Abstract
Cytochrome oxidase (COX) is a hetero-oligomeric complex of the mitochondrial inner membrane that reduces molecular oxygen to water, a reaction coupled to proton transfer from the mitochondrial matrix to the intermembrane space. In the yeast Saccharomyces cerevisiae, COX is composed of 11–13 different polypeptide subunits. Here, using pulse labeling of mitochondrial gene products in isolated yeast mitochondria, combined with purification of tagged COX subunits and ancillary factors, we studied the Cox2p assembly intermediates. Analysis of radiolabeled Cox2p obtained in pulldown assays by native gel electrophoresis revealed the existence of several assembly intermediates, the largest of which had an estimated mass of 450–550 kDa. None of the other known subunits of COX were present in these Cox2p intermediates. This was also true for the several ancillary factors having still undefined functions in COX assembly. In agreement with earlier evidence, Cox18p and Cox20p, previously shown to be involved in processing and in membrane insertion of the Cox2p precursor, were found to be associated with the two largest Cox2p intermediates. A small fraction of the Cox2p module contained Sco1p and Coa6p, which have been implicated in metalation of the binuclear copper site on this subunit. Our results indicate that following its insertion into the mitochondrial inner membrane, Cox2p assembles as a stand-alone protein with the compositionally more complex Cox1p and Cox3p modules.
Keywords: mitochondria, cytochrome c oxidase (complex IV), yeast genetics, mitochondrial respiratory chain complex, mitochondrial DNA (mtDNA), cytochrome oxidase assembly, Cox2p module, biogenesis, Saccharomyces cerevisiae
Introduction
Mitochondrial cytochrome oxidase (COX)3 is a hetero-oligomeric complex of the inner membrane that uses the electrons of ferrocytochrome c to reduce molecular oxygen to water, a reaction that is coupled to the transfer of protons from the internal matrix compartment to the intermembrane space (1). In Saccharomyces cerevisiae COX is composed of 11 different polypeptides of which 3, encoded in mitochondrial DNA, form the catalytic core with the heme A and copper prosthetic groups. The remaining 8 subunits are gene products of nuclear DNA that physically surround the 3 core subunit. These structural subunits do not appear to participate in either the electron or proton transferring activities of COX and are thought to shield and extend the half-life of this complex. Eleven COX subunits have homologues in bovine COX (1). Recently, two new subunits, Cox26p and Cox16p, have been reported to be physically associated with yeast COX. Cox26p is stoichiometric with the other subunits of COX in the supercomplexes (2, 3). Cox16p, a mitochondrial protein previously thought to be an assembly factor (4), was recently shown to be physically associated with yeast COX (5).
There has been a surge of interest in the past few years on the part of several laboratories to clarify the mechanism by which mitochondria biosynthesize this important respiratory complex (6–10). Pulse-chase analysis of COX intermediates in isolated mitochondria have revealed that the core Cox1p and Cox3p subunits assemble independently with their own proprietary set of nuclear-encoded subunits before the resultant modules assemble with each other. Most, if not all, structural subunits have been found to be associated with Cox1p and Cox3p (11, 12). In addition to the structural subunits, the Cox1p module also contains translational regulators Mss51p, Cox14p, and Coa3p as well as several other factors involved in the maturation of the protein (13–19).
In the present study we have analyzed the composition of the Cox2p module. Pulse-chase labeling of mitochondria disclosed the existence of several Cox2p assembly intermediates, the largest of which has an apparent mass of 450–550 kDa. Pulldown assays of tagged subunits of COX and of accessory factors known to be required for COX assembly, some with still poorly understood functions, were not found to be present in the Cox2p intermediates. Cox20p and Cox18p, with functions in membrane insertion and processing of the Cox2p precursor, have been shown to interact with Cox2p in yeast and human mitochondria (20, 21). Our evidence indicates that Cox18 and Cox20 interact sequentially with the two largest Cox2p intermediates. Based on these results we propose that unlike Cox1p and Cox3p that acquire specific subsets of nuclear-encoded subunits before assembling into COX, Cox2p interacts with the Cox1p and Cox3p assembly modules as a stand-alone core protein.
Results
Properties of strains expressing tagged Cox2p
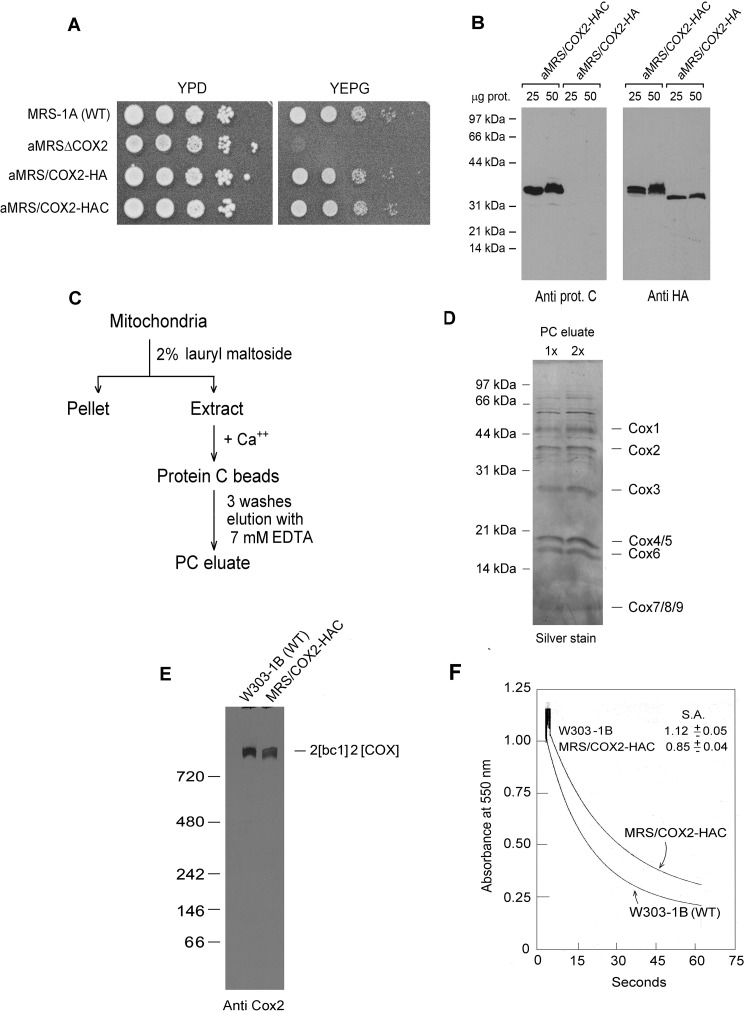
To study assembly of Cox2p in yeast, we fused the COX2 gene to two different sequences, one coding for the hemagglutinin tag (HA) and the other for a tandem HA followed by a protein C tag (HAC). Substitution of the fusion genes (COX2-HA and COX2-HAC) for the cox2 null allele of aMRSΔCOX2 restored growth of this mutant on nonfermentable carbon sources (Fig. 1A). Strains containing the fusion genes in their mitochondrial DNA were confirmed by Western blot analysis to express Cox2p with the corresponding tags (Fig. 1B).
Figure 1.
Properties of yeast expressing HA- and HAC-tagged Cox2p. A, overnight cultures of the parental WT MRS-1A, aMRSΔCOX2, aMRS/COX2-HA, and aMRS/COX2-HAC were serially diluted and spotted on rich glucose (YPD) and rich glycerol/ethanol (YEPG) plates. The photograph was taken after 2 days' incubation at 30 °C. B, mitochondria were prepared from aMRS/COX2-HA and aMRS/COX2-HAC. The indicated amounts of mitochondria were applied to a 12% polyacrylamide gel and separated by SDS-PAGE. Western blots were incubated with polyclonal antibody against the protein C epitope followed by secondary peroxidase-coupled antibody against rabbit γ globulin. Proteins were visualized with SuperSignal chemiluminescent substrate kit (Pierce). C, protocol used to purify HAC-tagged cytochrome oxidase. D, two different concentrations of cytochrome oxidase purified as in C were depolymerized in Laemmli sample buffer (43) and separated by SDS-PAGE on a 12% polyacrylamide gel. Proteins were stained with silver. The subunits of cytochrome oxidase are identified in the right-hand margin. E, mitochondria from the indicated strains were extracted and separated by BN-PAGE. Proteins were transferred to a PVDF membrane, reacted with a polyclonal antibody against Cox2p and further processed as in B. F, mitochondria of the WT strain W303–1B and MRS/COX2-HAC expressing tagged Cox2p were assayed for oxidation of ferrocytochrome c at 550 nm. S. A. refers to specific activity expressed as micromoles of cytochrome c oxidized per minute per milligram protein.
