Abstract
Biomolecular condensates are two- and three-dimensional compartments in eukaryotic cells that concentrate specific collections of molecules without an encapsulating membrane. Many condensates behave as dynamic liquids and appear to form through liquid-liquid phase separation driven by weak, multivalent interactions between macromolecules. In this review, we discuss current models and data regarding the control of condensate composition and we describe our current understanding of the composition of representative condensates including PML nuclear bodies, P-bodies, stress granules, the nucleolus, and two-dimensional membrane localized LAT and nephrin clusters. Specific interactions, such as interactions between modular binding domains, weaker interactions between intrinsically disorder regions and nucleic acid base pairing, and nonspecific interactions, such as electrostatic interactions and hydrophobic interactions, influence condensate composition. Understanding how specific condensate composition is determined is essential to understanding condensates as biochemical entities and ultimately discerning their cellular and organismic functions.
Biomolecular condensates are two- and three-dimensional compartments in eukaryotic cells that concentrate specific collections of molecules without an encapsulating membrane. Condensate formation has emerged as a fundamental mechanism for the organization of biomolecules within the nucleus and cytosol, and at membranes [1–4]. Many condensates behave as dynamic liquid, and appear to form through liquid-liquid phase separation (LLPS) driven by weak, multivalent interactions between macromolecules. There are numerous manifestations of this multivalency, including arrays of modular protein domains [5–10], distributed weakly adhesive motifs in intrinsically disordered regions (IDRs) of proteins [11–16], and repetitive base-pairing elements in RNA and DNA [17,18]. Recent reviews have focused on how these features of biomolecules drive LLPS, enable regulation of the process, dictate condensate material properties, and potentially give rise to new biochemical and cellular functions [1–4]. Here we will discuss current models and data regarding a less well-understood aspect of biomolecular condensates—their compositions. That is, for any given condensate, which molecules are concentrated within the structure and which are depleted (and to what quantitative degree for both), what physical factors determine concentration/depletion, and how can the collection of molecules be regulated? Such information is essential to understanding condensates as biochemical entities and ultimately discerning their cellular and organismic functions.
Models for compositional control of condensates
Individual condensates can contain hundreds of distinct molecular components. For example, PML bodies can contain over 200 unique proteins [19], the nucleolus can contain over 4500 unique proteins [20], and stress granules can contain over 100 proteins as well as over 1,000 RNA transcripts [21–24]. Some of these components are unique to a specific condensate, but others can be shared between different types, particularly among the various RNA-containing structures [18,23–26]. Composition can vary dramatically under different cellular conditions [23,24,26,27] and can rapidly change in response to signals [24,28]. Even in the absence of stimuli, most (but not all) residents rapidly exchange between the condensate and the surrounding cytoplasm or nucleoplasm [14,29–32].
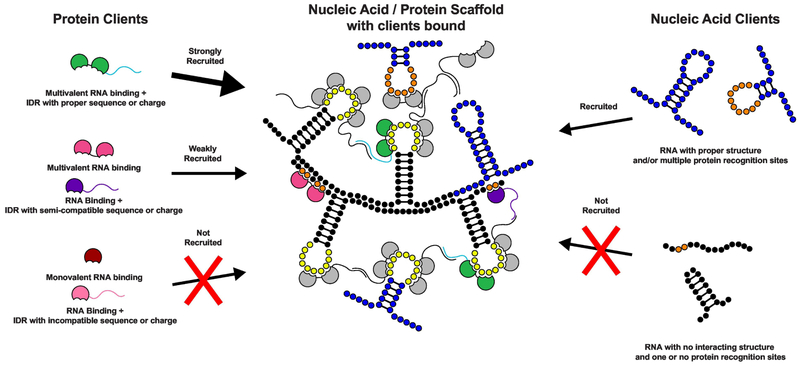
Where studied, only a few residents are typically necessary to form the condensate. Deletion or depletion of these molecules decreases the size and/or number of the structures in a cell, and sometimes overexpression has the opposite effect [33–36]. We refer to these elements as scaffolds (For reference, see the black and yellow nucleic acids and the gray proteins with black disordered tails bound to nucleic acids in the schematic in Figure 1). An example of a scaffold is PML; knocking out PML abolishes PML nuclear body formation [34,37] while increasing PML expression results in an increased number of PML nuclear bodies [38]. Other condensate residents are concentrated within the structure, often by direct interactions with scaffolds, but are not required for condensate formation. We refer to these elements as clients (For reference, see the colored nucleic acids and proteins that bind to the nucleic acid / protein scaffold in Figure 1). Examples of clients include PML nuclear body proteins Sp100 and BLM; knocking out either protein does not ablate PML nuclear body formation [34,39]. Since there are few scaffolds and generally many more clients, scaffolds should be present at a higher concentration than clients within condensates. In this context, the question of compositional specificity becomes one of understanding, for a given set of scaffolds, what clients are recruited and to what degree. In the engineered model condensates described below, the distinction between scaffold and client was stark, by design. But in natural condensates, the distinction between scaffold and client may be blurred for some molecules and could vary with cellular conditions. Nevertheless, the scaffold-client distinction is useful in considering condensate composition.
Figure 1.
A unified model of condensate composition
A model condensate formed from a protein scaffold with multivalent RNA binding domains (RBDs - grey) linked to an IDR (black) and an RNA scaffold with multiple RBD recognition elements (yellow) and specific secondary structure elements. Client protein recruitment is dependent on both the number of RBDs and the sequence of the IDR. Different IDR sequences are represented by the different colors of the client proteins. Client RNA recruitment is dependent on the RNA secondary structure and valency of RBD recognition elements (orange). For more detailed description of model, see the text.
Modular domains
Using a two scaffold model system, Banani and colleagues [40] combined biochemical and cellular experiments with computational modeling to provide an initial conceptual framework to explain how specific clients can be recruited to phase separated droplets generated by modular protein domain interactions. The biochemistry was based largely on engineered scaffolds composed of multiple repeats (5-10) of modular binding domains, which underwent LLPS when mixed with complementary scaffolds composed of repeated (5-10) interaction motifs. The resulting condensates then recruited clients containing a lower valency (1-3 repeats) of the same domains or motifs.
Several systems were examined: small ubiquitin like modifier (SUMO) domains binding to SUMO interacting motifs (SIMs); Src Homology (SH) 3 domains binding to proline rich motifs (PRMs); and RNA recognition motif (RRM) domains binding to the RNA element, UCUCU. In all cases, when the total solution concentration of the first scaffold exceeded that of the second, the resulting condensates recruited only clients that would bind the first (e.g. when polySUMO > polySIM, droplets recruited only SIM-containing clients). When the scaffold concentrations were reversed, the opposite client was recruited. Thus, clients partition into condensates only when their cognate scaffold is present in stoichiometric excess. Mathematical modeling based on simple mass action showed that this result is less obvious than it might seem at first, since it required concentration of both scaffolds into the droplets to equal degrees. If the two scaffolds are concentrated to slightly different degrees, the model predicted client partitioning to show non-intuitive behavior in some regions of the phase diagram (e.g. extremely high client partitioning near stoichiometric equality of the scaffolds), as was observed experimentally. In addition to scaffold ratio, the valency of the clients also dictated their recruitment. Higher valency of SIM or SUMO in clients increased their recruitment into polySUMO-polySIM droplets, likely due to avidity effects producing higher affinity. In general, mass action would predict that clients with a higher affinity for scaffolds should be recruited into droplets to a greater degree.
A final aspect of the framework is that molecules that bind to the scaffolds through regions that are not involved in scaffold-scaffold contacts (e.g. at other discrete binding sites or through non-specific interactions) should be recruited similarly across the entire phase diagram without regard for scaffold ratio. The combination of relevant parameters—scaffold ratio, client valency, client affinity, interactions with different domains, and non-specific interactions—provides a rich space of compositions achievable for a condensate generated by a modest number of scaffolds. As described below, this framework was borne out in studies of mammalian PML nuclear bodies and yeast P-bodies.
Intrinsically Disordered Regions
Like multivalent modular domains, protein IDRs can act as scaffolds to generate condensates and/or specify client recruitment. Many groups have shown that IDRs of some RNA binding proteins can undergo phase separation at high concentrations or when mixed with scaffold RNA [11,14,16,41,42]; whether this is a general principle that applies to the IDRs of all RNA binding proteins has not been determined. A variety of sidechain interaction types can promote IDR phase separation, including pi-pi (e.g. aromatic-aromatic), cation-pi (e.g. arginine-aromatic), polar-polar (e.g. poly-glutamine) and charge-charge [43,44]. Amyloid-like β-strand interactions of the polypeptide backbone also play important roles [41,45,46]. Additionally, other IDRs contain specific multivalent binding sequences, such as RGG motifs on Fus and Ews that bind Tudor domains when methylated on arginines [47]. Together, these interactions provide the weak, distributed adhesions necessary to promote phase separation of IDRs. The properties of an IDR scaffold that cause it to recruit specific clients should be related to those promoting phase separation, but have been less well studied. As in the modular domain systems described above, the valency of interaction motifs in IDRs appears to play an important role in client recruitment. For example, Kato and colleagues demonstrated that the N-terminal low complexity IDR of FUS forms hydrogels through a series of tyrosine motifs that promote formation of amyloid-like fibers [41]. The authors examined the rate of client loss from hydrogels following replacement of the surrounding buffer to learn how the number of tyrosine residues in the FUS IDR affected its retention by hydrogel. They observed that the half-life of FUS IDRs was correlated with the number of tyrosine residues in the IDR; IDRs that contained a full compliment of tyrosine residues (fraction of mutated tyrosine residues = 0 / 27) were not depleted following buffer replacement, IDRs in which 5 tyrosines (5 / 27) were mutated to serine had a halflife of ~16 s, IDRs in which 9 tyrosines (9 / 27) were mutated to serine had a half-life of ~9 s, IDRs in which 15 tyrosines (15 / 27) were mutated to serine had a half-life of ~7 s, and IDRs in which all tyrosines (27 / 27) were mutated to serine had a half-life of ~3 s. Similar observations were made in cells, where the number of tyrosines in the N-terminal IDR of a panel of FUS mutants correlated with recruitment of the proteins into stress granules; mutants with 27 or 22 tyrosines (0 / 27 or 5 / 27, respectively) partitioned efficiently, a mutant with 18 tyrosines (9 / 27) moderately partitioned, and those containing 12 or 0 tyrosines (15 / 27 or 27 / 27, respectively) did not measurably partition.
However, other experiments by Kato and colleagues demonstrated that the recruitment of IDRs into condensates is not simply determined by the number of tyrosine residues within a given client. Using hydrogel depletion assays to test whether hydrogels differentially retain client IDRs derived from other RNA binding proteins, they reported that the clients RBM3 and hnRNPA2 had half-lives in FUS hydrogels >10 s, CPEB2 and Tia1 had half-lives ~5 s, and hnRNPA1 had half-life <5 s. It is difficult to rationalize these retention times based simply on interactions between client tyrosine motifs and those of the FUS scaffold, as RMB3, hnRNPA2, and hnRNPA1 IDRs are tyrosine-rich while CPEB2 and Tia1 IDRs are composed of polyQ elements. Thus, straightfoward correlations of like-recruits-like may be more difficult to make when comparing between different proteins.
