Abstract
Background
Heat stress is a severe environmental stress that affects plant growth and reduces yield. Bax inhibitor-1 (BI-1) is a cytoprotective protein that is involved in the response to biotic and abiotic stresses. The Arabidopsis (Arabidopsis thaliana) BI-1 mutants atbi1–1 and atbi1–2 are hypersensitive to heat stress, and AtBI-1 overexpression rescues thermotolerance deficiency in atbi1 plants. Nevertheless, the mechanism of BI-1 in plant thermotolerance is still unclear.
Results
We identified a wheat (Triticum aestivum L.) BI-1 gene, TaBI-1.1, which was highly upregulated in an RNA sequencing (RNA-seq) analysis of heat-treated wheat. The upregulation of TaBI-1.1 under heat stress was further demonstrated by real time quantitative PCR (qRT-PCR) and β-glucuronidase (GUS) staining. Compared with the wild type Col-0, the atbi1–2 mutant is hypersensitive to heat stress, and constitutive expression of TaBI-1.1 in atbi1–2 (35S::TaBI-1.1/ atbi1–2) rescued the deficiency of atbi1–2 under heat stress. Furthermore, we identified TaFKBP62 as a TaBI-1.1-interacting protein that co-localized with TaBI-1.1 on the endoplasmic reticulum (ER) membrane and enhanced heat stress tolerance. Additionally, HSFA2, HSFB1, ROF1, HSP17.4B, HSP17.6A, HSP17.8, HSP70B, and HSP90.1 expression levels were suppressed in atbi1–2 plants under heat stress. In contrast, 35S::TaBI-1.1/atbi1–2 relieved the inhibitory effect of AtBI-1 loss of function.
Conclusions
TaBI-1.1 interacted with TaFKBP62 and co-localized with TaFKBP62 on the ER membrane. Both TaBI-1.1 and AtBI-1 regulated the expression of heat-responsive genes and were conserved in plant thermotolerance.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1485-0) contains supplementary material, which is available to authorized users.
Keywords: Bax inhibitor-1, Heat stress, TaFKBP62, RNA-seq, Heat-responsive genes
Background
Heat stress is a major environmental factor that affects almost all aspects of plant growth, development, reproduction, and yield. High temperature affects the stability of multiple proteins and cytoskeletal structures and alters the efficiency of enzymatic reactions, and exposure to sustained high temperature can even result in plant death [1].
Plants have evolved a variety of mechanisms to resist heat stress, including the maintenance of membrane stability, ROS scavenging, antioxidant production, the accumulation and adjustment of compatible solutes, the induction of protein kinase cascades, chaperone signalling, and transcriptional activation [2]. Heat shock proteins (HSPs) are important components of the response to heat stress that can improve photosynthesis, assimilate partitioning, water and nutrient use efficiency, and membrane stability [3–5]. HSPs are induced through the action of heat-stress transcription factors (HSFs), which mediate the synthesis of HSPs by binding to heat-stress elements in the promoters of heat-responsive genes.
Bax inhibitor-1 (BI-1) is a cell death suppressor that is conserved in plants and mammals [6, 7]. BI-1 was first identified in mammals but was functionally verified in yeast and shown to suppress Bax-induced cell death [8]. Programmed cell death (PCD) is a conserved process in eukaryotes induced by stress stimuli or during development [7]. Numerous studies have revealed that BI-1 inhibits cell death induced by biotic and abiotic stresses. Overexpressing Arabidopsis (Arabidopsis thaliana) AtBI-1 in tobacco BY-2 cells suppresses Bax-, H2O2- or salicylic acid (SA)-mediated cell death [9]. Overexpressing TaBI-1 in tobacco blocks Bax-induced cell death, and silencing TaBI-1 in wheat (Triticum aestivum L.) enhances the susceptibility to Puccinia striiformis [10]. HvBI-1 overexpression increases susceptibility to biotrophic Blumeria graminis f.sp. hordei but enhances resistance to Fusarium graminearum (Fg) and inhibits cell death induced by mammalian BAX expression [11]. Enhanced barley BI-1 expression suppresses penetration resistance to B. graminis f. sp. tritici [12]. AtBI-1 overexpression in sugarcane increases tolerance to long-term water deficit [13]. Recent studies suggest that BI-1 is involved in the response to heat stress. MrBI-1 partially rescues Bax-induced cell death in yeast, and deleting MrBI-1 impairs heat tolerance [14]. Pepper CaBI-1 is upregulated by high temperature [15]. Two AtBI-1 mutants (atbi1–1 and atbi1–2) exhibit hypersensitivity to tunicamycin (TM)-induced PCD progression and increased sensitivity to heat shock-induced cell death. In contrast, AtBI-1 overexpression rescues thermotolerance deficiency in atbi1 plant [16, 17]. However, the mechanism of BI-1 in thermotolerance is unclear.
FK506-binding proteins (FKBPs) are a superfamily of peptidyl prolyl cis-trans isomerases (PPIases) that are characterized by their enzymatic activity [18, 19]. The catalytic activity of FKBP is inhibited upon binding of the immunosuppressive drug FK506 [20]. FKBP family members are found in multiple subcellular locations, including the endoplasmic reticulum (ER) [21], cytosol [22], nucleus [23], and mitochondria [24]. FKBP isoforms are distinguished by their molecular weights, which range from 12 kDa to over 77 kDa [25–27]. Large FKBPs contain additional domains that enable them to function as chaperones, such as AtFKBP62 (ROF1) and AtFKBP65 (ROF2). ROF1 and ROF2 share similar domains and high sequence identity. These proteins contain three peptidylprolyl cis/trans isomerases (PPIase) domains, a tetratricopeptide repeat motif (TPR) domain, and a calmodulin-binding domain [28]. The first PPIase domain possesses PPIase activity and an FK506 binding site, while the remaining two PPIase-like domains exhibit only partial identity to the functional domain [27]. The TPR domain is necessary for ROF1 to interact with HSP90 [29]. The expression levels of both ROF1 and ROF2 increase under heat stress, but ROF2 is detected only after heat treatment [29]. ROF1 plays a role in prolonging thermotolerance by sustaining the levels of the small HSPs that are necessary under heat stress [30]. Nevertheless, the functions of large FKBPs in other species are largely unknown.
During the late growth stage of wheat, dry hot winds can substantially reduce grain filling. Currently, no studies have investigated the involvement of wheat BI-1 in the response to heat stress, and little is known about the function of AtBI-1 in plant thermotolerance. In our previous work, we identified the ER-resident protein TaBI-1.1 from an RNA sequencing (RNA-seq) analysis of Fg-treated wheat. Here, we demonstrated that TaBI-1.1, a TaFKBP62-interacting protein, is conserved with AtBI-1 in the response to heat stress and that both TaBI-1.1 and AtBI-1 positively regulate heat-responsive genes expression.
Results
TaBI-1.1 is upregulated under heat stress
We analysed the RNA-seq data from heat-treated wheat to investigate the biological mechanism of the heat stress response. A highly upregulated gene, TaBI-1.1 (12.15-fold increase, TRIAE_CS42_U_TGACv1_644608_AA2140670), was identified from the RNA-seq data (Additional file 1: Table S1). To determine whether TaBI-1.1 was upregulated under heat stress, we monitored TaBI-1.1 expression using real time quantitative PCR (qRT-PCR). TaBI-1.1 mRNA accumulated during heat treatment, reaching a peak of ~ 6-fold at 8 h (Fig. 1a). We analysed the histological β-glucuronidase (GUS) activity via GUS staining of PBI::GUS transgenic Arabidopsis to investigate the spatial expression pattern of TaBI-1.1. Higher levels of GUS protein accumulation were observed in the mature leaves of heat-treated plants than in those of control plants (Fig. 1b and c). GUS mRNA levels were also obviously increased, as shown by qRT-PCR, confirming the upregulation of TaBI-1.1 under heat stress (Fig. 1d).
Fig. 1.
