was observed between CPNE1 expression and other clinicopathological features, including age, sex, tumor size and tumor differentiation in the present study. In addition to the above, CPNE1 protein expression were detected in 5 NSCLC cell lines and BEAS-2B cells by western blot analysis (Fig. 1B). Therefore, it was demonstrated that CPNE1 expression was frequently increased in NSCLC tissues and cell lines compared with the control.
Figure 1.
CPNE1 is frequently overexpressed in lung adenocarcinoma tissues and cell lines. (A) A total of 128 formalin-fixed and paraffin-embedded NSCLC tissues were subjected to immunohistochemical analyses of the CPNE1 protein. Representative images are presented of CPNE1 antibody staining in (a and b) NSCLC tissues (c) and normal tissues, at ×100 magnification, and at ×200 magnification (d-f). (B) The level of CPNE1 in human NSCLC cells was detected by western blotting. (C) Lentiviral transfection efficiency was detected using fluorescence microscopy at ×100 magnification to observe cells expressing green fluorescent protein at 5 days following transfection. (D) CPNE1 mRNA and protein levels in stable A549 and H1299 cells. Each cell line was divided into the following two groups: The sh-NC group (cells infected with Lv-si-CTRL) and the sh-CPNE1 group (cells infected with Lv-si-CPNE1). **P<0.01 vs. sh-NC. CPNE1, Copine 1; NSCLC, non-small cell lung cancer; sh-NC, negative control cells; sh-CPNE1, CPNE1-silenced cells.
Table I.
Distribution of CPNE1 status in NSCLC according to clinicopathological characteristics.
| Characteristic | CPNE1 expression
|
P-value | ||
|---|---|---|---|---|
| Low | High | χ2 | ||
| Sex | ||||
| Male | 25 (37.31%) | 42 (62.69%) | 0.056 | 0.813 |
| Female | 24 (39.34%) | 37 (60.66%) | ||
| Age, years) (mean ± SD) | 57.98±11.12 | 61.27±8.88 | −1.845 | 0.067 |
| Tumor size, cm | ||||
| <3 | 26 (48.15%) | 28 (51.85%) | 5.082 | 0.079 |
| ≥3, <5 | 18 (35.29%) | 33 (64.71%) | ||
| ≥5 | 5 (21.74%) | 18 (78.26%) | ||
| Differentiation | ||||
| Low | 22 (33.85%) | 43 (66.15%) | 1.099 | 0.294 |
| Medium, high | 27 (42.86%) | 36 (57.14%) | ||
| Nodal status | ||||
| N0 | 32 (50.00%) | 32 (50.00%) | 8.986 | 0.011 |
| N1 | 9 (36.00%) | 16 (64.00%) | ||
| N2/N3 | 8 (20.51%) | 31 (79.49%) | ||
| Distant metastasis | ||||
| No | 46 (41.82%) | 64 (58.18%) | 4.142 | 0.042 |
| Yes | 3 (16.67%) | 15 (83.33%) | ||
| TNM stage | ||||
| I–II | 40 (57.97%) | 29 (42.03%) | 24.563 | <0.001 |
| III–V | 9 (15.25%) | 50 (84.75%) | ||
Data are presented as n (percentage) unless otherwise stated. CPNE1, Copine 1; NSCLC, non-small cell lung cancer; SD, standard deviation; TNM, tumor, node and metastasis.
Lentivirus-mediated RNA interference efficiently inhibits CPNE1 expression in lung adenocarcinoma cells
In order to evaluate the association between CPNE1 and lung adenocarcinoma, lentivirus-delivered CPNE1-siRNA and scr-siRNA vector were successfully constructed. The results of transfection demonstrated that >90% of the A549 and H1299 cells exhibited green fluorescence indicating successful infection (Fig. 1C). To determine the silencing efficiency, the mRNA and protein expression levels of CPNE1 were detected by RT-qPCR and western blot analysis, respectively. The results demonstrated that the mRNA and protein expression levels of CPNE1 were significantly decreased in the sh-CPNE1 group compared with the sh-NC group (P<0.01; Fig. 1D). These data suggest that CPNE1 silencing may effectively downregulate CPNE1 expression.