The presence of the HAC tag on Cox1p and Cox3p was used previously to affinity purify COX intermediates and supercomplexes containing the fully assembled enzyme (10, 12). The accessibility of the tag of Cox2p-HAC to the protein C antibody and the extent of purification attained by a single affinity step was examined by extracting mitochondria from aMRS/COX2-HAC with lauryl maltoside and purifying the extract on the protein C antibody beads (Fig. 1C). The purity of COX in the fraction eluted from the protein antibody beads was assessed by silver staining of proteins separated by SDS-PAGE on a 17.5% polyacrylamide gel. With the exception of a few high molecular contaminants, the other proteins in this fraction corresponded to known subunits of COX (Fig. 1D). Immunochemical (Fig. 1E) and enzymatic (Fig. 1F) assays of COX indicated that the HAC tag on Cox2p reduced both COX assembly and activity by ∼20%.
Detection of Cox2p intermediates in mitochondria
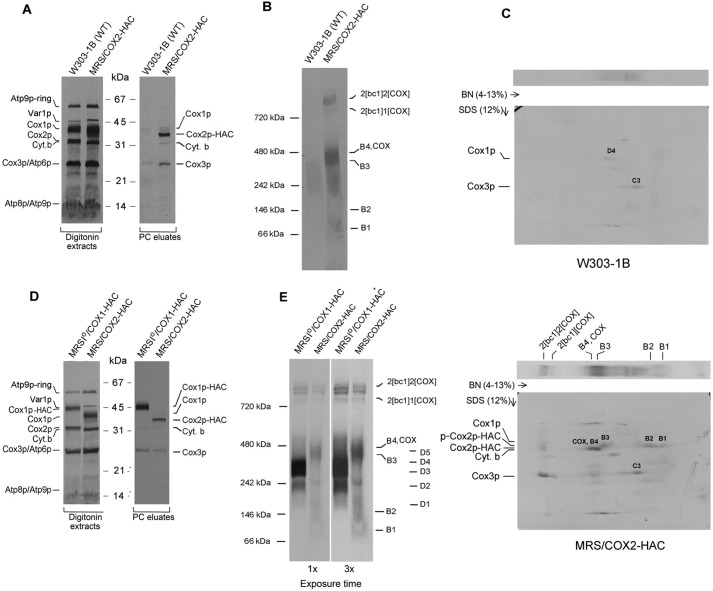
Mitochondria from the parental WT, MRS/COX2-HAC, and MRSIO/COX1-HAC, the latter two expressing Cox2p-HAC and Cox1p-HAC, respectively, were radiolabeled with [35S]methionine plus [35S]cysteine. The mitochondria were extracted with digitonin and purified on protein C antibody beads. Total mitochondrial proteins and the purified fraction eluted from the antibody beads with EDTA were separated by SDS-PAGE on a 12% polyacrylamide gel, transferred to nitrocellulose, and exposed to X-ray film. The autoradiograph disclosed that the EDTA eluate obtained from the MRS/COX2-HAC mitochondria contained predominantly Cox2p-HAC4 and less prominently Cox1p, Cox3p, and cytochrome b bands (Fig. 2A). The presence of the latter two subunits is consistent with the results of the blue native gel, which indicated co-purification on the protein C antibody beads of supercomplexes with 1 and 2 units of COX per 2 units of bc1 complex (Fig. 2B). Most of the Cox3p in the purified fraction is contributed by COX, some of which is associated with the supercomplexes. Some tag-independent Cox3p, however, is adsorbed to the antibody beads (10). The lack of stoichiometry of Cox1p with the other two COX subunits was observed previously in studies of Cox1p and Cox3p intermediates (10, 12) and can be explained by dilution of newly synthesized radiolabeled Cox1p by the large steady-state pool of the mitochondrial Cox1p (D3 and D4) intermediates (10).
Figure 2.
In organello labeling of Cox2p intermediates. A, mitochondria were isolated from respiratory competent haploid strains W303–1B and MRS/COX2-HAC. Mitochondria (250 μg protein) were labeled in vitro with [35S]methionine plus [35S]cysteine and extracted with digitonin as described previously (10). The digitonin extract (5% of total) and the fraction purified on protein C antibody beads as in Fig. 1D (15% of total) were separated by SDS-PAGE on a 12% polyacrylamide gel, transferred to nitrocellulose and exposed to X-ray film. B, the fraction purified on the protein C beads was separated by BN-PAGE on a 4–13% polyacrylamide gel, transferred to a PVDF membrane, and exposed to X-ray. C, the purified PC eluates from WT and from MRS/COX2-HAC were separated by BN-PAGE on a 4–13% polyacrylamide gel in the first dimension and by SDS-PAGE in the second dimension. The background bands detected in the autoradiogram of the WT control (2D gel of top panel) correspond to the D3 and D4 intermediates of the Cox1p module and the C3 intermediate of Cox3p module. The radiolabeled bands pulled down by Cox2p are the B1, B2, and B3 intermediates. The Cox2p precursor (p-Cox2p), the B4 intermediate and COX with mature Cox2p and the nonspecific adsorbed C3 intermediate with Cox3p (2D gel of lower panel). The migration of Cox1p, Cox2p-HAC, and the Cox2p precursor are marked in the margins. D and E, same as A and B, except that MRSIO/COX1-HAC, a strain expressing Cox1p with an HAC tag was used for comparison. The Western blotting of the native gel in the right panel was exposed three times longer.
Two bands (B1, B2) detected by BN-PAGE migrated as a single diffuse band on a 4–13% native gel. These bands migrated differently from Cox1p intermediates D1–D5 (Fig. 2E) (10). The presence of Cox2p in the B1 and B2 intermediates was confirmed by SDS-PAGE electrophoresis in the second dimension (Fig. 2C).
The third prominent region of radioactivity consisted of a broad band that migrated in the region of the COX dimer, slightly below the 480 kDa marker (Fig. 2, B and E). Our data suggest that this region includes two intermediates (B3 and B4) and that the latter overlaps with the COX dimer. This is supported by pulldown experiments with strains expressing either Cox18p or Cox20p with HAC tags (see below).
Consistent with its lower methionine and cysteine content, labeling of Cox2p is significantly less than of Cox1p and Cox3p (10, 12). This is evident by comparing labeling of Cox1p and Cox2p-HAC in digitonin extracts separated by SDS-PAGE (Fig. 2D) and of affinity purified Cox1p intermediates D3 and D4 with the Cox2p intermediates B3 and B4 separated by BN-PAGE (Fig. 2E). Because of the lower level of Cox2p labeling, longer exposure times to X-ray film were required, resulting in a higher background from nonspecific adsorption of radiolabeled proteins to the beads. This is especially true of the abundant Cox1p and Cox3p intermediates that have a tendency to adsorb nonspecifically to the protein C antibody beads (Fig. 2C, top panel).
