ABSTRACT
Dynamin-related protein 1 (Drp1) is a dynamin superfamily GTPase, which drives membrane constriction during mitochondrial division. To mediate mitochondrial division, Drp1 is recruited to the mitochondrial outer membrane and is assembled into the division machinery. We previously showed that Drp1 interacts with phosphatidic acid (PA) and saturated phospholipids in the mitochondrial membrane, and this interaction restrains Drp1 in initiating the constriction of mitochondria. Here, we show that the role of saturated acyl chains of phospholipids is independent of their contribution to the membrane curvature or lipid packing suggesting their direct interaction with Drp1. We further show that an unstructured loop in the stalk domain of Drp1 is critical for interaction with unsaturated PA. Our data significantly advance our understanding of this unique protein-lipid interaction involved in mitochondrial division.
KEYWORDS: dynamin-related protein 1, GTPase, lipid binding, liposomes, mitochondria, organelle dynamics, phosphatidic acid
Mitochondria are essential organelles and function in many important processes including energy production, metabolism, signal transduction and cell death.1-4 To execute these diverse functions, mitochondria divide and fuse and regulate size, distribution and turnover. Unregulated mitochondrial division and fusion have been associated with a wide range of human diseases including cancers, metabolic syndromes and neurological disorders such as Alzheimer disease, Parkinson disease and Charcot-Marie-Tooth diseases.5-7
Mitochondrial division depends on dynamin-related protein 1 (Drp1), which belongs to the dynamin GTPase superfamily.8 Demonstrating the direct link between Drp1 defects and human diseases, mutations in Drp1 result in severe abnormality in brain development, neurodegeneration, epilepsy and postnatal death.9-11 Further supporting the physiological importance of Drp1, whole body deletion of Drp1 in mouse models causes embryonic lethality.12,13 Neuron-specific deletion of Drp1 leads to mitochondrial dysfunction and neurodegeneration in the brain, while cardiomyocyte-specific deletion of Drp1 causes heart failure.14-18 At the cellular level, the loss of Drp1 enlarges mitochondria due to unopposed mitochondrial fusion in neurons and cardiomyocytes and makes mitochondria resistant to autophagic degradation ultimately leading to accumulation of dysfunctional mitochondria.15
Drp1 is a soluble cytosolic enzyme and is recruited to the mitochondrial outer membranes by Drp1 receptor proteins such as Mff, Mid49, Mid41 and Fis1. On the mitochondria, Drp1 oligomerizes into helical structures that can wrap around mitochondria. GTP-regulated conformational changes of these helices are thought to drive the constriction of mitochondria during division. At late stages of mitochondrial division, another GTPase (dynamin-2) works with Drp1 to further constrict mitochondria and complete the membrane fission process. In addition to these protein components, studies have shown that phospholipids in the mitochondrial membrane also play key roles in mitochondrial division. A mitochondria-specific phospholipid, cardiolipin, is synthesized in the mitochondrial inner membrane, and a small fraction of cardiolipin is transported to the outer membrane. On the mitochondrial outer membrane, cardiolipin promotes mitochondrial division likely by enhancing the oligomerization of Drp1 and the oligomerization-stimulated GTPase activity of Drp1.19-23 As a binding site for cardiolipin, the variable domain of Drp1 has been proposed because mutations in this domain interfere with interactions of Drp1 with cardiolipin.24
We recently reported that a signaling phospholipid, phosphatidic acid (PA), binds to Drp1 and regulates mitochondrial division.25 Interaction of Drp1 with PA is unique in 2 ways (Fig. 1). First, this protein-lipid interaction involves both the head group and acyl chains of PA. More specifically, Drp1 only binds to PA that contains saturated acyl chains with physiological length (C16–18). Second, in this interaction, the head group of PA and saturated acyl chains can be separated into 2 phospholipid molecules. Drp1 binds to liposomes that contain unsaturated PA and saturated phosphatidylcholine (PC). In contrast, Drp1 does not associate with liposomes that contain only unsaturated PA or saturated PC. These data suggest that Drp1 can separately recognize the head group of PA and saturated acyl chains of another phospholipid. Domain analysis has revealed that Drp1 has 2 saturated PA binding domains—the stalk and variable domains both of which lack a known lipid-binding motif.25
Figure 1.
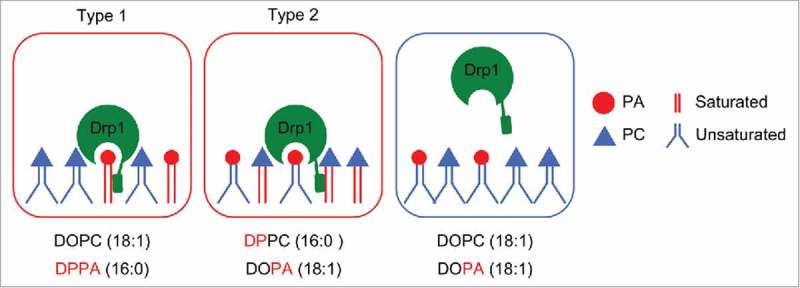
Coincident interaction of Drp1 with PA. In type 1 binding mode, Drp1 binds to saturated PA. In type 2 binding mode, Drp1 simultaneously binds to unsaturated PA and saturated non-PA phospholipids such as PC.
