Abstract
Variant interpretation depends on accurate annotations using biologically relevant transcripts. We have developed a systematic strategy for designating primary transcripts and have applied it to 109 hearing loss–associated genes that were divided into three categories. Category 1 genes (n = 38) had a single transcript; category 2 genes (n = 33) had multiple transcripts, but a single transcript was sufficient to represent all exons; and category 3 genes (n = 38) had multiple transcripts with unique exons. Transcripts were curated with respect to gene expression reported in the literature and the Genotype-Tissue Expression Project. In addition, high-frequency loss-of-function variants in the Genome Aggregation Database and disease-causing variants in ClinVar and the Human Gene Mutation Database across the 109 genes were queried. These data were used to classify exons as clinically significant, insignificant, or of uncertain significance. Interestingly, 6% of all exons, containing 124 reportedly disease-causing variants, were of uncertain significance. Finally, we used exon-level next-generation sequencing quality metrics generated at two clinical laboratories and identified a total of 43 technically challenging exons in 20 different genes that had inadequate coverage and/or homology issues that might lead to false-variant calls. We have demonstrated that transcript analysis plays a critical role in accurate clinical variant interpretation.
CME Accreditation Statement: This activity (“JMD 2018 CME Program in Molecular Diagnostics”) has been planned and implemented in accordance with the accreditation requirements and policies of the Accreditation Council for Continuing Medical Education (ACCME) through the joint providership of the American Society for Clinical Pathology (ASCP) and the American Society for Investigative Pathology (ASIP). ASCP is accredited by the ACCME to provide continuing medical education for physicians.
The ASCP designates this journal-based CME activity (“JMD 2018 CME Program in Molecular Diagnostics”) for a maximum of 18.0 AMA PRA Category 1 Credit(s)™. Physicians should claim only credit commensurate with the extent of their participation in the activity.
CME Disclosures: The authors of this article and the planning committee members and staff have no relevant financial relationships with commercial interests to disclose.
With the rapid growth of genomic testing and the decreasing cost of sequencing, proper analysis of genetic variants is becoming increasingly critical for patient care. The American College of Medical Genetics and Genomics and the Association for Molecular Pathology have set forth guidelines for the interpretation of sequence variants.1 However, use of the guidelines requires an understanding of the transcriptional architecture of each gene. There can be several mRNA transcripts for each gene, and, currently, each laboratory individually determines which transcript to use when annotating, interpreting, and reporting variants in any gene. Human transcripts are being designated and annotated by multiple groups. The most commonly used sets of coding transcripts are currently available from GENCODE [https://www.gencodegenes.org; mainly Ensembl and HAVANA (https://www.ensembl.org), the Consensus CDS (CCDS; https://www.ncbi.nlm.nih.gov/projects/CCDS/CcdsBrowse.cgi), and the Locus Reference Genomic (LRG; https://www.lrg-sequence.org) databases], University of California, Santa Cruz, reference sequence (https://genome.ucsc.edu; mainly the NCBI Reference Sequence Database, RefSeq, https://www.ncbi.nlm.nih.gov/refseq, and LRG), RefSeqGene, a subset of RefSeq defining reference sequences for well-characterized genes (https://www.ncbi.nlm.nih.gov/refseq/rsg), and the NCBI AceView genes (https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly).2, 3, 4, 5, 6, 7, 8 Each group annotates transcripts with a combination of computational and manual literature curation. Although current Human Genome Variation Society standards mention that describing variants in the context of exons and introns is optional,9 in many genes there are exon-specific factors that influence interpretation, and tracking these data on an exon level is important.
In addition to the above annotation challenges, technical limitations of next-generation sequencing (NGS) can also lead to inaccurate variant calls. Several genes contain coding sequences that can pose several technical problems, including sequences with high homology to other genomic regions, sequences with high GC content, and repetitive sequences. If a gene has significant sequence overlap with another gene or a pseudogene, it can be difficult to align the short NGS reads to the right genomic location, leading to false-negative and/or false-positive variant calls. DNA with high GC content is not easily amplified, and highly repetitive DNA is prone to sequencing and/or alignment errors. All such regions should be systematically investigated in the targeted genes of interest to address test limitations and design necessary ancillary assays.10
On passing sequencing quality metrics and filtration cutoffs, a rare variant would then be evaluated on the basis of the most biologically relevant transcript for the disease of interest. It is common to choose the longest transcript for sequencing pipelines. However, variants are often evaluated in the context of this transcript, which does not necessarily encompass all essential exons and can also contain nonbiologically functional exons. Thus, choosing a medically relevant transcript is essential for variant interpretation and for understanding the molecular consequence of a variant on the gene's function.
Herein, we provide a framework for transcript curation and selection using a combination of tissue expression and genomic data sets, protein functional domains, and published work from animal and human studies. We apply this framework to hearing loss, a relatively common condition that affects 1 in 300 infants, half of whom have a genetic cause.11 Because of the complexity of the auditory system, it is also highly heterogeneous, with >100 genes causative for nonsyndromic hearing loss alone.12 We also use clinical NGS data sets generated at two different diagnostic laboratories to systematically highlight technically challenging regions across the hearing loss genes. We demonstrate the utility of our framework and its impact on variant annotation and interpretation. Although our analysis was limited to hearing loss, we recommend that this guidance be used for all genes that are definitively associated with fully penetrant diseases.
Materials and Methods
Transcript Curation Process
A total of 109 hearing loss–associated genes, largely from the OtoGenome Test (GTR000509148.8) at the Laboratory for Molecular Medicine (LMM), were included for transcript curation. All known (NM) RefSeq transcripts in these genes were curated with respect to function, tissue specificity, and temporal expression from published literature (Figure 1). Exon-specific expression data were extracted from the Genotype-Tissue Expression Project (GTEx, http://www.gtexportal.org, last accessed January 15, 2018). GTEx was supported by the Common Fund of the Office of the Director of the NIH, and by the National Cancer Institute, the National Human Genome Research Institute, the National Heart, Lung, and Blood Institute, the National Institute on Drug Abuse, the National Institute of Mental Health, and the National Institute of Neurological Disorders and Stroke. In addition, high allele frequency (>0.3%) predicted loss-of-function (LoF) variants (nonsense, frameshift, and ±1, 2 splice site), were queried from the Genome Aggregation Database (gnomAD, http://gnomad.broadinstitute.org, last accessed January 15, 2018),13 and exons containing such LoF variants were flagged (Figure 1). An allele frequency of 0.3% was chosen because variants in hearing loss genes that are above this frequency can be considered likely benign, as defined by Duzkale et al.14
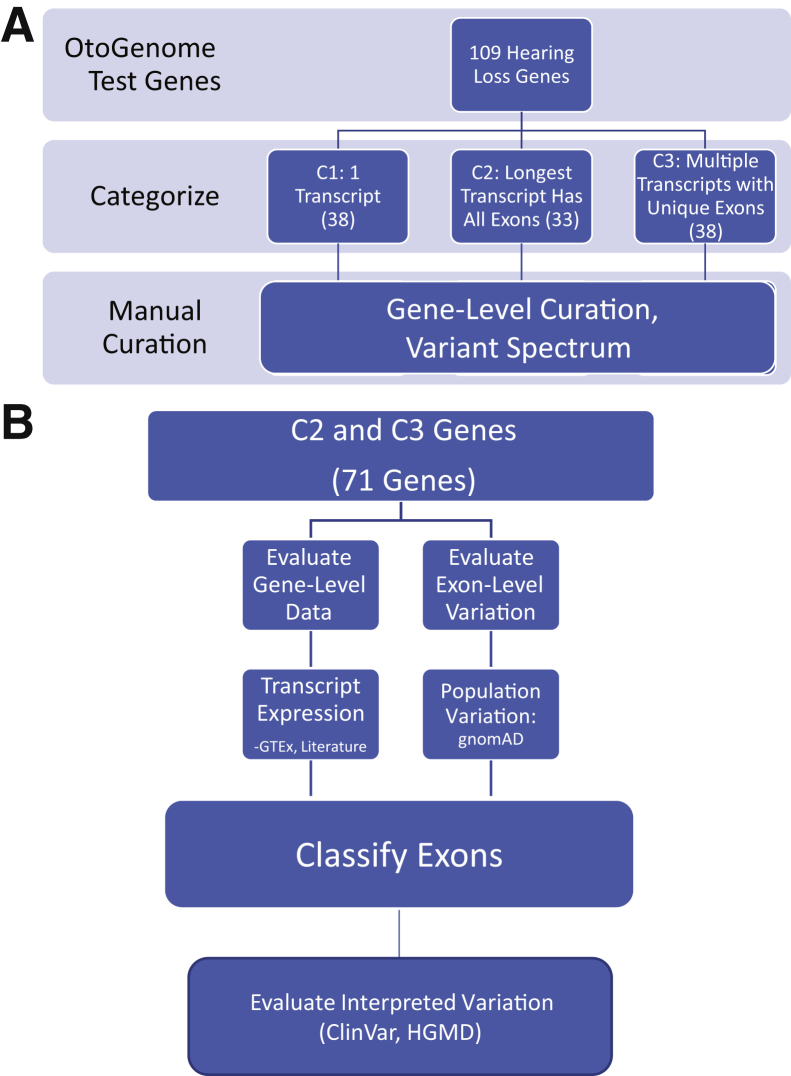
Figure 1.
