Significance
Understanding the thermodynamic factors responsible for enhanced stability of thermophilic cytochrome P450 enzymes is significant in their development as efficient catalysts for high-temperature monooxygenation reactions. These factors are usually inferred from structural comparison with mesophilic P450s and invoke rigidifying enthalpic interactions as primary thermodynamic determinants for thermostability. Using the thermophilic P450 CYP119 as an example, the present work, however, questions this enthalpy-driven notion of P450 thermostability and provides strong experimental evidence that their thermostability may actually be entropy-driven due to increased flexibility in the folded state relative to mesophilic P450s and that this flexibility is partitioned effectively between functional activity and thermal stability. This represents a major paradigm shift in understanding the basis of thermostability in thermophilic P450s.
Keywords: thermophilic protein, cytochrome p450, entropy-driven, thermostability, flexibility
Abstract
The enhanced thermostability of thermophilic proteins with respect to their mesophilic counterparts is often attributed to the enthalpy effect, arising from strong interactions between protein residues. Intuitively, these strong interresidue interactions will rigidify the biomolecules. However, the present work utilizing neutron scattering and solution NMR spectroscopy measurements demonstrates a contrary example that the thermophilic cytochrome P450, CYP119, is much more flexible than its mesophilic counterpart, CYP101A1, something which is not apparent just from structural comparison of the two proteins. A mechanism to explain this apparent contradiction is that higher flexibility in the folded state of CYP119 increases its conformational entropy and thereby reduces the entropy gain during denaturation, which will increase the free energy needed for unfolding and thus stabilize the protein. This scenario is supported by thermodynamic data on the temperature dependence of unfolding free energy, which shows a significant entropic contribution to the thermostability of CYP119 and lends an added dimension to enhanced stability, previously attributed only to presence of aromatic stacking interactions and salt bridge networks. Our experimental data also support the notion that highly thermophilic P450s such as CYP119 may use a mechanism that partitions flexibility differently from mesophilic P450s between ligand binding and thermal stability.
Cytochrome P450s are ubiquitous hemeprotein monooxygenases that catalyze various biochemical reactions constituting drug metabolism, lipid and steroid biosynthesis, carcinogenesis, and degradation of pollutants in higher organisms (1–5). The functional activity of these enzymes relies on conformational flexibility of the binding regions to recognize substrates with different physicochemical properties and convert them to monooxygenated products with high regioselectivity and stereo selectivity. Further development of P450 enzymes as practical catalysts in medical and industrial applications would benefit from this flexibility, but would also require them to be highly stable to enable efficient usage in a bioreactor-type setting for such applications. While most regular (mesophilic) P450 enzymes denature around 50 °C to 60 °C, thermally stable (thermophilic) P450s are adapted to meet this requirement, as they are structurally stable and function optimally at temperatures of 80 °C or above (6), and thus can be particularly advantageous in catalyzing reactions such as oxidation of unactivated C−H bonds, for example in lipophilic substrates, often feasible only at very high temperatures. In addition, the enhanced structural stability has great potential utility in carrying out mechanistic investigations of the P450 catalytic cycle that are difficult to carry out with the mesophilic enzymes (7, 8). Thus, understanding the microscopic factors governing the thermostability of these enzymes is of significance in designing new enzymes that can work at extreme environment for even a wider variety of defined substrates (9–13).
Discerning the basis of thermostability, in principle, can potentially be accomplished from a detailed structural and thermodynamic study of a large number of thermophilic P450s and then comparison with mesophilic homologs within a superfamily to identify the relationships between structural elements/interactions and thermodynamic factors that impart thermostability. Elucidation of the specific determinants of thermostability in thermophilic P450s so far have relied mainly on structural analysis of four thermophilic P450s with available crystal structures: CYP119, CYP175A1, CYP231A2, and P450st (14–17). Structural comparison with mesophilic P450 analogs suggests varying structural features such as shorter loops, more compact hydrophobic core, aromatic side-chain stacking, and/or salt bridge networks that may primarily contribute to the enhanced stability of thermophilic P450s, with no single universal feature believed to be responsible for thermostability (15, 18–21). For example, by comparing the crystal structures of CYP119 with CYP101A1, considered to be its mesophilic counterpart based on closest sequence and structural homology (22), it is suggested that the extensive clustering of aromatic residues and salt bridge interaction networks enhances the thermal stability of CYP119. On the other hand, CYP175A1 lacks such aromatic clustering and other factors such as salt bridge interactions, hydrogen bonds, and shorter loops have been invoked to account for its thermostability (15, 22). Such comparisons have also been extended to other thermophilic P450s identified so far and used to interpret the thermodynamics basis of their thermal stability (14–17).
Based on such comparisons, a prevailing view is that the thermostability of thermophilic proteins is primarily enthalpic in origin and the entropic contribution to the thermal stability [∆G(T)] of a folded protein (Eq. 1) is generally deemed to be negligible in such cases.
| [1] |
Intuitively, the strong attractive forces inside the protein molecule, such as hydrogen bonds, salt bridges, aromatic stacking interactions, and nonpolar van der Waals interactions that are enthalpic in nature, will not only stabilize the structure but also rigidify the macromolecule in the folded state to protect it from denaturing at high temperature (23–25). However, a wealth of experimental data accumulated on flexibility of various mesophilic P450s suggest that these proteins are highly dynamic entities which use their conformational dynamics as an integral feature to facilitate substrate recognition and catalysis (26). Structures of thermophilic P450s with various ligands indicate a similar trend, with the flexibility especially high within the active site regions involving the F/G helices and the F/G loop region, where distinct changes allow each thermophilic P450 to adapt to specific ligands with appropriate physicochemical properties, similar to what is observed in mesophilic P450s (27–29). Recent experimental data on various mesophilic P450s by several groups, including ours, indicate that the protein dynamics is not only restricted to the active site regions but also involves regions remote from the catalytic site (27, 30–34). For example, in P450-BM3, mutations done at remote sites affected ligand binding, likely due to change in active site dynamics (35). Also, in CYP101A1, collective dynamic modes on the picosecond to nanosecond timescales were detected to be prevalent throughout the protein, and not just the active site, that create access to the active site for the substrate (26, 36). Even in the case of other thermophilic proteins such as rubredoxin and glutamate dehydrogenase, increased fluctuations were detected throughout the protein by hydrogen/deuterium exchange NMR spectroscopy and molecular dynamics (MD) simulations (37–39). These motions were correlated to a broadened energy landscape exhibiting more conformational states than their mesophilic counterparts, which was then related to an increased entropic reservoir to explain the observed thermodynamic properties of the proteins. Results from these studies therefore raise an inevitable question as to whether the thermal resilience of thermophilic P450s arises primarily from the rigidity resulting from enthalpic interactions or whether there is an important previously unobserved role for the inherent flexibility of these proteins that would provide a thermodynamic explanation for the increased thermal stability.
In the present study, we reconcile this flexibility/rigidity notional discrepancy by providing evidence from solution NMR and neutron scattering measurements on CYP119 and its mesophilic counterpart CYP101A1, which shows that the thermophilic CYP119 exhibits increased global flexibility and conformational disorder in the folded state relative to its mesophilic counterpart CYP101A1, while, at the same time, retaining its enthalpic interactions and thermostability. CYP119 is an ideal candidate for such dynamic and thermodynamic characterization since it was the first thermophilic P450 discovered from the thermophilic organism Sulfolobus acidocaldarius with a melting temperature of ∼90 °C compared with a value of ∼50 °C for its mesophilic counterpart CYP101A1 and has been the most experimentally characterized from the group of thermophilic P450s in terms of mechanistic and structural features (22, 40). By performing detailed measurements of thermodynamic parameters for CYP119 using a fluorescence-based technique and comparing them to similar parameters in the mesophilic counterpart CYP101A1, we show that increased dynamics of CYP119 in the folded state can lower the entropy gain and thus increase the free energy of unfolding, making an important entropic contribution to the protein’s thermal stability.
