Abstract
The cellular abundance of proteins can vary even between isogenic single cells. This variability between single-cell protein levels can have regulatory roles, such as controlling cell fate during apoptosis induction or the proliferation/quiescence decision. Here, we review examples connecting protein levels and their dynamics in single cells to cellular functions. Such findings were made possible by the introduction of antibodies, and subsequently fluorescent proteins, for tracking protein levels in single cells. However, in heterogeneous cell populations, such as tumors or differentiating stem cells, cellular decisions are controlled by hundreds, even thousands of proteins acting in concert. Characterizing such complex systems demands measurements of thousands of proteins across thousands of single cells. This demand has inspired the development of new methods for single-cell protein analysis, and we discuss their trade-offs, with an emphasis on their specificity and coverage. We finish by highlighting the potential of emerging mass-spec methods to enable systems-level measurement of single-cell proteomes with unprecedented coverage and specificity. Combining such methods with methods for quantitating the transcriptomes and metabolomes of single cells will provide essential data for advancing quantitative systems biology.
Introduction
Early experimental investigations of cellular heterogeneity focussed on isogenic bacterial populations. Despite being isogenic and growing in the same culture, individual bacteria varied in persistence, λ phage burst size, β-galactosidase production, and chemotactic behaviour [1–4]. These pioneering studies used elegant approaches to investigate heterogeneity and its functional consequences but were limited by the technology at the time, having no means of detecting gene expression in single cells. In 1994 a new technology, GFP, was introduced [5] which allowed researchers to measure and dynamically track protein levels in single cells. This technological innovation enabled the accurate measurement of protein levels and their variability across thousands of isogenic cells [6]. The measurements revealed unexpected variability in the levels of proteins expressed from the same promoter, which the authors interpreted as biochemical noise comprising two components: intrinsic, inherent to the biochemical process of transcription and translation, and extrinsic, dominated by external environmental fluctuations.
Regulation and functions of single-cell protein variability
While these first studies focussed on clonal cells and attributed the variability of a protein to noise in gene expression, in many cases the differences in the abundance of a protein across single cells reflects different cellular states that may lead to different functional outcomes [7]. For instance, in single mitotically cycling MCF10A cells, the level of p21, a cyclin-dependent kinase 2 (CDK2) inhibitor, determines whether a cell enters a quiescent or proliferative state [8]. If p21 is present above a threshold at the end of mitosis, it inhibits CDK2 and the cell enters quiescence. Conversely, if the level of p21 is below the threshold, CDK2 remains active and the cell continues to proliferate. By making measurements of single cells, the authors also found that modulating p21 levels altered the proportion of quiescent or proliferative cells, and that different cell lines exhibited different inherent proportions of each. Thus, the level of a single protein affects the proportion of cells in a quiescent or proliferative state.
In other cases, experiments have demonstrated that changes in genetic parameters can tune the variability in gene expression, and cells can exploit this variability to respond dynamically to environmental changes. To study the effect of genetic parameters on gene expression noise, the relative contributions of transcription and translation to phenotypic noise in B. subtilis were quantitated at various rates of transcription and translation [9]. The authors demonstrated that the efficiency of either process, and the resulting noise profile, could be altered by mutating the promoter, which affected transcription [10] or ribosomal binding, which affected translation [11]. Subsequently, a different group introduced both cis- and trans-acting mutations that changed the expression noise profile of a given gene [12], providing further evidence of how gene expression noise can be biochemically encoded and evolved. These studies indicated that gene expression variability is a selectable trait, evolved to suit the gene and its particular function.
Spencer et al. [13] provided an example of how this evolved, inherent variability in protein levels between cells could lead to graded cellular responses across the population, and confer an overall survival advantage. They monitored HeLa and MCF10 cells on their path toward TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis and observed highly variable outcomes between single cells: most cells died, doing so at an exponentially decaying rate, but a small subpopulation always survived altogether and continued growing. After measuring the protein-level distributions of five apoptotic regulators, the authors found that the measured inherent variability in the levels of these proteins was enough to account for the variability in cellular response time between induction and apoptosis itself. Thus, inherent distributed protein levels can lead to graded responses to stress at the population level, and can improve the chances that a small population of cells survives a particular stress. Similarly, variable response to stress as a bet-hedging strategy was theoretically predicted [14] and later experimentally demonstrated in yeast [15], where it was shown that more stochastic expression of MSN2/4 target genes increased the population survival rate under stress by 20%. The examples above demonstrate that protein expression noise plays a regulatory role in population-level co-operation, co-ordination, and survival. Discovering and understanding such regulatory mechanisms requires single-cell measurements.
New technology can enable new biology
Just as the seminal work finding gene expression variability [6] depended on a new technology, so did current efforts to understand and control cell function and fate. Studying regulation across heterogeneous cellular systems, such as human tissues, cancers, or differentiating cells, demands technologies that can measure gene expression at the systems level. Toward this end, single-cell RNA-seq methods have made much progress at measuring single-cell transcriptomes [16]. Over the last decade, the technology has progressed rapidly from identifying transcripts in single cells [17] to droplet-based sequencing of tens of thousands of cells [18]. This progress has led to its widespread use for classifying subpopulations of cells, often uncovering previously undetected but biologically relevant rare cell subpopulations [19,20].
While single-cell RNA-seq techniques are steadily advancing our knowledge of heterogeneous cell systems, they can only capture a portion of the transcriptional gene expression profile in any given cell. Since sequence reads are split amongst all analyzed cells, increasing the number of analyzed cells per experiment decreases the number of sequence reads per cell. This trend increases the number of missing data points, which were present and confounded the analysis even of lower throughput methods [21]. Since single-cell RNA-seq methods usually captured less than 20% of the mRNAs, they incur substantial sampling error, especially given the low average mRNA copy number per cell (median: ~17). Proteins have >1000-fold higher average copy numbers per cell (median: ~50000), and thus single-cell proteomics has an opportunity to alleviate the uncertainty incurred by sampling error [22].
