Significance
Here, we demonstrate that the antirecombinase Srs2 is unable to dismantle recombination intermediates containing the meiosis-specific recombinase Dmc1. Our work defines a function of Dmc1 as an inhibitor of the prototypical antirecombinase Srs2. Based on these findings, we propose that the presence of Dmc1 may help to prevent Srs2 from disruption of early meiotic recombination intermediates, which may in turn be anticipated to favor the formation of cross-over recombination products.
Keywords: Srs2, Rad51, Dmc1, meiosis, homologous recombination
Abstract
Cross-over recombination products are a hallmark of meiosis because they are necessary for accurate chromosome segregation and they also allow for increased genetic diversity during sexual reproduction. However, cross-overs can also cause gross chromosomal rearrangements and are therefore normally down-regulated during mitotic growth. The mechanisms that enhance cross-over product formation upon entry into meiosis remain poorly understood. In Saccharomyces cerevisiae, the Superfamily 1 (Sf1) helicase Srs2, which is an ATP hydrolysis-dependent motor protein that actively dismantles recombination intermediates, promotes synthesis-dependent strand annealing, the result of which is a reduction in cross-over recombination products. Here, we show that the meiosis-specific recombinase Dmc1 is a potent inhibitor of Srs2. Biochemical and single-molecule assays demonstrate that Dmc1 acts by inhibiting Srs2 ATP hydrolysis activity, which prevents the motor protein from undergoing ATP hydrolysis-dependent translocation on Dmc1-bound recombination intermediates. We propose a model in which Dmc1 helps contribute to cross-over formation during meiosis by antagonizing the antirecombinase activity of Srs2.
Homologous recombination (HR) is necessary for maintaining genome integrity across all domains of life, and HR is also required to repair programmed double strand breaks (DSBs) during meiosis (1–5). Meiosis is a broadly conserved eukaryotic cellular program that yields haploid progeny from diploid precursors and is essential for sexual reproduction (4, 6, 7). HR plays a central role in meiosis, and, in most organisms, the chromosomal changes that take place during meiosis are dependent on recombination (4, 6, 7). Importantly, the transition from mitotic repair to meiotic repair is denoted by a marked difference in cross-over vs. non–cross-over HR outcomes. Cross-overs are considered less desirable in mitotic cells because they can lead to loss of heterozygosity and other chromosomal rearrangements (5, 8). In contrast, cross-overs are favored during meiosis because they are essential for chromosome segregation and they also help enhance genetic diversity among progeny (4, 6, 7). The molecular basis for the differential regulation of cross-overs during mitosis and meiosis remains poorly understood.
The DNA transactions that take place during HR are catalyzed by the broadly conserved Rad51/RecA family of recombinases, which are ATP-dependent DNA-binding proteins that form extended helical filaments on ssDNA (2, 3). Most eukaryotes require two recombinases, Rad51, which catalyzes DNA strand exchange during mitosis, and Dmc1, which catalyzes strand exchange during meiosis (1, 3, 4). Rad51 and Dmc1 are ∼46% identical and arose from a gene-duplication event early in eukaryotic evolution coinciding with the emergence of meiosis (1, 3, 4, 9). However, it remains unknown why eukaryotes require a specialized recombinase for meiosis, and the relationship between recombinase identity and the regulation of cross-over vs. non–cross-over recombination outcomes remains unknown (1–5).
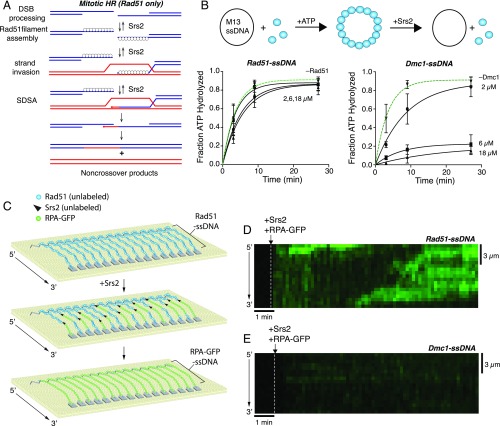
Srs2 is an ATP-dependent 3′→5′ ssDNA motor protein that plays a central role in minimizing cross-overs during mitosis (2, 10–17). Srs2 functions by dismantling Rad51-ssDNA and D-loop intermediates, thereby channeling HR intermediates through the synthesis-dependent strand annealing (SDSA) pathway, which exclusively yields non–cross-over recombination products (Fig. 1A) (2, 10–16). Consequently, srs2Δ mutants have a mitotic hyperrecombination phenotype characterized by frequent cross-overs and gross chromosomal rearrangements (15, 16, 18–20). Srs2 antirecombinase activity is counterbalanced by the Rad51 paralog complex Rad55/57, the Shu complex (composed of the Rad51 paralogs Psy3 and Csm2 and the SWIM-domain proteins Shu1 and Shu2), and Rad52, all of which enhance assembly or stability of early HR intermediates (Fig. 1A) (21–24). The role of Srs2 in helping to minimize cross-overs during mitosis is well established, but its functions during meiosis remain poorly understood (15). Srs2 expression is up-regulated during meiosis (25), but srs2Δ mutants have relatively mild meiotic phenotypes (15, 26). However, mechanistic interpretation of the molecular defects underlying srs2Δ phenotypes are complicated by the fact that Srs2 also participates in HR-independent processes related to genome integrity (reviewed in refs. 16 and 27), in particular with respect to replication through DNA secondary structure (28–30), replication checkpoint activation (31, 32), preventing ssDNA gaps present at stalled replication forks from being used to initiate recombination (33, 34), and Top1-mediated removal of misincorporated ribonucleotides (35).
Fig. 1.
Srs2 cannot remove Dmc1 from ssDNA. (A) Model for mitotic HR highlighting the regulatory role of Srs2 in disruption of Rad51-containing intermediates. (B) Srs2 ATP hydrolysis assays with Rad51 or Dmc1. Data points represent the mean ± SD of three experiments. (C) Schematic of ssDNA curtain assay for Srs2. (D) Kymograph showing unlabeled Srs2 (500 pM) disruption of Rad51-ssDNA as revealed by RPA-GFP (100 pM) rebinding (shown in green). (E) Kymograph showing that unlabeled Srs2 (500 pM) does not dismantle Dmc1-ssDNA.
Interestingly, Srs2 overexpression in meiosis results in the disruption of Rad51 repair foci, but does not disrupt Dmc1 repair foci (36), leading to the suggestion that Srs2 may not play a major role in regulating HR in meiosis (15). However, the activities of Srs2 on meiotic HR intermediates have not been explored in vitro, and it remains unknown whether this differential response of Rad51 and Dmc1 foci to Srs2 overexpression is caused by an inherent difference between the two recombinases, or whether it might reflect the effects of another protein component of the meiotic presynaptic complex (reviewed in ref. 3), such as the Dmc1-specific mediator protein complex Mei5/Sae3 (37), the Dmc1 accessory factors Hop2/Mnd1 (38, 39) or Rdh54 (40), or perhaps the ZMM proteins (including, Zip1, Zip2, Zip3, Zip4/Spo22, Mer3, Msh4, and Msh5), which help to coordinate recombination with synaptonemal complex formation and have also been implicated as inhibitors of Sgs1 (41).