Results
Far-UV Circular Dichroism Spectra and Thermal Melting Curves of CYP101A1 and CYP119.
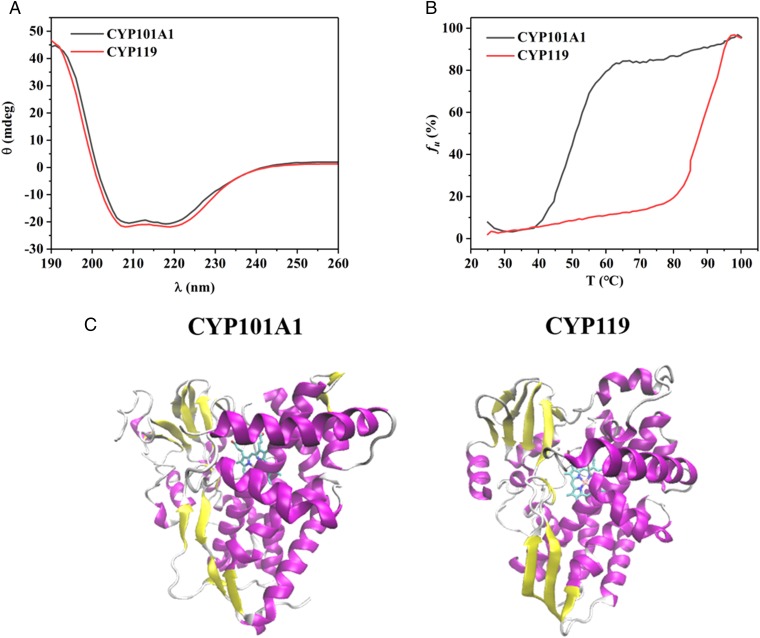
Far-UV circular dichroism (CD) spectra of CYP101A1 and CYP119 were acquired at 25 °C (Fig. 1A) and indicate that both proteins are well folded and contain a mix of alpha-helices and beta-sheets in terms of secondary structure, similar to what is observed in the ligand-free crystal structures of the two proteins (14, 40), as shown in Fig. 1C. Thermal melting curves which monitor the unfolding of protein as a function of temperature were also obtained for the two proteins. Unfolding profile for the secondary structure in each protein was measured at a wavelength of 222 nm as a function of increasing temperature until the protein unfolded. The unfolded fraction, fu, was calculated using
| [2] |
where CT is the observed CD value (millidegrees) of CYP101A1 or CYP119 at a given temperature T, and Cmax and Cmin are the maximum and minimum CD values (millidegrees) of CYP101A1 or CYP119 during the thermally induced unfolding process. By fitting the melting curves presented in Fig. 1B with a Boltzmann function, the melting temperature (Tm) for each protein was calculated. From the resulting values, it is evident that CYP119 used in our studies is much more thermostable than its mesophilic counterpart CYP101A1, since its Tm value (89.0 ± 1.2 °C) is significantly higher than that of CYP101A1 (50 ± 1.0 °C), in agreement with previously reported results (41).
Fig. 1.
CD spectra and thermal melting curves of ligand-free CYP101A1 and ligand-free CYP119. (A) Comparison of far-UV CD spectra of CYP101A1 (black) and CYP119 (red). Spectra were acquired at 25 °C on a sample of each protein with a concentration of 5 µM in 50 mM potassium phosphate, 50 mM potassium chloride buffer, pH 7.4. (B) Comparison of thermal unfolding curves of CYP101A1 (black) and CYP119 (red). Unfolding of the secondary structure of each protein was monitored at a fixed wavelength of 222 nm as a function of increasing temperature, where fu is the unfolding fraction. Sample conditions were similar to those used in measurement of far-UV CD spectra in A. Experimentally measured ellipticity values were converted to percentage unfolded fraction fu as described in the text. (C) Crystal structures of (Left) ligand-free CYP101A1 (PDB ID code 3L61) and (Right) ligand-free CYP119 (PDB ID code 1IO7).
Comparison of Thermodynamic Parameters Between CYP101A1 and CYP119 Based on Free Energy Stability Curves.
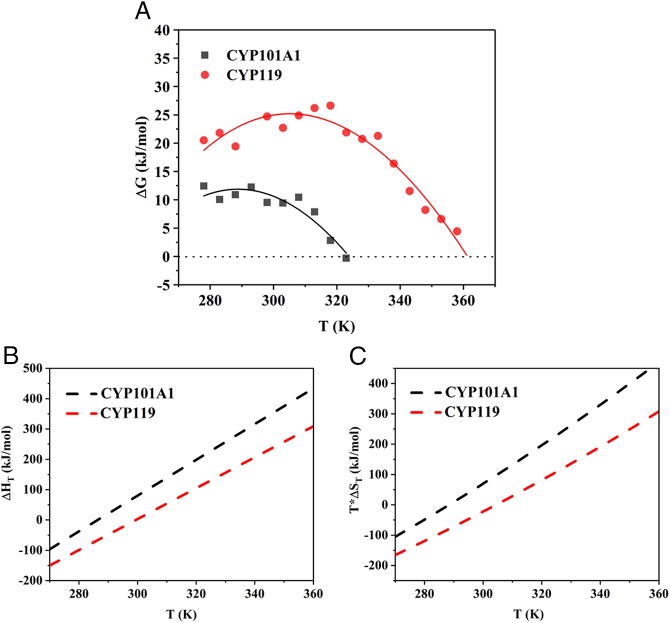
Free energy stability curves were determined for both CYP101A1 and CYP119 via the chemical denaturation method using guanidinium hydrochloride (GdnHCl). The temperature dependence of GdnHCl-induced unfolding of each protein was measured as described in the experimental section to determine the free energy of unfolding ∆G and also the free energy stability curve (Fig. 2A). A fluorescence-based technique was utilized to monitor the unfolding of the protein which relies on use of the fluorescent dye, SYPRO Orange. SYPRO Orange is known to bind to hydrophobic regions of a protein with high affinity and has been used previously to study ligand binding, protein stability, and unfolding of proteins to determine Tm values using differential scanning fluorimetry or thermal shift assays (42, 43). The fluorescent intensity of the dye increases dramatically when it binds to exposed hydrophobic regions of the protein as the protein unfolds, making this method a very useful quantitative monitor of protein unfolding. We have adapted this fluorescence-based method in a manner to determine protein free energy stability curves for thermodynamic parameter measurements by systematically measuring the protein unfolding process as a function of increasing amounts of chemical denaturant, which, to our knowledge, has not been demonstrated previously for any protein. A major advantage of using fluorescence as a means of detection rather than traditional CD spectroscopic methods for such thermodynamic measurements is that it can be done with greater speed and requires lower amounts of samples. This is because the method is amenable to setup in a 96-well or 384-well format in a real-time PCR instrument, where, due to availability of a temperature-gradient feature and compatible fluorescence filter sets (λex 470 nm/λem 570 nm), the entire stability curve comprising free energy measurements at different temperatures can be acquired in a single run, reducing the overall measurement time from days to a few hours.
Fig. 2.