Lin and colleagues performed similar experiments to examine how adding the FUS N-terminal low complexity sequence to a modular, multivalent scaffold could affect recruitment of client IDRs to condensates [11]. Polypyrimidine tract-binding protein (PTB) contains four RNA recognition motifs (RRMs) and forms liquid droplets when mixed with RNA containing five RRM binding elements in vitro. Fusing the FUS IDR to PTB (PTB-FUS) enhanced the recruitment of certain client IDRs into these protein-RNA droplets. Recruitment of FUS and Tia1 IDRs was appreciably enhanced (approximately 2-fold), recruitment of the Lsm4 IDR was slightly enhanced (approximately 1.1-fold) and recruitment of the Pub1 and eIF4GII IDRs was unchanged. As in the FUS hydrogel recruitment above, the differences here do not simply reflect like-recruiting-like, as the FUS and eIF4GII IDRs are tyrosine-rich, Lsm4 is asparagine-rich, while the Tia1 and Pub1 IDRs are glutamine-rich. The differences in recruitment pattern enhancement may be related to the pI of each IDR. The theoretical pI of the Fus and Tia1 IDRs is 4.7 and 5.8, respectively, while the theoretical pIs of Lsm4 IDR, Pub1 IDR, and eIF4GII IDR range from 9.6 to 11.5. Thus the enhanced recruitment does appear to favor acidic clients, although this is not straightforward to reconcile with the acidic nature of the FUS IDR in the FUS-PTB scaffold. Together, these data suggest that IDR scaffolds can differentially recruit IDR clients, although the relationship between scaffold sequence, or amino acid composition, and client sequence/composition remains to be understood.
Some work has been done to understand the recruitment of nucleic acids into IDR-based condensates. Ddx4, a scaffold found in nuage and germ granules, contains disordered regions that are composed of alternating positively and negatively charged groups of residues as well as cation-pi interactions between arginine and phenylalanine residues. Interactions between these charge blocks promote LLPS of Ddx4 [12]. The Ddx4 droplets recruit ssDNA while excluding dsDNA, an effect the authors suggest may arise from pairing of arginine sidechains with the bases of flexible single-stranded polynucleotides [12]. In work to further investigate nucleic acid partitioning into Ddx4 condensates, Nott and colleagues observed that short, stem-loop structured RNA and DNA, as well as unstructured RNA and ssDNA, strongly partitioned into Ddx4 condensates while rigid dsDNA, dsRNA, and hybrid dsRNA/dsDNA oligonucleotides of longer than 20 bases were excluded [48]. They suggest that the interior of the Ddx4 condensates may act like an organic solvent that could destabilize and melt short dsDNA into ssDNA that could be stabilized by arginine – nucleotide interactions. The authors also posit that shorter, more compact nucleic acid structures are preferred over longer, rigid structures because smaller nucleic acids do not disrupt the structure of the condensate.
Despite a variety of data indicating that IDRs can promote phase separation in vitro and in cells, it was recently shown that phase separation of several IDRs can be inhibited by non-specific competitors such as BSA, lysozyme, and RNaseA in vitro [49]. These agents decrease the concentration of IDR within the droplet phase and also increase the critical threshold for droplet formation. Such observations suggest that the weak, non-specific interactions that promote LLPS of IDRs may be particularly susceptible to non-specific competition, and thus that IDRs may not strongly contribute to condensate composition in vivo, where competitors are abundant. Consistent with this idea, the IDR fragments of Lsm4, Dhh1, Pop2 and Ccr4 were not strongly recruited into yeast P-bodies, and only Pop2 showed a substantial decrease in recruitment when its IDR was deleted [49]. Note, however, that this study did not quantify protein concentrations in P-bodies, only the fraction of P-bodies that contained the proteins above an arbitrary threshold intensity. So it remains possible that deletion of the IDRs did decrease the strength of recruitment into P-bodies. Thus, although IDRs can contribute to recruitment specificity into phase separated structures in vitro, it is unclear how strongly they affect compositional specificity relative to modular domain interactions in vivo. Nevertheless, IDRs are clearly important to condensates, since some IDRs do phase separate to form novel structures in cells [12,48,50], and both in vitro and in vivo data have shown that when IDRs are fused to modular domains that drive LLPS through more specific interactions, the IDRs act cooperatively with the modular domains to promote phase separation in the presence of competitors [49,51].
RNA-based compositional control
While RNAs are important scaffolds and clients in many condensates, little was known until recently about how RNA might regulate the specific molecular composition of condensates. Langdon and colleagues [18] recently observed that secondary structure controls complimentary base pairing in mRNA molecules and dictates the specificity of RNA incorporation into condensates in vitro and in vivo. This study provides a framework for understanding how RNA-RNA interactions can specifically modulate condensate composition.
Whi3 is a Poly-Q containing RNA binding protein that forms RNA-containing liquid-like condensates in fungal cells. Both RNA and Whi3 act as scaffolds and are required for proper condensate formation in vivo [42,52]. However, Whi3 can assemble into spatially and compositionally distinct condensates within the same cell. Polarity Whi3 condensates located at cell tips contain BNI1 and SPA2 mRNA transcripts, while perinuclear Whi3 condensates lack BNI1 and SPA2 but contain CLN3 mRNA transcripts that regulate cell cycle progression. In addition to having distinct molecular identities, these two distinct Whi3 condensates also have different biophysical properties in vivo. Polarity condensates are more dynamic, less viscous, and have a higher protein to RNA ratio than perinuclear condensates [42].
To determine how the same protein scaffold (Whi3) could form distinct condensates with different RNA scaffolds (SPA2 versus CLN3), Langdon et al. reconstituted Whi3 condensates in vitro. Droplets were formed with BNI1 and Whi3, and then SPA2 or CLN3 mRNA was added to solution. While SPA2 was rapidly incorporated into BNI1 droplets, CLN3 was not recruited to BNI1 droplets. Rather, CLN3 competed Whi3 away from BNI1 to form a distinct class of droplets, reducing the number and size of BNI1 droplets. In the absence of Whi3, the mRNA transcripts were able to phase separate in the presence of spermine, analogous to recent observations with RNAs generated in nucleotide-repeat expansion diseases [17]. Moreover, they showed a similar pattern of self-organization as the Whi3-containing droplets: CLN3 only localized with itself, while SPA2 and BNI1 colocalized in the same gel-like condensates. Thus, intermolecular RNA-RNA interactions were sufficient to sort mRNA into distinct phase separated compartments. Finally, using several different perturbations Langdon and colleagues showed that secondary structure was the key feature that determined whether RNA transcripts would interact and coassemble in Whi3 droplets, since disruption of secondary structure eliminated segregation of CLN3 from BNI1. Thus, RNA secondary structure plays a critical role in determining the specific molecular composition of RNA-containing condensates. An interesting inference from this work is that the inability of Whi3-RNA interactions to recruit both CLN3 and BNI1 (which both contain Whi3-binding elements) into both droplet classes suggests that RNA oligomers/aggregates may be kinetically trapped in the droplets in which they initially assemble (otherwise free RNA binding sites on the Whi3 in one class of droplet should be able to recruit at least some of the other RNA). Such non-equilibrium behavior could play an important role in cells, where local transcription and oligomerization of certain founder RNAs could produce specific structures, whose identities are defined by those RNAs and maintained by their extremely slow kinetics of dissociation.
A unified model of condensate composition
Figure 1A shows a model that brings together our current understanding of how interactions of modular domain, IDRs and RNA molecules may promote formation of an RNA and protein containing condensate and define its composition. The figure shows a model condensate formed from a protein scaffold with multivalent RNA binding domains (RBDs) linked to an IDR and an RNA scaffold with multiple RBD recognition elements and specific secondary structure elements. The protein and RNA scaffolds interact through specific protein:RNA interactions (and also through potentially less specific IDR:RNA interactions, not shown in the figure). The protein scaffolds can also further interact through their IDRs. Thus, both modular domain interactions and IDRs contribute to phase separation. The resulting condensate can recruit client proteins or client RNAs depending on the specific molecular features of the clients. Client RNA with compatible sequence and secondary structure partitions into the condensate by complementary base pairing with scaffold RNA, while RNA lacking the compatible sequence and secondary structure does not. Some RNA:RNA interactions may be kinetically trapped, keeping the system out of equilibrium. RNA clients can also be recruited via multivalent interactions with the protein scaffold. Several molecular features promote client protein partitioning into the condensates. Client proteins with higher RBD valency partition more than monovalent clients due to increased interactions with the RNA scaffold. Client proteins that contain an IDR with certain sequence or charge features will preferentially partition over client proteins with IDRs lacking these sequence features. The effects of modular binding domains and IDRs on client recruitment are cooperative, such that clients containing both multivalent RBDs and a compatible IDR will most strongly partition to the condensate. Neither monovalent RNA-binding domains, nor incompatible IDRs partition strongly into the model condensate. Composition can be tuned by modifying either scaffolds or clients (Figure 2). Combined, these models can serve as a conceptual framework to understand how the composition of biomolecular condensates is defined and can be dynamically regulated (see below).
Figure 2.
Compositional control of condensates formed from scaffolds with modular domains
(A) Model of client recruitment with different scaffold stoichiometries. Client recruitment is dependent on the availability of binding sites on the scaffolds and the valency of the clients. (B) Droplets were formed with 6 μM of a (SUMO)9-(SIM)8 scaffold containing Ulp1 cleavage sites after only the two N-terminal SUMOs and were equilibrated with 50 nM of GFP-(SIM)2 (Client 1, green) and RFP-(SUMO)2 (Client 2, magenta). At time 0, 10 nM of Ulp1 was added. Pseudocolored images of the merged fluorescent signals from the two clients are shown. Figure reproduced with permission from [40].
Dynamic regulation of condensate composition
Recent studies that used either model engineered systems or specific cellular examples have suggested several mechanisms by which condensate composition can be dynamically altered. Some of these hinge on dynamic regulation of scaffold or client valency through changes in post-translational modifications that mediate scaffold-client interactions. For example, Banani and colleagues [40] showed that a multivalent scaffold composed of both SUMO and SIM modules, with the former in excess, will recruit SIM-containing clients, but not SUMO-containing clients. However, after removal of some SUMO modules by addition of the deSUMOylating enzyme ULP-1, the SUMO:SIM ratio in the scaffold is inverted, and SIM-containing clients leave the condensate while SUMO-containing clients are recruited to it (Figure 2B). Thus, by altering the relative valency, post-translational modification of multivalent scaffolds can reverse client composition, a behavior that should be general for condensates based on molecular interactions with post-translational modifications. In such systems, addition or loss of post-translational modifications from clients should also increase or decrease their partitioning, respectively, in dynamic fashion.
The post-translational modification of IDRs can also modulate condensate composition through changing chemical properties and interactions of the polypeptide chain. For example, phosphorylation can decrease self-association of IDRs [53,54], causing dissolution of IDR-based condensates [46,53,54] and/or release of IDRs from condensates [45]. Similarly, arginine methylation of the IDR of Ddx4 [12] and Fus [55] can also attenuate condensate formation.
Finally, the work of Langdon and colleagues suggests additional RNA-based mechanisms of regulation. These authors show that RNA secondary structure can be regulated by protein binding. For example, when Whi3 binds to CLN3, the transcript extends. Whi3 binding to BLN1 increases the conformational dynamics of the RNA. These changes could in turn influence recruitment of additional RNAs or proteins, altering condensate composition and function [18].
Compositions of natural condensates
In the sections below we describe our current understanding of the composition of representative condensates—PML nuclear bodies, P-bodies, stress granules, the nucleolus, and two-dimensional membrane localized LAT and nephrin clusters—through the lens of the models above.