Interactions with PA do not stimulate the GTPase activity of Drp1.25 Rather, this coincident PA interaction involves the hydrophilic head and hydrophobic tail to inhibit the function of Drp1 in mitochondrial division. In cells, increasing PA levels inhibit mitochondrial division and leads to the accumulation of Drp1 oligomers on mitochondria suggesting that PA inhibits mitochondrial division after Drp1 is oligomerized on mitochondria.25 This PA-mediated inhibition of mitochondrial division also requires saturation of acyl chains as decreasing levels of saturated acyl chains block the inhibitory effect of PA on mitochondrial division.25 Therefore, PA along with saturated phospholipids provide a novel mechanism that controls mitochondrial division.
Furthermore, we found that Drp1 also interacts with MitoPLD,25 a phospholipase which produces PA from cardiolipin in the mitochondrial outer membrane.26-29 This protein-protein interaction suggests that MitoPLD might provide Drp1 with the relatively rare lipid PA and create a PA-enriched lipid environment around the division machinery. In the current study, we further characterized the role of acyl chains in this coincident lipid interaction using liposome flotation assays to define this novel mechanism of Drp1-PA/saturated phospholipid interactions. We also identified a region in the stalk domain of Drp1 that is important for this interaction.
To determine the role of acyl chains in Drp1-PA/saturated phospholipid interactions, we first tested whether association of Drp1 with membranes depends on the concentration of saturated acyl chains using a liposome flotation assay (Fig. 2). We generated three types of liposomes with different ratios of saturated PC (DPPC) and unsaturated PC (DOPC): 0:84, 42:42 and 84:0 (% mol). We held the amount of PA constant (15%) in these liposomes. The 1% rhodamine-phosphatidylethanolamine (rhodamine-PE) was included to detect liposomes. Purified recombinant His6-Drp1 was incubated with liposomes at 4°C for 60 min. Drp1-liposome samples was mixed with sucrose to make the final concentration of sucrose 1.73 M. This was placed at the bottom of discontinuous sucrose gradient (1.25 and 0.25 M) (Fig. 2A). After ultracentrifugation, the majority of liposomes floated to the top half of the gradient based on the fluorescence intensity of rhodamine-PE.25 We collected the upper half of the gradient (2.5 ml) as a liposome-bound fraction and a lower half (2.5 ml) as an unbound fraction (Fig. 2A). These fractions were analyzed by SDS-PAGE and sliver staining (Fig. 2B and C). We found that the amounts of Drp1 associated with liposomes increased as the amounts of saturated PC increased in liposomes. These data also show that 42% of saturated acyl chains are not saturated in terms of binding to Drp1 in the flotation assay.
Figure 2.
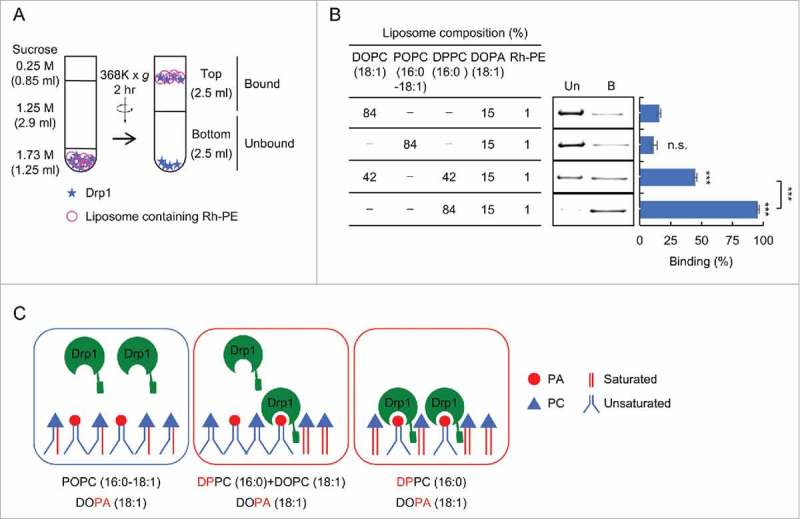
Two saturated acyl chains in single phospholipids are necessary for interactions of Drp1 with liposomes. (A) Liposome flotation assay. Recombinant His6-Drp1 was incubated with liposomes at 4°C for 1 hour and analyzed by sucrose density gradient. The majority of liposomes floated to the upper half of the gradient. (B) Liposome flotation assay was performed using full length His6-Drp1 and the indicated liposomes. Equal amounts of the bottom (unbound, Un) and top (bound, B) were analyzed by SDS-PAGE and silver staining. Band intensity was quantified, and relative amounts of Drp1 in the bound fraction are shown (Mean ± SEM; n = 3). (C) Two saturated acyl chains must be present in the same molecule to mediate Drp1 interactions. Drp1 does not bind to PA that contains one saturated acyl chain and one unsaturated acyl chain. Student's t-test: ***p < 0.001.
This led us to wonder about the role of acyl chain in Drp1-membrane interactions. The role of acyl chains in phospholipids may be to change the membrane curvature or lipid packing through saturation and unsaturation. If this model is correct, then interactions of Drp1 with liposomes depend on the total amount of saturated acyl chains in PC in our liposome flotation assay. In contrast, acyl chains may play a more direct role through interactions with Drp1 independent of these biophysical properties of the lipid bilayer.