(A) Free energy stability curves for ligand-free CYP101A1 (black) and ligand-free CYP119 (red) determined by fluorescence dye method as described in Methods. The solid lines were fitted to the ΔG(T) points obtained at various temperatures by using Eq. 3, where ΔG(T) is the free energy of unfolding determined via chemical denaturation using GdnHCl as described in Methods and SI Appendix. (B) Enthalpy change (∆HT) and (C) entropy change (T*∆ST) during unfolding versus temperature for CYP101A1 (black) and CYP119 (red). Values of ∆HT and T*∆ST were derived from Eqs. 4 and 5, respectively. The dashed lines in the plot are illustrative lines for the enthalpy change and entropy change parameters for each protein as a function of temperature.
For comparison, the free energy stability curve of CYP119 obtained by the traditional CD spectroscopy method is presented in SI Appendix, Fig. S2 and agrees quite well with the fluorescence measurements. We envision that this method will find general applicability in similar thermodynamic measurements for other thermophilic P450s, although due diligence must be performed for factors such as solubility of the protein, exposed hydrophobic patches, and background fluorescence from intrinsically fluorescent groups that may complicate the accuracy of the measurement and interpretation of results. In our case, these factors were not an issue for either protein based on structural analysis and control measurements performed (see experimental section). The free energy stability curve for CYP119 is much higher than that for CYP101A1 at all temperatures below the melting temperature of CYP101A1 (Fig. 2A), demonstrating that CYP119 is thermally more stable. It is also found that CYP119 is maximally stable at 35 °C with a maximum stabilization free energy of ∼25 kJ/mol, which is considerably higher than that for CYP101A1, which is ∼12 kJ/mol at its maximal stability temperature of 15 °C.
Next, we determined the enthalpic and entropic contribution to the free energy of unfolding (Eq. 1) at any given temperature via the modified Gibbs−Helmholtz equation,
| [3] |
where Tm is the melting temperature, ∆G is the unfolding free energy, ∆Cp is the change in heat capacity, and ∆Hm and ∆Sm are the enthalpy penalty and entropy gain of unfolding at the melting temperature. All of these thermal parameters can be determined by fitting Eq. 3 to the free energy stability curves (Fig. 2A), and the results are presented in Table 1. From the fit, the values of Tm obtained are ∼361 K (CYP119) and ∼323 K (CYP101A1), in general agreement with the values derived from the thermal unfolding experiments obtained by circular dichroism (Fig. 1B). The change in heat capacity ∆Cp was also obtained from the fit and shows that CYP119 has smaller ∆Cp (∼5.1 kJ⋅mol−1⋅K−1) compared with CYP101A1 (∼5.9 kJ⋅mol−1⋅K−1), which is reflected in the broadening of the CYP119 stability curve relative to that of CYP101A1 (Fig. 2A). It is also observed that the ∆Hm value for CYP119 (314 kJ⋅mol−1) is about 45% higher than that of CYP101A1 (216 kJ⋅mol−1), which would usually imply that the enthalpic interactions play a major role in stabilizing CYP119 relative to CYP101A1. Additionally, the ∆Sm value for CYP119 (869 J⋅mol−1⋅K−1) is found to be about 30% higher than the ∆Sm value for CYP101A1 (668 J⋅mol−1⋅K−1), suggesting that there is a larger entropy change for CYP119 at its unfolding temperature. Thus, it would seem that the favorable enthalpic change for CYP119 is solely responsible for determining the thermostability of CYP119 while the entropic contribution would not be of any significance, as has also been suggested by previous thermodynamic studies with other P450s such as CYP175A1 (15). However, this conclusion changes if the comparison is done at similar temperatures rather than the respective unfolding temperatures for the two proteins which are significantly different.
Table 1.
Thermodynamic parameters of CYP101A1 and CYP119 determined from the thermal stability curves
| Protein | Tm, K | ∆Hm, kJ⋅mol−1 | ∆Cp, kJ⋅mol−1⋅K−1 | ∆Sm, J⋅mol−1⋅K−1 |
| CYP101A1 | 323 ± 1.7 | 216 ± 25 | 5.9 ± 0.6 | 668 ± 73 |
| CYP119 | 361 ± 1.4 | 314 ± 22 | 5.1 ± 0.6 | 869 ± 60 |
Tm, ∆Hm, and ∆Cp were determined by fitting the stability curve for each protein to the Gibbs−Helmholtz equation (Eq. 3). ∆Sm was calculated from ∆Hm and Tm values. Errors represent SEM.
Such a comparison can be made by determining the individual enthalpic and entropic contributions to unfolding free energy at any given temperature other than Tm using the relationships
| [4] |
| [5] |
The resulting plots of ∆HT and ∆ST as a function of temperature are presented in Fig. 2 B and C. A striking trend observed from these plots is that both the enthalpy and entropy changes for CYP119 are actually much lower than those for CYP101A1 at all temperatures. Thus, even though CYP119 seems thermally more stable than CYP101A1 at their respective unfolding temperatures due to higher enthalpic contribution, when the comparison is made at the same temperature, the relative enthalpic contribution is actually lower, while the entropic contribution to thermal stability is much more significant for CYP119 due to a smaller entropy gain during unfolding, which reduces the free energy of unfolding and thus thermally stabilizes CYP119. In other words, at the same temperature, CYP119 is actually less stable compared with CYP101 if only the enthalpic term is taken into account, and the reason to render CYP119 thermally more stable is that it has much less entropy gain during unfolding. Therefore, assessment of thermostability solely based on crystal structures could sometimes be misleading, as structural analysis normally addresses the enthalpic contribution while ignoring the entropic contribution.
Comparison of Inherent Protein Flexibility Between CYP101A1 and CYP119.
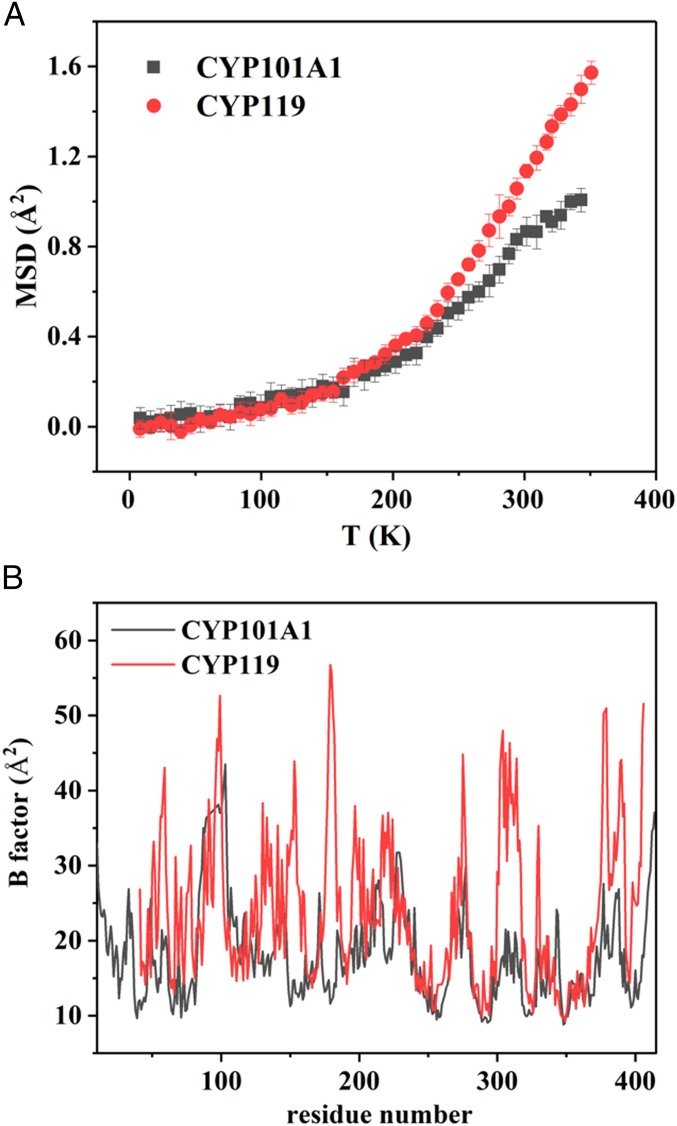
To compare the internal dynamics in CYP101A1 and CYP119, we performed neutron scattering experiments on protein powders hydrated with D2O, and derived the mean squared atomic displacements (MSDs), <x2(∆t)>, of CYP101A1 and CYP119 as a function of temperature as outlined in the experimental section. Use of neutron scattering is inherently advantageous in this case due to its ability to readily observe change in protein flexibility on a global scale over a wide temperature range and, in particular, at very high temperatures that are typically not accessible to crystallographic methods or solution methods such as NMR. Analysis of the MSD results (Fig. 3A) from neutron scattering reveals a clear dynamic transition, i.e., change in the slope of MSD at ∼200 K, for both proteins. This dynamic transition is coupled to the thermal activation of higher-order, large-scale collective protein motions and is thus thought to be important for specific protein function in proteins (36, 44). Below this transition temperature, protein dynamics consists of mostly harmonic vibrations and methyl rotations, which are essentially the same for both CYP101A1 and CYP119, as expected. Above the transition temperature, anharmonic large-scale collective dynamic motions set in, and the MSD comparison between the two proteins demonstrates that these dynamical processes are clearly more abundant in CYP119 than in CYP101A1 at all temperatures up to their melting temperatures. Hence, it can be concluded that CYP119 is, overall, more dynamic than CYP101A1 at any given temperature, especially in terms of anharmonic, collective atomic motions.
