Abstract
Brown adipose tissue (BAT) is a crucial regulator of energy expenditure. Emerging evidence suggests that n-3 PUFA potentiate brown adipogenesis in vitro. Since the pregnancy and lactation is a critical time for brown fat formation, we hypothesized that maternal supplementation of n-3 PUFA promotes BAT development in offspring. Female C57BL/6 mice were fed a diet containing n-3 PUFA (3%) derived from fish oil (FO), or an isocaloric diet devoid of n-3 PUFA (Cont) during pregnancy and lactation. Maternal n-3 PUFA intake was delivered to the BAT of neonates significantly reducing the n-6/n-3 ratio. The maternal n-3 PUFA exposure was linked with upregulated brown-specific gene and protein profiles and the functional cluster of brown-specific miRNAs. In addition, maternal n-3 PUFA induced histone modifications in the BAT evidenced by 1) increased epigenetic signature of brown adipogenesis, i.e., H3K27Ac and H3K9me2, 2) modified chromatin-remodeling enzymes, and 3) enriched the H3K27Ac in the promoter region of Ucp1. The offspring received maternal n-3 PUFA nutrition exhibited a significant increase in whole-body energy expenditure and better maintenance of core body temperature against acute cold treatment. Collectively, our results suggest that maternal n-3 PUFA supplementation potentiates fetal BAT development via the synergistic action of miRNA production and histone modifications, which may confer long-lasting metabolic benefits to offspring.
Keywords: BAT, n-3 PUFA, fish oil, thermogenesis, maternal nutrition
1. Introduction
Brown adipose tissue (BAT) is a specialized fat that dissipates excess energy into heat (non- shivering thermogenesis) through mitochondrial uncoupling protein 1 (UCP1) [1]. Current research renews the metabolic function of BAT by revealing BAT as a crucial regulator in maintaining energy balance by increasing thermogenic energy expenditure. A significant amount of BAT is found in healthy adults as well as most children and adolescents [2–4], but not in the obese adults, suggesting that loss of active BAT depots is a contributing factor to obesity. Childhood obesity predisposes adults with metabolic susceptibility to obesity and type 2 diabetes (T2D) [5–7]. Therefore, identifying early regulatory factors to prevent childhood obesity is critical to combat the current obesity epidemic. The fetal and neonatal stages are critical for fetal BAT development, which are expected to have long-term impacts on offspring BAT function [8]. However, a limited amount of studies have been conducted regarding the effects of maternal nutrition on prenatal BAT development. Despite the well-established physiological relevance of BAT in obesity outcome, it is unclear whether the amount of BAT at birth or the rate of BAT loss (either by degeneration or by transdifferentiation into WAT) are associated with susceptibility to obesity in later life. In particular, it is poorly understood whether prenatal BAT development through dietary intervention could be a manageable target to attenuate the risk for childhood obesity.
Human BAT depots are found in the deeper neck (cervical), supraclavicular, paravertebral, perirenal, and axillary areas [9–11] and possesses 50-times greater respiratory activities than white adipose tissue (WAT) [12]. These BAT depots are comprised of 1.5% of total body mass (roughly 5% of total fat mass), and up to 90% depots could be activated BAT [13]. An image-guided mapping of rodent adipose depots reveals the topological analogy of BAT between rodents and humans [14], which provides the feasibility to use rodent in studying human BAT. Also, the comparable functional analysis demonstrated that human supraclavicular BAT features functional similarity with rodent interscapular BAT regarding mitochondrial activity and thermogenic potential [12]. Regarding the timeline for BAT development, late pregnancy (at the last trimester) is the critical time for human BAT formation [15, 16]. The human brown adipocytes in the interscapular are rapidly lost after birth via either degeneration and replacement with white adipocyte, or transdifferentiation into white adipocytes. However, brown adipocytes reside in the deeper neck and supraclavicular regions, remain active to adolescence and adulthood until they lost thermogenic potential with the progression of obesity, type 2 diabetes, or aging [6]. Given these developmental similarities, the regulation of interscapular BAT in rodents during pregnancy seems to be translatable to BAT around the neck and supraclavicular in humans.
BAT development is modulated by epigenetic modifications that are heritable and reversible changes in gene expression occur without altering DNA sequences through DNA methylation, chromatin histone remodeling, and noncoding RNAs such as miRNAs [17–19]. Nearly a dozen miRNAs have been identified in promoting the transcriptional program of brown adipogenesis [20]. On the other hand, the site-specific acetylation (Ac) and methylation (Me) status on the lysine (K) residues of histone tails, especially at H3K9 and H3K27, play essential roles in adipogenesis by controlling gene activation or repression[21, 22]. Maternal nutrition is a key epigenetic modulator for fetus development. However, it is largely unknown whether the BAT epigenome is a viable target for obesity control through maternal nutrition.
Accumulating evidence has supported that n-3 PUFA promotes brown adipogenesis and adaptive thermogenesis [23–27]. We have previously demonstrated that eicosapentaenoic acids (EPA) promote the brown adipogenic program through a miRNA-dependent epigenetic mechanism in the murine primary brown adipocytes [28], suggesting that maternal n-3 PUFA nutrition may be effective in reinforcing embryonic BAT developmental program during pregnancy. This study aimed to investigate the impact of early n-3 PUFA exposure through maternal nutrition on the fetal BAT development. Here, we demonstrate that maternal n-3 PUFA intake during pregnancy and lactation enhanced brown transcriptional programming through miRNA and histone modification-mediated epigenetic regulations. These results open a new research avenue emphasizing that ‘boosting prenatal BAT development’ could be a novel therapeutic target for attenuating childhood obesity through thermogenic energy expenditure.
2. Material and methods
2.1. Animals
All animal experiments were conducted according to the protocols approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Nebraska-Lincoln. C57BL/6 male and female mice were purchased at 8–10 weeks of age from the Jackson Laboratory. After 3 days of acclimatization period, mice were put into the breeding colony. Female mice were randomly assigned to two diet groups receiving either a diet containing 3% of n-3 PUFA from the fish oil (FO) or a diet devoid of n-3 PUFA (Cont). The AIN-93G rodent formulation was modified for the fat composition containing 15% of total calorie from fat either 10% palm oil (Cont) or fish oil (FO), the other 5% of total calorie from soybean oil as a source of essential FA. Gas Chromatography (GC) was used to analyze fatty acid profiles of each diet and dietary composition in Supplement Table 1. The same diets were maintained throughout gestation and lactation. The pregnancy of the female mice last for 19–21 days, and the pups (n=16 each group) were weaned (3 weeks postpartum). Necropsy was conducted at weaning, to collect blood, liver, interscapular BAT (iBAT), and inguinal (iWAT) and epididymal WAT (eWAT). Tissue samples were snap-frozen in liquid nitrogen and kept at −80 °C for further analysis.
2.2. Energy expenditure by metabolic cages
To measure the effect of maternal n-3 PUFA intake on energy expenditure, pups (n=5 each group) were individually placed into the metabolic cage (TSE systems) for six days (two days of adaptation and four days of measurement). Indirect oxygen consumption (VO2) and carbon dioxide production (VCO2) were used to calculate metabolic parameters. Energy expenditure (EE) and respiratory exchange rate (RER=VCO2/VO2) were calculated and obtained from the TSE systems software and plotted into a figure with hourly time point.
2.3. Cold exposure, measurement of rectal temperature and heat release
To measure the long-term effect of maternal n-3 PUFA supplementation on the offspring’s thermogenic potential, male pups from both maternal fish oil or control diet (n=6 per group) were switched to a standard AIN-93G diet (no additional n-3 PUFA). At week 11, mice were exposed to cold temperature (6°C) acutely (1–3 hour) or for 24 hours (See Fig 5A study design). To measure the core body temperature, a rectal thermometer (Kent Scientific Corp) was used. The probe was positioned into the anal ducts of the mice and three readings of each time point were recorded. Infrared (IR) camera (A655sc, FLIR Systems) was used to detect thermal release and to capture images of the surface body temperature. FLIR Research IR program software was used to display surface heat release via color palette representing temperatures between 22 and 34 °C.
Figure 5.