A: Transcript curation workflow: 109 hearing loss–associated genes, predominantly from the OtoGenome test (GTR000509148.7), were categorized. Genes were divided into three categories using National Center for Biotechnology Information (NCBI) reference sequence transcripts. Category 1 (C1) contained genes that had a single transcript; category 2 (C2) genes had multiple transcripts, but the longest transcript encompassed all exons; and category 3 (C3) genes had multiple transcripts with unique exons. B: Category 2 and 3 curation process: category 2 and 3 genes were manually curated. Exon-specific expression data were obtained from the Genotype-Tissue Expression Project (GTEx). Literature searches were performed for information about functional domains; additional expression data, such as tissue-specific transcript expression; and temporal expression. To evaluate population variation, loss-of-function variants were obtained from the Genome Aggregation Database (gnomAD). To evaluate interpreted variation, likely pathogenic/pathogenic variants were retrieved from our internal database (also in ClinVar), and ClinVar and disease-causing mutation variants were obtained from the Human Gene Mutation Database (HGMD).
Exon Numbering and Classification
All coding and noncoding exons in the primary transcript and minor transcripts were numbered sequentially, consistent with RefSeqGene annotations. Transcripts were aligned and viewed using Alamut version 2.6.1 (Interactive Biosoftware, Rouen, France). To distinguish primary and secondary transcripts, each secondary transcript was given a different letter that was added after the exon number. For example, if there were two transcripts with unique alternate exon 1, they were numbered as 1A and 1B. To define a minimal curated transcript list, unique exons were listed in the minimal number of secondary transcripts, and the designated longest transcript contained the most coding bases (Table 1 and Supplemental Tables S1, S2, and S3).15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80
Table 1.
Curated Transcripts for Category 1, 2, and 3 Genes
Transcript numbers were taken from RefSeqGene: https://www.ncbi.nlm.nih.gov/refseq/rsg. The minimal curated transcript set, with all unique exons for categories 1, 2, and 3, is listed. Category 2 genes, in which the curated transcript is not the longest one, are in bold.
Longest transcript.
Primary transcript.
Exons were classified as a clinically significant exon, an exon of uncertain significance, or a clinically insignificant exon (Table 2). Exons were considered clinically significant if there was no evidence they were alternatively spliced, they did not contain high-frequency exonic LoF variants, or they were supported by tissue-specific inner ear expression in the literature. Exons of uncertain significance were spliced out of major transcripts, had no expression data, and, for some, contained one high-frequency LoF variant. Finally, clinically insignificant exons were noncoding, had nonsupporting human or animal tissue expression data, or had multiple high-frequency LoF variants.
Table 2.
Exons Were Categorized as Clinically Significant, Uncertain Significance, Clinically Insignificant, or Noncoding on the Basis of the Pieces of Evidence Listed
| Exon designation (in genes with a clinical validity of at least moderate) | Evidence used for designation |
|---|---|
| Clinically significant exon | • Evidence that exon is required for biological function (eg, literature, absence of high frequency LoF variant in general population) |
| AND/OR | |
| • No alternative splicing | |
| Exon of uncertain significance | • No expression data in the literature |
| AND | |
| • Alternative splicing | |
| AND/OR | |
| • 1 High-frequency LoF variant in gnomAD (>0.3%) | |
| Clinically insignificant exon | • Literature confirms exon is not expressed in tissue of clinical relevance |
| AND/OR | |
| • >1 High-quality, high-frequency LoF variant in gnomAD (>0.3%) | |
| Noncoding exon | • Exon does not code for protein |
gnomAD, Genome Aggregation Database; LoF, loss of function.
Variant Counts
Pathogenic, likely pathogenic, benign, and likely benign variants and variants of uncertain clinical significance in the ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar, last accessed January 15, 2018),81 in addition to disease-causing mutations in the Human Gene Mutation Database Professional version 2018.1 (Qiagen, Redwood City, CA),82 were counted across all uncertain and insignificant exons (Figure 1). Each variant was evaluated on the basis of transcript location and predicted molecular consequence to the gene.
Technically Challenging Regions
Exon-level NGS quality metrics, including average mapping quality and average minimum depth of coverage, were calculated across all 109 genes using exome sequencing data. Exome targets were captured using the Agilent Clinical Research Exome V5 kit (Agilent, Santa Clara, CA), after which 2 × 100 bp paired-end sequencing was performed on the Illumina HiSeq platform (Illumina, San Diego, CA).
Exome sequencing using the above conditions was performed at the Children's Hospital of Philadelphia83 and LMM with an overall average coverage of approximately 100× and 180×, respectively. Poor-quality regions, as defined in Results, were compared between the two sites. In addition, 87 of the 109 genes were targeted at the LMM (in the previous version of the hearing loss panel12) using a custom-based capture kit (Agilent), followed by 2 × 150 bp paired-end sequencing to an average coverage of approximately 600×. Exon-level NGS quality metrics from this capture-based sequencing approach were calculated and compared with those from exome sequencing.
Results
Transcript Curation
A total of 109 genes on LMM's hearing loss panel were curated for clinically relevant transcripts, as outlined in Figure 1 and Table 2. These genes had between 1 and 17 National Center for Biotechnology Information reference sequence RefSeq transcripts and between 1 and 72 unique exons, for a total of 340 unique transcripts and 2146 unique exons across all genes (Figure 2A). Genes were divided into three categories using RefSeq transcripts (https://www.ncbi.nlm.nih.gov/refseq/rsg, last accessed January 15, 2018).5 All genes with only one RefSeq transcript were classified as category 1 genes. Genes with multiple transcripts were considered category 2 genes if the longest transcript included all annotated exons, and those with multiple transcripts and mutually exclusive exons were grouped under category 3 (C3) (Figure 1).
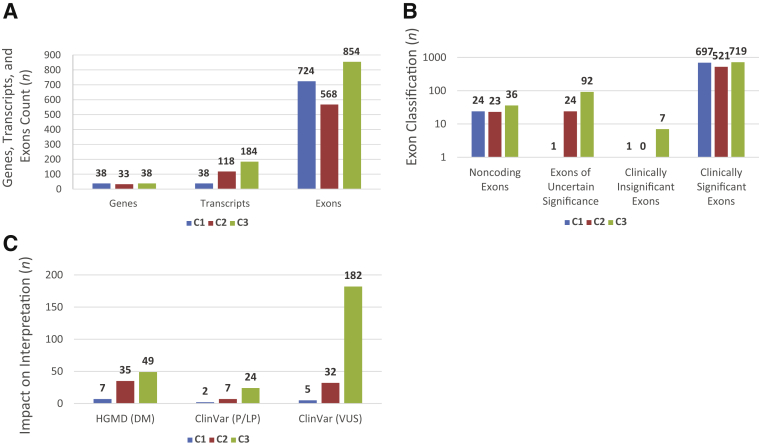
Figure 2.
A: Gene, exon, and transcript counts: genes were categorized [category 1 (C1), category 2 (C2), and category 3 (C3)], and transcripts and exons were counted. B: Exon counts across categories: for each of the three categories, exons were classified as Clinically significant, uncertain significance, clinically insignificant, or noncoding, as per the definitions in Figure 1C. C: Impact on interpretation: variant counts in uncertain and insignificant exons for each category were collected from the Human Gene Mutation Database (HGMD) and ClinVar. DM, disease-causing mutation; LP, likely pathogenic; P, pathogenic; VUS, variant of uncertain significance.
Category 1 Genes
Of the 109 genes evaluated, 38 had a single RefSeq transcript, each with an average of 19 exons (total of 38 transcripts and 724 exons across 38 genes) (Figure 2A and Supplemental Table S1). Because each gene had only one known RefSeq transcript, category 1 transcripts had minimal curation, and almost all exons, except for noncoding ones (n = 24), were critical for inclusion in diagnostic testing and variant interpretation. Interestingly, however, on the basis of exon-level counts of high allele frequency LoF variants in the general population (Materials and Methods), there was one insignificant exon (MYO15A, NM_016239.3, exon 26) and another of uncertain significance (ATP6V1B1, NM_001692.3, exon 1) in this category.