Fig. 3.
(A) Comparison of MSD of ligand-free CYP101A1 (black) and ligand-free CYP119 (red) as a function of temperature from 4 K to 353 K determined by neutron scattering spectroscopy. Error bars represent one standard deviation. The detailed procedures for preparing the samples for neutron scattering, data acquisition, and analysis are provided in Methods. (B) Comparison of crystallography B factors of ligand-free CYP101A1 (PDB ID code 3L61) and ligand-free CYP119 (PDB ID code 1IO7) at 100 K.
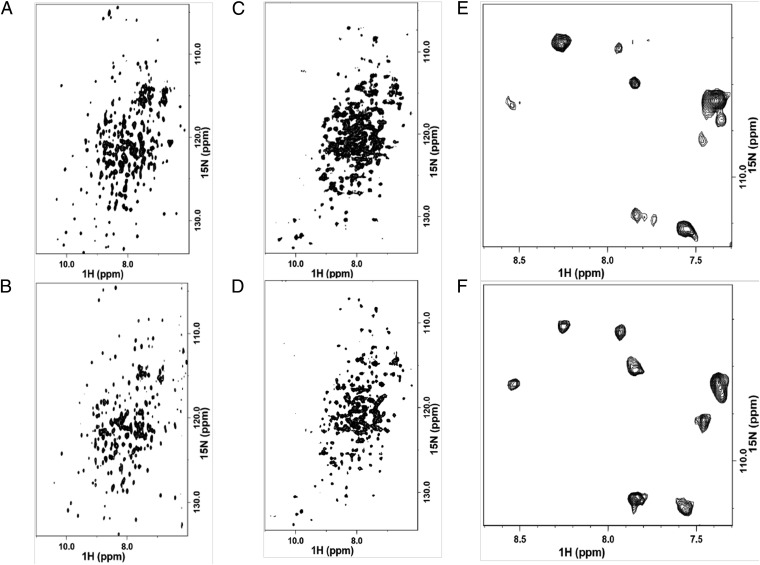
The inherent protein dynamics in CYP101A1 and CYP119 was also studied by solution NMR spectroscopy to obtain residue-level comparison of protein dynamics throughout the protein. The 2D 1H-15N transverse relaxation-optimized spectroscopy (TROSY) NMR spectra of CYP101A1 and CYP119 were acquired under identical conditions at 30 °C, and peak patterns in the two spectra were compared. As can be seen from the NMR spectra of ligand-free CYP101A1 and CYP119 in Fig. 4 A, C, and E, extensive line-broadening and splitting of peaks into multiplets was observed for CYP119 resonances throughout the spectra relative to the resonances for CYP101A1. The increased NMR line-broadening in CYP119 relative to CYP101A1 is unexpected since it is a smaller protein than CYP101A1 (368 vs. 414 residues), which indicates that CYP119 is sampling multiple conformations that may be in intermediate or slow exchange, unlike CYP101A1, which exhibits a smaller amount of such characteristics. The presence of peak clusters instead of individual peaks combined with extensive line-broadening clearly implies the presence of higher conformational disorder as well as increased dynamics in CYP119 compared with CYP101A1, consistent with the results obtained from neutron scattering experiments (Fig. 3A). Interestingly, the conformational disorder still persists to a significant extent in CYP119 even in the presence of a high-affinity ligand such as 4-phenylimidazole, although the overall conformational dynamics is reduced (Fig. 4 D and F). Similar reduction in dynamics is also observed for 4-phenylimidazole−bound CYP101A1 (Fig. 4B) compared with 4-phenylimidazole−bound CYP119. This observation supports our hypothesis that the inherent flexibility of CYP119 thus may not be entirely utilized for binding ligands as is generally deduced from analysis of ligand-bound and ligand-free crystal structures of CYP119, but may also likely make an important entropic contribution to its enhanced thermostability. Presence of large conformational flexibility in CYP119 in the F/G loop region has been previously inferred from crystal structures of the ligand−protein complexes (14) in the form of conformational changes, from previous MD simulations, and also via 2D NMR spectroscopy of 13C-p-methoxyphenylalanine insertions or 15N selectively labeled Phe residues in CYP119 (45, 46), which all suggest that CYP119 samples a range of predetermined conformational states in solution and ligand binding selects for the appropriate conformation(s). However, the observations from these studies have been restricted primarily to the ligand binding regions that require such flexibility for diverse substrate recognition, similar to several mesophilic P450s (47). None of these previous studies have been able to detect the extent of temperature-dependent dynamic disorder observed from neutron scattering and NMR spectra in our study. Given that a protein encounters a hierarchy of states in its conformational landscape, dynamic fluctuations can occur on several timescales and length scales. Thus, it may not be possible to assess various forms of protein flexibility from MD simulations on the small range of timescales employed in these studies or from limited NMR snapshots of the protein with selectively labeled amino acids, as the contributing fluctuation modes may be partitioned in different proportions over the protein matrix and do not represent the entire protein dynamics.
Fig. 4.
Comparison of 2D 1H-15N TROSY-HSQC (heteronuclear single-quantum coherence) NMR spectra of (A) ligand-free CYP101A1, (B) 4-phenylimidazole-bound CYP101A1, (C) ligand-free CYP119, and (D) 4-phenylimidazole-bound CYP119 at 30 °C. (E and F) Comparison of zoomed-in example regions of ligand-free CYP119 and 4-phenylimidazole-bound CYP119 full spectra in C and D to better show increased conformational disorder relative to CYP101A1 and dynamic changes upon ligand binding in CYP119. Spectra were acquired using samples of 15N uniformly labeled protein with and without the ligands at a concentration of 150 μM in 50 mM potassium phosphate and 50 mM potassium chloride, pH 7.4.
The presence of increased overall flexibility of CYP119 relative to CYP101A1 is also evident from comparison of B factors of crystal structures of ligand-free CYP101A1 [Protein Data Bank (PDB) ID code 3L61] and ligand-free CYP119 (PDB ID code 1IO7), which shows that CYP119 is globally more dynamic than CYP101A1, as shown in Fig. 3B, and not just the active site regions, which is in agreement with the neutron scattering and NMR experimental results.