Single-cell RNA-seq techniques have been transformative [19,20] and continue to advance RNA-based biological research, but mRNA levels alone are insufficient for characterizing, understanding, and controlling biological systems. Even at the superficial descriptive level, single-cell RNA-seq cannot capture post-translational modifications (PTMs). Well-characterized regulatory mechanisms, such as long-lived proteins [23] and translational regulation [24] make mRNAs poor surrogates for functional proteins. Munsky et al. [25] simulated the distributions of mRNA and protein levels for a number of gene regulatory motifs, and showed that depending on the motif, mRNA levels were not necessarily correlated to the corresponding protein’s level. The different time scales of mRNAs and proteins could account for differences between mRNA and protein levels. Globally, mRNA levels can explain differences between the abundance of different proteins, but only poorly explain the relative changes of individual proteins across human tissues [26]. These relative differences implicate post-transcriptional regulation as an important regulatory mechanism that shapes tissue-specific proteomes. In order to characterize the molecular and signaling mechanisms controlling cell function, new technologies for quantitating all proteoforms in single cells will be needed. A proteoform is defined as the set of all molecular forms of a protein produced from one gene [27]. Different proteoforms of the same protein can have different functions and need to be quantitated distinctly [28]. Ultimately, comprehensive quantitation of proteoforms across thousands of single cells can reduce the number of assumptions in signaling network modelling, and even enable causal inference [22].
Although quantitating proteins in single cells is necessary for systems-level analysis, it is also insufficient. Additional measurements of other layers of biological regulation can capture important information upstream of translation, furnishing a more complete understanding of a regulatory motif or pathway. To characterize the interplay between various layers of regulation at a systems level [29], new single-cell technologies and studies should strive toward ‘multiomics’ methods [30] that enable simultaneous systems-level measurement of proteins, metabolites, mRNAs, and DNA modifications.
Methods for quantifying protein levels in single cells
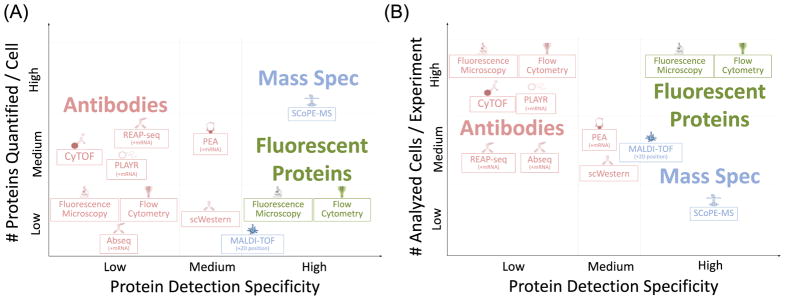
There are three major modalities for identifying and quantitating proteins in single cells: (i) genetically engineered fluorescent proteins, (ii) antibodies, and (iii) mass spectrometry (MS). The first two have so far dominated cellular protein research, and have enabled many discoveries, including those reviewed above. The third method shows the greatest promise for increasing both the specificity and the throughput of single-cell protein analysis, Figure 1. Below we discuss the distinctive advantages and weaknesses of the current methods, which are summarized in Table 1.
Figure 1. Classification of single-cell protein analysis methods based on their specificity, proteome coverage, and cell throughput.
(A) Antibody-based methods, in red, are widely utilized and generally applicable to intermediate number of proteins at once. Their specificity depends on the antibody and can be rather low. The specificity of antibodies can be increased by electrophoretic separation (scWestern) or using multiple antibodies per protein (PEA). Fluorescent protein-based methods, in green, are highly specific and facilitate monitoring protein levels over time, but are limited to quantitating only a few proteins per cell because of spectral overlap. MS can increase both specificity and depth of coverage. MALDI-TOF has been used to study single cells and spatial questions for decades, but it offers only medium specificity and low proteome coverage. SCoPE-MS enables simultaneous identification and quantitation of hundreds of proteins from single cells, and demonstrates one path toward comprehensive quantitation of proteins in single cells. (B) Flow cytometry-based and automated imaging techniques, encompassing many antibody and fluorescent protein-based methods, can robustly assay tens of thousands of cells per experiment, lighting the way for statistical analysis of single cell data. For the remaining techniques of higher specificity antibodies or MS, there is generally a trade-off between specificity and cellular throughput. All values are applicable for a typically sized mammalian cell, with a diameter of ~15 μm and ~500 pg of total protein. Abbreviations: PEA, proximity extension assay; SCoPE-MS, single cell proteomics by MS; scWestern, single-cell Western blot.
Table 1.