The MYO15A exon 26 contained a nonsense variant (c.5925G>A; p.Trp1975*) that was present in 1.3% (316/23,712) South Asian alleles, including three homozygotes in the gnomAD. Interestingly, RNA sequencing data from the GTEx study showed that this exon is not expressed across all tested tissues. This exon was, therefore, considered to be clinically insignificant, and the variants identified therein were likely to be benign. The ATP6V1B1 exon 1 contained a start loss variant (c.2T>C; p.M1?) that was present in 40% of total alleles in gnomAD, including 23,280 homozygotes. It is possible that the exon start is erroneously annotated or that reinitiation might occur elsewhere, including at any of the two downstream methionines in this exon. However, there are currently no functional data to support either possibility or to rule out potential reinitiation at downstream exons. This exon was, therefore, classified as uncertain clinical significance, wherein sequence variants should be carefully interpreted.
Category 2 Genes
This category included 33 genes, each with an average of four transcripts and 18 unique exons (Figure 2A and Supplemental Table S2). A total of 118 transcripts, including 568 unique exons, were curated in this category. Although the longest transcript represented all annotated RefSeq exons and was presumed to be the major transcript for category 2 genes, the shorter (minor) transcripts were curated for potentially identifying nonbiologically or nonclinically relevant exons in the longer transcript. Apart from the 23 noncoding exons in these genes, there were 24 coding exons not contained in minor transcripts, thus questioning their clinical relevance (Figure 2B). Of those exons, seven were not expressed in any tissue in the GTEx database, including four exons (CCDC50 exon 6, DFNA5 exon 2, EDN3 exon 4, and ILDR1 exon 6) harboring high allele frequency LoF variants in gnomAD (Supplemental Table S2). Of the 17 exons that showed expression in the GTEx database, 5 (CEP78 exons 1, 2, and 16, DFNA5 exon 6, and KARS exon 15) also contained high-frequency LoF variants in gnomAD (Supplemental Table S2), whereas the remaining 12 did not, although more information is needed to clarify their biological or clinical relevance.
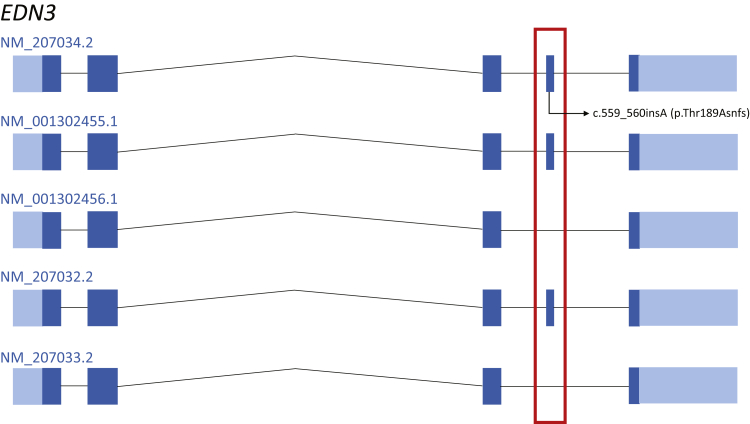
An illustrative example in this category is the EDN3 gene known to cause Waardenburg syndrome (WS), type 4.84, 85 This gene has five RefSeq transcripts sharing coding exons 1 to 3 and 5 but differing in the inclusion of coding exon 4. Specifically, the NM_001302456.1 and NM_207033.2 transcripts do not contain the fourth coding exon shared by the three other transcripts (Figure 3). Interestingly, a frameshift variant (NM_207034.2: c.559_560insA; p.Thr189Asnfs) in this exon was present in 0.6% (157/25,790) Finnish European alleles in gnomAD, with a high-quality variant score, including two homozygotes (Figure 3), supporting the clinical insignificance of this exon. In addition, exon 4 is predicted to be spliced out in most tissues in the GTEx expression database, further calling in to question the pathogenicity of the two variants present in exon 4 that were reported as disease-causing mutations in the Human Gene Mutation Database.
Figure 3.
Visualization of category 2 example, EDN3. Transcript view of the EDN3 gene. EDN3 has five reference sequence transcripts sharing coding exons 1 to 3 and 5 but differing in the inclusion of coding exon 4. A frameshift variant (NM_207034.2: c.559_560insA; p.Thr189Asnfs) in this exon was present in 0.6% (157/25,790) Finnish European alleles in the Genome Aggregation Database, with a high-quality variant score, including two homozygotes. This high-frequency loss-of-function variant is in the alternatively spliced exon (boxed area).
Category 3 Genes
There were 38 genes in this category, each with an average of 5 transcripts and 22 unique exons (Figure 2A and Supplemental Table S3). In total, C3 genes had 184 RefSeq transcripts and 854 unique exons. Given the multiple transcripts with mutually exclusive exons in this category, a thorough curation was performed to select the most clinically relevant transcript for each C3 gene (Supplemental Table S3). Published human and/or animal tissue expression studies supported transcript selection for 32 C3 genes; the longest transcript was supported in 25 genes, whereas a shorter isoform was most relevant in 7 genes. There were no expression data to guide selection of the most biologically relevant transcript for six C3 genes for which the longest transcript was defaulted to. Overall, 92 coding exons in the C3 genes met our criteria for uncertain significance, whereas 7 coding exons were classified as clinically insignificant (Materials and Methods) (Figure 2B).
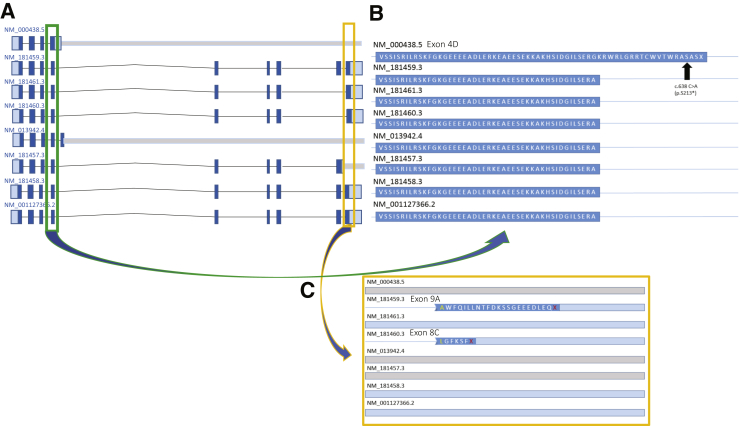
A C3 example is the PAX3 gene, which is a common cause of WS, type 1.86, 87, 88 This gene has eight RefSeq transcripts with varied tissue and temporal expression58, 59, 60 and with significant alternative splicing; a transcript can include 4, 5, 8, 9, or 10 exons. Certain exons use alternate splice junctions that can also change the reading frame for the terminal exon (Figure 4A). Interestingly, one putative LoF variant (c.638C>A; p.S213*) in exon 4D of the NM_000438.5 transcript was found in 0.54% (166/30,592) South Asian alleles, including one homozygous individual. This allele frequency is inconsistent with the estimated disease prevalence for autosomal dominant WS of approximately 1:40,000.88, 89 Exon 4D is only 20 amino acids longer than exon 4 on all other transcripts. Human expression data in the GTEx database strongly support use of the exon 4 (and not 4D) splice donor site, suggesting that exon 4D is not biologically relevant (Figure 4B).
Figure 4.
Visualization of category 3 example, PAX3. A: Transcript view of the PAX3 gene. PAX3 has eight reference sequence transcripts; a transcript can include 4, 5, 8, 9, or 10 exons. B: A close-up of the putative loss-of-function variant (c.638C>A; p.S213*) in exon 4D of the NM_000438.5 transcript, which was found in 0.54% (166/30,592) South Asian alleles in the Genome Aggregation Database, including one homozygous individual. C: A close-up of the two uncertain exons in PAX3: 9A (NM_181459.3) and 8C (NM_181460.3).
Similarly, exon 8C in the NM_181460.3 transcript of the PAX3 gene is unlikely to be biologically relevant, as supported by lack of its expression (GTEx database) and the presence of a putative LoF splicing variant (c.1024+1G>C) affecting this exon in 0.15% (47/30,764) South Asian alleles in gnomAD. This same variant has a missense effect (p.Arg402Pro) on the NM_181461.3 transcript, further highlighting the importance of appropriate transcript selection for variant annotation and interpretation (Figure 4C).
Impact on Clinical Testing and Interpretation
Transcript selection can significantly alter variant interpretation because variants can have differing molecular consequences on each transcript. For example, pathogenic missense and nonsense variants in the MITF gene, encoding a transcription factor critical for melanocyte development, are known to cause WS, type 2.88, 90 This gene has 13 curated RefSeq transcripts. On 4 of 13 transcripts, a particular pathogenic variant (which segregated with disease in >10 members of a family with WS91) is annotated as a variant in a +1 canonical splice donor site (c.33+1G>A). However, this nucleotide change is a deep intronic variant in the other nine transcripts (eg, NM_001184967.1: c.199-1066G>A) and could easily be misclassified if only the deep intronic consequence was interpreted.