Discussion
Increased Protein Flexibility as a Source of Entropic Contribution to Thermostability of CYP119.
Various enthalpic factors that rely on interactions between individual amino acid residues have been proposed to account for the thermostability of thermophilic proteins (48). In the case of thermophilic P450s such as CYP119, based on comparison of crystal structures, the enhanced thermostability of the protein relative to the mesophilic counterparts such as CYP101A1 has been suggested to arise primarily from an increased network of salt bridge interactions and extensive clustering of aromatic rings in CYP119, both of which are deemed to contribute to the enthalpy penalty toward unfolding (14, 22). Mutations involving these interactions are known to lower Tm in the range of ∼1 °C to 10 °C (49, 50). This indicates that, while the aromatic stacking interactions and salt bridge interaction networks are important in contributing to the thermostability of CYP119, they, by themselves, do not account entirely for the observed thermostability of CYP119, the melting temperature of which is more than 35 °C higher than that of CYP101A1. Recently, nonpolar interactions within the Cys ligand loop in CYP119 were also implicated in stabilization of the CYP119 fold based on lowering of Tm by up to ∼16 °C for mutations made in this region (27). Other factors suggested to augment thermostability of CYP119 include stronger internal hydrogen bonding, a smaller, more compact structure with shorter β-turns compared with CYP101A1, and an increased number of branched amino acids such as isoleucine leading to greater hydrophobic interactions in the core of the protein. However, the contribution of these factors to thermostability of CYP119 was not deemed to be significant, based on statistical analysis of different P450 structures that found no large deviations for these factors between CYP119 and mesophilic P450s (14, 22).
A striking characteristic feature of the above factors, especially the aromatic stacking and salt bridge interactions, is that they all mostly contribute to the enthalpy term of the free energy (Eq. 1) (51). Intuitively, these stronger attractive interactions between residues would rigidify the protein molecule, and therefore one would expect CYP119 to be more rigid compared with CYP101A1. However, this is contrary to the results obtained here using neutron scattering, NMR, and comparison of crystallography B factors, which all clearly indicate that CYP119 is globally more dynamic than CYP101A1 (Figs. 3 and 4). To explain this discrepancy, it is important to consider the entropy term in understanding the thermostability of CYP119. As is observed from the plot of enthalpy and entropy terms as a function of temperature, while the enthalpy of unfolding for CYP119 clearly makes a negative contribution to the free energy stability curve compared with CYP101A1 at every temperature, the enhanced thermal stability, i.e., higher unfolding free energy of the thermophilic enzyme, results instead from its much smaller entropy gain during unfolding. The reduction in entropy gain will definitely benefit from the protein being more flexible and disordered in the folded state, as shown in Figs. 3 and 4. Such entropic driving forces are not unusual for stability of proteins, and it has been argued that the enhanced thermostability of many thermophilic proteins are indeed entropy-driven, with the thermostable proteins, on average, exhibiting a lower value of conformational entropy change (per amino acid) of unfolding (52, 53). In fact, enhancement of protein stability by site-directed mutations to reduce conformational entropy gain of unfolding has been demonstrated quite early on in bacteriophage T4 lysozyme, where Gly and Pro substitutions at key positions in the lysozyme structure enabled entropic stabilization of the protein (54). A recent study of thermal stability of Taq DNA polymerase similarly proposed a reduced entropic folding penalty as the basis for enhanced thermal stabilization for this enzyme upon analysis of thermodynamic data on thermophilic Taq and mesophilic Escherichia coli DNA polymerases (55). When the analysis was extended to available thermodynamic data for 17 other thermophilic−mesophilic protein pairs, analogous entropic patterns were observed in seven of these systems, which suggests that this mechanism of stabilizing thermophilic proteins may be more common than previously thought (11, 55). More recently, thermodynamic studies carried out on another thermophilic P450, CYP175A1, also support the same conclusion that the thermostability is entropy-driven, when their thermodynamic data are reanalyzed in a manner similar to ours (SI Appendix, Fig. S3) (15). Since the original analysis of thermodynamic data on CYP175A1 and mesophilic CYP101A1 in that publication was carried out at their respective unfolding temperatures, the entropic effects were found to be less prominent for the thermostability of the protein. However, if the comparison is carried out at the same temperature to assess entropy values, the results are actually opposite and more in line with that for CYP119 (SI Appendix, Fig. S3). This would also explain why CYP175A1 is highly thermostable, similar to CYP119, even though its structure is missing the characteristic aromatic stacking interactions observed in CYP119. Accounting for entropic effects is an important consideration that is often missed when the analysis is solely based on invoking enthalpic interactions by simply comparing the static crystal structures.
It is interesting to trace the origin of this entropic contribution toward thermostability in CYP119. The neutron scattering data clearly indicate significantly higher MSDs for CYP119 compared with CYP101A1 at all temperatures up to the melting temperature of CYP119, pointing to the presence of greater dynamic and conformational heterogeneity in CYP119. However, it is not readily obvious from this data what the nature of this dynamic difference is and whether this occurs throughout the protein. Analysis of NMR spectra of the two proteins offers further clues in this regard, since the NMR resonances can be analyzed in a residue-wise manner and give a dynamic assessment of the entire protein on all timescales. Upon comparison of Fig. 4 A and C, considerable conformational disorder and dynamic differences are observed throughout the spectrum of CYP119, whereas only a small percentage of residues (<30%) are affected in CYP101A1 spectrum to the extent that they are in CYP119. This indicates that multiple conformations on a variety of timescales are being sampled by CYP119 across most of the protein, and thus we deduce that the entire structure of CYP119 protein is flexible and this flexibility is not completely abolished in CYP119 even when a strong affinity ligand such as 4-phenylimidazole binds to the active site, indicating that the purpose of the high inherent flexibility in CYP119 is not solely to accomplish the function of ligand binding. Additionally, the increased global flexibility of CYP119 is reconfirmed by the comparison of the crystallography B factors of these two proteins (Fig. 3B).
Such increased global flexibility is not unprecedented for thermophilic proteins. Enhanced flexibility for the thermophilic variants over the mesophilic counterparts on the millisecond to second timescale were detected for rubredoxin and α-amylase by hydrogen/deuterium exchange experiments (20, 37). Also, a study of backbone dynamics on multiple timescales of thermophilic and mesophilic ribonuclease enzymes by 15N NMR relaxation measurements found that the extent of flexibility for thermophilic ribonuclease is significantly more than the mesophilic protein on certain timescales (56). Larger dynamic fluctuations on the picosecond timescale at a global level have also been reported for thermophilic malate dehydrogenase and dihydrofolate reductase by elastic incoherent neutron scattering (57, 58). The above studies clearly dispel the notion that thermophilic proteins are more rigid than their mesophilic counterparts, at least on certain timescales. These dynamic fluctuations throughout the protein may cause the low-level conformational disorder observed throughout the protein, creating a large number of accessible conformational microstates that can be occupied as the temperature increases and contribute to the conformational entropy of the folded state, thereby increasing its thermostability. In contrast, the mesophilic counterpart can initiate unfolding at earlier temperatures if it has fewer of these accessible states, especially conformations corresponding to the native folded state. It is interesting to note that another previous MD simulation on CYP119 predicted a coupled outer shell which had different dynamic characteristics than the active site that is not observed in CYP101A1 (29). Also, another recent simulation on CYP119 found hotspots remote from the active site which formed a coupled network of residues to the Cys ligand loop that, when disrupted, leads to rapid unfolding of the protein (27). Mutation of certain aromatic residues in these hotspots was found to lower the thermostability up to 16 °C (27). It is therefore plausible that a dynamic network is pervasive throughout the entire protein which has distinct microstates modulated on various timescales to impart additional thermal stability via an entropic contribution, evidence of which is visible in the differential linewidths and/or peak splittings observed throughout the protein backbone in our NMR spectra (Fig. 4B). This change in backbone flexibility can be small and lead to entropic effects without causing major changes in structure that would disrupt enthalpic interactions on a global scale, and would therefore mask the entropic contribution when analyzing crystal structures of thermophilic proteins.