Comparison of methods for single cell-protein analysis
| Method name | Method type | Specificity | # Quantified proteins/cell | # Analyzed cells | Cell fixation | PTM quantification | Examples of applications |
|---|---|---|---|---|---|---|---|
| Microscopy | Fluorescent protein | High | Low | High | Compatible | No | Protein dynamics [6,31] |
| Flow cytometry | Fluorescent protein | High | Low | High | Required | No | Gene expression variability [9,15] |
| Microscopy | Antibody | Low | Low | High | Compatible | Yes | Spatial mapping [43] |
| Flow Cytometry | Antibody | Low | Low | High | Required | Yes | Cell subtype heterogeneity [13,54] |
| CyTOF | Antibody | Low | Medium | High | Required | Yes | Immune cell subtype heterogeneity [52] |
| PLAYR | Antibody (+mRNA) | Low | Medium | High | Required | Yes | mRNA and protein discrepancies [80] |
| Abseq | Antibody (+mRNA) | Low | Low | Medium | Compatible | Yes | Surface marker heterogeneity [53] |
| REAP-seq | Antibody (+mRNA) | Low | Medium | Medium | Compatible | Yes | Immune cell subtype heterogeneity [48] |
| scWestern | Antibody | Medium | Low | Medium | Compatible | Yes | Resolve non-specific probe signal [55,56] |
| PEA | Antibody (+mRNA) | Medium | Medium | Medium | Compatible | Yes | Drug response heterogeneity [57,81] |
| MALDI-TOF | Mass Spec | Medium/High | Medium/High | Medium | Compatible | Yes | Spatial mapping of molecules [61,62] |
| SCoPE-MS | Mass Spec | High | High | Low | Limited compatibility | Likely | Stem cell population heterogeneity [70] |
Methods for protein quantitation in single cells exhibit an inverse relationship between detection specificity and number of analyzed cells per experiment. Most methods are compatible with fixed cells, some require it, and MS-based methods are compatible with some fixatives, e.g. methanol, but poorly compatible with others, e.g. formaldehyde. All antibody-based methods can detect PTMs, and while MS can accomplish this for larger samples, its potential to do so at the single-cell level remains unproven. Most recent methods have emphasized increases in depth and throughput to allow exploring global gene expression variability at the protein level, in order to address questions of functional heterogeneity in diverse cell populations. Abbreviations: CyTOF, mass cytometry; PEA, proximity extension assay; REAP-seq, RNA expression and protein sequencing; SCoPE-MS, single cell proteomics by MS; scWestern, single cell Western blot.
Fluorescent proteins enable quantifying protein dynamics
Since the discovery and cloning of GFP [5], the community has engineered many fluorescent proteins with substantially enhanced functions, such as different spectral characteristics, fast folding and maturation, increased and decreased resistance to photobleaching, and FRET [31,32]. Fluorescent proteins allow dynamic measurements of protein levels and location over time, Table 1. Such measurements have been instrumental for discovering biological functions that depend not merely on the levels of a protein, but also on its dynamics [33–36]. For example, different dynamics of p53 induce the transcription of different sets of genes and different cell fates [37]. Indeed, important cellular functions are regulated by dynamic signaling mechanisms [38,39], and thus measuring protein dynamics is essential to understand biological systems. However, the number of proteins that can be quantitated per cell using fluorescent proteins remains capped at ~12, limited by the fluorophores’ spectral overlap [40]. Furthermore, fluorescent proteins have limited utility with systems that cannot be genetically engineered, such as clinical samples. Engineering new suites of fluorescent proteins for each new biological question requires much time and effort, and is often prohibitive for systems level measurements.
Antibody-based methods
Antibodies can target some protein portion of many cellular pathways in single cells, for instance enabling studies of emergent cancer resistance [41], and can be applied over a broad dynamic range [42]. Immunohistochemistry enables visualization of tissue sections with single-cell resolution, while immunocytochemistry does the same for monolayer cell cultures [43]. Antibodies are frequently incorporated in flow cytometry, characterizing patterns of a few proteins across tens of thousands of cells [44]. Additionally, antibodies have a long history of detecting and quantitating PTMs. However, detecting multiple PTMs per protein, especially PTMs in close proximity, with antibodies can be challenging. Antibodies face two primary hurdles for more comprehensive protein coverage: specificity and scalability [45–48]. Indeed, a recent study tested 1124 antibodies and validated only 354 (~31%) antibodies [49]. New methods are attempting to overcome these challenges by increasing protein multiplexing while maintaining or improving antibody specificity for protein quantitation in single cells. See Figure 1 and Table 1.
Conjugating fluorophores to antibodies was the first widely used readout of antibody binding and continues to be used widely in conjunction with microscopy and flow cytometry (Figure 1 and Table 1). These methods allow quantitating many cells per unit time, and microscopy provides spatial information on protein localization. A primary limitation of antibody-conjugated fluorophores is the overlap between the spectra of distinct fluorophores. This overlap allows relatively few antibody-conjugated fluorophores to be assayed at a time (Figure 1 and Table 1). More recent antibody-based methods aim to relieve this limitation by using antibody readouts that allow for assaying more analytes per cell.
Such readouts, used by mass cytometry (CyTOF) [50], are transition element isotopes conjugated to antibodies. Since these metals are normally absent from biology, the non-specific background is minimal. CyTOF starts by labeling fixed cells with antibodies conjugated to transition metals. Then droplets containing single cells are isolated, the cells vaporized, and the remaining transition metals analyzed by a TOF mass spectrometer. CyTOF is a mature technology that can routinely analyze tens of thousands of cells per hour, detect and quantitate very lowly abundant epitopes such as PTMs, and has been multiplexed to simultaneously process 60 samples [51]. Bendall et al. [52] employed CyTOF to quantitate the immune response of thousands of single cells from healthy human bone marrow samples. They developed two panels of antibodies designed to interrogate different aspects of the immune response. From the 13 proteins common to both panels, the authors created a map of phenotypically and presumably functionally linked immune cell populations. The authors then overlaid the data from the 18 remaining, panel-specific proteins to refine their map, uncovering further heterogeneity within numerous subsets of the larger, annotated cell populations. Although CyTOF begins to probe the pathway level for thousands of cells at a time, it relies on single antibodies, and is limited to fewer than 40 epitopes per cell, Table 1.