PML Nuclear Bodies
The Promyelocytic leukemia protein (PML) forms biomolecular condensates termed PML nuclear bodies (PML NBs) in the nuclei of mammalian cells. As described earlier, PML is the only protein known to be required to form PML NBs [34]. PML is a member of the tripartite motif (TRIM) family of proteins, and as such contains an N-terminal Ring domain, two B-box domains and a coiled-coil region. There are numerous isoforms of PML, which are distinguished by distinct C-terminal elements. PML condensate formation is driven by PML:PML interactions involving at least the first B-box domain [56] and the coiled-coil domain [57], PML is also SUMOylated at three main positions and several minor sites [58,59]. These modifications, as well as a C-terminal SIM element found in most isoforms, contribute to the formation and ultrastructure of the bodies [60,61]. Approximately 200 PML NB client proteins have been reported [19] and body composition changes under different conditions [28,62,63]. Posttranslational modification of both the PML scaffold and clients can regulate client recruitment to PML NBs. SUMOylation of PML is essential for recruitment of numerous cellular SIM-containing clients including Daxx, Sp100, HIPK2, BLM, and IKKε [28,34,64,65]. Similarly, engineered clients composed of repeated SIMs localize to PML NBs formed by wild-type PML but not to bodies formed by a mutant PML that cannot be SUMOylated. Thus, the SUMOylation of the PML scaffold promotes the recruitment of clients containing SIMs. Interestingly, engineered clients composed of repeated SUMO domains do not localize to PML NBs formed by wild-type PML, but do localize to bodies generated by a mutant PML lacking the three main SUMOylation sites [40]. The combined data suggest that wild type PML NBs lie on the SUMO-excess side of the SUMO-SIM phase diagram and that mutation of the main SUMOylation sites in PML moves the bodies to the SIM-excess side, inverting their recruitment profile. Posttranslational modification of client SIMs can increase SIM affinity for SUMO [66,67]. For instance, phosphorylation of the Daxx SIM increases its affinity for SUMO-1 [66] and, presumably, SUMOylated PML. Together, these data illustrate how SUMOylation of PML and SUMOylation or phosphorylation of clients can regulate the composition of PML NBs through modulating scaffold-client interactions.
The composition of PML NBs changes as cells progress through the cell cycle or differentiate [62,63,68]. Dellaire and colleagues [62] investigated the cell-cycle dependent recruitment of two PML NB clients, Sp100 and Daxx. During G1 phase, most PML NBs are enriched with both Sp100 and Daxx. During metaphase, both Sp100 and Daxx rapidly disperse from PML NBs so that by the time cells enters prometaphase, very few PML NBs contain Sp100 or Daxx. SUMO-1 levels within PML NBs also decrease during prometaphase, indicating that PML is actively deSUMOylated as the cell moves from prophase to prometaphase. Thus, as a cell progresses through the cell cycle, the SUMOylation state of the PML scaffold is altered and this may modulate scaffold:client interactions. While the scope of this study was limited to only two clients during the cell cycle, it may provide a lens to understand how the overall composition of PML NBs is either maintained or altered at any given time.
Much work needs to be done to fully understand how the composition of PML NBs is regulated. For instance, although engineered polySUMO clients do not strongly partition into interphase PML NBs [40], all three SUMO isoforms are highly concentrated in the structures [57]. Does this SUMO consist of long polySUMO chains attached to PML, as has been suggested [57,60,69,70], or if not, what are the SUMOylated clients and how do they concentrate in the bodies while an engineered polySUMO client does not? What clients remain in PML NBs during mitosis, when many SIM-containing clients depart? How are the activities of SUMO ligases and SUMO proteases, which SUMOylate and deSUMOylate targets respectively and are both concentrated within PML NBs [71,72], balanced dynamically to control the SUMOylation status of PML and other components of the bodies? Although the biochemical functions of PML NBs remain unclear, changes in composition during the cell cycle could alter such functions. Future work to address these questions will likely reveal additional levels of PML body compositional control and promote additional studies to understand how other modifications, such as phosphorylation, acetylation, and ubiquitinylation [73] also regulate PML body dynamics and composition.
P-bodies
P-bodies (short for “processing bodies”) are RNP granules found in the cytoplasm of eukaryotic cells that concentrate translationally repressed mRNA and mRNA processing enzymes (see review [74]). P-bodies serve as sites of both RNA degradation and RNA storage, and thus are thought to have an important role in regulating translation [75]. Inhibiting translation initiation with cycloheximide inhibits P-body formation, and treating partially purified P-bodies with RNAase is sufficient to dissolve them [76]. Thus, mRNA is an important scaffold in P-bodies. Genetic screens to identify the protein scaffolds of yeast P-bodies have shown that among the numerous residents of P-bodies (see below) only deletion of Edc3, Dcp2, Dhh1, Pat1 or Lsm4 causes substantial decreases in P-body size, number, and client recruitment [35,36,51]. In addition to Dhh1, Lsm1, and Lsm4, GW182, a protein not found in yeast, was identified in an RNAi screen for proteins required to form P-bodies in human U20S cells [77]. There appears to be redundancy between the protein scaffolds, since loss of a single scaffold does not completely abolish P-body formation, but reduces their number and size [35]. The deletion of multiple scaffolds, such as Edc3 and Lsm4, abolishes P-body formation in yeast (i.e. no condensates containing Dcp1 or Dhh1 are observed), but even this can be rescued by overexpressing additional proteins with multivalent protein and RNA binding domains, including Psp2, Pby1, and Dhh1 [36]. Recent in vitro reconstitutions have also provided evidence for the importance of multivalent protein and RNA scaffolds in P-bodies [6]. Minimal LLPS droplets can be formed in vitro with Edc3, Dcp1, and Dcp2, but adding either the multivalent protein Pdc1 or RNA with a length of greater than 30 nucleotides greatly reduces the critical concentration for phase separation [78]. Thus, although P-body scaffolds are redundant, multivalent interactions between protein scaffolds and RNAs are essential for P-body assembly [36].
In addition to these scaffolds, there are numerous protein clients that localize to P-bodies. While previous studies have identified dozens of P-body components including Lsm1-7, Xrn1, and Pop2 [51], recent proteomics studies provide a more comprehensive list of P body components [24,79]. In one study, P-bodies were purified from human epithelial cells (HEK293T) using a method involving cell disruption followed by isolation of GFP-Lsm14a-labeled particles with a novel flow cytometry approach [79]. Mass spectrometry analysis of the collected particles revealed 125 proteins enriched in Lsm14a-labeled particles including translation repression and mRNA decay factors (4E-T, LSM14A, LSM14B, and IGF2BP2), components of the miRNA pathway (AGO1, AGO2, MOV10, and ZCCHC3), decapping complex components (DCP1A, DCP1B, DCP2, and EDC4), nonsense-mediated mRNA decay factors (UPF1 and SMG7), and, unexpectedly, myosin motors (MYO1C, MYO6, and MYH10). In another study, proximity-dependent biotinylation (BioID) was performed to label P-body components in human epithelial cells (HEK293T) using either BirA*-PatL1 or BirA*-Dcp2a as baits. Biotinylated proteins were immunoprecipitated and identified with mass spectrometry. Over 100 proteins were identified, including many proteins involved in mRNA decapping (DCP1A, DCP1B, PNRC1, EDC3, and EDC4), components of the miRNA pathway (AGO1, AGO2, AGO3, TRNC6A, and ZCCHC3), nonsense-mediated mRNA decay factors (SMG5 and SMG7), and, unexpectedly, proteins involved in COP-II vesicle budding (SEC16A, SEC23B, SEC24B) and mitotic spindle organization (CEP192 and WDR62). Both proteomics studies found that P-bodies only share 10-25% of their components with stress granules, suggesting that the two condensates are compositionally and functionally distinct [24,79].
Only seven proteins (Ago1, Ago2, Dcp1b, Ddx6, Edc4, EIF4ENIF1, and SMG7) were identified in all 3 proteomics screens (i.e. isolation of Lsm14a particles, and BioID with PatL1 and Dcp2). These proteins are likely constitutive components of mammalian P-bodies, and all of them are capable of binding RNA. 21 proteins were identified in 2 or more screens, while over 100 proteins were only identified in a single screen. However, all screens returned some subset of proteins involved in mRNA decapping, miRNA processing, and nonsense-mediated mRNA decay.
The discrepancy between the mass spectrometry results could be due to the different techniques or bait proteins used. Partial purification of P-bodies likely introduces some artifacts, similar to artifacts observed during partial purification of stress granules [22]. In cells, P-bodies are liquid-like and many components rapidly exchange with the cytoplasm. By isolating P-bodies from the cytoplasm, many of the dynamic components are likely lost, resulting in a bias towards purification of more solid-like particles. Therefore, this approach is more likely to identify components that reside stably in the P-body. In contrast, proximity-dependent biotinylation is excellent for identifying the more dynamic components of P-bodies. However, with this approach, it is more difficult to differentiate between interactions that specifically occur in P-bodies from those that occur in the cytoplasm. Furthermore, BirA*-PatL1 and BirA*-Dcp2a shared less than 40% of interacting components, suggesting that no one bait is sufficient to identify the complete P-body interactome with BioID.
While RNA is required for P-body formation, it is unclear which mRNA transcripts act as scaffolds and which act as clients, or whether there is a clear distinction for these in P-bodies. Several studies have demonstrated that P-bodies are enriched with translationally repressed mRNA [76,79]. Transcriptomic analysis of the Lsm14a-labeled, partially purified P-bodies identified thousands of mRNA transcripts in P-bodies [79]. While 89% of P-body enriched transcripts are protein-coding RNAs, they are poorly translated and exhibit very heterogeneous Poly(A) tail length, two characteristics of translationally repressed mRNA. P-bodies are enriched with mRNA transcripts that encode regulatory processes, such as chromatin remodeling, RNA processing, cell division, and morphogenesis, but are generally lacking in mRNA transcripts encoding housekeeping functions, such as histones, translation machinery, and catabolic and metabolic pathways. Furthermore, transcripts for protein complexes, such as the cohesion complex, are enriched together. The authors speculate that regulatory mRNA transcripts are temporarily stored in P-bodies to repress translation, and that release of transcripts from P-bodies allows for posttranscriptional regulation of protein expression. However, this model remains to be experimentally tested.
Since P-bodies are enriched with specific classes of mRNA, the targeting of mRNAs to P-bodies must be a carefully regulated process. However, the mechanisms that regulate mRNA localization to P-bodies remain poorly understood. The RNA-targets of P-body enriched RNA binding proteins were significantly enriched in P-bodies [79], suggesting that RNA-protein interactions regulate RNA composition. However, it is likely more complicated than RNA binding proteins simply recognizing the primary sequence on mRNA transcripts. For example, mRNAs bound to Ago2 in their UTR were enriched in P-bodies, while those bound to Ago2 in their coding sequence were not enriched in P-bodies [79]. As described above, RNA-RNA interactions, RNA sequence, and secondary structure can determine the ability of RNAs to phase separate and partition into RNP condensates [17,18]. Thus, many features of RNA, including sequence, length, and secondary structure could influence the propensity for transcripts to partition into P-bodies. Since P-bodies are also enriched with mRNA helicases, such as UPF1 and DDX6, RNA secondary structure or length is likely actively changed within the P-bodies [79]. Consistent with the idea of active RNA processing within condensates, RNAs that form static, solid-like structures in vitro are able to form dynamic, liquid droplets in vivo, whose dynamic properties depend on cellular ATP levels [17].
While recent proteomics and transcriptomics studies have contributed to a more complete understanding of P-body composition, it remains poorly understood how this composition is regulated. P-bodies disassemble during mitosis, reform after cytokinsesis, and then increase in number and size during S phase [80]. However, it is not known how P-body composition might change throughout the cell cycle. During stress, P-body size and number increase and their composition changes [24,77]. For example, P-bodies become enriched with ubiquitination-related proteins under arsenite stress [24]. However, specific mechanisms that regulate client recruitment under these different conditions are currently unknown.
