Significance
Understanding roles and mechanisms of protein posttranslational modifications (PTMs) would greatly impact multiple scientific domains, from bioengineering to biomedical science. PTMs are known to interfere with drug action and influence biochemical networks of engineered organisms. Many PTM sites have been identified, but it remains unclear under which conditions these sites are modified. Furthermore, there is a need to understand how the cell utilizes PTMs to increase fitness. Here, we approach this challenge by integrating tools from molecular biology, biochemistry, and systems biology to unravel mechanisms through which PTMs regulate enzymes throughout Escherichia coli metabolism and demonstrate how these individual PTMs further regulate pathways and ultimately cell phenotypes. This workflow could be applied to study PTMs and their roles across species.
Keywords: systems biology, posttranslational modifications, metabolism, protein chemistry, omics data
Abstract
Understanding the complex interactions of protein posttranslational modifications (PTMs) represents a major challenge in metabolic engineering, synthetic biology, and the biomedical sciences. Here, we present a workflow that integrates multiplex automated genome editing (MAGE), genome-scale metabolic modeling, and atomistic molecular dynamics to study the effects of PTMs on metabolic enzymes and microbial fitness. This workflow incorporates complementary approaches across scientific disciplines; provides molecular insight into how PTMs influence cellular fitness during nutrient shifts; and demonstrates how mechanistic details of PTMs can be explored at different biological scales. As a proof of concept, we present a global analysis of PTMs on enzymes in the metabolic network of Escherichia coli. Based on our workflow results, we conduct a more detailed, mechanistic analysis of the PTMs in three proteins: enolase, serine hydroxymethyltransferase, and transaldolase. Application of this workflow identified the roles of specific PTMs in observed experimental phenomena and demonstrated how individual PTMs regulate enzymes, pathways, and, ultimately, cell phenotypes.
The confluence of genomic analyses and computational power is rapidly changing the types of questions that can now be addressed in the biological and medical sciences. Current genomic, proteomic, and metabolomic datasets enable quantitative tracking of RNA transcripts, proteins, and metabolites in unprecedented detail (1–5). On the other hand, computational methods are limited in their capacity to address this increasingly diverse span of experimental data types (6). Addressing these and other challenges brought forth by these advancements requires creating interdisciplinary frameworks upon which disparate biological data types can be analyzed and interpreted.
Here, we take advantage of several synergistic domains of science—systems biology, biochemistry, and synthetic biology—to develop a workflow that reconciles systems-level, multiomics analysis and genome-scale modeling with all-atom molecular dynamics (MD) simulations. Bringing these disparate domains together enables us to address the multilayered challenge of characterizing posttranslational modifications (PTMs) of proteins. To this end, the confluence of these technologies addresses the questions: “What does each PTM do?” and “How does the cell use PTMs to regulate itself during changes in environmental conditions?”
It is widely accepted that PTMs are central to the elaborate control mechanisms that regulate bacterial metabolism as nutritional sources change (7–9). However, a mechanistic understanding of how PTMs on metabolic enzymes influence cellular fitness remains unclear. On one hand, great efforts have been made to understand how enzyme activity is controlled through PTMs through decades of biochemistry research. On the other hand, mechanisms of cellular adaptation are less clear, as applying omics data to study PTMs remains a challenge. Hundreds of potentially functional PTM sites have been identified in bacteria (10). While some may be spurious and low-stoichiometry chemical modifications (11), many are high stoichiometry and likely to regulate bacterial metabolism (12–16). However, it remains difficult to unravel the physiological roles of the PTMs in a high-throughput manner.
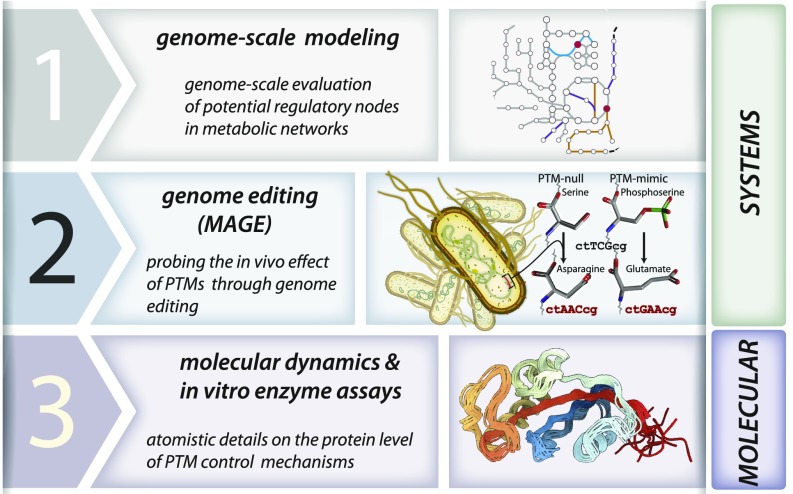
Studying PTMs is a multilayered challenge, in which one must first, demonstrate how modifications of protein residues influence protein activity; and second, understand if the changes in proteins influence pathways and cell physiology. To address this multifaceted question, our workflow consists of three stages (Fig. 1). In the first stage, we use genome-scale metabolic modeling to identify a subset of enzymes likely to require regulation during changes in environmental conditions. The second stage characterizes the cellular effect of probing experimentally measured PTM sites in the subset of enzymes through genome-editing techniques. The third stage utilizes all-atom MD simulations to understand the detailed mechanisms of the specific PTMs that are demonstrated to have an effect on cellular fitness. We apply this framework to study PTMs in Escherichia coli and their influence across multiple nutrient conditions. We demonstrate that this strategy is capable of clarifying complex changes in metabolic network usage, identifying key regulatory nodes, and elucidating the complex interplay between environment and the PTM state of the metabolic proteome.
