Abstract
Omics technologies have made it easier and cheaper to evaluate thousands of biological molecules at once. These advances have led to novel therapies approved for use in the clinic, elucidated the mechanisms behind disease-associated mutations, led to increased accuracy in disease subtyping and personalized medicine, and revealed novel uses and treatment regimes for existing drugs through drug repurposing and pharmacology studies. In this review, we summarize some of these milestones and discuss the potential of integrative analyses that combine multiple data types for further advances.
Introduction
The “omics revolution” that has been sweeping biological research since the advent of genomic sequencing has generated an incredible amount of data, and given birth to technologies that make it ever easier and cheaper to measure biological molecules en masse. The task of translating those data into actionable therapeutic knowledge, however, remains an area of active research. We briefly review omics data and technologies, discuss the types of questions translational researchers might ask using omics datasets, and highlight important translational advances and accomplishments from the last few years.
The vast promise of omics technologies
“Omics” assays are those that attempt to interrogate an entire layer of molecular activity in a cell or sample. The omics revolution was set off by genomic arrays, which contained hundreds of probes for selected variants in predetermined regions of the genome. Now, omics technologies have expanded to include more unrestricted approaches, such as assays based on next-generation sequencing and mass spectrometry. There are customized assays for each layer of molecular activity, from genomes to metabolomes. A scientist can choose to measure genomics (e.g. whole genome or whole exome sequencing), transcriptomics (e.g. RNA-seq), epigenomics (e.g. bisulfide sequencing, ChIP-seq for histone modifications, ATAC-Seq for open chromatin), the three-dimensional arrangement of the genome (e.g. Hi-C or ChIA-PET), proteomics or phosphoproteomics, and metabolomics (most commonly by mass spectrometry). Each layer’s assay comes with its own technical requirements and caveats, but each can give rich, detailed information about the choreography of molecules in a sample.
Increasingly, researchers are recognizing the value of skillful integration of multiple layers of omics data, termed multi-omic studies. Modeling and discovering the interplay between different omic layers can lead to important functional and clinical discoveries [1,2]. A recent example of multi-omic studies successfully leading to translational impact comes from the study of IDH mutations in cancer. A 2008 genomics study found common mutations in the IDH1 gene in glioblastoma that were associated with increased survival [3], and subsequent studies found this mutation in other cancers as well [4]. However, it wasn’t until combined genomic and epigenomic studies that the full implications of this mutation were discovered. The mutation in IDH1 and a similar mutation in the IDH2 gene produce altered forms of the encoded enzymes with a gain of function that leads to metabolic, epigenetic, and transcriptomic changes that block differentiation of cancerous cells [5*,6]. Last year, less than 10 years after the studies that identified the mutations, a drug that targets mutated IDH2 was approved for the treatment of acute myeloid leukemia [7,8], and new drugs targeting these enzymes are being developed for other indications.
What types of clinical insights can omics data provide?
There are several distinct types of questions one could ask with omics data that would be useful for translational research. Here, we split them into five categories and give recent examples of each.
Disease-altered molecules, therapeutic targets, and biomarkers
A straightforward result of omics studies is a list of molecules that are altered in a disease, or correlated with disease severity. While omics approaches are sometimes derided as “hypothesis free science,” in reality these lists of molecules are the necessary step of observation from which hypotheses can be generated systematically. The lists of molecules point to pathways that could contain new therapeutic targets, biomarkers, or lead to functional insights into the disease.
Prioritizing these often very long lists of altered molecules is critical. It may seem natural to focus on molecules that are supported by interesting functions known in the literature. However, such an approach ends up reinforcing prior beliefs at the expense of novel discovery. Alternative approaches include focusing on the network or pathways that are enriched in the observed molecules [9].
Functional insights
Omics data can also help researchers come to a better understanding of the mechanism of disease. In the case of genome-wide association studies (GWAS), for example, mechanistic insights are often vital once genomic variants have been statistically associated with a disease. For example, GWAS have found strong association of mutations in the region of the gene FTO with obesity [10,11]. A recent study used further omics data to show that a causal variant in this region leads to derepression of important bioenergetic genes [12**]. This work represents an exciting move towards understanding the mechanism behind heritable obesity.
