Abstract
Controlled drug delivery systems have been utilized to enhance the therapeutic effects of many drugs by delivering drugs in a time-dependent and sustained manner. Here, with the aid of 3D printing technology, drug delivery devices were fabricated and tested using a model drug (fluorophore: rhodamine B). Poly(ethylene glycol) dimethacrylate (PEGDMA) devices were fabricated using a two-photon polymerization (TPP) system and rhodamine B was homogenously entrapped inside the polymer matrix during photopolymerization. These devices were printed with varying porosity and morphology using varying printing parameters such as slicing and hatching distance. The effects of these variables on drug release kinetics were determined by evaluating device fluorescence over the course of one week. These PEGDMA-based structures were then investigated for toxicity and biocompatibility in vitro, where MTS assays were performed using a range of cell types including induced pluripotent stem cells (iPSCs). Overall, tuning the hatching distance, slicing distance, and pore size of the fabricated devices modulated the rhodamine B release profile, in each case presumably due to resulting changes in the motility of the small molecule and its access to structure edges. In general, increased spacing provided higher drug release while smaller spacing resulted in some occlusion, preventing media infiltration and thus resulting in reduced fluorophore release. The devices had no cytotoxic effects on human embryonic kidney cells (HEK293), bone marrow stromal stem cells (BMSCs) or iPSCs. Thus, we have demonstrated the utility of two-photon polymerization to create biocompatible, complex miniature devices with fine details and tunable release of a model drug.
Keywords: 3D Printing, Two-photon Polymerization, Controlled Drug Release, PEGDMA
Introduction
Recent advances in 3D printing, including the ability to construct complex structures with fine precision, open the door to rapid and facile production of controlled release devices (Acosta-Velez et al., 2018; Clark et al., 2017; Do et al., 2015; Fina et al., 2018; Goole and Amighi, 2016; Hollander et al., 2018; Khaled et al., 2018; Martinez et al., 2017; Zhang et al., 2017). Newly developed technologies such as two-photon polymerization (TPP) enable researchers to prototype micro- and nanostructures with high resolution. Compared to typical fused deposition modeling printing (50-200 μm resolution) and stereolithography (~20 μm resolution), TPP can be used to create structures with features of the order of approximately 100 nm resolution (Melchels et al., 2010). With such high resolution and precision, TPP enables the fabrication of complex nanoscale devices (Timashev et al., 2016; Worthington et al., 2017).
While historically this technique has been used primarily to create optical devices, micromachines and microfluidics, such high resolution and precision enables the fabrication of complex nanoscale devices that could directly influence cellular growth, differentiation and behavior. Several groups, including our own, have applied this ability to biological systems, namely to study cell motility and mechanical response,(Klein et al., 2010) and to encourage infiltration and alignment within TPP scaffolds (Timashev et al., 2016; Worthington et al., 2017).
Owing to its tendency to hinder non-specific protein absorption, poly(ethylene glycol) (PEG) has been used extensively to increase drug stability and retention time of biopharmaceuticals (Ivens et al., 2015; Turecek et al., 2016). Furthermore, methacrylated PEG (e.g., PEGDMA), which can undergo photocrosslinking, is one of the most widely studied platforms in tissue engineering, especially when co-crosslinked or otherwise functionalized with integrin binding peptides such as RGD (Bryant and Anseth, 2003; Clapper et al., 2008). Many groups, including our own, have demonstrated that photocrosslinked PEGDMA and its close relatives are compatible with a wide variety of cell and tissue types (Adiguzel et al., 2017; Clapper et al., 2008; Xu et al., 2013; Zhou et al., 2016). PEGDMA hydrogels, whose architecture can be tightly controlled using photopolymerization (Bryant and Anseth, 2003), have also been used for a number of drug delivery applications, including stimuli-responsive systems (Aimetti et al., 2009; Lin and Anseth, 2009). Yet, the traditional photopolymerization methods used in these approaches are inherently limited in their ability to create micron-scale features with high precision. TPP, capable of overcoming this challenge, has been used to create simple PEGDMA scaffolds with some success (Ulasan et al., 2015), but this powerful ability has not been fully characterized or realized in the context of pharmaceutics, controlled release, or tissue engineering.
In this study, we investigated the effect of manipulating a number of parameters during the TPP 3D printing process to determine if such changes could be used to control the release of a model drug, rhodamine B. Using TPP, PEGDMA devices were printed with varying parameters and their drug release kinetics were assessed according to five traditional drug delivery models.. PEGDMA was chosen due to its biocompatible properties and its ability to be used as a photopolymer for TPP printing (Ulasan et al., 2015). We also tested for any cytotoxic effects of the devices on a range of cell lines that are widely used for biomedical testing. We hypothesized that the 3D printing parameters of slicing, hatching, and pore size would affect drug release profiles of printed structures. The results obtained from this study provide a valuable foundation that connects TPP based 3D printing settings to controllable drug release. To the best of our knowledge, this is the first study to characterize the effects of TPP printing parameters on (model) drug release kinetics.
Materials and Methods
Device Design
Stereolithographic files of printable devices were designed and generated using AutoCAD software (Autodesk, San Francisco, California). The general design consisted of either a cuboid or a "woodpile" structure, which contained multiple layers of regularly-spaced cylinders, each layer lying in the x-y plane but rotated 90° compared to the previous layer. The overall dimensions of each device was 105 × 105 × 60 μm (LxWxH), unless otherwise noted. For woodpile structures, cylinder diameter and spacing were each varied according to the parameters listed in Table 1. When 3D models are translated into a series of commands for line-by-line two-photon polymerization, the user must determine the line density. The 3D solid is first split into a series of horizontal layers in a process known as slicing. The distance between each of these layers is known as the slicing distance. Each of these layers is then split into a series of parallel lines in a process known as hatching. As with slicing, the distance between each line is known as the hatching distance. In our experiments, the slicing and hatching distances were each varied for cuboid and woodpile structures, with all other parameters held constant (Table 1).
Table 1.