Tissue-specific transcript expression can also significantly alter variant interpretation. Variants in TBC1D24, a GTPase-activating protein, are associated with nonsyndromic hearing loss, DOORS (Deafness, Onychodystrophy, Osteodystrophy, mental Retardation, and Seizures) syndrome, or a spectrum of epilepsy conditions.30, 89, 90, 91, 92, 93, 94, 95 This gene has two curated RefSeq transcripts: NM_001199107.1, the longest which contains eight exons and is most abundant in mouse neurons; and NM_020705.2, which is missing exon 3, contains only seven exons, and is expressed in mouse cochlea and nonneuronal tissues.30 NM_001199107.1:c.969_970delGT (p.Ser324Thrfs), a frameshift variant in exon 3, is not present in the shorter transcript and was identified in the homozygous state in five members of a consanguineous family with severe lethal epileptic encephalopathy but no hearing loss, thereby supporting the tissue-specific expression of the longest transcript.93
On the basis of our comprehensive curation of all transcripts, there were 125 coding exons with no or uncertain clinical significance, constituting 6% of all 2089 coding exons across all 109 genes (Figure 2B). Because of the limited evidence supporting those exons' clinical relevance, variants therein should be carefully interpreted because they can be a source of false-positive diagnoses. Interestingly, there are 124 variants that are labeled as disease causing (91 disease-causing mutations and 33 pathogenic/likely pathogenic) in disease databases (the Human Gene Mutation Database and ClinVar, respectively), in addition to 224 variants of uncertain clinical significance across those exons (Figure 2C). These variants interpreted as clinically significant will all require further assessment to ensure sufficient evidence is present to implicate them in hearing loss.
Technical Assessment
Because of the genetic heterogeneity of hearing loss, most clinical genetics laboratories use targeted or exome-based panels to sequence a comprehensive set of genes known to cause hearing loss. Although robust, such approaches have limitations inherent to the NGS technology, including the inability to reliably capture and sequence low-complexity and/or high-homology genomic regions.
Regions were identified in a set of 109 hearing loss genes that are technically challenging to sequence using current short-read (100 to 150 bp) NGS in clinical laboratories. Clinical exome sequencing data generated in two different sites (Children's Hospital of Philadelphia and LMM) were used to calculate exon-level quality metrics across all exons in the 109 hearing loss genes. An average mapping quality and an average depth of coverage cutoff of 20 and 15, respectively, are strong indicators of poor-quality regions.83 Forty-three well-baited (approximately 90% baited bases) exons were identified in 20 genes with the above cutoffs, despite being exome sequenced to an overall average coverage of up to 180× at the two clinical laboratories (Supplemental Table S4) (Materials and Methods). Of those exons, 31 were sequenced to an overall coverage of approximately 600× using a different targeted capture (average percentage baited exons, 95%) and longer reads (150 bp), but still had low-quality metrics (Supplemental Table S4).
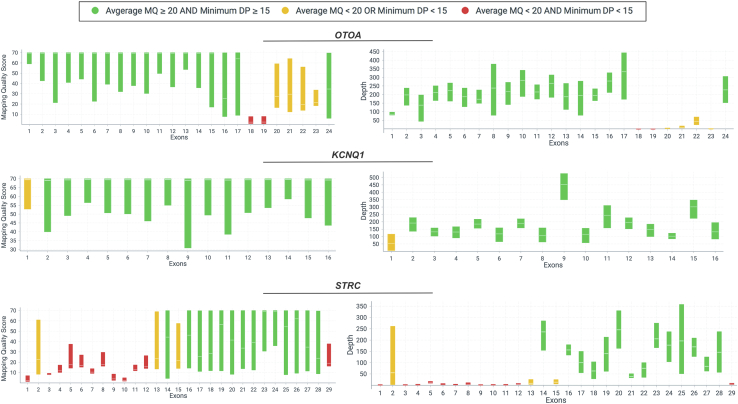
The 43 regions included exons with high homology to other genomic sequences (n = 21 exons in STRC and OTOA) or exons that have GC-rich or repeat sequences (n = 22; eg, KCNQ1, MYO15A, and TPRN) (Figure 5). It is unlikely that sequence variants in all 43 exons will be reliably detected using available NGS chemistries and, therefore, false-positive and/or false-negative variant calls in those exons should be highly expected.
Figure 5.
Visualization of three genes with known technically challenging regions. Exons in otoancorin (OTOA) or stereocilin (STRC), with high homology to other genomic sequences, and GC-rich first exon in the potassium channel, KCNQ1. Mapping quality (MQ) and depth of coverage (DP) plots are displayed for OTOA, KCNQ1, and STRC. Green bars indicate exons with both average MQ ≥ 20 and minimum DP ≥ 15; yellow bars, either average MQ < 20 or minimum DP < 15; red bars, poor exons, where both average MQ < 20 and minimum DP < 15. Each bar in MQ and DP plots shows minimum and maximum range for each exon, and average is shown by a tick mark in the middle of each bar.
Discussion
Transcript selection is critical for determining DNA variants' potential effects on RNA and/or protein expression, function, and stability. This annotation, in turn, significantly affects variant interpretation. Although most clinical laboratories use one set of coding transcripts (commonly RefSeq) for variant annotation, any set might contain multiple transcripts for each gene; some of these are true clinically relevant isoforms, whereas others can be false annotations. Even for those genes with multiple isoforms, deciphering the relevant isoform for a given disease—often on the basis of tissue-specific expression data—is necessary for interpretation. In the absence of uniform guidelines for transcript selection, each laboratory applies different internal rules for identifying the most appropriate transcript(s) for interpretation and reporting.
Herein, we provide a comprehensive evidence-based framework for transcript curation and selection. We apply this framework to 109 hearing loss genes and illustrate its utility in transcript selection and variant annotation and interpretation. These efforts are in line with our collaboration with the LRG effort to standardize the use of RefSeq transcripts for these 109 hearing loss genes. Of all coding (RefSeq) exons in these genes, 6% were shown to have no or questionable clinical validity, rendering them a potential source of false-variant calls irrespective of their predicted protein effect (eg, missense or LoF). The utility of this analysis process was confirmed by retrospectively analyzing 828 hearing loss patients tested at the LMM, and it was found that four likely pathogenic/pathogenic variants in C3 genes across 13 cases were present on alternate transcripts and would have been misinterpreted had transcript information not been evaluated.
A challenge with this approach is that it requires significant manual curation of a defined set of genes; such curation is essential for accurate interpretation and is arguably more effective if performed ahead of testing, and not retrospectively, to minimize analysis and wet bench burdens. This may be a challenge for exome or genome analysis; transcript-specific information should be evaluated at the variant level in this case. However, with the growth of disease expert groups, through international efforts like the Clinical Genome Resource (ClinGen) and others, accurate transcript curations can be leveraged across multiple diseases and gene panels and then shared with the larger community, with the hope of generating a compendium of clinically curated transcripts as a resource for more comprehensive clinical use.
Another challenge is that it is highly dependent on availability of human and/or animal expression data in the relevant disease tissue—the inner ear in this current work. However, leveraging existing large human genomic population (gnomAD) and transcriptome (GTEx) sequencing data as well as high-quality variant databases (ClinVar) can support the selection of clinically relevant transcripts in our genes. Although most genes associated with genetic hearing loss have LoF as the proposed disease mechanism, there are exceptions. LoF should be the proposed disease mechanism in the relevant gene before calling an exon insignificant based on high allele frequency LoF variants in the general population.
Finally, exon-level NGS quality metrics were used to highlight regions that are inaccessible to sequencing and/or accurate variant calling, especially with short-read (100- to 150-bp) chemistries that are mostly used in clinical and research laboratories. It is possible that some of those regions can be recovered with longer reads and improved bioinformatics pipelines. Until then, however, it is highly important that different ancillary assays, such as Sanger sequencing, be validated to accurately capture sequence variants in those regions.
In summary, we recommend that our transcript selection framework and exon classification system be used in other disease areas for more efficient and accurate variant interpretation, and to avoid erroneous annotations and, potentially, misdiagnoses. We hope to expand this analysis to disorders of reduced penetrance and complex diseases as guidelines for gene curation and variant classification are developed in these areas.
Footnotes
Supported in part by the National Human Genome Research Institute, in conjunction with additional funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development under award U41HG006834 (H.L.R.).
Disclosures: None declared.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Supplemental material for this article can be found at https://doi.org/10.1016/j.jmoldx.2018.06.005.