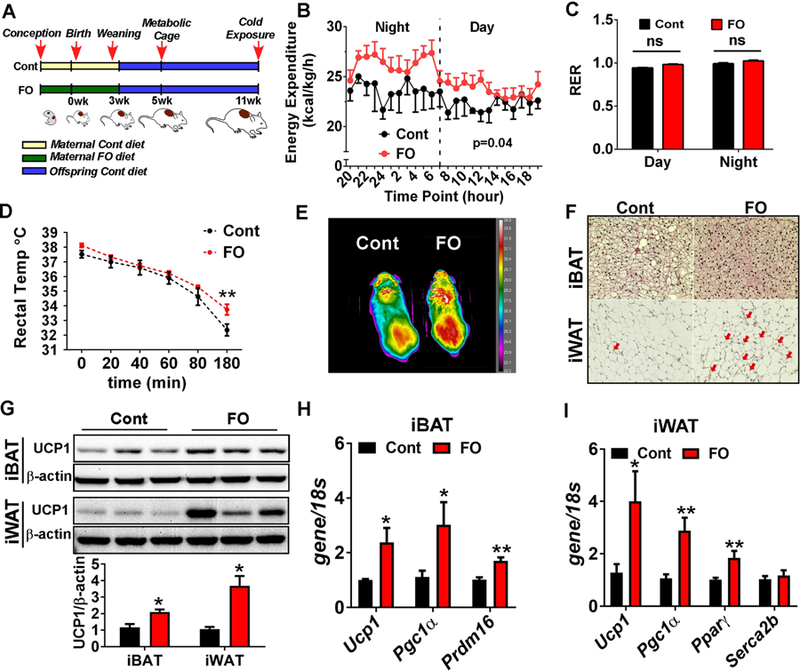
Maternal n-3 PUFA supplementation was associated with metabolic improvement in the later life of offspring. A. Schematic presentation of experimental design. The pups received maternal control diet (Cont)- or FO-diet (FO) were weaned at 3 week-old, and maintained until 11 week-old with no additional dietary modification. B. Average energy expenditure for 4 days measured by metabolic cages (n=5) C. Average RER (VO2/VCO2) values for 4 days measured by using metabolic cages (n=5). D. Core body temperature measured by rectal temperature upon exposure to cold temperature (6°C) for 3 hours (n=6). E. Heat release captured by IR camera upon 3-hour cold exposure. F. Representative images with H&E staining of iBAT and iWAT after 24-hour exposure to cold temperature (6°C). Red arrows indicate the emergence of brown-like structure within iWAT. G. Western blot pattern of UCP1 in the iBAT and iWAT after 24-hour cold exposure (left). Relative UCP1 levels were quantified by Image J (right). H. Brown signature gene expressions of Ucp1, Pgc1α, and Prdm16 in the IBAT after 24-hour cold exposure (n=6), I. Gene expression levels of Ucp1, Pgc1α, PPARγ and Scerca2b (a responsible gene for calcium- cycling dependent thermogenesis) in the iWAT after 24-hour cold exposure (n=6). All data are expressed as mean ± SEM. *p< 0.05, **p< 0.01, ***p<0.001 by Student’s t-test.
2.4. Gas Chromatography (GC) for fatty acid analysis
To determine FA profiles in the red blood cells, whole blood was collected, and red blood cells were precipitated by centrifugation (6000×g for 15 minutes). A 200μl of the packed volume of red blood cells were transferred to a fresh glass vial and total lipids were extracted. They were then subjected to FA methylation by 14 % boron trifluoride (BF3)-methanol reagent (Sigma, USA) at 100 °C for 1 hour to form fatty acid methyl ester (FAME). Agilent Technologies HP-88 column (100m × 0.25mm × 0.2 μm film thickness) was used. The individual FA peak was identified by comparing its relative retention times with the commercial mixed-FA standard (NuCheck PreP), and the area percentages for all resolved peaks were analyzed using the GC Chemstation software.
2.5. Blood Chemistry
To measure plasma glucose, insulin, and cAMP levels, immunoassays were conducted by using mouse glucose assay (Crystal Chem), ultra-sensitive mouse insulin ELISA kit (Crystal Chem), and mouse cAMP parameter assay kit (R&D Systems), respectively, in accordance to the manufacturer’s protocol.
2.6. qPCR of mRNA and microRNA analysis
Total RNA was extracted using Trizol® reagent (Invitrogen) from homogenized tissues. RNA was purified using DNase treatment & removal kit (Invitrogen), and 2 μg of RNA was converted into cDNA (iScript, BioRad) via reverse transcription. Relative gene expressions were determined based on the 2−ΔΔCT method with normalization of the raw Ct value to 18s. For miRNA analysis, miRNA was converted to cDNA using the miScript reverse transcription kit (Qiagen) according to the manufacturer’s instructions. MiRNA was measured by using the commercial miScript Universal Primer, with the miScript primer assay kit (Qiagen). Primers of miR-30b, −193b, −365, and RNU6–2 were purchased from Qiagen. The Ct values were normalized to RNU6–2 (U6 small nuclear).
2.7. Western blot analysis
Total protein was extracted from tissue by homogenizer using RIPA buffer along with a protease inhibitor and phosphatase inhibitor. Proteins were separated by SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membrane (0.45 μm, Thermogenic Scientific) using a wet-transfer method. Antibody targeting UCP1 (14670), PPARγ (2444) and β-actin (4967) were purchased from Cell Signaling Technology. PRDM16 (sc-55697) was purchased from Santa Cruz Biotechnology. Blots were visualized with a FluorChem™E imaging system (Protein Simple).
2.8. Chromatin immunoprecipitation assay (ChIP)
The murine brown adipocyte cell-line (HIB1B) was a generous gift from Dr. Johannes Klein (University of Lubeck, Lubeck, Germany), who has generated the cell line from brown fat of newborn or 6–8 week old of C57BL/6 or FVB mice [29]. HIB1B cells were differentiated and treated with EPA (100 μM), as we described previously [28]. Chromatin immunoprecipitation assay was performed using CHIP-IT® Express kit according to the manufacturer’s instructions (Active Motif, USA). Cells were fixed with 1% formaldehyde to cross-link DNA and proteins. Chromatins were isolated and DNA was sheared into 200–1000 bp length. Fifty μl of each sample was removed as the input control. The samples were incubated with magnetic beads-AcH3K27 antibody complex (Cell Signaling Technology, USA) overnight. Next, magnetic beads were washed, and chromatins were eluted and reversely cross-linked. After treated with Proteinase K, DNA was ready to be used immediately for real-time qPCR analysis using SYBR Green (QuantStudio 6, life technology). The Ucp1 and Pgc1α promoter regions were assessed in acetylation histone 3 at lysine 27 (H3K27) ChIP samples. Fold of enrichment was calculated and presented according to the methods provided by the manufacturer. Primer sequence: Ucp1 proximal promoter, 5’-CCCACTAGCAGCTCTTTGGA-3’ and 5’-CTGTGGAGCAGCTCAAAGGT-3’; Pgc1 α CRE region, 5’-CAAAGCTGGCTTCAGTCACA-3’ and 5’-AAAAGTAGGCTGGGCTGTCA-3’ [30].
2.9. Statistical Analysis
All data are presented as mean ± SEM. Independent samples from control group and treatment group were analyzed and compared using two-tailed Student’s t-test, with * P < 0.05, ** P < 0.01, *** P <0.001. All statistical analyses were conducted by Graph Pad Prism 7 (Version 7.03).
3. Results
3.1. Maternal n-3 PUFA supplementation decreased WAT mass and n-6/n-3 PUFA ratio
To investigate the effects of n-3 PUFA on BAT development on offspring, C57BL/6 female were fed either a diet containing with 3% of n-3 PUFA from fish oil (FO) or an isocaloric diet absent of additional n-3 PUFA (Cont) during pregnancy and lactation. Both male and female pups were weaned at 3 weeks postpartum. There was a significant decrease in body weight in FO compared to Cont in both genders. Although there was no difference in liver weight, WAT was significantly reduced in both genders, indicating that maternal n-3 PUFA intake decreased white fat accumulation. A similar trend was observed in iBAT mass, although the differences did not reach a statistical significance (Fig 1A–C).
Figure 1.
