Coxiella burnetii is an obligate intracellular bacterium and the etiological agent of Q fever. Successful host cell infection requires the Coxiella type IVB secretion system (T4BSS), which translocates bacterial effector proteins across the vacuole membrane into the host cytoplasm, where they manipulate a variety of cell processes.
KEYWORDS: Coxiella burnetii, IL-17, type 4 secretion, innate immunity, macrophage
ABSTRACT
Coxiella burnetii is an obligate intracellular bacterium and the etiological agent of Q fever. Successful host cell infection requires the Coxiella type IVB secretion system (T4BSS), which translocates bacterial effector proteins across the vacuole membrane into the host cytoplasm, where they manipulate a variety of cell processes. To identify host cell targets of Coxiella T4BSS effector proteins, we determined the transcriptome of murine alveolar macrophages infected with a Coxiella T4BSS effector mutant. We identified a set of inflammatory genes that are significantly upregulated in T4BSS mutant-infected cells compared to mock-infected cells or cells infected with wild-type (WT) bacteria, suggesting that Coxiella T4BSS effector proteins downregulate the expression of these genes. In addition, the interleukin-17 (IL-17) signaling pathway was identified as one of the top pathways affected by the bacteria. While previous studies demonstrated that IL-17 plays a protective role against several pathogens, the role of IL-17 during Coxiella infection is unknown. We found that IL-17 kills intracellular Coxiella in a dose-dependent manner, with the T4BSS mutant exhibiting significantly more sensitivity to IL-17 than WT bacteria. In addition, quantitative PCR confirmed the increased expression of IL-17 downstream signaling genes in T4BSS mutant-infected cells compared to WT- or mock-infected cells, including the proinflammatory cytokine genes Il1a, Il1b, and Tnfa, the chemokine genes Cxcl2 and Ccl5, and the antimicrobial protein gene Lcn2. We further confirmed that the Coxiella T4BSS downregulates macrophage CXCL2/macrophage inflammatory protein 2 and CCL5/RANTES protein levels following IL-17 stimulation. Together, these data suggest that Coxiella downregulates IL-17 signaling in a T4BSS-dependent manner in order to escape the macrophage immune response.
INTRODUCTION
The intracellular bacterium Coxiella burnetii is the etiological agent of Q fever, a zoonotic infectious disease. Initially, Q fever manifests as an acute self-limited flu-like illness. However, patients can develop chronic disease that can be life threatening due to serious clinical manifestations, such as endocarditis (1). Furthermore, the current therapy recommended for chronic Q fever requires at least 18 months of doxycycline and hydroxychloroquine treatment (2). An effective vaccine (Q-Vax) has been developed for humans but is currently licensed only in Australia due to adverse effects, especially when administered to previously infected populations (3). In addition, Q fever outbreaks have occurred in several countries, including the Netherlands (4), the United States (5), Spain (6), Australia (7), Japan (8), and Israel (9), exemplifying how expansive C. burnetii infection is worldwide and the need for novel therapeutic targets.
Human infection occurs primarily by inhaling contaminated dust or aerosols, often from close contact with livestock. In the lungs, C. burnetii displays tropism for alveolar macrophages, where it forms a phagolysosome-like parasitophorous vacuole (PV) necessary to support bacterial growth (10, 11). C. burnetii's ability to survive and replicate inside the PV, an inhospitable environment for most bacteria, is a unique feature essential for C. burnetii pathogenesis. C. burnetii exploits the acidic PV pH for metabolic activation (12) and actively manipulates PV fusogenicity and maintenance (13). PV establishment requires the translocation of bacterial proteins into the host cell cytoplasm by the C. burnetii Dot/Icm (defect in organelle trafficking/intracellular multiplication) type IVB secretion system (T4BSS), closely related to the Dot/Icm T4BSS of Legionella pneumophila (14). T4BSS effector proteins manipulate not only host vesicular trafficking during PV development but also other cellular processes, such as lipid metabolism, host gene expression, apoptosis, host translation, iron transport, ubiquitination, autophagy, and immunity (15, 16). Based on in silico prediction, there are more than 100 putative C. burnetii T4BSS effector proteins (17–19), but functional data are lacking for the majority of these proteins. In particular, the role of T4BSS effector proteins in manipulating the innate immune response is poorly understood. Recently, the C. burnetii T4BSS effector protein IcaA was found to inhibit caspase 11-mediated, noncanonical activation of the nucleotide binding domain and the leucine-rich repeat-containing protein (NLRP3) inflammasome during C. burnetii infection (20). Since cytosolic lipopolysaccharide (LPS) is known to activate noncanonical inflammasomes (21, 22), it is possible that C. burnetii LPS triggers this pathway and that the bacterium utilizes T4BSS effectors, such as IcaA, to block this innate immune response. Given the low infectious dose (<10 organisms) (23), C. burnetii certainly inhibits several immediate host cell responses in order to establish infection.
In order to identify new immune response pathways manipulated by C. burnetii T4BSS effector proteins, we compared the transcriptome of alveolar macrophages infected with wild-type (WT) or T4BSS mutant C. burnetii. We identified a set of inflammatory genes downregulated by C. burnetii T4BSS effector proteins, with the interleukin-17 (IL-17) signaling pathway being one of the top targeted host cell pathways. As IL-17 is a proinflammatory cytokine that plays a role in the protective response against a variety of bacterial infections, including those caused by the pulmonary intracellular pathogens Mycoplasma pneumonia, Mycobacterium tuberculosis, Francisella tularensis, and Legionella pneumophila (24–27), we further investigated the role of IL-17 during C. burnetii infection. Our data revealed that stimulation of the macrophage IL-17 signaling pathway leads to C. burnetii killing in a dose-dependent manner, with the T4BSS mutant displaying increased sensitivity compared to WT bacteria. Finally, our findings demonstrate that C. burnetii downregulates the IL-17 signaling pathway in macrophages through T4BSS effector proteins.
RESULTS
Differentially expressed genes in C. burnetii-infected macrophages.
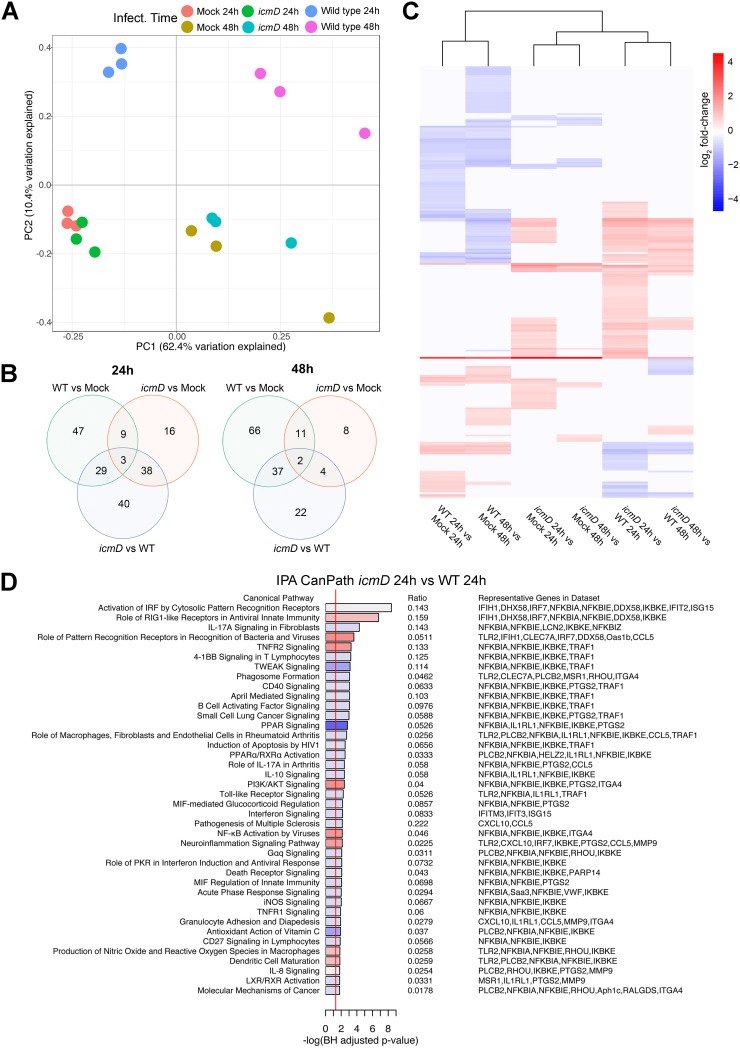
In order to identify T4BSS-dependent changes in the expression of host genes, we determined the whole transcriptome of murine alveolar (MH-S) macrophages infected with either wild-type (WT) C. burnetii or a C. burnetii mutant lacking icmD, an essential component of the T4BSS (14). We previously found minimal differences in PV size and bacterial replication between the WT and a C. burnetii T4BSS mutant during the first 48 h of infection of MH-S macrophages (28). Thus, to avoid changes in host cell gene expression that could occur due to PV expansion and bacterial replication after 48 h and because C. burnetii T4BSS effector protein secretion occurs by 4 h postinfection (hpi) (29), we analyzed gene expression at 24 and 48 hpi. By principal components analysis (PCA), global transcription in T4BSS mutant-infected cells more closely resembled that in mock-infected cells than that in WT-infected cells, suggesting that the active T4BSS in WT bacteria drastically alters the host cell response to C. burnetii infection (Fig. 1A). The number of differentially expressed genes (DEGs) was determined for each comparison at 24 or 48 hpi, using an absolute fold change threshold of >1.5 and false discovery rate (FDR) of <5% (see Data Set S1 in the supplemental material). The largest differences in gene expression at 24 hpi and 48 hpi were between icmD mutant-infected cells versus WT-infected cells (110 DEGs at 24 hpi) and WT-infected cells versus mock-infected cells (116 DEGs at 48 hpi), respectively (Fig. 1B; Data Set S1). Unsupervised hierarchical cluster analysis of fold change values for DEGs (absolute log2 fold change, >0.585; FDR, <5%) across all six possible two-way comparisons revealed that the majority of DEGs were upregulated in icmD mutant-infected cells compared to their expression in WT-infected cells (Fig. 1C, icmD versus WT). In contrast, the majority of DEGs were downregulated in WT-infected versus mock-infected cells (Fig. 1C, WT versus mock). Overall, there were fewer downregulated genes in the icmD mutant-infected cells than in the mock-infected cells (Fig. 1C, icmD versus mock). This provides evidence that C. burnetii T4BSS effector proteins may play a role in the downregulation of host cell genes during the initial stages of infection.