Fig. 1.
A workflow for bridging systems and molecular science with multiomics data, genome-scale models of E. coli metabolism, and MD simulations. Our workflow involves a staging of computational and experimental methods: (1) computational genome-scale modeling to predict which enzymes require regulation across specific nutrient shifts; (2) genome editing through MAGE to probe the effect of changing PTM sites on cellular fitness; and (3) MD and in vitro enzyme assays to probe the effect of modifying a residue that is a known PTM site on protein configuration.
Results and Discussion
Stage One: Identify Regulatory Nodes in the E. coli Metabolic Network.
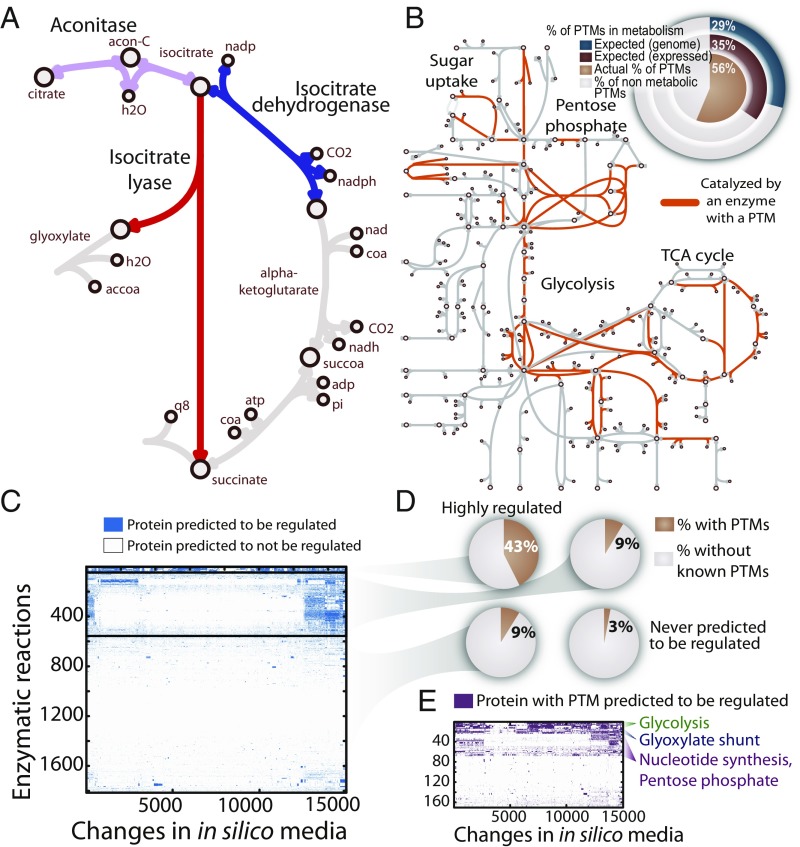
Stage one of the workflow (Fig. 1) identifies key enzymes in E. coli metabolism that require regulation (Fig. 2A). Since PTMs are enriched in metabolic pathways and may regulate them (Fig. 2B and SI Appendix, Fig. S1), this stage further demonstrates that PTMs are preferentially localized to these enzymes. First, using constraint-based modeling (17–19), we predict metabolic pathway usage, which estimates the steady-state flux through all metabolic pathways in a cell (19) (SI Appendix, Fig. S2). Second, we extend this framework to predict which enzymes require regulation in a sudden nutritional shift (to ensure optimal cell fitness). To do this, we use a method called Regulated Metabolic Branch Analysis (RuMBA) (SI Appendix, Fig. S3) to predict which metabolic enzymes, if regulated, are positioned to rapidly reroute flux to more favorable pathways. Lastly, we compare our findings with published lists (20–23) of metabolic enzymes with measured PTMs. With this framework, thousands of metabolic enzymes can be systematically assessed across thousands of nutrient shifts to understand which enzymes are likely modified during environmental shifts.
Fig. 2.
Metabolically regulated enzymes can be predicted in silico. (A) Metabolic regulation rebalances metabolism often at metabolic branch points, such as the flux split at isocitrate, which divides flux between the TCA cycle and glyoxylate shunt during the glucose–acetate diauxie. (B) PTMs are found on many enzymes in central metabolism and other pathways. We found that 56% of proteins with PTMs are in E. coli metabolism, which is ∼2× expected (29%) when considering protein-coding genes (or 35% when considering only expressed genes). (C) Clustering analysis of all enzymes predicted to require regulation for at least one of the 15,051 media shifts. (D) We found that 43% of the enzymes in the highly regulated cluster have one or more PTM, while ∼9% in the less regulated clusters have PTMs. Only 3% of the enzymes that never require regulation have PTMs. (E) Clusters of enzymes with PTMs reveal pathways that the PTMs will regulate more frequently during nutritional shifts.