Stress Granules
Stress granules are cytoplasmic RNP granules that form in response to diverse cellular stresses including heat shock, hypoxia, starvation, and viral infection (see [81] for an in depth review). Stress granule formation is required for cellular survival during stress [82–84], although how stress granules specifically promote cell survival is not completely understood. Recent proteomics studies reveal that stress granule composition changes under different stresses (as discussed below), suggesting that stress granule composition and function is tuned depending on the specific cellular stress [23,24]. Yeast stress granules are more solid-like than the liquid-like yeast P bodies, while both mammalian stress granules and mammalian P bodies behave like liquid droplets [85]. Mammalian stress granules are composed of a stable, solid-like core surrounded by a dynamic, liquid-like shell [22]. ATP is required to maintain the liquid-like properties of the shell, suggesting that the localization of ATPases, such as RNA helicases, to stress granules could promote the turnover of components [22]. However, the drop in cytoplasmic pH that occurs when ATP is depleted in yeast might also explain these observations [86]. ATP depletion rapidly acidifies the cell, which causes the cytoplasm to solidify and reduces protein mobility [86].
Like P-bodies, stress granules are organized around both protein and RNA scaffolds. Non-translating mRNAs are thought to be an important scaffold for stress granule formation, since stress granules contain Poly(A) binding protein (PABP), stain positive for Poly(A) mRNA, and form when translation initiation is inhibited [87,88]. Transcriptomics of purified cores found that mRNA make up over 80% of the RNA in stress granules and suggests that mRNAs with longer coding regions and 3’ UTRs are more likely to localize to stress granules [21], In addition to mRNA, stress granules also require different protein scaffolds depending on the type of stress. Phosphorylated eukaryotic initiation factor (elF)2a is required for mammalian stress granules to form in response to arsenite [89], since cells fail to form elF3/TIAR/G3BP positive foci when the WT elF2a allele is replaced with a non-phosphorylatable mutant. Additionally, deleting either UBAP2L or CSDE1 with CRISPR/CAS9 significantly reduced the number of G3BP foci formed during arsenite stress [24]. Deletion, truncation, or cleavage of G3BP prevents stress granule formation during arsenite stress [90] and viral infection [91], but not during osmotic stress or heat shock [92]. Thus, like P-bodies, there appears to be some redundancy between the protein scaffolds required for stress granule formation. Previous cell biology and genetics studies have identified numerous stress granule clients including translation initiation factors, aa tRNA-synthases, and RNA binding proteins, such as TIA1, Fus, and Ews [89,93,94].
Recent proteomics studies have contributed to a more complete understanding of stress granule composition [22–24]. The stable, stress granule cores of yeast and mammalian cells treated with arsenite were isolated and analyzed by mass spectrometry. Both yeast and mammalian stress granules were enriched with tRNA sythases, ribosome biogenesis factors, protein chaperones, and RNA/DNA helicases [22]. Proximity labeling approaches using G3BP1 as bait found that many G3BP1 interactions were stress independent [23,24], suggesting that many of the identified interactions were not specifically happening in the stress granule but also occurring in the unstressed cytoplasm. Stress granule proteins identified by proximity labeling include those involved in regulating RNA stability, translation initiation, viral translation, and RNA splicing [23,24]. However, as with P-body component analysis, proteomics results from purified stress granules versus proximity labeling are not expected to be exactly the same. Because stress granules are composed of a stable core and dynamic shell, many rapidly exchanging shell components will be lost during the purification process leaving stress granules that are enriched in only stable core proteins for mass spectrometry analysis. Proximity-labeling, on the other hand, does not distinguish between interactions that occur in stress granules from those that occur in the surrounding cytoplasm. Additionally, stress granule composition was found to vary between different cell types and different stresses, making direct comparisons between different proteomics studies difficult [23]. Furthermore, additional protein baits may be required to visualize the entire stress granule interactome with proximity labeling.
There is evidence that stress granule composition changes depending on the type of stress, suggesting that stress granule scaffolds and clients are stress specific [90,92,93,95,96]. For example, unlike yeast stress granules formed during glucose starvation, yeast stress granules formed in response to arsenite contain components associated with later stages of translation initiation such as eIF3, eIF4A/B, eIF5B and eIF1A [97]. Recent screens have begun to provide a more comprehensive view of these composition changes. Comparing stress granules formed with arsenite to those formed with heat shock revealed that 23% of components are stress specific [23]. Arsenite stress granules recruit additional proteins involved in translation initiation (EIF4G3, ABCF1, EIF3A), RNA splicing (DAZAP1), and RNA binding (Fus and SLBP), while heat shock stress granules recruit additional proteins involved in RNA silencing (DDX4 and EIF2C1) and RNA splicing (SF1). However, more comprehensive studies are required to understand how stress granule composition changes under other cellular stresses such as glucose starvation, hypoxia, and viral infection.
Although stress granule composition is known to be both cell type and stress dependent [23], the mechanisms that regulate client recruitment are not clear. Both yeast and mammalian stress granules are enriched with proteins that add or remove post-translational modifications, including kinases, phosphatases, E3 ligases, and methyl-transferases [22], and there is some evidence that stress granule composition can be regulated by changes in post-translational modifications. For example, phosphorylation of G3BP impairs stress granule assembly [90], while O-Glc-NAc glycosylation of proteins enhances stress granule assembly [77]. The methylation of RGG motifs on RNA binding proteins, such as Fus and Ews, can recruit Tudor domain containing proteins, such as TDRD3, to stress granules [47]. NEDD8 (neural precursor cell expressed developmentally downregulated protein 8) is a small ubiquitin-like protein that is covalently conjugated to Lys residues on protein substrates [98]. Inhibiting NEDDylation prevents stress granule assembly in response to arsenite treatment, and NEDDylation of the stress granule component SRSF3 is required for its interaction with TIA1 but not G3BP1. Thus, NEDDylation could regulate specific protein-protein interactions at stress granules. Polymers of ADP-ribose are covalently attached to proteins in a process called PolyADP-ribosylation (PARylation). Poly(ADP-ribose) polymerases localize to stress granules, and Ago2, G3BP1 and TIA-1 are all PARylated on their RNA binding domains upon stress with pateamine A [99]. Whether this alters their ability to bind RNA is not clear.
There is also evidence that the post-translational modification of stress granule components can be regulated in a stress specific manner. For example, eIF4A2 is SUMOylated in response to arsenite and ionising radiation, but not in response to heat shock or hippuristanol, and elF4A2 SUMOylation correlates with its localization to stress granules (through a still unknown mechanism) [100]. Additionally, different elF2a kinases are required for mammalian cells to form G3BP1 foci when exposed to arsenite, heat shock, thapsigargin, or MG132 [101], suggesting that different signaling pathways are used to initiate stress granule formation in response to different stresses. While these are a few examples of how specific clients may be recruited to stress granules under certain conditions, we lack a comprehensive understanding of how stress granule composition is regulated. Understanding how stress granule composition changes in response to specific stresses is important and may even reveal general principles of how RNP granules are able to adjust their composition to meet cellular needs.
The Nucleolus
The nucleolus is an RNP condensate that self-organizes around tandemly repeated ribosomal DNA sequences in the nucleus and functions to guide ribosome biogenesis [20]. Nucleoili are composed of three distinct subregions: the fibrillar center (FC), the dense fibrillar components (DFCs), and the granular components (GCs). Each subregion of the nucleolus contains specific proteins that regulate subsequent stages of ribosomal RNA production, processing, and assembly [102]. RNA polymerase I localizes to repeated rDNA elements to initiate transcription of rRNA in the interior FC [103]. rRNA transcripts are spliced and modified by small nucleolar ribonucleoproteins (snoRNPs) within the middle DFC and then assembled into mature ribosomal subunits in the outer GC [104]. Processed ribosomal subunits exit the nucleolus and are assembled into the ribosome in the cytoplasm.
The nucleolus exhibits properties consistent with a condensate formed through LLPS [29,105–107]. Solubilized nucleoli components can re-assemble and self-organize into nucleolar-like structures in vitro [108]. Purified fibrillarin (FIB1), a component of the DFC, and nucleophosmin (NPM1), a component of the GC, can each undergo LLPS in vitro at physiologic concentrations when combined with physiologic concentrations of salt and ribosomal RNA (rRNA) [107,109]. However, FIB1 droplets and NPM1 droplets each exhibit distinct biophysical properties. Droplets of fluorescently-labeled FIB1 recover more slowly following photobleaching than those of fluorescently-labeled NPM1, similar to the dynamics of these proteins within Xenopus laevis nucleoli [107]. When combined in the same in vitro assay at appropriate relative concentrations, the two proteins form multilayered droplets. In these, FIB1 is enriched in a central compartment and NPM1 is enriched in an outer layer, as in Xenopus laevis nucleoli. This organization arises from the different surface tensions of the two subcompartments [107]. Although the FC has not been reconstituted, neighboring FCs have been observed to undergo fusion during G1 phase, suggesting that the FC is a third liquid-like phase within the nucleolus [105]. Thus, within each nucleolus, the phase separated FC, DFC, and GC form from different components, are maintained by biophysical properties that emerge as a result of their ability to phase separate, and perform different functions in ribosome biogenesis [102].
Recently, potential nucleolar scaffolds have been identified, including both proteins and nucleic acids. The FC contains concentrated RNA polymerase I organized around repeated rDNA elements, similar to transcription condensates seen elsewhere in the nucleus [110,111]. Thus, rDNA may be a scaffold for the FC. rRNA and non-coding RNAs are likely nucleic acid scaffolds in the DFC and GC [112]. The nucleolus is enriched in RNA polymerase II-generated, non-coding RNAs that are rich in Alu elements [112]. Antisense-oligo-mediated depletion of Alu-containing RNA or RNA polymerase II inhibition disrupts the nucleolus and inhibits rRNA transcription, suggesting that Alu-containing RNAs function as a scaffold within the nucleolus. In addition to nucleic acids, WDR46 and NPM1 have been identified as potential protein scaffolds of the GC. WDR46 localizes to the GC subregion and contains an IDR that facilitates interactions with other IDR-containing proteins [113]. Knockdown of WDR46 to 40% of wild-type levels results in the mis-localization of potential client proteins nucleolin and DDX21 from the GC to the outer surface of the nucleolus. Other binding partners, NOP2 and EBP2, localize to the GC regardless of WDR46 expression levels. NPM1 also localizes to the GC subregion and is composed of an N-terminal pentamerization domain, an amphiphilic IDR and a folded C-terminal nucleic acid binding domain (NBD) [114]. The pentamerization domain and the IDR each contain acidic elements that bind to proteins containing arginine-rich motifs (R-motifs) [109]. Deletion of NPM1 results in a disruption of nucleolar morphology and rearrangement of perinucleolar heterochromatin when compared with cells expressing NPM1. NPM1 also regulates the localization of viral [115], tumor suppressor [116], and ribosomal proteins [117] within the nucleolus. R-motifs within nucleolar proteins have been identified as nucleolus-targeting sequences, and NPM1 is likely the scaffold for these R-motif clients [109,118,119]. Thus, WDR46 and NPM1 are two protein scaffolds localized within the GC that can recruit IDR-containing or R-motif-containing clients, respectively. More research is required to determine additional protein scaffolds within each of the nucleolar subregions. Each subregion could even contain redundant protein scaffolds, similar to P bodies and stress granules.