Another approach for increasing the antibody readouts per cell relies on conjugating antibodies with DNA oligonucleotides (barcodes), and a number of recent methods, including Abseq, cellular indexing of transcriptome and epitope by sequencing (CITE-seq), and RNA expression and protein sequencing (REAP-seq), have employed this strategy [48,53,54] (see Figure 1 and Table 1). With Abseq, single cells are incubated with barcoded antibodies recognizing cell-surface proteins; the method is limited to surface proteins. The incubation is done in a high-throughput microfluidic device, then the barcodes are amplified by PCR and the resulting DNA sequenced. The number of reads per barcode is interpreted as a surrogate for the protein level. CITE-seq and REAP-seq also employ droplet microfluidics, and subsequently generate protein and RNA level readouts by integrating oligonucleotide-tagged antibodies into established single cell transcriptomic workflows. The microfluidic medium enables parallel processing of thousands of single cells, fewer than flow-based technologies such as CyTOF. Although there is no practical limit to the number of unique identifiers that can be chemically conjugated to antibodies of choice, these methods are inherently limited by the number of available antibodies, their specificity, the epitope availability, and the number of antibodies that can be introduced per cell before molecular crowding becomes a limiting factor. These limitations have so far circumscribed the number of quantitated proteins to two surface markers for Abseq and ~80 proteins for REAP-seq.
Approaches for increasing the specificity of antibody-based methods
The above methods depend critically on the specificity of a single antibody and will perform very poorly in the presence of non-specific binding. To alleviate such concerns, two strategies have been developed to increase the specificity of antibody-based single-cell methods as described below:
First, single-cell Western blotting (scWestern) increases the specificity of antibodies by physically separating proteins from single-cell lysates. This is accomplished by electrophoresis [55] or isoelectric focussing of proteins [56], and the added dimension of separation helps single-antibody probing resolve non-specific signals. Hughes et al. [55] used in-house fabricated open-microwell arrays to separate proteins with at least 50% different masses from single cells. In the open-microwell format, the authors simultaneously applied scWestern to 5040 single-cell samples and obtained quantitative measurements for 1608 of these samples in just over an hour. However, the method introduces additional protein loss of ~40% for each single cell, and introduces a limit of detection of 27000 molecules, while the median protein has ~50000 copies per cell, e.g. per murine fibroblast. Although the same blot can be stripped and reprobed for different proteins upward of nine times, the number of proteins simultaneously measurable by scWestern is fundamentally the number of re-blotting cycles and the number of high-quality antibodies.
The second approach to increasing signal specificity for antibody-based methods is the proximity extension assay (PEA). PEA increases specificity by requiring the binding of two different oligonucleotide-tagged antibodies to the same protein before a signal can be generated [57,58]; see Figure 1 and Table 1. Two antibodies bound to the same protein carry overlapping sequences that ligate upon binding, allowing subsequent extension, amplification, digestion, and quantitation of a few dozen proteins by microfluidic qPCR, which can be performed on plates of FACS-sorted single cells, yielding a throughput of ~100 cells per hour. Requiring two antibodies reduces background signal compared with single-antibody probing, and the less stringent specificity requirements permit the use of a wider range of antibodies. However, not all of these additional antibodies are necessarily applicable; epitope availability and molecular crowding inhibit how comprehensively PEA can be applied, since any given protein must have two antibody binding sites that are amenable to oligonucleotide overlap.
Single-cell antibody methods will continue to scale as efforts to retain or improve antibody specificity evolve (Table 1).
MS-based methods
Proteins can be identified and quantitated by MS. Indeed, MS applied to bulk samples comprising millions of cells can already measure thousands of proteins at once with high specificity [59,60], including important PTMs that affect cell function and dynamics, such as phosphorylation [52]. Although most MS methods remain bound to bulk samples, new methods are enabling the simultaneous analysis of more complete proteomes of increasing numbers of single cells.
A combination of matrix assisted laser desorption/ionization (MALDI) with time-of-flight (TOF) MS, commonly abbreviated as MALDI-TOF, has been applied to single cells for approximately two decades [61,62], enabling identification and spatial mapping of dozens of molecules, metabolites, and peptides, Figure 1. However, the variability in the fraction of peptides ionized across samples limits the quantitative accuracy of measurements relying on MALDI ionization. Furthermore, since peptides are not separated and enter the instrument at the same time, the acquired spectra comprise a mixture of the spectra of many peptides. These complex spectra are hard to interpret and relatively few peptide sequences can be confidently identified. Thus, while MALDI has been employed widely in the spatial mapping of neuropeptides in single neurones [63] or proteins in tissue samples [64], it remains bound to biological questions of localization rather than quantitation.
The MS method that has allowed for well-controlled and accurate measurements of tens of thousands of proteins is liquid chromatography (LC) combined with electrospray ionization (ESI) and tandem MS, usually abbreviated as LC-MS/MS. Ideally, LC-MS/MS can be applied to single-cell lysates to give the deep and accurate quantitation of proteins and proteoforms that it has afforded with bulk samples. However, large losses during sample preparation and delivery for LC-MS/MS and relatively low sensitivity have limited its application to very small samples.
Very sensitive workflows have begun to apply LC-MS/MS to small samples comprising hundreds of human cells [60] and even to large single cells, such as oocytes [65,66] and muscle fibers [67,68]. These applications developed and applied very sensitive methods to quantitate between 450 and 800 proteins in single human oocytes, and ~2100 proteins in single muscle fibers. Yet the typical single mammalian cell contains orders of magnitude less protein than oocytes and muscle fibers [69] and poses even greater challenges. Our early attempts at using LC-MS/MS to identify and quantify proteins from typically sized single mammalian cells, dubbed single-cell proteomics by MS (SCoPE-MS) [70], aimed to increase the efficiency of delivering proteins to the MS instrument and the confidence of peptide sequencing by combining two strategies.