Supplemental Data
References
- 1.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., Voelkerding K., Rehm H.L. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casper J., Zweig A.S., Villarreal C., Tyner C., Speir M.L., Rosenbloom K.R., Raney B.J., Lee C.M., Lee B.T., Karolchik D., Hinrichs A.S., Haeussler M., Guruvadoo L., Navarro Gonzalez J., Gibson D., Fiddes I.T., Eisenhart C., Diekhans M., Clawson H., Barber G.P., Armstrong J., Haussler D., Kuhn R.M., Kent W.J. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 2018;46:D762–D769. doi: 10.1093/nar/gkx1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenbloom K.R., Sloan C.A., Malladi V.S., Dreszer T.R., Learned K., Kirkup V.M., Wong M.C., Maddren M., Fang R., Heitner S.G., Lee B.T., Barber G.P., Harte R.A., Diekhans M., Long J.C., Wilder S.P., Zweig A.S., Karolchik D., Kuhn R.M., Haussler D., Kent W.J. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thierry-Mieg D., Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7 Suppl 1:S12.11–S12.14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pruitt K.D., Harrow J., Harte R.A., Wallin C., Diekhans M., Maglott D.R. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aken B.L., Achuthan P., Akanni W., Amode M.R., Bernsdorff F., Bhai J. Ensembl 2017. Nucleic Acids Res. 2017;45:D635–D642. doi: 10.1093/nar/gkw1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.den Dunnen J.T., Dalgleish R., Maglott D.R., Hart R.K., Greenblatt M.S., McGowan-Jordan J., Roux A.F., Smith T., Antonarakis S.E., Taschner P.E. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37:564–569. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- 10.Roy S., Coldren C., Karunamurthy A., Kip N.S., Klee E.W., Lincoln S.E., Leon A., Pullambhatla M., Temple-Smolkin R.L., Voelkerding K.V., Wang C., Carter A.B. Standards and guidelines for validating next-generation sequencing bioinformatics pipelines: a Joint Recommendation of the Association for Molecular Pathology and the College of American Pathologists. J Mol Diagn. 2018;20:4–27. doi: 10.1016/j.jmoldx.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Alford R.L., Arnos K.S., Fox M., Lin J.W., Palmer C.G., Pandya A., Rehm H.L., Robin N.H., Scott D.A., Yoshinaga-Itano C. American College of Medical Genetics and Genomics guideline for the clinical evaluation and etiologic diagnosis of hearing loss. Genet Med. 2014;16:347–355. doi: 10.1038/gim.2014.2. [DOI] [PubMed] [Google Scholar]
- 12.Abou Tayoun A.N., Al Turki S.H., Oza A.M., Bowser M.J., Hernandez A.L., Funke B.H., Rehm H.L., Amr S.S. Improving hearing loss gene testing: a systematic review of gene evidence toward more efficient next-generation sequencing-based diagnostic testing and interpretation. Genet Med. 2018;18:545–553. doi: 10.1038/gim.2015.141. [DOI] [PubMed] [Google Scholar]
- 13.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duzkale H., Shen J., McLaughlin H., Alfares A., Kelly M.A., Pugh T.J., Funke B.H., Rehm H.L., Lebo M.S. A systematic approach to assessing the clinical significance of genetic variants. Clin Genet. 2013;84:453–463. doi: 10.1111/cge.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Modamio-Hoybjor S., Mencia A., Goodyear R., del Castillo I., Richardson G., Moreno F., Moreno-Pelayo M.A. A mutation in CCDC50, a gene encoding an effector of epidermal growth factor-mediated cell signaling, causes progressive hearing loss. Am J Hum Genet. 2007;80:1076–1089. doi: 10.1086/518311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Namburi P., Ratnapriya R., Khateb S., Lazar C.H., Kinarty Y., Obolensky A., Erdinest I., Marks-Ohana D., Pras E., Ben-Yosef T., Newman H., Gross M., Swaroop A., Banin E., Sharon D. Bi-allelic truncating mutations in CEP78, encoding centrosomal protein 78, cause cone-rod degeneration with sensorineural hearing loss. Am J Hum Genet. 2016;99:777–784. doi: 10.1016/j.ajhg.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robertson N.G., Resendes B.L., Lin J.S., Lee C., Aster J.C., Adams J.C., Morton C.C. Inner ear localization of mRNA and protein products of COCH, mutated in the sensorineural deafness and vestibular disorder, DFNA9. Hum Mol Genet. 2001;10:2493–2500. doi: 10.1093/hmg/10.22.2493. [DOI] [PubMed] [Google Scholar]
- 18.Robertson N.G., Skvorak A.B., Yin Y., Weremowicz S., Johnson K.R., Kovatch K.A., Battey J.F., Bieber F.R., Morton C.C. Mapping and characterization of a novel cochlear gene in human and in mouse: a positional candidate gene for a deafness disorder, DFNA9. Genomics. 1997;46:345–354. doi: 10.1006/geno.1997.5067. [DOI] [PubMed] [Google Scholar]
- 19.Van Laer L., Huizing E.H., Verstreken M., van Zuijlen D., Wauters J.G., Bossuyt P.J., Van de Heyning P., McGuirt W.T., Smith R.J., Willems P.J., Legan P.K., Richardson G.P., Van Camp G. Nonsyndromic hearing impairment is associated with a mutation in DFNA5. Nat Genet. 1998;20:194–197. doi: 10.1038/2503. [DOI] [PubMed] [Google Scholar]
- 20.Delmaghani S., del Castillo F.J., Michel V., Leibovici M., Aghaie A., Ron U., Van Laer L., Ben-Tal N., Van Camp G., Weil D., Langa F., Lathrop M., Avan P., Petit C. Mutations in the gene encoding pejvakin, a newly identified protein of the afferent auditory pathway, cause DFNB59 auditory neuropathy. Nat Genet. 2006;38:770–778. doi: 10.1038/ng1829. [DOI] [PubMed] [Google Scholar]
- 21.Cheng J., Zhu Y., He S., Lu Y., Chen J., Han B., Petrillo M., Wrzeszczynski K.O., Yang S., Dai P., Zhai S., Han D., Zhang M.Q., Li W., Liu X., Li H., Chen Z.Y., Yuan H. Functional mutation of SMAC/DIABLO, encoding a mitochondrial proapoptotic protein, causes human progressive hearing loss DFNA64. Am J Hum Genet. 2011;89:56–66. doi: 10.1016/j.ajhg.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grifa A., Wagner C.A., D'Ambrosio L., Melchionda S., Bernardi F., Lopez-Bigas N., Rabionet R., Arbones M., Monica M.D., Estivill X., Zelante L., Lang F., Gasparini P. Mutations in GJB6 cause nonsyndromic autosomal dominant deafness at DFNA3 locus. Nat Genet. 1999;23:16–18. doi: 10.1038/12612. [DOI] [PubMed] [Google Scholar]
- 23.Borck G., Ur Rehman A., Lee K., Pogoda H.M., Kakar N., von Ameln S. Loss-of-function mutations of ILDR1 cause autosomal-recessive hearing impairment DFNB42. Am J Hum Genet. 2011;88:127–137. doi: 10.1016/j.ajhg.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santos-Cortez R.L., Lee K., Azeem Z., Antonellis P.J., Pollock L.M., Khan S., Irfanullah, Andrade-Elizondo P.B., Chiu I., Adams M.D., Basit S., Smith J.D., University of Washington Center for Mendelian Genomics. Nickerson D.A., McDermott B.M., Jr., Ahmad W., Leal S.M. Mutations in KARS, encoding lysyl-tRNA synthetase, cause autosomal-recessive nonsyndromic hearing impairment DFNB89. Am J Hum Genet. 2013;93:132–140. doi: 10.1016/j.ajhg.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beisel K.W., Rocha-Sanchez S.M., Morris K.A., Nie L., Feng F., Kachar B., Yamoah E.N., Fritzsch B. Differential expression of KCNQ4 in inner hair cells and sensory neurons is the basis of progressive high-frequency hearing loss. J Neurosci. 2005;25:9285–9293. doi: 10.1523/JNEUROSCI.2110-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu T., Nie L., Zhang Y., Mo J., Feng W., Wei D., Petrov E., Calisto L.E., Kachar B., Beisel K.W., Vazquez A.E., Yamoah E.N. Roles of alternative splicing in the functional properties of inner ear-specific KCNQ4 channels. J Biol Chem. 2007;282:23899–23909. doi: 10.1074/jbc.M702108200. [DOI] [PubMed] [Google Scholar]