Labeling and assembly of COX and the bc1 complex in mitochondria from cells grown in the presence of chloramphenicol
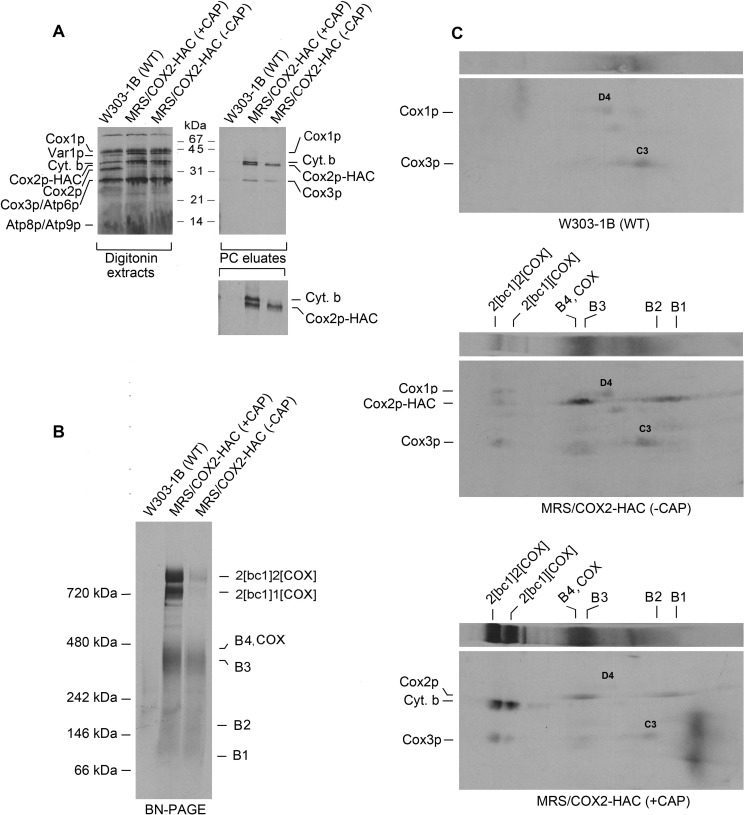
Growth of yeast in the presence of chloramphenicol, an inhibitor of mitochondrial but not cytoplasmic ribosomes, enhances translation of some mitochondrial gene products such as Atp9p, most likely by increasing the pool of nuclear encoded subunits that normally interact with their partners translated on mitochondrial ribosomes (23). Mitochondria of chloramphenicol-treated and nontreated cells did not show a significant difference in translation of either Cox1p or Cox2p. Labeling of cytochrome b, however, was significantly increased in mitochondria of cells that had been treated with chloramphenicol (Fig. 3A). The chloramphenicol treatment also elicited a significant increase in co-immunoadsorption with Cox2-HAC of the cytochrome b component of the bc1 complex (Fig. 3A). Blue native gels of affinity purified Cox2p-HAC reveal an increase of radiolabel in the COX/B3–B4 region and an even more significant increase of labeled supercomplexes (Fig. 3B). Two-dimensional (2D) BN-PAGE followed by SDS-PAGE indicated that the increase in radiolabeled supercomplexes was due largely to co-immunoadsorption of cytochrome b in the bc1 complex of the supercomplexes (Fig. 3C, lower panel).
Figure 3.
Effect of 2-h growth in the presence of chloramphenicol on translation of cytochrome b. A, mitochondria were isolated from the WT strain W303-1B and MRS/COX2-HAC, a strain expressing Cox2p-HAC, grown to early stationary phase in rich galactose (YPGal) medium without chloramphenicol (−CAP) and with an additional 2-h growth in YPGal containing 2 mg/ml chloramphenicol (+CAP). Mitochondria labeled for 20 min with [35S]methionine plus [35S]cysteine were extracted with digitonin and Cox2p-HAC purified on protein C antibody beads as described under “Materials and methods.” The digitonin extracts and purified fraction from the beads (PC eluates) were separated by SDS-PAGE (43) on a 17% polyacrylamide gel, transferred to nitrocellulose and exposed to an X-ray film. The radiolabeled mitochondrial gene products are identified in the margin. The migrations of Var1p relative to Cox1p and of Cox2p relative to cytochrome b are reversed at 17% compared with 12% acrylamide. The small inset panel of the PC eluates has been expanded to better show the separation of cytochrome b and Cox2p-HAC. B, the PC eluates of A were separated by BN-PAGE on a 4–13% polyacrylamide gel, transferred to a PVDF membrane and exposed to X-ray film. The bands corresponding to the supercomplexes, COX, and Cox2p intermediates are marked in the margin. C, the PC eluates of A were first separated by BN-PAGE as in B and by SDS-PAGE on a 12% polyacrylamide gel in the second dimension. The large area of radioactivity near the right edge of the lower gel is an artifact.
Pulse-chase of Cox2 intermediates
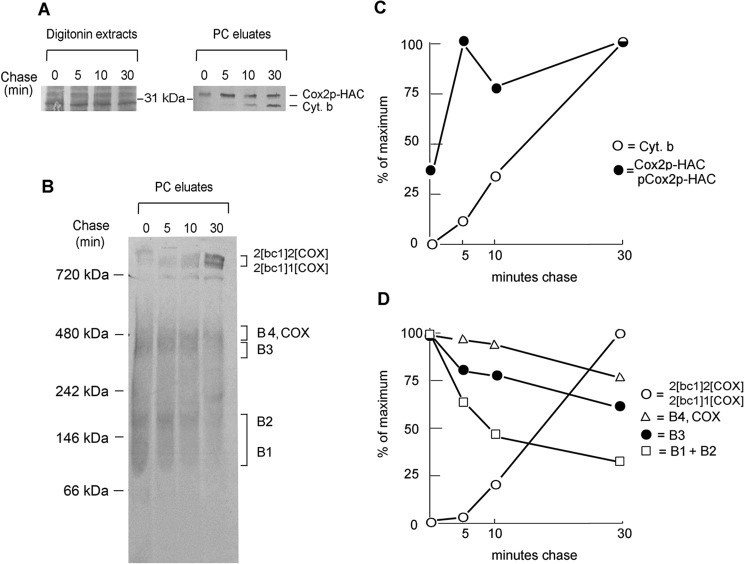
The precursor product relationship of the Cox2p assembly intermediates was studied by pulse-chase labeling of mitochondria expressing Cox2p-HAC. Mitochondria from cells that had been incubated in medium containing chloramphenicol for 2 h were pulsed for 5 min with radiolabeled methionine and cysteine and chased for different times following addition of puromycin and excess unlabeled methionine. The fractions obtained after extraction with digitonin and purification on protein C antibody beads were separated by SDS-PAGE and BN-PAGE. Quantitation of radioactivity in the supercomplexes and the B1 plus B2, the B3 and the B4/COX regions of the native gel indicated a time-dependent decrease of radiolabel from the intermediates and an increase of label in the supercomplexes (Fig. 4, B–D). It was not possible to distinguish what fraction of the decrease in the B1/COX region is contributed by the B1 intermediate and/or by COX as both would be expected to be incorporated into the supercomplexes.
Figure 4.
Pulse-chase analysis of mitochondria expressing Cox2p-HAC. A, mitochondria of MRS/COX2-HAC that were grown on YPGal with a subsequent incubation in the presence of chloramphenicol were labeled with [35S]methionine plus [35S]cysteine for 5 min. Translation was terminated by addition of puromycin and cold methionine and chased by further incubation for the indicated times. Mitochondria sampled at each time point were extracted with digitonin and Cox2p-HAC purified on protein C antibody beads (PC eluates). The digitonin extracts and PC eluates were separated by SDS-PAGE on a 12% polyacrylamide gel, transferred to nitrocellulose and exposed to X-ray. B, the PC eluates from A were separated by BN-PAGE on a 4–13% polyacrylamide gel, transferred to a PVDF membrane and exposed to X-ray film. C, radiolabeled Cox2p-HAC and cytochrome b on the Western blotting of A were quantified with a phosphorimager. D, the band of B corresponding to the radiolabeled supercomplexes, COX plus intermediates B4, B3, and B1 plus B2 were quantified with a phosphorimager.
The SDS-PAGE gel of the eluates from the protein C antibody beads shows an increase of radiolabeled cytochrome b during the chase (Fig. 4, A–C). This increase reflects the incorporation of cytochrome b into the bc1 complex that is incorporated into the supercomplexes and co-immunoadsorbed with Cox2p-HAC on the antibody beads.