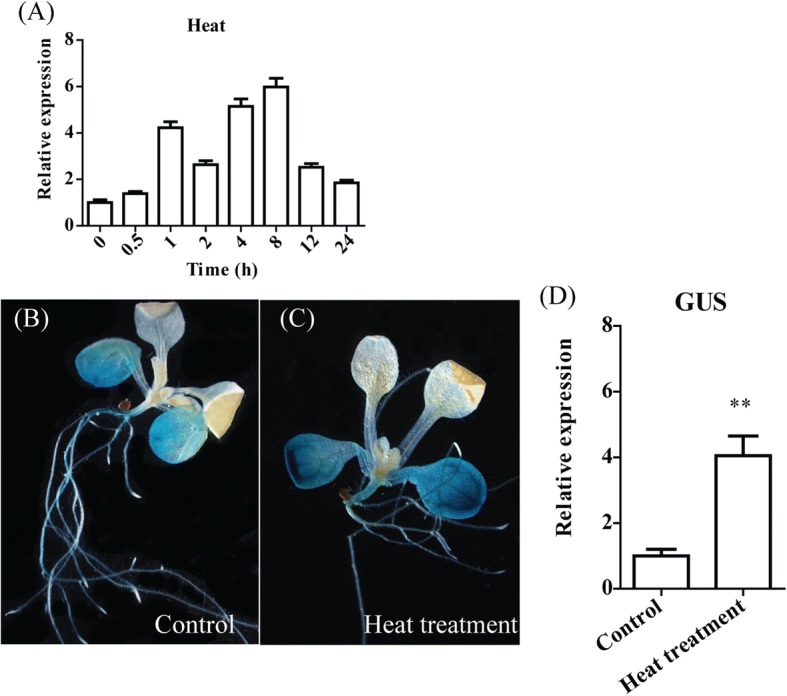
Relative and spatial expression patterns of TaBI-1.1. a Expression profile of TaBI-1.1 under heat treatment for 0, 0.5, 1, 2, 4, 8, 12, and 24 h. Wheat Actin was used as a reference. The vertical coordinates are the fold-changes, and the horizontal ordinates are the different time periods. b and c GUS activity of PBI::GUS transgenic Arabidopsis under normal conditions (b) and heat treatment (c) was detected via GUS histochemical staining. Seedlings grown under normal conditions were used as controls. d GUS expression levels in the PBI:GUS transgenic lines were monitored by qRT-PCR. Actin2 was used as a reference. The results are shown as the means±standard deviation (SD) of three biological replicates. Error bars indicate the SD. Asterisks (**) indicate the significant differences (P < 0.01) compared with Col-0 (Student’s t-test)
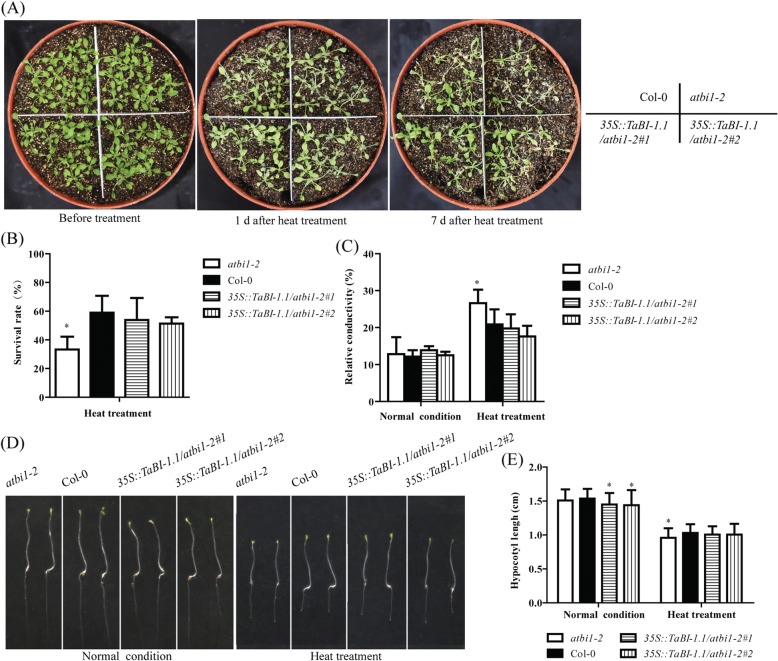
Constitutive expression of TaBI-1.1 in atbi1–2 fully rescues defective heat tolerance in atbi1–2 plants
BI-1 is known to be highly evolutionarily conserved. atbi1–2 mutants are hypersensitive to heat stress, and AtBI-1 overexpression can rescue this deficiency [17]. Based on the high upregulation of TaBI-1.1 under heat treatment, we surmised that TaBI-1.1 might play a similar role as AtBI-1 in the response to heat stress. To test this hypothesis, we generated transgenic lines that ectopically expressed TaBI-1.1 in the atbi1–2 background under the control of the cauliflower mosaic virus (CaMV) 35S promoter. Two homozygous lines with relatively high TaBI-1.1 expression levels (35S::TaBI-1.1/atbi1–2#1 and 35S::TaBI-1.1/atbi1–2#2) were selected for further analysis. Eighteen-day-old atbi1–2 and Col-0 plants, as well as the two transgenic lines, were exposed to 45 °C for 6 h, and the survival rates were tested after 7 days. The survival rates of Col-0, 35S::TaBI-1.1/atbi1–2#1, and 35S::TaBI-1.1/atbi1–2#2 plants were not significantly different, and all the three genotypes exhibited significantly higher survival rates than atbi1–2. Compared with the survival rates of atbi1–2 and Col-0, the two transgenic lines fully rescued the deficiency of atbi1–2 under heat stress (Fig. 2a and b). Furthermore, we examined ion leakage in the different genotypes. Under normal conditions, no differences in relative conductivity were observed between any of the genotypes. However, under heat treatment, atbi1–2 showed a significantly higher relative conductivity than Col-0 and the two transgenic lines, and no differences in relative conductivity were detected between Col-0 and the two transgenic lines, which confirmed that TaBI-1.1 fully rescued the deficiency of atbi1–2 in response to heat stress, indicating the conserved function of BI-1 between wheat and Arabidopsis (Fig. 2c).
Fig. 2.
Constitutive TaBI-1.1 expression in atbi1–2 rescues the deficiency of atbi1–2 in heat tolerance. a The phenotypes of atbi1–2, Col-0, 35S::TaBI-1.1/atbi1–2#1, and 35S::TaBI-1.1/atbi1–2#2 under heat stress. b The survival rates of atbi1–2, Col-0, 35S::TaBI-1.1/atbi1–2#1, and 35S::TaBI-1.1/atbi1–2#2 under heat stress. The results are shown as the means±SD of three biological replicates. Error bars indicate the SD. c The relative conductivity of atbi1–2, Col-0, 35S::TaBI-1.1/atbi1–2#1, and 35S::TaBI-1.1/atbi1–2#2 after heat treatment. The results are shown as the means±SD of six biological replicates. Error bars indicate the SD. d Hypocotyl elongation in atbi1–2, Col-0, 35S::TaBI-1.1/atbi1–2#1, and 35S::TaBI-1.1/atbi1–2#2 after heat treatment. e Statistical analysis of hypocotyl elongation in atbi1–2, Col-0, 35S::TaBI-1.1/atbi1–2#1, and 35S::TaBI-1.1/atbi1–2#2 after heat treatment. The results are shown as the means±SD of 36 biological replicates. Error bars indicate the SD. All the asterisks in the figure (* and **) indicate significant differences (P < 0.05 and P < 0.01, respectively) compared with Col-0 (Student’s t-test)
Hypocotyl elongation is known to be inhibited by heat stress [31]. To elucidate the role of BI-1 during hypocotyl elongation under heat stress, plants from all four genotypes were grown in the dark for 3 days and then exposed to 45 °C for 2 h. Hypocotyl elongation was tested after 3 days of recovery. Six-day-old plants grown in the dark were used as the control. In the control group, no differences in hypocotyl elongation were observed between atbi1–2 and Col-0, while the two transgenic lines displayed significantly shorter hypocotyls than Col-0. In contrast, atbi1–2 exhibited less hypocotyl elongation than the two transgenic lines and Col-0 under heat treatment (Fig. 2d and e). These results suggested that hypocotyl elongation in atbi1–2 is hypersensitive under heat stress and that TaBI-1.1 rescues the hypersensitive phenotype of atbi1–2.