To test these models, we generated another type of liposome that contains 84% POPC, which contains one saturated acyl chain and one unsaturated acyl chains. We compared this liposome to liposomes that contain 42% saturated PC (DPPC, both chains saturated) and 42% unsaturated PC (DOPC, both chains unsaturated). In these 2 types of liposomes, the total amount of acyl chains are same; however, saturated and unsaturated acyl chains are paired in the first liposome while 2 saturated acyl chains are present in the same PC molecule in the second liposome. Intriguingly, the results showed that the POPC liposome only poorly interacts with Drp1 similar to negative control liposomes containing only an unsaturated PC (Fig. 2B and C). These data rule out the model that saturated acyl chains facilitate Drp1-membrane interactions by modulating the membrane curvature or lipid packing. It appears that 2 saturated acyl chains must be present in the same PC molecule to mediate Drp1 interactions.
To further test whether Drp1 binds to liposomes independently of the membrane curvature, we generated liposomes with 2 different diameters. We chose 50 and 400 nm because the diameter of the mitochondria in cells is typically 300–400 nm. In addition, a previous study using cryoelectron microscopy of purified a yeast homolog of Drp1 has shown that Drp1 forms spiral structures whose inner diameter is approximately 90 nm30 We therefore reasoned that the diameter of 50 and 400 nm would cover the reasonable range of size that are relevant to mitochondrial division. We first tested 50 and 400 nm liposomes that contain unsaturated PA (DOPA) and saturated PC (DPPC) in our flotation assay. We found that Drp1 similarly associate with these 2 liposomes (Fig. 3A and B). We then generated liposomes that contain saturated PA (DPPA). Again, we observed similar association of Drp1 with liposomes regardless of their diameter (Fig. 3C and D). Therefore, these results further confirm the model that recruitment of Drp1 to membrane thorough interactions with PA is independent of the membrane curvature of the liposomes.
Figure 3.
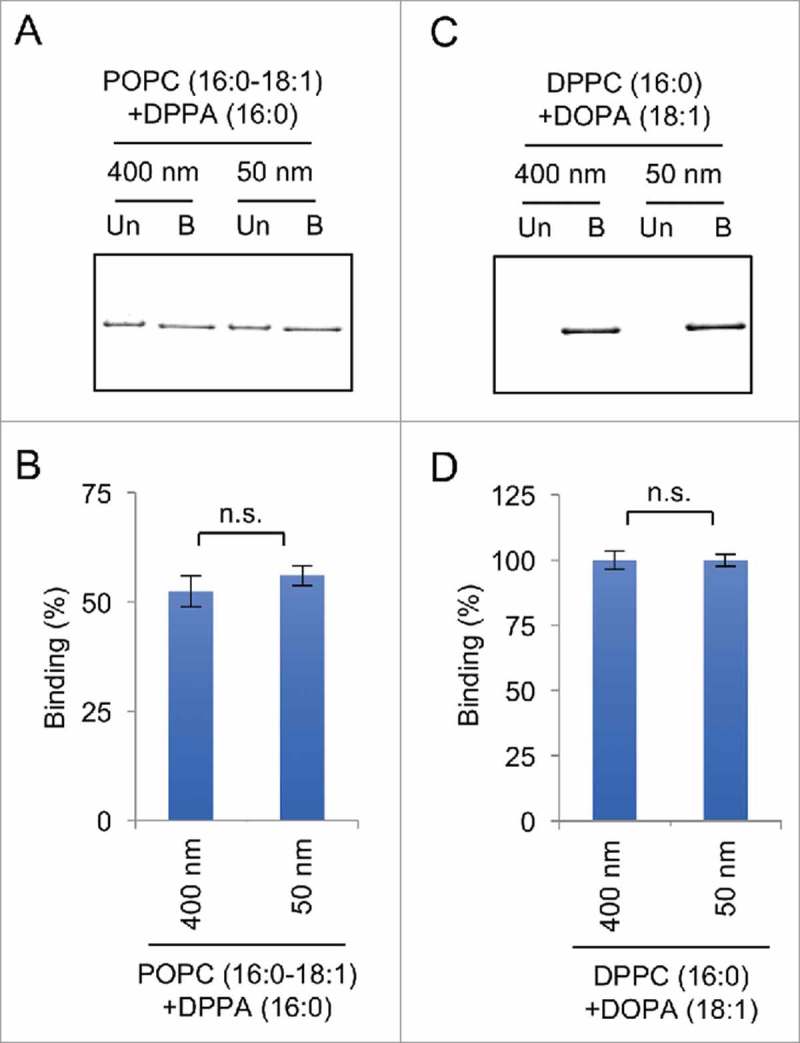
Drp1 binds to liposomes regardless of their diameter in liposome flotation assays. (A and C) Flotation assays were performed using His6-Drp1 and liposomes that contain saturated PA (DPPA) in (A) and unsaturated PA (DOPA) and saturated PC (DPPC) in (C). To change the diameter, we used 2 different nanopore membranes with a pore size of 50 or 400 nm. The upper and lower fractions were collected and analyzed by SDS-PAGE and silver staining. (B and D) The band intensity was quantified, and the relative amounts of Drp1 in the bound fraction are shown (Mean ± SEM; n = 3).