FIG 1.
C. burnetii infection alters the gene expression profile of alveolar macrophages in a T4BSS-dependent manner. The transcriptomes of MH-S macrophages infected with wild-type (WT) or icmD T4SS mutant C. burnetii were analyzed at 24 or 48 hpi. (A) Principal components analysis (PCA) of genome-wide expression across all RNA-seq samples after normalization of raw data. (B) Venn diagram of differentially expressed genes for each comparison for each time point using an absolute log2 fold change (logFC) of >0.585 (1.5-fold in linear space) and a false discovery rate (FDR) of <0.05. (C) Unsupervised hierarchical clustering heat map of log2 fold change values for all six comparisons using the ward.D2 clustering method and Euclidean distance. Red intensity indicates increased expression in the first group relative to that in the second group, whereas blue intensity indicates decreased expression. (D) Ingenuity Pathway Analysis (IPA) using differentially expressed genes for the comparison between WT-infected cells and icmD mutant-infected cells at 24 hpi. Red shading indicates increased pathway activity in icmD mutant-infected cells, whereas blue shading indicates increased pathway activity in WT-infected cells, based on IPA activity Z-scores. White shading indicates that no activity pattern was predicted or could be determined. TNFR2, tumor necrosis factor receptor 2; PPAR, peroxisome proliferator-activated receptor; PI3K, phosphatidylinositol 3-kinase; MIF, migration inhibition factor; PKR, protein kinase R.
To identify the biological pathways targeted by C. burnetii T4BSS effector proteins, we used two methods: gene set enrichment via CERNO testing (30) using Gene Ontology (GO) annotations as gene sets (Fig. S1) and Ingenuity Pathways Analysis (IPA) using DEGs with an absolute log2 fold change of >0.585 and an FDR of <5% as inputs (Fig. 1D and S2). Both methods revealed differential expression of several immune and inflammatory pathways, including pathogen recognition and activation of the interferon regulatory factor (IRF) by cytosolic pattern recognition receptors (PRRs) and transmembrane PRRs, signaling pathways induced by the proinflammatory cytokines IL-1α and IL-1β, chemokine activity, T cell migration, and nuclear factor κB (NF-κB) phosphorylation. In addition, our data indicate that the C. burnetii T4BSS significantly downregulates the macrophage type I interferon (IFN) response. This finding supports published data that C. burnetii does not induce a robust type I IFN response in macrophages (31). In addition, IL-17 signaling was among the top three overrepresented canonical pathways between mutant and WT infection (Fig. 1D), with upstream regulator analysis predicting activation of IL-17 signaling in icmD mutant-infected cells relative to mock-infected cells (Fig. S2). Given that IL-17 is known to be an important proinflammatory cytokine against several pulmonary pathogens, we specifically tested for the differential expression of IL-17-related genes (32) using self-contained gene set testing. The IL-17 gene set was overexpressed in icmD mutant-infected macrophages relative to WT-infected macrophages (Table S1), suggesting that the C. burnetii T4BSS downregulates IL-17 signaling in macrophages.
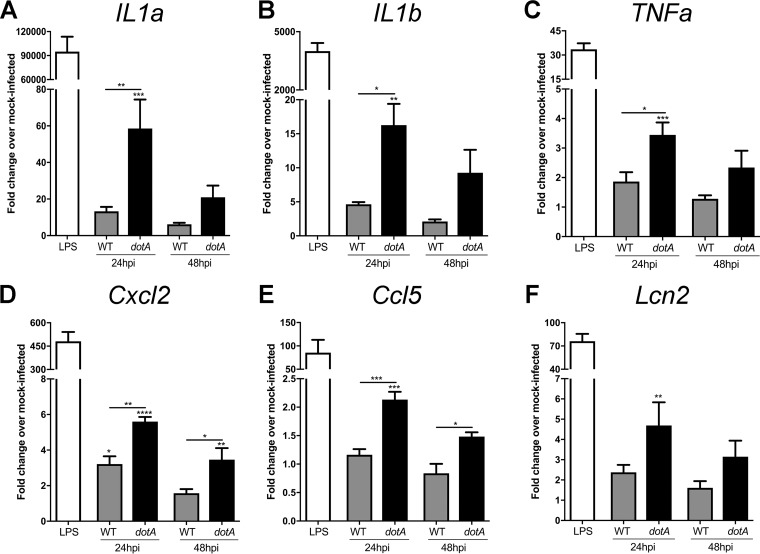
To validate the transcriptome analysis, we used quantitative reverse transcription-PCR (qRT-PCR) of infected macrophages to test the expression of the IL-17 pathway and other proinflammatory genes. RNA was isolated from MH-S macrophages infected with either WT C. burnetii or a C. burnetii mutant lacking dotA, another essential component of the T4BSS (33). Like the icmD mutant, the dotA mutant does not translocate T4BSS effector proteins, allowing us to confirm that gene expression changes are indeed T4BSS dependent. Lipopolysaccharide (LPS), a potent stimulator of the inflammatory response (34), served as a positive control. Between the WT- and dotA mutant-infected macrophages, we observed a significant difference in expression of the proinflammatory genes Il1a, Il1b, and Tnfa (Fig. 2A to C), as well as the IL-17 signaling pathway chemokines Cxcl2/Mip2 and Ccl5/Rantes (Fig. 2D and E) and the antimicrobial protein Lipocalin-2 (Lcn2) (Fig. 2F). These genes were upregulated in the dotA mutant-infected macrophages compared to WT-infected macrophages, with more significant differences being found at 24 hpi than at 48 hpi. The gene for IL-17A itself was not differentially regulated in either our RNA sequencing (RNA-seq) data or qRT-PCR data (data not shown), which is not surprising, given that macrophages produce very little IL-17 (35). These data suggest that, during the early stages of macrophage infection, the C. burnetii T4BSS may target the IL-17 pathway in order to downregulate expression of several proinflammatory genes.
FIG 2.
C. burnetii T4BSS effector proteins downregulate expression of the IL-17 pathway. Quantitative RT-PCR gene expression analysis of IL-17 pathway genes in macrophages infected with wild-type (WT) or dotA mutant C. burnetii at 24 or 48 hpi. Mock-infected macrophages treated with LPS (100 ng/ml) for 24 h were used as a positive control. The expression of individual genes was normalized to that of Gapdh, and the fold change in expression over that in mock-infected cells was determined. Compared to their levels of expression in WT-infected cells, macrophages infected with dotA mutant C. burnetii had higher levels of expression of the IL-17 pathway genes IL1a (A), IL1b (B), Tnfa (C), Cxcl2 (D), Ccl5 (E), and Lcn2 (F). Error bars show the average ± SEM from three independent experiments, performed in biological duplicate with three technical replicates. P values were determined by one-way ANOVA with Tukey's post hoc test and represent statistically significant differences compared to mock-infected cells or between WT and dotA mutant-infected cells. *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001.