To gain insights into the molecular mechanisms governing cross-over regulation during meiosis, here, we investigated the ability of a central regulator of mitotic cross-over formation, the Srs2 antirecombinase, to act upon meiotic recombination intermediates. By using a combination of biochemical and single-molecule imaging assays, we show that the meiosis-specific recombinase Dmc1 is a highly potent inhibitor of Srs2 activity. Dmc1 acts by inhibiting the ATP hydrolysis by Srs2, which in turn prevents Srs2 from undergoing ATP-dependent translocation on ssDNA. Our results are consistent with a model in which direct physical contact with the 3′ ends of the Dmc1 filaments halts Srs2 motor activity, suggesting the possibility that this inhibition occurs through an allosteric regulatory mechanism. We propose a model in which meiotic recombination intermediates containing Dmc1 are protected from disruption by the antirecombinase activity of Srs2, and therefore the ability of Dmc1 to inhibit Srs2 may help to promote cross-over recombination outcomes during meiosis.
Results
Dmc1 Is a Potent Inhibitor of Srs2 ATP Hydrolysis Activity.
To examine potential functions for Srs2 regulation during meiosis, we sought to determine whether Srs2 could dismantle Dmc1-ssDNA filaments. Srs2 uses ATP-dependent translocation to remove Rad51 from ssDNA; therefore ATP hydrolysis activity is a good proxy for Srs2 antirecombinase activity (10, 11). As expected, Srs2 retained vigorous ATP hydrolysis activity in the presence of increasing concentrations of Rad51 (Fig. 1B). In striking contrast, ATP hydrolysis by Srs2 was inhibited with increasing concentrations of Dmc1 (Fig. 1B). These bulk biochemical data strongly suggest that Srs2 was unable to remove the meiosis-specific recombinase Dmc1 from ssDNA.
Rad51I345T Does Not Strongly Inhibit Srs2 ATP Hydrolysis Activity.
Rad55/57 and Rad52 have been implicated as potential regulators of Srs2 activity, albeit through poorly understood mechanisms (21, 22). Interestingly, Rad55/57 and Rad52 have no impact on Srs2 ATP hydrolysis rates (22, 24, 42), and Srs2 readily removes Rad52 from RPA–ssDNA complexes (42). Rad51I345T is a suppressor mutation that bypasses the need for Rad55/57 (22, 43). To gain insight into how Rad51I345T functions, we tested the ability of this rad51 mutant to inhibit ATP hydrolysis by Srs2. These results revealed that Rad51I34T has only a moderate impact on Srs2 ATPase hydrolysis activity (SI Appendix, Fig. S1 A and B). We have previously shown that Srs2 removes Rad51I345T from ssDNA at a reduced velocity (∼85 nt⋅s−1) compared with the velocity observed with WT Rad51 (∼140 nt⋅s−1) (44), indicating that Rad51I345T slows but does not stop the movement of Srs2, which is consistent with the observed reduction in Srs2-mediated ATP hydrolysis in the presence of Rad51I345T. Together, these findings suggest that Rad55/57 (22), Rad52 (24, 42), and Rad51I345T (present study and ref. 44) do not function by inhibiting Srs2 ATP hydrolysis activity. These results are in striking contrast to our findings for Dmc1, which strongly inhibits ATP hydrolysis by Srs2 (Fig. 1B). Our data suggest that Dmc1 may act as a direct inhibitor of Srs2, which affects Srs2 through a mechanism that may be distinct from all of the aforementioned regulatory factors.
Dmc1 Blocks Srs2 Translocation on ssDNA.
We have established ssDNA curtain assays for visualizing the behaviors of Srs2 on HR intermediates in real time (Fig. 1C) (42, 44, 45). In brief, long ssDNA substrates (≥50 kilonucleotides) are generated by rolling circle replication using a 5′ biotinylated primer (45–47). The resulting 5′ biotinylated ssDNA is tethered to a supported lipid bilayer on the surface of a microfluidic sample chamber through a biotin–streptavidin linkage (45–47). The 5′ ends of the ssDNA are then aligned at nanofabricated Cr barriers to lipid diffusion, and the downstream ends of the ssDNA are attached to Cr pedestals, which are deposited onto the fused silica by electron-beam lithography (45–47). Addition of GFP- or mCherry-RPA allows the ssDNA to be extended by hydrodynamic force, and also provides means of visualizing the ssDNA by total internal reflection fluorescence microscopy (45–47). In these assays, unlabeled Srs2 disrupts Rad51-ssDNA, allowing for the rapid replacement of Rad51 by RPA-GFP, and single Srs2 complexes can translocate over distances spanning thousands of nucleotides (Fig. 1D) (44). In striking contrast, we found no evidence of Srs2 translocation on Dmc1-ssDNA even in assays containing fivefold more Srs2 (500 pM) relative to typical assays with Rad51-ssDNA (Fig. 1E) (44). Importantly, Srs2 overexpression disrupts Rad51 foci in vivo while leaving Dmc1 foci unaffected (36). Thus, cell-based assays and our biochemical and single-molecule analysis all suggest that Dmc1-containing HR intermediates are impervious to Srs2.