- 27.Zazo Seco C., Serrao de Castro L., van Nierop J.W., Morin M., Jhangiani S., Verver E.J., Schraders M., Maiwald N., Wesdorp M., Venselaar H., Spruijt L., Oostrik J., Schoots J., Baylor-Hopkins Center for Mendelian G., van Reeuwijk J., Lelieveld S.H., Huygen P.L., Insenser M., Admiraal R.J., Pennings R.J., Hoefsloot L.H., Arias-Vasquez A., de Ligt J., Yntema H.G., Jansen J.H., Muzny D.M., Huls G., van Rossum M.M., Lupski J.R., Moreno-Pelayo M.A., Kunst H.P., Kremer H. Allelic mutations of KITLG, encoding KIT ligand, cause asymmetric and unilateral hearing loss and Waardenburg syndrome type 2. Am J Hum Genet. 2015;97:647–660. doi: 10.1016/j.ajhg.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riazuddin S., Ahmed Z.M., Fanning A.S., Lagziel A., Kitajiri S., Ramzan K., Khan S.N., Chattaraj P., Friedman P.L., Anderson J.M., Belyantseva I.A., Forge A., Riazuddin S., Friedman T.B. Tricellulin is a tight-junction protein necessary for hearing. Am J Hum Genet. 2006;79:1040–1051. doi: 10.1086/510022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahituv N., Sobe T., Robertson N.G., Morton C.C., Taggart R.T., Avraham K.B. Genomic structure of the human unconventional myosin VI gene. Gene. 2000;261:269–275. doi: 10.1016/s0378-1119(00)00535-7. [DOI] [PubMed] [Google Scholar]
- 30.Rehman A.U., Santos-Cortez R.L., Morell R.J., Drummond M.C., Ito T., Lee K., Khan A.A., Basra M.A., Wasif N., Ayub M., Ali R.A., Raza S.I., University of Washington Center for Mendelian G., Nickerson D.A., Shendure J., Bamshad M., Riazuddin S., Billington N., Khan S.N., Friedman P.L., Griffith A.J., Ahmad W., Riazuddin S., Leal S.M., Friedman T.B. Mutations in TBC1D24, a gene associated with epilepsy, also cause nonsyndromic deafness DFNB86. Am J Hum Genet. 2014;94:144–152. doi: 10.1016/j.ajhg.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adato A., Lefevre G., Delprat B., Michel V., Michalski N., Chardenoux S., Weil D., El-Amraoui A., Petit C. Usherin, the defective protein in Usher syndrome type IIA, is likely to be a component of interstereocilia ankle links in the inner ear sensory cells. Hum Mol Genet. 2005;14:3921–3932. doi: 10.1093/hmg/ddi416. [DOI] [PubMed] [Google Scholar]
- 32.Mathur P.D., Zou J., Zheng T., Almishaal A., Wang Y., Chen Q., Wang L., Vashist D., Brown S., Park A., Yang J. Distinct expression and function of whirlin isoforms in the inner ear and retina: an insight into pathogenesis of USH2D and DFNB31. Hum Mol Genet. 2015;24:6213–6228. doi: 10.1093/hmg/ddv339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J., Liu X., Zhao Y., Adamian M., Pawlyk B., Sun X., McMillan D.R., Liberman M.C., Li T. Ablation of whirlin long isoform disrupts the USH2 protein complex and causes vision and hearing loss. PLoS Genet. 2010;6:e1000955. doi: 10.1371/journal.pgen.1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebrahim S., Ingham N.J., Lewis M.A., Rogers M.J.C., Cui R., Kachar B., Pass J.C., Steel K.P. Alternative splice forms influence functions of whirlin in mechanosensory hair cell stereocilia. Cell Rep. 2016;15:935–943. doi: 10.1016/j.celrep.2016.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wayne S., Robertson N.G., DeClau F., Chen N., Verhoeven K., Prasad S., Tranebjarg L., Morton C.C., Ryan A.F., Van Camp G., Smith R.J. Mutations in the transcriptional activator EYA4 cause late-onset deafness at the DFNA10 locus. Hum Mol Genet. 2001;10:195–200. doi: 10.1093/hmg/10.3.195. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y., Liu Y., Nie H., Ma X., Xu Z. Alternative splicing of inner-ear-expressed genes. Front Med. 2016;10:250–257. doi: 10.1007/s11684-016-0454-y. [DOI] [PubMed] [Google Scholar]
- 37.Lagziel A., Overlack N., Bernstein S.L., Morell R.J., Wolfrum U., Friedman T.B. Expression of cadherin 23 isoforms is not conserved: implications for a mouse model of Usher syndrome type 1D. Mol Vis. 2009;15:1843–1857. [PMC free article] [PubMed] [Google Scholar]
- 38.Riazuddin S., Belyantseva I.A., Giese A.P., Lee K., Indzhykulian A.A., Nandamuri S.P., Yousaf R., Sinha G.P., Lee S., Terrell D., Hegde R.S., Ali R.A., Anwar S., Andrade-Elizondo P.B., Sirmaci A., Parise L.V., Basit S., Wali A., Ayub M., Ansar M., Ahmad W., Khan S.N., Akram J., Tekin M., Riazuddin S., Cook T., Buschbeck E.K., Frolenkov G.I., Leal S.M., Friedman T.B., Ahmed Z.M. Alterations of the CIB2 calcium- and integrin-binding protein cause Usher syndrome type 1J and nonsyndromic deafness DFNB48. Nat Genet. 2012;44:1265–1271. doi: 10.1038/ng.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilcox E.R., Burton Q.L., Naz S., Riazuddin S., Smith T.N., Ploplis B., Belyantseva I., Ben-Yosef T., Liburd N.A., Morell R.J., Kachar B., Wu D.K., Griffith A.J., Riazuddin S., Friedman T.B. Mutations in the gene encoding tight junction claudin-14 cause autosomal recessive deafness DFNB29. Cell. 2001;104:165–172. doi: 10.1016/s0092-8674(01)00200-8. [DOI] [PubMed] [Google Scholar]
- 40.Adato A., Vreugde S., Joensuu T., Avidan N., Hamalainen R., Belenkiy O., Olender T., Bonne-Tamir B., Ben-Asher E., Espinos C., Millan J.M., Lehesjoki A.E., Flannery J.G., Avraham K.B., Pietrokovski S., Sankila E.M., Beckmann J.S., Lancet D. USH3A transcripts encode clarin-1, a four-transmembrane-domain protein with a possible role in sensory synapses. Eur J Hum Genet. 2002;10:339–350. doi: 10.1038/sj.ejhg.5200831. [DOI] [PubMed] [Google Scholar]
- 41.Vastinsalo H., Jalkanen R., Dinculescu A., Isosomppi J., Geller S., Flannery J.G., Hauswirth W.W., Sankila E.M. Alternative splice variants of the USH3A gene Clarin 1 (CLRN1) Eur J Hum Genet. 2011;19:30–35. doi: 10.1038/ejhg.2010.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lynch E.D., Lee M.K., Morrow J.E., Welcsh P.L., Leon P.E., King M.C. Nonsyndromic deafness DFNA1 associated with mutation of a human homolog of the Drosophila gene diaphanous. Science. 1997;278:1315–1318. [PubMed] [Google Scholar]
- 43.Sanchez-Mejias A., Fernandez R.M., Lopez-Alonso M., Antinolo G., Borrego S. New roles of EDNRB and EDN3 in the pathogenesis of Hirschsprung disease. Genet Med. 2010;12:39–43. doi: 10.1097/GIM.0b013e3181c371b0. [DOI] [PubMed] [Google Scholar]
- 44.Cui L., Wong E.H., Cheng G., Firmato de Almeida M., So M.T., Sham P.C., Cherny S.S., Tam P.K., Garcia-Barcelo M.M. Genetic analyses of a three generation family segregating Hirschsprung disease and iris heterochromia. PLoS One. 2013;8:e66631. doi: 10.1371/journal.pone.0066631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grillet N., Schwander M., Hildebrand M.S., Sczaniecka A., Kolatkar A., Velasco J., Webster J.A., Kahrizi K., Najmabadi H., Kimberling W.J., Stephan D., Bahlo M., Wiltshire T., Tarantino L.M., Kuhn P., Smith R.J., Muller U. Mutations in LOXHD1, an evolutionarily conserved stereociliary protein, disrupt hair cell function in mice and cause progressive hearing loss in humans. Am J Hum Genet. 2009;85:328–337. doi: 10.1016/j.ajhg.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmed Z.M., Masmoudi S., Kalay E., Belyantseva I.A., Mosrati M.A., Collin R.W., Riazuddin S., Hmani-Aifa M., Venselaar H., Kawar M.N., Tlili A., van der Zwaag B., Khan S.Y., Ayadi L., Riazuddin S.A., Morell R.J., Griffith A.J., Charfedine I., Caylan R., Oostrik J., Karaguzel A., Ghorbel A., Riazuddin S., Friedman T.B., Ayadi H., Kremer H. Mutations of LRTOMT, a fusion gene with alternative reading frames, cause nonsyndromic deafness in humans. Nat Genet. 2008;40:1335–1340. doi: 10.1038/ng.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hershey C.L., Fisher D.E. Genomic analysis of the Microphthalmia locus and identification of the MITF-J/Mitf-J isoform. Gene. 2005;347:73–82. doi: 10.1016/j.gene.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 48.Chen L., Guo W., Ren L., Yang M., Zhao Y., Guo Z. A de novo silencer causes elimination of MITF-M expression and profound hearing loss in pigs. BMC Biol. 2016;14:52. doi: 10.1186/s12915-016-0273-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmed Z.M., Yousaf R., Lee B.C., Khan S.N., Lee S., Lee K., Husnain T., Rehman A.U., Bonneux S., Ansar M., Ahmad W., Leal S.M., Gladyshev V.N., Belyantseva I.A., Van Camp G., Riazuddin S., Friedman T.B., Riazuddin S. Functional null mutations of MSRB3 encoding methionine sulfoxide reductase are associated with human deafness DFNB74. Am J Hum Genet. 2011;88:19–29. doi: 10.1016/j.ajhg.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kelley P.M., Weston M.D., Chen Z.Y., Orten D.J., Hasson T., Overbeck L.D., Pinnt J., Talmadge C.B., Ing P., Mooseker M.S., Corey D., Sumegi J., Kimberling W.J. The genomic structure of the gene defective in Usher syndrome type Ib (MYO7A) Genomics. 1997;40:73–79. doi: 10.1006/geno.1996.4545. [DOI] [PubMed] [Google Scholar]