Knockdown of CPNE1 inhibits in vitro cell growth, cell cycle progression and migration of NSCLC cells
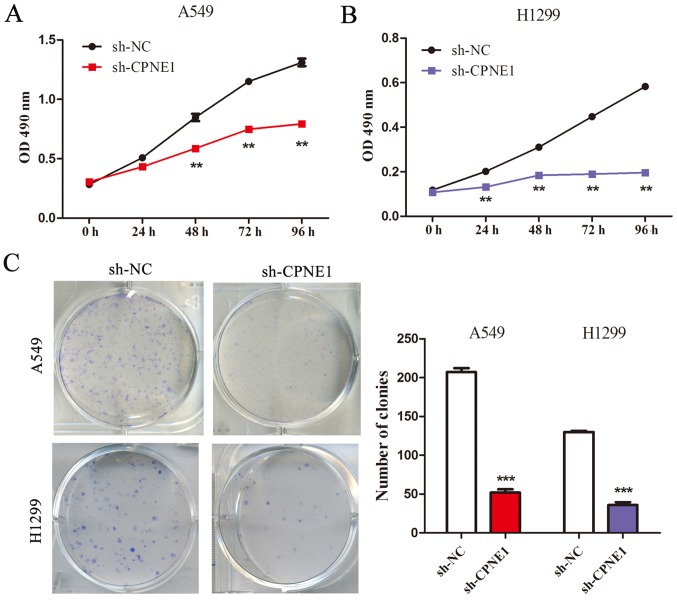
The CCK-8 assay demonstrated that compared with the control cells, cell growth was significantly inhibited in cells with stable knockdown of CPNE1 at 48 and 72 h following transfection (P<0.01; Fig. 2A and B). These results were also confirmed by performing a clonogenic assay (P<0.001; Fig. 2C). These results indicate that CPNE1 can promote cell growth via its effects on the cell cycle and apoptosis in NSCLC cells.
Figure 2.
Silencing of CPNE1 inhibits NSCLC cell proliferation and clone formation. (A and B) Cell Counting Kit-8 assay of cell proliferation in NSCLC cell lines; cell viability was determined at 24, 48 and 72 h. (C) Representative images and quantitative clonogenic analysis of plate colony formation in NSCLC cells. All experiments were performed in triplicate **P<0.01 and ***P<0.001 vs. sh-NC. CPNE1, Copine 1; NSCLC, non-small cell lung cancer; OD, optical density; sh-NC, negative control cells; sh-CPNE1, CPNE1-silenced cells.
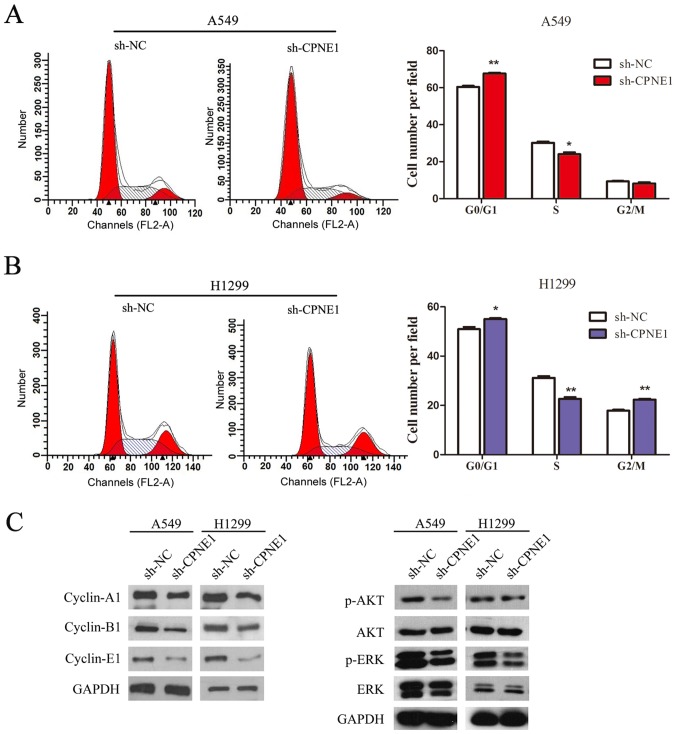
Furthermore, flow cytometric results indicated that transfection with sh-CPNE1 in NSCLC cells had an effect on the cell cycle, as the proportion of cells in the S phase was significantly decreased and the proportion of cells in the G0/G1 phase was significantly increased in CPNE1-silenced cells compared with the control cells (P<0.05; Fig. 3A and B).