The first strategy sought to minimize sample loss by optimizing a ‘clean’ sample processing pipeline that required no chemical cleanup; rather, it employed mechanical cell lysis by sonication and only LC- and MS-compatible reagents. This obviated the losses typically incurred by chemical cleanup and pipetting. The second strategy increased both the number of quantitated cells per run and the confidence in peptide identification by using isobaric mass tags [71]. These tags covalently bond with peptides, allowing peptides from different samples to be distinguished from one another. We used these tags to label the cell lysates of single cells as well as the lysate of 200 cells, termed carrier cells, and combined the labeled single cells and the carrier cells into a single sample. Incorporating the carrier cells into the workflow conferred two benefits: (i) it reduced the peptide loss experienced by the single cells during sample preparation and nanoLC separation, and (ii) it improved identification confidence by increasing the total number of ions delivered to the mass spectrometer. Combining these strategies minimized sample loss in the processing pipeline, which then delivered enough single-cell sample for confident identification. This allowed reliable relative quantitation of ~600 proteins in any given single cell, and over 1000 proteins across hundreds of differentiating mouse embryonic stem cells.
We have outlined specific strategies that can increase throughput – both number of quantitated proteins and number of analyzed cells – by orders of magnitude [22]. Such improvements are required for MS to become a powerful platform for single cell protein analysis, Figure 1. Though current MS-based methods are limited in cell throughput to ~10 cells/h per instrument, incorporating existing automation technologies and increasing cellular multiplexing can increase cell throughput, a critical determinant of statistical power [22]. Reducing sample volumes from microliters to nanoliters can significantly alleviate protein adsorption, as demonstrated recently by lysing cells in hundreds of nanoliters using a custom chip dubbed ’nanodroplet processing in one pot for trace sample’ (nanoPOTS) [72]. Another avenue for improvement is peptide separation. It can be achieved not only by LC but also by capillary electrophoresis (CE), and CE-MS/MS [66] can offer some advantages over LC-MS/MS for very limited complex samples, such as the proteomes of single cells, since it allows reduced flow rates and thus improved ionization of molecules. Furthermore, CE-MS/MS is the most promising method for quantitating proteins without having to digest them in what is commonly referred to as top-down MS. Critically, the LC-MS/MS strategy also has the potential to measure PTMs, such as phosphorylation and glycosylation [73–75]. The development of such methods is just beginning and although unpublished results from our laboratory demonstrate their feasibility, much further development is needed to make single-cell PTM analysis routine. Finally, technical improvements in parallel ion accumulation and sampling efficiency will improve the sensitivity, accuracy, and depth of quantitation across the analyzed cells [22].
Many of the analytical methods developed for bulk MS data can be applied to single-cell MS data as well, but single-cell data pose additional challenges that will motivate the development of new methods. Some of the challenges for single-cell proteomics are similar to those for scRNA-seq, e.g. significant measurement noise and missing data (i.e. not all proteins will be quantitated in all cells). We believe that the fraction of missing data can be reduced by using targetted MS approaches, while the noise can be reduced by improved ion sampling, e.g. by increasing the ion accumulation time [22]. These aspects of the data can also benefit from methods developed for scRNA-seq data [21]. Other challenges, such as reliable sequence identification, are specific to single-cell proteomics and will necessitate new analytical methods, including improved analysis of MS spectra and the use of additional informative features, such as retention time [22].
More excitingly, we believe that quantitating all relevant proteoforms across thousands of single cells can enable powerful data-driven models of biological networks. Empirically estimated joint and conditional distributions can obviate assumptions about the functional form of interactions between proteins and afford distinguishing direct from indirect effects in biological networks [22]. While the inference of probabilistic graphical models has received much attention in biology [76,77], it has always been limited by a lack of sufficient observations across conditions/cells, by missing variables (e.g. only mRNAs are measured while key proteins and PTMs that mediate signals are not), and by the confounding effects of population averages. We believe high-throughput single-cell MS has the potential to overcome these data limitations and will motivate the development of new inference algorithms that can accommodate loopy networks and afford efficient computation based on empirical distributions. If successful, these approaches will transcend mere molecular descriptions and empower models that enable rational control of cells, e.g. efficient directed differentiation, and hopefully uncover new principles of emergent biological behavior.
Simultaneous quantitation of proteins and RNAs in single cells
While quantitating protein levels in single cells can be powerful alone, it is even more powerful when combined with the quantitated transcriptomes of the same single cells. Thus, an important direction in the advancement of single-cell proteomics methods is making them compatible with single-cell transcriptomic methods, which are generally more mature [30], and single-cell metabolomics methods, many of which already employ MS [78,79].
In initial attempts, both CyTOF and PEA, described above, have been used to quantitate some transcripts and their corresponding proteins in the same single cell. To enable quantitating mRNAs by CyTOF, Frei et al. [80] developed and integrated a proximity ligation assay for RNA (PLAYR) into the CyTOF workflow. The PLAYR method is compatible with flow cytometry and CyTOF, and the authors used both to quantitate transcripts alone, in addition to simultaneously quantitating ten transcripts and corresponding proteins in single primary human peripheral blood mononuclear cells (PBMCs). Darmanis et al. [81] extended the use of PEA to simultaneously measure ~22 mRNAs and corresponding proteins by performing TaqMan and PEA on the split lysate of single neural stem cells undergoing BMP4-induced differentiation. Their comparative analysis of the predictive power of mRNA and/or protein levels for assigning single cells to a treatment group demonstrated that proteins were better predictors for the functional response to BMP4-treatment than RNA, though both proteins and mRNA levels contributed unique information; the combined data predicted treatment group more effectively than mRNA or protein alone.