Based on the conserved function of TaBI-1.1 and AtBI-1 under heat stress, as well as their high sequence conservation, we analysed the number of BI-1 members in some important species by screening the Ensembl plant database and constructed a phylogenetic tree of the BI-1 family (Additional file 2: Figure S1B). Aegilops tauschii, Arabidopsis thaliana, Brachypodium distachyon, Hordeum vulgare, and Oryza sativa each contained only one BI-1. Additionally, only three members were identified in Triticum aestivum and Zea mays (Additional file 2: Figure S1A). These results showed that several BI-1 members are present in plant species, revealing the pivotal role of BI-1 in plants.
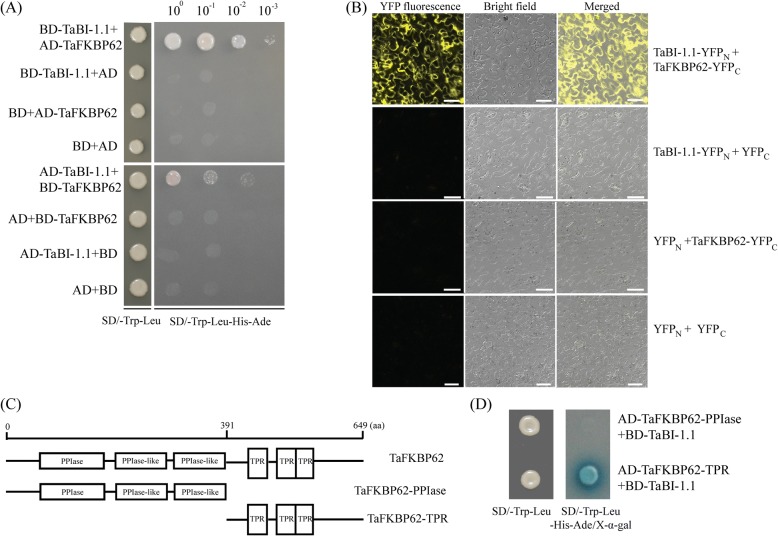
TaBI-1.1 interacts with the TPR domain of TaFKBP62
To further explore the cellular mechanisms of TaBI-1.1 in heat tolerance, we performed yeast two-hybrid assays using TaBI-1.1 expressed from the pGBKT7 (BD) vector as the bait protein to screen a wheat cDNA library. One candidate interacting partner, the ROF1 homologue TaFKBP62, which shares 59.23% amino acid sequence identity with ROF1, was obtained. Both proteins contain three PPIase domains and one TPR domain and belong to the FKBP62 family. Their TPR domains share 70.94% identity, indicating that the TPR domain of FKBP62 is conserved between wheat and Arabidopsis.
Yeast two-hybrid and bimolecular fluorescence complementation (BiFC) assays were used to explore the interaction between TaBI-1.1 and TaFKBP62. A BD vector containing TaBI-1.1 (BD-TaBI-1.1) and a pGADT7 (AD) vector containing TaFKBP62 (AD-TaFKBP62) were constructed for the yeast two-hybrid analysis. Four groups, BD-TaBI-1.1 + AD-TaFKBP62, BD-TaBI-1.1 + AD, BD + AD-TaFKBP62, and BD + AD, were co-transformed into yeast cells. Only yeast cells transformed with BD-TaBI-1.1 and AD-TaFKBP62 were able to grow on selective medium lacking Trp, Leu, His, and Ade (SD/−Trp-Leu-Ade-His). Conversely, co-transformants expressing BD-TaBI-1.1 + AD, BD + AD-TaFKBP62 or BD + AD did not grow on SD/−Trp-Leu-Ade-His medium. Similar results were obtained when we switched the TaBI-1.1 and TaFKBP62 vectors, indicating that TaBI-1.1 interacted with TaFKBP62 in yeast cells (Fig. 3a). This interaction was further demonstrated using BiFC assay. Four vector groups, TaBI-1.1–YFPN (N-terminal fragment of yellow fluorescent protein) and TaFKBP62–YFPC (C-terminal fragment of yellow fluorescent protein), TaBI-1.1–YFPN and YFPC, YFPN and TaFKBP62–YFPC, and YFPN and YFPC, were injected into Nicotiana benthamiana leaves. YFP fluorescence was only detected in leaf epidermal cells injected with TaBI-1.1–YFPN and TaFKBP62–YFPC (Fig. 3b), confirming the interaction between TaBI-1.1 and TaFKBP62.
Fig. 3.
The interaction between TaBI-1.1 and TaFKBP62. a The interaction between TaBI-1.1 and TaFKBP62 measured using yeast two-hybrid assays. b The interaction between TaBI-1.1 and TaFKBP62 measured via BiFC assays in Nicotiana benthamiana epidermal cells. Bars = 50 μm. c The two fragments, TaFKBP62-PPIase and TaFKBP62-TPR, corresponding to the PPIase and TPR domain, respectively, of TaFKBP62. d Interactions between TaBI-1.1 and the two TaFKBP62 fragments detected via yeast two-hybrid assay
To determine which region of TaFKBP62 interacted with TaBI-1.1, the TaFKBP62 sequence was divided to two fragments, TaFKBP62-PPIase (N-terminal and three PPIase domains) and TaFKBP62-TPR (TPR domain), according to the protein domains (Fig. 3c). The two fragments were cloned into an AD vector and co-transformed with BD-TaBI-1.1 into yeast cells. Both AD-TaFKBP62-PPIase + BD-TaBI-1.1 and AD-TaFKBP62-TPR + BD-TaBI-1.1 cells grew on SD/−Trp-Leu medium, whereas only transformants expressing AD-TaFKBP62-TPR and BD-TaBI-1.1 grew on SD/−Trp-Leu-His-Ade/X-α-gal medium (Fig. 3d). These results suggested that TaBI-1.1 specifically interacts with the TPR domain of TaFKBP62.
Given the conserved TPR domain between TaFKBP62 and ROF1, the sequence conservation between TaBI-1.1 and AtBI-1, and the interaction between TaFKBP62 and TaBI-1.1, we also tested the interaction between AtBI-1 and ROF1, as well as AtBI-1 and the TPR domain of ROF1. However, AtBI-1 did not interact with ROF1 or the TPR domain of ROF1 in yeast cells (Additional file 3: Figure S2).
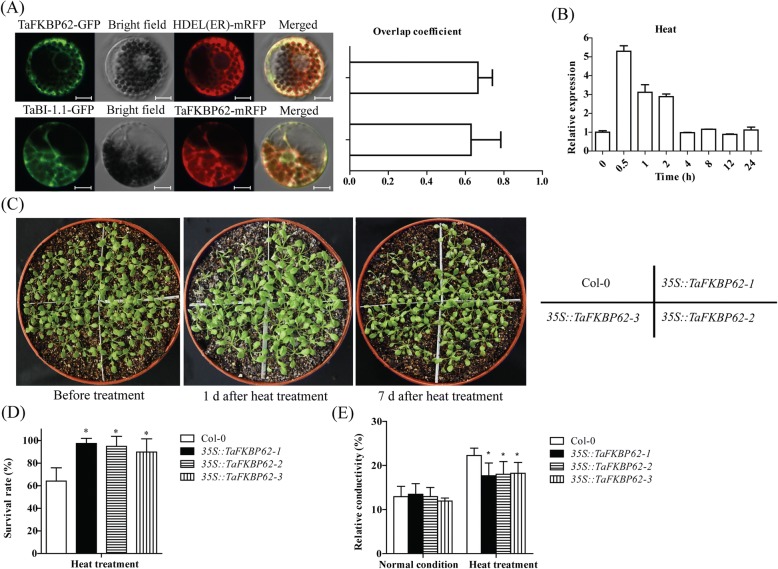
TaFKBP62 co-localizes with TaBI-1.1 on the ER membrane and enhances the heat stress tolerance in Arabidopsis
We previously showed that TaBI-1.1 localizes to the ER membrane [32]. In view of the interaction between TaBI-1.1 and TaFKBP62, we constructed recombinant TaFKBP62-GFP and TaFKBP62-mRFP vectors to detect the subcellular localization of TaFKBP62 and to determine whether TaBI-1.1 co-localizes with TaFKBP62 at the ER membrane. Two groups, TaFKBP62-GFP + mRFP-HDEL (an ER marker) and TaBI-1.1-GFP + TaFKBP62-mRFP, were co-transformed into wheat protoplasts. The overlap coefficient for TaFKBP62-GFP and mRFP-HDEL fluorescence was 0.67, indicating that TaFKBP62 localized to the ER membrane in the wheat protoplasts (Fig. 4a). The overlap coefficient for TaBI-1.1-GFP and TaFKBP62-mRFP fluorescence was 0.63, suggesting that TaBI-1.1 co-localized with TaFKBP62 at the ER membrane (Fig. 4a). ROF1 localizes to the cytoplasm under normal conditions and translocates into the nucleus under heat treatment [30]. However, we did not observe the nuclear translocation of either TaFKBP62 or TaBI-1.1 under heat stress (Additional file 4: Figure S3).