C. burnetii downregulates CXCL2/MIP-2 and CCL5/RANTES expression in a T4BSS-dependent manner.
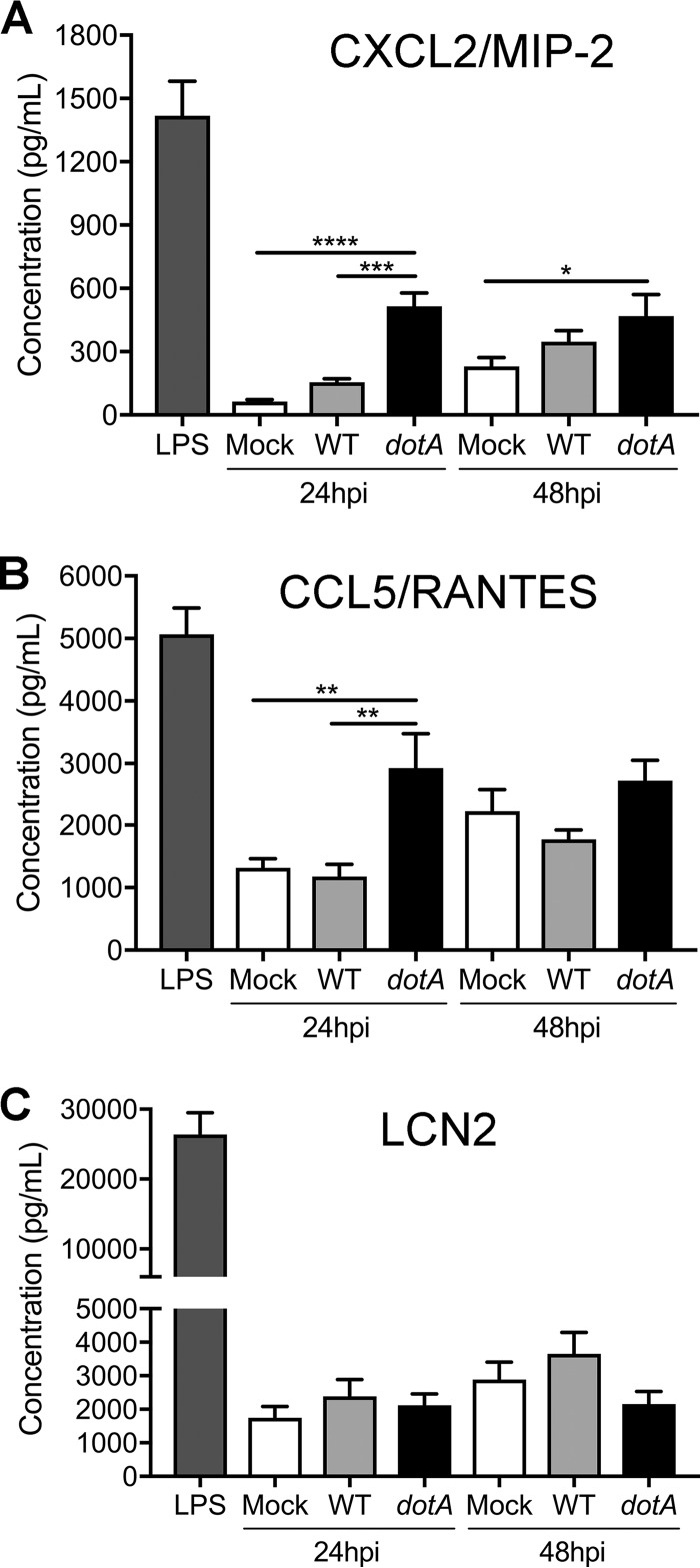
A number of studies have shown that IL-17 plays an important role in the innate immune response against bacteria by stimulating secretion of multiple chemokines. Within the context of infection, these chemokines recruit macrophages, neutrophils, and lymphocytes to the infection site, thereby enhancing inflammation. To validate the gene expression changes between macrophages infected with the WT or the T4BSS mutant, we measured the secretion of CXCL2/macrophage inflammatory protein 2 (MIP-2) and CCL5/RANTES at 24 or 48 hpi using an enzyme-linked immunosorbent assay (ELISA). We observed a significant difference in CXCL2/MIP-2 and CCL5/RANTES protein levels between the WT- and dotA mutant-infected macrophages, with a 3-fold increase of both cytokines in the dotA mutant-infected macrophages (Fig. 3A and B), confirming the gene expression data. While the CXCL2/MIP-2 level was significantly higher at both 24 and 48 hpi, we detected a difference in CCL5/RANTES expression only at 24 hpi (Fig. 3B).
FIG 3.
The C. burnetii T4BSS decreases the secretion of CXCL2/MIP-2 and CCL5/RANTES in infected MH-S macrophages. ELISA quantitation of the CXCL2/MIP-2, CCL5/RANTES, and Lipocalin-2 (LCN2) proteins in the supernatant of macrophages infected with WT or dotA mutant C. burnetii at 24 or 48 hpi. Cells treated with LPS (100 ng/ml) were used as a positive control. Compared to the levels of secretion in mock-infected and WT-infected macrophages at 24 hpi, dotA mutant-infected macrophages had increased secretion of CXCL2/MIP-2 (A) and CCL5/RANTES (B) but not LCN2 (C). Shown are the means ± SEM from three independent experiments done in duplicate. P values were determined by one-way ANOVA with Tukey's post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001.
LCN2 expression is strongly induced by IL-17 and blocks catecholate-type siderophores of Gram-negative bacteria, preventing the bacteria from scavenging the free iron required for bacterial growth (36, 37). While Lcn2 gene expression was differentially regulated (Fig. 2F), we did not observe a significant difference in secreted LCN2 protein levels between the WT- and dotA mutant-infected macrophages at either 24 or 48 hpi (Fig. 3C). These conflicting data may be due to the posttranslational regulation of LCN2 (38). However, our data do suggest that the C. burnetii T4BSS downregulates macrophage secretion of the chemokines CXCL2/MIP-2 and CCL5/RANTES during infection.
C. burnetii T4BSS effector proteins impair IL-17-stimulated CXCL2/MIP-2 and CCL5/RANTES secretion.
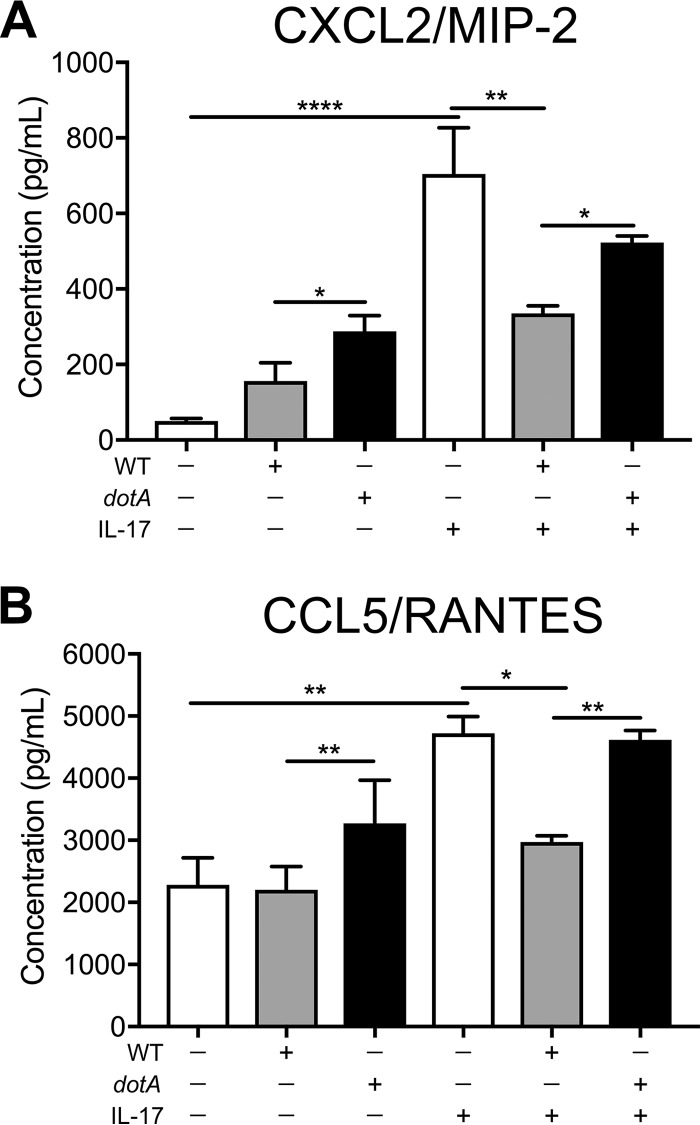
The chemoattractant CXCL2/MIP-2 is typically secreted by monocytes and macrophages and recruits neutrophils that are required for pathogen clearance (39). Furthermore, CCL5/RANTES, which is secreted by lymphocytes, macrophages, and endothelial cells, also recruits and activates leukocytes (40). We first confirmed that IL-17 upregulates CXCL2/MIP-2 and CCL5/RANTES in MH-S alveolar macrophages by treating uninfected macrophages with recombinant mouse IL-17A and analyzing the cell-free supernatant by ELISA. In uninfected macrophages, the levels of CXCL2/MIP-2 and CCL5/RANTES increased 14-fold and 2-fold, respectively, following IL-17A treatment for 24 h (Fig. 4A and B). To test if C. burnetii T4BSS effector proteins block IL-17-stimulated chemokine secretion, WT- or dotA mutant-infected macrophages were treated with IL-17A for 24 h. In IL-17-stimulated macrophages infected with WT bacteria, CXCL2/MIP-2 levels decreased 2.2-fold compared to those in stimulated mock-infected macrophages, while CCL5/RANTES levels decreased 1.6-fold (Fig. 4A and B), suggesting that WT C. burnetii blocks IL-17-induced chemokine secretion. Further, dotA mutant C. burnetii did not block IL-17A-stimulated CXCL2/MIP-2 and CCL5/RANTES (Fig. 4A and B). These data suggest that the C. burnetii T4BSS impairs IL-17 signaling in macrophages, including secretion of CXCL2/MIP-2 and CCL5/RANTES.
FIG 4.
C. burnetii T4BSS blocks IL-17-stimulated secretion of CXCL2/MIP-2 and CCL5/RANTES. The results of ELISA quantitation of CXCL2/MIP-2 (A) and CCL5/RANTES (B) protein levels after IL-17A (100 ng/ml) treatment of mock-infected, WT-infected, or dotA mutant-infected macrophages at 24 hpi are shown. The means ± SEM from three individual experiments performed in duplicate are shown. P values were determined by one-way ANOVA with Tukey's post hoc test and represent statistically significant differences compared to mock-infected cells or between WT and dotA mutant-infected cells. *, P < 0.05; **, P < 0.01; ****, P < 0.001.
Triggering the macrophage IL-17 pathway is bactericidal.