Most GWAS findings, however, tend to be common genomic variants with very small effects on the probability of disease. A recent model proposed by Boyle et. al. suggests that variants in almost any gene expressed in disease-relevant cells may contribute to disease, and that these small effects add up to account for most of the heritability of diseases [13]. This hypothesis, which they call the omnigenic model, could be true because of the highly interconnected nature of genes and other molecules in the cell; expression changes of nearly any set of genes can work through these interactions to affect important disease pathways. Their findings emphasize the need for detailed integrative models to uncover functional insights in cases where the mechanisms of disease-driving variants or pathways are not obvious.
Disease classification and prediction
Omics data can lead to further subdivision beyond a binary classification of healthy vs. diseased that can prove to be hugely beneficial in the clinic. Such approaches can lead to better treatment for patients based on the actual biology of their specific disease, by placing patients within subtypes or along a spectrum of their disease. Pirhaji et al. recently showed that even relatively crude ordinal classification of disease severity can be used effectively to find disease-related pathways [14*]. Finding the best methods for subtyping [15] and for improving results by integrating different kinds of molecular data [16] are extremely active areas of research.
The subtyping of breast cancer has been especially well-studied [17]. Classical subtypes rely on gene expression of specific markers, and those subtypes are associated with different treatments and prognoses. A recent study by Vazquez et. al. directly showed the advantage of adding multi-omics information into models predicting the progression of breast cancer [18**].
Personalized medicine
Although related to subtyping and disease classification, the exciting potential of omics studies to contribute to personalized and precision medicine deserves special attention. The increasing availability of omics technologies in the clinic could lead to decisions and treatments being tailored to an individual patient [19,20].
In 2012, a model for this approach was put forward in the form of an “integrative Personal Omics Profile (iPOP)”, where researchers performed multiple omics analysis on the blood of a healthy individual over several months [21]. One of the important results of this study was the high level of variability in molecular activity such as mRNA and miRNA expression for the same person over time. Further studies in a large number and of diverse subjects would be needed to determine the level of “background” variability expected in healthy and diseased individuals before monitoring like this could be widely implemented.
Personalized medicine is also a promising application for analysis of the gut microbiome. Sequencing genomic material from intestinal bacteria leads to estimations of which species colonize the gut and their abundance. The microbiome in the gut of each person is distinct, and has important implications for their health [22*]. The vast potential that microbiomes could contribute to personalized medicine is reviewed in [23].
Drug repurposing & pharmacology
Along with identifying new therapeutic targets and disease phenotypes – which could lead to novel drugs – omics studies promise to help identify new uses for existing drugs. Such repurposing of approved drugs could save years and tens or hundreds of millions of dollars into research. Omics studies have the potential to discover new purposes for drugs in an unbiased manner, without prior hypotheses about which drugs and diseases might go together [24].
One popular approach to unbiased drug repurposing involves comparing transcriptomic profiles of cell lines treated with a library of approved drugs to the transcriptomic profile from disease samples. In particular, the goal is to find drugs that raise the expression of genes that have lowered expression in the disease, and vice versa. The Connectivity Map (CMAP) provides a public repository of such gene expression data for this purpose [25,26]. In one such study, researchers found such a transcriptomic connection between small cell lung cancer and tricyclic antidepressants [27,28*]. Identifying connections between approved or investigational drugs and new diseases could have tremendous impact. However, there are significant commercial barriers to repurposing, which may determine whether or not this approach is ever broadly adopted.
Finally, omics data can help us to further understand existing or developing drugs by providing insights into pharmacology [29], or to track pharmacodynamics in real time [30]. In this way, data like these can help researchers develop better drugs and treatment regimes for patients.
Public databases and comparative studies
One of the great advantages of omics data is that they can remain easily accessible for further analysis long after the initial study is finished. Many of the examples above reanalyzed data that were made public as part of large collaborative efforts. These projects invested the necessary resources to make sure that the experiments were well-documented through extensive metadata, thus ensuring that future users would be able to interpret them [26,31–37].
Putting omics data in public databases allows for many sets of eyes on the same dataset, maximizing the useful findings that might be wrung from the data. It also allows for meta-analyses where interesting data can be directly compared between studies. These comparative studies require careful work and expertise to contend with batch effects and variable data collection methods, but they can be powerful ways to determine if findings are consistent across a field [38]. Of course, they can also highlight reproducibility problems across omics studies [39].