Figure 3.
Knockdown of CPNE1 decelerates the cell cycle in NSCLC cells. (A and B) Flow cytometric analysis of A549 and H1299 cells. Cells were harvested and stained with propidium iodide. The percentage of cells in each cell cycle phase is presented in the inset of each panel, in which the values represent the mean ± standard deviation of three measurements. (C) Signaling molecules were detected, and the data demonstrated that p-AKT, p-ERK, cyclin-A1, cyclin-B1 and cyclin-E1 levels were markedly decreased in sh-CPNE1 compared with those in sh-NC. *P<0.05 and **P<0.01 vs. sh-NC. CPNE1, Copine 1; NSCLC, non-small cell lung cancer; p, phosphorylated; AKT, protein kinase B; ERK, extracellular signal regulated kinase; sh-NC, negative control cells; sh-CPNE1, CPNE1-silenced cells.
Furthermore, the molecular alterations and associated pathways in CPNE1-silenced cells were measured by western blotting. As illustrated in Fig. 3C, the results of the present study demonstrated that cell cycle associated proteins (cyclin-A1, cyclin-B1 and cyclin-E1), p-AKT and p-ERK levels were markedly decreased in the CPNE1-silenced cells compared with the control cells.
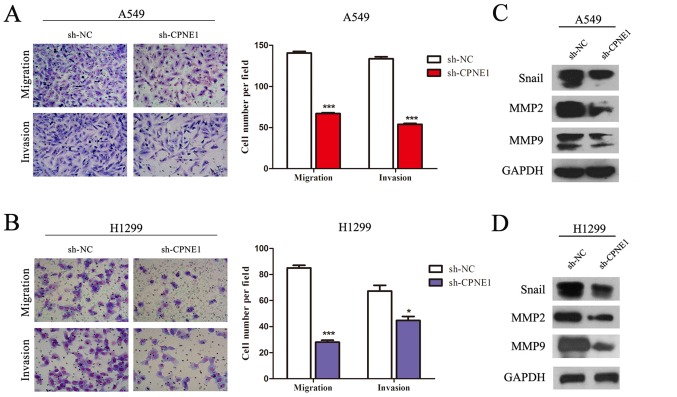
In addition, as presented in Fig. 4, a Transwell assay of the NSCLC stable cells lines further indicated that the migratory ability of NSCLC cells was significantly suppressed as a result of the loss of CPNE1 (P<0.001). In line with it, it was observed that Snail, MMP2, MMP9 were markedly decreased in the CPNE1-silenced cells compared with those in the control cells (Fig. 4B and D).
Figure 4.
Silencing of CPNE1 inhibits the migratory and invasive abilities of NSCLC cells, and its associated pathways. (A and B) CPNE1 silencing inhibits invasion and migration of NSCLC cells. Assays were performed in a Transwell plate. The cells that had migrated were stained with crystal violet and counted in at least three microscopic fields (magnification, ×100). Subsequently, the cells were treated as above and allowed to invade through the Matrigel-coated membrane in Transwell plates. Invasive cells were stained with crystal violet and counted under a light microscope (magnification, ×100). (C and D) Snail, MMP2, MMP9 were markedly decreased in sh-CPNE1 compared with sh-NC. *P<0.05 and ***P<0.001 vs. sh-NC. CPNE1, Copine 1; NSCLC, non-small cell lung cancer; MMP, matrix metalloproteinase; sh-NC, negative control cells; sh-CPNE1, CPNE1-silenced cells.