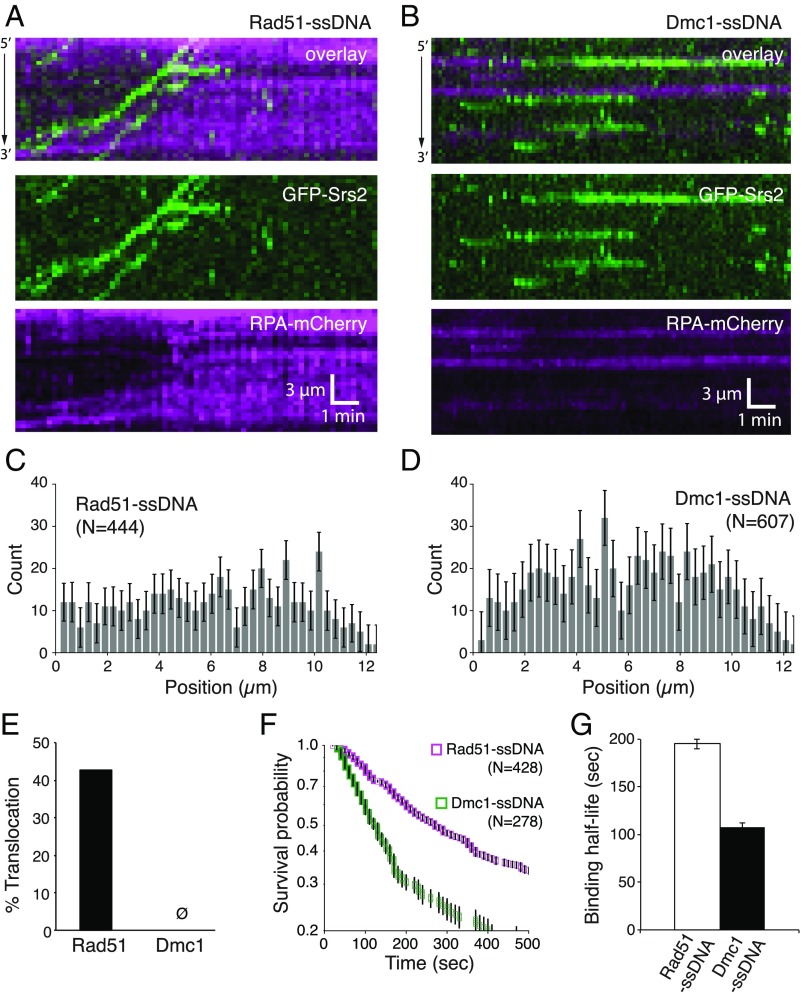
We considered two mechanisms for Dmc1-mediated inhibition of Srs2: (i) Dmc1 might prevent Srs2 from binding to the presynaptic complex or (ii) Dmc1 might allow Srs2 binding but block its motor activity. We used GFP-tagged Srs2 to distinguish between these models (Fig. 2) (42, 44). As previously demonstrated, GFP-Srs2 is targeted to clusters of RPA interspersed between Rad51 filaments and then rapidly dismantles the Rad51 filaments (Fig. 2A) (44). GFP-Srs2 recruitment was much less efficient on Dmc1-ssDNA relative to Rad51-ssDNA, as evidenced by the need for ∼fivefold more GFP-Srs2 to attain similar levels of binding (Fig. 2 A and B). GFP-Srs2 was targeted to RPA within Dmc1 filaments (Fig. 2B), and the overall GFP-Srs2 binding distributions were similar for both recombinases (Fig. 2 C and D). However, there was no evidence for GFP-Srs2 translocation on the Dmc1-ssDNA complexes (Fig. 2 B and E). Interestingly, GFP-Srs2 bound to the Dmc1-ssDNA filaments with a mean lifetime of 108 ± 4.0 s (n = 278), compared with 195 ± 5.0 s (n = 428) for Rad51-ssDNA, indicating that Srs2 dissociates approximately twofold faster from Dmc1-ssDNA compared with Rad51-ssDNA (Fig. 2 F and G). These results demonstrate that Srs2 can bind to Dmc1-ssDNA, albeit at reduced levels, but Srs2 is unable to translocate on the ssDNA in the presence of the meiosis-specific recombinase Dmc1. Thus, our ssDNA curtain measurements experiments reveal a crucial mechanistic difference between Srs2 assays with Rad51I345T and Srs2 assays conducted with Dmc1: namely, Rad51I345T merely slows Srs2 translocation, whereas Dmc1 completely halts it.
Fig. 2.
Srs2 binds to Dmc1 filaments but cannot translocate. (A) Kymographs showing GFP-Srs2 (150 pM; green) translocation on Rad51-ssDNA in the presence of RPA-mCherry (magenta). (B) Kymographs of GFP-Srs2 (500 pM) binding to Dmc1-ssDNA in the presence of RPA-mCherry (100 nM; magenta). (C) Binding distribution histograms for GFP-Srs2 on Rad51-ssDNA (n = 444). (D) Binding distribution histograms for GFP-Srs2 on Dmc1-ssDNA (n = 607). Error bars in C and D indicate 95% CIs generated by bootstrap analysis. (E) Percentage of bound Srs2 complexes that exhibit translocation activity on Rad51-ssDNA (n = 444) or Dmc1-ssDNA (n = 607). (F) Survival probabilities for GFP-Srs2 bound to Rad51-ssDNA (n = 428) or Dmc1-ssDNA (n = 278). Error bars indicate 95% CI generated by bootstrap analysis. (G) GFP-Srs2 binding t1/2 on Rad51-ssDNA (n = 428) or Dmc1-ssDNA (n = 278). Error bars indicate 95% CIs obtained for the fit to the survival probability plots.
Dmc1 Prevents Heteroduplex Joint Disruption by Srs2.
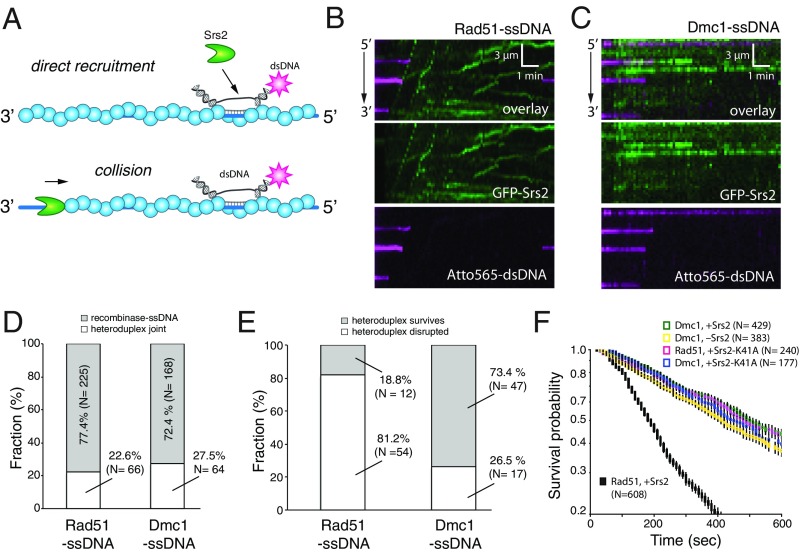
Srs2 has the ability to disrupt heteroduplex DNA intermediates, and this activity plays a central role in the ability of Srs2 to contribute to genome integrity (13, 15, 18, 19, 27). Similarly, we have shown that Srs2 can disrupt short heteroduplex DNA joints made with Rad51-ssDNA and 70-bp dsDNA fragments harboring a 15-nt tract of homology complementary to the presynaptic ssDNA (44). Srs2 disrupts these intermediates through two distinct pathways: through direct recruitment to the heteroduplex joint or through collisional encounters while undergoing 3′→5′ translocation along the Rad51-ssDNA (Fig. 3 A and B). Srs2 cannot translocate on Dmc1-ssDNA; therefore, the collisional pathway for heteroduplex disruption is not operational in the presence of Dmc1 (Fig. 3C). GFP-Srs2 is recruited to heteroduplex DNA joints in the presence of Dmc1, but these intermediates remained resistant to Srs2-mediated disruption (Fig. 3 C–E), and the lifetimes of the heteroduplex joints were substantially longer for Dmc1-ssDNA (430 ± 15 s) compared with Rad51-ssDNA (127 ± 2.1 s; Fig. 3F). Indeed, the lifetimes of the heteroduplex joints were similar for reactions that used (i) Srs2 and Dmc1-ssDNA (430 ± 15 s), (ii) the ATP hydrolysis-deficient mutant Srs2K41A and Rad51-ssDNA (343 ± 5.0 s), or (iii) Dmc1-ssDNA in the absence of Srs2 (405 ± 10 s; Fig. 3F). These results suggest that Srs2 does not strongly affect the stability of heteroduplex DNA joints in the presence of Dmc1.
Fig. 3.