Early research into the composition of the nucleolus used quantitative mass spectrometry and quantitative western blotting to analyze isolated, intact nucleoli from cultured HeLa cells [120]. Real-time fluorescence microscopy corroborated mass spectrometry and western blot results. Through the combination of these techniques, the authors found that the inhibition of rRNA transcription causes some dead box helicases, such as DDX3X, DDX59, and DDX17, to accumulate in the nucleolus while other dead box helicases, such as DDX 18, DDX51, and DDX54 disperse out of the nucleolus. rRNA processing proteins, such as Upt1 1, Upt1 4, and Upt7, and RNA Polymerase I also disperse from the nucleolus upon inhibition of rRNA transcription. While mass spectrometry studies can provide a list of nucleolar components, they do not provide insight into subnucleolar localization of these proteins. Future studies should link specific proteins to specific subregions. For instance, RNA Pol I subunits localize to the FC, fibrillarin and other snoRNPs localize to the DFC, and NPM1 and WDR46 localize to the GC. Similar localization for other identified nucleolar proteins would improve compositional and functional understanding of the condensate.
Given that the nucleolus is a multi-layered condensate, each subregion likely contains distinct scaffolds and clients, and much work is required to understand how the composition of each subregion might be regulated through RNA processing or post-translational modification of proteins. The mass spectrometry experiments described above show that the nucleolus composition can fluctuate, but it is unknown how composition is regulated in vivo. In addition to ribosome biogenesis, the nucleolus is involved in cell-cycle control [121,122], genome stability [123], and the stress response [124,125], and it is unclear how the composition of each subregion regulates these processes. Infection of cells with virus also induces compositional changes in the nucleolus, however the mechanism by which these changes occur is not understood [126,127]. More work is required to understand how compositional changes in the nucleolus are regulated, whether each subregion is individually regulated or whether changes in one subregion lead to changes in all subregions, and how compositional changes alter nucleolar function.
Membrane Clusters
Many transmembrane receptors, including growth factor receptors, adhesion receptors, and immune receptors, form higher-order clusters that are essential for downstream signaling [128–130]. There is growing evidence that many of these clusters are membrane-associated condensates that form through LLPS. Currently, two analogous signaling systems have been shown to form membrane-associated condensates through LLPS [5,7,9]. Nephrin is a disordered transmembrane receptor that regulates cell-cell adhesion in podocyte cells of the kidney [131]. The protein Linker for the Activation of T cells (LAT) is a disordered transmembrane protein that regulates signaling in activated T cells [132]. Both Nephrin and LAT are phosphorylated on multiple tyrosine residues in response to upstream stimuli [133,134]. Their phosphorylated cytoplasmic tails then interact with collections of cytoplasmic, multivalent adaptor proteins containing SH2 domains, SH3 domains, and proline-rich motifs (PRMs). Multivalency in these molecules enables their assembly and concomitant phase separation, forming micron-scale signaling clusters on supported lipid bilayers (SLBs) in vitro and at the plasma membrane of cells [7,9]. The formation of these clusters is correlated with downstream signaling events such as actin polymerization and the phosphorylation of Erk [9]. Thus, it is important to understand how the composition of membrane-associated condensates is regulated to tune downstream signaling activities.
Phosphorylated nephrin will phase separate when combined with fellow scaffolds Nek and N-WASP [5,7]. Unlike nephrin clusters, LAT clusters have redundant scaffolds. Phosphorylated LAT will phase separate when mixed with Grb2 and Sos1, Gads and SLP-76, or combinations of these four scaffolds [9], Thus, the stoichiometry of Grb2, Gads, Sos1, and SLP-76 potentially varies within any given LAT cluster. Whether the stoichiometry of the scaffolds within LAT clusters is regulated by local cellular conditions is unknown. In LAT clusters, unlike in nephrin clusters, Nek and WASP family proteins act as clients that are not required for phase separation but are recruited through phosphotyrosine sites on SLP-76, which bind Nck. Recruitment of Nck, N-WASP and Arp2/3 complex then promotes actin assembly at LAT clusters, likely in a manner similar to that observed at nephrin, Nck, and N-WASP clusters.
In addition to specific binding, electrostatic sorting of client proteins is another mechanism by which LAT condensates tune their composition. Following phase separation of the scaffolds LAT, Grb2, and Sos1 on supported lipid bilayers, engineered client proteins that are negatively charged, such as the intracellular domain of CD45, the native phosphatase for LAT, and engineered SNAP protein fused to polyglutamate, were excluded from clusters. Conversely, positively charged model clients, such as SNAP fused with polyarginine or modified GFP [135], are enriched within clusters [9]. In this system, the size of client proteins does not appear to be a factor controlling partitioning into condensates (at least up to ~106 kDa). Thus, while not yet examined in cells, electrostatic sorting is likely a significant mechanism by which cluster composition is regulated. It remains to be determined whether analogous charge effects play significant roles in defining the composition of three-dimensional condensates.
Recent work has demonstrated that in the nephrin and LAT systems, the composition of membrane-associated clusters has important influence on biochemical and cellular functions, respectively [136]. In both systems, clusters locally generate actin filaments through the adaptor protein Nck, its ligand N-WASP, and the Arp2/3 complex, which nucleates new actin filaments from the sides of existing mother filaments [137]. Moreover, LLPS increases the specific activity of molecules in the pathways toward the Arp2/3 complex [9] (L.B. Case, M.K. Rosen, unpublished). The mechanism of this increase has recently been elucidated, and has novel implications for cluster composition (L.B. Case, M.K. Rosen, unpublished). Grb2 and Sos1 (in LAT clusters), and N-WASP (in nephrin clusters) all have both increased density and increased membrane dwell time within phase separated condensates on supported lipid bilayers in vitro [138] (L.B. Case, M.K. Rosen, unpublished). In nephrin clusters, the specific activity of N-WASP toward the Arp2/3 complex positively correlates with the membrane dwell time of N-WASP, but is independent of N-WASP density. Thus, LLPS increases Arp2/3-dependent actin assembly by increasing the membrane dwell time of upstream signaling proteins. Theoretical analyses of the LAT-Grb2-Sos1 pathway suggest that this effect is likely to be generally observed for signaling pathways that, like the Arp2/3 and Sos1 pathways, are slow, multi-step and driven out of equilibrium by irreversible steps [138]. Additionally, since membrane dwell time is strongly influenced by the multivalent connectivity within clusters, and connectivity is determined by the relative stoichiometry of the components, dwell time is controlled by stoichiometry. In turn, specific activity of the pathway is controlled by stoichiometry. Thus, the biochemical function of condensates is not only dependent on the types of molecules that they concentrate, but also on the stoichiometries of those components. This mechanism of regulating protein activity is unique to biomolecular condensates, which can form across a range of component stoichiometries, unlike molecular machines, which assemble with specific compositions.
The composition of LAT clusters has significant consequences on the cell biology of T cell activation [132,139]. In Jurkat T cells, following engagement of the T cell receptor by an MHC-peptide complex on an antigen-presenting cell, LAT clusters form at the edge of the cell-cell junction, termed the immune synapse (IS; [140]). Over time, the LAT clusters migrate toward the center of the IS through the actions of cortical actomyosin. During this process the clusters traverse two distinct actin networks, a branched network at the periphery of the IS that moves in a radial direction, and a more central network of actin arcs that appear to move in a telescoping fashion with both radially and circularly directed components [141,142]. We recently showed that the composition of LAT clusters may control their interactions with and movement by these two actin networks [136]. Nck is enriched in LAT clusters when they are in the peripheral region of the IS. However, Nck dissipates as the clusters move across the boundary between the two distinct actin networks. Through in vitro biochemical reconstitution of LAT clustering in motile actomyosin networks on supported lipid bilayers, we found that LAT clusters containing Nck and WASP family proteins bind to and are dragged by motile actin filaments. In contrast, LAT clusters lacking these proteins do not bind filaments and are instead pushed (less efficiently) by physical collisions with moving actin filaments. Together the in vitro and in vivo observations led to a model in which LAT clusters in the periphery bind the branched actin network tightly through Nck and WASP, and move with its radial movement. Near the junction with the actin arcs, Nck (and likely WASP) is released from the clusters, weakening their binding to actin and enabling them to be moved by the radial component of the arc movement, while the randomly directed circular component averages away. Thus, changes in composition enable LAT clusters to migrate radially through differential engagement with the two differently moving actin filament networks found at the IS. Future work to determine the mechanisms that regulate LAT cluster composition in different regions of the IS and how composition determines downstream signaling outputs will be essential toward understanding function of LAT clusters in the broader context of T cell activation.
Future Perspectives
Here we have reviewed recent advances in our understanding of condensate composition, and proposed a unified framework to explain how the protein and RNA components of condensates may be defined and dynamically regulated. However, there remains much that we do not understand in this area, and this framework will need to be updated as the field advances.
Most of the data we have presented show how specific intermolecular interactions can control condensate composition. However, nonspecific electrostatic interactions have also been shown to significantly impact composition in biochemical reconstitutions [9,12,48]. The relative importance of specific binding and non-specific electrostatics has not been examined in detail, and may be different in simplified in vitro model systems from the more complex cellular environment [49]. Moreover, quantitative models relating binding affinity between clients and scaffolds to the degrees of client concentration in condensates are lacking.
Current proteomic studies of condensate composition have necessarily probed average behaviors across many individual condensates in large numbers of cells [23,24]. However, these approaches cannot address the possibility that condensates within the same cytoplasm could have heterogeneous compositions that are regulated by their local, subcellular environment. Such non-equilibrium behavior is indeed demonstrated in fungal cells, where perinuclear condensates formed by the Whi3 protein scaffold contain different RNA clients than peripheral polarity condensates formed by the same Whi3 scaffold [18,42]. Similarly, TDP-43 RNA neuronal granules in a single axon exhibited differences in material properties depending on their location relative to the cell body [25]. LAT clusters also have different compositions depending on their position in the IS [136]. It seems likely that other condensates, such as P bodies or stress granules, could exhibit changes in composition depending on subcellular localization or local environmental fluctuations. Careful, quantitative live-cell imaging of condensate composition will be necessary to understand the level of heterogeneity found within cells.
Addressing these issues will require quantitative imaging of in vitro model systems and natural condensates in live cells, additional proteomics analyses and deep mechanistic studies of individual molecules, as well as complementary development of theories to explain both passive and active compositional control. Ultimately, a predictive understanding of condensate composition will provide the foundation for understanding the biochemical and cellular functions of these fascinating cellular compartments.
Highlights.