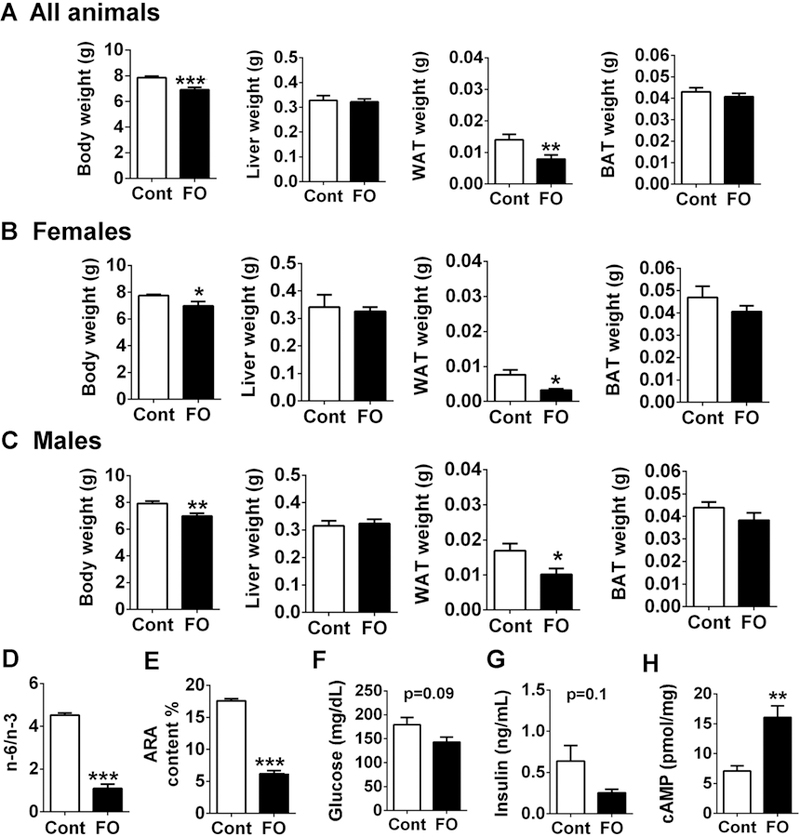
Maternal n-3 PUFA supplementation decreased offspring’s WAT accumulation and altered blood lipid profile at the time of weaning. A-C. Effects of maternal n-3 PUFA supplementation on body weight, liver weight, epididymal WAT, and interscapular BAT weight from all pups (A, n=18 each group), female pups (B, n=8) and male pups (C, n=10). D. n-6/n-3 PUFA ratio by GC analysis. E. Arachidonic acid content (%). F. Plasma glucose level. G. Insulin concentration by ELISA. H. cAMP concentration. All data was expressed as mean ± SEM.* p<0.05, **p< 0.01, and ***P < 0.001 by Student’s t-test.
To investigate the impact of maternal n-3 PUFA intake in the pups, FA profile was determined in the red blood cells of the weaned pups. There was a significant decrease in n-6/n-3 PUFA ratio in the maternal n- 3 PUFA received group (FO) compared to the control, indicating that maternal n-3 PUFA was effectively delivered from mothers to the pups (Fig 1D). Concomitantly, the content of arachidonic acid (ARA), a precursor for pro-inflammatory eicosanoids, was significantly decreased in the maternal n-3 PUFA group (Fig 1E). Also, there was a trend towards a decrease in plasma glucose (p=0.09) and insulin concentrations (p=0.1) in the maternal n-3 PUFA group (Fig 1F, G), indicating that maternal n-3 PUFA intake may have an impact on the glucose tolerance and insulin sensitivity in pups at weaning. Adrenaline, released via the action of the sympathetic nervous system, plays a vital role in brown and beige adipose tissue thermogenic program [31]. Our results also indicated that there was a significant increase in cAMP levels (p<0.01) in the plasma of FO group (Fig. 1H), which is a downstream target upon β3-adrenergic receptor (ADRB3) activation, thereby regulating lipolysis and UCP1-mediated thermogenic responses [32].
3.2. Maternal n-3 PUFA supplementation promoted the fetal BAT development
Despite there were no differences in iBAT mass, maternal n-3 PUFA intake was linked with a darker in color in the picture of BAT taken upon sacrifice (Fig 2A), and lower lipid accumulation in the H&E staining image (Fig 2B). Similar to the results from red blood cells, maternal FO intake significantly altered the lipid profile in the iBAT with a 5-fold reduction in n-6/n-3 ratio and a 2-fold decrease in ARA content compared to control (Fig 2C). To identify the effect of maternal n-3 PUFA supplementation on BAT activities, we measured the brown-specific gene expression levels in the iBAT. There was a significant increase in Ucp1, Cidea, Prdm16, and Pgc1α, along with the Gpr120, a membrane receptor of long chain fatty acid receptor in both gender pups (Fig. 2D-F). In accordance with the increased brown- specific transcripts, protein expressions of UCP1, PRDM16, and GRP120 were higher in the maternal FO-fed group (Fig 2G). The mitochondrial DNA content showed a trend towards an increase in the maternal n-3 PUFA group, but it was not statistically significant (p=0.08) (Fig 2H).
Figure 2.
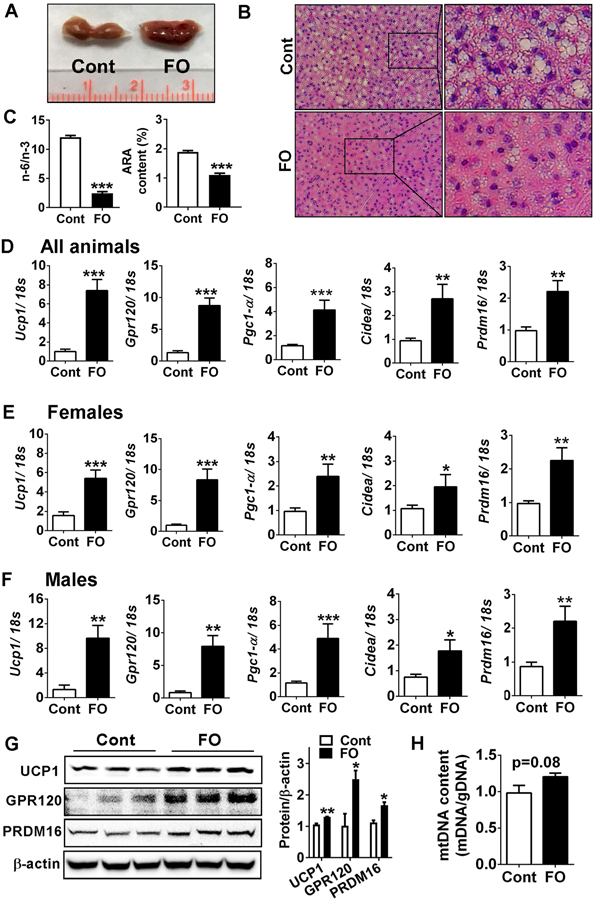
Maternal n-3 PUFA supplementation enhanced BAT development in offspring at the time of weaning. A. Gross image of iBAT without (Cont) or with (FO) maternal n-3 PUFA supplementation. B. Representative microscopic images with H&E staining of iBAT. C. n-6/n-3 PUFA ratio and arachidonic acid (ARA) content (%) in the iBAT (left) by GC analysis. D-F. Brown signature gene expressions of Ucp1, Gpr120, Pgc1-α, Cidea, and Prdm16 in all pups (D), female pups (E) and male pups (F) by qPCR. G. Western blot pattern of UCP1, GPR120, and PRDM16 in the iBAT (left). Relative protein intensities to β-actin quantified by Image J (right). H. Relative mitochondrial DNA to genomic DNA in the iBAT (n=6). All data are shown as mean ± SEM. *p < 0.05, **p < 0.01 and ***p < 0.001 by Student’s t-test.
3.3. Maternal n-3 PUFA supplementation modulated epigenetic factors for BAT developments
We have previously demonstrated that n-3 PUFA potentiate brown adipogenesis in the primary murine brown precursor cells via miRNA-mediated epigenetic mechanisms [28]. After confirming that maternal n-3 PUFA upregulated BAT transcriptional programs of the offspring (Fig 2), we sought to determine whether the similar epigenetic mechanisms are involved in this process. The functional cluster of miRNAs of miR-30b, miR-193b, and miR-365 was significantly increased in maternal FO received pups (Fig 3A), recapitulating our in vitro results [28]. Intriguingly, the BAT with maternal n-3 PUFA exposure showed a substantial increase of Drosha, an RNA double-strand ribonuclease and critical component of the microprocessor for initiating the cleavage of pri-miRNA into stem-loop pre-miRNA (Fig 3B), in both gene and protein expression (Fig 3C, D). These results suggest that nuclear processing of pri-miRNA might be upregulated by early exposure of FO during fetal BAT development.
Figure 3.