Fig. 4.
The co-localization between TaBI-1.1 and TaFKBP62 and TaFKBP62 expression patterns under stress treatments. a The subcellular localization of TaFKBP62 and the co-localization between TaBI-1.1 and TaFKBP62 in wheat protoplasts. The left panel shows the fluorescence of GFP, mRFP and the merge. Bars = 10 μm. The right panel shows the co-localization levels calculated from the overlap coefficients obtained from at least ten individual protoplasts. b TaFKBP62 expression patterns after heat treatment for 0, 0.5, 1, 2, 4, 8, 12, and 24 h. Wheat Actin was used as a reference. The vertical coordinates represent the fold-changes, and the horizontal ordinates represent the different time periods. The results are shown as the means±SD of three biological replicates. Error bars indicate the SD. c The phenotypes of Col-0, 35S::TaFKBP62–1, 35S::TaFKBP62–2, and 35S::TaFKBP62–3 after heat stress. d The survival rates of Col-0, 35S::TaFKBP62–1, 35S::TaFKBP62–2, and 35S::TaFKBP62–3 under heat stress. The results are shown as the means±SD of three biological replicates. Error bars indicate the SD. e The relative conductivity of Col-0, 35S::TaFKBP62–1, 35S::TaFKBP62–2, and 35S::TaFKBP62–3 after heat treatment. The results are shown as the means±SD of six biological replicates. Error bars indicate the SD
To determine the possible role of TaFKBP62 under heat stress, we monitored TaFKBP62 expression patterns using qRT-PCR and found that TaFKBP62 obviously accumulated during heat treatment, reaching a peak point of ~ 5.3-fold at 0.5 h (Fig. 4b). TaFKBP62 upregulation was also detected in the RNA-seq data from heat-treated wheat (Additional file 5: Table S2). We generated transgenic Arabidopsis plants that constitutively expressed TaFKBP62 under the control of the CaMV 35S promoter. Three homozygous lines, 35S::TaFKBP62–1 35S::TaFKBP62–2 and 35S::TaFKBP62–3, were selected for further analysis. Eighteen-day-old Col-0 and the three transgenic lines were exposed to 45 °C for 6 h, and survival rates were tested after 7 days. The survival rates of the three transgenic lines were significantly higher than those of Col-0 (Fig. 4c and d). Ion-leakage assays showed that the three transgenic lines exhibited significantly lower relative conductivity than Col-0 under heat stress, indicating that TaFKBP62 enhanced heat stress tolerance in Arabidopsis (Fig. 4e).
TaBI-1.1 is conserved with AtBI-1 in regulating heat-responsive gene expression
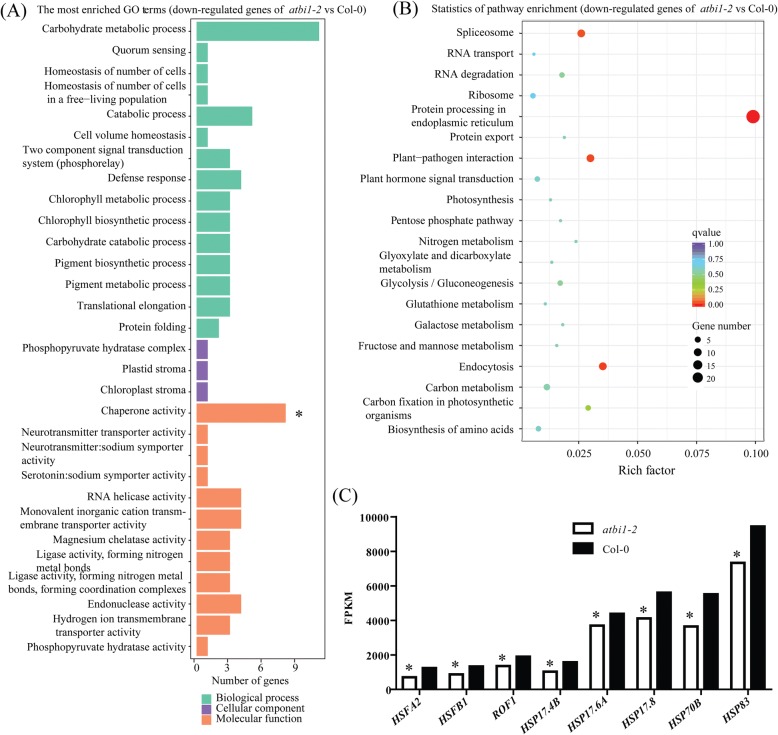
We conducted an RNA-seq analysis of atbi-1 and Col-0 under heat treatment to further investigate the mechanism of BI-1 in response to heat stress. Thirty-five upregulated genes and 80 downregulated genes were identified (Additional file 6: Figure S4). The gene identity (ID) numbers and fold-changes are shown in Additional file 5: Table S2. Most of the HSPs were positively regulated under heat stress. Therefore, we analysed the downregulated genes in atbi-1 versus (vs) Col-0. The top 30 enriched Gene Ontology (GO) terms and the top 20 enriched Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways among the downregulated genes are shown in Fig. 5a and b. Only the term chaperone activity was significant among the 30 enriched GO terms. Of the top 20 enriched pathways, the greater enrichment factor represented a higher degree of enrichment. The enriched pathway protein processing in the ER had the highest enrichment factor, indicating that most of the downregulated genes were involved in the protein processing pathway in the ER. Eight differentially expressed genes, HSFA2, HSFB1, ROF1, HSP17.4B, HSP17.6A, HSP17.8, HSP70B, and HSP90.1, were selected to generate a histogram for visual analysis based on their fragments per kilobase of transcript per million mapped reads (FPKM) in atbi-1 and Col-0. The FPKM of these genes was significantly lower in atbi-1 than in Col-0 (Fig. 5c), revealing that atbi1–2 deficiency might be caused by the downregulation of these genes under heat stress and that the loss of AtBI-1 function might affect protein processing and chaperone activity under heat stress.
Fig. 5.
GO term enrichment and KEGG enrichment analyses were conducted using the RNA-seq data. a GO term enrichment analysis of the downregulated genes. The vertical coordinates are the enriched GO terms, and the horizontal coordinates are the numbers of the downregulated genes in these GO terms. The green columns represent the biological process GO terms; the purple columns represent the cellular component GO terms; the orange columns represent the molecular function GO terms. “*” indicates a significantly enriched GO term. b KEGG enrichment analysis of the downregulated genes. The vertical coordinates are the enriched pathways, and the horizontal coordinates are the rich factors. The size of each point represents the number of downregulated genes in the pathway, and the colour of the point represents the q-value. c FPKM values of the eight downregulated genes. Asterisks (*) indicate significant differences (padj< 0.005) compared with Col-0
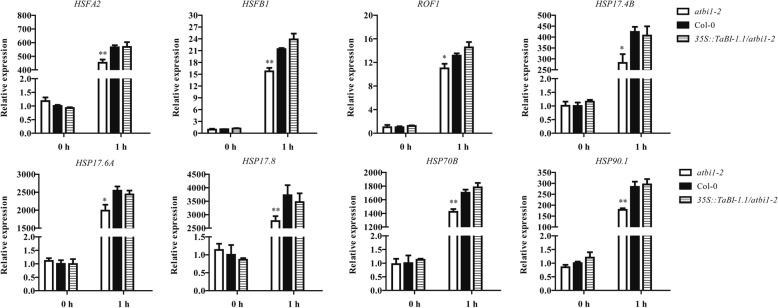
To verify the accuracy of the RNA-seq analysis and investigate whether TaBI-1.1 can rescue the downregulation of these genes in atbi1–2, qRT-PCR analyses were performed using the aforementioned eight genes. Under normal conditions (0 h), no significant differences in gene expression levels were detected among the three genotypes. After heat treatment for 1 h, all eight genes showed obvious upregulation, indicating that they were heat-responsive genes. The expression levels of these genes were significantly lower in atbi1–2 than in Col-0. In contrast, no significant differences in gene expression were observed between Col-0 and 35S::TaBI-1.1/atbi1–2 (Fig. 6). These results suggested that TaBI-1.1 fully rescued the downregulation of these heat-responsive genes in atbi1–2.