Using the above schema, we applied RuMBA to study E. coli’s canonical diauxic shift from glucose to acetate metabolism. The metabolic network is used to simulate the metabolic flux through all pathways, thus enabling the computation of changes in flux splits at each metabolite node when shifting between two nutrient conditions (Fig. 2A). Here, we tested whether the flux through metabolic branch points changed significantly upon nutrient shift from one reaction to another reactions in the same branch point (SI Appendix, Fig. S3 B–D). Our model predicted a significant diversion of flux at the branch point between the TCA cycle and the glyoxylate shunt (7, 24) during the shift from glucose to acetate metabolism, suggesting that this branch point is likely regulated (P << 1 × 10−5; SI Appendix, Fig. S3E and Dataset S1). Consistent with these predictions, isocitrate dehydrogenase is used predominantly during glucose metabolism, while growth on acetate uses isocitrate lyase to support anaplerosis. To this end, RuMBA-predicted enzymes are likely to be situated at key points in the network for regulation to divert flux to key pathways.
We compared our predictions for the glucose–acetate diauxie to several experimentally validated cases to show that RuMBA accurately identifies key regulatory nodes in metabolism. First, the predicted regulators significantly overlap with enzymes that are known to be regulated by small molecules (using 1,219 experimentally validated cases; SI Appendix, Figs. S3F and S5 and Dataset S2), especially for enzymes that are regulated allosterically by metabolites (SI Appendix, Fig. S3G). Second, analysis of transcriptomics data indicates that regulation of these proteins is eventually reinforced through transcriptional regulation (P = 3 × 10−10; SI Appendix, Fig. S6). In other words, proteins are regulated transiently through small molecules and may be sustained on longer time scales by transcriptional changes. Third, we find that PTMs [including serine, threonine, or tyrosine phosphorylation, lysine acetylation, or succinylation sites (20–23)] are enriched among proteins that are predicted to require regulation (SI Appendix, Fig. S4). Furthermore, several of these PTMs significantly change in abundance between glucose and acetate media (Dataset S17) (16).
We further investigated changes at metabolic branch points under diverse changes in environmental conditions, beyond the glucose–acetate diauxie. We simulated changes in metabolism for 15,051 shifts between pairs of 174 different media (Dataset S4) and predicted regulation in each nutritional change. Across all conditions, known PTMs are enriched among the regulated set of enzymes in 92% of the simulated nutrient shifts [hypergeometric test; false discovery rate (FDR) < 0.01; SI Appendix, Fig. S4C]. We found that proteins with experimentally measured PTMs required regulation more than those without PTMs (Wilcoxon P = 6 × 10−6). RuMBA-predicted enzymes were clustered by nutrient shift (Fig. 2C), and our findings indicate that far fewer enzymes have experimentally measured PTM sites in clusters of enzymes that require occasional regulation (9%) or enzymes not requiring regulation (3%) (Fig. 2D). In contrast, 43% of the enzymes in the most highly regulated cluster had PTMs, which is enriched in glycolytic enzymes and the glyoxylate shunt (Fig. 2E and SI Appendix, Fig. S7). Thus, in E. coli, PTMs are effectively localized to enzymes that have the greatest potential to regulate metabolism under different growth conditions.
Stage Two: Characterize the Cellular Impact of Probing PTM Sites.
We hypothesize that regulation of the RuMBA-predicted enzymes (from stage one) is critical to rapidly force flux from one branch to another immediately following a change in nutrient condition. To test this hypothesis, we used genome editing to study the in vivo effect of mimicking modification as well as disabling the cell’s ability to modify specific PTM sites within this subset of enzymes during nutrient shifts. The main understanding gained from this stage is identification of PTM sites (among the subset of RuMBA-predicted enzymes), that elicit changes in cellular fitness under specific nutrient shifts.
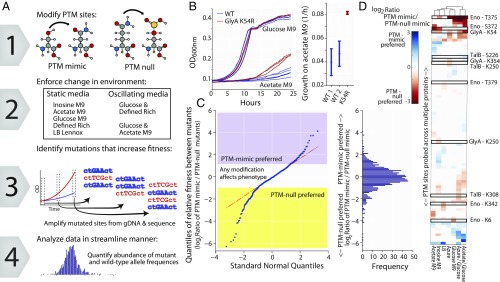
We assessed global changes in cell fitness after using multiplexed automated genome engineering (MAGE) (25) to perturb PTM status in a pooled screen approach (Fig. 3A) (26). PTM sites were selected from the RuMBA-predicted enzymes (268 modifications to 134 known PTM sites in 61 proteins, covering all proteins with PTMs and known protein structures at the time of the experimental design) and studied across five media conditions at multiple time points using MAGE-Sequencing (MAGE-Seq) (27) (Datasets S5 and S6). Codons were changed to (i) mimic a PTM (i.e., replacing S/T/Y residues with glutamate to mimic the negative charge of phosphorylation or asparagine to mimic the lack of charge in acetylation; referred to as “PTM-mimic”); or (ii) remove the propensity for PTM addition (i.e., replacing S/T/Y with asparagine which cannot be phosphorylated to mimic the size and polarity of a S/T/Y or lysine with arginine, which cannot be acetylated but retains the positive charge; referred to as “PTM-null”).