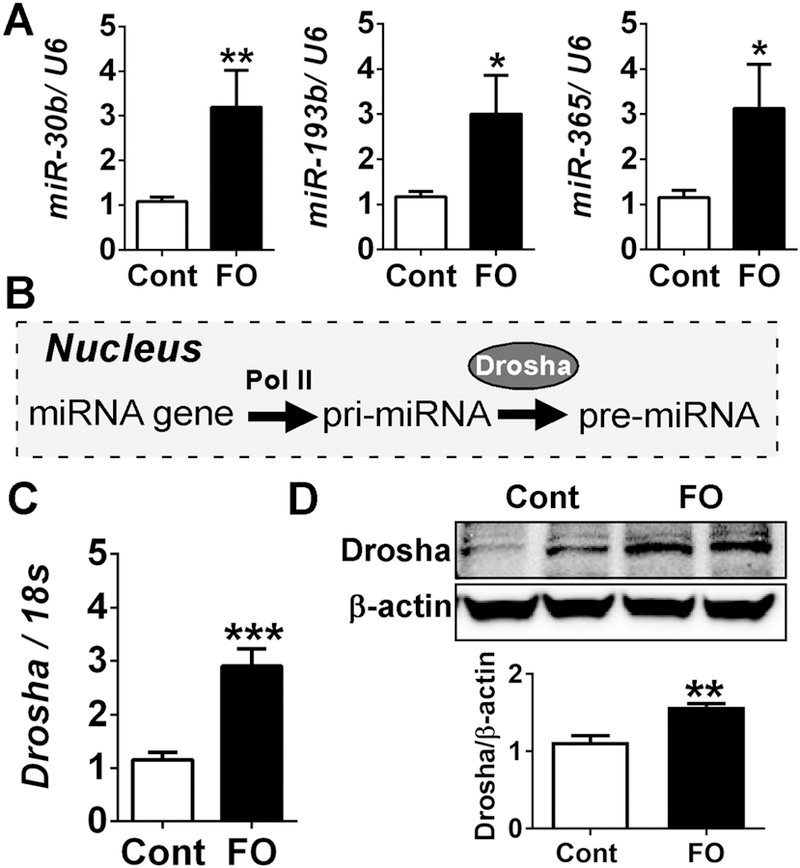
Maternal n-3 PUFA supplementation promoted brown adipogenesis via miRNA biogenesis at the time of weaning. A. qPCR analysis of miRNAs of miR-30b, 193b, and 365 in all pups (n=18), B. Schematic diagram of the nuclear processing of pri-miRNA by Drosha. C. Drosha mRNA expression (n=8) by qPCR, D. Protein levels of Drosha in iBAT (upper), and its relative intensity to β-actin by Image J (lower). All data are shown as mean ± SEM. *p < 0.05, **p < 0.01 and ***p < 0.001 by Student’s t-test.
Next, we examined the role of maternal n-3 PUFA intake on histone modifications. There was a remarkable decrease in histone deacetylase 1 (HDAC1). The reduced HADC1 levels were linked with an increase of acetylation (Ac), but a decrease of tri-methylation (me3), at histone H3 lysine 27 (H3K27) site (Fig 4A), a critical post-translational modifications (PTM) mark for brown adipogenesis[22]. The decrease H3K27me3 was consistent with decreased gene expression of JmjC domain-containing protein 3 (Jmjd3), an H3K27-specific demethylase [33], in FO group compared to control (Fig 4B). Intriguingly, no specific increase in acetylation levels at H3K9, another critical regulatory site for brown adipogenesis. However, there was a distinct increase in methylation status (H3K9me2) with maternal n-3 PFUA intake (Fig 4A). Consistently, the expression of euchromatic histone N-lysine methyltransferase 1 (Ehmt1), a methyltransferase to H3K9[34], was significantly increased without altering expression levels of Jumonji Domain Containing 1A (Jmjd1a, same with Kdm3a or Jmdm2a), an H3K9-specific demethylase (Fig 4B). These site-specific epigenetic modulations by maternal n-3 PUFA were linked with increased brown adipocyte differentiation evidenced by increased protein expression of PPARγ, fatty acid synthase (FAS) and fatty acid binding protein aP2 in the iBAT compared to control (Fig 4C).
Figure 4.
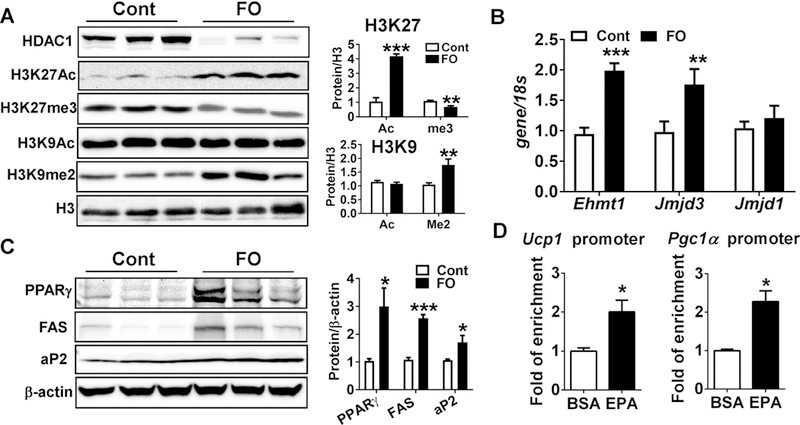
Maternal n-3 PUFA supplementation promoted brown adipogenesis via histone modifications at the time of weaning. A. Western blot pattern of epigenetic markers of HDAC1, H3K27Ac, H3K27Me3, H3K9Ac, H3K9me2 (left). Image J was used to determine the relative status of histone acetylation and methylation at the H3K27 and H3K9 (right). B. qPCR analysis of epigenetic enzymes of histone methyltransferase Ehmt1, and demethylases of Jmjd3 and Jmjd1. C. Western blot pattern of adipocyte proteins of PPARγ, FAS, and aP2 (left). The membrane intensity was quantified by Image J (right). D. Enrichment of H3K27Ac at the promoter region of Ucp1 and Pgc1α by ChIP assay in the HIB1B cells that were differentiated with either vehicle (BSA) or EPA (n=4). All data are expressed as mean ± SEM. *p< 0.05, **p< 0.01, ***p<0.001 by Student’s t-test.
To further ascertain whether the modulation of H3K27Ac occurs at the promoter/enhancer region of Ucp1 and Pgc1α we conducted chromatin immunoprecipitation (ChIP) assay by pulling down the DNA with the antibody targeting to H3K27Ac in the murine HIB1B brown adipocyte cell lines that are treated with or without EPA treatment (100 μM). There was more than a 2-fold enrichment at H3K27Ac in the n-3 PUFA treated group compared with the control both in the Ucp1 enhancer region and Pgc1α-cAMP response element (CRE) binding region (Fig 4D). Taken together, these results suggest that maternal FO exposure alters the epigenetic signature of histone acetylation and methylation and miRNA abundance in the fetal BAT, thereby enhancing brown-specific transcriptional program.
3.4. Maternal n-3 PUFA supplementation conferred long-lasting thermogenic benefits to offspring
To gain an insight as to whether augmented BAT activity by maternal FO intake exerts extended metabolic benefits, we conducted additional experiments. The weaned mice with or without maternal n- 3 PUFA intake were kept in the same diet without additional n-3 PUFA until they were used for measurement of energy expenditure (at 5-week of age) and cold resistance (at 11-week of age) (Fig 5A). The pups that received maternal n-3 PUFA showed significantly higher (p=0.04) energy expenditure especially at night (Fig 5B). There were no significant differences in RER (p=0.3), implicating no difference in the substrate use between groups (Fig 5C). Subsequently, upon mice reaching 11 weeks of age (8 weeks after maternal nutrition), mice were subjected to acute cold treatment (6°C), and rectal temperature was recorded. At the 3-hour time point, core body temperature was significantly higher in the maternal n-3 PUFA fed group compared to control group (Fig 5D). Additionally, under the IR camera, pups from maternal FO group presented a higher heat release on the surface than control mice (Fig 5E). After 24-hour cold exposure, adipocyte morphology was visualized by H&E staining. In iBAT, fat accumulation was lower in maternal FO-received mice than Cont, indicating the higher BAT activity (Fig 5F). Similarly, cold exposure induced the massive brown-like adipocyte morphology in the inguinal WAT (iWAT) in the FO group, but not in the Cont group (Fig 5F). Consistently, UCP1 protein expression was higher in both iBAT (2-fold) and iWAT (3.5-fold) of the FO group than control (Fig 5G). The brown-specific gene expression levels, i.e., Ucp1, Pgc1α and Prdm16, were maintained higher in the iBAT of the FO group than control (Fig 5H). In addition, the browning related gene expressions of Ucp1, Pparγ, and Pgc1α were significantly higher in the iWAT of FO group than the control (Fig 5I). However, the transcript levels of sarcoplasmic/endoplasmic reticulum calcium-ATPase (Serca2b), a responsive gene for calcium-dependent thermogenesis[35], was not different between groups (Fig 5I). In addition, the two gene expressions that are responsible for another UCP1-independent thermogenic program via creatine-driven substrate cycle, i.e., Ckmt1, Ckmt2, were not detectable in both groups (data not shown). These results suggest that maternal FO supplementation not only maintains stronger thermogenic potential in the iBAT but also links with enhanced adaptive thermogenesis (UCP1-dependent) in the iWAT. Taken together, augmented BAT formation by maternal FO may also mediate extended metabolic benefits in later life.