Fig. 6.
The expression levels of eight genes in atbi1–2, Col-0 and 35S::TaBI-1.1/atbi1–2 detected by qRT-PCR after heat treatment. Gene expression levels in Col-0 at 0 h were set to “1.” The results are shown as the means±SD of three biological replicates. Error bars indicate the SD. Asterisks (* and **) indicate significant differences (P < 0.05 and P < 0.01, respectively) compared with Col-0 (Student’s t-test)
Discussion
TaBI-1.1 is conserved with AtBI-1 in response to heat stress
BI-1 is an ER-resident cell death suppressor that is highly evolutionarily conserved across species. Numerous studies have shown that plant BI-1 functions as an attenuator of cell death induced by biotic and abiotic stresses such as pathogens, heat, cold, drought, and salt. Human BI-1 blocks cell death induced by the ectopic expression of the mouse Bax [8]. The overexpression of rice and Arabidopsis BI-1 genes in yeast and plant cells also suppresses cell death induced by mammalian Bax [32, 33]. Arabidopsis BI-1 is upregulated under heat stress [17]. The two mutants, atbi1–1 and atbi1–2, exhibit increased sensitivity to heat stress, and BI-1 overexpression rescues the deficiency of the mutants [17]. In the present study, we demonstrated that TaBI-1.1 is a heat-responsive gene and that overexpressing TaBI-1.1 in atbi1–2 rescues the hypersensitive phenotype of atbi1–2 under heat stress (Figs. 1 and 2). Eight heat-responsive genes, HSFA2, HSFB1, ROF1, HSP17.4B, HSP17.6A, HSP17.8, HSP70B, and HSP90.1, were significantly downregulated in atbi1–2 compared with their levels in Col-0 under heat stress, whereas the downregulated expression levels were fully supplemented in 35S::TaBI-1.1/atbi1–2 (Figs. 5c and 6), indicating that TaBI-1.1 is conserved with AtBI-1 in response to heat stress.
The sequence of BI-1 is highly conserved in plants, and only a few BI-1 members were identified in each plant species (Additional file 2: Figure S1), revealing a crucial role of BI-1 in plants. In view of the conserved function of BI-1 between wheat and Arabidopsis in response to heat stress, we surmised that BI-1 might also be involved in thermotolerance in other plant species.
The interaction between TaBI-1.1 and TaFKBP62 may be involved in heat tolerance
TPR-containing proteins exhibit a large degree of sequence diversity, but structural comparison reveals three conserved helical coiled structures formed by the concatenation of ~ 34-residue helical hairpins [34]. Most identified HSP90 co-chaperones contain a TPR domain that binds to the MEEVD motif at the C-terminus of HSP90 [35, 36]. Human TPR-containing immunophilins FKBP52 and FKBP38 bind HSP90 through their TPR domains [37, 38]. Arabidopsis ROF1 binds HSP90.1 via its TPR domain [29]. The ROF1–HSP90.1 complex translocates into the nucleus and interacts with HSFA2 upon exposure to heat stress. However, HSFA2 only interacts with HSP90.1, but not with ROF1 [30]. Therefore, ROF1 mediates the interaction between HSP90.1 and HSFA2 through its TPR domain in response to heat stress [30]. In our study, TaBI-1.1 interacted with the TPR domain of TaFKBP62, and the TPR domain of FKBP62 was conserved between wheat and Arabidopsis (Fig. 3). Overexpressing TaBI-1.1 in atbi1–2 upregulated ROF1 expression under heat treatment, indicating that TaBI-1.1 positively regulates ROF1 expression under heat stress (Fig. 6). Constitutive TaFKBP62 expression increased tolerance to heat stress in Arabidopsis (Fig. 4). Thus, we hypothesized that the interaction between TaBI-1.1 and TaFKBP62 might be involved in thermotolerance.
Despite the high degree of sequence conservation between TaBI-1.1 and AtBI-1 and the conserved TPR domain between TaFKBP62 and ROF1, neither ROF1 nor the TPR domain of ROF1 interacted with AtBI-1 (Additional file 3: Figure S2). The ROF1–HSP90.1 complex localizes to the cytoplasm under normal conditions and translocates to the nucleus following exposure to heat stress [30]. In the present study, TaFKBP62 was found to be an ER-resident protein (Fig. 4a) and did not translocate to the nucleus under heat stress (Additional file 4: Figure S3), implying that the mechanisms of TaFKBP62 and ROF1 in plant thermotolerance were different. We hypothesized that TaFKBP62 may act as a TaBI-1.1 chaperonin through its TPR domain on the ER membrane in response to heat stress.
BI-1 might respond to heat stress by regulating heat-responsive gene expression
HSFA2 acts as a positive regulator in the response to heat stress [39, 40]. HSFB proteins were suggested to be coactivators of HSFAs [41]. HSPs function in protein folding, the maintenance of protein stability, and the activation and maturation of cellular proteins through cooperation with client proteins and co-chaperones to regulate cellular processes [42–44]. Both TaBI-1.1 and AtBI-1 regulated the expression of HSFA2, HSFB1, ROF1, HSP17.4B, HSP17.6A, HSP17.8, HSP70B, and HSP90.1 under heat stress (Figs. 5c and 6). Therefore, we hypothesized that the involvement of BI-1 in thermotolerance might be related to the regulation of heat-responsive genes.
In the RNA-seq analysis of heat-treated atbi-1 and Col-0 plants, the most enriched GO term and pathway were chaperone activity and protein processing in the ER, respectively (Fig. 5a and b). AtBI-1, as an ER-resident protein, modulates ER stress-mediated PCD [7]. The loss of AtBI-1 function in atbi1–2 negatively regulated HSFA2, HSFB1, ROF1, HSP17.4B, HSP17.6A, HSP17.8, HSP70B, and HSP90.1 expression (Figs. 5c and 6). The ROF1–HSP90.1 complex interacts with HSFA2 upon exposure to heat stress to regulate the accumulation of small HSPs [30]. Therefore, we hypothesized that AtBI-1 regulates ROF1 and HSP90.1 protein processing at the ER membrane to affect the stability of the ROF1–HSP90.1–HSFA2 complex, resulting in the accumulation of small HSPs following exposure to heat stress.
Conclusions
Compared with Col-0, atbi1–2 mutants are hypersensitive to heat stress, and constitutive TaBI-1.1 expression in atbi1–2 plants rescued the deficiency of atbi1–2 under heat stress. TaBI-1.1 interacted with the TPR domain of TaFKBP62. TaFKBP62 was found to be an ER-resident protein that co-localized with TaBI-1.1. Additionally, both TaBI-1.1 and AtBI-1 regulated the expression of ROF1 in response to heat stress. Because constitutive TaFKBP62 expression enhanced heat-stress tolerance in Arabidopsis, we hypothesized that the interaction between TaFKBP62 and TaBI-1.1 may be involved in the response to heat stress. Our results revealed the role of TaBI-1.1 and AtBI-1 in response to heat stress and the possible relationship between BI-1 and FKBP62 in Arabidopsis and wheat. Nevertheless, additional studies are needed to fully elucidate the mechanism of BI-1 in plant thermotolerance.
Methods
Plant materials and gene cloning
The wheat cultivar Xiaobaimai and the Arabidopsis mutant atbi1–2 (CS323793) and wild-type Columbia-0 (Col-0) were used in this study. TaBI-1.1 (TRIAE_CS42_U_TGACv1_644608_AA2140670) was identified in a previous study [32]. The TaFKBP62 (TRIAE_CS42_2DS_TGACv1_178636_AA0598780) coding sequence was amplified and inserted into the pLB vector (TIANGEN, China) (for primer sequences, see Additional file 7: Table S3). DNAMAN 6.0 software was used to determine amino acid sequence identity (Lynnon Biosoft, U.S.). Phylogenetic analyses were conducted using Molecular Evolutionary Genetics Analysis (MEGA 5.0, U.S.) software.