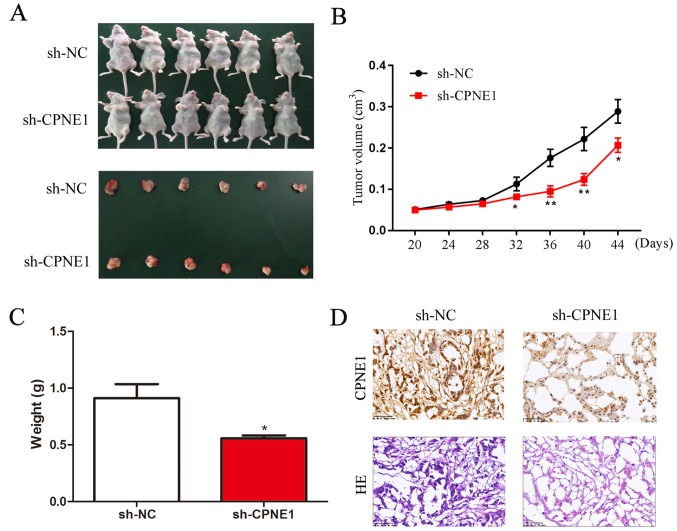
Knockdown of CPNE1 inhibits tumor growth in vivo
To further assess the effect of CPNE1-silenced on NSCLC cells in vivo, a xenograft model was used to determine the role of CPNE1 in the tumorigenicity and development of A549 cells. As demonstrated in Fig. 5A and B, tumor growth in the sh-CPNE1 group was significantly decreased compared with the sh-NC group. The final tumor volume at 44 days was 0.222±0.059 cm3 in the CPNE1-siRNA group, whereas the volume in the scr-siRNA group was 0.400±0.142 cm3 (P<0.01). The mean tumor weight in the CPNE1-siRNA group was significantly decreased compared with the scr-siRNA group (0.60±0.10 g vs. 0.90±0.30 g; Fig. 5C). Consequently, these data demonstrate that silencing CPNE1 expression may suppress tumor growth of human lung adenocarcinoma cells in nude mice. The tissues resected from the xenograft tumors were analyzed to verify CPNE1 expression using hematoxylin and immunohistochemistry (Fig. 5D).
Figure 5.
Knockdown of CPNE1 inhibits tumor growth in vivo. (A) CPNE1-silenced A549 cell xenografts in nude mice (n=6) at the experimental endpoint; tumors were dissected and images were captured as presented. (B) Tumor growth curves in mice (n=6 in each group) inoculated with the indicated cells at the indicated days. (C) Each tumor formed was weighed and the mean weight in each group was calculated. (D) Immunohistochemical staining for CPNE1 was quantified based on staining intensity. *P<0.05 and **P<0.01 vs. sh-NC. CPNE1, Copine 1; sh-NC, negative control cells; sh-CPNE1, CPNE1-silenced cells; HE, hematoxylin and eosin.
Discussion
Despite improvements in therapeutic strategies, the survival and outcome of patients with NSCLC have not been markedly affected (4). The majority of patients have progressed to the advanced stage of the disease when the diagnosis is confirmed, which is beyond the optimal treatment period (18). Therefore, it is essential to elucidate the pathogenesis of NSCLC and to identify novel treatment targets.
It has been reported that Copine 1, encoded by CPNE1, is a soluble membrane-binding protein, which includes two tandem C2 domains at the N-terminus and an A domain at the C-terminus (6,9). Copine 1 can bind with different intracellular proteins via its A domain at the C-terminus (7). Further study has demonstrated that this A domain has been identified in several integrins and extracellular matrix proteins and may serve as a protein-binding domain (10). The C2 domain was demonstrated to be important for calcium and phospholipid binding (19,20). In addition, multiple proteins have been identified as Copine targets, and a number of these are proteins associated with intracellular signalling pathways including dual specificity mitogen-activated protein kinase kinase 1/ERK, protein phosphatase 5, and CDC42-regulated protein kinase (7,8). Notably, Park et al (9) previously reported that CPNE1 serves a vital role in regulating neuronal differentiation of HiB5 cells, which may be associated with activating AKT signalling via phosphorylating on the residue 473 (S473) of AKT. Recently, another study demonstrated that CPNE1 may promote the development and progression of prostate cancer via its C2 domain (16). Although CPNE1 was demonstrated to bind several intracellular proteins with diverse biological functions, the role of CPNE1 in regulating biological processes is not well understood.