Fig. 3.
Enforcing PTM states with MAGE demonstrates that PTMs are functionally relevant and impact fitness. (A) Codons in the MAGE-generated mutant population were edited at PTM sites to mimic the PTM or disallow the PTM (PTM-null). A mutant pool harboring 268 different mutations to 134 codons on 61 genes was obtained and screened on diverse media conditions. Mutant abundance was quantified by amplicon-sequencing MAGE sites (MAGE-Seq). The mutant pool was screened on five static media conditions and two oscillating conditions to identify PTM sites that, when forced in a PTM-mimic or -null state, had altered cellular fitness in specific conditions. (B) Individual mutants showed media-specific responses (e.g., enhanced growth on acetate M9 media for the GlyA K54R mutant; Right). (C) PTM-mimic and -null modifications for each growth condition and time point were compared. PTM sites on 35 proteins exhibited a significant preference for the PTM-mimic or -null states. (D) Most mimic/null preferences were condition-specific, with oscillating media showing more extensive preferences (example PTM sites are highlighted; see SI Appendix, Fig. S9C for all sites with significant preferences for PTM-mimic or -null modifications).
Targeted mutations impacted organism fitness under specific media conditions, as exemplified by the PTM-null mutation at position K54 of serine hydroxymethyltransferase (glyA). For example, this mutation doubled the growth rate on acetate M9 minimal medium, but did not have a considerable impact on growth on glucose M9 minimal medium (Fig. 3B). However, following glucose depletion, the K54R mutant showed an increased growth rate during the brief shift to residual fermentation products (SI Appendix, Fig. S8).
Computational analysis of the MAGE data indicate that modification of 88% of the sites significantly increased or decreased in fitness in at least one condition (Fig. 3D and SI Appendix, Fig. S9 A and C). Furthermore, 35 genes had PTM sites showing significantly different impacts between PTM-mimic and -null variants in at least one condition (FDR < 0.05; Fig. 3C). This suggests that many PTM states are preferred in specific nutrient conditions.
An analysis of all screen data points to global patterns: PTMs influence cellular fitness by regulating critical proteins at key positions in dynamic nutrient shifts. We quantified the properties of the modified proteins that contributed the most to fitness changes using a generalized estimating equation (GEE) (28). First, our analysis showed that the enzymes that are critical for survival are prime targets for regulation to gain fitness advantages (SI Appendix, Fig. S9B). Second, modifications impacting fitness often occur at positions in proteins that impact enzyme activity (e.g., salt bridge residues or near active site residues, P << 1 × 10−10 and P = 9 × 10−7, respectively). Lastly, mutations more significantly impact cellular fitness when nutrients oscillate (e.g., shifting between glucose to acetate minimal medium and back to glucose; P = 3 × 10−3). Thus, PTMs are positioned on the enzymes that directly regulate growth, and when proteins are forced to remain in a single PTM state (thereby preventing transient control at these sites), they more significantly impact cellular fitness.
Stage Three: Elucidate Control Mechanisms of PTMs.
Atomistic simulations provide a contextual basis for the analysis of how PTMs modulate protein structure and function. To unravel the specific mechanisms by which the PTMs regulate the proteins, we studied three E. coli proteins. Classical MD simulations and in vitro biochemical assays showed that the PTMs (i) modulate interactions at protein interfaces in serine hydroxymethyltransferase (GlyA), (ii) manipulate binding site conformations in transaldolase, and (iii) control catalytic residues in enolase.
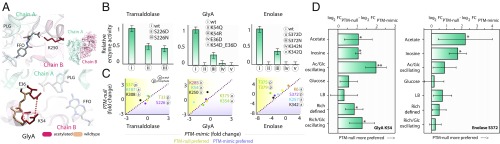
First, we found that PTMs can influence enzyme activity by modulating protein interaction. Specifically, MD simulations, in vitro enzymatic assays, and MAGE for GlyA all suggest that acetylation at the dimer interface disrupts activity, thus providing a transient control mechanism (Fig. 4A and SI Appendix, Fig. S10). For GlyA, which is regulated in 12% of the 15,051 simulated substrate shifts, acetylation takes place at positions K54, K250, and K354, near the dimer interface, which also form part of the substrate binding domain (Fig. 4A). Analysis of a 100-ns MD trajectory suggests that acetylation of K250 and K354 impacts the structure of the N-terminal domain of monomeric GlyA [average root mean squared deviation (rmsd) of 3.5 Å relative to the crystallographic structure; SI Appendix, Fig. S11] in addition to cofactor binding (tetrahydrofolate). Site-directed mutation of these residues to mimic acetylation ultimately influenced in vitro enzymatic activity (Fig. 4B) as well as organism fitness for the PTM-mimic in vivo (Fig. 4C). Similarly, acetylation of K54 was disruptive to cellular fitness, as it disrupted a salt bridge interaction with E36 of the neighboring subunit, increasing the intermolecular interaction by 2–4 Å, relative to WT protein (Fig. 4A and SI Appendix, Fig. S12). These findings are consistent with in vitro assays that mimic the loss of this interaction (Fig. 4B), which demonstrate nearly a complete loss in enzymatic activity. In vivo, the K54N PTM-mimic modification significantly decreased fitness, while the K54R mutant was preferred (Fig. 4C). This response was strongest for growth on gluconeogenic substrates, in which the PTM could block their catabolism when glycolytic substrates were available. Furthermore, modifying the PTM state was especially disruptive to the cell when nutrient condition oscillated between acetate and glucose (Fig. 4D).