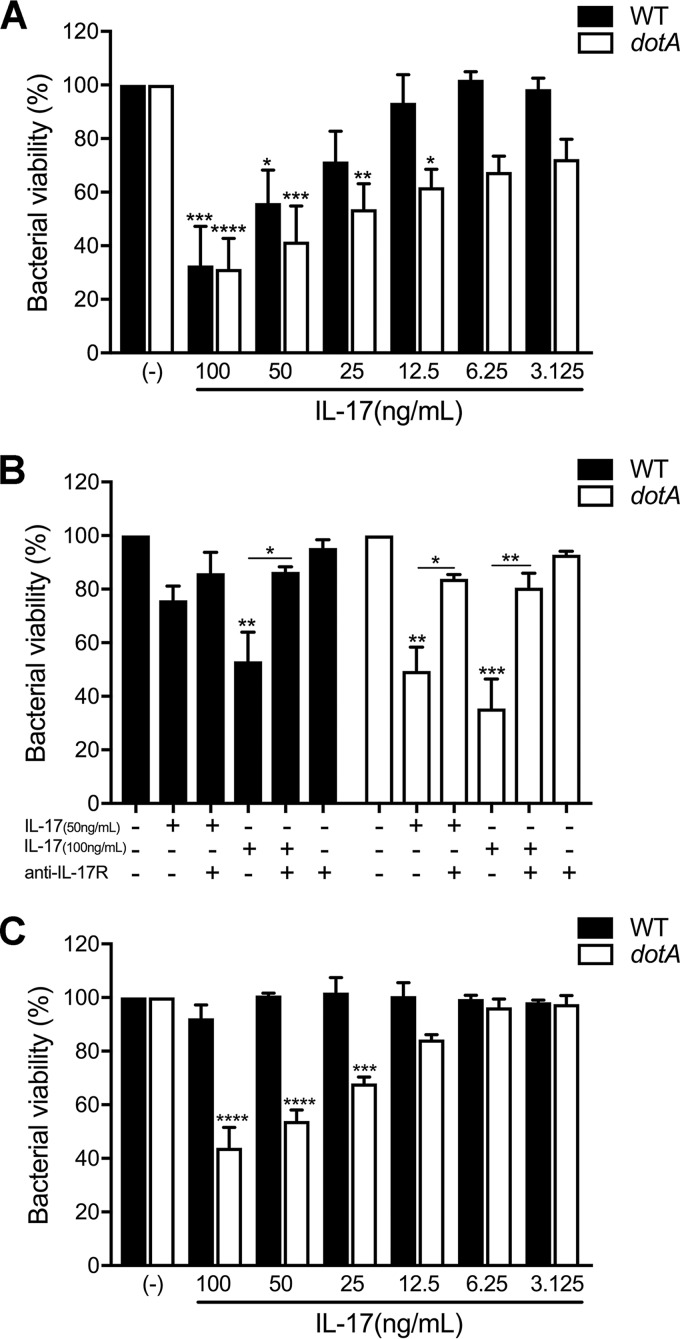
Several studies have demonstrated that IL-17 plays a protective role for the host during bacterial infections (32, 41–43). To evaluate if IL-17 affects C. burnetii viability in macrophages, we treated infected macrophages at 24 or 48 hpi with recombinant mouse IL-17A and enumerated the viable bacteria 24 h later using a fluorescent infectious focus-forming unit (FFU) assay in Vero cells (44). IL-17 stimulation decreased C. burnetii viability at 24 and 48 hpi in a dose-dependent manner, with an ∼40% decrease being found at the highest concentration (Fig. S3). IL-17A treatment did not affect macrophage viability (data not shown). Interestingly, C. burnetii appeared to be more resistant to IL-17 stimulation at 48 hpi than at 24 hpi, as low concentrations of IL-17 (50 ng/ml) led to a significant loss of bacterial viability only at 24 hpi (Fig. S3A and B). To determine if the T4BSS is related to bacterial susceptibility to IL-17, we infected macrophages with either WT or dotA mutant C. burnetii, stimulated with IL-17A, and measured bacterial viability by a CFU assay on agarose plates (45). We observed a stronger bactericidal effect of IL-17 on dotA mutant C. burnetii than on WT C. burnetii, as the presence of 25 and 12.5 ng/ml of IL-17 led to a 47% and 39% loss of dotA mutant C. burnetii viability, respectively, but did not affect WT C. burnetii viability (Fig. 5A and S4). To further assess the specificity of IL-17 activity, we treated infected cells with IL-17 (50 or 100 ng/ml) in the presence or absence of an antibody that blocks the IL-17 receptor (IL-17R). The IL-17 bactericidal effect was significantly neutralized by blocking the IL-17 receptor, as cotreatment with IL-17 and the anti-IL-17 receptor antibody rescued over 30% of the bacterial viability compared to that of IL-17-treated infected cells (Fig. 5B and S4).
FIG 5.
Activating the macrophage IL-17 pathway can kill C. burnetii, but the T4BSS effector proteins play a protective role. (A and B) MH-S cells were infected for 24 h, followed by treatment for 24 h with either IL-17 alone (A) or IL-17 and an IL-17A receptor-blocking antibody (2 μg/ml) (B). (C) hMDMs were infected for 24 h and treated with human IL-17A for 24 h. Viable bacteria were quantitated using an agarose-based CFU assay, and the loss of bacterial viability was calculated by dividing the number of WT or T4SS mutant bacteria in treated samples by the number in their respective untreated samples (−). The dotA T4BSS mutant is more sensitive to IL-17 than WT bacteria in both MH-S and hMDMs, and the viability of both WT and dotA mutant C. burnetii could be recovered by blocking IL-17 receptor signaling. Error bars indicate the mean ± SEM from four individual experiments. P values were determined by one-way ANOVA with Dunnett's post hoc test and represent statistically significant differences compared to the untreated controls. *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001.
In order to validate the bactericidal effect of IL-17 in primary cells, human monocyte-derived macrophages (hMDMs) were infected with the WT or the C. burnetii dotA mutant and treated at 24 hpi with recombinant human IL-17A, and bacterial viability was measured after 24 h by the CFU assay. Confirming our results obtained in MH-S cells, the dotA T4BSS mutant viability decreased with IL-17 treatment (Fig. 5C and S4), with a 50% decrease in the number of viable bacteria being seen at 100 ng/ml. However, IL-17 had no effect on WT C. burnetii in primary hMDMs. Together, these data suggest that activation of the IL-17 signaling pathway in macrophages kills intracellular C. burnetii bacteria, with the C. burnetii T4BSS playing a protective role.
DISCUSSION
The innate immune response relies on pathogen detection by pattern recognition receptors, which activate signaling pathways and trigger an inflammatory response (46). While the innate immune response is essential to protect the host, pathogens such as C. burnetii have evolved strategies to overcome the host innate immune response (47, 48). Despite being sequestered in a growth-permissive vacuole, C. burnetii T4BSS effector proteins manipulate a variety of host cell signaling processes, including the innate immune responses of inflammasome-mediated pyroptotic and apoptotic cell death (20, 49–51). To identify potential targets of C. burnetii T4BSS effector proteins, we compared the transcriptomes of murine alveolar macrophages infected with either WT or T4BSS mutant C. burnetii. We identified several inflammatory pathways downregulated by C. burnetii T4BSS effector proteins, including IL-17 signaling. Previous studies demonstrated that IL-17 plays a protective role against several pathogens, including L. pneumophila, the closest pathogenic relative of C. burnetii (25–27, 41, 42). We found that C. burnetii downregulates the macrophage IL-17 signaling pathway in a T4BSS-dependent manner, protecting the bacteria from IL-17-mediated killing by the macrophage and blocking the secretion of proinflammatory chemokines. To our knowledge, this is the first demonstration of a pathogenic bacterium directly downregulating intracellular macrophage IL-17 signaling.
Previous studies demonstrated that C. burnetii infection leads to secretion of the proinflammatory cytokines tumor necrosis factor alpha (TNF-α) and gamma interferon (IFN-γ), with both cytokines playing critical roles in restricting C. burnetii replication (52–54). In our studies, gene expression analysis during the early stages of infection revealed striking differences in the immunological response to WT and T4BSS mutant C. burnetii, with C. burnetii T4BSS mutant-infected macrophages having a stronger proinflammatory response. For example, the proinflammatory genes Il1a, Il1b, and Tnfa are expressed at higher levels in macrophages infected with T4BSS mutant C. burnetii than in WT-infected macrophages. Bacterium-driven downregulation of these and other proinflammatory cytokines would benefit the bacteria in establishing infection. In support of our data, C. burnetii infection of primary macrophages does not activate caspase 1 (20), an enzyme required for the production of the proinflammatory cytokines IL-1β and IL-1α (55, 56). Interestingly, C. burnetii does not directly inhibit caspase 1 activation but appears to interfere with upstream signaling events, including blocking TNF-α signaling (20, 57). However, a recent study did not detect significant differences in TNF-α production in murine bone marrow-derived macrophages infected with WT C. burnetii or icmL mutant C. burnetii, a mutant with a nonfunctional T4BSS (31). These apparently conflicting data may be explained by the use of C57BL/6 mice in the latter study; C57BL/6 mice, in contrast to other inbred mouse strains, are not permissive for intracellular C. burnetii replication due to the large amount of TNF-α produced upon Toll-like receptor (TLR) stimulation (31, 58–60). Further experimentation is required to elucidate the mechanism(s) behind C. burnetii T4BSS-mediated downregulation of the macrophage proinflammatory response.
Pathogen-associated molecular patterns (PAMPs) are sensed by different PRRs, which activate IRFs and initiate key inflammatory responses, including the transcription of type I interferons (IFN) and IFN-inducible genes (61, 62). Type I IFN can be induced by many intracellular bacterial pathogens either via recognition of bacterial surface molecules, such as LPS, or through stimulatory ligands released by the bacteria via specialized bacterial secretion systems (63). Our transcriptome analysis revealed C. burnetii T4BSS-mediated downregulation of macrophage IRF activation by cytosolic and transmembrane PRRs. A recent study found that C. burnetii does not trigger cytosolic PRRs or induce robust type I IFN production in mouse macrophages (31). Additionally, IFN-α receptor-deficient (IFNAR−/−) mice were protected from C. burnetii infection, suggesting that type I IFNs are not required to restrict bacterial replication (31, 64). However, delivery of recombinant IFN-α to the lungs of C. burnetii-infected mice protected against bacterial replication, revealing a potential role of type I IFN in the control of C. burnetii infection in the lung (64). Interestingly, type I IFN is induced during L. pneumophila infection and plays a key role in macrophage defense by restricting intracellular bacterial replication (65, 66). However, to counteract this host immune response, the L. pneumophila type IV secretion system (T4SS) effector protein SdhA suppresses the induction of IFN through an unknown mechanism (67). Similarly, our data suggest that C. burnetii T4BSS effector proteins negatively modulate the type I IFN response in alveolar macrophages, most likely as a bacterial immune evasion mechanism.