Composition plays an essential role in the function of biomolecular condensates
Condensate components can be roughly classified as scaffolds or clients
Some aspects of client recruitment by scaffolds are known, but much remains unknown
Composition is increasingly understood through proteomics and mechanistic studies
Acknowledgements:
Work in the Rosen lab is supported by the Howard Hughes Medical Institute. This work was additionally supported by a National Research Service Award from NIDDK (F32 DK101188 to J.A.D.) and a Howard Hughes Medical Institute Collaborative Innovation Award. L.B.C. is a Robert Black Fellow of the Damon Runyon Cancer Research Foundation (DRG-2249-16).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hyman AA, Weber CA, Julicher F, Liquid-liquid phase separation in biology, Annu Rev Cell Dev Biol 30 (2014) 39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- [2].Banani SF, Lee HO, Hyman AA, Rosen MK, Biomolecular condensates: organizers of cellular biochemistry, Nat Rev Mol Cell Biol 18 (2017) 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shin Y, Brangwynne CP, Liquid phase condensation in cell physiology and disease, Science (80-. ). 357 (2017). doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- [4].Alberti S, Phase separation in biology, Curr. Biol 27 (2017) R1097–R1102. doi: 10.1016/j.cub.2017.08.069. [DOI] [PubMed] [Google Scholar]
- [5].Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, V Hollingsworth J, King DS, Banani SF, Russo PS, Jiang QX, Nixon BT, Rosen MK, Phase transitions in the assembly of multivalent signalling proteins, Nature. 483 (2012) 336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fromm SA, Kamenz J, Nöldeke ER, Neu A, Zocher G, Sprangers R, In vitro reconstitution of a cellular phase-transition process that involves the mRNA decapping machinery., Angew. Chem. Int. Ed. Engl 53 (2014) 7354–9. doi: 10.1002/anie.201402885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Banjade S, Rosen MK, Phase transitions of multivalent proteins can promote clustering of membrane receptors, Elife. 3 (2014) e04123. doi: 10.7554/eLife.04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zeng M, Shang Y, Araki Y, Guo T, Huganir RL, Zhang M, Phase Transition in Postsynaptic Densities Underlies Formation of Synaptic Complexes and Synaptic Plasticity, Cell. 166 (2017) 1163–1175.e12. doi: 10.1016/j.cell.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, Vale RD, Phase separation of signaling molecules promotes T cell receptor signal transduction Science, 352 (2016) 595–599. doi: 10.1126/science.aad9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sun D, Wu R, Zheng J, Li P, Yu L, Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation, Cell Res 28 (2018) 405–415. doi: 10.1038/s41422-018-0017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lin Y, Protter DSW, Rosen MK, Parker R, Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins, Mol. Cell 60 (2015) 208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, Baldwin AJ, Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles, Mol Cell. 57 (2015) 936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, Pozniakovski A, Poser I, Maghelli N, Royer LA, Weigert M, Myers EW, Grill S, Drechsel D, Hyman AA, Alberti S, A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation, Cell. 162 (2015) 1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- [14].Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP, Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization, Cell. 163 (2015) 123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Murakami T, Qamar S, Lin JQ, Schierle GSK, Rees E, Miyashita A, Costa AR, Dodd RB, Chan FTS, Michel CH, Kronenberg-Versteeg D, Li Y, Yang S-P, Wakutani Y, Meadows W, Ferry RR, Dong L, Tartaglia GG, Favrin G, Lin W-L, Dickson DW, Zhen M, Ron D, Schmitt-Ulms G, Fraser PE, Shneider NA, Holt C, Vendruscolo M, Kaminski CF, St George-Hyslop P, ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function, Neuron. 88 (2015) 678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, McKnight SL, The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei, Cell. 163 (2015) 829–839. doi: 10.1016/j.cell.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jain A, Vale RD, RNA phase transitions in repeat expansion disorders, Nature, advance on (2017). doi: 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Langdon EM, Qiu Y, Ghanbari Niaki A, McLaughlin GA, Weidmann C, Gerbich TM, Smith JA, Crutchley JM, Termini CM, Weeks KM, Myong S, Gladfelter AS, mRNA structure determines specificity of a polyQ-driven phase separation, Science (80-. ). (2018). doi: 10.1126/science.aar7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Van Damme E, Laukens K, Hai Dang T, Van Ostade X, A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics, J Int Biol Sci 6 (2010) 51–67. doi: 10.7150/ijbs.6.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ahmad Y, Boisvert F-M, Gregor P, Cobley A, Lamond AI, NOPdb: Nucleolar Proteome Database—2008 update, Nucleic Acids Res 37 (2009) D181–D184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R, The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules, Mol. Cell 68 (2017) 808–820.e5. doi: 10.1016/j.molcel.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R, ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure., Cell. 164 (2016) 487–98. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo E-C, Krach F, Yang D, Sen A, Fulzele A, Wozniak JM, Gonzalez DJ, Kankel MW, Gao F-B, Bennett EJ, Lecuyer E, Yeo GW, Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules., Cell. 172 (2018) 590–604.e13. doi: 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Youn J, Dunham WH, Hong SJ, Fabian M, Knight JDR, Bashkurov M, Chen G.l., Bagci H, Rathod B, MacLeod G, Eng SWM, Angers S, Morris Q, Fabian M, Côté J-F, Gingras A-C, High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies, Mol. Cell 69 (2018) 517–532. doi: 10.1016/j.molcel.2017.12.020. [DOI] [PubMed] [Google Scholar]
- [25].Gopal PP, Nirschl JJ, Klinman E, Holzbaur ELF, Amyotrophic lateral sclerosis-linked mutations increase the viscosity of liquid-like TDP-43 RNP granules in neurons, Proc. Natl. Acad. Sci 114 (2017) E2466–E2475.doi: 10.1073/pnas.1614462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Buchan JR, Parker R, Eukaryotic Stress Granules: The Ins and Outs of Translation, Mol. Cell 36 (2009) 932–941.doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Fong K, Li Y, Wang W, Ma W, Li K, Qi RZ, Liu D, Songyang Z, Chen J, Whole-genome screening identifies proteins localized to distinct nuclear bodies, J. Cell Biol 203 (2013) 149 LP–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Weidtkamp-Peters S, Lenser T, Negorev D, Gerstner N, Hofmann TG, Schwanitz G, Hoischen C, Maul G, Dittrich P, Hemmerich P, Dynamics of component exchange at PML nuclear bodies, J. Cell Sci 121 (2008) 2731 LP–2743. doi:doi: 10.1242/jcs.031922. [DOI] [PubMed] [Google Scholar]
- [29].Brangwynne CP, Mitchison TJ, Hyman AA, Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes, Proc. Natl.Acad. Sci 108 (2011) 4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, Hyman AA, The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin, Cell. 169 (2017) 1066–1077.e10.doi: 10.1016/j.cell.2017.05.028. [DOI] [PubMed] [Google Scholar]
- [31].Schwarz-Romond T, Metcalfe C, Bienz M, Dynamic recruitment of axin by Dishevelled protein assemblies, J. Cell Sci 120 (2007) 2402 LP–2412. [DOI] [PubMed] [Google Scholar]
- [32].Dundr M, Misteli T, Functional architecture in the cell nucleus, Biochem. J 356 (2001) 297 LP–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB, An Architectural Role for a Nuclear Noncoding RNA: NEAT1 RNA Is Essential for the Structure of Paraspeckles, Mol. Cell 33 (2009) 717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ETH, Strauss JF, Maul GG, Pml Is Critical for Nd10 Formation and Recruits the Pml-lnteracting Protein Daxx to This Nuclear Structure When Modified by Sumo-1, J. Cell Biol 147 (1999) 221 LP–234.doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Teixeira D, Parker R, Analysis of P-body assembly in Saccharomyces cerevisiae., Mol. Biol. Cell 18 (2007) 2274–87. doi: 10.1091/mbc.E07-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Rao BS, Parker R, Numerous interactions act redundantly to assemble a tunable size of P bodies in Saccharomyces cerevisiae, Proc Natl Acad Sci USA. 114 (2017) E9569–E9578. doi: 10.1073/pnas.1712396114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhong S, Müller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP, Role of SUMO-1-modified PML in nuclear body formation, Blood. 95 (2000) 2748 LP–2752. [PubMed] [Google Scholar]
- [38].de Stanchina E, Querido E, Narita M, V Davuluri R, Pandolfi PP, Ferbeyre G, Lowe SW, PML Is a Direct p53 Target that Modulates p53 Effector Functions, Mol. Cell 13 (2004) 523–535. doi: 10.1016/S1097-2765(04)00062-0. [DOI] [PubMed] [Google Scholar]
- [39].Zhong S, Hu P, Ye T-Z, Stan R, Ellis NA, Pandolfi PP, A role for PML and the nuclear body in genomic stability, Oncogene. 18 (1999) 7941. [DOI] [PubMed] [Google Scholar]
- [40].Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK, Compositional Control of Phase-Separated Cellular Bodies, Cell. 166 (2016) 651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, Grishin NV, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, Eisenberg D, Tycko R, McKnight SL, Cell-free Formation of RNA Granules: Low Complexity Sequence Domains Form Dynamic Fibers within Hydrogels, Cell. 149 (2012) 753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS, RNA Controls PolyQ Protein Phase Transitions, Mol. Cell 60 (2015) 220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Brangwynne CP, Tompa P, Pappu RV, Polymer physics of intracellular phase transitions, Nat. Phys 11 (2015) 899. doi: 10.1038/nphys3532. [DOI] [Google Scholar]
- [44].Vernon RM, Chong PA, Tsang B, Kim TH, Bah A, Farber P, Lin H, Forman-Kay JD, Pi-Pi contacts are an overlooked protein feature relevant to phase separation, Elife. 7 (2018) e31486. doi: 10.7554/eLife.31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL, Phosphorylation-Regulated Binding of RNA Polymerase II to Fibrous Polymers of Low-Complexity Domains, Cell. 155 (2013) 1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Murray DT, Kato M, Lin Y, Thurber KR, Hung I, McKnight SL, Tycko R, Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains, Cell. 171 (2017) 615–627.e16. doi: 10.1016/j.cell.2017.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Goulet I, Boisvenue S, Mokas S, Mazroui R, Cote J, TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules., Hum. Mol. Genet 17 (2008) 3055–74. doi: 10.1093/hmg/ddn203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nott TJ, Craggs TD, Baldwin AJ, Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters, Nat. Chem 8 (2016) 569. doi: 10.1038/nchem.2519. [DOI] [PubMed] [Google Scholar]