We showed that both stalk and variable domains of Drp1 interact with saturated PA.25 The stalk domain contains about 300 amino acids and mainly consists of α helices. In contrast, the variable domain consists largely of an unstructured loop with about 100 amino acids. Previous studies have suggested that cardiolipin binds to the variable domain. We showed that cardiolipin and saturated PA binds to Drp1 through different mechanisms. We therefore were interested in further mapping the regions that are involved in saturated PA interactions in the stalk domain.
We thought that Drp1 likely penetrates the membrane because Drp1 recognizes the acyl chains of phospholipids. It has been shown that some peripheral lipid-binding proteins carrying a pleckstrin homology, FYVE, or C2 domain are inserted into the hydrophobic core of the bilayer, which is typically ∼3–5 nm away from the headgroup.31,32 Membrane insertion is often mediated by an unstructured loop with hydrophobic amino acids. For example, the FYVE domain of the EEA1 protein penetrates the hydrophobic core via an insertion loop consisting of 10 amino acids (total length) and 2–3 hydrophobic residues that are sufficient to reach acyl chains in one leaflet.33 We therefore reasoned that a loop in the stalk domain, which contains hydrophobic residues, is important for its interactions with saturated PA.
A structural analysis of the stalk domain predicted 4 unstructured loops. The loop corresponding to amino acids TAKYIETSEL 351–360 (Fig. 4A, putative insertion loop) carries 4 amino acids with hydrophobic side-chains (underlined. We hypothesized that this loop is important for interactions of the stalk domain with saturated PA. We collectively mutated these hydrophobic residues to glycine in the His6-stalk-domain and measured the association of the mutant recombinant protein (His6-stalk-domain4G) with liposomes containing saturated PA (DPPA) (Fig. 4B). Supporting our model, the His6-stalk4G showed significant decreases in interaction with saturated PA liposomes compared with a wildtype His6-stalk-domain (Fig. 4C and D).
Figure 4.
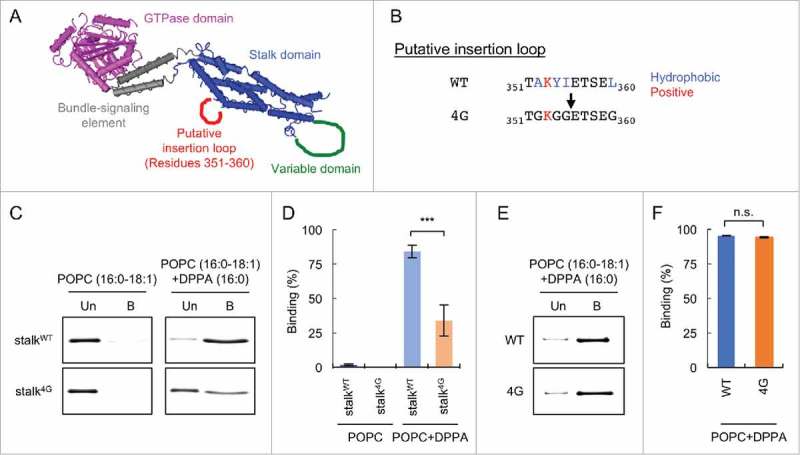
Identification of a loop that is critical for interactions with saturated PA. (A) The 3D structure of Drp136. A putative membrane insertion loop is shown (red). (B) Amino acid sequence of the putative insertion loop. Four hydrophobic residues (blue) are substituted by glycine in the 4G mutant. (C) Liposome flotation assay using His6-stalk and His6-stalk4G. Liposomes containing saturated PA (DPPA) were used. As a negative control, liposomes that lack DPPA were used. (D) Band intensity was quantified, and relative amounts of Drp1 in the bound fraction are shown (Mean ± SEM; n = 3). (E) Liposome flotation assay using His6-Drp1 and His6-Drp14G. (D) Relative amounts of Drp1 in the bound fraction are shown (Mean ± SEM; n = 3). Student's t-test: ***p < 0.001.
These data prompted us to further test whether these mutations affect interactions of full length Drp1 with saturated PA-containing liposomes. We introduced the 4G mutations into the full length Drp1 and purified His6-Drp14G. When we performed liposome flotation assays using His6-Drp1 and His6-Drp14G, we found that these proteins similarly bound to saturated PA-containing liposomes (Fig. 4E and F). These data suggest that the stalk and variable domains are functionally redundant in association with saturated PA.
It has been suggested that the stalk domain mediates the oligomerization of Drp1.34 We were therefore interested in testing whether the association of Drp1 with saturated PA depends on the oligomerization of Drp1. To address this question, we introduced a substitution (G350D) that blocks the oligomerization of Drp1.35 We confirmed that wildtype His6-Drp1, but not His6-Drp1G350D, oligomerizes in the presence of GTPγS and could be pelleted by ultracentrifugation in a sedimentation assay consistent with previous studies (Fig. 5A and B). Although we currently do not know the reason, a small fraction of His6-Drp1G350D was consistently pelleted in the absence of GTPγS. We then tested their interaction with liposomes containing saturated PA or unsaturated PA and saturated PC. We found that wildtype and mutant proteins similarly associate with these liposomes (Fig. 5C and D). Therefore, interactions of Drp1 with saturated PA do not require Drp1 oligomerization.