Design parameters for two-photon polymerized PEGDMA woodpile and cuboid devices.
| Structure Type |
Cylinder Diameter |
Clyinder Spacing |
Slicing Distance |
Hatching Distance |
|
|---|---|---|---|---|---|
| Varying Cylinder Size | Woodpile | 5 μm | 5 μm | 0.10 μm | 0.10 μm |
| Woodpile* | 10 μm | 10 μm | 0.10 μm | 0.10 μm | |
| Woodpile | 15 μm | 15 μm | 0.10 μm | 0.10 μm | |
| Varying Cylinder Spacing | Woodpile | 5 μm | 5 μm | 0.10 μm | 0.10 μm |
| Woodpile | 5 μm | 9.29 μm | 0.10 μm | 0.10 μm | |
| Woodpile | 5 μm | 15 μm | 0.10 μm | 0.10 μm | |
| Varying Slicing Distance | Woodpile* | 10 μm | 10 μm | 0.05 μm | 0.10 μm |
| Woodpile* | 10 μm | 10 μm | 0.15μm | 0.10 μm | |
| Woodpile* | 10 μm | 10 μm | 0.151μm | 0.10 μm | |
| Varying Hatching Distance | Woodpile* | 10 μm | 10 μm | 0.10 μm | 0.05 μm |
| Woodpile* | 10 μm | 10 μm | 0.101μm | 0.10 μm | |
| Woodpile* | 10 μm | 10 μm | 0.10 μm | 0.15 μm | |
| Varying Slicing Distance | Cuboid | N/A | N/A | 0.051μm | 0.10 μm |
| Cuboid | N/A | N/A | 0.10 μm | 0.10 μm | |
| Cuboid | N/A | N/A | 0.151μm | 0.10 μm | |
| Varying Hatching Distance | Cuboid | N/A | N/A | 0.10 μm | 0.05 μm |
| Cuboid | N/A | N/A | 0.101μm | 0.10 μm | |
| Cuboid | N/A | N/A | 0.10 μm | 0.15 mm |
These devices were each 110 × 110 × 60 μm
Device Fabrication
Based on previous work and preliminary results, the optimal formulation to create repeatable TPP poly(ethylene glycol) structures containing sufficient fluorophore for detection was found to be: 88.3 % wt PEGDMA (Mn of 575, viscosity of ~57 cP, Sigma-Aldrich, St. Louis, MO), 2.7 % wt Irgacure 369 (photoinitiator, BASF, Germany), and 10 % wt rhodamine B (Sigma-Aldrich) mixture (10 mg/mL in water) or water only (no fluorophore) as the control. To facilitate adhesion of the printed structure to the substrate, we functionalized glass coverslips with polymerizable groups prior to their use as two-photon polymerization substrates (Worthington et al., 2017). Briefly, we exposed glass substrates to oxygen plasma (Plasma Cleaner equipped with PlasmaFlo gas flow control, Harrick Plasma, Ithaca, NY) at an oxygen flow rate of 22.5 mL/min at 30 W radio frequency power for three minutes. The substrates were then submerged in a 1% solution of 3-(trimethoxysilyl)propyl methacrylate (Sigma-Aldrich) in hexanes (Fisher Scientific, Waltham, MA) overnight. We then rinsed the glass substrates in hexanes, dried them and stored them in an airtight container at room temperature until ready for use.
For each set of devices, the substrate was placed in the sample holder and a droplet of the formulation was placed in the center of the substrate which was a 30 mm ½” glass coverslip (CS-30R, Warner Instrument, Hamden, CT). The structures were created using a two-photon lithography system (780 nm laser, Nanoscribe GmbH) using a 25X objective (NA=0.8). Laser power (100%), scanning speed (50,000 um/s) and all other lithography parameters were held constant for all experiments. After fabrication, the structures were removed from the sample holder, submerged in deionized water for five minutes and dried at room temperature overnight in the dark.
Device Morphology
The morphology of the printed devices was examined using scanning electron microscopy (SEM). Samples were mounted on an aluminum stub using double-sided carbon tape. These samples were then dried overnight in ambient air for 24 h prior to being coated with gold-palladium using an argon beam K550 sputter coater (Emitech Ltd., Kent, England). Once coated, samples were imaged using a Hitachi S-4800 SEM (Hitachi High-Technologies, Tokyo, Japan) at an accelerating voltage of 1 kV with the sample stage tilted at 30°.
Release of Rhodamine B
Samples were initially imaged with an EVOS FL fluorescence microscope (Thermo Fisher, Waltham, MA) and used as the starting reference point for fluorophore release (starting concentration of 100%: Supplemental Figure 1A). Samples were imaged at the lowest brightness settings: 10% intensity at an exposure rate of 15 ms. Devices were then submerged in 3 mL of nanopure water and incubated in a shaking incubator set at 300 rpm and 37°C. Samples were collected at the following time points: 1, 3, 6, 12, 24, 48, 72, 96, 168 h. At each designated time point, the water was aspirated and the devices were left to dry for 5 minutes prior to imaging. Images of the devices were collected using a fluorescence microscope at the settings previously mentioned, then analyzed using ImageJ as described previously (see example in Supplemental Figure 1B-C) (Gavet and Pines, 2010; McCloy et al., 2014). In order to generate this standard curve, PEGDMA discs (4 mm × 50 μm) were fabricated using UV polymerization. Briefly, laminate molds were constructed using two standard glass slides with two layers of heavy duty aluminum foil (approximately 0.5 in. × 1 in.; total thickness ~5 μm) on each end as spacers. These layers were clamped on each end using binder clips and each mold was filled with 60 mL of PEGDMA with rhodamine. The pre-polymerized formulations were the same as described above and to each other except for the amount of rhodamine B added, which ranged from 0 mg/mL to 2 mg/mL. Each sample was photopolymerized by exposing the mold to high intensity UV light (Omnicure Series 2000 equipped with 8 mm liquid light guide, Excelitas Technologies, Waltham, MA) at a distance of 2 inches from the end of the light guide and an intensity of 6 W/cm2 (measured at the source) for 50 seconds. After polymerization, each mold was carefully deconstructed, and the resulting films were rinsed by rapid submersion in excess deionized water three times to remove trace amounts of unreacted prepolymer. The films were then blotted with a laboratory napkin and a 4 mm biopsy punch was used to create uniform discs (n > 5) from the periphery of the film, where photobleaching had not occurred. After imaging as described above, our results confirmed that the amount of added rhodamine B was within the linear range of the standard curve and therefore the structures were not saturated with the fluorophore (Supplemental Figure 2).