Another exciting avenue involves comparing data across different models of the same disease. Studies like these can be used to validate new model systems, find pathways common across several models of disease, or identify pathways that are only altered in one model system, which may not translate to humans. For example, comparison of the transcriptome [40] and epigenome [41] between mouse models and human tissue in Alzheimer’s disease showed several disease-related pathways are conserved between the two, such as immune response alterations, while pathways such as glial-neuron interactions may only be altered in human disease. These studies can help us understand the strengths and weaknesses of preclinical models, and, in particular, whether findings in these models are likely to translate to humans.
Tools for integrative multi-omics studies
One simple, but powerful tool for multi-omics studies is correlation analysis. Identifying a correlation between distinct types molecules can be an effective way to generate new hypotheses – as in the finding of a mutation that correlates with a specific epigenetic state [5]. As multi-omic studies get more complicated, however, more sophisticated tools that integrate multi-omic studies are increasingly important. There are several distinct types of methods proposed for mathematical integration of omics data [42].
One promising type of methods used to integrate omics layers is based on networks and pathways [43]. Databases of known biological pathways and gene ontologies are important resources for the community, and can be used to map omics hits to known functions [44–49]. However, such approaches are inherently limited by the incomplete knowledge of molecular pathways.
A new class of tools is emerging that do not rely on previously known functional pathways, and instead infer networks and connections among the data. Tools like these may use different types of networks as their underlying model [50]. They can focus on how networks may differ between states, such as disease and control [51]. Tools like Omics Integrator [52,53**] and PIUMet [54*] are recent examples of methodological advances that meet these challenges. In both cases, the underlying networks and input datatype(s) are flexible in order to suit diverse experimental situations, methods to determine the robustness and specificity of the network results are included, and user interfaces have been built so that computational biology expertise is not needed in order to run the tools.
A vision for the future
It is likely the most important contributions of multi-omic methods still lie in the future. At least two important advances are needed before these approaches routinely contribute to the discovery of disease mechanisms. First, most omic studies, especially in the clinic, are currently carried out on bulk tissue. However, most disease processes represent a complex interplay of different cell types and tissues. Recent advances in single-cell/nucleus omics assays have demonstrated the extremely diverse patterns of molecular activity present in healthy tissue, tumors, and other samples [55]. These assays so far have primarily measured gene expression in single cells, but recently tools are emerging to perform genomic, epigenomic, and small proteomic screens as well [55]. Advances in these types of omic data collection, coupled with temporal and spatial information, may someday provide the necessary data to understand the contributions of the complex, dynamic interactions of cell types to disease.
However, dramatic advances in measurement technology alone will not suffice. It will be essential to develop better methods for distinguishing correlated events from causal behavior. Current computational methods for causal modeling do not scale to the omics level, as they require vast quantities of data from large numbers of samples [56, 57]. Some types of data are more amenable to causal modeling, especially interventional experiments. In these experiments one systematically inactivates (or activates) each molecule of interest and monitors the effect on all other molecules. Obviously, it would be prohibitively expensive to carry out interventional experiments on a genome/proteome-wide scale. There are also critical practical considerations of choosing an experimental model in which to conduct such experiments, which cannot be conducted on biopsy or post-mortem samples.
The future for causal modeling almost certainly lies at the interface of computation and experiment. Computational methods, including some of the solutions we have described here, such as network optimization techniques, can shrink the scale of the problem. Once a focused list of genes/proteins have been identified, interventional experiments can be conducted on tens or hundreds of molecules rather than tens of thousands. But these experiments will necessarily be conducted in a model of the disease that can never fully capture the full complexity of the problem. Model organisms, immortalized cell lines, induced pluripotent stem cell (iPSC)-based approaches, and even 3D models of specific organs are being utilized to great advantage, but all involve tradeoffs [58]. The challenge will be to develop computational methods that translate findings across these models [59], and perhaps can ultimately discover from the data what aspects of the model are relevant to the human disease.