An additional factor that can add to this entropic contribution from the backbone is side-chain configurational entropy from the numerous salt bridge networks identified in CYP119. CYP119 has a smaller number of two-residue salt bridges but is known to have a higher number of three or more residue salt bridge networks (∼10) compared with CYP101A1 (∼6) (41). The multitude of favorable electrostatic interactions in these networks is believed to be an important source of thermostability for CYP119 (14, 22). Previous structural and computational studies suggest that salt bridge networks in thermophilic proteins generally are dynamic rather than static, with multiple configurations observed for salt bridge residue side chains (12). Incorporation of salt-bridges in networks overcomes the energetic cost of desolvation at high temperatures, due to favorable geometry and better electrostatic interactions resulting from shorter pair distances (an enthalpic effect), while, at the same time, the stochastic disorder due to increased thermal motions of the side chains at higher temperatures is easily accommodated in a network by increasing salt bridge occupation frequencies among the various charged side chains in the network (59). This makes the side-chain interactions long-lived, even though it reduces specific charged residue pair correlations and tends to increase the configurational entropy of both backbone and side-chain atoms in the folded form. This entropic effect would augment the enthalpic component of thermal stability imparted by the electrostatic effects. This was very elegantly demonstrated for a homotrimeric coiled coil LPP56 containing a well-packed hydrophobic core and a number of salt bridge networks on the surface (59). Analysis of salt bridge networks in ligand-free CYP119 by comparing them between two molecules present in the same asymmetric unit of the crystal structure similarly reveals presence of multiple configurations for several side chains in these networks (14). For example, in the salt- ridge network formed by Arg188/Glu184/Asp85, the side chain of Arg188 is strongly hydrogen-bonded to the side chains of both Glu184 and Asp85 in one of the molecules, but is only strongly hydrogen-bonded to the side chain of Glu184, since it is flipped in orientation and exhibits a much weaker hydrogen bonding to Asp85 in the second molecule. Another example is the side-chain network formed by Arg100/Glu96/Arg328/Glu332/Glu333, which shows different configurations in the two molecules, with differences in hydrogen bonding between Arg100 and Glu333 due to change in distance and orientation of the side chains and also in the hydrogen bonding between Arg100 and Glu332 side chains due to change in their orientation. Moreover, differences in side-chain configurations are also observed when comparing crystal structures of ligand-free CYP119 solved at two different temperatures (100 K and 270 K), with some of the largest differences in B factors between the two structures corresponding to residues incorporating the above side chains (14). The resulting side-chain configurational entropy seen in these salt bridge networks therefore sheds additional light on the role of salt bridge networks in enhancing thermal stability in thermophilic P450s, which is quite distinct from their typically enthalpic role. Since the overall entropy is distributed throughout the protein, it would also explain why individual single-site mutations in salt bridge networks only cause minor changes in Tm values, as they would result only in small amounts of entropic destabilization, but multisite mutations would likely cause larger changes due to cumulative entropic effects (50). Another worthwhile possibility to consider with regards to an alternate entropic role for side chains is aromatic residues, especially those involved in aromatic clusters in thermophilic proteins such as CYP119 that have been invoked as contributing to increased stability by virtue of their characteristic enthalpic interactions, such as aromatic stacking (41). Two prominent aromatic clusters, not found in other thermophilic P450s, are present in CYP119: one cluster formed by five residues—Tyr2, Trp4, Phe5, Phe24, and Trp281—and another cluster formed by seven residues—Phe225, Phe228, Trp231, Tyr250, Phe298, Phe334, and Phe338 (41). Apart from increasing thermal stability in CYP119 via aromatic interactions, such clusters could, in part, contribute favorably to the entropic term in these interactions via a hydrophobic effect that entails release of ordered water molecules from the surfaces of these hydrophobic aromatic side chains when they come together to form such clusters in the folded structure. This contribution, while important, is unlikely to be a major factor for enhanced stability, since these clusters are not found in other thermophilic P450s. Also, in CYP119, they are present close to the surface and exposed to water, which has the potential to form similar ordered shells around the exposed part of aromatic side chains, albeit to a reduced extent compared with the unfolded state. This would again explain the small changes in Tm values (<10 °C) observed upon site-specific mutations carried out in these clusters (49, 50).
The presence of an increased number of accessible dynamic microstates would not only contribute toward the conformational entropy but also in increasing the heat capacity of the folded state toward that of the denatured form. As a result, ∆Cp of unfolding is reduced in CYP119 similar to what is observed in other thermophilic proteins. For example, by measuring the backbone conformational dynamics of B1 domain of streptococcal protein G, reduced conformational heat capacity was calculated from backbone NMR order parameters and correlated to increased thermal stability (60). Assuming that the unfolded states for both CYP101A1 and CYP119 are similar in nature, this would explain the lower heat capacity change for CYP119 relative to CYP101A1 reflected in the broadening of the free energy stability curve for CYP119 with a shift of Tm to higher temperatures. However, it is important to note that other factors may also contribute to the smaller ∆Cp and ∆S, such as more rigid or compact structure of the denatured protein molecules, different nature of interactions, and/or smaller entropy gain of water molecules during denaturation. These factors are very difficult to quantify in the present work and would require further extensive investigation, and thus no firm conclusions for these factors can be drawn from experiment at this time. We note here that a high-temperature (500 K) MD simulation to study unfolding of CYP119 and CYP101A1 found only a marginal increase in the radius of gyration for CYP101A1 versus CYP119 upon unfolding (27), making it less likely that differences in denatured state of the two proteins are a major source of entropic differences between the two proteins.
Partitioning of Protein Flexibility Between Ligand Binding and Thermal Stability.
Finally, one may also consider the relative contribution of protein flexibility toward functional aspects such as ligand binding and catalytic activity vis-à-vis its role in thermostability. Our NMR data on the dynamic changes in CYP119 with and without ligand indicate preservation of a residual level of dynamics throughout the protein even in the presence of ligand, which points to a notion of flexibility partitioning where part of the flexibility is used for ligand binding/catalysis and part for imparting thermal stability, and that this partitioning may operate differently between thermophilic and mesophilic enzymes. This represents a previously unobserved paradigm for use of protein dynamics in two distinct biological roles within the same protein and, to our knowledge, has not been reported previously for any other thermophilic enzyme. A similar notion in context of thermal stability (but not ligand binding) has been put forward from rigidity analysis and H/D exchange studies on rubredoxin that is based upon a different way of flexibility partitioning between the mesophilic and thermophilic forms (37, 61, 62). It is possible that increased conformational fluctuations may be utilized at lower temperatures to reduce the entropic gain for thermal stability in thermophilic enzymes such as CYP119 which is not necessary in mesophilic enzymes, whereas flexibility related to activity is restricted at lower temperature and becomes more prominent at higher temperatures in these enzymes. It has been generally observed that the functional activity of thermophilic enzymes in terms of substrate turnover are several-fold lower than mesophilic enzymes around room temperature; however, activities of both types of enzymes become optimum much closer to their respective Tm values, which would support this conjecture. Further detailed experimental studies in terms of thermodynamic and dynamic measurements on multiple timescales (e.g., by NMR) are needed on CYP119 in various ligand-bound forms to characterize the extent of this partitioning as a function of temperature and delineate the entropic portion. It remains to be seen whether the significance of entropic contribution toward enhanced thermal stability manifested in CYP119 and CYP175A1 is a general trend in thermophilic P450s. If so, further characterization could potentially open up new avenues for engineering thermostability in P450 enzymes using entropic considerations.