In addition to proinflammatory cytokines and PRRs, we discovered an important role for the cytokine IL-17 during C. burnetii infection of macrophages. The protective role of IL-17 against extracellular bacteria has been extensively studied; additionally, IL-17 can be critical for the full immune response leading to the control of intracellular bacteria (32, 42, 43, 68). IL-17 is produced by T helper 17 (Th17) cells, γδ T cells, and invariant natural killer T (iNKT) cells (69). In the lung, γδ T cells have been implicated as a primary source of early IL-17 production in several in vivo models of infection (70), which may have implications for C. burnetii lung infection. Exogenous IL-17 binds the IL-17 receptor on the surface of the macrophage, triggering chemokine secretion, neutrophil recruitment, and a Th1 response, thus enhancing bacterial clearance (26, 27, 71, 72). By both gene expression and protein analysis, we found that C. burnetii downregulates IL-17-stimulated chemokine secretion in macrophages in a T4BSS-dependent manner. A previous study found that following C. burnetii aerosol infection in mice, neutrophils are not present in the airways until 7 days postinfection, though the mechanism of this delay remains unknown (73). Further, neutrophils play a critical role in inflammation and bacterial clearance following intranasal C. burnetii infection, but it is unknown whether neutrophils directly kill the bacteria or serve to enhance the immune response (74). Based on our findings in alveolar macrophages, we hypothesize that C. burnetii T4BSS effector proteins downregulate the IL-17 pathway to suppress chemokine secretion as a mechanism to avoid neutrophil recruitment at early stages of infection. This could be an important immune evasion strategy that enables the bacteria to establish long-term persistence. In addition to chemokines, the IL-17-stimulated protein LCN2 may also be downregulated by C. burnetii. LCN2 is a siderophore-binding antimicrobial protein that can limit bacterial growth by iron restriction.
A previous study demonstrated that C. burnetii-infected IL-17 receptor knockout mice had a bacterial burden in the spleen and lung similar to that in infected WT mice, suggesting that IL-17 does not play an essential role during C. burnetii infection (74). In contrast, our in vitro studies revealed that activating the IL-17 signaling pathway in macrophages can directly kill intracellular C. burnetii. Further, the C. burnetii T4BSS appears to play a protective role, presumably by blocking the intracellular signaling pathway triggered by IL-17 binding to IL-17R. Our data may explain the lack of phenotypic changes in IL-17 receptor knockout mice infected by WT C. burnetii, as the intracellular signaling pathway is not activated in the absence of the IL-17 receptor.
During acute C. burnetii infection in humans, the number of γδ T cells rises significantly in the peripheral blood of patients (75). Given that γδ T cells can secrete large amounts of IL-17 (76), it is possible that the downregulation of intracellular IL-17 signaling by T4BSS effector proteins might be an essential mechanism of immune evasion that allows C. burnetii persistence. IL-17 activates common downstream pathways in macrophages, including the nuclear factor κB (NF-κB) and mitogen-activated protein kinase (MAPK) pathways (77, 78). Our transcriptome data suggest that the C. burnetii T4BSS downregulates the genes involved in the IL-17 canonical NF-κB signaling pathway, including Il17ra, Il17rc, Traf6, Nfkb1, and Nfkb2. This hypothesis is consistent with a recent study that found that C. burnetii can modulate the NF-κB canonical pathway through the T4BSS (79). NF-κB activation correlates with enhanced expression of inducible nitric oxide synthase (iNOS) (80) and NADPH oxidase (NOX) (81), which generate nitric oxide (NO) and reactive oxygen species (ROS), respectively. Both NO and ROS are signature molecules for M1 macrophages (82), while C. burnetii-infected macrophages exhibit an M2 polarization that is unable to control bacterial replication (83). As IL-17 alters macrophage polarization (84), one potential mechanism is that IL-17 polarizes toward the M1 phenotype, triggering ROS and NO, leading to C. burnetii killing. In addition, as IFN-γ plays a clear role in C. burnetii killing (53, 54), the IL-17 bactericidal effect might be related to IFN-γ, as IL-17 can induce an IFN-γ response (85). Further experimentation is needed to identify not only the C. burnetii T4BSS effector protein modulating IL-17 signaling in macrophages but also how IL-17 leads to C. burnetii death inside macrophages.
In summary, this study suggests that C. burnetii employs the T4BSS to downregulate IL-17 signaling in macrophages during the early stages of infection. This has important implications in both controlling the proinflammatory response elicited by the macrophages and avoiding direct killing by the macrophage. Further studies identifying the bacterial T4BSS effector proteins involved in this mechanism and elucidating how IL-17 kills C. burnetii will give new insight into immune evasion by C. burnetii.
MATERIALS AND METHODS
Bacteria and mammalian cells.
Coxiella burnetii Nine Mile phase II (NMII; clone 4, RSA439) was purified from Vero cells (African green monkey kidney epithelial cells; CCL-81; American Type Culture Collection, Manassas, VA) and stored as previously described (86). For all experiments, the C. burnetii NMII wild type (WT), icmD mutant (14), and dotA mutant (33) were grown for 4 days in acidified citrate cysteine medium 2 (ACCM-2) at 37°C in 2.5% O2 and 5% CO2, washed twice with phosphate-buffered saline (PBS), and stored as previously described (87). Murine alveolar (MH-S) macrophages (CRL-2019; ATCC) were maintained in growth medium consisting of RPMI 1640 medium (Corning, New York, NY, USA) containing 10% fetal bovine serum (FBS; Atlanta Biologicals, Norcross, GA, USA) at 37°C in 5% CO2. The multiplicity of infection (MOI) for each bacterial stock was optimized for each bacterial stock and culture vessel for a final infection of approximately 1 internalized bacterium per cell. To obtain human monocyte-derived macrophages (hMDMs), peripheral blood mononuclear cells were isolated from buffy coats (Indiana Blood Center) using Ficoll-Paque (catalog number 17144002; GE Healthcare). Monocytes were isolated from lymphocytes by positive selection using CD14 magnetic beads (catalog number 11367D; Dynabeads FlowComp human CD14; Thermo Fisher Scientific). Following isolation, monocytes were cultured for 7 days with RPMI 1640 medium containing 10% FBS, 100 mg/ml penicillin-streptomycin, and 50 ng/ml human macrophage colony-stimulating factor (M-CSF; catalog number 14-8789-62; Thermo Fisher Scientific). At 24 h prior to infection, the medium containing antibiotics and M-CSF was replaced with RPMI 1640 medium containing 10% FBS.
RNA sequencing.
MH-S cells (4 × 105 cells per well of a 6-well plate) were mock infected or infected with WT or icmD mutant C. burnetii, with three replicates being used per condition. Total RNA was isolated at 24 and 48 hpi using an RNeasy Plus minikit (Qiagen). RNA samples had an RNA integrity number of >7, as determined on an Agilent 2100 bioanalyzer. RNA-seq libraries were prepared using a ScriptSeq Complete kit (Illumina, Inc.) according to the manufacturer's instructions. Libraries were sequenced at 30 million reads per sample on an Illumina NextSeq platform with read lengths of 75 bp by the Indiana University Bloomington Center for Genomics and Bioinformatics and mapped to the mouse reference genome mm10 by the Indiana University Center for Computational Biology and Bioinformatics. RNA processing and sequencing were performed as a single batch. The median library size (mapped reads) was 17.8 million reads, with the minimum being 13.4 million reads.
Gene expression analysis.
RNA-seq differential gene expression (DGE) analysis was performed using the edgeR package (version 3.16.5) in R (version 3.3.3). After filtering genes with low levels of expression across a majority of samples, trimmed mean of M (TMM) value normalization was applied to the remaining 9,400 genes. Expression data for these genes were converted to log counts per million (logCPM) for data visualization with principal components analysis (PCA) plots. DGE analysis was performed using the glmLRT function as 2-way comparisons between the three classes using the following model matrix formula: ∼0 + Infection_Time, where Infection_Time is a combined factor variable consisting of cell treatment and time point (six levels). The fold change in gene expression was determined by comparing wild-type- or icmD mutant-infected cells to mock-infected cells or each other at either 24 h or 48 h (six different comparisons). Differential expression of functional pathways was assessed by two methods using the list of differentially expressed genes (DEGs) for each comparison: (i) gene enrichment analysis using CERNO testing in the tmod package (version 0.31) (30) in R with Gene Ontology (GO; C5 in MSigDB [88]) annotations as gene sets and all DEGs without cutoff criteria but ranked by ascending P values and (ii) Ingenuity Pathways Analysis (version 42012434) using DEGs with a ∣log2 fold change∣ of >0.585 (1.5 in linear space) and an FDR of <5% as inputs. In addition, self-contained gene set testing for enrichment of IL-17-related genes (32) was also performed using the roast.DGElist function in the edgeR package and the following gene list: Ccl5, Il17rc, Lcn2, Traf6, Il17ra, Nfkb1, Nfkb2, Ccl2, and Ccl3.
Quantitative gene expression by real time-PCR (RT-qPCR).