PEGDMA Biocompatibilty
Human embryonic kidney 293 cells (HEK293, ATCC, Rockville, MD) and bone marrow stromal stem cells (BMSCs, ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco®, Life Technologies Corporations, Brooklyn NY) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA), 10 mM HEPES (Gibco®), 50 μg/mL gentamycin sulfate (Cellgro, Manassas, VA), 1 mM sodium pyruvate (Gibco®), and 1 mM Glutamax (Gibco®). Both cell types were incubated at 37°C and 5% CO2 in a humidified atmosphere. Murine induced pluripotent stem cells (MiPSCs) were generated as described previously (Tucker et al., 2013a; Tucker et al., 2013b; Worthington et al., 2016). Pluripotency media comprised Dulbecco’s modified Eagle’s medium: Nutrient Mixture F-12 (DMEM/F12; Life Technologies, Gibco, Carlsbad, CA) with 15% fetal bovine serum (Life Technologies), 1% 100X nonessential amino acids (NEAA; Life Technologies), 0.4 mM L-glutamine (Life Technologies), 0.1 mg/mL Primocin (InvivoGen, San Diego, CA), and 8.88 ng/mL 2-mercaptoethanol (Sigma-Aldrich). Just before use, 2 U/mL of mouse recombinant leukemia inhibitory factor (mLif, ESGRO; EMD Millipore, Billerica, MA) was added and media were warmed to 37 °C.
To test for cytotoxicity of the PEGDMA-based devices, 5 mL of complete media was added to 60 mm petri dishes that contained the PEGDMA devices and were incubated overnight at 37 °C. The supernatant was then harvested and used as growth media to feed each cell line. The “conditioned” media was incubated with the cells for 24 hours before cytotoxicity was analyzed. Cytotoxicity was assessed using an MTS assay (CellTiter 96®, Promega, Madison, WI) following manufacturer's instructions. In brief, cells were plated in a 96-well plate at a density of 1 × 104 cells/well in 100 μl of DMEM (+ supplements) for 24 h prior to treatments. The media was then aspirated and then the indicated supernatants (described above) were added at varying dilutions. The cultures were then incubated for 24 h, after which the media was removed, cells were washed with 1X PBS, and then replenished with fresh media. Then, 20 μL of MTS reagent was added and cells were incubated for a further 3 hours. Finally, the absorbance was measured at 490 nm using a SpectraMax plus 384 Microplate spectrophotometer (Molecular Devices, Sunnyvale, CA). Relative cell viability was analyzed using untreated cells as the control group.
Statistical Analysis
The loss of fluorescence was used as a means of quantifying drug release (Supplemental Figure 1) and release curves were fit to five drug release models: zero order, first order, Higuchi, Hixson-Crowell, and Korsmeyer-Peppas. Model accuracy was determined using R-squared and Sy.x values, with
| (1) |
where n is the number of data points in the set (typically 24: 3 measurements at each of 8 time points) and K is the number of parameters fitted in the model. The most accurate model and extra sum-of-squares F-tests were used to test the null hypothesis that all release profiles within a given group could be modeled using the same parameters. P-value was two-sided and a value of less than 0.05 was considered to be statistically significant. Data were presented as mean ± the standard error of the mean, unless stated otherwise.
Results and Discussion
In order to identify the potential connection between 3D printing settings and controlled drug release, we investigated the effects of varying printing parameters on controlled fluorophore release. Using a two-photon polymerization system, 3D printed devices were printed with varying slicing, hatching, spacing, and shape and were homogenously loaded with rhodamine B in order to assess controlled drug delivery functionality. Furthermore, the printed devices were analyzed for cytotoxic effects on a variety of cells in an effort to demonstrate the potentially wide applicability of this technology to multiple disease models.
Cylinder Diameter
In this experiment, rhodamine B was homogenously encapsulated in the material (PEG) and thus can be assumed to be distributed equally across the entire volume. Meanwhile, diffusion is known to be dependent on the surface area available for transport. Hence, a higher surface area per volume should equate to more rapid transport. For an individual cylinder, this relationship becomes:
| (2) |
Where r is the cylinder diameter and h is cylinder length. Thus, for a constant cylinder length, decreasing the diameter increases the surface area to volume ratio. Accordingly, we hypothesized that the rate of rhodamine B released from PEG “woodpile” structures would increase as the diameter of cylinders decreased.
The PEG structures created to test this first hypothesis matched the overall intended woodpile design (Figure 1A-F). It should be noted, however, that PEG structures are known to be hydrophilic and in this case, water accounts for roughly 10% of the mass. The majority, if not all, of this hydration can be presumed to have been lost upon dessication, which was required for SEM imaging. This drying effect likely contributed to slight deviations (e.g. shrinkage) in the appearance of the structures from their morphology prior to dessication (Figure 1D-F).
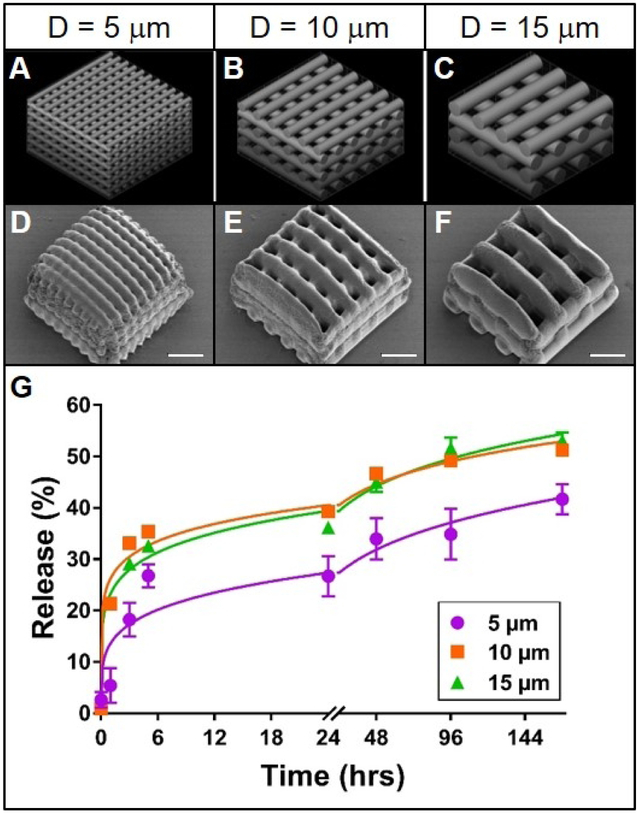
Figure 1. Effect of cylinder size on rhodamine release from TPP woodpile structures.