Figure 5.
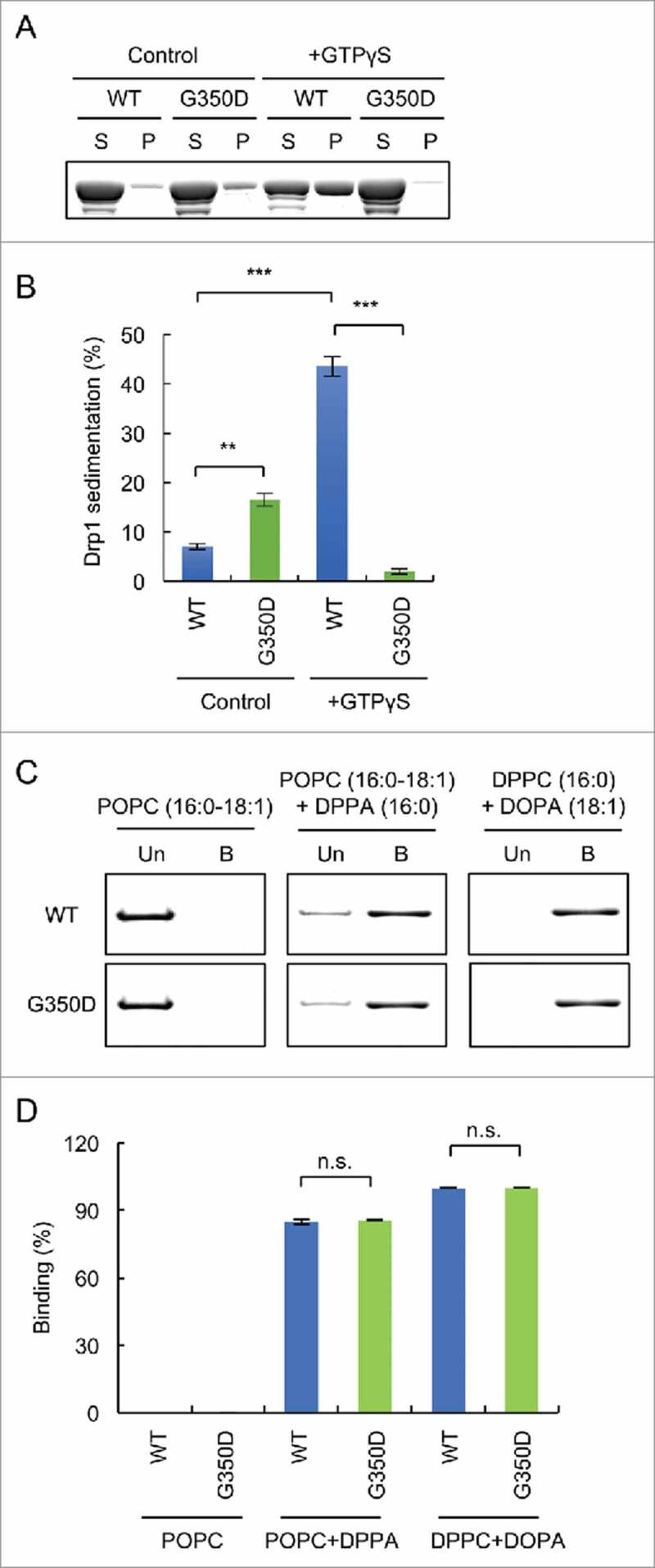
Drp1 binds to PA-containing liposomes independently of oligomerization. (A) His6-Drp1 and His6-Drp1G350D were incubated in the presence or absence of 2 mM GTPγS at room temperature for 30 min and ultracentrifuged at 100,000 x g for 20 min. The supernatant and pellet fractions were analyzed by SDS-PAGE and Coomassie brilliant blue staining. (B) The band intensity was quantified and relative amounts of Drp1 in the pellet fractions are shown (mean ± SEM; n = 3). (C) Liposome flotation assay using His6-Drp1 and His6-Drp1G350D. (D) Band intensity was quantified, and relative amounts of Drp1 in the bound fraction is shown (mean ± SEM; n = 3). Student's t-test: **p < 0.005; ***p < 0.001.
In this study, we showed that saturated acyl chains of phospholipids help Drp1 interact with the membrane together with the head group of PA independent of the membrane curvature or lipid packing. We also found that an unstructured loop in the stalk domain is important for interactions with saturated PA. It is important to further understand if this loop actually inserts into the membrane and directly interacts with the hydrophobic acyl chains of phospholipids in future studies. Studies have shown that the recruitment of Drp1 to mitochondria depends on its receptor proteins Mff, Mid49, Mid51 and Fis1. We therefore suggest that the PA/saturated phospholipids themselves do not control the recruitment of Drp1. Rather, these phospholipids regulate the function of Drp1 once it is recruited to mitochondria and assembled into the division machinery. We are also interested in studying the functional relationship between the Drp1 receptors and phospholipids by reconstituting these components together with Drp1 in vitro.