Interaction of structural subunits with the Cox2p module
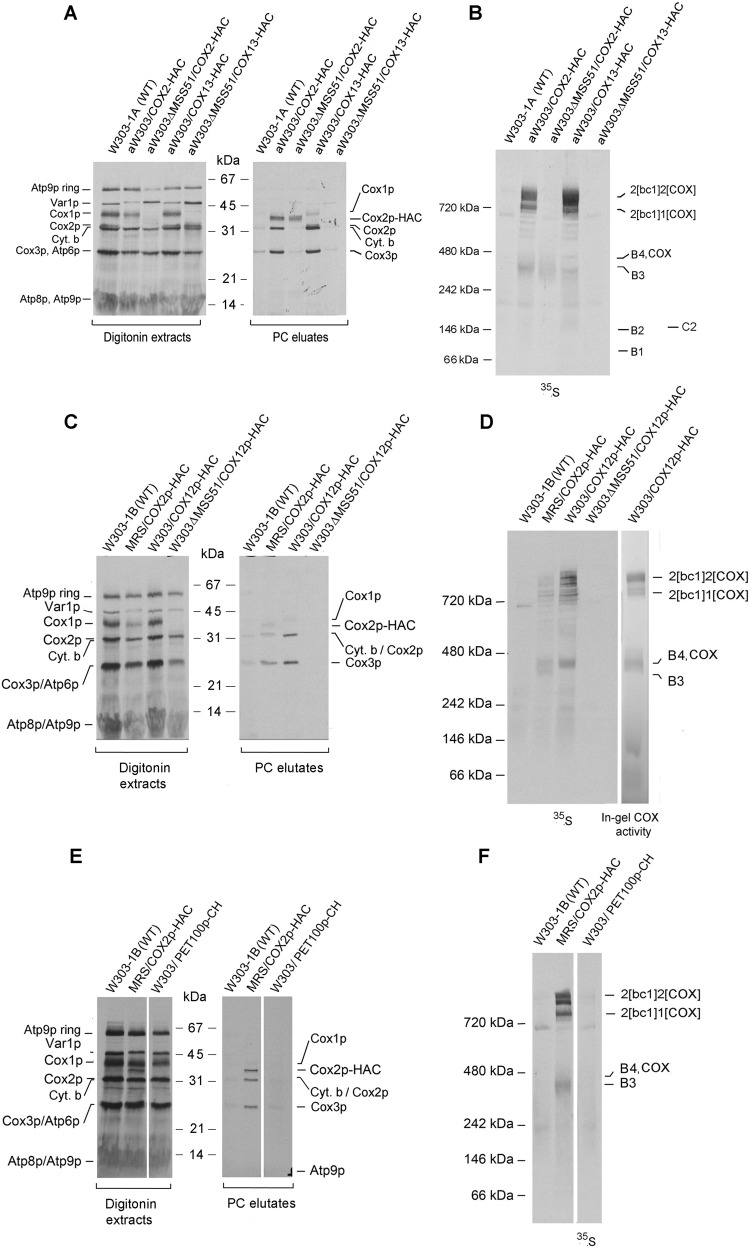
Subunits of COX imported into mitochondria from the cytoplasm were previously shown to be selectively associated with the Cox1p and Cox3p module (11, 12). The exception was Cox4p, which was present in both the Cox1p and Cox3p modules (12). To complete the analysis of this set of COX constituents, mitochondria were isolated from strains expressing Cox9p (subunit 7a), Cox12p (subunit 6b), and Cox16p (4, 5, 24) with an HAC or CH tag at their C termini. In prior studies all the other structural subunits had been assigned to either the Cox1p or Cox3p modules (10–12). Cox9p and Cox12p, in particular, were good candidates to be components of the Cox2p module as they both make direct contact with Cox2p in mature COX (25).
Co-immunoadsorption of Cox2p with the tagged proteins, including Cox2p as positive and Cox13p as negative controls, was also examined in an mss51 null mutant that lacks COX (22) and Cox1p intermediates (11). The presence of radiolabeled Cox2p-HAC was determined by SDS-PAGE, and the presence of Cox2p assembly intermediates was determined by BN-PAGE. As expected, the mss51 mutation did not affect pulldown on the protein C antibody beads of tagged Cox2p (Fig. 5, A and B). Significantly, radiolabeled Cox3p did not co-immunopurify with Cox2p-HAC in the mss51 mutant (Fig. 5F), suggesting the absence of any significant interaction of Cox2p with Cox3p in the absence of Cox1p.
Figure 5.
Pulldown assays of radiolabeled mitochondrial gene products of strains expressing Cox12p-HAC, Cox13p-HAC, and Pet100p-CH. A and B, the parental WT and strains expressing Cox2p-HAC and Cox13p-HAC with and without an mss51 null mutation were grown in YPGal medium and incubated for an additional 2 h in fresh YPGal containing 2 mg/ml chloramphenicol. Mitochondria were labeled for 20 min and the tagged proteins purified on protein C beads as in Fig. 3A. A and B, the fractions purified on protein C antibody beads were separated on a 12% polyacrylamide gel by SDS-PAGE (A) and on a 4–13% polyacrylamide gel by BN-PAGE (B). Following transfer to nitrocellulose (A) or PVDF (B), the membranes were exposed to an X-ray film. The positions of the radiolabeled mitochondrial products and assembly intermediates are marked in the margins of the autoradiograms. C and D, yeast strains expressing Cox12p-HAC were analyzed as in A and B. E and F, yeast strains expressing Pet100p-CH were analyzed as in A and B. In D, the in-gel COX activity was measured as described previously (10).
Cox13p was previously found to be associated with the Cox3p module (12). As expected, newly translated Cox2p in the mss51 mutant expressing Cox13p-HAC was not detected in the fraction eluted from the protein C antibody beads (Fig. 5, A and B), confirming that this subunit is not a component of the Cox2p intermediates. This was also true of Cox9p or Cox12p, neither of which was found to be in the Cox2 intermediates. Purification on protein C antibody beads of tagged Cox12p-HAC from W303/COX12-HAC displayed a radiolabeled band at the position of B4/COX but not B3 (Fig. 5D). Neither COX nor the supercomplexes were present in the mss51 mutant. The Cox12-HAC purified from this strain did not contain any radiolabeled COX subunits (Fig. 5C) nor were the B3 or B4 intermediates detected on the native gel (Fig. 5D). Similar results were obtained with strains expressing Cox9p-HAC (Fig. S1).
Interaction of COX assembly factors with the Cox2p module
The Cox1 and Cox3 modules have been shown to contain regulatory and assembly factors (14–18, 26–28). To ascertain if this is also true of the Cox2p module, pulldown assays were done with strains expressing Pet100p-CH (29, 30), Pet117p-CH (31, 32), Cox18p-HAC (33), Cox20p-HAC (34), Sco1p-CH (35), and Coa6p-CH (36). The functions of Pet100p or Pet117p are still poorly understood. Both are essential for COX assembly, but are not present in the mature complex. Purification on protein C beads of these assembly factors tagged with CH indicated that Pet100p (Fig. 5, E and F) and Pet117 (Fig. S1) are not associated with either the Cox2p or the other two COX modules.
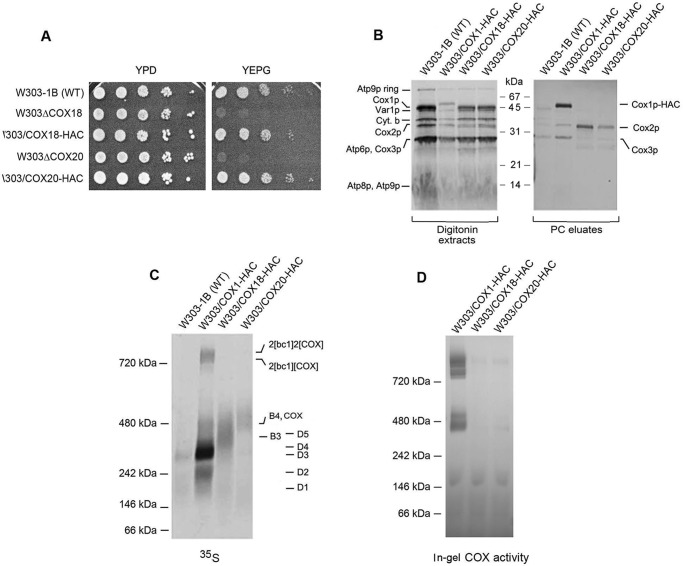
Two other factors, Cox18p and Cox20p, promote transfer of the C-terminal domain of Cox2p to the intermembrane side of the inner membrane (33, 34). Both factors physically interact with Cox2p in yeast (20) and in human mitochondria (21). The presence of yeast Cox18p and Cox20p in the Cox2p intermediates was examined in strains expressing each protein with C-terminal HAC tag. The presence of the tag did not compromise the function of either Cox18p or Cox20p as evidenced by the normal growth on nonfermentable carbon sources of strains expressing the tagged in lieu of the WT proteins (Fig. 6A).