Dmc1 prevents Srs2 from disrupting heteroduplex DNA joints. (A) Pathways for Srs2 recruitment to heteroduplex DNA joints. (B) Kymographs illustrating GFP-Srs2 (500 pM; green) disruption of heteroduplex DNA joints prepared with 70-bp Atto565-labeled dsDNA fragments (magenta) bound to Rad51-ssDNA. (C) Kymographs showing GFP-Srs2 (500 pM) acting upon the same dsDNA fragments (magenta) bound to Dmc1-ssDNA. (D) Fraction of GFP-Srs2 that is directly recruited to the heteroduplex DNA joints with Rad51-ssDNA or Dmc1-ssDNA. (E) Fraction of dsDNA that remains bound after injection of GFP-Srs2 (500 pM). (F) Survival probability plots for dsDNA bound to Rad51-ssDNA or Dmc1-ssDNA in the presence of GFP-Srs2 (500 pM), GFP-Srs2K41A (500 pM), or no Srs2 as indicated. Error bars indicate 95% CIs generated by bootstrap analysis.
Srs2 ATPase Activity Is Inhibited by Mixed Recombinase Filaments.
Meiotic presynaptic complexes contain Rad51 and Dmc1. However, Rad51 and Dmc1 do not form highly intermixed filaments in vivo, but instead are thought to segregate into side-by-side homotypic filaments (1, 3, 48, 49) (Fig. 4A). We have shown that this side-by-side homotypic filament organization can be recapitulated in vitro in assays containing mixtures of Rad51 and Dmc1, indicating that these two recombinases have an intrinsic capacity to form separate filaments on the same ssDNA molecules even in the absence of any other assembly factors (50) (Fig. 4 A and B). Therefore, we next asked how Srs2 behaved in reactions containing mixtures of Rad51 and Dmc1. Remarkably, bulk biochemical assays revealed that Srs2 ATP hydrolysis activity was markedly reduced in reactions containing 3:1 or 1:1 ratios of Rad51 to Dmc1, indicating that Srs2 ATP hydrolysis activity was down-regulated within the context of these mixed recombinase filaments (Fig. 4C).
Fig. 4.
Srs2 action on mixed recombinase filaments. (A) Schematic of Rad51 and Dmc1 filaments bound to the same ssDNA. (B) Images showing Rad51 only, Dmc1 only, and mixed recombinase filaments in the presence of Hed1-GFP (green). Dmc1 (dark) and Rad51-Hed1 (green) regions are highlighted. (C) Srs2 ATP hydrolysis assays with Rad51 only, Dmc1 only, or 3:1 and 1:1 mixtures of Rad51 to Dmc1. Data points represent the mean ± SD of three experiments. (D) Kymographs illustrating examples of complete disassembly, local disassembly, and no disassembly for reactions performed with Rad51 (1.5 µM) and Dmc1 (1.5 µM), Hed1-GFP (10 nM), RPA-mCherry (100 pM), and unlabeled Srs2 (500 pM). (E) Distribution of observed event types for Srs2 reactions performed with Rad51 only, 3:1 Rad51:Dmc1, 1:1 Rad51:Dmc1, or Dmc1 only as indicated.
The Rad51-Binding Factor Hed1 Does Not Affect Srs2.
During meiosis, Rad51 strand-exchange activity is down-regulated by the meiosis-specific Rad51-binding factor Hed1, which prevents Rad54 from associating with Rad51-ssDNA filaments (51–53). Importantly, we used GFP-tagged Hed1 in DNA curtain assays to identify the locations of Rad51 within mixed recombinase filaments containing Rad51 and Dmc1 (Fig. 4B) (52). Bulk biochemical assayed revealed that Hed1-GFP has no appreciable impact on Srs2 ATP hydrolysis activity (SI Appendix, Fig. S2A), and DNA curtain assays demonstrated that Srs2 readily disrupts Hed1-Rad51-ssDNA filaments, exhibiting velocity and processivity values similar to reactions without Hed1 (SI Appendix, Fig. S2 B–G) (44). These findings demonstrated that Hed1 does not alter the ability of Srs2 to remove Rad51 from ssDNA.
Srs2 Translocation Is Highly Restricted Within Mixed Recombinase Filaments.
We next sought to visualize the behavior of unlabeled Srs2 on meiotic presynaptic complexes containing both Rad51 and Dmc1, in which the locations of the Rad51 filaments were demarcated by GFP-tagged Hed1 and the removal of the recombinases was monitored with RPA-mCherry. These experiments revealed outcomes that could be categorized as (i) complete filament disassembly; (ii) local disassembly, in which a portion of the filament was disrupted; or (iii) no disassembly (Fig. 4D). At a 3:1 ratio of Rad51 to Dmc1, 57% of the presynaptic complexes exhibited no Srs2 activity, 19% were fully disassembled, and the remaining 23% showed local disassembly (Fig. 4E). At a 1:1 of Dmc1 to Rad51, 94% of the presynaptic complexes remained fully intact, 6% exhibited local disruption, and none of the filaments were completely disassembled (Fig. 4E). These findings with mixed Rad51 plus Dmc1 recombinase filaments contrast with control reactions for Rad51 only, in which all (100%) of the presynaptic complexes were completely disassembled by Srs2, and also contrasted with reactions containing only Dmc1, in which none (0%) of the presynaptic complexes were disassembled (Fig. 4E). We conclude that Srs2 activity is greatly restricted within mixed recombinase filaments containing Rad51 and Dmc1.
Interestingly, the local disassembly events observed on mixed recombinase filaments appeared to initiate primarily within the Rad51-Hed1-GFP sections of the filaments. To analyze these events in more detail, we conducted experiments at a 1:1 ratio of Rad51 to Dmc1 in the presence of Hed1-GFP and mCherry-Srs2 (SI Appendix, Fig. S3 A and B). Most of the mCherry-Srs2 (69%; n = 223) was recruited to Hed1-Rad51 within the mixed filaments (SI Appendix, Fig. S3 B and C), even though only ∼20% of mixed recombinase filaments are composed of Rad51 under these conditions (50). The majority of the mCherry-Srs2 (89%; n = 289) exhibited no detectable translocation activity, whereas a small population (1.8%; n = 6) translocated more than approximately 1,000 nt (SI Appendix, Fig. S3D). Interestingly, a somewhat larger population (8.2%; n = 28) appeared to translocate over shorter distances as evidenced by the replacement of the green fluorescence signal from Rad51-Hed1-GFP with the magenta signal from RPA-mCherry (no more than approximately 1,000 nt; SI Appendix, Figs. S3D and S4). Taken together, these findings suggest that, although Srs2 can bind to and remove Rad51-Hed1 from within the mixed recombinase filaments, it fails to progress beyond the 3′ ends of the adjacent Dmc1 filaments (Fig. 4E and SI Appendix, Fig. S4).
Discussion
Our work shows that the meiosis-specific recombinase Dmc1 prevents Srs2 from dismantling early meiotic recombination intermediates. Our data demonstrate that, although Srs2 can bind to presynaptic complexes containing Dmc1, it is unable to initiate ATP-dependent translocation on these Dmc1-bound substrates. These studies highlight a biochemical distinction between Rad51 and Dmc1, namely, Rad51 is highly susceptible to disruption by the antirecombinase activity of Srs2 whereas Dmc1 is not. We speculate that the ability of Dmc1 to inhibit Srs2 may help explain in part why eukaryotes utilize a specialized recombinase during meiosis.
Model for Dmc1 as an Inhibitor of Srs2.