Fig. 4.
PTMs affecting cell fitness modulate binding configurations, protein–protein interactions, and enzymatic activity. (A) Molecular interactions that take place in serine hydroxymethyl transferase (GlyA). Acetylation of K250 of GlyA disrupts a salt bridge, leading to decreased THF binding (Upper). K54 participates in a salt bridge with E36 of the other protein subunit (Lower), and acetylation increases the distance between K54 and E36 by >4 Å (SI Appendix, Fig. S12). (B) Enzymatic assays for transaldolase, GlyA, and enolase. (C) Average fitness for all significantly changing conditions is plotted for the PTM-mimic (x axis) and the PTM-null (y axis) MAGE mutants for transaldolase, GlyA, and enolase. Comparing B and C, we see that, for transaldolase, enzymatic assays show reduced activity of S226D and S226N mutants, but the open conformation rescues the reduced activity of the S226 MAGE PTM-mimic state. For GlyA, the K54 PTM-mimic state decreases catalytic activity and cellular fitness, while the K250 PTM-null state significantly enhances growth. For enolase, PTMs modifying two catalytic residues abolish enzyme activity, which is consistent with MAGE mutants preferring the PTM-null genotype. (D) Forcing the PTM-mimic mutation of K54 in GlyA and S372/K342 in enolase inhibits growth in gluconeogenic conditions, and especially deleterious to growth during oscillating conditions. FFO, 5-formyl tetrahydrofolate; PLG, N-glycine-[3-hydroxy-2-methyl-5-phosphono-oxymethyl-pyridin-4-yl-methane].
Second, our findings suggest that certain PTMs act as precise, yet transient, mechanochemical “switches” for tuning enzyme activity by impacting active site accessibility. For transaldolase, which is regulated in 22% of the 15,051 simulated substrate shifts, many experimentally detected PTMs localize to a shallow channel where metabolites pass to enter/exit the active site (SI Appendix, Fig. S10). The conformation of this channel is known to impact substrate binding and catalysis (29). We studied protein conformations of wild-type (WT) and five modified transaldolase proteins during a 1.05-μs MD trajectory (SI Appendix, Fig. S13) and found that PTMs on certain residues influence the accessibility of the channel, namely, S226 and K308. Phosphorylation of S226 promoted an opening of the channel, whereas acetylation strongly induced its closing. In vitro, modification of S226 reduced enzyme activity by 40% (Fig. 4B), yet the MAGE PTM-mimic state was preferred in specific conditions compared with the PTM-null state (Fig. 4C), presumably to maintain the opening of the substrate channel. Forcing the mimicking of acetylation at K308 significantly decreased organism fitness (Fig. 4C), by disrupting the accessibility of the binding pocket, which is upheld by a salt bridge between D305 and S37. Thus, for transaldolase, PTMs seem to be used to maintain binding-pocket accessibility, and this influences cellular fitness in specific conditions.
Third, modification of catalytic residues directly impact protein reactivity, as found for enolase. Enolase was a branch-point enzyme in the metabolic network that was predicted to be regulated in 48% of the 15,051 substrate shifts. An analysis of the protein structure showed that PTMs on active-site residues S372 and K341 destabilized the binding of two Mg2+ ions, which are required for catalysis (SI Appendix, Figs. S10 and S14). We further performed in vitro enzyme assays to study the effect of modifying K341 and S372 on enzyme activity (Fig. 4B and SI Appendix, Fig. S15). Consistent with a recent study (30), mutating K341 to asparagine or glutamine completely arrested catalysis. Mutating S372 to aspartate also completely arrested activity, as the phosphorylation mimic introduced a negative charge near Mg2+, while S372N only partially decreased activity, thus showing that the charge of the PTM is more responsible for switching the enzyme off. These changes also caused significant impacts on organism fitness in vivo across certain nutrient environments (Fig. 4C). Specifically, the PTM-null state was preferred for gluconeogenic substrates (Fig. 4D). These findings suggest that the direct modulation of catalytic residues provides a precise mechanism to regulate a highly sensitive branch point, such as for enolase.