- [49].Protter DSW, Rao BS, Van Treeck B, Lin Y, Mizoue L, Rosen MK, Parker R, Intrinsically Disordered Regions Can Contribute Promiscuous Interactions to RNP Granule Assembly, Cell Rep 22 (2018) 1401–1412. doi: 10.1016/j.celrep.2018.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Pak CW, Kosno M, Holehouse AS, Padrick SB, Mittal A, AN R, Yunus AA, Liu DR, Pappu RV, Rosen MK, Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein, Mol. Cell 63 (2016) 72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Decker CJ, Teixeira D, Parker R, Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae, J. Cell Biol 179 (2007) 437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lee C, Occhipinti P, Gladfelter AS, PolyQ-dependent RNA-protein assemblies control symmetry breaking, J. Cell Biol 208 (2015) 533 LP–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lin Y, Currie SL, Rosen MK, Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs, J. Biol. Chem 292 (2017) 19110–19120. doi: 10.1074/jbc.M117.800466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Monahan Z, Ryan VH, Janke AM, Burke KA, Rhoads SN, Zerze GH, O’Meally R, Dignon GL, Conicella AE, Zheng W, Best RB, Cole RN, Mittal J, Shewmaker F, Fawzi NL, Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity., EMBO J. (2017) e201696394. doi: 10.15252/embj.201696394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hofweber M, Hutten S, Bourgeois B, Spreitzer E, Niedner-Boblenz A, Schifferer M, Ruepp M-D, Simons M, Niessing D, Madl T, Dormann D, Phase Separation of FUS Is Suppressed by Its Nuclear Import Receptor and Arginine Methylation, Cell. 173 (2018) 706–719.e13. doi: 10.1016/j.cell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- [56].Huang S-Y, Naik MT, Chang C-F, Fang P-J, Wang Y-H, Shih H-M, Huang T, The B-box 1 dimer of human promyelocytic leukemia protein, J. Biomol. NMR 60 (2014) 275–281. doi: 10.1007/s10858-014-9869-4. [DOI] [PubMed] [Google Scholar]
- [57].Sahin U, Ferhi O, Jeanne M, Benhenda S, Berthier C, Jollivet F, Niwa-Kawakita M, Faklaris O, Setterblad N, de Thé H, Lallemand-Breitenbach V, Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins, J. Cell Biol 204 (2014) 931 LP–945. doi: 10.1083/jcb.201305148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kamitani T, Nguyen HP, Kito K, Fukuda-Kamitani T, Yeh ETH, Covalent Modification of PML by the Sentrin Family of Ubiquitin-like Proteins, J. Biol. Chem 273(1998) 3117–3120. doi: 10.1074/jbc.273.6.3117. [DOI] [PubMed] [Google Scholar]
- [59].Nisole S, Maroui MA, Mascle X, Aubry M, Chelbi-Alix M, Differential Roles of PML Isoforms, Front. Oncol 3 (2013) 125. doi: 10.3389/fonc.2013.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Shen TH, Lin H-K, Scaglioni PP, Yung TM, Pandolfi PP, The Mechanisms of PML-Nuclear Body Formation, Mol. Cell 24 (2006) 331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Li C, Peng Q, Wan X, Sun H, Tang J, C-terminal motifs in promyelocytic leukemia protein isoforms critically regulate PML nuclear body formation, J. Cell Sci 130 (2017) 3496 LP–3506. doi: 10.1242/jcs.202879. [DOI] [PubMed] [Google Scholar]
- [62].Dellaire G, Eskiw CH, Dehghani H, Ching RW, Bazett-Jones DP, Mitotic accumulations of PML protein contribute to the re-establishment of PML nuclear bodies in G1, J. Cell Sci 119 (2006) 1034 LP–1042. doi: 10.1242/jcs.02817. [DOI] [PubMed] [Google Scholar]
- [63].Salsman J, Rapkin LM, Margam NN, Duncan R, Bazett-Jones DP, Dellaire G, Myogenic differentiation triggers PML nuclear body loss and DAXX relocalization to chromocentres, Cell Death Dis 8 (2017) e2724. doi: 10.1038/cddis.2017.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wagenknecht N, Reuter N, Scherer M, Reichel A, Müller R, Stamminger T, Contribution of the Major ND10 Proteins PML, hDaxx and Sp100 to the Regulation of Human Cytomegalovirus Latency and Lytic Replication in the Monocytic Cell Line THP-1, Viruses. 7 (2015). doi: 10.3390/v7062751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Renner F, Moreno R, Schmitz ML, SUMOylation-Dependent Localization of IKKε in PML Nuclear Bodies Is Essential for Protection against DNA-Damage-Triggered Cell Death, Mol. Cell 37 (2010) 503–515.doi: 10.1016/j.molcel.2010.01.018. [DOI] [PubMed] [Google Scholar]
- [66].Chang C-C, Naik MT, Huang Y-S, Jeng J-C, Liao P-H, Kuo H-Y, Ho C-C, Hsieh Y-L, Lin C-H, Huang N-J, Naik NM, Kung CC-H, Lin S-Y, Chen R-H, Chang K-S, Huang T-H, Shih H-M, Structural and Functional Roles of Daxx SIM Phosphorylation in SUMO Paralog-Selective Binding and Apoptosis Modulation, Mol. Cell 42 (2011) 62–74. doi: 10.1016/j.molcel.2011.02.022. [DOI] [PubMed] [Google Scholar]
- [67].Anamika L Spyracopoulos, Molecular Basis for Phosphorylation-dependent SUMO Recognition by the DNA Repair Protein RAP80, J. Biol. Chem 291 (2016) 4417–4428. doi: 10.1074/jbc.M115.705061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Chen Y-CM, Kappel C, Beaudouin J, Eils R, Spector DL, Live Cell Dynamics of Promyelocytic Leukemia Nuclear Bodies upon Entry into and Exit from Mitosis, Mol. Biol. Cell 19 (2008) 3147–3162. doi: 10.1091/mbc.E08-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Condemine W, Takahashi Y, Zhu J, Puvion-Dutilleul F, Guegan S, Janin A, de Thé H, Characterization of Endogenous Human Promyelocytic Leukemia Isoforms, Cancer Res 66 (2006) 6192 LP–6198. doi: 10.1158/0008-5472.CAN-05-3792. [DOI] [PubMed] [Google Scholar]
- [70].Lallemand-Breitenbach V, de Thé H, PML nuclear bodies: from architecture to function, Curr. Opin. Cell Biol 52 (2018) 154–161. doi: 10.1016/j.ceb.2018.03.011. [DOI] [PubMed] [Google Scholar]
- [71].Liang Y-C, Lee C-C, Yao Y-L, Lai C-C, Schmitz ML, Yang W-M, SUM05, a Novel Poly-SUMO Isoform, Regulates PML Nuclear Bodies, Sci. Rep 6 (2016) 26509. doi: 10.1038/srep26509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Gao C, Ho C-C, Reineke E, Lam M, Cheng X, Stanya KJ, Liu Y, Chakraborty S, Shih H-M, Kao H-Y, Histone Deacetylase 7 Promotes PML Sumoylation and Is Essential for PML Nuclear Body Formation, Mol. Cell. Biol 28 (2008) 5658–5667. doi: 10.1128/MCB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Cheng X, Kao H-Y, Post-translational modifications of PML: consequences and implications, Front. Oncol 2 (2013) 210. doi: 10.3389/fonc.2013.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Luo Y, Na Z, Slavoff SA, P-Bodies: Composition, Properties, and Functions, Biochemistry. 57 (2018) 2424–2431. doi: 10.1021/acs.biochem.7b01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Aizer A, Kalo A, Kafri P, Shraga A, Ben-Yishay R, Jacob A, Kinor N, Shav-Tal Y, Quantifying mRNA targeting to P-bodies in living human cells reveals their dual role in mRNA decay and storage., J. Cell Sci 127 (2014) 4443–56. doi: 10.1242/jcs.152975. [DOI] [PubMed] [Google Scholar]
- [76].Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R, Processing bodies require RNA for assembly and contain nontranslating mRNAs, RNA. 11 (2005) 371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P, A functional RNAi screen links O-GIcNAc modification of ribosomal proteins to stress granule and processing body assembly., Nat. Cell Biol 10 (2008) 1224–31. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Schütz S, Nöldeke ER, Sprangers R, A synergistic network of interactions promotes the formation of in vitro processing bodies and protects mRNA against decapping, Nucleic Acids Res 45 (2017) 6911–6922. doi: 10.1093/nar/gkx353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hubstenberger A, Courel M, Bénard M, Souquere S, Ernoult-Lange M, Chouaib R, Yi Z, Morlot J-B, Munier A, Fradet M, Daunesse M, Bertrand E, Pierron G, Mozziconacci J, Kress M, Weil D, P-Body Purification Reveals the Condensation of Repressed mRNA Regulons., Mol. Cell 68 (2017) 144–157.e5. doi: 10.1016/j.molcel.2017.09.003. [DOI] [PubMed] [Google Scholar]
- [80].Aizer A, Kafri P, Kalo A, Shav-Tal Y, The P Body Protein Dcpla Is Hyper-phosphorylated during Mitosis, PLoS One. 8 (2013) e49783.doi: 10.1371/journal.pone.0049783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Protter DSW, Parker R, Principles and Properties of Stress Granules, Trends Cell Biol 26 (2016) 668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Mateju D, Franzmann TM, Patel A, Kopach A, Boczek EE, Maharana S, Lee HO, Carra S, Hyman AA, Alberti S, An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function, EMBO J. 36 (2017) 1669 LP–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kroschwald S, Munder MC, Maharana S, Franzmann TM, Richter D, Ruer M, Hyman AA, Alberti S, Different Material States of Pub1 Condensates Define Distinct Modes of Stress Adaptation and Recovery, Cell Rep 23 (2018) 3327–3339. doi: 10.1016/j.celrep.2018.05.041. [DOI] [PubMed] [Google Scholar]
- [84].Riback JA, Katanski CD, Kear-Scott JL, V Pilipenko E, Rojek AE, Sosnick TR, Drummond DA, Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response., Cell. 168 (2017) 1028–1040.e19.doi: 10.1016/j.cell.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Kroschwald S, Maharana S, Mateju D, Malinovska L, Nuske E, Poser I, Richter D, Alberti S, Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules, Elite. 4 (2015) e06807. doi: 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Munder MC, Midtvedt D, Franzmann T, Nuske E, Otto O, Herbig M, Ulbricht E, Muller P, Taubenberger A, Maharana S, Malinovska L, Richter D, Guck J, Zaburdaev V, Alberti S, A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy, Elite. 5 (2016) e09347. doi: 10.7554/eLife.09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P, Wickens MP, Evidence That Ternary Complex (elF2-GTP-tRNAi Met)-Deficient Preinitiation Complexes Are Core Constituents of Mammalian Stress Granules, Mol. Biol. Cell 13 (2002) 195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Dang Y, Kedersha N, Low W-K, Romo D, Gorospe M, Kaufman R, Anderson P, Liu JO, Eukaryotic Initiation Factor 2α-independent Pathway of Stress Granule Induction by the Natural Product Pateamine A, J. Biol. Chem 281 (2006) 32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- [89].Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P, Stress granules and processing bodies are dynamically linked sites of mRNP remodeling, J. Cell Biol 169 (2005) 871 LP–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J, The RasGAP-associated endoribonuclease G3BP assembles stress granules., J. Cell Biol 160 (2003) 823–31. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [91].White JP, Cardenas AM, Marissen WE, Lloyd RE, Inhibition of Cytoplasmic mRNA Stress Granule Formation by a Viral Proteinase, Cell Host Microbe 2 (2007) 295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- [92].Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, Mclnerney GM, Ivanov P, Anderson P, G3BP-Caprin1- USP10 complexes mediate stress granule condensation and associate with 40S subunits., J. Cell Biol 212 (2016) 845–60. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Cherkasov V, Grousl T, Theer P, Vainshtein Y, Glasser C, Mongis C, Kramer G, Stoecklin G, Knop M, Mogk A, Bukau B, Systemic control of protein synthesis through sequestration of translation and ribosome biogenesis factors during severe heat stress., FEBS Lett 589 (2015) 3654–64.doi: 10.1016/j.febslet.2015.10.010. [DOI] [PubMed] [Google Scholar]