Ultimately, we would like to comprehensively quantitate the transcriptome, the proteome, and the metabolome of the same single cell. RNA-seq is steadily advancing high-throughput single-cell transcriptomics toward this end, while MS offers the most promising method for high-throughput single-cell proteomics and metabolomics. In the short term, single-cell multiomics experiments will likely be performed on cell lysates divided in half, where one part is sent for nucleotide base sequencing and the other for MS-based proteomic and metabolic analysis. The losses incurred by such lysate splitting could be mitigated by first amplifying the nucleic acids or by developing techniques that allow nearly lossless separation of molecular types, an area in which we expect to see much progress in the near future.
Concluding remarks
Growing evidence elaborates on how protein-level cellular heterogeneity is intricately tied to cell fate in such varied biological contexts as cancer, differentiation, and mitosis. Transcriptomic studies provide an appreciation for the heterogeneous nature of the cells making up these systems, but cannot capture most PTMs, such as phosphorylation and glycosylation, which are critical layers of regulation. Ultimately, single cell proteomic measurements are required to characterize functional variability at the systems level. MS is poised to enable such measurements, and will continue to improve alongside parallel advances in instrumentation, automation, and computation. MS can also enable multilayer analyses of the same single cell beyond the protein level and begin to quantitate PTMs, metabolomes and transcriptomes simultaneously with proteomes. These high-powered, high-dimensional data will power systems-level measurement and characterization across many biological questions, and have an opportunity to set new standards of accuracy, applicability, and depth in quantitative systems biology.
Summary.
The levels of a protein can vary across single cells both because of stochastic influences, noise in gene expression due to low-copy-number molecules, and because of cellular responses and regulatory mechanisms.
Protein levels influence cellular functions and determine cell fate.
Current technologies enable the simultaneous study of a few proteins, but face challenges when scaled to pathway-level multiplexing.
The next generation of systems biology needs more powerful methods for quantitating proteins in single cells.
MS is poised to enable deep quantitation of single-cell proteomes for the next generation of systems biology.
Abbreviations
- BMP4
bone morphogenic protein 4
- CDK2
cyclin-dependent kinase 2
- CE
capillary electrophoresis
- CITE-seq
cellular indexing of transcriptome and epitope by sequencing
- CyTOF
mass cytometry
- ESI
electrospray ionization
- MALDI
matrix assisted laser desorption/ionization
- nLC
nano liquid chromatography
- PEA
proximity extension assay
- PLAYR
proximity ligation assay for RNA
- PTM
post-translational modification
- REAP-seq
RNA expression and protein sequencing
- scWestern
single cell Western blot
Footnotes
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Bigger J. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet. 1944;244:497–500. doi: 10.1016/S0140-6736(00)74210-3. [DOI] [Google Scholar]
- 2.Delbruck M. The burst size distribution in the growth of bacterial viruses (bacteriophages) J Bacteriol. 1945;50:131–135. doi: 10.1128/JB.50.2.131-135.1945. [DOI] [PubMed] [Google Scholar]
- 3.Novick A, Weiner M. Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci USA. 1957;43:553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spudich JL, Koshland DE. Non-genetic individuality: chance in the single cell. Nature. 1976;262:467–471. doi: 10.1038/262467a0. [DOI] [PubMed] [Google Scholar]
- 5.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 6.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. https://doi.org/0.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 7.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spencer SL, Cappell SD, Tsai FC, Overton KW, Wang CL, Meyer T. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell. 2013;155:369–383. doi: 10.1016/j.cell.2013.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 10.Ptashne M, Backman K, Humayun MZ, Jeffrey A, Maurer R, Meyer B, et al. Autoregulation and function of a repressor in bacteriophage lambda. Science. 1976;194:156–161. doi: 10.1126/science.959843. [DOI] [PubMed] [Google Scholar]
- 11.Coleman J, Inouye M, Nakamura K. Mutations upstream of the ribosome-binding site affect translational efficiency. J Mol Biol. 1985;181:139–143. doi: 10.1016/0022-2836(85)90332-8. [DOI] [PubMed] [Google Scholar]
- 12.Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thattai M, van Oudenaarden A. Stochastic gene expression in fluctuating environments. Genetics. 2004;167:523–530. doi: 10.1534/genetics.167.1.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart-Ornstein J, Weissman JS, El-Samad H. Cellular noise regulons underlie fluctuations in Saccharomyces cerevisiae. Mol Cell. 2012;45:483–493. doi: 10.1016/j.molcel.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saliba A-E, Westermann AJ, Gorski SA, Vogel J. Single-cell RNA-seq: advances and future challenges. Nucleic Acids Res. 2014;42:8845–8860. doi: 10.1093/nar/gku555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 18.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C, et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature. 2017;546:431–435. doi: 10.1038/nature22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stubbington MJT, Rozenblatt-Rosen O, Regev A, Teichmann SA. Single-cell transcriptomics to explore the immune system in health and disease. Science. 2017;358:58–63. doi: 10.1126/scienceaan6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicks SC, Townes FW, Teng M, Irizarry RA. Missing data and technical variability in single-cell RNA-sequencing experiments. Biostatistics. 2017 doi: 10.1093/biostatistics/kxx053. [DOI] [PMC free article] [PubMed]