Conclusions & challenges
Despite its many promises and applications, research that leverages omics data faces important open challenges. Among them is the task of accurate quantification and inter-lab reproducibility of high-throughput assays. Careful experimental design and transparent releases of data, meta-data, and computer code used for analysis should improve reproducibility [60]. Some fields, such as gene expression analysis, have very well-established and frequently used databases for sharing results. However, the situation is much less developed for other areas, such as metabolomics. Attention should also be paid to extraneous sources of variability that may have been ignored previously, such as the handling of tissue prior to data procurement [61].
As the mechanisms for accurately comparing datasets advance, the next big challenge will be to carry out such studies on clinical samples. Such studies need extra care to ensure that patient privacy is respected. Yet the success of The Cancer Genome Atlas, among others, proves that these problems can be overcome. So far, most studies have focused on collecting genomic, and to some extent transcriptional data. However, as other omic methods become more common, it is likely that more comprehensive studies will follow. These future studies may also be able to include richer data from patients’ electronic medical records, and perhaps even from wearable devices.
Beyond any technical challenges there is also an urgent need to make sure such studies provide benefit to all. The generation of omics data and its analysis take considerable resources, and those resources will increase immensely in personalized medicine paradigms. If these expensive approaches result, as hoped, in major advances in health care, the gaps in public health outcomes between first and third world countries will only increase [56]. As we develop and improve methods for translational omics research, we should also pay attention to economic issues and look for opportunities to bring the promise of omics data to less advantaged regions.
Finally, there is a need for smart, flexible, and easy to use methods for integrative omics data analyses to continue to be improved and developed. As biologists become more comfortable and knowledgeable about the methods and caveats associated with omics data, specialized tools for these professionals will be needed. Even tools that are currently created with non-programmers in mind, such as Omics Integrator and PIUMet, require some knowledge of how their respective parameters affect their results, and how their results should be interpreted. In addition, tools that can discover and implicate causal relationships between molecules and pathways would be a great boon to network-based tools. However, omics data is already making its mark on translational research, and ongoing work to improve its reproducibility, accessibility, and ease of use will continue to unlock its potential.
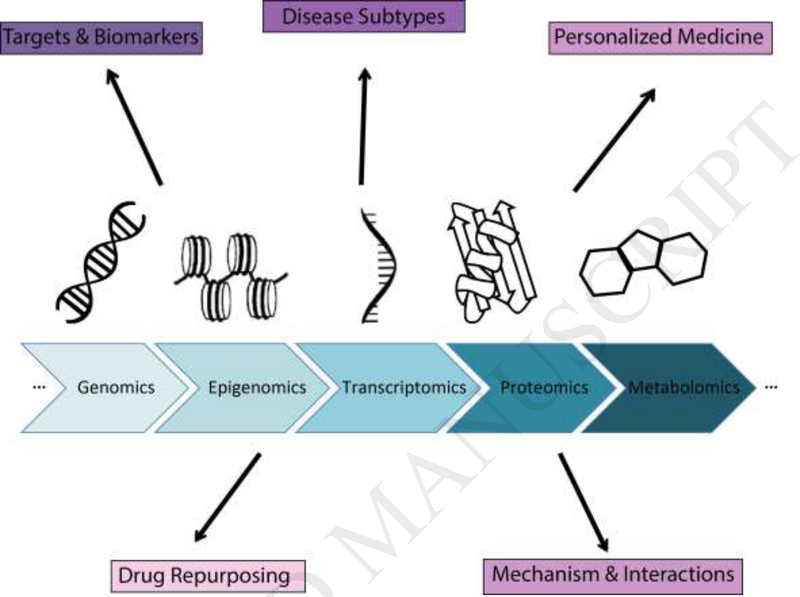
Figure 1.
Omics data measure entire layers of molecular activity. A few of the technologies are shown in the center. Integrating and analyzing these data can serve several important purposes for translational research.
Highlights.