- [94].Andersson MK, Stahlberg A, Arvidsson Y, Olofsson A, Semb H, Stenman G, Nilsson O, Aman P, The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response, BMC Cell Biol 9 (2008) 37. doi: 10.1186/1471-2121-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Wallace EWJ, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, Airoldi EM, Pan T, Budnik BA, Drummond DA, Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress., Cell. 162 (2015) 1286–98. doi: 10.1016/j.cell.2015.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Yang X, Shen Y, Garre E, Hao X, Krumlinde D, Cvijović M, Arens C, Nyström T, Liu B, Sunnerhagen P, Stress granule-defective mutants deregulate stress responsive transcripts., PLoS Genet 10 (2014) e1004763.doi: 10.1371/journal.pgen.1004763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Buchan JR, Yoon J-H, Parker R, Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae, J. Cell Sci 124 (2011) 228 LP–239. doi: 10.1242/jcs.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Jayabalan AK, Sanchez A, Park RY, Yoon SP, Kang G-Y, Baek J-H, Anderson P, Kee Y, Ohn T, NEDDylation promotes stress granule assembly, Nat. Commun 7 (2016) 12125. doi: 10.1038/ncomms12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Leung AKL, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P, Poly(ADP-Ribose) Regulates Stress Responses and MicroRNA Activity in the Cytoplasm, Mol. Cell 42 (2011) 489–99. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Jongjitwimol J, Baldock RA, Morley SJ, Watts FZ, Sumoylation of elF4A2 affects stress granule formation, J. Cell Sci 129 (2016) 2407–LP-2415. doi: 10.1242/jcs.184614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Aulas A, Fay MM, Lyons SM, Achorn CA, Kedersha N, Anderson P, Ivanov P, Stress-specific differences in assembly and composition of stress granules and related foci, J. Cell Sci 130 (2017) 927 LP–937. doi: 10.1242/jcs.199240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Boisvert F-M, van Koningsbruggen S, Navascués J, Lamond AI, The multifunctional nucleolus, Nat. Rev. Mol. Cell Biol 8 (2007) 574. [DOI] [PubMed] [Google Scholar]
- [103].Albert B, Léger-Silvestre I, Normand C, Ostermaier MK, Pérez-Fernández J, Panov K.l., Zomerdijk JCBM, Schultz P, Gadal O, RNA polymerase l–specific subunits promote polymerase clustering to enhance the rRNA gene transcription cycle, J. Cell Biol 192 (2011) 277 LP–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Smirnov E, Borkovec J, Kováčik L, Svidenská S, Schröfel A, Skalníková M, Švindrych Z, Křížek P, Ovesný M, Hagen GM, Juda P, Michalová K, Cardoso MC, Cmarko D, Raška I, Separation of replication and transcription domains in nucleoli, J. Struct. Biol 188 (2014) 259–266. doi: 10.1016/j.jsb.2014.10.001. [DOI] [PubMed] [Google Scholar]
- [105].Wang F, Ying C, Shang G, Jiao M, Hongfang Z, The new evidence of nucleolar ultrastructural dynamic change: Fibrillar centre (FC) fusion in G1 phase and regeneration in S phase, Micron. 49 (2013) 15–20.doi: 10.1016/j.micron.2013.02.006. [DOI] [PubMed] [Google Scholar]
- [106].Weber SC, Brangwynne CP, Inverse Size Scaling of the Nucleolus by a Concentration-Dependent Phase Transition, Curr. Biol 25 (2015) 641–646. doi: 10.1016/j.cub.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, V Pappu R, Brangwynne CP, Coexisting Liquid Phases Underlie Nucleolar Subcompartments, Cell. 165 (2016) 1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Trimbur GM, Walsh CJ, Nucleolus-like morphology produced during the in vitro reassociation of nucleolar components, J. Cell Biol 122 (1993) 753 LP–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, Nourse A, Deniz AA, Kriwacki RW, Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA, Elife. 5 (2016) e13571. doi: 10.7554/eLife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, V Zamudio A, Manteiga JC, Li CH, Guo YE, Day DS, Schuijers J, Vasile E, Malik S, Hnisz D, Lee T.l., Cisse II, Roeder RG, SharpS PA, Chakraborty AK, Young RA, Coactivator condensation at super-enhancers links phase separation and gene control, Science (80-. ). (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Cho W-K, Spille J-H, Hecht M, Lee C, Li C, Grube V, 1.1. Cisse, Mediator and RNA polymerase II clusters associate in transcription-dependent condensates, Science (80-. ). (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Caudron-Herger M, Pankert T, Seiler J, Németh A, Voit R, Grummt I, Rippe K, Alu element-containing RNAs maintain nucleolar structure and function, EMBO J. 34 (2015) 2758 LP–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Yuya H, Emilie L, Toshiyuki O, Masahiro K, Yuzo W, Tsuneyoshi H, Kunio T, Nucleolar scaffold protein, WDR46, determines the granular compartmental localization of nucleolin and DDX21, Genes to Cells 18 (2013) 780–797. doi:doi: 10.1111/gtc.12077. [DOI] [PubMed] [Google Scholar]
- [114].Holmberg Olausson K, Nistér M, Lindström MS, Loss of Nucleolar Histone Chaperone NPM1 Triggers Rearrangement of Heterochromatin and Synergizes with a Deficiency in DNA Methyltransferase DNMT3A to Drive Ribosomal DNA Transcription, J. Biol. Chem 289 (2014) 34601–34619. doi: 10.1074/jbc.M114.569244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Duan-Porter WD, Woods VL Jr., Maurer KD, Li S, Rosen A, Dynamic Conformations of Nucleophosmin (NPM1) at a Key Monomer-Monomer Interface Affect Oligomer Stability and Interactions with Granzyme B, PLoS One. 9 (2014) e115062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Bertwistle D, Sugimoto M, Sherr CJ, Physical and Functional Interactions of the Arf Tumor Suppressor Protein with Nucleophosmin/B23, Mol. Cell. Biol . 24 (2004) 985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Lindström MS, Elucidation of Motifs in Ribosomal Protein S9 That Mediate Its Nucleolar Localization and Binding to NPM1/Nucleophosmin, PLoS One. 7 (2012) e52476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Mitrea DM, Grace CR, Buljan M, Yun M-K, Pytel NJ, Satumba J, Nourse A, Park C-G, Madan Babu M, White SW, Kriwacki RW, Structural polymorphism in the N-terminal oligomerization domain of NPM1, Proc. Natl. Acad. Sci 111 (2014) 4466 LP–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Mitrea DM, Cika JA, Stanley CB, Nourse A, Onuchic PL, Banerjee PR, Phillips AH, Park C-G, Deniz AA, Kriwacki RW, Self-interaction of NPM1 modulates multiple mechanisms of liquid-liquid phase separation, Nat. Commun 9 (2018) 842. doi: 10.1038/s41467-018-03255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Andersen JS, Lam YW, Leung AKL, Ong S-E, Lyon CE, Lamond AI, Mann M, Nucleolar proteome dynamics, Nature. 433 (2005) 77. [DOI] [PubMed] [Google Scholar]
- [121].de los Santos-Velazquez AI, de Oya IG, Manzano-Lopez J, Monje-Casas F, Late rDNA Condensation Ensures Timely Cdc14 Release and Coordination of Mitotic Exit Signaling with Nucleolar Segregation, Curr. Biol 27 (2017) 3248–3263.e5. doi: 10.1016/j.cub.2017.09.028. [DOI] [PubMed] [Google Scholar]
- [122].Yuan F, Xu C, Li G, Tong T, Nucleolar TRF2 attenuated nucleolus stress-induced HCC cell-cycle arrest by altering rRNA synthesis, Cell Death Dis 9 (2018) 518. doi: 10.1038/s41419-018-0572-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Lindström MS, Jurada D, Bursae S, Orsolic I, Bartek J, Volarevic S, Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis, Oncogene. 37 (2018) 2351–2366. doi: 10.1038/s41388-017-0121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Boulon S, Westman BJ, Hutten S, Boisvert F-M, Lamond AI, The Nucleolus under Stress, Mol. Cell 40 (2010) 216–227.doi: https://doi.Org/10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Chen J, Lobb IT, Morin P, Novo SM, Simpson J, Kennerknecht K, von Kriegsheim A, Batchelor EE, Oakley F, Stark LA, Identification of a novel TIF-IA-NF-κΒ nucleolar stress response pathway, Nucleic Acids Res 46 (2018) 6188–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Jarboui MA, Bidoia C, Woods E, Roe B, Wynne K, Elia G, Hall WW, Gautier VW, Nucleolar Protein Trafficking in Response to HIV-1 Tat: Rewiring the Nucleolus, PLoS One. 7 (2012) e48702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Salvetti A, Greco A, Viruses and the nucleolus: The fatal attraction, Biochim. Biophys. Acta - Mol. Basis Dis 1842 (2014) 840–847. doi: 10.1016/j.bbadis.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Hiroshima M, Pack C, Kaizu K, Takahashi K, Ueda M, Sako Y, Transient Acceleration of Epidermal Growth Factor Receptor Dynamics Produces Higher-Order Signaling Clusters, J. Mol. Biol 430 (2018) 1386–1401.doi: 10.1016/j.jmb.2018.02.018. [DOI] [PubMed] [Google Scholar]
- [129].Rishita C, Michael S, Integrin and cadherin clusters: A robust way to organize adhesions for cell mechanics, BioEssays. 39 (2016) e201600123.doi: 10.1002/bies.201600123. [DOI] [PubMed] [Google Scholar]
- [130].Hui E, Cheung J, Zhu J, Su X, Taylor MJ, Wallweber HA, Sasmal DK, Huang J, Kim JM, Mellman I, Vale RD, T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition, Science (80-. ). 355 (2017) 1428 LP–1433. doi: 10.1126/science.aaf1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Patrakka J, Tryggvason K, Nephrin-a unique structural and signaling protein of the kidney filter, Trends Mol Med 13 (2007) 396–403. doi: 10.1016/j.molmed.2007.06.006. [DOI] [PubMed] [Google Scholar]
- [132].Zhang W, Sloan-Lancaster J, Kitchen J, Trible RP, Samelson LE, l_AT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation, Cell. 92 (1998) 83–92. doi: 10.1016/S0092-8674(00)80901-0. [DOI] [PubMed] [Google Scholar]
- [133].Verma R, Kovari I, Soofi A, Nihalani D, Patrie K, Holzman LB, Nephrin ectodomain engagement results in Src kinase activation, nephrin phosphorylation, Nek recruitment, and actin polymerization, J Clin Invest 116 (2006) 1346–1359. doi: 10.1172/JCI27414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Paz PE, Wang S, Clarke H, Lu X, Stokoe D, Abo A, Mapping the Zap-70 phosphorylation sites on LAT (linker for activation of T cells) required for recruitment and activation of signalling proteins in T cells, Biochem J. 356 (2001) 461–471. doi: 10.1042/bj3560461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].McNaughton BR, Cronican JJ, Thompson DB, Liu DR, Mammalian cell penetration, siRNA transfection, and DNA transfection by supercharged proteins, Proc. Natl. Acad. Sci 106 (2009) 6111 LP–6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Ditlev JA, Vega AR, V Köster D, Su X, Lakoduk A, Vale RD, Mayor S, Jaqaman K, Rosen MK, A Composition-Dependent Molecular Clutch Between T Cell Signaling Clusters and Actin, BioRxiv. (2018). doi: 10.1101/316414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Mullins RD, Heuser JA, Pollard TD, The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments, Proc Natl Acad Sci U S A. 95 (1998) 6181–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Huang WYC, Yan Q, Lin W-C, Chung JK, Hansen SD, Christensen SM, Tu H-L, Kuriyan J, Groves JT, Phosphotyrosine-mediated LAT assembly on membranes drives kinetic bifurcation in recruitment dynamics of the Ras activator SOS, Proc. Natl. Acad. Sci 113 (2016) 8218–8223. doi: 10.1073/pnas.1602602113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Shen S, Chuck M.l., Zhu M, Fuller DM, Yang CW, Zhang W, The importance of LAT in the activation, homeostasis, and regulatory function of T cells, J Biol Chem 285 (2010) 35393–35405. doi: 10.1074/jbc.M110.145052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Balagopalan L, Barr VA, Kortum RL, Park AK, Samelson LE, Cutting edge: cell surface linker for activation of T cells is recruited to microclusters and is active in signaling, J Immunol 190 (2013) 3849–3853. doi: 10.4049/jimmunol.1202760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Yi J, Wu XS, Crites T, Hammer JA 3rd, Actin retrograde flow and actomyosin II arc contraction drive receptor cluster dynamics at the immunological synapse in Jurkat T cells, Mol Biol Cell. 23 (2012) 834–852. doi: 10.1091/mbc.E11-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Murugesan S, Hong J, Yi J, Li D, Beach JR, Shao L, Meinhardt J, Madison G, Wu X, Betzig E, Hammer JA, Formin-generated actomyosin arcs propel T cell receptor microcluster movement at the immune synapse, J. Cell Biol 215 (2016) 383–399. doi: 10.1083/jcb.201603080. [DOI] [PMC free article] [PubMed] [Google Scholar]