Figure 6.
Cox18p and Cox20p are components of the Cox2p module. A, the parental WT, the cox18 and cox20 null mutants, and the null mutants expressing HAC tagged Cox18p and Cox20p were serially diluted and spotted on rich glucose (YPD) and rich ethanol plus glycerol (YEPG). The plates were incubated for 2 days at 30 °C. B, mitochondria of the WT and the stains expressing HAC-tagged Cox1p (MRSIO/COX1-HAC), Cox18p (W303/COX18-HAC), and Cox20p (W303/COX20-HAC) were grown in YPGal without additional growth in medium containing chloramphenicol. Mitochondria were prepared and labeled as in Fig. 3A. The digitonin extracts and eluates from the protein C antibody beads (PC eluates) were separated by SDS-PAGE on a 12% polyacrylamide gel. C, the fractions of B eluted from the PC beads were separated by BN-PAGE on a 4–13% polyacrylamide gel, transferred to a PVDF membrane and exposed to X-ray. D, same as C except that the gel was stained for COX activity.
Cox18p-HAC and Cox20p-HAC were adsorbed to protein C antibody beads from digitonin extracts of mitochondria that had been labeled with [35S]methionine and [35S]cysteine. SDS-PAGE of the fraction eluted from the beads indicated an enrichment of Cox2p but not Cox1p or Cox3p. This result confirms that the yeast Cox18p and Cox20p, like their human counterparts, are associated with newly translated Cox2p (Fig. 6B). Analysis of the same fractions on a native gel showed that Cox2p pulled down by Cox18p-HAC migrated with an apparent mass of 350 to 550 kDa and Cox20-HAC of 450 and 550 kDa (Fig. 6C), suggesting that Cox18p interacts with the B3 intermediate before Cox20p. As indicated earlier, the in-gel enzymatic assay indicates that intermediate B4 containing Cox20p co-migrates with dimeric COX (Fig. 6D).
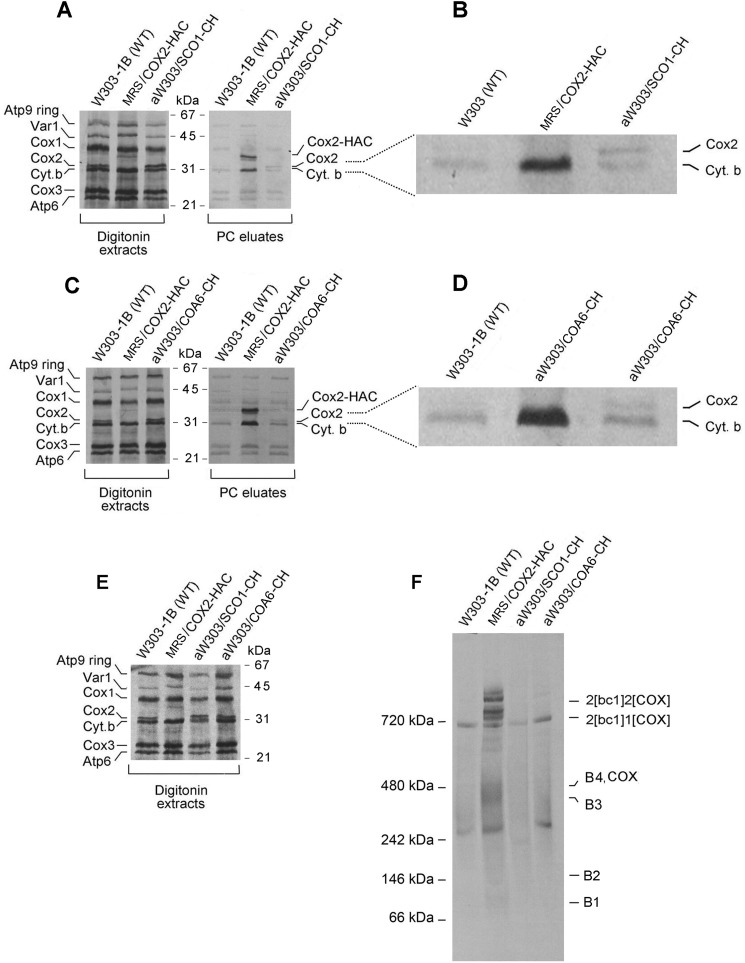
We also tested the presence of Sco1p and Coa6p in the Cox2 module. Both proteins have been implicated in metalation of the binuclear CuA center of Cox2p. Sco1p was shown to transiently bind to Cox2p (37) and has been implicated in maturation of the CuA center in the human subunit (35, 38). More recently, studies of patients deficient in COX have uncovered another protein, Coa6p, that interacts with Sco1p (39) and also appears to function in maturation of the CuA site (33, 40). Mitochondria of strains expressing Sco1p-CH and Coa6-CH were labeled in vitro and the two tagged proteins purified on protein C antibody beads. In both cases the protein fraction eluted from the beads was enriched in radiolabeled Cox2p (Fig. 7, A–D). Although the Cox2p signals were weak, they were consistently higher than the background of radiolabeled proteins nonspecifically bound to the beads (see WT in Fig. 7, B and D). The signal of newly translated Cox2p-HAC in MRS/COX2-HAC, most of which is associated with the intermediate, was significantly greater than that of the Cox2p enriched with Sco1p-CH and Coa6-CH. This suggests that only a small fraction of the Cox2 module contains these proteins. This is also supported by the blue native gel of the eluates from the protein C antibody beads, which indicated that very little of Cox2p intermediates were pulled down by Sco1p-CH and Coa6p-CH (Fig. 7, E and F). This circumstance preempted identifying the intermediate(s) containing Sco1p and Coa6p in strains with null mutations in genes coding for these proteins. The patterns of labeled Cox2p intermediates in the sco1 and coa6 null mutants did not differ from WT (not shown).
Figure 7.
Association of Sco1p and Coa6p with Cox2p. A, the different strains were grown to early stationary phase in 2% YPGal with a final 2-h growth in the presence of 2 mg/ml chloramphenicol. Mitochondria were labeled for 20 min and fractionated as in Fig. 2A. The digitonin extracts and eluates from the protein C antibody beads were separated by SDS-PAGE on a 12% polyacrylamide gel containing 6 m urea and 10% by weight glycerol. B, an enlargement of the Cox2p and cytochrome b region of the gel shown in A. C, the different strains of yeast were grown and mitochondria were labeled and fractionated, and the digitonin extracts and eluates from the protein C antibody beads were electrophoresed as in A. D, an enlargement of the Cox2p and cytochrome b region of the gel shown in C. E, the indicated strains were grown and mitochondria were labeled and fractionated, and the digitonin extracts electrophoresed as in A. F, the digitonin extracts of E were purified on protein C antibody beads and separated by BN-PAGE on a 4–13% polyacrylamide gel.
Discussion
Cox2p is one of the three subunits that make up the catalytic core of COX. It is translated on mitochondrial ribosomes with an N-terminal 15 amino acid long extension that is proteolytically removed during insertion of the protein into the inner membrane. Membrane insertion of Cox2p is co-translation and requires the IMP protease complex to process the Cox2p precursor (41) and the Cox2p-specific factors Cox18p and Cox20p to transfer the large hydrophilic C-terminal domain across the inner membrane (23, 34). Cox2p interacts directly with core subunits Cox1p and Cox3p, and with two structural subunits, Cox9p and Cox12p, that are encoded in nuclear DNA (25).