- 22.Specht H, Slavov N. Transformative opportunities for single cell proteomics. Journal of Proteome Research. 2018 doi: 10.1021/acs.jproteome.8b00257. [DOI] [PMC free article] [PubMed]
- 23.Thayer NH, Leverich CK, Fitzgibbon MP, Nelson ZW, Henderson KA, Gafken PR, et al. Identification of long-lived proteins retained in cells undergoing repeated asymmetric divisions. Proc Natl Acad Sci USA. 2014;111:14019–14026. doi: 10.1073/pnas.1416079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polymenis M, Schmidt EV. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munsky B, Neuert G, van Oudenaarden A. Using gene expression noise to understand gene regulation. Science. 2012;336:183–187. doi: 10.1126/science.1216379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franks A, Airoldi E, Slavov N. Post-transcriptional regulation across human tissues. PLoS Comput Biol. 2017;13:e1005535. doi: 10.1371/journal.pcbi.1005535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith LM, Kelleher NL, Linial M, Goodlett D, Langridge-Smith P. Proteoform: a single term describing protein complexity. Nat Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malioutov D, Chen T, Jaffe J, Airoldi E, Carr S, Budnik B, et al. Quantifying homologous proteins and proteoforms. bioRxiv. 2017 doi: 10.1101/168765. [DOI] [PMC free article] [PubMed]
- 29.Cheng Z, Otto GM, Powers EN, Keskin A, Mertins P, Carr SA, et al. Pervasive, coordinated protein-level changes driven by transcript isoform switching during meiosis. Cell. 2018;172:910–923. e16. doi: 10.1016/j.cell.2018.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macaulay IC, Ponting CP, Voet T. Single-cell multiomics: multiple measurements from single cells. Trends Genet. 2017;33:155–168. doi: 10.1016/j.tig.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 32.Cranfill PJ, Sell BR, Baird MA, Allen JR, Lavagnino Z, de Gruiter HM, et al. Quantitative assessment of fluorescent proteins. Nat Methods. 2016;13:557–562. doi: 10.1038/nmeth.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muzzey D, van Oudenaarden A. Quantitative time-lapse fluorescence microscopy in single cells. Annu Rev Cell Dev Biol. 2009;25:301–327. doi: 10.1146/annurev.cellbio.042308.113408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, et al. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- 35.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine JH, Lin Y, Elowitz MB. Functional roles of pulsing in genetic circuits. Science. 2013;342:1193–1200. doi: 10.1126/science.1239999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purvis JE, Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell. 2013;152:945–956. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slavov N, Carey J, Linse S. Calmodulint ransduces Ca2+ oscillations into differential regulation of its target proteins. ACS Chem Neurosci. 2013;4:601–612. doi: 10.1021/cn300218d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Autissier P, Soulas C, Burdo TH, Williams KC. Evaluation of a 12-color flow cytometry panel to study lymphocyte, monocyte, and dendritic cell subsets in humans. Cytom Part A. 2010:9999A. doi: 10.1002/cyto.a.20859. [DOI] [PMC free article] [PubMed]
- 41.Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C, et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature. 2017;546:431–435. doi: 10.1038/nature22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ullal AV, Peterson V, Agasti SS, Tuang S, Juric D, Castro CM, et al. Cancer cell profiling by barcoding allows multiplexed protein analysis in fine-needle aspirates. Sci Transl Med. 2014;6:219ra9. doi: 10.1126/scitranslmed.3007361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller MA, Weissleder R. Imaging of anticancer drug action in single cells. Nat Rev Cancer. 2017;17:399–414. doi: 10.1038/nrc.2017.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adan A, Alizada G, Kiraz Y, Baran Y, Nalbant A. Flow cytometry: basic principles and applications. Crit Rev Biotechnol. 2017;37:163–176. doi: 10.3109/07388551.2015.1128876. [DOI] [PubMed] [Google Scholar]
- 45.Michaud GA, Salcius M, Zhou F, Bangham R, Bonin J, Guo H, et al. Analyzing antibody specificity with whole proteome microarrays. Nat Biotechnol. 2003;21:1509–1512. doi: 10.1038/nbt910. [DOI] [PubMed] [Google Scholar]
- 46.Bumbaca D, Wong A, Drake E, Reyes AE, II, Lin BC, Stephan J-P, et al. Highly specific off-target binding identified and eliminated during the humanization of an antibody against FGF receptor 4. MAbs. 2011;3:376–386. doi: 10.4161/mabs.3.4.15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Persson H, Preger C, Marcon E, Lengqvist J, Graslund S. Antibody Validation by Immunoprecipitation Followed by Mass Spectrometry Analysis. Humana Press; New York, N.Y: 2017. pp. 175–187. [DOI] [PubMed] [Google Scholar]
- 48.Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, et al. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol. 2017;35:936–939. doi: 10.1038/nbt.3973. [DOI] [PubMed] [Google Scholar]
- 49.Marcon E, Jain H, Bhattacharya A, Guo H, Phanse S, Pu S, et al. Assessment of a method to characterize antibody selectivity and specificity for use in immunoprecipitation. Nat Methods. 2015;12:725–731. doi: 10.1038/nmeth.3472. [DOI] [PubMed] [Google Scholar]