A recent study demonstrated that CPNE3 is upregulated, and can enhance cell metastasis in NSCLC (21). Further study demonstrated that CPNE3 can activate downstream ErbB2 signalling and promote migration in SKBr3 breast cancer cells (22). In accordance with these findings, Heinrich et al (23) also demonstrated that CPNE3 can interact with ErbB2 and promote tumor cell migration. The AKT serine/threonine kinase serves essential roles in regulating cell growth, cell migration, invasion, survival, and glycolysis. Furthermore, aberrant activation of AKT signalling is associated with the pathogenesis of cancer and poor prognosis (24,25). Among the AKT feedback signalling molecules, ERK is generally activated with AKT in tumor cells and is pivotal for cell proliferation and evasion of cell apoptosis (26). In specific cases, AKT and ERK signalling pathways are compensatory for each other (27,28). Notably, it was demonstrated in the present study that p-AKT and p-ERK levels were decreased in the CPNE1-silenced cells compared with the control cells.
Cyclin B1 is a key regulator in the cell cycle progression from G2 to M phase. It has been demonstrated that cyclin B1 serves a pivotal role in tumorigenesis and tumor development: Deregulation of cyclin B1 can frequently lead to unrestricted cell-cycle progression and malignant transformation (29-31), and cyclin B1 overexpression has been detected in various types of human cancer (32,33). Cyclin E1 is a key regulator of the cell cycle and serves an important role in tumorigenesis and angiogenesis (34). Previous studies have demonstrated that overexpression of cyclin E1 was important in the growth of ovarian cancer cells and strongly associated with poor prognosis (35,36). In the present study, the results demonstrated that transfection with sh-CPNE1 in NSCLC cells had an effect on the cell cycle, and cyclin-A1, cyclin-B1 and cyclin-E1 levels were lower in the CPNE1-silenced cells than those in the control cells.
Metastasis and relapse is the major cause of mortality for lung cancer patients (37). Epithelial-mesenchymal transition is a critical step for morphogenesis during embryonic development and the conversion of early-stage tumors into invasive malignancies (38,39), which is marked by induction of Snail and MMPs (40,41). In the present study, it was also demonstrated that Snail, MMP2, MMP9 were decreased in the CPNE1-silenced cells compared with those in the control cells.
In conclusion, to the best of our knowledge, the present study reported for the first time that CPNE1 expression is upregulated in NSCLC and it was observed that increased expression of CPNE1 is associated with advanced TNM stage, lymph node metastasis and distant metastasis in lung adenocarcinoma. Furthermore, the function of CPNE1 in regulation of cell growth, migration and invasion was investigated, and it was demonstrated that knockdown of CPNE1 inhibits the cell cycle in NSCLC cells. Collectively, these data strongly suggest that CPNE1 is an oncogene in NSCLC and serves an important role in tumorigenesis of NSCLC progression.
Acknowledgments
Not applicable.
Funding
The present study was supported by grants from The National Natural Science Foundation of China (grant no. 81201575), The Science and Technology Plan Projects of Suzhou (grant no. SYS201612), Jiangsu Provincial Medical Youth Talent (grant no. QNRC2016746), Medicine and Technology Projects of Zhejiang province (grant no. 2017KY646), The Societal and Developmental Project of Suzhou (grant no. SS201630), The Suzhou Key Laboratory for Respiratory Medicine (grant no. SZS201617), The Clinical Medical Center of Suzhou (grant no. Szzx201502), Jiangsu Provincial Key Medical Discipline (grant no. ZDXKB2016007) and The Clinical Key Specialty Project of China.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
SLL, HCT, JJZ, HGD, YYZ, WWD, ZLD, PTS and YZ participated in data collection and analysis. SLL, JAH and ZYL participated in the design of the study. ZYL participated in the writing of the manuscript and data interpretation. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Experiments using tissue samples from human subjects were approved by the Ethics Committee of The First Affiliated Hospital of Soochow University (Suzhou, China). All participants provided written informed consent for the whole study. Experiments on animals were performed following approval from the Animal Ethical and Welfare Committee of The First Affiliated Hospital of Soochow University.