- 51.Haraksingh R.R., Jahanbani F., Rodriguez-Paris J., Gelernter J., Nadeau K.C., Oghalai J.S., Schrijver I., Snyder M.P. Exome sequencing and genome-wide copy number variant mapping reveal novel associations with sensorineural hereditary hearing loss. BMC Genomics. 2014;15:1155. doi: 10.1186/1471-2164-15-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zwaenepoel I., Mustapha M., Leibovici M., Verpy E., Goodyear R., Liu X.Z., Nouaille S., Nance W.E., Kanaan M., Avraham K.B., Tekaia F., Loiselet J., Lathrop M., Richardson G., Petit C. Otoancorin, an inner ear protein restricted to the interface between the apical surface of sensory epithelia and their overlying acellular gels, is defective in autosomal recessive deafness DFNB22. Proc Natl Acad Sci U S A. 2002;99:6240–6245. doi: 10.1073/pnas.082515999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi B.Y., Ahmed Z.M., Riazuddin S., Bhinder M.A., Shahzad M., Husnain T., Riazuddin S., Griffith A.J., Friedman T.B. Identities and frequencies of mutations of the otoferlin gene (OTOF) causing DFNB9 deafness in Pakistan. Clin Genet. 2009;75:237–243. doi: 10.1111/j.1399-0004.2008.01128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cohen-Salmon M., El-Amraoui A., Leibovici M., Petit C. Otogelin: a glycoprotein specific to the acellular membranes of the inner ear. Proc Natl Acad Sci U S A. 1997;94:14450–14455. doi: 10.1073/pnas.94.26.14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schraders M., Ruiz-Palmero L., Kalay E., Oostrik J., del Castillo F.J., Sezgin O., Beynon A.J., Strom T.M., Pennings R.J., Zazo Seco C., Oonk A.M., Kunst H.P., Dominguez-Ruiz M., Garcia-Arumi A.M., del Campo M., Villamar M., Hoefsloot L.H., Moreno F., Admiraal R.J., del Castillo I., Kremer H. Mutations of the gene encoding otogelin are a cause of autosomal-recessive nonsyndromic moderate hearing impairment. Am J Hum Genet. 2012;91:883–889. doi: 10.1016/j.ajhg.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Housley G.D., Kanjhan R., Raybould N.P., Greenwood D., Salih S.G., Jarlebark L., Burton L.D., Setz V.C., Cannell M.B., Soeller C., Christie D.L., Usami S., Matsubara A., Yoshie H., Ryan A.F., Thorne P.R. Expression of the P2X(2) receptor subunit of the ATP-gated ion channel in the cochlea: implications for sound transduction and auditory neurotransmission. J Neurosci. 1999;19:8377–8388. doi: 10.1523/JNEUROSCI.19-19-08377.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lynch K.J., Touma E., Niforatos W., Kage K.L., Burgard E.C., van Biesen T., Kowaluk E.A., Jarvis M.F. Molecular and functional characterization of human P2X(2) receptors. Mol Pharmacol. 1999;56:1171–1181. doi: 10.1124/mol.56.6.1171. [DOI] [PubMed] [Google Scholar]
- 58.Monsoro-Burq A.H. PAX transcription factors in neural crest development. Semin Cell Dev Biol. 2015;44:87–96. doi: 10.1016/j.semcdb.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 59.Tsukamoto K., Nakamura Y., Niikawa N. Isolation of two isoforms of the PAX3 gene transcripts and their tissue-specific alternative expression in human adult tissues. Hum Genet. 1994;93:270–274. doi: 10.1007/BF00212021. [DOI] [PubMed] [Google Scholar]
- 60.Blake J.A., Ziman M.R. Pax3 transcripts in melanoblast development. Dev Growth Differ. 2005;47:627–635. doi: 10.1111/j.1440-169X.2005.00835.x. [DOI] [PubMed] [Google Scholar]
- 61.Alagramam K.N., Miller N.D., Adappa N.D., Pitts D.R., Heaphy J.C., Yuan H., Smith R.J. Promoter, alternative splice forms, and genomic structure of protocadherin 15. Genomics. 2007;90:482–492. doi: 10.1016/j.ygeno.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alagramam K.N., Yuan H., Kuehn M.H., Murcia C.L., Wayne S., Srisailpathy C.R., Lowry R.B., Knaus R., Van Laer L., Bernier F.P., Schwartz S., Lee C., Morton C.C., Mullins R.F., Ramesh A., Van Camp G., Hageman G.S., Woychik R.P., Smith R.J. Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum Mol Genet. 2001;10:1709–1718. doi: 10.1093/hmg/10.16.1709. [DOI] [PubMed] [Google Scholar]
- 63.Ahmed Z.M., Riazuddin S., Ahmad J., Bernstein S.L., Guo Y., Sabar M.F., Sieving P., Riazuddin S., Griffith A.J., Friedman T.B., Belyantseva I.A., Wilcox E.R. PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum Mol Genet. 2003;12:3215–3223. doi: 10.1093/hmg/ddg358. [DOI] [PubMed] [Google Scholar]
- 64.Khan S.Y., Ahmed Z.M., Shabbir M.I., Kitajiri S., Kalsoom S., Tasneem S., Shayiq S., Ramesh A., Srisailpathy S., Khan S.N., Smith R.J., Riazuddin S., Friedman T.B., Riazuddin S. Mutations of the RDX gene cause nonsyndromic hearing loss at the DFNB24 locus. Hum Mutat. 2007;28:417–423. doi: 10.1002/humu.20469. [DOI] [PubMed] [Google Scholar]
- 65.Jin H., May M., Tranebjaerg L., Kendall E., Fontan G., Jackson J., Subramony S.H., Arena F., Lubs H., Smith S., Stevenson R., Schwartz C., Vetrie D. A novel X-linked gene, DDP, shows mutations in families with deafness (DFN-1), dystonia, mental deficiency and blindness. Nat Genet. 1996;14:177–180. doi: 10.1038/ng1096-177. [DOI] [PubMed] [Google Scholar]
- 66.Nakane T., Inada Y., Ito F., Itoh N., Tazawa S., Chiba S. Cloning and expression of mouse deafness dystonia peptide 1 cDNA. Biochem Biophys Res Commun. 2000;273:759–764. doi: 10.1006/bbrc.2000.3004. [DOI] [PubMed] [Google Scholar]
- 67.Scott H.S., Kudoh J., Wattenhofer M., Shibuya K., Berry A., Chrast R., Guipponi M., Wang J., Kawasaki K., Asakawa S., Minoshima S., Younus F., Mehdi S.Q., Radhakrishna U., Papasavvas M.P., Gehrig C., Rossier C., Korostishevsky M., Gal A., Shimizu N., Bonne-Tamir B., Antonarakis S.E. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat Genet. 2001;27:59–63. doi: 10.1038/83768. [DOI] [PubMed] [Google Scholar]
- 68.Verpy E., Leibovici M., Zwaenepoel I., Liu X.Z., Gal A., Salem N., Mansour A., Blanchard S., Kobayashi I., Keats B.J., Slim R., Petit C. A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat Genet. 2000;26:51–55. doi: 10.1038/79171. [DOI] [PubMed] [Google Scholar]