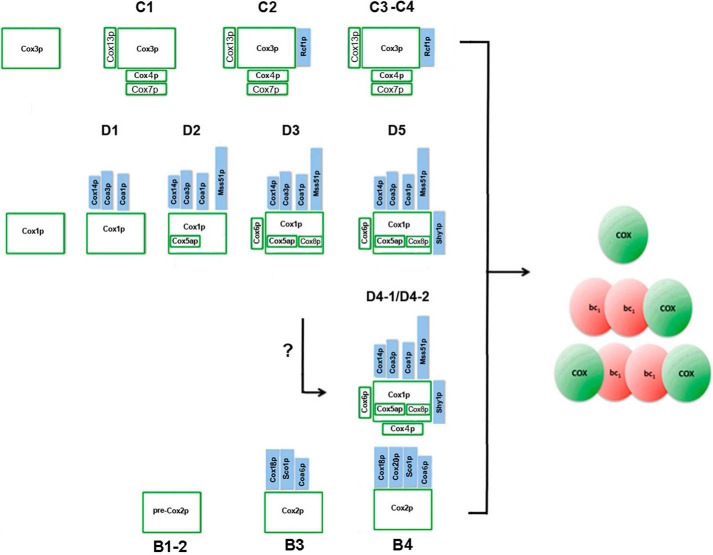
In previous studies we characterized two intermediates of yeast COX, one containing Cox1p and the other Cox3p. The Cox1p and Cox3p intermediates were shown to assemble independently into modules that interacted with each other to form COX (10, 12). Significantly, each module contained a specific subset of the nuclear-encoded structural subunits. The subunits associated with each module corresponded to the same ones that are associated with Cox1p and Cox3p subunits in mature COX (Fig. 8). In addition to the structural subunits, the modules contain regulatory and assembly factors that act on Cox1p or Cox3p (14–18, 26–28).
Figure 8.
The three cytochrome oxidase modules. The structural subunits and ancillary assembly factors present in each module are shown.
In this study we examined the composition of the Cox2p assembly intermediates by the approach used previously to characterize the Cox1p and Cox3p modules. Pulse labeling and pulse-chase analysis of mitochondria from a yeast strain expressing Cox2p with a C-terminal protein C plus HA double tag disclosed the existence of several intermediates ranging from 100 to 450 kDa. The largest intermediate (B4) migrated in the same region as dimeric COX but could be distinguished from the latter in a COX mutant.
Cox9p and Cox12p are known to make contact with Cox2p in the mature enzyme (25). As neither of these two subunits had previously been detected in the Cox1p or Cox3p modules, they were good candidates to be associated with newly translated Cox2p. This expectation, however, is not supported by the results of pulldown experiments. Cox2p did not co-immunopurify with tagged Cox9p or Cox12p of mitochondria in which the endogenous gene products had been pulse labeled with [35S]methionine and [35S]cysteine. This suggests that these subunits interact with Cox2p after it assembles with the Cox1p and Cox3p modules. However, we cannot exclude the alternative possibility that the interaction of Cox9p and Cox12p with Cox2p may be too weak to withstand the solubilization and purification protocol used in the pulldown assay.
Cox2p intermediates were tested for the presence of accessory factors Pet110p and Pet117p. Pulldown assays of tagged Pet100p and Pet117p indicated that neither assembly factor is present in the Cox2p intermediates. They were also not detected in the Cox1p and Cox3p modules, suggesting that their role is unrelated to either maturation or regulation of expression of the catalytic subunits. We also examined which Cox2p intermediates contain Cox18p, Cox20p, Sco1p, and Coa6p. Cox18p and Cox20p are involved in insertion of the Cox2p hydrophobic transmembrane sequences into the phospholipid bilayer, transfer of the hydrophilic domain with the CuA active site across the inner membrane, and proteolytic removal of the N-terminal presequence of the Cox2p precursor (33, 34). Sco1p and Coa6p have been implicated in maturation of the CuA site of Cox2p (36–40). The results of pulldown assays indicated Cox18p and Cox20p to be associated with the two largest Cox2p intermediates. This is consistent with earlier evidence that they interact with yeast (20) and human (21) Cox2p. The presence of Cox20p in B4 and of Cox18p in both B3 and B4 suggests that the interaction of Cox18p with the Cox2p precursor precedes that of Cox20p.
Yeast Sco1p and Coa6p, like their human counterparts, form complexes with Cox2p intermediates (39, 40). In both cases the interactions appear to be transient as only a small fraction of the newly translated Cox2p co-immunopurified with tagged Sco1p and Coa6p. Most of Cox2p pulled down by tagged Coa6p is associated with the B3 intermediate, suggesting the function of Sco1p and Coa6p is exercised at a late stage of Cox2p biogenesis.
In view of the absence of any of known COX structural subunits in the B4 intermediate, Cox2 probably interacts with the Cox1p and/or Cox3p modules without its direct partner subunits in COX (Cox9p and Cox12p). Shedding of Cox18p and Cox20p from the B4 intermediate must occur prior to assembly of Cox2p with its Cox1p and Cox3p partners. This is supported by the earlier finding that fully processed Cox2p is detected in a cox4 mutant defective in assembly of COX and with greatly reduced steady-state concentrations of Cox1p (33).
Materials and methods
Strains and growth media
The genotypes and sources of the S. cerevisiae strains used in this study are described in Table S1. The compositions of solid and liquid YPD, YPGal, and YEPG have been described previously (42).
Construction of hybrid genes with HA and tandem HA plus protein C tags
COX2 was first modified at the 3′ end to express Cox2p with an HA tag. The gene was PCR amplified with the forward primers cox2–5 starting 500 bp upstream of the gene and cox2–13, which contained the sequence complementary to the HA tag followed by the sequence complementary to the last eight codons of the gene. The PCR fragment was digested with a combination of SacI and BamH1 and was cloned into pUC19/COX3 (pUC19 with COX3 inserted at the HindIII site of the multiple cloning sequence). The plasmid obtained from this ligation (pCOX2/ST10) was fused to a BamH1-PstI fragment with 500 bp of COX2 3′-UTR obtained with primers cox2–11 and cox2–12. The COX2-HA gene in pCOX2/ST10 was used as the template for amplification of COX2-HAC with primers cox2–5 and HA-protC. The amplified sequence consisting of 500 bp 5′-UTR followed by COX2 plus the sequence coding for the HAC double tag, was substituted for the SacI-BamHI fragment of pCOX2/ST10 to obtain pCOX2/ST11 containing COX2-HAC plus 500 bp of 5′ and 3′ flanking sequences.
Plasmids pCOX2/ST10 and pCOX2/ST11 were each introduced into the kar1 ρ0 strain DFK160 by biolistic transformation. DFK160 is identical to DFS160 (43) except that it has an additional lys2 mutation. The hybrid COX2 genes in DFK160 transformants were substituted for the cox2 null allele of MRSΔCOX2 by recombination. The latter strain was obtained by transferring the mitochondrial genome of HMD22 with the cox2 mutation to MRS-3Bρ0. The sequences of the primers used for the constructs in this and subsequent sections are listed in Table S2.
Construction of a pet100 null allele
A plasmid (pG135/ST1) containing a BamHI fragment with PET100 and ∼500 bp of 5′- and 3′-UTR sequences in YEp352 (44) was digested with BglII and NheI to remove 26 nucleotides internal to the gene. The gapped plasmid was ligated to a 1 kb BamH1-XbaI fragment containing HIS3 to produce pG135/ST2. The pet100::HIS3 allele, obtained as a BamH1 fragment from pG135/ST2 and was introduced by homologous recombination (45) in the respiratory competent strains W303–1A and W303–1B. A similar procedure was used to obtain a pet117null mutant.
Construction of the COX18-HAC, COX20-HAC, COX16-CH, PET100-CH, PET117-CH, SCO1-CH, and COA6-CH fusion genes
The sequence coding for the HAC tag was added to these genes in two steps. First, each gene was modified to express a C-terminal HA tag by PCR amplification of ∼500 bp of 5′-UTR plus the coding sequence. The genes were amplified with a 5′ primer consisting of 18–21 nucleotides and reverse primer with sequences complementary to the HA tag followed by 18–21 terminal codons of the gene. The resultant product was ligated to either YIp351 containing LEU2 (44) or YIp349 with the TRP1 selectable gene. The plasmids obtained from this ligation were used as a template for amplification with the same 5′ primer and a 3′ primer consisting of a sequence complementary to the protein C tag followed by two glycine codons and ending with 21 nucleotides of a sequence complementary to the HA tag. The PCR products were ligated into the same integrative plasmids.