- 50.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 51.McCarthy RL, Mak DH, Burks JK, Barton MC. Rapid monoisotopic cisplatin based barcoding for multiplexed mass cytometry. Sci Rep. 2017;7:3779. doi: 10.1038/s41598-017-03610-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bendall SC, Simonds EF, Qiu P, Amir ED, Krutzik PO, Finck R, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shahi P, Kim SC, Haliburton JR, Gartner ZJ, Abate AR. Abseq: Ultrahigh-throughput single cell protein profiling with droplet microfluidic barcoding. Sci Rep. 2017;7:44447. doi: 10.1038/srep44447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hughes AJ, Spelke DP, Xu Z, Kang CC, Schaffer DV, Herr AE. Single-cell western blotting. Nat Methods. 2014;11:749–755. doi: 10.1038/nmeth.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tentori AM, Yamauchi KA, Herr AE. Detection of isoforms differing by a single charge unit in individual cells. Angew Chem Int Ed (Engl ) 2016;55:12431–12435. doi: 10.1002/anie.201606039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lundberg M, Eriksson A, Tran B, Assarsson E, Fredriksson S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011;39:e102. doi: 10.1093/nar/gkr424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Assarsson E, Lundberg M, Holmquist G, Björkesten J, Bucht Thorsen S, Ekman D, et al. Homogenous 96-Plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE. 2014;9:e95192. doi: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 60.Li S, Plouffe BD, Belov AM, Ray S, Wang X, Murthy SK, et al. An integrated platform for isolation, processing, and mass spectrometry-based proteomic profiling of rare cells in whole blood. Mol Cell Proteomics. 2015;14:1672–1683. doi: 10.1074/mcp.M114.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69:47511–4760. doi: 10.1020/ac970888i. [DOI] [PubMed] [Google Scholar]
- 62.Li L, Garden RW, Sweedler JV. Single-cell MALDI: a new tool for direct peptide profiling. Trends Biotechnol. 2000;18:151–160. doi: 10.1016/S0167-7799(00)01427-X. [DOI] [PubMed] [Google Scholar]
- 63.Hummon AB, Amare A, Sweedler JV. Discovering new invertebrate neuropeptides using mass spectrometry. Mass Spectrom Rev. 2006;25:77–98. doi: 10.1002/mas.20055. [DOI] [PubMed] [Google Scholar]
- 64.Schwartz SA, Reyzer ML, Caprioli RM. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J Mass Spectrom. 2003;38:699–708. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- 65.Virant-Klun I, Leicht S, Hughes C, Krijgsveld J. Identification of maturation-specific proteins by single-cell proteomics of human oocytes. Mol Cell Proteomics. 2016;15:2616–2627. doi: 10.1074/mcp.M115.056887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lombard-Banek C, Moody SA, Nemes P. Single-cell mass spectrometry for discovery proteomics: quantifying translational cell heterogeneity in the 16-cell frog (Xenopus) embryo. Angew Chem Int Ed (Engl ) 2016;55:2454–2458. doi: 10.1002/anie.201510411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Murgia M, Nagaraj N, Deshmukh AS, Zeiler M, Cancellara P, Moretti I, et al. Single muscle fiber proteomics reveals unexpected mitochondrial specialization. EMBO Rep. 2015;16:387–395. doi: 10.15252/embr.201439757. https://doi.org/10.15252/embr.201439757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murgia M, Toniolo L, Nagaraj N, Ciciliot S, Vindigni V, Schiaffino S, et al. Single muscle fiber proteomics reveals fiber-type-specific features of human muscle aging. Cell Rep. 2017;19:2396–2409. doi: 10.1016/j.celrep.2017.05.054. [DOI] [PubMed] [Google Scholar]
- 69.Milo R, Jorgensen P, Moran U, Weber G, Springer M. BioNumbers—the database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010;38:D750–D753. doi: 10.1093/nar/gkp889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Budnik B, Levy E, Harmange G, Slavov N. Mass-spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. bioRxiv. 2018 doi: 10.1101/102681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 72.Zhu Y, Piehowski PD, Zhao R, Chen J, Shen Y, Moore RJ, et al. Nanodroplet processing platform for deep and quantitative proteome profiling of 10–100 mammalian cells. Nat Commun. 2018;9:882. doi: 10.1038/s41467-018-03367-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Slavov N, Budnik B, Schwab D, Airoldi E, van Oudenaarden A. Constant growth rate can be supported by decreasing energy flux and increasing aerobic glycolysis. Cell Rep. 2014;7:705–714. doi: 10.1016/j.celrep.2014.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Steen H, Jebanathirajah JA, Rush J, Morrice N, Kirschner MW. Phosphorylation analysis by mass spectrometry myths, facts, and the consequences for qualitative and quantitative measurements*. Mol Cell Proteomics. 2005;5:172–181. doi: 10.1074/mcp.M500135-MCP200. [DOI] [PubMed] [Google Scholar]
- 75.Morelle W, Michalski J-C. Analysis of protein glycosylation by mass spectrometry. Nat Protoc. 2007;2:1585–1602. doi: 10.1038/nprot.2007.227. [DOI] [PubMed] [Google Scholar]
- 76.Koller D, Friedman N. Probabilistic Graphical Models: Principles and Techniques. MIT Press; 2009. p. 1233. [Google Scholar]
- 77.Slavov N. Inference of sparse networks with unobserved variables. Application to gene regulatory networks. Proc Machine Learning Res. 2010;9:757–764. [Google Scholar]
- 78.Zenobi R. Single-cell metabolomics: analytical and biological perspectives. Science. 2013;342:1243259. doi: 10.1126/science.1243259. [DOI] [PubMed] [Google Scholar]
- 79.Rubakhin SS, Lanni EJ, Sweedler JV. Progress toward single cell metabolomics. Curr Opin Biotechnol. 2013;24:95–104. doi: 10.1016/j.copbio.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frei AP, Bava FA, Zunder ER, Hsieh EWY, Chen SY, Nolan GP, et al. Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat Methods. 2016;13:269–275. doi: 10.1038/nmeth.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Darmanis S, Gallant CJ, Marinescu VD, Niklasson M, Segerman A, Flamourakis G, et al. Simultaneous multiplexed measurement of RNA and proteins in single cells. Cell Rep. 2016;14:380–389. doi: 10.1016/j.celrep.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]