MH-S cells (2 × 105 cells per well of a 6-well plate) were mock infected or infected with WT or dotA mutant C. burnetii in 0.5 ml growth medium for 2 h at 37°C in 5% CO2, washed extensively with PBS, and incubated in 2 ml of growth medium. Cells treated with LPS (100 ng/ml) from Escherichia coli O111:B4 (catalog number L4392; Sigma) were used as a positive control. RNA was isolated using an RNeasy Plus minikit at 24 and 48 hpi and analyzed for quantity and the A260/A280 ratio (Implen NanoPhotometer), and cDNA was generated using a SuperScript III first-strand synthesis system kit (Invitrogen). Real-time PCR using a Luminaris Color HiGreen quantitative PCR (qPCR) master mix (Thermo Fisher Scientific) was done on a Bio-Rad CFX Connect real-time system according to the manufacturer's instructions. Mouse-specific primers (5′ to 3′) were as follows: for Il1b, forward primer TGTAATGAAAGACGGCACACC and reverse primer TCTTCTTTGGGTATTGCTTGG; for Il1a, forward primer CGCTTGAGTCGGCAAAGAAAT and reverse primer ACAAACTGATCTGTGCAAGTCTC; for Tnfa, forward primer TTCTGTCTACTGAACTTCGGG and reverse primer GTATGAGATAGCAAATCGGCT; for CCL5/RANTES, forward primer ACTCCCTGCTGCTTTGCCTAC and reverse primer ACTTGCTGGTGTAGAAATACT; for CXCL2/MIP-2, forward primer CGCTGTCAATGCCTGAAGAC and reverse primer ACACTCAAGCTCTGGATGTTCTTG; for Lcn2, forward primer TTTCACCCGCTTTGCCAAGT and reverse primer GTCTCTGCGCATCCCAGTCA; and for Gapdh (glyceraldehyde-3-phosphate dehydrogenase), forward primer AAGGTCATCCCAGAGCTGAA and reverse primer CTGCTTCACCACCTTCTTGA. The relative levels of transcripts were calculated by the ΔΔCT threshold cycle (CT) method using Gapdh as the internal control. The relative levels of mRNA from the mock-infected samples were adjusted to 1 and served as the basal control value. Each experiment was done in biological duplicate, and qPCR was performed on three separate cDNA preparations from each RNA.
ELISA.
CXCL2-MIP-2, CCL5-RANTES, and Lipocalin-2 protein levels in cell-free supernatants were measured by ELISA (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions. In brief, MH-S cells (5 × 104 cells per well of a 24-well plate) were plated and allowed to adhere overnight. The cells were then mock infected or infected with WT or dotA mutant C. burnetii in 0.25 ml growth medium for 2 h at 37°C in 5% CO2, washed extensively with PBS, and incubated in 0.5 ml growth medium. To examine the IL-17 pathway expression in infected cells, the cells were pretreated with 100 ng/ml of IL-17A recombinant mouse protein (catalog number PMC0174; Thermo Fisher Scientific) for 24 h and then infected as described above. LPS-treated cells (100 ng/ml; catalog number L4391; Sigma) were used as a positive control. The cell supernatant was collected at 24 or 48 hpi, centrifuged at 20,000 × g for 10 min, and analyzed by ELISA. Each experiment was performed in biological duplicate with two technical replicates.
C. burnetii viability by fluorescent infectious FFU and CFU assays.
MH-S cells (5 × 104 cells per well of a 24-well plate) were plated and allowed to adhere overnight, while monocytes (1 × 105 cells per well of a 24-well plate) were plated and differentiated to hMDMs for 7 days. The cells were then mock infected or infected with WT or dotA mutant C. burnetii in 0.25 ml growth medium for 2 h at 37°C in 5% CO2, washed extensively with PBS, and incubated in 0.5 ml growth medium. At the indicated time points, the cells were treated with different concentrations of either recombinant mouse IL-17A or recombinant human IL-17A (catalog number 14-8179-62; Thermo Fisher Scientific) for 24 h. The cells were lysed in sterile water for 5 min and analyzed by a focus-forming unit (FFU) assay as previously described (89). For the CFU assay, the released bacteria were diluted 1:5 in ACCM-2 and plated in 2-fold serial dilutions onto 0.25% ACCM-2 agarose plates (45). The plates were incubated for 7 to 9 days at 37°C in 2.5% O2 and 5% CO2, and the number of colonies was counted to measure bacterial viability. Each experiment was performed in biological duplicate, and the bacteria were spotted in triplicate.
Antibody neutralization.
MH-S cells (5 × 104) were mock infected or infected with WT or dotA mutant C. burnetii in a 24-well plate. At 24 hpi, the cells were treated with 50 or 100 ng/ml of IL-17A in the presence or absence of 2 μg/ml of anti-IL-17Ra monoclonal antibody (catalog number MAB4481; Thermo Fisher Scientific) for 24 h. Bacteria were released by water lysis and analyzed by the CFU assay as described above. Each experiment was performed in biological duplicate, and the bacteria were spotted in triplicate.
Data analysis.
Statistical analyses were performed using ordinary one-way analysis of variance (ANOVA) with Dunnett's or Turkey's multiple-comparison test in Prism (version 7) software (GraphPad Software, Inc., La Jolla, CA).
Supplementary Material
ACKNOWLEDGMENTS
We thank Mark Kaplan and Dhritiman Samanta for helpful discussions and James Ford, Hongyu Gao, and Yunlong Liu for assistance with RNA sequencing and bioinformatics.
This research was supported by the National Institute of Allergy and Infectious Diseases, NIH (AI121786 to S.D.G.; 5K08AI125682 to T.M.T.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00532-18.
REFERENCES
- 1.Maurin M, Raoult D. 1999. Q fever. Clin Microbiol Rev 12:518–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mazokopakis EE, Karefilakis CM, Starakis IK. 2010. Q fever endocarditis. Infect Disord Drug Targets 10:27–31. doi: 10.2174/187152610790410918. [DOI] [PubMed] [Google Scholar]
- 3.Ackland JR, Worswick DA, Marmion BP. 1994. Vaccine prophylaxis of Q fever. A follow-up study of the efficacy of Q-Vax (CSL) 1985-1990. Med J Aust 160:704–708. [PubMed] [Google Scholar]
- 4.Kampschreur LM, Dekker S, Hagenaars JC, Lestrade PJ, Renders NH, de Jager-Leclercq MG, Hermans MH, Groot CA, Groenwold RH, Hoepelman AI, Wever PC, Oosterheert JJ. 2012. Identification of risk factors for chronic Q fever, the Netherlands. Emerg Infect Dis 18:563–570. doi: 10.3201/eid1804.111478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjork A, Marsden-Haug N, Nett RJ, Kersh GJ, Nicholson W, Gibson D, Szymanski T, Emery M, Kohrs P, Woodhall D, Anderson AD. 2014. First reported multistate human Q fever outbreak in the United States, 2011. Vector Borne Zoonotic Dis 14:111–117. doi: 10.1089/vbz.2012.1202. [DOI] [PubMed] [Google Scholar]
- 6.Alonso E, Lopez-Etxaniz I, Hurtado A, Liendo P, Urbaneja F, Aspiritxaga I, Olaizola JI, Pinero A, Arrazola I, Barandika JF, Hernaez S, Muniozguren N, Garcia-Perez AL. 2015. Q fever outbreak among workers at a waste-sorting plant. PLoS One 10:e0138817. doi: 10.1371/journal.pone.0138817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Archer BN, Hallahan C, Stanley P, Seward K, Lesjak M, Hope K, Brown A. 2017. Atypical outbreak of Q fever affecting low-risk residents of a remote rural town in New South Wales. Commun Dis Intell Q Rep 41:E125–E133. [PubMed] [Google Scholar]
- 8.Porter SR, Czaplicki G, Mainil J, Horii Y, Misawa N, Saegerman C. 2011. Q fever in Japan: an update review. Vet Microbiol 149:298–306. doi: 10.1016/j.vetmic.2010.11.017. [DOI] [PubMed] [Google Scholar]
- 9.Amitai Z, Bromberg M, Bernstein M, Raveh D, Keysary A, David D, Pitlik S, Swerdlow D, Massung R, Rzotkiewicz S, Halutz O, Shohat T. 2010. A large Q fever outbreak in an urban school in central Israel. Clin Infect Dis 50:1433–1438. doi: 10.1086/652442. [DOI] [PubMed] [Google Scholar]
- 10.Stein A, Louveau C, Lepidi H, Ricci F, Baylac P, Davoust B, Raoult D. 2005. Q fever pneumonia: virulence of Coxiella burnetii pathovars in a murine model of aerosol infection. Infect Immun 73:2469–2477. doi: 10.1128/IAI.73.4.2469-2477.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khavkin T, Tabibzadeh SS. 1988. Histologic, immunofluorescence, and electron microscopic study of infectious process in mouse lung after intranasal challenge with Coxiella burnetii. Infect Immun 56:1792–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hackstadt T, Williams JC. 1981. Biochemical stratagem for obligate parasitism of eukaryotic cells by Coxiella burnetii. Proc Natl Acad Sci U S A 78:3240–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howe D, Melnicakova J, Barak I, Heinzen RA. 2003. Maturation of the Coxiella burnetii parasitophorous vacuole requires bacterial protein synthesis but not replication. Cell Microbiol 5:469–480. doi: 10.1046/j.1462-5822.2003.00293.x. [DOI] [PubMed] [Google Scholar]
- 14.Beare PA, Gilk SD, Larson CL, Hill J, Stead CM, Omsland A, Cockrell DC, Howe D, Voth DE, Heinzen RA. 2011. Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. mBio 2:e00175-. doi: 10.1128/mBio.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Schaik EJ, Chen C, Mertens K, Weber MM, Samuel JE. 2013. Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat Rev Microbiol 11:561–573. doi: 10.1038/nrmicro3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu J, Luo ZQ. 2017. Legionella and Coxiella effectors: strength in diversity and activity. Nat Rev Microbiol 15:591–605. doi: 10.1038/nrmicro.2017.67. [DOI] [PubMed] [Google Scholar]