Conclusions
From a systems perspective, determining the molecular basis of a phenotype requires a complete delineation of the cell parts and pathway structure, and an understanding of how the parts and pathways interact with the cell environment. Our combined computational and experimental approach provides a means to reliably navigate a set of experimentally determined PTMs and probe promising functional roles considering specific environmental perturbations. These analyses open vistas in systems biology, empowering the systematization of biochemistry and shaping the study of PTMs. Indeed, approaches used here could be applied to study PTMs in other organisms (31). Perhaps most important is the approach taken to understand how a modification at specific sites in individual proteins can impact biological fitness, both on the molecular and physiological levels.
Ultimately, in this work, we demonstrated how PTMs fit into the regulation of E. coli metabolism and cell fitness. Specifically, our work suggests that many PTMs are poised to help cells respond to familiar extrinsic nutrient fluctuations and intrinsic expression noise as the nutrient environment fluctuates. When PTMs are deployed to suppress pathways that are suboptimal following an acute stress or nutritional change, it demands less energy and allows for an immediate response, compared with irreversible protein degradation or transcriptional and translational control mechanisms (13). Our work also clearly shows, from multiple angles, that PTMs can directly impact cellular fitness. Furthermore, we elucidated examples of how regulation of specific enzymes achieves this result. Indeed, by carrying out three stages of the workflow, we find that many PTMs are located at model-predicted branch points in the metabolic network, where they could control cell physiology. Furthermore, many PTMs occur at functionally relevant positions in the essential proteins, exactly where the most impactful regulation could be elicited (e.g., salt bridges and active sites), as we found through the analyses of enolase, transaldolase, and serine hydroxymethyltransferase (and many other proteins; SI Appendix, Figs. S3 and S4). Perhaps most importantly, we found that the modification of PTM states impact cellular fitness most when the primary nutrients change, which reinforces the notion that PTMs help the cells to adapt as nutrient conditions change.
Methods
PTMs and Data on Metabolic Regulation.
Lists of metabolic proteins with PTMs were obtained from proteomic studies on serine, threonine, or tyrosine phosphorylation (20), lysine acetylation (21, 22), or lysine succinylation (23). All reported occurrences of noncovalent metabolite-mediated metabolic regulation were obtained from EcoCyc (32) and are reported in Dataset S2.
Constraint-Based Modeling Analysis.
RuMBA computes the relative change in flux splits, based on model-simulated flux by using Markov chain Monte Carlo (MCMC) sampling. Simulations were conducted by using the COBRA Toolbox (33, 34). MCMC sampling of the metabolic flux provided the range and distribution of feasible steady-state fluxes for each reaction (35). In RuMBA, for each metabolite, all incoming and outgoing fluxes for each MCMC flux vector were summed. For each ith reaction, the fraction of total flux through the metabolite, vmet, was computed as follows:
where vi is the flux through reaction i and fi is the fraction of all flux passing through the metabolite of interest that is passing through reaction i. This is done for each feasible flux vector, to obtain a distribution of fi fractions for each reaction for the two growth conditions of interest. A P value is computed that measures the overlap of the fi values for that reaction under the given growth condition—that is, the probability of finding an fi value in the first growth condition that is equal to or more extreme than an fi value for the same reaction in the second growth condition. The P values are corrected for multiple hypotheses (FDR < 0.01). Since this method focuses on flux splits, rather than absolute flux, moderate variations in metabolite uptake rates have little effect on our predictions (SI Appendix, Fig. S16). See SI Appendix for more details on the method and its validation.
Oligonucleotide Design for MAGE.
A panel of phosphorylation and acetylation sites were identified from previous studies (20–22), and codons for the phosphorylation sites on serine and threonine or lysine acetylation were changed. Serine and threonine were changed to glutamate to mimic the phosphorylation and an asparagine to mimic the unphosphorylated residue. Lysine was converted to glutamine to mimic the acetylated state and arginine to inhibit acetylation. All 90-mer MAGE oligonucleotide sequences are provided for the subset of genes studied (Datasets S5 and S6). MAGE oligonucleotides were synthesized by Integrated DNA Technologies with standard purification.
MAGE.
MAGE was conducted as described (25). Four rounds of MAGE were conducted. Multiplex allele-specific colony PCR was used as described (36) to verify mutations and to identify specific mutants for phenotyping. See SI Appendix for further details.
Screen for PTM Mutation Fitness.
We tested multiple media conditions (e.g., LB, Azure-defined rich + glucose, Glucose M9, Acetate M9, and Inosine M9) at 30 °C as well as for two oscillating conditions (Azure and glucose M9 or glucose and acetate M9). The screens were sampled at two to four time points (Dataset S12), and allele frequencies were quantified by amplifying the genes with PTM sites from the genomic DNA and sequencing the amplicons with next-generation sequencing. To obtain the final pool with all MAGE mutants, MAGE was conducted in five batches, each with ∼46 different MAGE oligos. MAGE oligos were grouped to ensure that no two oligos targeted within 100 base pairs of each other, to avoid competition between oligos in any one pool. The batches of mutants were combined and subjected to phenotypic selections.
Measurements of the allele frequency were made at 3 h after electroporation and pooling and overnight storage at 4 °C. Cells pellets were subsequently washed with the medium used in the screen. Cells were maintained at 30 °C at exponential growth by serial dilution at regular intervals (about every three doublings; see Dataset S12 for values). Aliquots were saved at each dilution, and time points were selected for subsequent sequencing and analysis of allele frequencies at each PTM site.