A-C: 3D models of woodpile structures with designed cylinder diameters and cylinder spacing of 5, 10 or 15 μm, respectively. D-F: Representative scanning electron micrographs of the corresponding poly(ethylene glycol) woodpile structures. Scale bars represent 25 μm. G: Release profiles (data points) and best-fit Korsmeyer-Peppas regression curves (lines) of rhodamine B diffusion from the devices shown in A-F. Error bars represent standard error of the mean; n=3.
Release of rhodamine from the woodpile structures most closely followed a Korsmeyer-Peppas model; R2 values were higher and Sy.x values lower overall for this model than for the other four models (Supplemental Table 1). It is reasonable to anticipate that such pore obstruction in the 5 μm woodpile structure would inhibit release from the structure. Indeed, the initial rate of release of rhodamine B from the 5 μm woodpile structures was lower than that from the 10 or 15 μm structures (Figure 1G). Furthermore, the ultimate amount of rhodamine B released from the 5 μm structures (41%) was lower than the amount released from the 10 or 15 μm structures (51% and 53%, respectively; Figure 1G). These differences were further validated by comparing the mathematical parameters: the release rate constant (k) for the 5 μm structures was roughly half the value of its larger diameter counterparts, while the release rate exponent (n) was nearly 1.5 times greater. Indeed, the possibility that these three structures share a common release model is statistically unlikely (p < 0.0001). However, these data do not support the hypothesis that decreasing cylinder diameter increases release rate from woodpile structures. On the contrary, structures composed of the smallest cylinders released the model drug more slowly than structures with larger cylinders. This difference in release can likely be attributed to the occlusion of water due to the small pore size of the structures containing 5 μm cylinders. Potentially indicating that there are pore size limitations for water infiltration. Furthermore, differences in pore size (spaces between cylinders) in this experiment could have confounded the effect of cylinder size. With a larger pore size, more water would be more readily able to enter the device and thus increase the rate of fluorophore diffusion and release. Since cylinder diameter and spacing were directly linked in this experiment, the expected effect of increasing the cylinder diameter (decreasing the release rate) because of the decrease in surface area to volume ratio could have been offset by the concomitant expected effect of increasing the spacing between cylinders (increasing release rate).
Cylinder Spacing
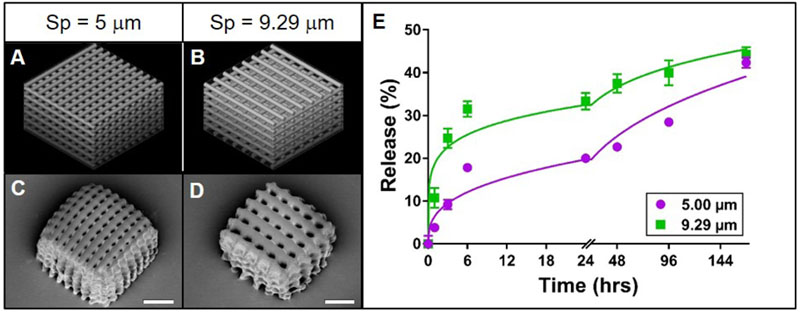
In order to gain more clarity into the effects of woodpile morphology on rhodamine B release rate, the effects of cylinder diameter and cylinder spacing were separated. In theory, increasing the space between cylinders in a woodpile structure will increase the concentration gradient, boosting the driving force for diffusion of rhodamine B out of the cylinder. In essence, smaller pores may occlude water infiltration while larger pores allow for higher rates of diffusion. Thus, we hypothesized that for PEG woodpile structures with constant cylinder diameters, increasing the spacing between cylinders would increase the release rate of encapsulated rhodamine B. As in the initial experiment described above, the PEG woodpile structures resembled their respective 3D models reasonably well (Figure 2A-D). While a structure with 15 μm spaces between cylinders was initially included in this experiment, these structures could not be reliably fabricated and were often absent from the sample surface, most likely because the large spacing prevented adequate intra-structural adherence of the cylinders to one another. Thus, this group was excluded from further analysis. Regardless, the initial rate of release of rhodamine B from the structures with larger spacing between cylinders (9.29 μm) was higher than that from the structures with smaller spacing (5 μm; Figure 1E). Meanwhile, the amount of rhodamine B that had been released at the end of the experiment was relatively similar between the structures with larger spacing (44%) compared to smaller spacing (42%; Figure 1E). Furthermore, both release profiles closely followed a Korsmeyer-Peppas model (Supplemental Table 2), and their regression parameters were significantly different from one another (p < 0.0001). In fact, the release constant for structures with larger spacing was approximately three times larger and their release exponent half the value of the smaller-spaced structure (Supplemental Table 2). These data support the hypothesis that increasing spacing between cylinders in a PEG woodpile device increases the release rate of encapsulated rhodamine B. Also, as was observed here, the planned porosity should be considered carefully, as pores that are too large can cause structural integrity issues of the printed device (Sabree et al., 2015).
Figure 2. Effect of cylinder spacing on rhodamine B release from TPP woodpile structures.
A-B: 3D models of woodpile structures with designed space between cylinders of 5 or 9.29 μm, respectively, with constant cylinder diameter of 5 μm. C-D: Representative scanning electron micrographs of the corresponding PEG woodpile structures. Scale bars represent 25 μm. E: Release profiles (data points) and best-fit Korsmeyer-Peppas regression curves (lines) of rhodamine B diffusion from the devices shown in A-D. Error bars represent standard error of the mean; n=3.