Materials and methods
Protein expression and purification
We expressed and purified recombinant full length Drp1 and the stalk domain as described previously.25 Rosetta 2(DE3) pLysS competent cells (Novagen) were transfected with pET15b vectors carrying Drp1 or the stalk domain. These cells were grown in LB-containing ampicillin and chloramphenicol overnight at 37°C. Then, 1 ml of the culture was diluted into 1 L of the same growth medium and continued to grow the culture for 3–5 hours at 37°C. The culture was cooled down on ice, and 0.1 mM of IPTG was added.
The expression of recombinant proteins was induced overnight at 16°C. The bacteria were washed twice with PBS by centrifugation at 4000 rpm for 15 min and harvested and froze at −80°C. The frozen cell pellets were resuspended in 40 ml of lysis buffer (10 mM imidazole, 1 mM MgCl2, 500 mM NaCl, 2 mM β-mercaptoethanol, 20 mM Hepes, pH 7.4) and sonicated on ice for 10 × 5 sec at setting 5 and then 10 × 5 sec at setting 2 using a Fisher Scientific Sonic dismembrator model 100. The cell homogenate was centrifuged at 4100 rpm for 15 min at 4°C, and the supernatant was further clarified at 15,000 rpm for 15 min at 4°C. After being passed through a membrane filter with a pore size of 0.45 µm, the lysate was incubated with 2 ml of pre-washed 50% Ni-NTA beads (His-Bind Resin, Novagen) overnight at 4°C and placed in a 15-ml column (Poly-prep chromatography column, Bio-Rad). The beads were washed with 3 ml of lysis buffer, 15 ml of lysis buffer, 3 ml of wash buffer (40 mM imidazole, 1 mM MgCl2, 500 mM NaCl, 2 mM β-mercaptoethanol, 20 mM Hepes, pH 7.4), and 15 ml of wash buffer.
The His6-tagged recombinant proteins were eluted from the column using 3 × 0.9 ml of elution buffer (250 mM imidazole, 1 mM MgCl2, 500 mM NaCl, 2 mM β-mercaptoethanol, 20 mM Hepes, pH 7.4) and collected it into 9 300-µl fractions. After confirming the purification of proteins using SDS-PAGE and Coomassie brilliant blue staining of each fraction, the peak fractions (typically fractions #3 and 4) were collected and diluted into 15 ml of lysis buffer without imidazole. The residual imidazole was removed by passing the solution 3 times through an Amicon Ultra Centrifugation Filter (a 50K filer for Drp1 and a 10K filter for the stalk domain) according to the manufacturer's instructions. The purified proteins were mixed with 20% DMSO, snap frozen in liquid nitrogen, and stored at −80°C.
Liposome flotation assay
We performed liposome floatation assays as described25 with some modifications. We obtained DPPA (830855), DOPA (840875), DPPC (850355), DOPC (850375), POPC (850457), and rhomamine-DPPE (810158) from Avanti Polar Lipids. The lipids were mixed and dried under a flow of nitrogen gas for 5 min and then dried in a SpeedVac overnight. The lipid film was resuspended at a concentration of 10 mM in 100 mM NaCl and 20 mM MES (pH 7.0), vortexed for 1 hour, and subjected to 5 freeze-thaw cycles using dry ice and a 42°C heat block. Unilamellar liposomes were generated via extrusion through a nanopore membrane with a pore size of 50 or 400 nm; this process was repeated 21 times. The liposomes (5 mM lipids) were mixed with His6-tagged recombinant proteins (5 µM) (final volume: 200 µl) and incubated at 4°C with gentle mixing for 1 hour. The protein-liposome mixture was diluted in 1.73 M sucrose and 20 mM MES (pH 7.0) (final volume: 1.25 ml) and placed at the bottom of tubes. We then overlaid 2.9 ml of 1.25 M sucrose/20 mM MES (pH7.0) and 0.85 ml of 0.25 M sucrose/20 mM MES (pH7.0). The sucrose gradient was centrifuged at 55,000 rpm for 2 hours at 4°C in an SW55Ti (Beckman). Two 2.5-ml fractions were collected from the top. The majority of liposomes were floated to the upper fractions, and both fractions were analyzed using SDS-PAGE and silver staining. The stained gels were scanned, and the band intensity was quantified using NIH ImageJ. The liposomes were detected based on the fluorescence intensity of rhodamine-PE.
Sedimentation assay
The oligomerization of Drp1 was assessed in a sedimentation assay as described.36 Purified His6-Drp1 and His6-Drp1G350D were resuspended at 15 µM in 100 µl of 2 mM MgCl2, 5 mM β-mercaptoethanol, and 20m M Hepes (pH 7.4), in the presence or absence of 2 mM GTPγS. After incubation for 30 min at room temperature, samples were ultracentrifuged at 100,000 x g for 20 min. Equivalent amounts of the pellet and supernatant fractions were analyzed by SDS-PAGE and Coomassie brilliant blue staining. The band intensity was quantified with NIH ImageJ software.
Abbreviations
- DOPA
dioleoyl phosphatidic acid
- DOPC
dioleoyl phosphatidylcholine
- DPPA
dipalmitoyl phosphatidic acid
- DPPC
dipalmitoyl phosphatidylcholine
- PA
phosphatidic acid
- PC
phosphatidylcholine
- POPC
palmitoyl-oleoylphosphatidylcholine
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank members of the Iijima and Sesaki laboratories for helpful discussion.