Performance evaluation under heat treatment
The fusion vector 35S::TaBI-1.1, in which the TaBI-1.1 coding sequence was cloned into the pCAMBIA1302 vector under the control of the CaMV 35S promoter, was described previously [32]. The 35S::TaBI-1.1 vector was transformed into atbi1–2 to generate 35S::TaBI-1.1/ atbi1–2 transgenic line via the floral-dipping method. The TaFKBP62 coding sequence was cloned into the pCAMBIA1302 vector under the control of the CaMV 35S promoter using an In-Fusion HD Cloning Kit (Clontech, United States) (for the primer sequences, see Additional file 7: Table S3). The 35S::TaFKBP62 fusion vector was transformed into Col-0 plants via the floral-dipping method. Homozygous T3 seeds were sterilized and germinated on MS medium [45] in a greenhouse at 22 °C under an 8-h light regime. For the phenotype analysis, 18-day-old plants were exposed to 45 °C for 6 h, and survival rates were measured after 7 days. Three biological replicates were performed.
For the ion leakage assay, 21-day-old plants were exposed to 45 °C for 3 h, and eight leaf discs (6 mm in diameter) were snipped from the leaves and placed in 5 mL of deionized water after heat treatment. After vacuuming for 30 min, the ion conductivity of the four genotypes was recorded as the initial conductivity, C1, and the ion conductivity of deionized water was recorded as C0. After incubating in boiling water for 30 min, we recorded the ion conductivity as C2, and the relative conductivity was calculated based on the formula:(C1-C0)/(C2-C0) × 100%. Ion conductivity was recorded using an FG3-B FiveGo™ conductivity meter (METTLER TOLEDO, Switzerland). Untreated plants were used as controls. Each treatment contained six independent replicates.
For hypocotyl-elongation assays, three-day-old plants grown in the dark were subjected to 45 °C for 2 h. After heat treatment, the plants were grown in the dark for three additional days, after which hypocotyl length was measured. The experiments included at least 36 seedlings for each genotype. Plants grown under normal conditions were used as controls.
qRT-PCR analysis
Ten-day-old wheat seedlings grown at 22 °C were used for the heat treatments. For heat treatment, seedlings were exposed to 38 °C. For analyses of heat-responsive gene expression, eight-day-old Col-0, atbi1–2, and the 35S::TaBI-1.1/ atbi1–2 seedlings were exposed to 37 °C for 0 and 1 h. Three biological replicates were performed. The samples were harvested after treatment and were rapidly frozen in liquid nitrogen. Total RNA was extracted from the seedlings using the RNAprep pure Plant Kit (TIANGEN, China). qRT-PCR was performed on an ABI7500 system (ABI, U.S.). Primers used for qRT-PCR are listed in Additional file 7: Table S3.
β-Glucuronidase (GUS) activity assay
The PBI::GUS fusion vector containing a 1.7 kb TaBI-1.1 promoter, as well as the corresponding transgenic Arabidopsis, were obtained previously [32]. For heat treatment, five-day-old seedlings were exposed to 37 °C for 3 h. Untreated seedlings were used as controls. GUS staining was examined using a GUS histochemical assay kit (Real-Times, Beijing) according to the manufacturer’s instructions. Image analysis was performed using a Leica M165 FC stereomicroscope (Wetzlar, Germany).
Yeast two-hybrid system
The BD-TaBI-1.1 and AD-TaBI-1.1 recombinant plasmids were obtained previously [32]. BD-TaBI-1.1 was used as a bait plasmid to screen a wheat cDNA library via yeast two-hybrid assays, which were performed according to the MATCHMAKER two-hybrid system user manual (Clontech, United States). The full-length TaFKBP62 coding sequence was amplified and cloned into pGBKT7 and pGADT7 to create BD-TaFKBP62 and AD-TaFKBP62, respectively. The TaFKBP62 fragments TaFKBP62-PPIase and TaFKBP62-TPR were cloned into pGADT7 to create AD-TaFKBP62-PPIase and AD-TaFKBP62-TPR recombinant plasmids (for the primer sequences, see Additional file 7: Table S3). The full-length coding sequences of AtBI-1 and ROF1 were amplified and cloned into pGBKT7 and pGADT7 to create BD-AtBI-1 and AD-ROF1, respectively. The TPR region of ROF1, ROF1-TPR, was cloned into pGADT7 to create the AD- ROF1-TPR recombinant plasmid (for the primer sequences, see Additional file 7: Table S3). To test the interactions, combinations BD-TaBI-1.1 + AD-TaFKBP62, AD-TaBI-1.1 + BD-TaFKBP62, AD-TaBI-1.1 + BD, BD-TaBI-1.1 + AD, AD + BD-TaFKBP62, BD + AD-TaFKBP62, AD + BD, AD-TaFKBP62-PPIase + BD-TaBI-1.1, AD-TaFKBP62-PPIase + BD, AD-TaFKBP62-TPR + BD-TaBI-1.1, AD-TaFKBP62-TPR + BD, BD-AtBI-1 + AD-ROF1, BD-AtBI-1 + AD, BD + AD-ROF1, BD-AtBI-1 + AD-ROF1-TPR, BD-AtBI-1 + AD, and BD + AD-ROF1-TPR were coexpressed in AH109 yeast cells and selected on SD/−Trp-Leu-His-Ade or SD/−Trp-Leu-His-Ade/X-α-gal medium for 5 days (Clontech, U.S.).
Subcellular localization assay
The coding region of TaFKBP62 was amplified without a stop codon from wheat cDNA via PCR and inserted into the p16318GFP and pcDNA3.1-mRFP vectors (for the primer sequences, see Additional file 7: Table S3). The TaBI-1.1-GFP recombinant plasmid was obtained previously [32]. TaFKBP62-GFP and mRFP-HDEL, as well as TaBI-1.1-GFP and TaFKBP62-mRFP, were introduced into wheat protoplasts via polyethylene glycol-mediated transformation [46]. After incubation for 12 h in the dark at 22 °C, fluorescence signals were detected using a confocal laser-scanning microscope. For the subcellular localization of TaFKBP62 or TaBI-1.1 under heat treatment, TaFKBP62-GFP or TaBI-1.1-GFP was transformed into wheat protoplasts and the fluorescence signals were observed after treatment at 37 °C for 3 h.
Bimolecular fluorescence complementation assay
The coding sequences of TaBI-1.1 and TaFKBP62 were amplified without stop codons via PCR using LA Taq DNA polymerase (Takara, Japan) and cloned into the medium vector pGWC-T according to the DNA Ligation Kit Ver.2.1 manual to generate pGWC-TaBI-1.1 and pGWC-TaFKBP62, respectively (for the primer sequences, see Additional file 7: Table S3). The sequences of TaBI-1.1 and TaFKBP62 from pGWC-TaBI-1.1 and pGWC-TaFKBP62 were further cloned into the destination vectors pEarlygate201-YN and pEarlygate202-YC, respectively, using gateway technology (Invitrogen, U.S.). The combined vectors were transformed into Agrobacterium tumefaciens strain GV3101 and introduced into Nicotiana benthamiana epidermal cells for transient expression as described previously [47]. YFP fluorescence was imaged using a confocal laser-scanning microscope after infiltration for 3 days.
RNA-seq analysis
Eight-day-old wheat seedlings were treated at 40 °C and isolated at 1 h and 3 h. The samples were mixed for RNA-seq analysis of heat-treated wheat (Novogene, Beijing). Six-day-old Col-0 and atbi1–2 plants were exposed to 37 °C for 1 h, and samples were collected for RNA-seq analysis of heat-treated Arabidopsis (Allwegene, Beijing). RNA-seq was performed using the HiSeq™2500 system (Illumina, U.S.).