4. Discussion
Emerging evidence suggests that n-3 PUFA potentiate thermogenic programs in brown and beige adipocytes [26, 36, 37]. Although pregnancy is a critical time frame for BAT development [8], it is largely unknown whether the availability of n-3 PUFA to the fetus could modulate embryonic BAT development. This study is designed to identify the effects of maternal n-3 PUFA intake on fetal BAT development, potential mechanisms, and long-term metabolic consequences. Here, we demonstrated that maternal n-3 PUFA intake altered the neonates’ FA profiles by reducing the n-6/n-3 PUFA ratio in the neonates, suggesting that n-3 PUFA is effectively delivered from mothers to the pups (Fig 1). The maternal n-3 PUFA intake (3%) was sufficient to potentiate transcriptional programming of the brown adipogenesis (Fig 2) via mechanisms involved in miRNA production (Fig 3) and histone modification (Fig 4). Furthermore, the augmented thermogenic activities endowed offspring with long-lasting metabolic benefits (Fig 5). Based on these results, we proposed that maternal n-3 PUFA intake is a molecular driver in promoting epigenetic liability of brown differentiation via a two-pronged mechanism through brown-specific miRNA biogenesis and histone modulation (Fig 6). To our knowledge, it is the first study to evaluate the potential of prenatal BAT as a new and feasible therapeutic target to promote thermogenic energy expenditure via maternal dietary intervention.
Figure 6.
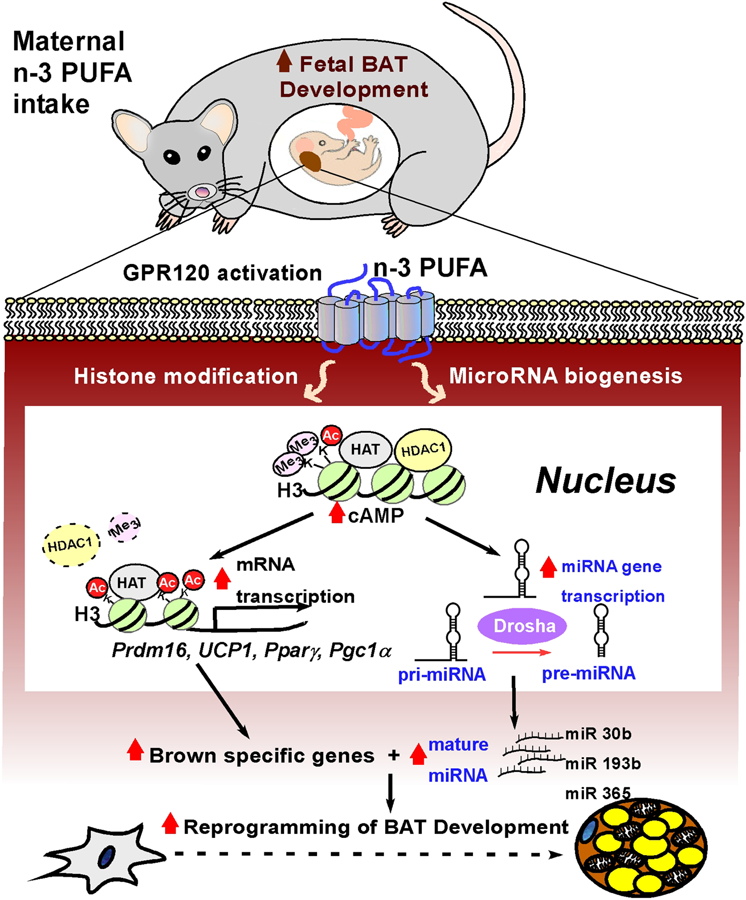
The proposed mechanism by which maternal n-3 PUFA supplementation potentiates fetal BAT development via epigenetic modulation. Maternal n-3 PUFA intake provides n-3 PUFA to the fetus across the placenta. In fetus, activation of GPR120, a membrane sensor for n-3 PUFA, triggers at least two epigenetic modulations, histone modifications, and miRNA biogenesis. Fetal availability of n-3 PUFA increases cAMP levels and alters PTM marks of H3K27Ac and H3K9me2, presumably with modulation of epigenetic enzymes including reduced HDAC1, resulting in enhanced transcriptional activation for brown specific genes, i.e., Ucp1, Prdm16, Pparγ, and Pgc1α. The augmented transcriptome by histone acetylation concurrently increases miRNA-gene transcription and Drosha expression, leading to enhanced nuclear pri-miRNA processing and mature miRNA biogenesis. With this concerted action of histone modification and miRNA regulation, early exposure to n-3 PUFA potentiates transcriptional program of the fetal BAT, which may mediate to long-lasting metabolic benefits in later life.
Regulation of fetal BAT development by maternal n-3 PUFA intake
The health benefits of maternal n-3 PUFA intake on offspring have received extensive attention [35, 38, 39]. Since the synthesis of n-3 PUFA in the fetus side is limited, the transfer of n-3 PUFA cross the placenta from the mother to the fetus is vital and necessary when it comes to maternal nutrition [40]. The maternal supplementation of n-3 PUFA increases the n-3 PUFA concentration in placenta and uterus cord blood, suggesting that maternal n-3 PUFA is effectively transferred to fetus prenatally [41]. Despite the multiple metabolic benefits of maternal n-3 PUFA intake to the offspring [42–45], the effects of maternal n-3 PUFA intake on modulating the fetal BAT development and thermogenic activities remain elusive. Indeed, per our knowledge, our study is the first to demonstrate maternal n-3 PUFA supplementation decreases n-6/n-3 ratio in the neonatal BAT (Fig 2C). Our results also provide scientific evidence that maternal n-3 PUFA supplementation promoted brown adipogenesis-related gene expressions including Ucp1, Pgc1α, Cidea, and Prdm16 (Fig 2). It is notable that activation of brown- specific genes in the iBAT relies on GPR120, a membrane sensor for n-3 PUFA. There was a robust increase in Gpr120 mRNA expression in iBAT in response to the maternal supply of n-3 PUFA in both genders. These results align with our previous finding that EPA-induced brown adipogenesis is dependent on GPR120 [28]. It is also compatible with the literature showing that knockout of GPR120 or treatment of GPR120 antagonist dampened the thermogenic activities [36]. Consistent with this notion, genetic ablation of GPR120 in mice results in hepatic steatosis and insulin resistance [46]. In humans, lack of GPR120 signaling activity due to a genetic mutation in the GPR120 gene (p.R2700H) is correlated with increased risk of obesity [46].
Intriguingly, augmented BAT activity by maternal n-3 PUFA was sustained for at least 8 weeks post-weaning (Fig 5); the pups received the maternal n-3 PUFA supplementation not only maintained the higher BAT activities in the iBAT but also exerted higher adaptive thermogenesis in the iWAT (Fig 5). Our results are consistent with the recent report that low perinatal n-6/n-3 ratio is important to attenuate susceptibility against diet-induced obesity in adult offspring [47]. Human studies also support an inverse correlation between maternal n-3 PUFA intake and prevalence of childhood obesity. Specifically, a higher risk of childhood obesity was correlated with a higher maternal n-6/n-3 fatty acid diet ratio, while maternal n-3 PUFA intake was linked with a lean body fat mass of children [48–51]. In contrast, there exist some inconsistent human studies that fail to observe the negative correlation between maternal n-3 PUFA and adiposity of the offspring [52–54]. These studies might be compounded by the variables such as supplementation periods, dose, and the ratio of DHA to EPA. Future human clinical trials with the long-term follow-up research design are warranted to establish the effect of maternal n-3 PUFA on thermogenesis and energy expenditure at the time of birth, childhood and adolescence.
Epigenetic regulation of BAT by early exposure to maternal n-3 PUFA
In terms of the underlying mechanism, we propose that maternal n-3 PUFA intake promotes brown transcriptional program of neonatal BAT via the synergistic action of miRNAs and histone modifications.