To construct genes expressing proteins with a C-terminal poly histidine and protein C tag, 500 bp of the 5′-UTRs plus the coding sequences of COX16, PET100, and PET117 (minus the termination codons) were ligated in-frame to the sequence coding for the CH tag in the integrative LEU2- or TRP1-bearing vectors YIp351-CH and YIp349-CH, respectively. The CH tag consists of the protein C epitope followed by three glycines and ending with six histidines. The modified genes were integrated into chromosomal DNA of the appropriate null mutant by homologous recombination (45) after linearization of their plasmids at LEU2 or TRP1. The strains expressing Sco1p-CH and Coa6p-CH were made the same way, except that the upstream primer started at 270 and 300 nucleotides ahead of the initiation codons, respectively. The sequences of the primers for these constructs are listed in Table S2.
Growth, isolation of mitochondria, labeling of mitochondrial gene products, and purification of tagged proteins
Yeast was grown in YPGal and mitochondria isolated as described (46). Small aliquots of mitochondria were frozen in liquid nitrogen and stored at −80 °C. Unless otherwise indicated mitochondria were labeled for 20 min at 25 °C with [35S]methionine plus [35S]cysteine (3000 Ci/mmol) (MP Biochemicals, Solon, OH) as described previously (10). The reaction was stopped with puromycin plus excess unlabeled methionine and further incubated for an additional 10 min. Digitonin extracts of the labeled mitochondria were purified on protein C antibody beads and analyzed by SDS-PAGE and BN-PAGE (10).
Miscellaneous procedures
Purification, ligation, and transformation of Escherichia coli were done under standard conditions (47). Yeast was transformed by the lithium acetate method (48). Proteins were separated by SDS-PAGE on 12 or 17% polyacrylamide gels run in Laemmli buffer (49). Proteins were separated by BN-PAGE on 4–13% polyacrylamide gels (50). Western blots were treated with monoclonal or polyclonal antibodies followed by a second reaction with anti-mouse or anti-rabbit IgG conjugated to horseradish peroxidase (Sigma) and proteins detected with SuperSignal chemiluminescent substrate kit (Pierce Biotechnology, Rockford, IL). Cytochrome oxidase activity was measured as described (51). The method of Lowry (52) was used to estimate protein concentration.
Author contributions
L. V. R. F., C. H. S., and A. T. conceptualization; L. V. R. F., C. H. S., G. P. M., and G. J. Y. data curation; L. V. R. F., C. H. S., and G. P. M. formal analysis; L. V. R. F., C. H. S., G. P. M., G. J. Y., and A. T. investigation; L. V. R. F., C. H. S., and A. T. writing-original draft; L. V. R. F., C. H. S., G. P. M., and A. T. writing-review and editing; G. P. M. and A. T. validation; A. T. supervision; A. T. funding acquisition; A. T. methodology; A. T. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Thomas Fox for the gift of the HMD22 strain of yeast.
This work was supported by National Institutes of Health Grant 5R01GM111864. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1 and S2 and Fig. S1.
The Cox2p and p-Cox2p (precursor) are very poorly separated in the Laemmli SDS-PAGE system. The band identified as Cox2p may include some p-Cox2p.
- COX
- cytochrome oxidase
- BN
- blue native
- HAC
- HA tag followed by protein C tag
- PC
- protein C antibody beads.
References
- 1. Yoshikawa S., Muramoto K., Shinzawa-Itoh K., Aoyama H., Tsukihara T., Ogura T., Shimokata K., Katayama Y., and Shimada H. (2006) Reaction mechanism of bovine heart cytochrome c oxidase. Biochim. Biophys. Acta 1757, 395–400 10.1016/j.bbabio.2006.04.028 [DOI] [PubMed] [Google Scholar]
- 2. Levchenko M., Wuttke J. M., Römpler K., Schmidt B., Neifer K., Juris L., Wissel M., Rehling P., and Deckers M. (2016) Cox26 is a novel stoichiometric subunit of the yeast cytochrome c oxidase. Biochim. Biophys. Acta. 1863, 1624–1632 10.1016/j.bbamcr.2016.04.007 [DOI] [PubMed] [Google Scholar]
- 3. Strecker V., Kadeer Z., Heidler J., Cruciat C. M., Angerer H., Giese H., Pfeiffer K., Stuart R. A., and Wittig I. (2016) Supercomplex-associated Cox26 protein binds to cytochrome c oxidase. Biochim. Biophys. Acta 1863, 1643–1652 10.1016/j.bbamcr.2016.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carlson C. G., Barrientos A., Tzagoloff A., and Glerum D. M. (2003) COX16 encodes a novel protein required for the assembly of cytochrome oxidase in Saccharomyces cerevisiae. J. Biol. Chem. 278, 3770–3775 10.1074/jbc.M209893200 [DOI] [PubMed] [Google Scholar]
- 5. Su C. H., and Tzagoloff A. (2017) Cox16 protein is physically associated with Cox1p assembly intermediates and with cytochrome oxidase. J. Biol. Chem. 292, 16277–16283 10.1074/jbc.M117.801811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mick D. U., Fox T. D., and Rehling P. (2011) Inventory control: Cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 12, 14–20 10.1038/nrm3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fox T. D. (2012) Mitochondrial protein synthesis, import, and assembly. Genetics. 192, 1203–1234 10.1534/genetics.112.141267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim H. J., Khalimonchuk O., Smith P. M., and Winge D. R. (2012) Structure, function, and assembly of heme centers in mitochondrial respiratory complexes. Biochim. Biophys. Acta 1823, 1604–1616 10.1016/j.bbamcr.2012.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Soto I. C., Fontanesi F., Liu J., and Barrientos A. (2012) Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim. Biophys. Acta 1817, 883–897 10.1016/j.bbabio.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McStay G. P., Su C. H., and Tzagoloff A. (2013) Modular assembly of yeast cytochrome oxidase. Mol. Biol. Cell 24, 440–452 10.1091/mbc.e12-10-0749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McStay G. P., Su C. H., Thomas S. M., Xu J. T., and Tzagoloff A. (2013) Characterization of assembly intermediates containing subunit 1 of yeast cytochrome oxidase. J. Biol. Chem. 288, 26546–26556 10.1074/jbc.M113.498592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Su C. H., McStay G. P., and Tzagoloff A. (2014) Assembly of the rotor component of yeast mitochondrial ATP synthase is enhanced when Atp9p is supplied by Atp9p-Cox6p complexes. J. Biol. Chem. 289, 31605–31616 10.1074/jbc.M114.602706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Perez-Martinez X., Broadley S. A., and Fox T. D. (2003) Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 22, 5951–5961 10.1093/emboj/cdg566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barrientos A., Zambrano A., and Tzagoloff A. (2004) Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 23, 3472–3482 10.1038/sj.emboj.7600358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pierrel F., Bestwick M. L., Cobine P. A., Khalimonchuk O., Cricco J. A., and Winge D. R. (2007) Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 26, 4335–4346 10.1038/sj.emboj.7601861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mick D. U., Vukotic M., Piechura H., Meyer H. E., Warscheid B., Deckers M., and Rehling P. (2010) Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J. Cell Biol. 191, 141–154 10.1083/jcb.201007026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fontanesi F., Clemente P., and Barrientos A. (2011) Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J. Biol. Chem. 286, 555–566 10.1074/jbc.M110.188805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mayorga J. P., Camacho-Villasana Y., Shingú-Vázquez M., García-Villegas R., Zamudio-Ochoa A., García-Guerrero A. E., Hernández G., and Pérez-Martínez X. (2016) A novel function of Pet54 in regulation of Cox1 synthesis in Saccharomyces cerevisiae mitochondria. J. Biol. Chem. 291, 9343–9355 10.1074/jbc.M116.721985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bareth B., Dennerlein S., Mick D. U., Nikolov M., Urlaub H., and Rehling P. (2013) The heme a synthase Cox15 associates with cytochrome c oxidase assembly intermediates during Cox1 maturation. Mol. Cell. Biol. 3, 4128–4137 10.1128/MCB.00747-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Elliott L. E., Saracco S. A., and Fox T. D. (2012) Multiple roles of the Cox20 chaperone in assembly of Saccharomyces cerevisiae cytochrome c oxidase. Genetics 190, 559–567 10.1534/genetics.111.135665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bourens M., and Barrientos A. (2017) Human mitochondrial cytochrome c oxidase assembly factor COX18 acts transiently as a membrane insertase within the subunit 2 maturation module. J. Biol. Chem. 292, 7774–7783 10.1074/jbc.M117.778514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Decoster E., Simon M., Hatat D., and Faye G. (1990) The MSS51 gene product is required for the translation of the COX1 mRNA in yeast mitochondria. Mol. Gen Genet. 224, 111–118 [DOI] [PubMed] [Google Scholar]