MD Simulations.
Classical MD simulations were performed starting from protein crystal structures, solvated with explicit solvent. Each structure was manually changed to study individual PTMs of interest. Parameters for phosphorylated amino acids were taken from previous studies (37). For each system, graphics processing unit-enabled PMEMD MD (38) was performed, by using the AMBER (39) 99sb force field (40, 41) for 50–120 ns per protein state (i.e., substrate-bound vs. substrate-free in WT and modified variant proteins). For more details, see SI Appendix.
Enzymatic Assays.
Enolase activity was assayed by measuring the conversion of 2-phospho-d-glycerate (2-PGE) to phosphoenolpyruvate (PEP) at 25 °C as described (42) with modifications. The reaction mixture contained 1 mM 2-PGE in reaction buffer (100 mM Hepes buffer, pH 8.5, 7.7 mM KCl, and 10 mM MgSO4, prewarmed to 25 °C), and enolase was added to initiate the reaction. The reaction was monitored spectrophotometrically by measuring absorbance at 240 nm for the production of PEP at 30-s intervals for 10 min.
For transaldolase, the reverse reaction catalyzed by transaldolase was tested at room temperature as described (43) with some modifications. The reaction mixture contained 5 mM d-fructose-6-phosphate, 0.2 mM erythrose-4-phosphate, 0.1 mM NADH, and 10 μg of α−glycerolphosphate dehydrogenase-triosephosphate isomerase (Sigma) in reaction buffer (40 mM triethanolamine, pH 7.6, and 5 mM EDTA), and transaldolase was added to initiate the reaction. The reaction was monitored spectrophotometrically by measuring absorbance at 340 nm at 30-s intervals for 10 min.
Serine hydroxymethyltransferase activity of THF-dependent cleavage was measured as described (44) with some modifications. The reaction mixture in a final volume of 75 μL consisted of 0.3 mM pyridoxal phosphate, 40 mM mercaptoethanol, 15 mM serine, and serine hydroxymethyltransferase in reaction buffer (10 mM potassium phosphate, pH 7.3, and 0.5 mM EDTA). After a 5-min incubation at 37 °C, 1 mM THF was added to initiate the reaction. The reaction was stopped after 2 min by the addition of 100 μL of pH 9.5 carbonate buffer. Twenty microliters of 2 mM NADP+ and enough methylene tetrahydrofolate dehydrogenase were then added to carry out the auxiliary reaction, and the increase in absorbance at 340 nm was followed to completion.
Supplementary Material
Acknowledgments
This work was supported by Swiss National Science Foundation Grant p2elp2_148961; the Gordon and Betty Moore Foundation GBMF 2550.04 Life Sciences Research Foundation postdoctoral fellowship; National Institutes of Health Grants R01-GM057089 and R35-GM119850; US Department of Energy Grant DE-FG02-02ER63445; and Novo Nordisk Foundation Center for Biosustainability Grant NNF10CC1016517. We also acknowledge National Energy Research Scientific Computing Center and XSEDE computer facilities (MCB140152).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811971115/-/DCSupplemental.
References
- 1.Kahn SD. On the future of genomic data. Science. 2011;331:728–729. doi: 10.1126/science.1197891. [DOI] [PubMed] [Google Scholar]
- 2.Gross M. Riding the wave of biological data. Curr Biol. 2011;21:R204–R206. doi: 10.1016/j.cub.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Metzker ML. Sequencing technologies—The next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 4.Fuhrer T, Zamboni N. High-throughput discovery metabolomics. Curr Opin Biotechnol. 2015;31:73–78. doi: 10.1016/j.copbio.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z, Wu S, Stenoien DL, Paša-Tolić L. High-throughput proteomics. Annu Rev Anal Chem (Palo Alto, Calif) 2014;7:427–454. doi: 10.1146/annurev-anchem-071213-020216. [DOI] [PubMed] [Google Scholar]
- 6.Berger B, Peng J, Singh M. Computational solutions for omics data. Nat Rev Genet. 2013;14:333–346. doi: 10.1038/nrg3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holms WH, Bennett PM. Regulation of isocitrate dehydrogenase activity in Escherichia coli on adaptation to acetate. J Gen Microbiol. 1971;65:57–68. doi: 10.1099/00221287-65-1-57. [DOI] [PubMed] [Google Scholar]
- 8.López-Maury L, Marguerat S, Bähler J. Tuning gene expression to changing environments: From rapid responses to evolutionary adaptation. Nat Rev Genet. 2008;9:583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- 9.Pisithkul T, Patel NM, Amador-Noguez D. Post-translational modifications as key regulators of bacterial metabolic fluxes. Curr Opin Microbiol. 2015;24:29–37. doi: 10.1016/j.mib.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Cain JA, Solis N, Cordwell SJ. Beyond gene expression: The impact of protein post-translational modifications in bacteria. J Proteomics. 2014;97:265–286. doi: 10.1016/j.jprot.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 11.Weinert BT, et al. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell. 2013;51:265–272. doi: 10.1016/j.molcel.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Baeza J, et al. Stoichiometry of site-specific lysine acetylation in an entire proteome. J Biol Chem. 2014;289:21326–21338. doi: 10.1074/jbc.M114.581843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chubukov V, Gerosa L, Kochanowski K, Sauer U. Coordination of microbial metabolism. Nat Rev Microbiol. 2014;12:327–340. doi: 10.1038/nrmicro3238. [DOI] [PubMed] [Google Scholar]