Funding
This work was supported by NIH grants to M.I. (GM084015) and H.S. (GM089853).
References
- [1].Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science 2012; 337:1062-5; PMID:22936770; https://doi.org/ 10.1126/science.1219855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pernas L, Scorrano L. Mito-Morphosis: Mitochondrial Fusion, Fission, and Cristae Remodeling as Key Mediators of Cellular Function. Annu Rev Physiol 2016; 78:505-31; PMID:26667075; https://doi.org/ 10.1146/annurev-physiol-021115-105011 [DOI] [PubMed] [Google Scholar]
- [3].Friedman JR, Nunnari J. Mitochondrial form and function. Nature 2014; 505:335-43; PMID:24429632; https://doi.org/ 10.1038/nature12985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shutt TE, McBride HM. Staying cool in difficult times: Mitochondrial dynamics, quality control and the stress response. Biochim Et Biophys Acta 2012; PMID:22683990; https://doi.org/ 10.1016/j.bbamcr.2012.05.024 [DOI] [PubMed] [Google Scholar]
- [5].Roy M, Reddy PH, Iijima M, Sesaki H. Mitochondrial division and fusion in metabolism. Curr Opin Cell Biol 2015; 33C:111-8; https://doi.org/ 10.1016/j.ceb.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Itoh K, Nakamura K, Iijima M, Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends Cell Biol 2013; 23:64-71; PMID:23159640; https://doi.org/ 10.1016/j.tcb.2012.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Serasinghe MN, Chipuk JE. Mitochondrial Fission in Human Diseases. Handb Exp Pharmacol 2017; PMID:28040850; https://doi.org/ 10.1007/164_2016_38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tamura Y, Itoh K, Sesaki H. SnapShot: Mitochondrial dynamics. Cell 2011; 145:1158, e1; https://doi.org/ 10.1016/j.cell.2011.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Vanstone JR, Smith AM, McBride S, Naas T, Holcik M, Antoun G, Harper ME, Michaud J, Sell E, Chakraborty P, et al.. DNM1L-related mitochondrial fission defect presenting as refractory epilepsy. Eur J Hum Genetics 2015; PMID:26604000; https://doi.org/ 10.1038/ejhg.2015.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Eng J Med 2007; 356:1736-41; PMID:17460227; https://doi.org/ 10.1056/NEJMoa064436 [DOI] [PubMed] [Google Scholar]
- [11].Yoon G, Malam Z, Paton T, Marshall CR, Hyatt E, Ivakine Z, Scherer SW, Lee KS, Hawkins C, Cohn RD. Lethal disorder of Mitochondrial fission caused by mutations in DNM1L. J Pediatr 2016; 171:313-6 e2; https://doi.org/ 10.1016/j.jpeds.2015.12.060 [DOI] [PubMed] [Google Scholar]
- [12].Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol 2009; 186:805-16; PMID:19752021; https://doi.org/ 10.1083/jcb.200903065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto Y, et al.. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol 2009; 11:958-66; PMID:19578372; https://doi.org/ 10.1038/ncb1907 [DOI] [PubMed] [Google Scholar]
- [14].Kageyama Y, Zhang Z, Roda R, Fukaya M, Wakabayashi J, Wakabayashi N, Kensler TW, Reddy PH, Iijima M, Sesaki H. Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage. J Cell Biol 2012; 197:535-51; PMID:22564413; https://doi.org/ 10.1083/jcb.201110034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kageyama Y, Hoshijima M, Seo K, Bedja D, Sysa-Shah P, Andrabi SA, Chen W, Höke A, Dawson VL, Dawson TM, et al.. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J 2014; 33:2798-813; PMID:25349190; https://doi.org/ 10.15252/embj.201488658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ishihara T, Ban-Ishihara R, Maeda M, Matsunaga Y, Ichimura A, Kyogoku S, Aoki H, Katada S, Nakada K, Nomura M, et al.. Dynamics of mtDNA nucleoids regulated by mitochondrial fission is essential for maintenance of homogeneously active mitochondria during neonatal heart development. Mol Cell Biol 2014; PMID:25348719; https://doi.org/ 10.1128/MCB.01054-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Song M, Mihara K, Chen Y, Scorrano L, Dorn GW 2nd.. Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab 2015; 21:273-85; PMID:25600785; https://doi.org/ 10.1016/j.cmet.2014.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ikeda Y, Shirakabe A, Maejima Y, Zhai P, Sciarretta S, Toli J, Nomura M, Mihara K, Egashira K, Ohishi M, et al.. Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ Res 2015; 116:264-78; PMID:25332205; https://doi.org/ 10.1161/CIRCRESAHA.116.303356 [DOI] [PubMed] [Google Scholar]