Conclusion
In conclusion, the thermophilic cytochrome P450, CYP119, shows higher flexibility throughout the protein than its mesophilic counterpart CYP101A1 as found by our experimental solution NMR and neutron scattering studies, which is not readily apparent only by their structural comparison. This higher flexibility in the folded state is not necessarily contradictory to enhanced thermal stability, but rather positively contributes to the latter through an entropic effect which can be reconciled by the fact that less entropy gain during denaturation will increase the unfolding free energy and thus stabilize the protein. This scenario is supported by thermodynamic measurements of the temperature dependence of unfolding free energy in the present work, which shows that the enhanced thermal stability of CYP119 relative to mesophilic CYP101A1 is indeed due to less entropy gain of the enzyme during unfolding. It is likely to result from an increased conformational entropy in the folded state that can arise from multiple sources, including backbone fluctuations on multiple timescales and dynamic disorder of side chains in salt bridge networks. As such, we propose that the higher flexibility of CYP119 may be partitioned effectively between functional activity and thermal stability and in a way that is different from mesophilic P450s.
Methods
Protein Expression and Purification.
CYP101A1 protein was expressed and purified in ligand-free form using methods described previously (26). The native CYP119 gene with a C-terminal His6-tag sequence was synthesized and subcloned into a pET29a vector (Genscript Inc). The resulting plasmid DNA was transformed into BL-21 pLysS (DE3) E. coli cells and placed on an LB/Agar plate containing 50 mg/mL of kanamycin for selection, followed by incubation overnight at 37 °C. A single isolated colony with kanamycin resistance from the resulting colonies was used to inoculate 50 mL of LB media containing 50 µg/mL of kanamycin and grown at 37 °C for 4 h to 5 h with shaking at 250 rpm until the culture reached an OD600 of roughly 0.6. The cells were then spun down, and the pellet was transferred to 1 L of defined M9 medium containing 50 µg/mL of kanamycin, trace metals, 4 g of glucose as the carbon source, and 1 g of ammonium sulfate as the nitrogen source, followed by growth for an additional 6 h to 7 h at 37 °C to a density of OD600 > 1. The heme precursor ∂-amino levulinic acid was added to the culture at this point for a final concentration of 300 µM, followed by induction with 1 mM isopropyl-D thiogalactopyranoside. Protein expression was then allowed overnight for 10 h to 12 h at 37 °C. Cell pellets were harvested in the morning by centrifugation and resuspended in buffer containing 50 mM potassium phosphate and 50 mM KCl at pH 7.4 (buffer A). The resulting solution was then subjected to sonication to disrupt the cell membranes, followed by high-speed centrifugation for 30 min to remove cell debris. The supernatant was loaded onto a nickel-charged agarose affinity resin column equilibrated with buffer A to allow binding of CYP119. The column was then washed with five column volumes of buffer A, and CYP119 was eluted with buffer A containing 150 mM imidazole. Next, the elute was loaded onto an anion exchange column, washed with five column volumes of buffer A, and the protein was again eluted with buffer A containing 250 mM KCl. The eluted protein was then concentrated and further purified by size exclusion chromatography on an ÄKTA-FPLC system (GE Healthcare Life Sciences), using an S-200 column (BioRad) equilibrated with buffer A. Protein fractions coming off from the size exclusion column were collected and checked for purity by measuring the (417/280)nm wavelength ratio using UV-visible spectroscopy. Fractions with the (417/280)nm ratio greater than 1.4 were judged to be pure and pooled together for storage at −80 °C until needed for further experiments.
Protein Secondary Structure Characterization and Thermal Melting Curves Using CD Spectroscopy.
CD measurements on protein secondary structure were conducted at 25 °C in a Jasco J-815 spectropolarimeter using a 1-mm-pathlength cell. For determination of the melting curves, signals in the far-UV CD region (222 nm) were monitored as a function of temperature. The ligand-free CYP101A1 and CYP119 protein concentrations were 5 μM in buffer A (pH 7.4) for the various measurements. Both proteins were heated in the range of 25 °C to 100 °C with a heating rate of 0.5 °C/min, equilibrated for at least 2 min at each temperature, and the CD data were collected at 0.5 °C intervals. Reversibility of thermal unfolding was confirmed by changing the temperature by a few degrees in either direction at each acquired data point and was observed to present no hysteresis.
Protein Chemical Denaturation Studies Using Fluorescence Spectroscopy.
For determination of the stability curves, samples of ligand-free CYP101A1 and ligand-free CYP119 were prepared with different guanidinium hydrochloride (GdnHCl) concentrations ranging from 0 M to 6 M. Chemical denaturation curves were collected corresponding to the various GdnHCl concentrations at temperatures ranging from 5 °C to 50 °C for CYP101A1 and 5 °C to 85 °C for CYP119. Samples containing 2 μM of each protein in buffer A were prepared in triplicate and added to PCR tubes (USA Scientific). Each sample was incubated at the set temperature for up to 24 h before the fluorescence measurement to achieve equilibrium unfolding conditions. SYPRO Orange dye available as 5,000× stock (Sigma-Aldrich) was added just before the measurement to the protein samples in an appropriate amount to achieve a final concentration of the dye of 5×. Chemical denaturation of the protein was monitored by exciting the SYPRO Orange dye at 470 nm and monitoring its fluorescence emission at 570 nm using a Chromo4 DNAEngine real-time 96-well PCR instrument. Controls included samples containing buffer only, protein plus buffer only, and dye plus buffer only, measurements from which were used for baseline correction using the Opticon Monitor software available on the PCR instrument. Normalized intensity values I0 used in the various chemical denaturation curves were obtained by normalizing the experimental fluorescence intensity values from 0 to 1 and plotted as a function of increasing GdnHCl concentration in the range 0 M to ∼6 M. The normalized fluorescence intensity values were plotted as a function of GdnHCl concentration and nonlinearly fitted to Eq. 6 to extract the unfolding free energy value ΔG(T) of CYP101A1 and CYP119 at zero denaturant concentration and a given temperature (63).
| [6] |
where I0 is the normalized fluorescence intensity of the SYPRO Orange dye bound to CYP101A1 or CYP119 at a given GdnHCl concentration, c. If and Iu represent the intercepts, while sf and su are the slopes of folded and unfolded baselines, respectively. R is the gas constant, T is the temperature, and s is the slope in the transition state. The unfolding free energy values ∆G(T) were then plotted versus temperature in the respective temperature range for each protein to obtain the protein stability curve for both CYP101A1 and CYP119 (Fig. 2A).
Elastic Incoherent Neutron Scattering Studies of CYP101A1 and CYP119.
Elastic incoherent neutron scattering spectra were collected on ligand-free CYP101A1 and CYP119 samples at h ≈ 0.4 (0.4 g water per gram protein) using the NG2 high-flux backscattering spectrometer at the Center for Neutron Research at National Institute of Standard and Technology with a fixed energy resolution of ∼0.8 μeV (corresponding to a time resolution of ∼1 ns). Elastic scans were performed for both protein samples in the temperature range of 4 K to 353 K at a heating rate of 1.0 K/min. No correction for multiple scattering is needed since the neutron transmission was over 0.9, and multiple scattering is thus negligible (64). The experimentally measured quantity is the so-called elastic intensity, i.e., the intensity of the peak in the dynamic structure factor, S(q, ∆t), as a function of temperature, where ∆t is the instrument resolution, which is 1 ns here. MSD, <x2(∆t)>, is derived using the widely used Gaussian approximation that S(q, ∆t) = exp(−1/6q2 <x2(∆t)>) in the q range from 0.36 Å−1 to 1.32 Å−1 (65). The obtained MSDs are plotted in Fig. 3A.