Patient consent for publication
All participants provided written informed consent for the whole study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Dai X, Guo G, Zou P, Cui R, Chen W, Chen X, Yin C, He W, Vinothkumar R, Yang F, et al. (S)-crizotinib induces apoptosis in human non-small cell lung cancer cells by activating ROS independent of MTH1. J Exp Clin Cancer Res. 2017;36:120. doi: 10.1186/s13046-017-0584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mulshine JL, Sullivan DC. Clinical practice. Lung cancer screening. N Engl J Med. 2005;352:2714–2720. doi: 10.1056/NEJMcp042630. [DOI] [PubMed] [Google Scholar]
- 5.Petschnigg J, Kotlyar M, Blair L, Jurisica I, Stagljar I, Ketteler R. Systematic Identification of Oncogenic EGFR Interaction Partners. J Mol Biol. 2017;429:280–294. doi: 10.1016/j.jmb.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Creutz CE, Tomsig JL, Snyder SL, Gautier MC, Skouri F, Beisson J, Cohen J. The copines, a novel class of C2 domain-containing, calcium-dependent, phospholipid-binding proteins conserved from Paramecium to humans. J Biol Chem. 1998;273:1393–1402. doi: 10.1074/jbc.273.3.1393. [DOI] [PubMed] [Google Scholar]
- 7.Tomsig JL, Snyder SL, Creutz CE. Identification of targets for calcium signaling through the copine family of proteins. Characterization of a coiled-coil copine-binding motif. J Biol Chem. 2003;278:10048–10054. doi: 10.1074/jbc.M212632200. [DOI] [PubMed] [Google Scholar]
- 8.Tomsig JL, Creutz CE. Biochemical characterization of copine: A ubiquitous Ca2+-dependent, phospholipid-binding protein. Biochemistry. 2000;39:16163–16175. doi: 10.1021/bi0019949. [DOI] [PubMed] [Google Scholar]
- 9.Park N, Yoo JC, Lee YS, Choi HY, Hong SG, Hwang EM, Park JY. Copine1 C2 domains have a critical calcium-independent role in the neuronal differentiation of hippocampal progenitor HiB5 cells. Biochem Biophys Res Commun. 2014;454:228–233. doi: 10.1016/j.bbrc.2014.10.075. [DOI] [PubMed] [Google Scholar]
- 10.Whittaker CA, Hynes RO. Distribution and evolution of von Willebrand/integrin A domains: Widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell. 2002;13:3369–3387. doi: 10.1091/mbc.e02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asamura H, Chansky K, Crowley J, Goldstraw P, Rusch VW, Vansteenkiste JF, Watanabe H, Wu YL, Zielinski M, Ball D, et al. The International Association for the Study of Lung Cancer Lung Cancer Staging Project: Proposals for the revision of the N descriptors in the forthcoming 8th edition of the TNM Classification for Lung Cancer. J Thorac Oncol. 2015;10:1675–1684. doi: 10.1097/JTO.0000000000000678. [DOI] [PubMed] [Google Scholar]
- 12.Tang H, Kirkness EF, Lippert C, Biggs WH, Fabani M, Guzman E, Ramakrishnan S, Lavrenko V, Kakaradov B, Hou C, et al. Profiling of short-tandem-repeat disease alleles in 12,632 human whole genomes. Am J Hum Genet. 2017;101:700–715. doi: 10.1016/j.ajhg.2017.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu J, Zeng Y, Li W, Qin H, Lei Z, Shen D, Gu D, Huang JA, Liu Z. CD73/NT5E is a target of miR-30a-5p and plays an important role in the pathogenesis of non-small cell lung cancer. Mol Cancer. 