Omics technologies allow evaluation of entire layers of molecular activity at once
Omics data can be used to answer a variety of questions in translational research
Integrating multiple layers of omics data leads to novel, important results
Tools for pathway-based and comparative studies will advance the field
Acknowledgements
This work was supported by NIH grant R01 NS089076.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hasin Y, Seldin M, Lusis A: Multi-omics approaches to disease. Genome Biol 2017, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang SSC, Fraenkel E: Integrating proteomic, transcriptional, and interactome data reveals hidden components of signaling and regulatory networks. Sci Signal 2009, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsons DW, Jones S, Zhang X, Lin JC-H, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu I-MM, Gallia GL, et al. : An integrated genomic analysis of human glioblastoma multiforme. Science (80- ) 2008, 321:1807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang H, Ye D, Guan KL, Xiong Y: IDH1 and IDH2 mutations in tumorigenesis: Mechanistic insights and clinical perspectives. Clin Cancer Res 2012, 18:5562–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. : Identification of a CpG Island Methylator Phenotype that Defines a Distinct Subgroup of Glioma. Cancer Cell 2010, 17:510–522. By testing for genomic variants and epigenomics in the same tumor samples, Noushmehr et. al. showed that the same subset of tumors which had IDH1 mutations also showed aberrant DNA methylation. This correlation led to the discovery of the mechanistic pathway between IDH1 and 2 mutations and cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AWM, Lu C, Ward PS, et al. : IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012, 483:479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stein EM, Dinardo CD, Pollyea DA, Fathi AT, Roboz GJ, Altman JK, Stone RM, Deangelo DJ, Levine RL, Flinn IW, et al. : Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood J 2017, 130:722–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amatangelo MD, Quek L, Shih A, Stein EM, Roshal M, David MD, Marteyn B, Farnoud NR, de Botton S, Bernard OA, et al. : Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood 2017, 130:732–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassani-Pak K, Rawlings C: Knowledge Discovery in Biological Databases for Revealing Candidate Genes Linked to Complex Phenotypes. J Integr Bioinform 2017, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frayling TM, Timpson NJ, Weedon MN, Freathy RM, Lindgren CM, Perry JRB, Katherine S, Lango H, Rayner NW, Shields B, et al. : A Common Variant in the FTO Gene Is Associated with Body Mass Index and Predisposes to Childhood and Adult Obesity. Science (80- ) 2007, 316:889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dina C, Meyre D, Gallina S, Durand E, Körner A, Jacobson P, Carlsson LMS, Kiess W, Vatin V, Lecoeur C, et al. : Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet 2007, 39:724–726. [DOI] [PubMed] [Google Scholar]
- 12.Claussnitzer M, Dankel SN, Kim K-H, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, et al. : FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med 2015, 373:895–907. Claussnitzer and colleagues used epigenomic chromatin state data across hundreds of tissue types to implicate the FTO region as an enhancer for two distant genes, IRX3 and IRX5 in adipose tissue, and then used transcriptomic data in adipocytes to suggest these genes may be important for mitochondrial function. They subsequently elucidated the regulatory pathway acting through the FTO region on IRX3 to affect thermogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyle EA, Li YI, Pritchard JK: An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 2017, 169:1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pirhaji L, Milani P, Dalin S, Wassie BT, Dunn DE, Fenster RJ, Avila-Pacheco J, Greengard P, Clish CB, Heiman M, et al. : Identifying therapeutic targets by combining transcriptional data with ordinal clinical measurements. Nat Commun 2017, 8 Authors propose a new method to examine the relationship between transcriptomic data and disease stages. By using ordinal regression to find genes whose expression is linked to progression of Huntington’s disease, they find new therapeutic targets for the disorder, and show that inhibition of the target is protective in a cell line model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saria S, Goldenberg A: Subtyping: What It is and Its Role in Precision Medicine. IEEE Intell Syst 2015, 30:70–75. [Google Scholar]