Additional files
Table S1. Differentially expressed genes in the RNA-seq analysis of heat-treated wheat. (XLS 10704 kb)
Figure S1. The number of BI-1 members in various plant species and phylogenetic analysis of the BI-1 family. (A) The number of BI-1 members in ten species. (B) Phylogenetic analysis of BI-1 family proteins. (TIF 1001 kb)
Figure S2. AtBI-1 did not interact with ROF1 or the TPR region of ROF1. (A) The sequence diagrams of the full-length ROF1 sequence and the TPR domain of the ROF1. (B) The interaction between AtBI-1 and ROF1, as well as AtBI-1 and the TPR domain of the ROF1 by yeast two-hybrid analysis. (TIF 2385 kb)
Figure S3. Subcellular localization of TaFKBP62 and TaBI-1.1 under heat stress. (TIF 6903 kb)
Table S2. Differentially expressed genes between atbi1–2 and Col-0 in the heat-treated RNA-seq analysis. (XLSX 22 kb)
Figure S4. Cluster analysis of the differentially expressed genes. (TIF 3543 kb)
Table S3. Primer sequences used in this study. (XLSX 13 kb)
Acknowledgements
We are grateful to Dr. Jian-Min Wan (Chinese Academy of Agricultural Sciences) for the vector of mRFP-HDEL.
Funding
This research was financially supported by the National Key Research and Development Program of China (2016YFD0100600) for designing and performing experiments, the National Transgenic Key Project of the Ministry of Agriculture of China (2018ZX0800909B and 2016ZX08002–002) for performing experiments and data analysis, the Open Research Fund Program of Beijing Key Lab of Plant Resource Research and Development, Beijing Technology and Business University, and the Fundamental Research Funds for the Central Universities (2014YB079) for writing and revising the manuscript.
Availability of data and materials
Wheat genes sequences in this article were downloaded from Ensembl Plant database (http://plants.ensembl.org/index.html); Arabidopsis genes sequences in this article were downloaded from The Arabidopsis Information Resource (TAIR) (http://www.arabidopsis.org/). The raw data of RNA-seq analysis are not publicly available, because they will be used in further researches, but they are available from the corresponding author on reasonable request.
Abbreviations
- ABA
abscisic acid
- ABRC
Arabidopsis Biological Resource Center
- AD
pGADT7
- BD
pGBKT7
- BI-1
Bax inhibitor-1
- BiFC
bimolecular fluorescence complementation
- CaMV
cauliflower mosaic virus
- ER
endoplasmic reticulum
- FKBP
FK506-binding protein
- FPKM
fragments per kilobase of transcript per million mapped reads
- GO
Gene Ontology
- GUS
β-glucuronidase
- HSF
heat stress transcription factor
- HSP
heat shock protein
- KEGG
Kyoto Encyclopaedia of Genes and Genomes
- PCD
programmed cell death
- PPIase
peptidyl prolyl cis-trans isomerase
- qRT-PCR
real time quantitative PCR
- RNA-seq
RNA sequencing
- SA
salicylic acid
- TM
tunicamycin
- TPR
tetratricopeptide repeat motif
- YFP
yellow fluorescent protein
Authors’ contributions
ZSX coordinated the project, conceived and designed experiments; PPL performed experiments and wrote the first draft; CTW modified and edited the manuscript, and performed supplementary experiments; WJZ and WYS conducted the bioinformatic work and performed experiments; JDF, MC, JC and YBZ contributed to acquisition of data and analysis; YJX edited and revised the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Pan-Pan Lu, Email: luckpanpan@yeah.net.
Wei-Jun Zheng, Email: zhengweijun@nwsuaf.edu.cn.
Chang-Tao Wang, Email: wangct@th.btbu.edu.cn.
Wen-Yan Shi, Email: shiwy12@126.com.
Jin-Dong Fu, Email: fujindong@caas.cn.
Ming Chen, Email: chenming02@caas.cn.
Jun Chen, Email: chenjun01@caas.cn.
Yong-Bin Zhou, Email: zhouyongbin@caas.cn.
Ya-Jun Xi, Email: xiyajun2002@126.com.
Zhao-Shi Xu, Email: xuzhaoshi@caas.cn.
References
- 1.Ruelland E, Zachowski A. How plants sense temperature. Environ Exp Bot. 2010;69:225–232. doi: 10.1016/j.envexpbot.2010.05.011. [DOI] [Google Scholar]
- 2.Wahid A, Gelani S, Ashraf M, Foolad M. Heat tolerance in plants: an overview. Environ Exp Bot. 2007;61:199–223. doi: 10.1016/j.envexpbot.2007.05.011. [DOI] [Google Scholar]
- 3.Camejo D, Rodríguez P, Morales MA, Dell’Amico JM, Torrecillas A, Alarcón JJ. High temperature effects on photosynthetic activity of two tomato cultivars with different heat susceptibility. J Plant Physiol. 2005;162:281–289. doi: 10.1016/j.jplph.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 4.Ahn YJ, Zimmerman JL. Introduction of the carrot HSP17.7 into potato (Solanum tuberosum L.) enhances cellular membrane stability and tuberization in vitro. Plant Cell Environ. 2006;29:95–104. doi: 10.1111/j.1365-3040.2005.01403.x. [DOI] [PubMed] [Google Scholar]
- 5.Momcilovic I, Ristic Z. Expression of chloroplast protein synthesis elongation factor, EF-Tu, in two lines of maize with contrasting tolerance to heat stress during early stages of plant development. J Plant Physiol. 2007;164:90–99. doi: 10.1016/j.jplph.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Hückelhoven R. BAX Inhibitor-1, an ancient cell death suppressor in animals and plants with prokaryotic relatives. Apoptosis. 2004;9:299–307. doi: 10.1023/B:APPT.0000025806.71000.1c. [DOI] [PubMed] [Google Scholar]
- 7.Ishikawa T, Watanabe N, Nagano M, Kawai-Yamada M, Lam E. Bax inhibitor-1: a highly conserved endoplasmic reticulum-resident cell death suppressor. Cell Death Differ. 2011;18:1271–1278. doi: 10.1038/cdd.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Q, Reed JC. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol Cell. 1998;1:337–346. doi: 10.1016/S1097-2765(00)80034-9. [DOI] [PubMed] [Google Scholar]
- 9.Kawai-Yamada M, Ohori Y, Uchimiya H. Dissection of Arabidopsis Bax inhibitor-1 suppressing Bax-, hydrogen peroxide-, and salicylic acid-induced cell death. Plant Cell. 2004;16:21–32. doi: 10.1105/tpc.014613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Tang C, Huang X, Li F, Chen X, Zhang G, Sun Y, Han D, Kang Z. Wheat BAX inhibitor-1 contributes to wheat resistance to Puccinia striiformis. J Exp Bot. 2012;63:4571–4584. doi: 10.1093/jxb/ers140. [DOI] [PubMed] [Google Scholar]
- 11.Babaeizad V, Imani J, Kogel KH, Eichmann R, Huckelhoven R. Over-expression of the cell death regulator BAX inhibitor-1 in barley confers reduced or enhanced susceptibility to distinct fungal pathogens. Theor Appl Genet. 2009;118:455–463. doi: 10.1007/s00122-008-0912-2. [DOI] [PubMed] [Google Scholar]
- 12.Eichmann R, Schultheiss H, Kogel KH, Hückelhoven R. The barley apoptosis suppressor homologue BAX inhibitor-1 compromises nonhost penetration resistance of barley to the inappropriate pathogen Blumeria graminis f. sp. tritici. Mol Plant Microbe In. 2004;17:484–490. doi: 10.1094/MPMI.2004.17.5.484. [DOI] [PubMed] [Google Scholar]
- 13.Ramiro DA, Melotto-Passarin DM, Barbosa Mde A, Santos FD, Gomez SG, Massola Junior NS, Lam E, Carrer H. Expression of Arabidopsis Bax Inhibitor-1 in transgenic sugarcane confers drought tolerance. Plant Biotechnol J. 2016;14:1826–1837. doi: 10.1111/pbi.12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Duan Z, Chen P, Shang Y, Wang C. The Bax inhibitor MrBI-1 regulates heat tolerance, apoptotic-like cell death, and virulence in Metarhizium robertsii. Sci Rep. 2015;5:10625. doi: 10.1038/srep10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isbat M, Zeba N, Kim SR, Hong CB. A BAX inhibitor-1 gene in Capsicum annuum is induced under various abiotic stresses and endows multi-tolerance in transgenic tobacco. J Plant Physiol. 2009;166:1685–1693. doi: 10.1016/j.jplph.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe N, Lam E. BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J Biol Chem. 2008;283:3200–3210. doi: 10.1074/jbc.M706659200. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe N, Lam E. Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. Plant J. 2006;45:884–894. doi: 10.1111/j.1365-313X.2006.02654.x. [DOI] [PubMed] [Google Scholar]