Slicing and Hatching Distance
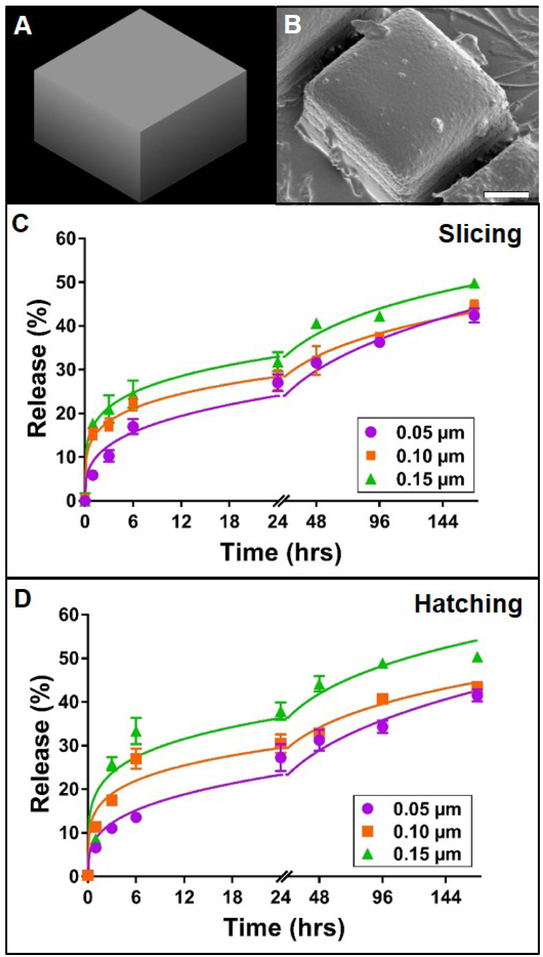
For any given material chemistry, the structural outcomes of TPP devices are highly dependent on selected TPP parameters, such as slicing and hatching distance. Given the impact of these subtle adjustments on final morphology, it was speculated that slicing and hatching distance may also play a role in the diffusion kinetics of encapsulated molecules from TPP devices. Slicing and hatching distance are each inversely correlated to line density within a structure. Thus, increasing either parameter should result in structures of decreasing density. In theory, this change would in turn enhance the mobility of encapsulated molecules and speed their diffusion out of the structure. Thus, we hypothesized that increasing slicing or hatching distance of PEG structures would increase the rate and ultimate amount of rhodamine B released from the device. This expectation is congruent not only with our second hypothesis (supported by Figure 2), but also with findings that suggest increased release rate of small molecules from highly porous mesoporous matrices (Horcajada et al., 2004), lidocaine from highly porous poly (lactic-coglycolic) microparticles (Klose et al., 2006), and vancomycin from calcium phosphate cement matrices (Schnieders et al., 2011). In order to separate the effects of slicing and hatching from those of cylinder size and spacing, as previously described, we first tested the slicing and hatching hypothesis using a simple cuboid structure. In all cases, the PEG cuboids were qualitatively similar to the originally designed model (Figure 3A-B) and to one another. Increasing slicing increased the initial rate of rhodamine B release from the cuboids (Figure 3C). Furthermore, the total amounts of rhodamine B released at the end of the experiment were 42, 44 and 50% for slicing distances of 0.05, 0.1 and 0.15 μm, respectively. Once again, all the release profiles followed the Korsmeyer-Peppas model (Supplemental Table 3), and significant differences were observed between their regression parameters (p < 0.0001). The modeled release rate constant increased with increasing slicing distance while the release rate exponent decreased or stayed the same (Supplemental Table 3). Increasing the hatching distance had a similar effect on rhodamine B release from PEG cuboids (Figure 3D). Release profiles followed a Korsmeyer-Peppas model and non-linear regression parameters were significantly different from one another (p < 0.0001) and responded to hatching distance in the same manner as slicing distance (Supplemental Table 3). These data support the hypothesis that increasing slicing or hatching distance would boost release from TPP PEG structures. Additionally, the release of rhodamine B from these cuboids was markedly slower than release from analogous woodpile structures. This difference can likely be attributed to the lower surface area and limited porosity of the cuboids compared to the woodpiles, which likely led to less water infiltration and thus decreased the rate of rhodamine B diffusion from the devices.
Figure 3. Effect of slicing and hatching distance on rhodamine B release from TPP cuboid structures.
A: 3D model of cuboid structure. B: Representative scanning electron micrograph of the corresponding poly(ethylene glycol) cuboid structure. Scale bar represents 25 μm, C-D: Release profiles (data points) and best-fit Korsmeyer-Peppas regression curves (lines) of rhodamine B diffusion from cuboid devices with varying slicing or hatching distance, respectively. Error bars represent standard error of the mean; n=3.
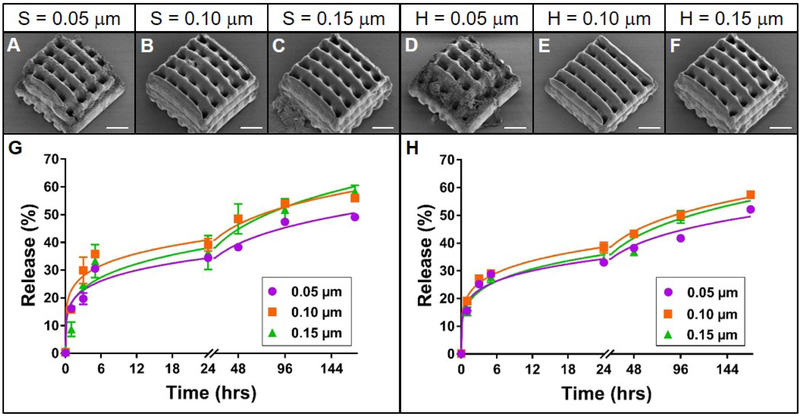
The effects of varying slicing and hatching distances were then investigated with woodpile structures described previously. While in all cases the resulting PEG woodpiles (Figure 4A-F) resembled the model (which is shown in Figure 1B), small slicing or hatching distances resulted in deterioration of the structure at the edges of the device (Figure 4A, D). Despite this structural difference, the profiles of rhodamine B release from these PEG devices still followed a Korsmeyer-Peppas model (Supplemental Table 4) and were remarkably similar to one another. The total amount of rhodamine B released from structures with slicing or hatching distances of 0.05 μm (49 or 52%, respectively) was slightly lower than that from 0.1 and 0.15 μm slicing or hatching (56 and 59, or 58 and 58%, respectively; Figure 4G-H). These subtle differences were reflected as significant differences (p < 0.05 and p < 0.0001, for varied slicing and hatching, respectively) between the regression parameters for these release profiles. However, absolute differences between the regression parameters were small compared to the effects of other parameters investigated (Supplemental Table 4), suggesting that at this scale, the kinetics of rhodamine B release from PEG woodpile structures are dictated mostly by structure geometry, with slicing and hatching distance playing a lesser but tangible role.