- 17.Chen C, Banga S, Mertens K, Weber MM, Gorbaslieva I, Tan Y, Luo ZQ, Samuel JE. 2010. Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc Natl Acad Sci U S A 107:21755–21760. doi: 10.1073/pnas.1010485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voth DE, Beare PA, Howe D, Sharma UM, Samoilis G, Cockrell DC, Omsland A, Heinzen RA. 2011. The Coxiella burnetii cryptic plasmid is enriched in genes encoding type IV secretion system substrates. J Bacteriol 193:1493–1503. doi: 10.1128/JB.01359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lifshitz Z, Burstein D, Peeri M, Zusman T, Schwartz K, Shuman HA, Pupko T, Segal G. 2013. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc Natl Acad Sci U S A 110:E707–E715. doi: 10.1073/pnas.1215278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cunha LD, Ribeiro JM, Fernandes TD, Massis LM, Khoo CA, Moffatt JH, Newton HJ, Roy CR, Zamboni DS. 2015. Inhibition of inflammasome activation by Coxiella burnetii type IV secretion system effector IcaA. Nat Commun 6:10205. doi: 10.1038/ncomms10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. 2013. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science 341:1250–1253. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszynski A, Forsberg LS, Carlson RW, Dixit VM. 2013. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 341:1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- 23.Brooke RJ, Kretzschmar ME, Mutters NT, Teunis PF. 2013. Human dose response relation for airborne exposure to Coxiella burnetii. BMC Infect Dis 13:488. doi: 10.1186/1471-2334-13-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Q, Martin RJ, Rino JG, Breed R, Torres RM, Chu HW. 2007. IL-23-dependent IL-17 production is essential in neutrophil recruitment and activity in mouse lung defense against respiratory Mycoplasma pneumoniae infection. Microbes Infect 9:78–86. doi: 10.1016/j.micinf.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gopal R, Monin L, Slight S, Uche U, Blanchard E, Fallert Junecko BA, Ramos-Payan R, Stallings CL, Reinhart TA, Kolls JK, Kaushal D, Nagarajan U, Rangel-Moreno J, Khader SA. 2014. Unexpected role for IL-17 in protective immunity against hypervirulent Mycobacterium tuberculosis HN878 infection. PLoS Pathog 10:e1004099. doi: 10.1371/journal.ppat.1004099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin Y, Ritchea S, Logar A, Slight S, Messmer M, Rangel-Moreno J, Guglani L, Alcorn JF, Strawbridge H, Park SM, Onishi R, Nyugen N, Walter MJ, Pociask D, Randall TD, Gaffen SL, Iwakura Y, Kolls JK, Khader SA. 2009. Interleukin-17 is required for T helper 1 cell immunity and host resistance to the intracellular pathogen Francisella tularensis. Immunity 31:799–810. doi: 10.1016/j.immuni.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimizuka Y, Kimura S, Saga T, Ishii M, Hasegawa N, Betsuyaku T, Iwakura Y, Tateda K, Yamaguchi K. 2012. Roles of interleukin-17 in an experimental Legionella pneumophila pneumonia model. Infect Immun 80:1121–1127. doi: 10.1128/IAI.05544-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulye M, Zapata B, Gilk SD. 2018. Altering lipid droplet homeostasis affects Coxiella burnetii intracellular growth. PLoS One 13:e0192215. doi: 10.1371/journal.pone.0192215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newton HJ, McDonough JA, Roy CR. 2013. Effector protein translocation by the Coxiella burnetii Dot/Icm type IV secretion system requires endocytic maturation of the pathogen-occupied vacuole. PLoS One 8:e54566. doi: 10.1371/journal.pone.0054566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiner J III, Domaszewska T. 2016. tmod: an R package for general and multivariate enrichment analysis. PeerJ Preprints 4:e2420v1 https://peerj.com/preprints/2420/. [Google Scholar]
- 31.Bradley WP, Boyer MA, Nguyen HT, Birdwell LD, Yu J, Ribeiro JM, Weiss SR, Zamboni DS, Roy CR, Shin S. 2016. Primary role for Toll-like receptor-driven tumor necrosis factor rather than cytosolic immune detection in restricting Coxiella burnetii phase II replication within mouse macrophages. Infect Immun 84:998–1015. doi: 10.1128/IAI.01536-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onishi RM, Gaffen SL. 2010. Interleukin-17 and its target genes: mechanisms of interleukin-17 function in disease. Immunology 129:311–321. doi: 10.1111/j.1365-2567.2009.03240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beare PA, Larson CL, Gilk SD, Heinzen RA. 2012. Two systems for targeted gene deletion in Coxiella burnetii. Appl Environ Microbiol 78:4580–4589. doi: 10.1128/AEM.00881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenfeld Y, Shai Y. 2006. Lipopolysaccharide (endotoxin)-host defense antibacterial peptides interactions: role in bacterial resistance and prevention of sepsis. Biochim Biophys Acta 1758:1513–1522. doi: 10.1016/j.bbamem.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 35.Ishigame H, Kakuta S, Nagai T, Kadoki M, Nambu A, Komiyama Y, Fujikado N, Tanahashi Y, Akitsu A, Kotaki H, Sudo K, Nakae S, Sasakawa C, Iwakura Y. 2009. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity 30:108–119. doi: 10.1016/j.immuni.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 36.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. 2004. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 37.Karlsen JR, Borregaard N, Cowland JB. 2010. Induction of neutrophil gelatinase-associated lipocalin expression by co-stimulation with interleukin-17 and tumor necrosis factor-alpha is controlled by IkappaB-zeta but neither by C/EBP-beta nor C/EBP-delta. J Biol Chem 285:14088–14100. doi: 10.1074/jbc.M109.017129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suk K. 2016. Lipocalin-2 as a therapeutic target for brain injury: an astrocentric perspective. Prog Neurobiol 144:158–172. doi: 10.1016/j.pneurobio.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 39.Ye P, Garvey PB, Zhang P, Nelson S, Bagby G, Summer WR, Schwarzenberger P, Shellito JE, Kolls JK. 2001. Interleukin-17 and lung host defense against Klebsiella pneumoniae infection. Am J Respir Cell Mol Biol 25:335–340. doi: 10.1165/ajrcmb.25.3.4424. [DOI] [PubMed] [Google Scholar]
- 40.Appay V, Rowland-Jones SL. 2001. RANTES: a versatile and controversial chemokine. Trends Immunol 22:83–87. doi: 10.1016/S1471-4906(00)01812-3. [DOI] [PubMed] [Google Scholar]
- 41.Cooper AM. 2009. IL-17 and anti-bacterial immunity: protection versus tissue damage. Eur J Immunol 39:649–652. doi: 10.1002/eji.200839090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khader SA, Gopal R. 2010. IL-17 in protective immunity to intracellular pathogens. Virulence 1:423–427. doi: 10.4161/viru.1.5.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guglani L, Khader SA. 2010. Th17 cytokines in mucosal immunity and inflammation. Curr Opin HIV AIDS 5:120–127. doi: 10.1097/COH.0b013e328335c2f6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coleman SA, Fischer ER, Howe D, Mead DJ, Heinzen RA. 2004. Temporal analysis of Coxiella burnetii morphological differentiation. J Bacteriol 186:7344–7352. doi: 10.1128/JB.186.21.7344-7352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vallejo Esquerra E, Yang H, Sanchez SE, Omsland A. 2017. Physicochemical and nutritional requirements for axenic replication suggest physiological basis for Coxiella burnetii niche restriction. Front Cell Infect Microbiol 7:190. doi: 10.3389/fcimb.2017.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medzhitov R. 2007. Recognition of microorganisms and activation of the immune response. Nature 449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 47.Baxt LA, Garza-Mayers AC, Goldberg MB. 2013. Bacterial subversion of host innate immune pathways. Science 340:697–701. doi: 10.1126/science.1235771. [DOI] [PubMed] [Google Scholar]
- 48.Cunha LD, Zamboni DS. 2013. Subversion of inflammasome activation and pyroptosis by pathogenic bacteria. Front Cell Infect Microbiol 3:76. doi: 10.3389/fcimb.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luhrmann A, Roy CR. 2007. Coxiella burnetii inhibits activation of host cell apoptosis through a mechanism that involves preventing cytochrome c release from mitochondria. Infect Immun 75:5282–5289. doi: 10.1128/IAI.00863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luhrmann A, Nogueira CV, Carey KL, Roy CR. 2010. Inhibition of pathogen-induced apoptosis by a Coxiella burnetii type IV effector protein. Proc Natl Acad Sci U S A 107:18997–19001. doi: 10.1073/pnas.1004380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klingenbeck L, Eckart RA, Berens C, Luhrmann A. 2013. The Coxiella burnetii type IV secretion system substrate CaeB inhibits intrinsic apoptosis at the mitochondrial level. Cell Microbiol 15:675–687. doi: 10.1111/cmi.12066. [DOI] [PubMed] [Google Scholar]
- 52.Zamboni DS, Campos MA, Torrecilhas AC, Kiss K, Samuel JE, Golenbock DT, Lauw FN, Roy CR, Almeida IC, Gazzinelli RT. 2004. Stimulation of Toll-like receptor 2 by Coxiella burnetii is required for macrophage production of pro-inflammatory cytokines and resistance to infection. J Biol Chem 279:54405–54415. doi: 10.1074/jbc.M410340200. [DOI] [PubMed] [Google Scholar]
- 53.Dellacasagrande J, Capo C, Raoult D, Mege JL. 1999. IFN-gamma-mediated control of Coxiella burnetii survival in monocytes: the role of cell apoptosis and TNF. J Immunol 162:2259–2265. [PubMed] [Google Scholar]