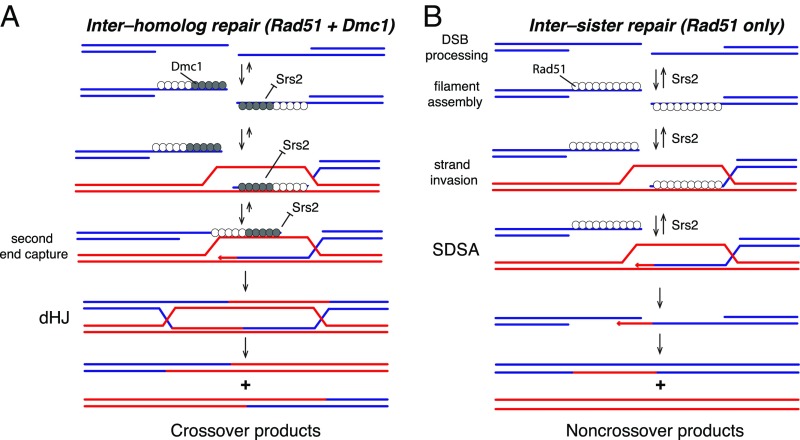
Srs2 readily removes Rad51 from ssDNA, and, during mitotic DSB repair, Srs2 is thought to promote SDSA by disrupting Rad51 filaments, by disrupting heteroduplex intermediates (e.g., D-loops), and/or by reducing the probability of second end capture (Fig. 5A) (10, 12, 15, 20). In striking contrast, our results demonstrate that Srs2 cannot remove the meiosis-specific recombinase Dmc1 from ssDNA (Fig. 5A). We propose that Dmc1 may help prevent premature dissolution of early meiotic recombination intermediates by blocking the antirecombinase activity of Srs2, which would in turn prevent these meiotic HR intermediates from being channeled through the SDSA pathway. Thus, the ability of Dmc1 to inhibit Srs2 may contribute to the elevated cross-over frequency that is the hallmark of meiosis. Mechanistically, Dmc1 acts by inhibiting Srs2 ATP hydrolysis, which in turn prevents Srs2 from translocating on ssDNA bound by Dmc1. The ability to shut down Srs2 translocation through inhibition of its ATP hydrolysis activity appears to distinguish Dmc1 from other known negative regulators of Srs2 antirecombinase activities, such as Rad55/5 or Rad52, which do not appear to affect the ssDNA-dependent ATP hydrolysis activity of Srs2 (22, 24, 42). Our findings suggest that this inhibition likely takes place as a result of direct physical contact between the advancing Srs2 motor proteins and the 3′ ends of Dmc1 filaments, suggesting a mechanism involving allosteric communication that turns off ATP hydrolysis by Srs2 (Fig. 5A). As a result, meiotic HR intermediates are rendered highly resistant to Srs2, which may favor cross-over product formation by increasing the likelihood of second strand capture and the formation of double Holliday junctions (Fig. 5A). Our results also provide a mechanistic explanation for the finding that Dmc1 foci are resistant to Srs2 overexpression in vivo (36), and pinpoint Dmc1 itself, as opposed to some other regulatory cofactor (e.g., Mei5/Sae3, Hop2/Mnd1, Rdh54, or the ZMM proteins), as the source of this resistance.
Fig. 5.
Models depicting the regulation of Srs2 on Rad51- and Dmc1-containing meiotic HR intermediates. (A) During meiosis, Srs2 is unable to effectively disrupt Dmc1-containing intermediates, which is anticipated to favor cross-over recombination outcomes. (B) Meiotic intermediates that are bound only by Rad51 would remain susceptible to the antirecombinase activity of Srs2 and would be channeled through the SDSA pathway.
The finding that Srs2 is inhibited by Dmc1 is remarkable given that Saccharomyces cerevisiae Rad51 and Dmc1 are ∼46% identical at the amino acid sequence level. Interestingly, there are many recombination events that take place during meiosis, which are independent of Dmc1 and are instead driven by the strand-exchange activity of Rad51 (54, 55). In addition, there are many intersister recombination events that take place during meiosis (56), and we speculate that these events may also be driven by Rad51. Importantly, our model for Dmc1-mediated inhibition of Srs2 would allow for two distinct modes of Srs2-mediated regulation during meiosis. Namely, Dmc1-mediated inhibition of Srs2 may selectively favor cross-over formation during interhomolog recombination (Fig. 5A), whereas Srs2 would still be able to act as an antirecombinase to promote Rad51-mediated intersister recombination via SDSA (Fig. 5B). Interestingly, Srs2 expression increases six to eightfold during meiosis (25). Although the reasons for the enhanced expression of Srs2 in meiosis remain unknown, one possibility is that it helps to ensure that there is sufficient Srs2 present to accommodate Rad51-mediated recombination events, or increased Srs2 levels may be necessary for its other replication-related functions as cells transition from 2n to 4n DNA content while the homologous chromosomes are being duplicated.
Conclusion.
Srs2 is considered a prototypical antirecombinase that dismantles Rad51-containing recombination intermediates. Similar antirecombinase functions have been ascribed to the S. cerevisiae RecQ helicase Sgs1, as well as several helicases that are important for dismantling HR intermediates in human cells, including FBH1, RECQ5, WRN, and BLM (57–62). Intriguingly, bulk biochemical assays have revealed that BLM can inhibit D-loop formation by human RAD51, but BLM does not inhibit D-loop formation by human DMC1 (63). Future work will be important to determine whether Dmc1 can inhibit other antirecombinase enzymes and whether this inhibition is conserved in higher eukaryotes.
Methods
Protein Purification.
The expression and purification of S. cerevisiae Rad51 and Dmc1 followed published procedures (64, 65). RPA, GFP-RPA, and mCherry-RPA were purified as described previously (42, 44). Srs2 proteins were also purified according to published procedures (42, 44). In brief, a pET11c vector encoding 9xHis-tagged Srs2 and pET15b vectors encoding GFP-Srs2898, mCherry-Srs2898, and GFP-Srs2K41A:898 were introduced into Escherichia coli Rosetta 2(DE3) cells (Novagen). Cells were grown in 3 L of Lysogeny Broth at 37 °C to an OD600 of 1–2. The temperature was reduced to 16 °C before addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 20 h of growth, cells were pelleted and frozen at −80 °C. The pellet was then resuspended in lysis buffer [40 mM NaHPO4, pH 7.5, 600 mM KCl, 5% glycerol, 10 mM imidazole, pH 7.8, 0.1 mM Tris (2-carboxyethyl)phosphine hydrochloride (TCEP), 0.05% Tween-20, 10 µM E-64, one pill per 100 mL of protease inhibitor mixture tablets, 1 mM benzamidine, 1 mM PMSF, 0.125% myo-inositol] and lysed by sonication on ice. The lysate was clarified by ultracentrifugation and incubated for 30 min with a Talon resin (Clontech) that was equilibrated with buffer Talon A (40 mM NaHPO4, pH 7.5, 300 mM KCl, 5% glycerol, 15 mM imidazole, 0.02% Tween-20, 1 mM benzamidine, 1 mM PMSF, 0.125% myo-inositol). Before elution, the Talon column was washed with buffer Talon A. The proteins were eluted with a step of buffer Talon A containing 100 mM imidazole (pH 7.8). The eluate was then dialyzed against heparin buffer (20 mM NaHPO4, pH 7.5, 100 mM KCl, 5% glycerol, 0.01% Tween-20, 1 mM TCEP, 2 mM EDTA, 0.125% myo-inositol) during a period of 2 h. The eluate was then loaded onto a 1-mL HiTrap heparin column (GE Lifesciences) equilibrated with heparin buffer, and proteins were eluted with a step of heparin buffer containing 500 mM KCl. The purified fraction was applied to a Superdex 200 size-exclusion column equilibrated with storage buffer (40 mM NaHPO4, pH 7.5, 300 mM KCl, 10% glycerol, 0.01% Tween-20, 1 mM TCEP, 0.5 mM EDTA, 0.125% myo-inositol). Fractions corresponding to monomeric Srs2 were pooled, concentrated, flash-frozen in liquid nitrogen, and stored at −80 °C.