Figure 4. Effect of slicing and hatching distance on rhodamine B release from TPP woodpile structures.
A-F: Representative scanning electron micrographs of PEG woodpile structures with 10 μm cylinders created using a slicing distance of 0.05, 0.1 or 0.15 μm at constant hatching distance or a hatching distance of 0.05, 0.1 or 0.15 μm at a constant slicing distance, respectively. Scale bars represent 25 μm. G-H: Release profiles (data points) and best-fit Korsmeyer-Peppas regression curves (lines) of rhodamine B diffusion from the devices shown in A-F with varying slicing (G) or hatching (H) distances. Error bars represent standard error of the mean; n=3.
At the end-points of the release studies (one week) presented in Figures 1 - 4, the devices did not completely release their payloads (maximum cumulative release was ~55%), while release appeared to have slowed substantially. There are a number of possible explanations as to why this may have occurred. First, small amounts of rhodamine B may still have continued to be released past the final time point, but the incremental change in fluorescence was not great enough to be detected. Second, since rhodamine B is photoactive, a proportion of it may have acted as a second photoinitiator during two-photon polymerization, thus becoming part of the crosslinked polymer via covalent bonding (Wagner, 1971). However, given the relatively high concentration of rhodamine B included in the formulation and the presence of the primary photoinitiator, which is more sensitive and efficient, we presume that rhodamine B activation during fabrication occurred to only a minor degree. Furthermore, even though this event may prevent the full release of the rhodamine B from the devices, it is a variable that is expected to be consistent between all groups. Finally, the saturation effect could also be due in part to the photodegradation properties of rhodamine B. Since the photodegradation of rhodamine B is relatively low (about 10% loss over 168 h) (Do et al., 2017), we believe that this contribution was also minimal. Several well-known groups suggest that only the first 60% of release should be used for mathematical modeling of drug delivery systems (Bruschi, 2015; Ritger and Peppas, 1987), so conclusions about general release behavior can confidently be drawn from the type of data we present here.
All of our release profiles followed a Korsmeyer-Peppas model, which enables the release rate exponent, n, to be used to identify the release mechanisms at play. Typically, values around 0.5 indicate Fickian diffusion and those higher than 0.5 signifying some degree of polymer relaxation effects, including swelling and crazing (Bruschi, 2015). The values we report for TPP PEGDMA structures are between 0.14 and 0.35, suggesting that release kinetics are likely dictated by diffusion rather than relaxation within the polymer network. However, even the highest rate exponent we found, 0.35, is quite low compared to the 0.5 “gold standard” of Fickian behavior. This discrepancy could be due to a number of factors, including sample geometry and covalent drug binding. First, distinct values of n that correspond to Fickian kinetics are known to depend on sample geometry, with cylindral and spherical samples having Fickian n-values of 0.45 and 0.43, respectively (Ritger and Peppas, 1987). Since the structures we created comprise a series of overlapping polymer lines on the sub-micron scale and overlapping cylindrical structures on the microscale, the geometry at hand is quite complex compared to the geometric systems used to form historical assumptions about n-values and Fickian behavior. Thus, it is plausible that n-values in the range of 0.15 to 0.35 are characteristic of release behavior that follows Fick’s law for these intricate structrures. Future studies should focus on controlling various parameters in order to test this hypothesis and to determine characteristic n-values for various release mechanisms. Secondly, a limited amount of covalent incorporation of the model drug is likely to have occurred during two-photon polymerization as a result of unavoidable Type II photoinitiation and associated hydrogen abstraction described above. Because release kinetics were inferred based on fluorescence of the structures, which included these presumptively bound molecules, the apparent release kinetics could have been dampened by the phenomenon. Although Type II photoinitiation has not been studied extensively in the context of two-photon polymerization, it usually plays only a minor role in traditional photopolymerization reactions and thus, we do not expect its effects to be dominant here (Lee et al., 2004).
PEGDMA Biocompatibility
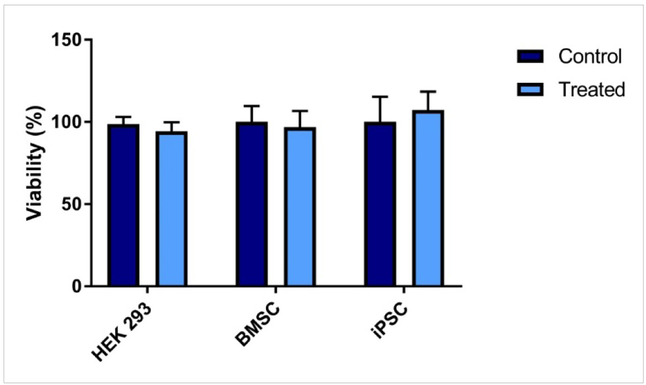
In order to be clinically translatable, the devices described here must not only be effective in delivering the intended therapeutic, but also show evidence of being biocompatible. With photopolymerization in particular, the presence of free radicals is at least one concern in terms of cytotoxicity (Platel et al., 2008). While exposure to photoinitiators and high intensity light during photopolymerization certainly causes cellular death to varying degrees depending on the initiator used (Williams et al., 2005), fabrication of the two-photon drug delivery systems we describe here occurs independently of cells and tissue, protecting them from this particular risk. Still, some photoinitiator fragments likely remain immediately after fabrication, so we sought to validate the compatibility of these in the context of our model system. We did so for prototype TPP devices using a range of cell types, including cells that may be used for tissue engineering purposes, such as iPSCs and BMSCs. In our experience, stem cell lines like these are especially sensitive to small changes in their culture conditions, qualifying their use at the 24-hour time point for determining overall biocompatibility. This acute response is likely due to stem cells’ characteristically high rate of self-renewal, which can reasonably be assumed to accelerate the effects of any cytotoxic media components. Media conditioned with presumptive leached products from the PEGDMA material had no significant cytotoxic effect when incubated with these various cell types (Figure 5). These results build upon what others have also noted; crosslinked PEGDMA has been shown to have no cytotoxic effects on other cell lines such as NIH-3T3 fibroblasts (Adiguzel et al., 2017; Clapper et al., 2008; Ulasan et al., 2015; Xu et al., 2013; Zhou et al., 2016). The lack of cytotoxic effects of the printed PEGDMA material towards stem cells is particularly important when considering these PEGDMA-based devices for tissue engineering purposes, as it demonstrates their versatility for a wide range of applications and tissue types. Further validation with more precise cell and tissue types of interest, as well as in an animal model, will certainly be required before two-photon polymerized drug delivery systems can be employed for any specific application.