MiRNAs are short non-coding RNAs associated with post-transcriptional regulation of gene expression, which play a critical role in BAT differentiation [20, 55–57]. The blockage of miRNA biogenesis by adipocyte-specific deletion of dicer significantly impairs BAT development [58]. The miR-193b/365 was first identified as a brown-specific miRNA cluster to promote brown adipogenesis by suppressing the myogeneic-lineage differentiation in the Myf5+ precursor cells[59]. Also, miR-30b was reported to induce brown-specific gene expressions and increased mitochondrial respiration through degradation of the transcriptional corepressor receptor-interacting protein 140 (Rip140) [28, 60]. Previously, we demonstrated the cell-autonomous function of n-3 PUFA in promoting brown adipogenesis in the primary brown precursor cells, leading to an increase of clusters of brown-specific miRNAs including miR-30b and miR-193b/365 [28]. In agreement with our in vitro study, n-3 PUFA exposure during pregnancy recapitulated the involvement of the same profile of miRNAs including miR-30b, and miR-193/365 cluster (Fig 3A). Interestingly, augmented BAT thermogenesis is correlated with an increase of the Drosha gene and protein expression (Fig 3C, D). Drosha is a ribonuclease and a critical component of the microprocessor for initiating the cleavage of pri-miRNA into stem-loop pre-miRNA [61]. It is conceivable that n-3 PUFA availability may alter microprocessor activity resulting in pre-miRNA biogenesis in the iBAT, thereby augmenting the microbiome of iBAT. In supporting this idea, deletion of fat-specific Dgcr8, a subunit of the microprocessor complex that recognizes and mediates the processing of pri-miRNA into pre-miRNA, resulted in defective BAT formation and severe cold intolerance [62]. We are currently investigating whether maternal n-3 PUFA intake alters the miRNA processor activities, thereby increasing the pri-miRNAs processing rate in the nucleus.
Histone modifications represent another important mode of epigenetic regulation of transcription in directing cell-lineage specification and tissue-specific gene expression. Accumulating evidence suggests that BAT development is governed by dynamic changes in post-translation modification via acetylation and methylation on the N-terminal tail region of histones[22]. In general, histone acetylation of lysine (K) side chains decreases positive changes and reduces chromatin compactness, thereby contributing to transcriptional activation. Histone acetylation is regulated by the balance between histone acetyltransferase (HAT) and histone deacetylases (HDACs). Jin et al. showed that Gcn5/PCAF, a HAT enzyme, facilitates brown adipogenesis [63]. The inhibition of HDAC1 [30] or HDAC9 is linked with transcriptional activation of brown adipogenesis [32, 39]. In contrast, deacetylase activity of HDAC3 is required for thermogenic adipose program [64]. Histone methylation occurs on lysine (K) or arginine (R) residue, thereby altering the DNA accessibility to the binding of chromatin modifiers and transcription factors or epigenetic readers. It is important to note that site-specific H3K9 methylation by G9a represses brown differentiation [38], while methyl transfer by EHMT1 promotes BAT-selective thermogenic program [34]. Controversially, deletion of demethylase JMJD1a, demethylase at H3K9, is linked with obesity [65, 66]. Another critical epigenetic site for histone methylation is H3K27. The reduction of H3K27me3 by demethylase enzymes of UTX [30] or JMJD3 [33] is essential for brown adipogenesis. Recently, the dynamic interplay between histone acetylation and methylation at H3K27was revealed during brown adipogenesis. The β3-adrenergic stimulation induced dissociation of HDAC1, resulting in increased H3K27 acetylation followed by H3K27me3 demethylation thus allowing transcription in the isolated brown precursor cells [30].
Most aforementioned epigenetic histone modifications during brown differentiation were derived from the animals or cells with specific deletions of epigenetic writers (enzymes that catalyze the addition of epigenetic marks) or erasers (enzymes that catalyze the removal of epigenetic marks). To our surprise, little information is available about how environmental factors such as diet modify epigenetic markers for brown adipogenesis. The transcriptional activation of the fetal brown thermogenic program by n-3 PUFA were linked with altered PTMs, i.e., enhanced H3K27Ac and diminished H3K27me3, through the coordinated modulation of epigenetic erasers between HDAC1 and JMJD3. In addition, the enhanced H3K9me2 status by n-3 PUFA intake was associated with an increase of epigenetic writer EHMT1 without altering the demethylase JMJD1 (Fig 4A, B). These results unanimously suggest that prenatal n-3 PUFA exposure serves as an epigenetic modulator, thereby potentiating transcriptional brown thermogenic program.
Limitation and future studies
One caveat of our study design is that we cannot distinguish the effects of prenatal n-3 PUFA exposure from the n-3 PUFA supply through lactation. Aware of this limitation, we plan to conduct new experiments to address the separate role of prenatal BAT development and lactation by collecting BAT at birth or swapping the pups during the lactation period. To ascertain the long-term metabolic benefits, we are currently investigating whether maternal n-3 PUFA intake confers the resistance against high fat diet- induced obesity and insulin resistance in adulthood. Despite numerous indications in experimental animals, our current understanding regarding the thermogenic function of n-3 PUFA in humans is inconsistent [47–54]. In particular, the role of maternal n-3 PUFA supplementation on thermogenicactivities in offspring, presumably through epigenetic modifications, has yet to be established. As pregnant women are recommended to take approximately 600~800 mg of n-3 PUFA daily during pregnancy [67, 68], the exact role of n-3 PUFA on fetal BAT development and its long-term metabolic benefits through enhanced brown thermogenic potential should be addressed in humans. Here, we presented initial evidence that fetal BAT development could be a feasible therapeutic target to attenuate childhood obesity via maternal n-3 PUFA supplementation.
Supplementary Material
Highlights.
Maternal n-3 PUFA intake potentiates the transcriptional program of the fetal BAT.
Maternal n-3 PUFA intake promotes the brown-specific miRNA biogenesis.
Maternal n-3 PUFA intake alters site-specific histone PTM marks in the fetal BAT (i.e., H3K27Ac and H3K9me2).
Maternal n-3 PUFA intake increases energy expenditure in offspring
Maternal n-3 PUFA intake may result in long-lasting metabolic benefits in offspring.
Acknowledgments
This work was supported in part by National Institutes of Health, Grant 1P20GM104320 (Project 5 to S.C.). It is also supported by Research Council Faculty seed grant at the University of Nebraska, and Nebraska EPSCoR Food for Health Initiative Grant (awarded to S.C). R.F. received Inaugural Robert and Leslie Lewinter-Suskind Pediatric Nutrition Student Travel Award to attend Nutrition 2018 conference.
Abbreviations used:
- ARA
arachidonic acid
- Cidea
cell death-inducing DNA fragmentation factor α- like effector A
- DHA
docosahexaenoic acids
- EHMT1
euchromatic histone N-lysine methyltransferase 1
- EPA
eicosapentaenoic acids
- eWAT
epididymal white adipose tissue
- FO
fish oil
- GPR120
G- protein-coupled receptor 120
- HF
high fat
- iBAT
interscapular brown adipose tissue
- iWAT
inguinal white adipose tissue
- Jmjd1
JmjC domain-containing protein 1
- Jmjd3
JmjC domain-containing protein 3
- miRNAs
microRNAs
- Prdm16
PR domain containing 16
- n-3 PUFA
omega-3 polyunsaturated fatty acids
- Pgc1-α
peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- PTM
post- translational modifications
- sWAT
subcutaneous white adipose tissue
- UCP1
uncoupling protein 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflict of interest.