GST-Hed1-6xHis-mCherry, GST-Hed1-6xHis, and GST-Hed1-6xHis-GFP were purified as described previously (50, 52). pGEX plasmids were transformed into E. coli Rosetta(DE3) cells (Novagen). Cell were grown to an OD of 0.6–0.8 at 37 °C, and cultures were then shifted to 16 °C and induced overnight with 0.1 mM IPTG. After overnight expression, cells were harvested and resuspended in 20 mL⋅L−1 cell lysis buffer [50 mM Tris⋅Cl, pH 7.5, 700 mM KCl, 1 mM EDTA, 10 mM β-mercaptoethanol (β-ME), protease inhibitor mixture (cat. no. 05892988001; Roche), 10% glycerol, and 1 mM PMSF]. Cells were lysed with lysozyme and sonicated. The lysate was clarified by ultracentrifugation for 45 min at 90,000 × g. Clarified extract was incubated in batch with glutathione resin (cat. no. 17-0756-01; GE Healthcare) for 1 h at 4 °C. After 1 h, the supernatant was removed and the resin was washed with 10 column volumes with buffer K1000 (20 mM Tris⋅Cl, pH 7.5, 1 M KCl, 10 mM β-ME, 1 mM PMSF, 10% glycerol, 2.5 mM imidazole). Resin was then washed with buffer K300 (20 mM Tris⋅Cl, pH 7.5, 0.3 M KCl, 10 mM β-ME, 1 mM PMSF, 10% glycerol, 2.5 mM imidazole). Protein was eluted with buffer K300 plus 25 mM glutathione. Peak fractions were bound to cOmplete nickel resin (cat. no. 05893682001; Roche) for 1 h at 4 °C. Resin was then washed with five column volumes of buffer K1000, followed by washing with five column volumes of buffer K300. Protein was then eluted with buffer K300 plus 100 mM imidazole. Peak fractions were pooled and dialyzed against buffer K 150 (20 mM Tris⋅Cl, pH 7.5, 150 mM KCl, 10 mM β-ME, 1 mM PMSF, 10% glycerol). Proteins were quantified by absorbance at 280 nm, and, in the case of GFP and mCherry protein concentrations, were quantified by measuring the absorbance of the chromophores at 488 nM (ε488 nm = 55,000 cm−1⋅M−1) or 587 nm (ε587 nm = 72,000 cm−1⋅M−1), respectively. Samples were flash-frozen and stored at −80 °C.
Single-Molecule Experiments.
All experiments were conducted with a prism-type total internal reflection fluorescence microscope (Nikon) equipped with a 488-nm laser (200 mW; Coherent Sapphire) and a 561-nm laser (200 mW; Coherent Sapphire). For all two-color images, we used a custom-built shuttering system to avoid the bleed-through from the green into the red channel during image acquisition. The lasers were each set to 50 mW output power, yielding illumination powers at the sample of ∼8.8 mW/mm2 for the 488-nm laser and 14.7 mW/mm2 for the 561-nm laser. With this system, images from the green (GFP) and the red (mCherry) channels are recorded independently, and these recordings are offset by 100 ms such that, when one camera records the red channel image, the green laser is shuttered off, and vice versa (42, 45).
Flowcells and ssDNA curtains were prepared as previously described (42, 44, 50). In brief, lipid bilayers were prepared with 91.5% DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), 0.5% biotinylated PE, and 8% mPEG 2000-DOPE [1,2-dioeoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (ammonium salt)]. The ssDNA substrate was generated by using a 5′ biotinylated primer and a circular M13 ssDNA template and rolling circle replication with phi29 DNA polymerase (66). The 5′ biotinylated ssDNA was injected into the sample chamber and attached to the bilayer through a biotin–streptavidin linkage. The flow cell was then attached to a microfluidic system, and buffer was delivered to the sample chambers by using a syringe pump (KD Scientific).
Rad51 and Dmc1 Filament Assembly.
The 5′ biotinylated ssDNA molecules were aligned along the diffusion barriers at a flow rate of 0.5 mL⋅min−1 in Dmc1 buffer plus RPA (30 mM Tris⋅Cl, pH 7.5, 100 mM KCl, 2 mM MgCl2, 1.5 mM CaCl2, 0.2 mg⋅mL−1 BSA, 1 mM DTT, 0.1 nM RPA-GFP, RPA-mCherry, or unlabeled RPA). When molecules had been aligned, the flow rate was adjusted to 1.0 mL⋅min−1 and 0.5 mL of 7 M urea was injected into the flow cell to disrupt any remaining secondary structure. The sample chamber was then flushed with Dmc1 buffer plus RPA-GFP or RPA-mCherry (0.1 nM) at 1.0 mL/min for 10 min. After 5 min, Dmc1 buffer plus ATP (30 mM Tris⋅Cl, pH 7.5, 100 mM KCl, 2 mM MgCl2, 1.5 mM CaCl2, 0.2 mg⋅mL−1 BSA, 1 mM DTT, plus 2.5 mM ATP) flowed at 1.0 mL⋅min−1 for 3 min. Rad51 (2 µM), Dmc1 (2 µM), or mixed ratios of the two (3 µM total Rad51 + Dmc1) were injected into the flow cell, buffer flow was terminated, and the reaction was incubated at 30 °C for 20 min to allow filament assembly. The RPA fluorescence signal was then monitored to verify filament assembly. In experiments in which unlabeled RPA was used, the reaction was allowed to incubate for 20 min without monitoring. Following a 20-min incubation, free recombinase was flushed from the sample chamber with Dmc1 buffer plus ATP, and experiments proceeded as described later.
Srs2 Translocation Assays.
All translocation measurements were conducted in Dmc1 buffer at 30 °C. After recombinase exchange, the flow cell was washed with 1 mL of Dmc1 buffer to remove free recombinase. Samples containing GFP-Srs2 with 0.1 pM RPA-mCherry, Srs2 with 0.1 pM RPA-GFP, or GFP-Srs2 with 0.1 pM RPA-mCherry were injected into the flow cell at a rate of 1 mL/min. For experiments in which Rad51 was labeled with GFP-Hed1 (10 nM), RFP-Srs2 was used with unlabeled RPA. Flow was stopped after 10 s, corresponding to the peak injection period, and the activity of Srs2 was monitored for 10–15 min. All data were collected at one frame per 10 s with 100 ms integration time, and the laser was shuttered between each acquired image to minimize photo-bleaching. Raw TIFF images were imported as image stacks into ImageJ, and kymographs were generated from the image stacks by defining a 1-pixel-wide region of interest along the long axis of the individual ssDNA molecules. From the kymographs, the lifetime, number, position, and translocation of Srs2 was quantified and used for further data analysis. Survival probabilities were calculated from binding lifetime measurements (Fig. 2F), and the data were fit by a constrained exponential decay function to determine the t1/2 (Fig. 2G). The error bars were generated by bootstrapping the data by using a custom python script. When indicted, the global association and dissociation rate of RPA or Hed1 was quantified by integrating the total signal intensity over entire ssDNA molecules, and the resulting data were fit to an exponential decay curve or scored for extent of disassembly.