2017;16:34. doi: 10.1186/s12943-017-0591-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Tomsig JL, Creutz CE. Copines: A ubiquitous family of Ca(2+)-dependent phospholipid-binding proteins. Cell Mol Life Sci. 2002;59:1467–1477. doi: 10.1007/s00018-002-8522-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang J, Zhang J, Ruan J, Mi Y, Hu Q, Wang Z, Wei B. CPNE1 is a useful prognostic marker and is associated with TNF receptor-associated factor 2 (TRAF2) expression in prostate cancer. Med Sci Monit. 2017;23:5504–5514. doi: 10.12659/MSM.904720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu C, Chen K, Zheng H, Guo X, Jia W, Li M, Zeng M, Li J, Song L. Overexpression of astrocyte elevated gene-1 (AEG-1) is associated with esophageal squamous cell carcinoma (ESCC) progression and pathogenesis. Carcinogenesis. 2009;30:894–901. doi: 10.1093/carcin/bgp064. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y, Ju Q, Qian B, Zhang F, Shi H. The effectiveness of PD-1 inhibitors in non-small cell lung cancer (NSCLC) patients of different ages. Oncotarget. 2017;9:7942–7948. doi: 10.18632/oncotarget.23678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nalefski EA, Falke JJ. The C2 domain calcium-binding motif: Structural and functional diversity. Protein Sci. 1996;5:2375–2390. doi: 10.1002/pro.5560051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rizo J, Südhof TC. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- 21.Lin HC, Zhang FL, Geng Q, Yu T, Cui YQ, Liu XH, Li J, Yan MX, Liu L, He XH, et al. Quantitative proteomic analysis identifies CPNE3 as a novel metastasis-promoting gene in NSCLC. J Proteome Res. 2013;12:3423–3433. doi: 10.1021/pr400273z. [DOI] [PubMed] [Google Scholar]
- 22.Choi HY, Park N, Na JB, Ko ES, Park JY, Yoo JC. Direct binding of Copine3 with Jab1 activates downstream ErbB2 signaling and motility in SKBr3 breast cancer cells. Oncol Rep. 2016;35:1147–1152. doi: 10.3892/or.2015.4472. [DOI] [PubMed] [Google Scholar]
- 23.Heinrich C, Keller C, Boulay A, Vecchi M, Bianchi M, Sack R, Lienhard S, Duss S, Hofsteenge J, Hynes NE. Copine-III interacts with ErbB2 and promotes tumor cell migration. Oncogene. 2010;29:1598–1610. doi: 10.1038/onc.2009.456. [DOI] [PubMed] [Google Scholar]
- 24.Ciruelos Gil EM. Targeting the PI3K/AKT/mTOR pathway in estrogen receptor-positive breast cancer. Cancer Treat Rev. 2014;40:862–871. doi: 10.1016/j.ctrv.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Miller TW, Rexer BN, Garrett JT, Arteaga CL. Mutations in the phosphatidylinositol 3-kinase pathway: Role in tumor progression and therapeutic implications in breast cancer. Breast Cancer Res. 2011;13:224. doi: 10.1186/bcr3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Z, Yang H, Sutton MN, Yang M, Clarke CH, Liao WS, Bast RC., Jr ARHI (DIRAS3) induces autophagy in ovarian cancer cells by downregulating the epidermal growth factor receptor, inhibiting PI3K and Ras/MAP signaling and activating the FOXo3a-mediated induction of Rab7. Cell Death Differ. 2014;21:1275–1289. doi: 10.1038/cdd.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saini KS, Loi S, de Azambuja E, Metzger-Filho O, Saini ML, Ignatiadis M, Dancey JE, Piccart-Gebhart MJ. Targeting the PI3K/AKT/mTOR and Raf/MEK/ERK pathways in the treatment of breast cancer. Cancer Treat Rev. 2013;39:935–946. doi: 10.1016/j.ctrv.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Britten CD. PI3K and MEK inhibitor combinations: Examining the evidence in selected tumor types. Cancer Chemother Pharmacol. 2013;71:1395–1409. doi: 10.1007/s00280-013-2121-1. [DOI] [PubMed] [Google Scholar]