- 23. Rak M., Gokova S., and Tzagoloff A. (2011) Modular assembly of yeast mitochondrial ATP synthase. EMBO J. 30, 920–930 10.1038/emboj.2010.364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aich A., Wang C., Chowdhury A., Ronsör C., Pacheu-Grau D., Richter-Dennerlein R., Dennerlein S., and Rehling P. (2018) COX16 promotes COX2 metallation and assembly during respiratory complex IV biogenesis. Elife 7, e32572. 10.7554/eLife.32572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., and Yoshikawa S. (1996) The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272, 1136–1144 10.1126/science.272.5265.1136 [DOI] [PubMed] [Google Scholar]
- 26. Perez-Martinez X., Butler C. A., Shingu-Vazquez M., and Fox T. D. (2009) Dual functions of Mss51 couple synthesis of Cox1 to assembly of cytochrome c oxidase in Saccharomyces cerevisiae mitochondria. Mol. Biol. Cell 20, 4371–4380 10.1091/mbc.e09-06-0522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fontanesi F., Soto I. C., Horn D., and Barrientos A. (2010) Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol. Cell. Biol. 30, 245–259 10.1128/MCB.00983-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Strogolova V., Furness A., Robb-McGrath M., Garlich J., and Stuart R. A. (2012) Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol. Cell. Biol. 32, 1363–1373 10.1128/MCB.06369-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Church C., Chapon C., Poyton and RO. (1996) Cloning and characterization of PET100, a gene required for the assembly of yeast cytochrome c oxidase. J. Biol. Chem. 271, 18499–18507 10.1074/jbc.271.31.18499 [DOI] [PubMed] [Google Scholar]
- 30. Church C., Goehring B., Forsha D., Wazny P., and Poyton R. O. (2005) A role for Pet100p in the assembly of yeast cytochrome c oxidase: Interaction with a subassembly that accumulates in a pet100 mutant. J. Biol. Chem. 280, 1854–1863 10.1074/jbc.M410726200 [DOI] [PubMed] [Google Scholar]
- 31. McEwen J. E., Hong K. H., Park S., and Preciado G. T. (1993) Sequence and chromosomal localization of two PET genes required for cytochrome c oxidase assembly in Saccharomyces cerevisiae. Curr. Genet. 23, 9–14 [DOI] [PubMed] [Google Scholar]
- 32. Taylor N. G., Swenson S., Harris N. J., Germany E. M., Fox J. L., Khalimonchuk O. (2017) The assembly factor Pet117 couples heme a synthase activity to cytochrome oxidase assembly. J. Biol. Chem. 292, 1815–1825 10.1074/jbc.M116.766980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saracco S. A., and Fox T. D. (2002) Cox18p is required for export of the mitochondrially encoded Saccharomyces cerevisiae Cox2p C-tail and interacts with Pnt1p and Mss2p in the inner membrane. Mol. Biol. Cell 13, 1122–1131 10.1091/mbc.01-12-0580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hell K., Tzagoloff A., Neupert W., and Stuart R. A. (2000) Identification of Cox20p, a novel protein involved in the maturation and assembly of cytochrome oxidase subunit 2. J. Biol. Chem. 275, 4571–4578 10.1074/jbc.275.7.4571 [DOI] [PubMed] [Google Scholar]
- 35. Glerum D. M., Shtanko A., and Tzagoloff A. (1996) SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 271, 20531–20535 [DOI] [PubMed] [Google Scholar]
- 36. Baertling F., van den Brand M. A. M., Hertecant J. L., Al-Shamsi A., van den Heuvel P. L., Distelmaier F., Mayatepek E., Smeitink J. A., Nijtmans L. G., and Rodenburg R. J. (2015) Mutations in COA6 cause cytochrome c oxidase deficiency and neonatal hypertrophic cardiomyopathy. Hum. Mutat. 36, 34–38 10.1002/humu.22715 [DOI] [PubMed] [Google Scholar]
- 37. Lode A., Kuschel M., Paret C., and Rödel G. (2000) Mitochondrial copper metabolism in yeast: interaction between Sco1p and Cox2p. FEBS Lett. 485, 19–24 10.1016/S0014-5793(00)02176-1 [DOI] [PubMed] [Google Scholar]
- 38. Nittis T., George G. N., and Winge D. R. (2001) Yeast Sco1, a protein essential for cytochrome c oxidase function is a Cu(I)-binding protein. J. Biol. Chem. 276, 42520–42526 10.1074/jbc.M107077200 [DOI] [PubMed] [Google Scholar]
- 39. Aich A., Wang C., Chowdhury A., Ronsör C., Pacheu-Grau D., Richter-Dennerlein R., Dennerlein S., and Rehling P. (2018) COX16 promotes COX2 metallation and assembly during respiratory complex IV biogenesis. eLife 7, e32572 10.7554/eLife.32572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ghosh A., Pratt A. T., Soma S., Theriault S. G., Griffin A. T., Trivedi P. P., and Gohil V. M. (2016) Mitochondrial disease genes COA6, COX6B and SCO2 have overlapping roles in COX2 biogenesis. Hum. Mol. Genet. 25, 660–671 10.1093/hmg/ddv503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Esser K., Pratje E., and Michaelis G. (1996) SOM1, a small new gene required for mitochondrial inner membrane peptidase function in Saccharomyces cerevisiae. Mol. Gen. Genet. 252, 437–445 [DOI] [PubMed] [Google Scholar]
- 42. Myers A. M., Pape L. K., and Tzagoloff A. (1985) Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 4, 2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bonnefoy N., and Fox T. D. (2007) Directed alteration of Saccharomyces cerevisiae mitochondrial DNA by biolistic transformation and homologous recombination. Methods Mol. Biol. 372, 153–166 10.1007/978-1-59745-365-3_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hill J. E., Myers A. M., Koerner T. J., and Tzagoloff A. (1986) Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2, 163–167 10.1002/yea.320020304 [DOI] [PubMed] [Google Scholar]
- 45. Rothstein R. J. (1983) One-step gene disruption in yeast. Methods Enzymol. 101, 202–211 10.1016/0076-6879(83)01015-0 [DOI] [PubMed] [Google Scholar]
- 46. Herrmann J. M., Foelsch H., Neupert W., and Stuart R. A. (1994) Isolation of yeast mitochondria and study of mitochondrial protein translation. in Cell Biology: A Laboratory Handbook (Celis J. E., ed) Vol. 1, pp. 538–544, Academic Press, San Diego, CA [Google Scholar]
- 47. Sambrook J., Fritsch E. F., and Maniatis T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 48. Schiestl R. H., and Gietz R. D. (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16, 339–346 10.1007/BF00340712 [DOI] [PubMed] [Google Scholar]
- 49. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 50. Wittig I., Braun H. P., and Schägger H. (2006) Blue native PAGE. Nat. Protoc. 1, 418–428 10.1038/nprot.2006.62 [DOI] [PubMed] [Google Scholar]
- 51. Wharton D. C., and Tzagoloff A. (1967) Cytochrome oxidase from beef heart mitochondria. Methods Enzymol. 10, 245–250 10.1016/0076-6879(67)10048-7 [DOI] [Google Scholar]
- 52. Lowry O. H., Rosebrough N. J., Farr A. L., and Randall R. J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.