Figure 5. Biocompatibility of photopolymerized poly(ethylene glycol).
The viability of human embryonic kidney cells (HEK 293), bone marrow stromal stem cells (BMSC) or mouse induced pluripotent stem cells (iPSC) was analyzed through an MTS assay after 24h of incubation in “conditioned” media. The cells were incubated with “conditioned” media that was made by incubating photopolymerized poly(ethylene glycol) dimethacrylate samples in cell culture media for 24 h. Error bars represent standard deviation; n=4.
Conclusions
In this study, we demonstrated that the manipulation of TPP based 3D printing parameters can be used to control drug release from printed constructs. Specifically, device design, porosity, and material density all play a role in the kinetics of small molecule release from two-photon polymerized structures. Since these can be tuned independently of material chemistry, this method of control has potential cost and time saving benefits over traditional controlled release methods. Additional future studies aimed at utilizing other drug candidates such as proteins and lipophilic drugs would further demonstrate the versatility of this approach.
Supplementary Material
Acknowledgements
We acknowledge financial support from the National Institutes of Health (1 R01 024605–01, 1 DP2 OD007483-01, 5P30CA086862), the Lyle and Sharon Bighley Chair of Pharmaceutical Sciences, The University of Iowa Graduate College and the Roy J. Carver Charitable Trust (Grant #18-5045). Scanning electron microscopy (NIH Shared Instrumentation Grant 1 S10 RR022498-01) was performed in the University of Iowa Central Microscopy Facility, a core resource supported by the Vice President for Research and Economic Development, the Holden Comprehensive Cancer Center, and the Carver College of Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acosta-Velez GF, Zhu TZ, Linsley CS, Wu BM, 2018. Photocurable poly(ethylene glycol) as a bioink for the inkjet 3D pharming of hydrophobic drugs. Int J Pharm 546, 145–153. [DOI] [PubMed] [Google Scholar]
- Adiguzel Z, Sagnic SA, Aroguz AZ, 2017. Preparation and characterization of polymers based on PDMS and PEG-DMA as potential scaffold for cell growth. Mater Sci Eng C Mater Biol Appl 78, 942–948. [DOI] [PubMed] [Google Scholar]
- Aimetti AA, Machen AJ, Anseth KS, 2009. Poly(ethylene glycol) hydrogels formed by thiolene photopolymerization for enzyme-responsive protein delivery. Biomaterials 30, 6048–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruschi ML, 2015. 5 - Mathematical models of drug release, in: Bruschi ML (Ed.), Strategies to Modify the Drug Release from Pharmaceutical Systems. Woodhead Publishing, pp. 63–86. [Google Scholar]
- Bryant SJ, Anseth KS, 2003. Controlling the spatial distribution of ECM components in degradable PEG hydrogels for tissue engineering cartilage. J. Biomed. Mater. Res. Part A 64A, 70–79. [DOI] [PubMed] [Google Scholar]
- Clapper JD, Pearce ME, Guymon CA, Salem AK, 2008. Biotinylated biodegradable nanotemplated hydrogel networks for cell interactive applications. Biomacromolecules 9, 1188–1194. [DOI] [PubMed] [Google Scholar]
- Clark EA, Alexander MR, Irvine DJ, Roberts CJ, Wallace MJ, Sharpe S, Yoo J, Hague RJM, Tuck CJ, Wildman RD, 2017. 3D printing of tablets using inkjet with UV photoinitiation. Int J Pharm 529, 523–530. [DOI] [PubMed] [Google Scholar]
- Do A-V, Akkouch A, Green B, Ozbolat I, Debabneh A, Geary S, Salem AK, 2017. Controlled and Sequential Delivery of Fluorophores from 3D Printed Alginate-PLGA Tubes. Annals of Biomedical Engineering 45, 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do AV, Khorsand B, Geary SM, Salem AK, 2015. 3D Printing of Scaffolds for Tissue Regeneration Applications. Adv Healthc Mater 4, 1742–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fina F, Goyanes A, Madla CM, Awad A, Trenfield SJ, Kuek JM, Patel P, Gaisford S, Basit AW, 2018. 3D printing of drug-loaded gyroid lattices using selective laser sintering. Int J Pharm 547, 44–52. [DOI] [PubMed] [Google Scholar]
- Gavet O, Pines J, 2010. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Developmental cell 18, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goole J, Amighi K, 2016. 3D printing in pharmaceutics: A new tool for designing customized drug delivery systems. Int J Pharm 499, 376–394. [DOI] [PubMed] [Google Scholar]
- Hollander J, Hakala R, Suominen J, Moritz N, Yliruusi J, Sandler N, 2018. 3D printed UV light cured polydimethylsiloxane devices for drug delivery. Int J Pharm 544, 433–442. [DOI] [PubMed] [Google Scholar]
- Horcajada P, Ramila A, Perez-Pariente J, Vallet R, x M, 2004. Influence of pore size of MCM-41 matrices on drug delivery rate. Microporous and Mesoporous Materials 68, 105–109. [Google Scholar]
- Ivens IA, Achanzar W, Baumann A, Brandli-Baiocco A, Cavagnaro J, Dempster M, Depelchin BO, Irizarry Rovira AR, Dill-Morton L, Lane JH, Reipert BM, Salcedo T, Schweighardt B, Tsuruda LS, Turecek PL, Sims J, 2015. PEGylated Biopharmaceuticals: Current Experience and Considerations for Nonclinical Development. Toxicologic Pathology 43, 959–983. [DOI] [PubMed] [Google Scholar]
- Khaled SA, Alexander MR, Wildman RD, Wallace MJ, Sharpe S, Yoo J, Roberts CJ, 2018. 3D extrusion printing of high drug loading immediate release paracetamol tablets. Int J Pharm 538, 223–230. [DOI] [PubMed] [Google Scholar]