- 69.Khateb S., Zelinger L., Ben-Yosef T., Merin S., Crystal-Shalit O., Gross M., Banin E., Sharon D. Exome sequencing identifies a founder frameshift mutation in an alternative exon of USH1C as the cause of autosomal recessive retinitis pigmentosa with late-onset hearing loss. PLoS One. 2012;7:e51566. doi: 10.1371/journal.pone.0051566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bahloul A., Michel V., Hardelin J.P., Nouaille S., Hoos S., Houdusse A., England P., Petit C. Cadherin-23, myosin VIIa and harmonin, encoded by Usher syndrome type I genes, form a ternary complex and interact with membrane phospholipids. Hum Mol Genet. 2010;19:3557–3565. doi: 10.1093/hmg/ddq271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Santos-Cortez R.L., Lee K., Giese A.P., Ansar M., Amin-Ud-Din M., Rehn K., Wang X., Aziz A., Chiu I., Hussain Ali R., Smith J.D., University of Washington Center for Mendelian Genomics. Shendure J., Bamshad M., Nickerson D.A., Ahmed Z.M., Ahmad W., Riazuddin S., Leal S.M. Adenylate cyclase 1 (ADCY1) mutations cause recessive hearing impairment in humans and defects in hair cell function and hearing in zebrafish. Hum Mol Genet. 2014;23:3289–3298. doi: 10.1093/hmg/ddu042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nyegaard M., Rendtorff N.D., Nielsen M.S., Corydon T.J., Demontis D., Starnawska A., Hedemand A., Buniello A., Niola F., Overgaard M.T., Leal S.M., Ahmad W., Wikman F.P., Petersen K.B., Cruger D.G., Oostrik J., Kremer H., Tommerup N., Frodin M., Steel K.P., Tranebjaerg L., Borglum A.D. A novel locus harbouring a functional CD164 nonsense mutation identified in a large danish family with nonsyndromic hearing impairment. PLoS Genet. 2015;11:e1005386. doi: 10.1371/journal.pgen.1005386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gagnon L.H., Longo-Guess C.M., Berryman M., Shin J.B., Saylor K.W., Yu H., Gillespie P.G., Johnson K.R. The chloride intracellular channel protein CLIC5 is expressed at high levels in hair cell stereocilia and is essential for normal inner ear function. J Neurosci. 2006;26:10188–10198. doi: 10.1523/JNEUROSCI.2166-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Diaz-Horta O., Subasioglu-Uzak A., Grati M., DeSmidt A., Foster J., 2nd, Cao L., Bademci G., Tokgoz-Yilmaz S., Duman D., Cengiz F.B., Abad C., Mittal R., Blanton S., Liu X.Z., Farooq A., Walz K., Lu Z., Tekin M. FAM65B is a membrane-associated protein of hair cell stereocilia required for hearing. Proc Natl Acad Sci U S A. 2014;111:9864–9868. doi: 10.1073/pnas.1401950111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao B., Wu Z., Muller U. Murine Fam65b forms ring-like structures at the base of stereocilia critical for mechanosensory hair cell function. Elife. 2016;5:e14222. doi: 10.7554/eLife.14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thoenes M., Zimmermann U., Ebermann I., Ptok M., Lewis M.A., Thiele H., Morlot S., Hess M.M., Gal A., Eisenberger T., Bergmann C., Nurnberg G., Nurnberg P., Steel K.P., Knipper M., Bolz H.J. OSBPL2 encodes a protein of inner and outer hair cell stereocilia and is mutated in autosomal dominant hearing loss (DFNA67) Orphanet J Rare Dis. 2015;10:15. doi: 10.1186/s13023-015-0238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grati M., Shin J.B., Weston M.D., Green J., Bhat M.A., Gillespie P.G., Kachar B. Localization of PDZD7 to the stereocilia ankle-link associates this scaffolding protein with the Usher syndrome protein network. J Neurosci. 2012;32:14288–14293. doi: 10.1523/JNEUROSCI.3071-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schneider E., Marker T., Daser A., Frey-Mahn G., Beyer V., Farcas R., Schneider-Ratzke B., Kohlschmidt N., Grossmann B., Bauss K., Napiontek U., Keilmann A., Bartsch O., Zechner U., Wolfrum U., Haaf T. Homozygous disruption of PDZD7 by reciprocal translocation in a consanguineous family: a new member of the Usher syndrome protein interactome causing congenital hearing impairment. Hum Mol Genet. 2009;18:655–666. doi: 10.1093/hmg/ddn395. [DOI] [PubMed] [Google Scholar]
- 79.Baig S.M., Koschak A., Lieb A., Gebhart M., Dafinger C., Nurnberg G., Ali A., Ahmad I., Sinnegger-Brauns M.J., Brandt N., Engel J., Mangoni M.E., Farooq M., Khan H.U., Nurnberg P., Striessnig J., Bolz H.J. Loss of Ca(v)1.3 (CACNA1D) function in a human channelopathy with bradycardia and congenital deafness. Nat Neurosci. 2011;14:77–84. doi: 10.1038/nn.2694. [DOI] [PubMed] [Google Scholar]
- 80.Schultz J.M., Khan S.N., Ahmed Z.M., Riazuddin S., Waryah A.M., Chhatre D., Starost M.F., Ploplis B., Buckley S., Velasquez D., Kabra M., Lee K., Hassan M.J., Ali G., Ansar M., Ghosh M., Wilcox E.R., Ahmad W., Merlino G., Leal S.M., Riazuddin S., Friedman T.B., Morell R.J. Noncoding mutations of HGF are associated with nonsyndromic hearing loss, DFNB39. Am J Hum Genet. 2009;85:25–39. doi: 10.1016/j.ajhg.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J., Jang W., Katz K., Ovetsky M., Riley G., Sethi A., Tully R., Villamarin-Salomon R., Rubinstein W., Maglott D.R. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A., Cooper D.N. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Niazi R, Gonzalez MA, Balciuniene J, Evans P, Sarmady M, Abou Tayoun AN:ExomeSlicer: a resource for the development and validation of exome-based clinical panels. bioRxiv 2018, doi: 10.1101/248906. [DOI] [PubMed]
- 84.Edery P., Attie T., Amiel J., Pelet A., Eng C., Hofstra R.M., Martelli H., Bidaud C., Munnich A., Lyonnet S. Mutation of the endothelin-3 gene in the Waardenburg-Hirschsprung disease (Shah-Waardenburg syndrome) Nat Genet. 1996;12:442–444. doi: 10.1038/ng0496-442. [DOI] [PubMed] [Google Scholar]
- 85.Hofstra R.M., Osinga J., Tan-Sindhunata G., Wu Y., Kamsteeg E.J., Stulp R.P., van Ravenswaaij-Arts C., Majoor-Krakauer D., Angrist M., Chakravarti A., Meijers C., Buys C.H. A homozygous mutation in the endothelin-3 gene associated with a combined Waardenburg type 2 and Hirschsprung phenotype (Shah-Waardenburg syndrome) Nat Genet. 1996;12:445–447. doi: 10.1038/ng0496-445. [DOI] [PubMed] [Google Scholar]
- 86.Baldwin C.T., Hoth C.F., Amos J.A., da-Silva E.O., Milunsky A. An exonic mutation in the HuP2 paired domain gene causes Waardenburg's syndrome. Nature. 1992;355:637–638. doi: 10.1038/355637a0. [DOI] [PubMed] [Google Scholar]
- 87.Tassabehji M., Read A.P., Newton V.E., Harris R., Balling R., Gruss P., Strachan T. Waardenburg's syndrome patients have mutations in the human homologue of the Pax-3 paired box gene. Nature. 1992;355:635–636. doi: 10.1038/355635a0. [DOI] [PubMed] [Google Scholar]
- 88.Song J., Feng Y., Acke F.R., Coucke P., Vleminckx K., Dhooge I.J. Hearing loss in Waardenburg syndrome: a systematic review. Clin Genet. 2016;89:416–425. doi: 10.1111/cge.12631. [DOI] [PubMed] [Google Scholar]
- 89.Shi Y., Li X., Ju D., Li Y., Zhang X., Zhang Y. A novel mutation of the MITF gene in a family with Waardenburg syndrome type 2: a case report. Exp Ther Med. 2016;11:1516–1518. doi: 10.3892/etm.2016.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tassabehji M., Newton V.E., Read A.P. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet. 1994;8:251–255. doi: 10.1038/ng1194-251. [DOI] [PubMed] [Google Scholar]
- 91.Azaiez H., Booth K.T., Bu F., Huygen P., Shibata S.B., Shearer A.E., Kolbe D., Meyer N., Black-Ziegelbein E.A., Smith R.J. TBC1D24 mutation causes autosomal-dominant nonsyndromic hearing loss. Hum Mutat. 2014;35:819–823. doi: 10.1002/humu.22557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Falace A., Filipello F., La Padula V., Vanni N., Madia F., De Pietri Tonelli D., de Falco F.A., Striano P., Dagna Bricarelli F., Minetti C., Benfenati F., Fassio A., Zara F. TBC1D24, an ARF6-interacting protein, is mutated in familial infantile myoclonic epilepsy. Am J Hum Genet. 2010;87:365–370. doi: 10.1016/j.ajhg.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guven A., Tolun A. TBC1D24 truncating mutation resulting in severe neurodegeneration. J Med Genet. 2013;50:199–202. doi: 10.1136/jmedgenet-2012-101313. [DOI] [PubMed] [Google Scholar]
- 94.Campeau P.M., Kasperaviciute D., Lu J.T., Burrage L.C., Kim C., Hori M. The genetic basis of DOORS syndrome: an exome-sequencing study. Lancet Neurol. 2014;13:44–58. doi: 10.1016/S1474-4422(13)70265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang L., Hu L., Chai Y., Pang X., Yang T., Wu H. A dominant mutation in the stereocilia-expressing gene TBC1D24 is a probable cause for nonsyndromic hearing impairment. Hum Mutat. 2014;35:814–818. doi: 10.1002/humu.22558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.