- 29.Niméus-Malmström E, Koliadi A, Ahlin C, Holmqvist M, Holmberg L, Amini RM, Jirström K, Wärnberg F, Blomqvist C, Fernö M, et al. Cyclin B1 is a prognostic proliferation marker with a high reproducibility in a population-based lymph node negative breast cancer cohort. Int J Cancer. 2010;127:961–967. doi: 10.1002/ijc.25091. [DOI] [PubMed] [Google Scholar]
- 30.Kedinger V, Meulle A, Zounib O, Bonnet ME, Gossart JB, Benoit E, Messmer M, Shankaranarayanan P, Behr JP, Erbacher P, et al. Sticky siRNAs targeting survivin and cyclin B1 exert an antitumoral effect on melanoma subcutaneous xenografts and lung metastases. BMC Cancer. 2013;13:338. doi: 10.1186/1471-2407-13-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthess Y, Raab M, Sanhaji M, Lavrik IN, Strebhardt K. Cdk1/cyclin B1 controls Fas-mediated apoptosis by regulating caspase-8 activity. Mol Cell Biol. 2010;30:5726–5740. doi: 10.1128/MCB.00731-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soria JC, Jang SJ, Khuri FR, Hassan K, Liu D, Hong WK, Mao L. Overexpression of cyclin B1 in early-stage non-small cell lung cancer and its clinical implication. Cancer Res. 2000;60:4000–4004. [PubMed] [Google Scholar]
- 33.Zhou L, Li J, Zhao YP, Cui QC, Zhou WX, Guo JC, You L, Wu WM, Zhang TP. The prognostic value of Cyclin B1 in pancreatic cancer. Med Oncol. 2014;31:107. doi: 10.1007/s12032-014-0107-4. [DOI] [PubMed] [Google Scholar]
- 34.Wang F, Fu XD, Zhou Y, Zhang Y. Down-regulation of the cyclin E1 oncogene expression by microRNA-16-1 induces cell cycle arrest in human cancer cells. BMB Rep. 2009;42:725–730. doi: 10.5483/BMBRep.2009.42.11.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakayama N, Nakayama K, Shamima Y, Ishikawa M, Katagiri A, Iida K, Miyazaki K. Gene amplification CCNE1 is related to poor survival and potential therapeutic target in ovarian cancer. Cancer. 2010;116:2621–2634. doi: 10.1002/cncr.24987. [DOI] [PubMed] [Google Scholar]
- 36.Schraml P, Bucher C, Bissig H, Nocito A, Haas P, Wilber K, Seelig S, Kononen J, Mihatsch MJ, Dirnhofer S, et al. Cyclin E overexpression and amplification in human tumours. J Pathol. 2003;200:375–382. doi: 10.1002/path.1356. [DOI] [PubMed] [Google Scholar]
- 37.Gupta GP, Massagué J. Cancer metastasis: Building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 38.Liu RY, Zeng Y, Lei Z, Wang L, Yang H, Liu Z, Zhao J, Zhang HT. JAK/STAT3 signaling is required for TGF-β-induced epithelial-mesenchymal transition in lung cancer cells. Int J Oncol. 2014;44:1643–1651. doi: 10.3892/ijo.2014.2310. [DOI] [PubMed] [Google Scholar]
- 39.Kang Y, Massagué J. Epithelial-mesenchymal transitions: Twist in development and metastasis. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 40.Ren F, Tang R, Zhang X, Madushi WM, Luo D, Dang Y, Li Z, Wei K, Chen G. Overexpression of MMP family members functions as prognostic biomarker for breast cancer patients: A systematic review and meta-analysis. PLoS One. 2015;10:e0135544. doi: 10.1371/journal.pone.0135544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ying TH, Lee CH, Chiou HL, Yang SF, Lin CL, Hung CH, Tsai JP, Hsieh YH. Knockdown of Pentraxin 3 suppresses tumorigenicity and metastasis of human cervical cancer cells. Sci Rep. 2016;6:29385. doi: 10.1038/srep29385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.