- 14.Hansen A-M, et al. The Escherichia coli phosphotyrosine proteome relates to core pathways and virulence. PLoS Pathog. 2013;9:e1003403. doi: 10.1371/journal.ppat.1003403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kochanowski K, Sauer U, Noor E. Posttranslational regulation of microbial metabolism. Curr Opin Microbiol. 2015;27:10–17. doi: 10.1016/j.mib.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Lim S, Marcellin E, Jacob S, Nielsen LK. Global dynamics of Escherichia coli phosphoproteome in central carbon metabolism under changing culture conditions. J Proteomics. 2015;126:24–33. doi: 10.1016/j.jprot.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 17.Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bordbar A, Monk JM, King ZA, Palsson BO. Constraint-based models predict metabolic and associated cellular functions. Nat Rev Genet. 2014;15:107–120. doi: 10.1038/nrg3643. [DOI] [PubMed] [Google Scholar]
- 19.Lewis NE, Nagarajan H, Palsson BO. Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods. Nat Rev Microbiol. 2012;10:291–305. doi: 10.1038/nrmicro2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macek B, et al. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol Cell Proteomics. 2008;7:299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, et al. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics. 2009;8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu BJ, Kim JA, Moon JH, Ryu SE, Pan J-G. The diversity of lysine-acetylated proteins in Escherichia coli. J Microbiol Biotechnol. 2008;18:1529–1536. [PubMed] [Google Scholar]
- 23.Zhang Z, et al. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinar G, Rabinowitz JD, Alon U. Robustness in glyoxylate bypass regulation. PLoS Comput Biol. 2009;5:e1000297. doi: 10.1371/journal.pcbi.1000297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haimovich AD, Muir P, Isaacs FJ. Genomes by design. Nat Rev Genet. 2015;16:501–516. doi: 10.1038/nrg3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelsic ED, et al. RNA structural determinants of optimal codons revealed by MAGE-seq. Cell Syst. 2016;3:563–571.e6. doi: 10.1016/j.cels.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shults J, Ratcliffe SJ. GEEQBOX: A MATLAB toolbox for generalized estimating equations and quasi-least squares. J Stat Softw. 2008;25:1–14. [Google Scholar]
- 29.Lehwess-Litzmann A, et al. Twisted Schiff base intermediates and substrate locale revise transaldolase mechanism. Nat Chem Biol. 2011;7:678–684. doi: 10.1038/nchembio.633. [DOI] [PubMed] [Google Scholar]
- 30.Nakayasu ES, et al. Ancient regulatory role of lysine acetylation in central metabolism. MBio. 2017;8:e01894-17. doi: 10.1128/mBio.01894-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barber KW, et al. Encoding human serine phosphopeptides in bacteria for proteome-wide identification of phosphorylation-dependent interactions. Nat Biotechnol. 2018;36:638–644. doi: 10.1038/nbt.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keseler IM, et al. EcoCyc: A comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schellenberger J, et al. Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox v2.0. Nat Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heirendt L, et al. 2017. Creation and analysis of biochemical constraint-based models: The COBRA Toolbox v3.0. arXiv:1710.04038. Preprint, posted October 11, 2017.
- 35.Schellenberger J, Palsson BØ. Use of randomized sampling for analysis of metabolic networks. J Biol Chem. 2009;284:5457–5461. doi: 10.1074/jbc.R800048200. [DOI] [PubMed] [Google Scholar]
- 36.Wang HH, et al. Genome-scale promoter engineering by coselection MAGE. Nat Methods. 2012;9:591–593. doi: 10.1038/nmeth.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Homeyer N, Horn AHC, Lanig H, Sticht H. AMBER force-field parameters for phosphorylated amino acids in different protonation states: Phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine. J Mol Model. 2006;12:281–289. doi: 10.1007/s00894-005-0028-4. [DOI] [PubMed] [Google Scholar]
- 38.Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, Walker RC. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J Chem Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 39.Case DA, et al. 2014 Amber 14. Available at https://orbilu.uni.lu/handle/10993/16614. Accessed July 8, 2015.
- 40.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem. 2000;21:1049–1074. [Google Scholar]
- 41.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu H, et al. Steinernema glaseri surface enolase: Molecular cloning, biological characterization, and role in host immune suppression. Mol Biochem Parasitol. 2012;185:89–98. doi: 10.1016/j.molbiopara.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Huang H, et al. The crystal structure and identification of NQM1/YGR043C, a transaldolase from Saccharomyces cerevisiae. Proteins. 2008;73:1076–1081. doi: 10.1002/prot.22237. [DOI] [PubMed] [Google Scholar]
- 44.Schirch LV. [180] Serine transhydroxymethylase (rabbit•liver) Methods Enzymol. 1971;17:335–340. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.