- 16.Nguyen T, Tagett R, Diaz D, Draghici S: A novel approach for data integration and disease subtyping. Genome Res 2017, 27:2025–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kittaneh M, Montero AJ, Glück S: Molecular profiling for breast cancer: a comprehensive review. Biomark Cancer 2013, 5:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vazquez AI, Veturi Y, Behring M, Shrestha S, Kirst M, Resende MFR, de los Campos G: Increased Proportion of Variance Explained and Prediction Accuracy of Survival of Breast Cancer Patients with Use of Whole-Genome Multiomic Profiles. Genetics 2016, 203:1425–1438. The authors show that using their model, they were able to predict cancer survival better using whole genome gene expression data than with currently used clinical information (i.e. cancer subtype and stage) alone. Further, they showed that using clinical information, gene expression, and DNA methylation information together outperformed other predictors. This is direct evidence of the added power of multiomics information for prognoses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen R, Snyder M: Promise of personalized omics to precision medicine. Wiley Interdiscip Rev Syst Biol Med 2013, 5:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman AA, Letai A, Fisher DE, Flaherty KT: Precision medicine for cancer with next-generation functional diagnostics. Nat Rev Cancer 2015, 15:747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HYK, Chen R, Miriami E, Karczewski KJ, Hariharan M, Dewey FE, et al. : Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 2012, 148:1293–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M, et al. : Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015, 163:1079–1095. Zeevi et. al. showed that different people respond differently to the same meals. By procuring metagenomic data in each person - measuring their gut bacteria - they could make personalized diet recommendations to lower blood glucose after meals. [DOI] [PubMed] [Google Scholar]
- 23.Kashyap PC, Chia N, Nelson H, Segal E, Elinav E: Microbiome at the Frontier of Personalized Medicine. Mayo Clin Proc 2017, 92:1855–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker NC, Ekins S, Williams AJ, Tropsha A: A bibliometric review of drug repurposing. Drug Discov Today 2018, doi: 10.1016/j.drudis.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. : The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science (80- ) 2006, 313:1929–1935. [DOI] [PubMed] [Google Scholar]
- 26.Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, et al. : A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171:1437–1452.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jahchan NS, Dudley JT, Mazur PK, Flores N, Yang D, Palmerton A, Zmoos AF, Vaka D, Tran KQT, Zhou M, et al. : A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov 2013, 3:1364–1377. Transcriptomic approaches showed that gene expression signatures in lung cancer were reversed in cell lines by tricyclic antidepressants. In mouse models, they inhibited tumor growth. Future studies into the connection between these common and inexpensive drugs and tumor growth in humans could prove to have important implications for these patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zingone A, Brown D, Bowman ED, Vidal OM, Sage J, Neal J, Ryan BM: Relationship between anti-depressant use and lung cancer survival. Cancer Treat Res Commun 2017, 10:33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao S, Iyengar R: Systems Pharmacology: Network Analysis to Identify Multiscale Mechanisms of Drug Action. Annu Rev Pharmacol Toxicol 2012, 52:505–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bordbar A, McCloskey D, Zielinski DC, Sonnenschein N, Jamshidi N, Palsson BO: Personalized Whole-Cell Kinetic Models of Metabolism for Discovery in Genomics and Pharmacodynamics. Cell Syst 2015, 1:283–292. [DOI] [PubMed] [Google Scholar]
- 31.Tomczak K, Czerwińska P, Wiznerowicz M: The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Poznań, Poland) 2015, 19:A68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keenan AB, Jenkins SL, Jagodnik KM, Koplev S, He E, Torre D, Wang Z, Dohlman AB, Silverstein MC, Lachmann A, et al. : The Library of Integrated Network-Based Cellular Signatures NIH Program: System-Level Cataloging of Human Cells Response to Perturbations. Cell Syst 2017, doi: 10.1016/j.cels.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feingold EA, Good PJ, Guyer MS, Kamholz S, Liefer L, Wetterstrand K, Collins FS, Gingeras TR, Kampa D, Sekinger EA, et al. : The ENCODE (ENCyclopedia of DNA Elements) Project. Science (80- ) 2004, 306:636–640. [DOI] [PubMed] [Google Scholar]
- 34.Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. : Integrative analysis of 111 reference human epigenomes. Nature 2015, 518:317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thiele I, Swainston N, Fleming RMT, Hoppe A, Sahoo S, Aurich MK, Haraldsdottir H, Mo ML, Rolfsson O, Stobbe MD, et al. : A community-driven global reconstruction of human metabolism. Nat Biotechnol 2013, 31:419–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N, et al. : The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013, 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. : NCBI GEO: Archive for functional genomics data sets - Update. Nucleic Acids Res 2013, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vasaikar SV, Straub P Wang J Zhang B LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018, 46:D956–D963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dupae J, Bohler S, Noben JP, Carpentier S, Vangronsveld J, Cuypers A: Problems inherent to a meta-analysis of proteomics data: A case study on the plants’ response to Cd in different cultivation conditions. J Proteomics 2014, 108:30–54. [DOI] [PubMed] [Google Scholar]