6. References
- [1].Harms M, Seale P, Brown and beige fat: development, function and therapeutic potential, Nature medicine, 19 (2013) 1252–1263. [DOI] [PubMed] [Google Scholar]
- [2].Gilsanz V, Hu HH, Kajimura S, Relevance of brown adipose tissue in infancy and adolescence, Pediatric research, 73 (2013) 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cypess AM, Kahn CR, The role and importance of brown adipose tissue in energy homeostasis, Current opinion in pediatrics, 22 (2010) 478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, Kolodny GM, Kahn CR, Identification and importance of brown adipose tissue in adult humans, The New England journal of medicine, 360 (2009) 1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chechi K, Nedergaard J, Richard D, Brown adipose tissue as an anti-obesity tissue in humans, Obesity reviews : an official journal of the International Association for the Study of Obesity, 15 (2014) 92–106. [DOI] [PubMed] [Google Scholar]
- [6].Merkestein M, Cagampang FR, Sellayah D, Fetal programming of adipose tissue function: an evolutionary perspective, Mammalian genome, 25 (2014) 413–423. [DOI] [PubMed] [Google Scholar]
- [7].Symonds ME, Pope M, Budge H, Adipose tissue development during early life: novel insights into energy balance from small and large mammals, Proceedings of the Nutrition Society, 71 (2012) 363– 370. [DOI] [PubMed] [Google Scholar]
- [8].Symonds ME, Pope M, Sharkey D, Budge H, Adipose tissue and fetal programming, Diabetologia, 55 (2012) 1597–1606. [DOI] [PubMed] [Google Scholar]
- [9].Lee P, Werner CD, Kebebew E, Celi FS, Functional thermogenic beige adipogenesis is inducible in human neck fat, International journal of obesity (2005), 38 (2014) 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zingaretti MC, Crosta F, Vitali A, Guerrieri M, Frontini A, Cannon B, Nedergaard J, Cinti S, The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue, FASEB J, 23 (2009) 3113–3120. [DOI] [PubMed] [Google Scholar]
- [11].Cypess AM, White AP, Vernochet C, Schulz TJ, Xue R, Sass CA, Huang TL, Roberts- Toler C, Weiner LS, Sze C, Chacko AT, Deschamps LN, Herder LM, Truchan N, Glasgow AL, Holman AR, Gavrila A, Hasselgren PO, Mori MA, Molla M, Tseng YH, Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat, Nature medicine, 19 (2013) 635–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Porter C, Herndon DN, Chondronikola M, Chao T, Annamalai P, Bhattarai N, Saraf MK, Capek KD, Reidy PT, Daquinag AC, Kolonin MG, Rasmussen BB, Borsheim E, Toliver- Kinsky T, Sidossis LS, Human and Mouse Brown Adipose Tissue Mitochondria Have Comparable UCP1 Function, Cell metabolism, 24 (2016) 246–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Leitner BP, Huang S, Brychta RJ, Duckworth CJ, Baskin AS, McGehee S, Tal I, Dieckmann W, Gupta G, Kolodny GM, Pacak K, Herscovitch P, Cypess AM, Chen KY, Mapping of human brown adipose tissue in lean and obese young men, Proceedings of the National Academy of Sciences of the United States of America, 114 (2017) 8649–8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang F, Hao G, Shao M, Nham K, An Y, Wang Q, Zhu Y, Kusminski CM, Hassan G, Gupta RK, Zhai Q, Sun X, Scherer PE, Oz OK, An Adipose Tissue Atlas: An Image-Guided Identification of Human-like BAT and Beige Depots in Rodents, Cell metabolism, 27 (2018) 252– 262.e253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Symonds ME, Pope M, Budge H, The ontogeny of brown adipose tissue, Annual review of nutrition, 35 (2015) 295–320. [DOI] [PubMed] [Google Scholar]
- [16].Symonds ME, Brown adipose tissue growth and development, Scientifica, 2013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Campos EI, Reinberg D, Histones: annotating chromatin, Annu. Rev. Genet, 43 (2009) 559–599. [DOI] [PubMed] [Google Scholar]
- [18].Fedorova E, Zink D, Nuclear architecture and gene regulation, Biochim. Biophys. Acta, 1783 (2008) 2174–2184. [DOI] [PubMed] [Google Scholar]
- [19].Handy DE, Castro R, Loscalzo J, Epigenetic modifications: basic mechanisms and role in cardiovascular disease, Circulation, 123 (2011) 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen Y, Pan R, Pfeifer A, Regulation of brown and beige fat by microRNAs, Pharmacology & therapeutics, 170 (2017) 1–7. [DOI] [PubMed] [Google Scholar]
- [21].Lecoutre S, Petrus P, Rydén M, Breton C, Transgenerational Epigenetic Mechanisms in Adipose Tissue Development, Trends in Endocrinology & Metabolism, (2018). [DOI] [PubMed]
- [22].Sambeat A, Gulyaeva O, Dempersmier J, Sul HS, Epigenetic regulation of the thermogenic adipose program, Trends in Endocrinology & Metabolism, 28 (2017) 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Okla M, Kim J, Koehler K, Chung S, Dietary Factors Promoting Brown and Beige Fat Development and Thermogenesis, Advances in nutrition (Bethesda, Md.), 8 (2017) 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bargut TC, Silva-e-Silva AC, Souza-Mello V, Mandarim-de-Lacerda CA, Aguila MB, Mice fed fish oil diet and upregulation of brown adipose tissue thermogenic markers, European journal of nutrition, 55 (2016) 159–169. [DOI] [PubMed] [Google Scholar]
- [25].Pahlavani M, Razafimanjato F, Ramalingam L, Kalupahana NS, Moussa H, Scoggin S, Moustaid-Moussa N, Eicosapentaenoic acid regulates brown adipose tissue metabolism in high-fat-fed mice and in clonal brown adipocytes, The Journal of nutritional biochemistry, 39 (2017) 101–109. [DOI] [PubMed] [Google Scholar]
- [26].Zhao M, Chen X, Eicosapentaenoic acid promotes thermogenic and fatty acid storage capacity in mouse subcutaneous adipocytes, Biochemical and biophysical research communications, 450 (2014) 1446–1451. [DOI] [PubMed] [Google Scholar]
- [27].Fan R, Koehler K, Chung S, Adaptive thermogenesis by dietary n-3 polyunsaturated fatty acids: Emerging evidence and mechanisms, Biochimica et biophysica acta, (2018). [DOI] [PMC free article] [PubMed]
- [28].Kim J, Okla M, Erickson A, Carr T, Natarajan SK, Chung S, Eicosapentaenoic Acid Potentiates Brown Thermogenesis through FFAR4-dependent Up-regulation of miR-30b and miR-378, The Journal of biological chemistry, 291 (2016) 20551–20562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Klein J, Fasshauer M, Klein HH, Benito M, Kahn CR, Novel adipocyte lines from brown fat: a model system for the study of differentiation, energy metabolism, and insulin action, Bioessays, 24 (2002) 382–388. [DOI] [PubMed] [Google Scholar]
- [30].Li F, Wu R, Cui X, Zha L, Yu L, Shi H, Xue B, Histone Deacetylase 1 (HDAC1) Negatively Regulates Thermogenic Program in Brown Adipocytes via Coordinated Regulation of Histone H3 Lysine 27 (H3K27) Deacetylation and Methylation, The Journal of biological chemistry, 291 (2016) 4523–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ng M, Fleming T, Gakidou E, Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013, Lancet (London, England), 384 (2014) 766–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Galmozzi A, Mitro N, Ferrari A, Gers E, Gilardi F, Godio C, Cermenati G, Gualerzi A, Donetti E, Rotili D, Inhibition of class I histone deacetylases unveils a mitochondrial signature and enhances oxidative metabolism in skeletal muscle and adipose tissue, diabetes, (2012) DB_120548. [DOI] [PMC free article] [PubMed]
- [33].Pan D, Huang L, Zhu LJ, Zou T, Ou J, Zhou W, Wang Y-X, Jmjd3-mediated H3K27me3 dynamics orchestrate brown fat development and regulate white fat plasticity, Developmental cell, 35 (2015) 568–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S, EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex, Nature, 504 (2013) 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Beckford RC, Howard SJ, Das S, Farmer AT, Campagna SR, Yu J, Hettich RL, Wilson JL, Voy BH, Maternal consumption of fish oil programs reduced adiposity in broiler chicks, Scientific reports, 7 (2017) 13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Quesada-Lopez T, Cereijo R, Turatsinze JV, Planavila A, Cairo M, Gavalda-Navarro A, Peyrou M, Moure R, Iglesias R, Giralt M, Eizirik DL, Villarroya F, The lipid sensor GPR120 promotes brown fat activation and FGF21 release from adipocytes, Nature communications, 7 (2016) 13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Laiglesia LM, Lorente-Cebrian S, Prieto-Hontoria PL, Fernandez-Galilea M, Ribeiro SM, Sainz N, Martinez JA, Moreno-Aliaga MJ, Eicosapentaenoic acid promotes mitochondrial biogenesis and beige-like features in subcutaneous adipocytes from overweight subjects, The Journal of nutritional biochemistry, 37 (2016) 76–82. [DOI] [PubMed] [Google Scholar]