ATP Hydrolysis Assays.
ATP hydrolysis was performed in Dmc1 buffer (30 mM Tris⋅Cl, pH 7.5, 100 mM KCl, 2 mM MgCl2, 1.5 mM CaCl2, 1 mM DTT, 0.2 mg⋅mL−1 BSA) in the presence of 2 mM cold ATP and trace amounts of γ–[32P]ATP. All reactions were performed at 30 °C, and contained 2.5 µM (in nucleotides) of M13 ssDNA. Aliquots were removed at specified time points and quenched with 25 mM EDTA and 1% SDS. The quenched reactions were spotted on TLC plates (cat. no. HX71732079; Millipore) and resolved in 0.5 M LiCl plus 1 M formic acid. Dried TLC plates were exposed to a phosphorimaging screen and scanned with a Typhoon platform (GE Healthcare).
Single-Molecule Joint Molecule Disruption Assays.
Heteroduplex joint disruption experiments were performed in Dmc1 buffer plus 5 mM MgCl2 (30 mM Tris⋅Cl, pH 7.5, 100 mM KCl, 5 mM MgCl2, 1.5 mM CaCl2, 1 mM DTT, 0.2 mg⋅mL−1 BSA) as described here earlier. All reactions were performed at 30 °C, and recombination filaments were reconstituted as described here earlier by using unlabeled RPA to extend the ssDNA. Recombinase exchange was allowed to proceed for 20 min, and then free recombinase was flushed away. A 70-bp DNA substrate labeled with Atto565 on the 5′ end of the complementary strand containing 15 bases of homology was then injected at concentrations of 1 nM (for reactions with Dmc1) and 10 nM (for reactions with Rad51) and incubated for 10 min as described previously (44). Unbound dsDNA was then flushed away, and data collection was initiated at one frame per 10 s; after 30 s, GFP-Srs2 (500 pM) was injected and monitored for 10–15 min. Data analysis was conducted as described here earlier. Survival probabilities were calculated from the dsDNA binding lifetimes and fit by an exponential decay function, and error bars were generated by bootstrapping by using a custom Python script.
Supplementary Material
Acknowledgments
We thank Douglas Bishop, Michael Lichten, Akira Shinohara, Neil Hunter, and Hannah Klein for helpful discussions and for sharing unpublished results; and members of the laboratories of E.C.G. and P.S. for comments on the manuscript. This research was funded by National Institutes of Health Grants R35GM118026 (to E.C.G.), R01ES007061 (to P.S.), R01ES015632 (to P.S.), and P01CA92584 (to P.S. and E.C.G.). J.B.C. is the Mark Foundation for Cancer Research Fellow for the Damon Runyon Cancer Research Foundation Grant (DRG 2310–17).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810457115/-/DCSupplemental.
References
- 1.Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kowalczykowski SC. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2015;7:a016410. doi: 10.1101/cshperspect.a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown MS, Bishop DK. DNA strand exchange and RecA homologs in meiosis. Cold Spring Harb Perspect Biol. 2014;7:a016659. doi: 10.1101/cshperspect.a016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunter N. Meiotic recombination: The essence of heredity. Cold Spring Harb Perspect Biol. 2015;7:a016618. doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pâques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keeney S, Lange J, Mohibullah N. Self-organization of meiotic recombination initiation: General principles and molecular pathways. Annu Rev Genet. 2014;48:187–214. doi: 10.1146/annurev-genet-120213-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zickler D, Kleckner N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol. 2015;7:a016626. doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Symington LS, Rothstein R, Lisby M. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics. 2014;198:795–835. doi: 10.1534/genetics.114.166140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin Z, Kong H, Nei M, Ma H. Origins and evolution of the recA/RAD51 gene family: Evidence for ancient gene duplication and endosymbiotic gene transfer. Proc Natl Acad Sci USA. 2006;103:10328–10333. doi: 10.1073/pnas.0604232103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antony E, et al. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol Cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veaute X, et al. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 12.Dupaigne P, et al. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: Implications for crossover incidence during mitotic recombination. Mol Cell. 2008;29:243–254. doi: 10.1016/j.molcel.2007.11.033. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, et al. Srs2 promotes synthesis-dependent strand annealing by disrupting DNA polymerase δ-extending D-loops. eLife. 2017;6:22195. doi: 10.7554/eLife.22195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasianovich Y, et al. Unloading of homologous recombination factors is required for restoring double-stranded DNA at damage repair loci. EMBO J. 2017;36:213–231. doi: 10.15252/embj.201694628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorenz A. Modulation of meiotic homologous recombination by DNA helicases. Yeast. 2017;34:195–203. doi: 10.1002/yea.3227. [DOI] [PubMed] [Google Scholar]
- 16.Niu H, Klein HL. Multifunctional roles of Saccharomyces cerevisiae Srs2 protein in replication, recombination and repair. FEMS Yeast Res. 2017;17:fow111. doi: 10.1093/femsyr/fow111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robert T, Dervins D, Fabre F, Gangloff S. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 2006;25:2837–2846. doi: 10.1038/sj.emboj.7601158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elango R, et al. Break-induced replication promotes formation of lethal joint molecules dissolved by Srs2. Nat Commun. 2017;8:1790. doi: 10.1038/s41467-017-01987-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piazza A, Wright WD, Heyer WD. Multi-invasions are recombination byproducts that induce chromosomal rearrangements. Cell. 2017;170:760–773.e15. doi: 10.1016/j.cell.2017.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burgess RC, et al. Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo. J Cell Biol. 2009;185:969–981. doi: 10.1083/jcb.200810055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, et al. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature. 2011;479:245–248. doi: 10.1038/nature10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernstein KA, et al. The Shu complex, which contains Rad51 paralogues, promotes DNA repair through inhibition of the Srs2 anti-recombinase. Mol Biol Cell. 2011;22:1599–1607. doi: 10.1091/mbc.E10-08-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma E, et al. Rad52-Rad51 association is essential to protect Rad51 filaments against Srs2, but facultative for filament formation. eLife. 2018;7:e32744. doi: 10.7554/eLife.32744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heude M, Chanet R, Fabre F. Regulation of the Saccharomyces cerevisiae Srs2 helicase during the mitotic cell cycle, meiosis and after irradiation. Mol Gen Genet. 1995;248:59–68. doi: 10.1007/BF02456614. [DOI] [PubMed] [Google Scholar]
- 26.Hong S, Kim KP. Shu1 promotes homolog bias of meiotic recombination in Saccharomyces cerevisiae. Mol Cells. 2013;36:446–454. doi: 10.1007/s10059-013-0215-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marini V, Krejci L. Srs2: The “odd-job man” in DNA repair. DNA Repair (Amst) 2010;9:268–275. doi: 10.1016/j.dnarep.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharyya S, Lahue RS. Saccharomyces cerevisiae Srs2 DNA helicase selectively blocks expansions of trinucleotide repeats. Mol Cell Biol. 2004;24:7324–7330. doi: 10.1128/MCB.24.17.7324-7330.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhattacharyya S, Lahue RS. Srs2 helicase of Saccharomyces cerevisiae selectively unwinds triplet repeat DNA. J Biol Chem. 2005;280:33311–33317. doi: 10.1074/jbc.M503325200. [DOI] [PubMed] [Google Scholar]