- 40.Miller JA, Horvath S, Geschwind DH: Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci 2010, 107:12698–12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gjoneska E, Pfenning AR, Mathys H, Quon G, Kundaje A, Tsai LH, Kellis M: Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature 2015, 518:365–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang S, Chaudhary K, Garmire LX: More is better: Recent progress in multi-omics data integration methods. Front Genet 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim TY, Kim HU, Lee SY: Data integration and analysis of biological networks. Curr Opin Biotechnol 2010, 21:78–84. [DOI] [PubMed] [Google Scholar]
- 44.Tanabe M, Kanehisa M: Using the KEGG database resource. Curr Protoc Bioinforma 2012, doi: 10.1002/0471250953.bi0112s38. [DOI] [PubMed] [Google Scholar]
- 45.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, et al. : The Reactome Pathway Knowledgebase. Nucleic Acids Res 2018, 46:D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD: PANTHER version 10: Expanded protein families and functions, and analysis tools. Nucleic Acids Res 2016, 44:D336–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z: GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 2009, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kutmon M, Riutta A, Nunes N, Hanspers K, Willighagen EL, Bohler A, Mélius J, Waagmeester A, Sinha SR, Miller R, et al. : WikiPathways: Capturing the full diversity of pathway knowledge. Nucleic Acids Res 2016, 44:D488–D494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP: Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27:1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Habermann B, Villaveces J, Koti P: Tools for visualization and analysis of molecular networks, pathways, and -omics data. Adv Appl Bioinforma Chem 2015, 8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ideker T, Krogan NJ: Differential network biology. Mol Syst Biol 2012, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tuncbag N, Gosline SJC, Kedaigle A, Soltis AR, Gitter A, Fraenkel E: Network-Based Interpretation of Diverse High-Throughput Datasets through the Omics Integrator Software Package. PLoS Comput Biol 2016, 12 Omics Integrator is a flexible tool for finding pathways and subnetworks enriched with your omics data, without mapping to previously known pathways. There is an easy-touse webtool available so that non-computational biologists can take advantage of it. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kedaigle AJ, Fraenkel E: Discovering Altered Regulation and Signaling Through Network-based Integration of Transcriptomic, Epigenomic, and Proteomic Tumor Data In Cancer Systems Biology; Methods in Molecular Biology. Springer Nature; 2018:13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pirhaji L, Milani P, Leidl M, Curran T, Avila-Pacheco J, Clish CB, White FM, Saghatelian A, Fraenkel E: Revealing disease-associated pathways by network integration of untargeted metabolomics. Nat Methods 2016, 13:770–776. This work introduces a network-based tool for integrating omics data that includes untargeted metabolomics screens. The tool is built to help identify metabolites by looking at connections of potential metabolite identities to other layers of omics data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanay A, Regev A: Scaling single-cell genomics from phenomenology to mechanism. Nature 2017, 541:331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alyass A, Turcotte M, Meyre D: From big data analysis to personalized medicine for all: Challenges and opportunities. BMC Med Genomics 2015, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pearl Judea: Causality – Models, Reasoning, and Inference. Cambridge University Press; 2009. [Google Scholar]
- 58.Shi Y, Inoue H, Wu J, Yamanaka S: Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discovery 2017, 16:115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuhrana V, Peng J, Chung CY, Auluck PK, Fanning S, Tardiff DF, Bartels T, Koeva M, Eichhorm SW, Benyamini H, et al. : Genome-Scale Networks Link Neurodegenerative Disease Genes to a-Synuclein through Specific Molecular Pathways. Cell Systems 2017, 4:157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kolker E, Özdemir V, Martens L, Hancock W, Anderson G, Anderson N, Aynacioglu S, Baranova A, Campagna SR, Chen R, et al. : Toward More Transparent and Reproducible Omics Studies Through a Common Metadata Checklist and Data Publications. Omi A J Integr Biol 2014, 18:10–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson SM, Craven RA, Nirmalan NJ, Harnden P, Selby PJ, Banks RE: Impact of pre-analytical factors on the proteomic analysis of formalin-fixed paraffinembedded tissue. Proteomics - Clin Appl 2013, 7:241–251. [DOI] [PubMed] [Google Scholar]