- Klein F, Striebel T, Fischer J, Jiang Z, Franz CM, von Freymann G, Wegener M, Bastmeyer M, 2010. Elastic fully three-dimensional microstructure scaffolds for cell force measurements. Adv Mater 22, 868–871. [DOI] [PubMed] [Google Scholar]
- Klose D, Siepmann F, Elkharraz K, Krenzlin S, Siepmann J, 2006. How porosity and size affect the drug release mechanisms from PLGA-based microparticles. International Journal of Pharmaceutics 314, 198–206. [DOI] [PubMed] [Google Scholar]
- Lee TY, Guymon CA, Jönsson ES, Hoyle CE, 2004. The effect of monomer structure on oxygen inhibition of (meth)acrylates photopolymerization. Polymer 45, 6155–6162. [Google Scholar]
- Lin CC, Anseth KS, 2009. PEG Hydrogels for the Controlled Release of Biomolecules in Regenerative Medicine. Pharm. Res 26, 631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez PR, Goyanes A, Basit AW, Gaisford S, 2017. Fabrication of drug-loaded hydrogels with stereolithographic 3D printing. Int J Pharm 532, 313–317. [DOI] [PubMed] [Google Scholar]
- McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A, 2014. Partial inhibition of Cdk1 in G2 phase overrides the SAC and decouples mitotic events. Cell Cycle 13, 1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchels FPW, Feijen J, Grijpma DW, 2010. A review on stereolithography and its applications in biomedical engineering. Biomaterials 31, 6121–6130. [DOI] [PubMed] [Google Scholar]
- Platel RH, Hodgson LM, Williams CK, 2008. Biocompatible Initiators for Lactide Polymerization. Polymer Reviews 48, 11–63. [Google Scholar]
- Ritger PL, Peppas NA, 1987. A simple equation for description of solute release I. Fickian and non-fickian release from non-swellable devices in the form of slabs, spheres, cylinders or discs. Journal of Controlled Release 5, 23–36. [PubMed] [Google Scholar]
- Sabree I, Gough JE, Derby B, 2015. Mechanical properties of porous ceramic scaffolds: Influence of internal dimensions. Ceramics International 41, 8425–8432. [Google Scholar]
- Schnieders J, Gbureck U, Vorndran E, Schossig M, Kissel T, 2011. The effect of porosity on drug release kinetics from vancomycin microsphere/calcium phosphate cement composites. Journal of Biomedical Materials Research Part B: Applied Biomaterials 99B, 391–398. [DOI] [PubMed] [Google Scholar]
- Timashev PS, Vedunova MV, Guseva D, Ponimaskin E, Deiwick A, Mishchenko TA, Mitroshina EV, Koroleva AV, Pimashkin AS, Mukhina IV, Panchenko VY, Chichkov BN, Bagratashvili VN, 2016. 3D in vitro platform produced by two-photon polymerization for the analysis of neural network formation and function. Biomedical Physics & Engineering Express 2, 035001. [Google Scholar]
- Tucker BA, Anfinson KR, Mullins RF, Stone EM, Young MJ, 2013a. Use of a Synthetic Xeno-Free Culture Substrate for Induced Pluripotent Stem Cell Induction and Retinal Differentiation. Stem Cells Translational Medicine 2, 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BA, Mullins RF, Streb LM, Anfinson K, Eyestone ME, Kaalberg E, Riker MJ, Drack AV, Braun TA, Stone EM, 2013b. Patient-specific iPSC-derived photoreceptor precursor cells as a means to investigate retinitis pigmentosa. eLife 2, e00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turecek PL, Bossard MJ, Schoetens F, Ivens IA, 2016. PEGylation of Biopharmaceuticals: A Review of Chemistry and Nonclinical Safety Information of Approved Drugs. Journal of Pharmaceutical Sciences 105, 460–475. [DOI] [PubMed] [Google Scholar]
- Ulasan M, Yavuz E, Bagriacik EU, Cengeloglu Y, Yavuz MS, 2015. Biocompatible thermoresponsive PEGMA nanoparticles crosslinked with cleavable disulfide-based crosslinker for dual drug release. Journal of Biomedical Materials Research Part A 103, 243–251. [DOI] [PubMed] [Google Scholar]
- Wagner PJ, 1971. Type II photoelimination and photocyclization of ketones. Accounts of Chemical Research 4, 168–177. [Google Scholar]
- Williams CG, Malik AN, Kim TK, Manson PN, Elisseeff JH, 2005. Variable cytocompatibility of six cell lines with photoinitiators used for polymerizing hydrogels and cell encapsulation. Biomaterials 26, 1211–1218. [DOI] [PubMed] [Google Scholar]
- Worthington KS, Green BJ, Rethwisch M, Wiley LA, Tucker BA, Guymon CA Salem AK., 2016. Neuronal Differentiation of Induced Pluripotent Stem Cells on Surfactant Templated Chitosan Hydrogels. Biomacromolecules 17, 1684–1695. [DOI] [PubMed] [Google Scholar]
- Worthington KS, Wiley LA, Kaalberg EE, Collins MM, Mullins RF, Stone EM, Tucker BA., 2017. Two-photon polymerization for production of human iPSC-derived retinal cell grafts. Acta biomaterialia 55, 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Cantu DA, Fu Y, Kim J, Zheng X, Hematti P, Kao WJ, 2013. Thiol-ene Michael-type formation of gelatin/poly(ethylene glycol) biomatrices for three-dimensional mesenchymal stromal/stem cell administration to cutaneous wounds. Acta Biomater 9, 8802–8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Feng X, Patil H, Tiwari RV, Repka MA, 2017. Coupling 3D printing with hot-melt extrusion to produce controlled-release tablets. Int J Pharm 519, 186–197. [DOI] [PubMed] [Google Scholar]
- Zhou W, Stukel JM, Cebull HL, Willits RK, 2016. Tuning the Mechanical Properties of Poly(Ethylene Glycol) Microgel-Based Scaffolds to Increase 3D Schwann Cell Proliferation. Macromol Biosci 16, 535–544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.