- 54.Dellacasagrande J, Ghigo E, Raoult D, Capo C, Mege JL. 2002. IFN-gamma-induced apoptosis and microbicidal activity in monocytes harboring the intracellular bacterium Coxiella burnetii require membrane TNF and homotypic cell adherence. J Immunol 169:6309–6315. doi: 10.4049/jimmunol.169.11.6309. [DOI] [PubMed] [Google Scholar]
- 55.Fettelschoss A, Kistowska M, LeibundGut-Landmann S, Beer HD, Johansen P, Senti G, Contassot E, Bachmann MF, French LE, Oxenius A, Kundig TM. 2011. Inflammasome activation and IL-1beta target IL-1alpha for secretion as opposed to surface expression. Proc Natl Acad Sci U S A 108:18055–18060. doi: 10.1073/pnas.1109176108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mariathasan S, Monack DM. 2007. Inflammasome adaptors and sensors: intracellular regulators of infection and inflammation. Nat Rev Immunol 7:31–40. doi: 10.1038/nri1997. [DOI] [PubMed] [Google Scholar]
- 57.Furuoka M, Ozaki K, Sadatomi D, Mamiya S, Yonezawa T, Tanimura S, Takeda K. 2016. TNF-alpha induces caspase-1 activation independently of simultaneously induced NLRP3 in 3T3-L1 cells. J Cell Physiol 231:2761–2767. doi: 10.1002/jcp.25385. [DOI] [PubMed] [Google Scholar]
- 58.Zamboni DS, Mortara RA, Freymuller E, Rabinovitch M. 2002. Mouse resident peritoneal macrophages partially control in vitro infection with Coxiella burnetii phase II. Microbes Infect 4:591–598. doi: 10.1016/S1286-4579(02)01577-0. [DOI] [PubMed] [Google Scholar]
- 59.Yoshiie K, Matayoshi S, Fujimura T, Maeno N, Oda H. 1999. Induced production of nitric oxide and sensitivity of alveolar macrophages derived from mice with different sensitivity to Coxiella burnetii. Acta Virol 43:273–278. [PubMed] [Google Scholar]
- 60.Zamboni DS. 2004. Genetic control of natural resistance of mouse macrophages to Coxiella burnetii infection in vitro: macrophages from restrictive strains control parasitophorous vacuole maturation. Infect Immun 72:2395–2399. doi: 10.1128/IAI.72.4.2395-2399.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 62.Zhao GN, Jiang DS, Li H. 2015. Interferon regulatory factors: at the crossroads of immunity, metabolism, and disease. Biochim Biophys Acta 1852:365–378. doi: 10.1016/j.bbadis.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 63.Monroe KM, McWhirter SM, Vance RE. 2010. Induction of type I interferons by bacteria. Cell Microbiol 12:881–890. doi: 10.1111/j.1462-5822.2010.01478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hedges JF, Robison A, Kimmel E, Christensen K, Lucas E, Ramstead A, Jutila MA. 2016. Type I interferon counters or promotes Coxiella burnetii replication dependent on tissue. Infect Immun 84:1815–1825. doi: 10.1128/IAI.01540-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Plumlee CR, Lee C, Beg AA, Decker T, Shuman HA, Schindler C. 2009. Interferons direct an effective innate response to Legionella pneumophila infection. J Biol Chem 284:30058–30066. doi: 10.1074/jbc.M109.018283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schiavoni G, Mauri C, Carlei D, Belardelli F, Pastoris MC, Proietti E. 2004. Type I IFN protects permissive macrophages from Legionella pneumophila infection through an IFN-gamma-independent pathway. J Immunol 173:1266–1275. doi: 10.4049/jimmunol.173.2.1266. [DOI] [PubMed] [Google Scholar]
- 67.Monroe KM, McWhirter SM, Vance RE. 2009. Identification of host cytosolic sensors and bacterial factors regulating the type I interferon response to Legionella pneumophila. PLoS Pathog 5:e1000665. doi: 10.1371/journal.ppat.1000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Curtis MM, Way SS. 2009. Interleukin-17 in host defence against bacterial, mycobacterial and fungal pathogens. Immunology 126:177–185. doi: 10.1111/j.1365-2567.2008.03017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jin W, Dong C. 2013. IL-17 cytokines in immunity and inflammation. Emerg Microbes Infect 2:e60. doi: 10.1038/emi.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu J, Qu H, Li Q, Ye L, Ma G, Wan H. 2013. The responses of gammadelta T-cells against acute Pseudomonas aeruginosa pulmonary infection in mice via interleukin-17. Pathog Dis 68:44–51. doi: 10.1111/2049-632X.12043. [DOI] [PubMed] [Google Scholar]
- 71.Chen J, Liao MY, Gao XL, Zhong Q, Tang TT, Yu X, Liao YH, Cheng X. 2013. IL-17A induces pro-inflammatory cytokines production in macrophages via MAPKinases, NF-kappaB and AP-1. Cell Physiol Biochem 32:1265–1274. doi: 10.1159/000354525. [DOI] [PubMed] [Google Scholar]
- 72.Barin JG, Baldeviano GC, Talor MV, Wu L, Ong S, Quader F, Chen P, Zheng D, Caturegli P, Rose NR, Cihakova D. 2012. Macrophages participate in IL-17-mediated inflammation. Eur J Immunol 42:726–736. doi: 10.1002/eji.201141737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Elliott A, Peng Y, Zhang G. 2013. Coxiella burnetii interaction with neutrophils and macrophages in vitro and in SCID mice following aerosol infection. Infect Immun 81:4604–4614. doi: 10.1128/IAI.00973-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Elliott A, Schoenlaub L, Freches D, Mitchell W, Zhang G. 2015. Neutrophils play an important role in protective immunity against Coxiella burnetii infection. Infect Immun 83:3104–3113. doi: 10.1128/IAI.00042-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schneider TJH, Liesenfeld O, Steinhoff D, Riecken EO, Zeitz M, Ullrich R. 1997. The number and proportion of Vgamma9 Vdelta2 T cells rise significantly in the peripheral blood of patients after the onset of acute Coxiella burnetii infection. Clin Infect Dis 24:261–264. doi: 10.1093/clinids/24.2.261. [DOI] [PubMed] [Google Scholar]
- 76.Papotto PH, Ribot JC, Silva-Santos B. 2017. IL-17(+) gammadelta T cells as kick-starters of inflammation. Nat Immunol 18:604–611. doi: 10.1038/ni.3726. [DOI] [PubMed] [Google Scholar]
- 77.Sonder SU, Saret S, Tang W, Sturdevant DE, Porcella SF, Siebenlist U. 2011. IL-17-induced NF-kappaB activation via CIKS/Act1: physiologic significance and signaling mechanisms. J Biol Chem 286:12881–12890. doi: 10.1074/jbc.M110.199547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hata K, Andoh A, Shimada M, Fujino S, Bamba S, Araki Y, Okuno T, Fujiyama Y, Bamba T. 2002. IL-17 stimulates inflammatory responses via NF-kappaB and MAP kinase pathways in human colonic myofibroblasts. Am J Physiol Gastrointest Liver Physiol 282:G1035–G1044. doi: 10.1152/ajpgi.00494.2001. [DOI] [PubMed] [Google Scholar]
- 79.Mahapatra S, Gallaher B, Smith SC, Graham JG, Voth DE, Shaw EI. 2016. Coxiella burnetii employs the Dot/Icm type IV secretion system to modulate host NF-kappaB/RelA activation. Front Cell Infect Microbiol 6:188. doi: 10.3389/fcimb.2016.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hatano E, Bennett BL, Manning AM, Qian T, Lemasters JJ, Brenner DA. 2001. NF-kappaB stimulates inducible nitric oxide synthase to protect mouse hepatocytes from TNF-alpha- and Fas-mediated apoptosis. Gastroenterology 120:1251–1262. doi: 10.1053/gast.2001.23239. [DOI] [PubMed] [Google Scholar]
- 81.Anrather J, Racchumi G, Iadecola C. 2006. NF-kappaB regulates phagocytic NADPH oxidase by inducing the expression of gp91phox. J Biol Chem 281:5657–5667. doi: 10.1074/jbc.M506172200. [DOI] [PubMed] [Google Scholar]
- 82.Jablonski KA, Amici SA, Webb LM, Ruiz-Rosado JDD, Popovich PG, Partida-Sanchez S, Guerau-de-Arellano M. 2015. Novel markers to delineate murine M1 and M2 macrophages. PLoS One 10:e0145342. doi: 10.1371/journal.pone.0145342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Benoit M, Barbarat B, Bernard A, Olive D, Mege JL. 2008. Coxiella burnetii, the agent of Q fever, stimulates an atypical M2 activation program in human macrophages. Eur J Immunol 38:1065–1070. doi: 10.1002/eji.200738067. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Q, Atsuta I, Liu S, Chen C, Shi S, Shi S, Le AD. 2013. IL-17-mediated M1/M2 macrophage alteration contributes to pathogenesis of bisphosphonate-related osteonecrosis of the jaws. Clin Cancer Res 19:3176–3188. doi: 10.1158/1078-0432.CCR-13-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eid RE, Rao DA, Zhou J, Lo SF, Ranjbaran H, Gallo A, Sokol SI, Pfau S, Pober JS, Tellides G. 2009. Interleukin-17 and interferon-gamma are produced concomitantly by human coronary artery-infiltrating T cells and act synergistically on vascular smooth muscle cells. Circulation 119:1424–1432. doi: 10.1161/CIRCULATIONAHA.108.827618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cockrell DC, Beare PA, Fischer ER, Howe D, Heinzen RA. 2008. A method for purifying obligate intracellular Coxiella burnetii that employs digitonin lysis of host cells. J Microbiol Methods 72:321–325. doi: 10.1016/j.mimet.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Omsland A, Cockrell DC, Howe D, Fischer ER, Virtaneva K, Sturdevant DE, Porcella SF, Heinzen RA. 2009. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc Natl Acad Sci U S A 106:4430–4434. doi: 10.1073/pnas.0812074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. 2011. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mulye M, Samanta D, Winfree S, Heinzen RA, Gilk SD. 2017. Elevated cholesterol in the Coxiella burnetii intracellular niche is bacteriolytic. mBio 8:e02313-16. doi: 10.1128/mBio.02313-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.