- [38].Wang L, Xu S, Lee JE, Baldridge A, Grullon S, Peng W, Ge K, Histone H3K9 methyltransferase G9a represses PPARγ expression and adipogenesis, The EMBO journal, 32 (2013) 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Basford JE, Knoll E, Tong WS, Blanco V, Blomkalns AL, Rudich S, Lentsch AB, Hui DY, Weintraub NL, HDAC9 knockout mice are protected from adipose tissue dysfunction and systemic metabolic disease during high fat feeding, Diabetes, (2013) DB_131148. [DOI] [PMC free article] [PubMed]
- [40].Larque E, Krauss-Etschmann S, Campoy C, Hartl D, Linde J, Klingler M, Demmelmair H, Cano A, Gil A, Bondy B, Koletzko B, Docosahexaenoic acid supply in pregnancy affects placental expression of fatty acid transport proteins, The American journal of clinical nutrition, 84 (2006) 853– 861. [DOI] [PubMed] [Google Scholar]
- [41].Ikeda K, Kang Q, Yoneshiro T, Camporez JP, Maki H, Homma M, Shinoda K, Chen Y, Lu X, Maretich P, UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis, Nature medicine, 23 (2017) 1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sanchez Meza K, Tene Perez CE, Sanchez Ramirez CA, Muniz Valencia R, Del Toro Equihua M, Levels of eicosapentaenoic acid in obese schoolchildren with and without insulin resistance, Nutricion hospitalaria, 31 (2014) 1102–1108. [DOI] [PubMed] [Google Scholar]
- [43].Kiecolt-Glaser JK, Belury MA, Andridge R, Malarkey WB, Hwang BS, Glaser R, Omega-3 supplementation lowers inflammation in healthy middle-aged and older adults: a randomized controlled trial, Brain, behavior, and immunity, 26 (2012) 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mori TA, Burke V, Puddey IB, Watts GF, O’Neal DN, Best JD, Beilin LJ,Purified eicosapentaenoic and docosahexaenoic acids have differential effects on serum lipids and lipoproteins, LDL particle size, glucose, and insulin in mildly hyperlipidemic men, The American journal of clinical nutrition, 71 (2000) 1085–1094. [DOI] [PubMed] [Google Scholar]
- [45].Itariu BK, Zeyda M, Hochbrugger EE, Neuhofer A, Prager G, Schindler K, Bohdjalian A, Mascher D, Vangala S, Schranz M, Krebs M, Bischof MG, Stulnig TM, Long-chain n-3 PUFAs reduce adipose tissue and systemic inflammation in severely obese nondiabetic patients: a randomized controlled trial, The American journal of clinical nutrition, 96 (2012) 1137–1149. [DOI] [PubMed] [Google Scholar]
- [46].Ichimura A, Hirasawa A, Poulain-Godefroy O, Bonnefond A, Hara T, Yengo L, Kimura I, Leloire A, Liu N, Iida K, Dysfunction of lipid sensor GPR120 leads to obesity in both mouse and human, (2012). [DOI] [PubMed]
- [47].Rudolph MC, Jackman MR, Presby DM, Houck JA, Webb PG, Johnson GC, Soderborg TK, de la Houssaye BA, Yang IV, Friedman JE, MacLean PS, Low Neonatal Plasma N-6/N-3 Pufa Ratios Regulate Offspring Adipogenic Potential and Condition Adult Obesity Resistance, Diabetes, (2017). [DOI] [PMC free article] [PubMed]
- [48].Vidakovic AJ, Gishti O, Voortman T, Felix JF, Williams MA, Hofman A, Demmelmair H, Koletzko B, Tiemeier H, Jaddoe VW, Gaillard R, Maternal plasma PUFA concentrations during pregnancy and childhood adiposity: the Generation R Study, Am J Clin Nutr, 103 (2016) 1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Donahue SM, Rifas-Shiman SL, Gold DR, Jouni ZE, Gillman MW, Oken E, Prenatal fatty acid status and child adiposity at age 3 y: results from a US pregnancy cohort, The American journal of clinical nutrition, 93 (2011) 780–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Moon RJ, Harvey NC, Robinson SM, Ntani G, Davies JH, Inskip HM, Godfrey KM, Dennison EM, Calder PC, Cooper C, Maternal plasma polyunsaturated fatty acid status in late pregnancy is associated with offspring body composition in childhood, J Clin Endocrinol Metab, 98 (2013) 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Foster BA, Escaname E, Powell TL, Larsen B, Siddiqui SK, Menchaca J, Aquino C, Ramamurthy R, Hale DE, Randomized Controlled Trial of DHA Supplementation during Pregnancy: Child Adiposity Outcomes, Nutrients, 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Muhlhausler BS, Yelland LN, McDermott R, Tapsell L, McPhee A, Gibson RA, Makrides M, DHA supplementation during pregnancy does not reduce BMI or body fat mass in children: follow- up of the DHA to Optimize Mother Infant Outcome randomized controlled trial, 2, The American journal of clinical nutrition, 103 (2016) 1489–1496. [DOI] [PubMed] [Google Scholar]
- [53].Meldrum S, Dunstan JA, Foster JK, Simmer K, Prescott SL, Maternal fish oil supplementation in pregnancy: a 12 year follow-up of a randomised controlled trial, Nutrients, 7 (2015) 2061–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Rytter D, Bech BH, Halldorsson T, Christensen JH, Schmidt EB, Danielsen I, Henriksen TB, Olsen SF, No association between the intake of marine n-3 PUFA during the second trimester of pregnancy and factors associated with cardiometabolic risk in the 20-year-old offspring, The British journal of nutrition, 110 (2013) 2037–2046. [DOI] [PubMed] [Google Scholar]
- [55].Trajkovski M, Lodish H, MicroRNA networks regulate development of brown adipocytes, Trends in endocrinology and metabolism: TEM, 24 (2013) 442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Arner P, Kulyte A, MicroRNA regulatory networks in human adipose tissue and obesity, Nature reviews. Endocrinology, 11 (2015) 276–288. [DOI] [PubMed] [Google Scholar]
- [57].Karbiener M, Scheideler M, MicroRNA Functions in Brite/Brown Fat - Novel Perspectives towards Anti-Obesity Strategies, Computational and structural biotechnology journal, 11 (2014) 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Mori MA, Thomou T, Boucher J, Lee KY, Lallukka S, Kim JK, Torriani M, Yki-Järvinen H, Grinspoon SK, Cypess AM, Altered miRNA processing disrupts brown/white adipocyte determination and associates with lipodystrophy, The Journal of clinical investigation, 124 (2014) 3339–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sun L, Xie H, Mori MA, Alexander R, Yuan B, Hattangadi SM, Liu Q, Kahn CR, Lodish HF, Mir193b–365 is essential for brown fat differentiation, Nature cell biology, 13 (2011) 958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hu F, Wang M, Xiao T, Yin B, He L, Meng W, Dong M, Liu F, miR-30 promotes thermogenesis and the development of beige fat by targeting RIP140, Diabetes, 64 (2015) 2056– 2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Ha M, Kim VN, Regulation of microRNA biogenesis, Nat Rev Mol Cell Biol, 15 (2014) 509–524 [DOI] [PubMed] [Google Scholar]
- [62].Kim H-J, Cho H, Alexander R, Patterson HC, Gu M, Lo KA, Xu D, Goh VJ, Nguyen LN, Chai X, MicroRNAs are required for the feature maintenance and differentiation of brown adipocytes, Diabetes, 63 (2014) 4045–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Jin Q, Wang C, Kuang X, Feng X, Sartorelli V, Ying H, Ge K, Dent SY, Gcn5 and PCAF regulate PPARγ and Prdm16 expression to facilitate brown adipogenesis, Molecular and cellular biology, (2014) MCB. 00622–00614. [DOI] [PMC free article] [PubMed]
- [64].Emmett MJ, Lim H-W, Jager J, Richter HJ, Adlanmerini M, Peed LC, Briggs ER, Steger DJ, Ma T, Sims CA, Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge, Nature, 546 (2017) 544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Tateishi K, Okada Y, Kallin EM, Zhang Y, Role of Jhdm2a in regulating metabolic gene expression and obesity resistance, Nature, 458 (2009) 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Inagaki T, Tachibana M, Magoori K, Kudo H, Tanaka T, Okamura M, Naito M, Kodama T, Shinkai Y, Sakai J, Obesity and metabolic syndrome in histone demethylase JHDM2a-deficient mice, Genes to Cells, 14 (2009) 991–1001. [DOI] [PubMed] [Google Scholar]
- [67].Dietary Guidelines for Americans, in, Government Printing Office, Washington, DC, U.S., 2010. [Google Scholar]
- [68].Swanson D, Block R, Mousa SA, Omega-3 fatty acids EPA and DHA: health benefits throughout life, Advances in nutrition, 3 (2012) 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