- 30.Dhar A, Lahue RS. Rapid unwinding of triplet repeat hairpins by Srs2 helicase of Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:3366–3373. doi: 10.1093/nar/gkn225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liberi G, et al. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J. 2000;19:5027–5038. doi: 10.1093/emboj/19.18.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yasuhira S. Redundant roles of Srs2 helicase and replication checkpoint in survival and rDNA maintenance in Schizosaccharomyces pombe. Mol Genet Genomics. 2009;281:497–509. doi: 10.1007/s00438-009-0426-x. [DOI] [PubMed] [Google Scholar]
- 33.Papouli E, et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 35.Potenski CJ, Niu H, Sung P, Klein HL. Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms. Nature. 2014;511:251–254. doi: 10.1038/nature13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sasanuma H, Furihata Y, Shinohara M, Shinohara A. Remodeling of the Rad51 DNA strand-exchange protein by the Srs2 helicase. Genetics. 2013;194:859–872. doi: 10.1534/genetics.113.150615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrari SR, Grubb J, Bishop DK. The Mei5-Sae3 protein complex mediates Dmc1 activity in Saccharomyces cerevisiae. J Biol Chem. 2009;284:11766–11770. doi: 10.1074/jbc.C900023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao W, et al. Mechanistic insights into the role of Hop2-Mnd1 in meiotic homologous DNA pairing. Nucleic Acids Res. 2014;42:906–917. doi: 10.1093/nar/gkt924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho HR, Kong YJ, Hong SG, Kim KP. Hop2 and Sae3 are required for Dmc1-mediated double-strand break repair via homolog bias during meiosis. Mol Cells. 2016;39:550–556. doi: 10.14348/molcells.2016.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nimonkar AV, et al. Saccharomyces cerevisiae Dmc1 and Rad51 proteins preferentially function with Tid1 and Rad54 proteins, respectively, to promote DNA strand invasion during genetic recombination. J Biol Chem. 2012;287:28727–28737. doi: 10.1074/jbc.M112.373290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jessop L, Rockmill B, Roeder GS, Lichten M. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet. 2006;2:e155. doi: 10.1371/journal.pgen.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Tullio L, et al. Yeast Srs2 helicase promotes redistribution of single-stranded DNA-bound RPA and Rad52 in homologous recombination regulation. Cell Rep. 2017;21:570–577. doi: 10.1016/j.celrep.2017.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fortin GS, Symington LS. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. EMBO J. 2002;21:3160–3170. doi: 10.1093/emboj/cdf293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaniecki K, et al. Dissociation of Rad51 presynaptic complexes and heteroduplex DNA joints by tandem assemblies of Srs2. Cell Rep. 2017;21:3166–3177. doi: 10.1016/j.celrep.2017.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Tullio L, Kaniecki K, Greene EC. Single-stranded DNA curtains for studying the Srs2 helicase using total internal reflection fluorescence microscopy. Methods Enzymol. 2018;600:407–437. doi: 10.1016/bs.mie.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibb B, Silverstein TD, Finkelstein IJ, Greene EC. Single-stranded DNA curtains for real-time single-molecule visualization of protein-nucleic acid interactions. Anal Chem. 2012;84:7607–7612. doi: 10.1021/ac302117z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma CJ, Steinfeld JB, Greene EC. Single-stranded DNA curtains for studying homologous recombination. Methods Enzymol. 2017;582:193–219. doi: 10.1016/bs.mie.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown MS, Grubb J, Zhang A, Rust MJ, Bishop DK. Small Rad51 and Dmc1 complexes oftenco-occupy both ends of a meiotic DNA double strand break. PLoS Genet. 2015;11:e1005653. doi: 10.1371/journal.pgen.1005653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cloud V, Chan YL, Grubb J, Budke B, Bishop DK. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science. 2012;337:1222–1225. doi: 10.1126/science.1219379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crickard JB, Kaniecki K, Kwon Y, Sung P, Greene EC. Spontaneous self-segregation of Rad51 and Dmc1 DNA recombinases within mixed recombinase filaments. J Biol Chem. 2018;293:4191–4200. doi: 10.1074/jbc.RA117.001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Busygina V, et al. Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev. 2008;22:786–795. doi: 10.1101/gad.1638708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crickard JB, et al. Regulation of Hed1 and Rad54 binding during maturation of the meiosis-specific presynaptic complex. EMBO J. 2018;37:e98728. doi: 10.15252/embj.201798728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsubouchi H, Roeder GS. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 2006;20:1766–1775. doi: 10.1101/gad.1422506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Argunhan B, et al. Fundamental cell cycle kinases collaborate to ensure timely destruction of the synaptonemal complex during meiosis. EMBO J. 2017;36:2488–2509. doi: 10.15252/embj.201695895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prugar E, Burnett C, Chen X, Hollingsworth NM. Coordination of double strand break repair and meiotic progression in yeast by a Mek1-Ndt80 negative feedback loop. Genetics. 2017;206:497–512. doi: 10.1534/genetics.117.199703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goldfarb T, Lichten M. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 2010;8:e1000520. doi: 10.1371/journal.pbio.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu Rev Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Branzei D, Szakal B. Building up and breaking down: Mechanisms controlling recombination during replication. Crit Rev Biochem Mol Biol. 2017;52:381–394. doi: 10.1080/10409238.2017.1304355. [DOI] [PubMed] [Google Scholar]
- 59.Brosh RM., Jr DNA helicases involved in DNA repair and their roles in cancer. Nat Rev Cancer. 2013;13:542–558. doi: 10.1038/nrc3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chu WK, Hickson ID. RecQ helicases: Multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 61.Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem. 2014;83:519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simandlova J, et al. FBH1 helicase disrupts RAD51 filaments in vitro and modulates homologous recombination in mammalian cells. J Biol Chem. 2013;288:34168–34180. doi: 10.1074/jbc.M113.484493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bugreev DV, Mazina OM, Mazin AV. Bloom syndrome helicase stimulates RAD51 DNA strand exchange activity through a novel mechanism. J Biol Chem. 2009;284:26349–26359. doi: 10.1074/jbc.M109.029371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Busygina V, et al. Functional attributes of the Saccharomyces cerevisiae meiotic recombinase Dmc1. DNA Repair (Amst) 2013;12:707–712. doi: 10.1016/j.dnarep.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Komen S, Macris M, Sehorn MG, Sung P. Purification and assays of Saccharomyces cerevisiae homologous recombination proteins. Methods Enzymol. 2006;408:445–463. doi: 10.1016/S0076-6879(06)08028-1. [DOI] [PubMed] [Google Scholar]
- 66.Qi Z, et al. DNA sequence alignment by microhomology sampling during homologous recombination. Cell. 2015;160:856–869. doi: 10.1016/j.cell.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.