- [19].Stepanyants N, Macdonald PJ, Francy CA, Mears JA, Qi X, Ramachandran R. Cardiolipin's propensity for phase transition and its reorganization by dynamin-related protein 1 form a basis for mitochondrial membrane fission. Mol Biol Cell 2015; 26:3104-16; PMID:26157169; https://doi.org/ 10.1091/mbc.E15-06-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bustillo-Zabalbeitia I, Montessuit S, Raemy E, Basanez G, Terrones O, Martinou JC. Specific interaction with cardiolipin triggers functional activation of Dynamin-Related Protein 1. PLoS One 2014; 9:e102738; PMID:25036098; https://doi.org/ 10.1371/journal.pone.0102738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ugarte-Uribe B, Muller HM, Otsuki M, Nickel W, Garcia-Saez AJ. Dynamin-related protein 1 (Drp1) promotes structural intermediates of membrane division. J Biol Chem 2014; 289:30645-56; PMID:25237193; https://doi.org/ 10.1074/jbc.M114.575779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Stepanyants N, Macdonald PJ, Francy CA, Mears JA, Qi X, Ramachandran R. Cardiolipin's propensity for phase transition and its reorganization by dynamin-related protein 1 form a basis for mitochondrial membrane fission. Mol Biol Cell 2015; 26:3104-16; PMID:26157169; https://doi.org/ 10.1091/mbc.E15-06-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Macdonald PJ, Stepanyants N, Mehrotra N, Mears JA, Qi X, Sesaki H, Ramachandran R. A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission. Mol Biol Cell 2014; 25:1905-15; PMID:24790094; https://doi.org/ 10.1091/mbc.E14-02-0728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Montessuit S, Somasekharan SP, Terrones O, Lucken-Ardjomande S, Herzig S, Schwarzenbacher R, Manstein DJ, Bossy-Wetzel E, Basañez G, Meda P, et al.. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell 2010; 142:889-901; PMID:20850011; https://doi.org/ 10.1016/j.cell.2010.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Adachi Y, Itoh K, Yamada T, Cerveny KL, Suzuki TL, Macdonald P, Frohman MA, Ramachandran R, Iijima M, Sesaki H. Coincident Phosphatidic acid interaction restrains Drp1 in mitochondrial division. Mol Cell 2016; 63:1034-43; PMID:27635761; https://doi.org/ 10.1016/j.molcel.2016.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, Frohman MA. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol 2006; 8:1255-62; PMID:17028579; https://doi.org/ 10.1038/ncb1487 [DOI] [PubMed] [Google Scholar]
- [27].Huang H, Gao Q, Peng X, Choi SY, Sarma K, Ren H, Morris AJ, Frohman MA. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell 2011; 20:376-87; PMID:21397848; https://doi.org/ 10.1016/j.devcel.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Baba T, Kashiwagi Y, Arimitsu N, Kogure T, Edo A, Maruyama T, Nakao K, Nakanishi H, Kinoshita M, Frohman MA, et al.. Phosphatidic acid (PA)-preferring phospholipase A1 regulates mitochondrial dynamics. J Biol Chem 2014; 289:11497-511; PMID:24599962; https://doi.org/ 10.1074/jbc.M113.531921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhang Y, Liu X, Bai J, Tian X, Zhao X, Liu W, Duan X, Shang W, Fan HY, Tong C, et al.. Mitoguardin regulates mitochondrial fusion through MitoPLD and is required for neuronal homeostasis. Mol Cell 2016; 61:111-24; PMID:26711011; https://doi.org/ 10.1016/j.molcel.2015.11.017 [DOI] [PubMed] [Google Scholar]
- [30].Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, Hinshaw JE. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Structural Mol Biol 2011; 18:20-6; https://doi.org/ 10.1038/nsmb.1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mashaghi A, Partovi-Azar P, Jadidi T, Nafari N, Maass P, Tabar MR, Bonn M, Bakker HJ. Hydration strongly affects the molecular and electronic structure of membrane phospholipids. J Chem Phys 2012; 136:114709; PMID:22443792; https://doi.org/ 10.1063/1.3694280 [DOI] [PubMed] [Google Scholar]
- [32].Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol 2008; 9:99-111; PMID:18216767; https://doi.org/ 10.1038/nrm2328 [DOI] [PubMed] [Google Scholar]
- [33].Kutateladze TG, Capelluto DG, Ferguson CG, Cheever ML, Kutateladze AG, Prestwich GD, Overduin M. Multivalent mechanism of membrane insertion by the FYVE domain. J Biol Chem 2004; 279:3050-7; PMID:14578346; https://doi.org/ 10.1074/jbc.M309007200 [DOI] [PubMed] [Google Scholar]
- [34].Strack S, Cribbs JT. Allosteric modulation of Drp1 mechanoenzyme assembly and mitochondrial fission by the variable domain. J Biol Chem 2012; 287:10990-1001; PMID:22334657; https://doi.org/ 10.1074/jbc.M112.342105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chang CR, Manlandro CM, Arnoult D, Stadler J, Posey AE, Hill RB, Blackstone C. A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division. J Biol Chem 2010; 285:32494-503; PMID:20696759; https://doi.org/ 10.1074/jbc.M110.142430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Frohlich C, Grabiger S, Schwefel D, Faelber K, Rosenbaum E, Mears J, Rocks O, Daumke O. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J 2013; 32:1280-92; PMID:23584531; https://doi.org/ 10.1038/emboj.2013.74 [DOI] [PMC free article] [PubMed] [Google Scholar]