- 18.Galat A. A note on clustering the functionally-related paralogues and orthologues of proteins: a case of the FK506-binding proteins (FKBPs) Comput Biol Chem. 2004;28:129. doi: 10.1016/j.compbiolchem.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Romano P, Gray J, Horton P, Luan S. Plant immunophilins: functional versatility beyond protein maturation. New Phytol. 2005;166:753–769. doi: 10.1111/j.1469-8137.2005.01373.x. [DOI] [PubMed] [Google Scholar]
- 20.Schreiber SL. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science. 1991;251:283. doi: 10.1126/science.1702904. [DOI] [PubMed] [Google Scholar]
- 21.Luan S, Kudla J, Gruissem W, Schreiber SL. Molecular characterization of a FKBP-type immunophilin from higher plants. P Natl Acad Sci USA. 1996;93:6964–6969. doi: 10.1073/pnas.93.14.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dwivedi RS, Breiman A, Herman EM. Differential distribution of the cognate and heat-stress-induced isoforms of high Mr cis-trans prolyl peptidyl isomerase (FKBP) in the cytoplasm and nucleoplasm. J Exp Bot. 2003;54:2679–2689. doi: 10.1093/jxb/erg307. [DOI] [PubMed] [Google Scholar]
- 23.Jin YJ, Burakoff SJ, Bierer BE. Molecular cloning of a 25-kDa high affinity rapamycin binding protein. FKBP25 J Biol Chem. 1992;267:10942–10945. [PubMed] [Google Scholar]
- 24.Breiman A, Fawcett TW, Ghirardi ML, Mattoo AK. Plant organelles contain distinct petidylprolyl cis, trans-isomerases. J Biol Chem. 1992;267:21293–21296. [PubMed] [Google Scholar]
- 25.Faure JD, Gingerich D, Howell SH. An Arabidopsis immunophilin, AtFKBP12, binds to AtFIP37 (FKBP interacting protein) in an interaction that is disrupted by FK506. Plant J. 1998;15:783–789. doi: 10.1046/j.1365-313X.1998.00248.x. [DOI] [PubMed] [Google Scholar]
- 26.Standaert RF, Galat A, Verdine GL, Schreiber SL. Molecular cloning and overexpression of the human FK506-binding protein FKBP. Nature. 1990;346:671–674. doi: 10.1038/346671a0. [DOI] [PubMed] [Google Scholar]
- 27.Kurek I, Aviezer K, Erel N, Herman E, Breiman A. The wheat peptidyl prolyl cis-trans-isomerase FKBP77 is heat induced and developmentally regulated. Plant Physiol. 1999;119:693–703. doi: 10.1104/pp.119.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meiri D, Tazat K, Cohen-Peer R, Farchi-Pisanty O, Aviezer-Hagai K, Avni A, Breiman A. Involvement of Arabidopsis ROF2 (FKBP65) in thermotolerance. Plant Mol Biol. 2010;72:191–203. doi: 10.1007/s11103-009-9561-3. [DOI] [PubMed] [Google Scholar]
- 29.Aviezer-Hagai K, Skovorodnikova J, Galigniana M, Farchi-Pisanty O, Maayan E, Bocovza S, Efrat Y, von Koskull-Doring P, Ohad N, Breiman A. Arabidopsis immunophilins ROF1 (AtFKBP62) and ROF2 (AtFKBP65) exhibit tissue specificity, are heat-stress induced, and bind HSP90. Plant Mol Biol. 2007;63:237–255. doi: 10.1007/s11103-006-9085-z. [DOI] [PubMed] [Google Scholar]
- 30.Meiri D, Breiman A. Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 2009;59:387–399. doi: 10.1111/j.1365-313X.2009.03878.x. [DOI] [PubMed] [Google Scholar]
- 31.Queitsch C, Hong S-W, Vierling E, Lindquist S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell. 2000;12:479–492. doi: 10.1105/tpc.12.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu PP, Yu TF, Zheng WJ, Chen M, Zhou YB, Chen J, Ma YZ, Xi YJ, Xu ZS. The wheat Bax Inhibitor-1 protein interacts with an aquaporin TaPIP1 and enhances disease resistance in Arabidopsis. Front Plant Sci. 2018;9:20. doi: 10.3389/fpls.2018.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawai-Yamada M, Uchimiya H. Mammalian Bax-induced plant cell death can be down-regulated by overexpression of Arabidopsis Bax Inhibitor-1 (AtBI-1) P Natl Acad Sci USA. 2001;98:12295–12300. doi: 10.1073/pnas.211423998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawai M, Pan L, Reed JC, Uchimiya H. Evolutionally conserved plant homologue of the Bax inhibitor-1 (BI-1) gene capable of suppressing Bax-induced cell death in yeast. FEBS Lett. 1999;464:143–147. doi: 10.1016/S0014-5793(99)01695-6. [DOI] [PubMed] [Google Scholar]
- 35.Das AK, Cohen PT, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein–protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med. 2003;228:111. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 37.Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 38.Riggs DL, Roberts PJ, Chirillo SC, Cheung-Flynn J, Prapapanich V, Ratajczak T, Gaber R, Picard D, Smith DF. The Hsp90-binding peptidylprolyl isomerase FKBP52 potentiates glucocorticoid signaling in vivo. EMBO J. 2003;22:1158. doi: 10.1093/emboj/cdg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edlich F, Erdmann F, Jarczowski F, Moutty MC, Weiwad M, Fischer G. The Bcl-2 regulator FKBP38-calmodulin-Ca2+ is inhibited by Hsp90. J Biol Chem. 2007;282:15341–15348. doi: 10.1074/jbc.M611594200. [DOI] [PubMed] [Google Scholar]
- 40.Busch W, Wunderlich M, Schöffl F. Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J. 2005;41:1. doi: 10.1111/j.1365-313X.2004.02272.x. [DOI] [PubMed] [Google Scholar]
- 41.Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 2006;48:535–547. doi: 10.1111/j.1365-313X.2006.02889.x. [DOI] [PubMed] [Google Scholar]
- 42.Bharti K, Koskulldöring PV, Bharti S, Kumar P, Tintschlkörbitzer A, Treuter E, Nover L. Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein Ortholog HAC1. Plant Cell. 2004;16:1521. doi: 10.1105/tpc.019927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caplan AJ, Jackson S, Smith D. Hsp90 reaches new heights. EMBO Rep. 2003;4:126. doi: 10.1038/sj.embor.embor742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- 45.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plantarum. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 46.Marion J, Bach L, Bellec Y, Meyer C, Gissot L, Faure JD. Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J. 2008;56:169–179. doi: 10.1111/j.1365-313X.2008.03596.x. [DOI] [PubMed] [Google Scholar]
- 47.Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc. 2006;1:2019. doi: 10.1038/nprot.2006.286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Differentially expressed genes in the RNA-seq analysis of heat-treated wheat. (XLS 10704 kb)
Figure S1. The number of BI-1 members in various plant species and phylogenetic analysis of the BI-1 family. (A) The number of BI-1 members in ten species. (B) Phylogenetic analysis of BI-1 family proteins. (TIF 1001 kb)
Figure S2. AtBI-1 did not interact with ROF1 or the TPR region of ROF1. (A) The sequence diagrams of the full-length ROF1 sequence and the TPR domain of the ROF1. (B) The interaction between AtBI-1 and ROF1, as well as AtBI-1 and the TPR domain of the ROF1 by yeast two-hybrid analysis. (TIF 2385 kb)
Figure S3. Subcellular localization of TaFKBP62 and TaBI-1.1 under heat stress. (TIF 6903 kb)
Table S2. Differentially expressed genes between atbi1–2 and Col-0 in the heat-treated RNA-seq analysis. (XLSX 22 kb)
Figure S4. Cluster analysis of the differentially expressed genes. (TIF 3543 kb)
Table S3. Primer sequences used in this study. (XLSX 13 kb)
Data Availability Statement
Wheat genes sequences in this article were downloaded from Ensembl Plant database (http://plants.ensembl.org/index.html); Arabidopsis genes sequences in this article were downloaded from The Arabidopsis Information Resource (TAIR) (http://www.arabidopsis.org/). The raw data of RNA-seq analysis are not publicly available, because they will be used in further researches, but they are available from the corresponding author on reasonable request.