NMR Studies of CYP101A1 and CYP119.
For solution NMR spectroscopy experiments on CYP101A1 and CYP119, 15N labeled samples of both proteins were prepared in a similar fashion to that described earlier in Protein Expression and Purification, except that 15N-labeled ammonium sulfate was used as a nitrogen source during cell growth. The final protein concentration used for the NMR experiments on both proteins was ∼0.15 mM. For the 4-phenylimidazole-bound form, 4-phenylimidazole was prepared as a 20-mM stock solution in methanol and added to the protein in small increments to achieve a final 1:1 relative molar concentration. The 1H-15N TROSY 2D spectra (Fig. 4) were acquired using these samples in ligand-free and 4-phenylimidazole-bound forms for both CYP101A1 and CYP119 in buffer containing 50 mM potassium phosphate and 50 mM KCl at pH 7.4 at 30 °C. The resulting data were processed and analyzed with Felix2000 software (Accelrys).
Supplementary Material
Acknowledgments
We thank Miranda Pratt for help with thermodynamic measurements using CD spectroscopy and Qin Xu for helpful discussions. The Shanghai Jiao Tong University (SJTU) group acknowledges support for Circular Dichroism Spectroscopy from Instrumental Analysis Center of SJTU. This work was supported by the National Science Foundation of China Grants 11504231 and 3163002. Access to High Flux Backscattering Source was provided by the Center for High Resolution Neutron Scattering, a partnership between the National Institute of Standards and Technology and the National Science Foundation under Agreement DMR-1508249.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807473115/-/DCSupplemental.
References
- 1.Sligar SG. Chemistry. Glimpsing the critical intermediate in cytochrome P450 oxidations. Science. 2010;330:924–925. doi: 10.1126/science.1197881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coon MJ. Cytochrome P450: Nature’s most versatile biological catalyst. Annu Rev Pharmacol Toxicol. 2005;45:1–25. doi: 10.1146/annurev.pharmtox.45.120403.100030. [DOI] [PubMed] [Google Scholar]
- 3.Schuler MA, Sligar SG. The Ubiquitous Roles of Cytochrome P450 Proteins. Wiley; New York: 2007. Diversities and similarities in P450 systems: An introduction; pp. 1–26. [Google Scholar]
- 4.Guengerich FP. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol. 2001;14:611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- 5.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Cowan DA. Thermophilic proteins: Stability and function in aqueous and organic solvents. Comp Biochem Physiol A Physiol. 1997;118:429–438. doi: 10.1016/s0300-9629(97)00004-2. [DOI] [PubMed] [Google Scholar]
- 7.Kellner DG, Hung S-C, Weiss KE, Sligar SG. Kinetic characterization of compound I formation in the thermostable cytochrome P450 CYP119. J Biol Chem. 2002;277:9641–9644. doi: 10.1074/jbc.C100745200. [DOI] [PubMed] [Google Scholar]
- 8.Sivaramakrishnan S, et al. A novel intermediate in the reaction of seleno CYP119 with m-chloroperbenzoic acid. Biochemistry. 2011;50:3014–3024. doi: 10.1021/bi101728y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fágáin CÓ. Understanding and increasing protein stability. Biochim Biophys Acta. 1995;1252:1–14. doi: 10.1016/0167-4838(95)00133-f. [DOI] [PubMed] [Google Scholar]
- 10.Taylor TJ, Vaisman II. Discrimination of thermophilic and mesophilic proteins. BMC Struct Biol. 2010;10:S5. doi: 10.1186/1472-6807-10-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Razvi A, Scholtz JM. Lessons in stability from thermophilic proteins. Protein Sci. 2006;15:1569–1578. doi: 10.1110/ps.062130306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar S, Nussinov R. How do thermophilic proteins deal with heat? Cell Mol Life Sci. 2001;58:1216–1233. doi: 10.1007/PL00000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urlacher VB, Girhard M. Cytochrome P450 monooxygenases: An update on perspectives for synthetic application. Trends Biotechnol. 2012;30:26–36. doi: 10.1016/j.tibtech.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Park S-Y, et al. Thermophilic cytochrome P450 (CYP119) from Sulfolobus solfataricus: High resolution structure and functional properties. J Inorg Biochem. 2002;91:491–501. doi: 10.1016/s0162-0134(02)00446-4. [DOI] [PubMed] [Google Scholar]
- 15.Behera RK, Mazumdar S. Thermodynamic basis of the thermostability of CYP175A1 from Thermus thermophilus. Int J Biol Macromol. 2010;46:412–418. doi: 10.1016/j.ijbiomac.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 16.Ho WW, Li H, Nishida CR, de Montellano PR, Poulos TL. Crystal structure and properties of CYP231A2 from the thermoacidophilic archaeon Picrophilus torridus. Biochemistry. 2008;47:2071–2079. doi: 10.1021/bi702240k. [DOI] [PubMed] [Google Scholar]
- 17.Oku Y, et al. Structure and direct electrochemistry of cytochrome P450 from the thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7. J Inorg Biochem. 2004;98:1194–1199. doi: 10.1016/j.jinorgbio.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Colombo G, Merz KM. Stability and activity of mesophilic subtilisin E and its thermophilic homolog: Insights from molecular dynamics simulations. J Am Chem Soc. 1999;121:6895–6903. [Google Scholar]
- 19.Dalhus B, et al. Structural basis for thermophilic protein stability: Structures of thermophilic and mesophilic malate dehydrogenases. J Mol Biol. 2002;318:707–721. doi: 10.1016/S0022-2836(02)00050-5. [DOI] [PubMed] [Google Scholar]
- 20.Fitter J, Heberle J. Structural equilibrium fluctuations in mesophilic and thermophilic α-amylase. Biophys J. 2000;79:1629–1636. doi: 10.1016/S0006-3495(00)76413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beadle BM, Baase WA, Wilson DB, Gilkes NR, Shoichet BK. Comparing the thermodynamic stabilities of a related thermophilic and mesophilic enzyme. Biochemistry. 1999;38:2570–2576. doi: 10.1021/bi9824902. [DOI] [PubMed] [Google Scholar]
- 22.Yano JK, et al. Crystal structure of a thermophilic cytochrome P450 from the archaeon Sulfolobus solfataricus. J Biol Chem. 2000;275:31086–31092. doi: 10.1074/jbc.M004281200. [DOI] [PubMed] [Google Scholar]
- 23.Vihinen M. Relationship of protein flexibility to thermostability. Protein Eng. 1987;1:477–480. doi: 10.1093/protein/1.6.477. [DOI] [PubMed] [Google Scholar]
- 24.Radestock S, Gohlke H. Exploiting the link between protein rigidity and thermostability for data-driven protein engineering. Eng Life Sci. 2008;8:507–522. [Google Scholar]
- 25.Chan C-H, et al. Relationship between local structural entropy and protein thermostability. Proteins. 2004;57:684–691. doi: 10.1002/prot.20263. [DOI] [PubMed] [Google Scholar]
- 26.Miao Y, et al. Coupled flexibility change in cytochrome P450cam substrate binding determined by neutron scattering, NMR, and molecular dynamics simulation. Biophys J. 2012;103:2167–2176. doi: 10.1016/j.bpj.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meharenna YT, Poulos TL. Using molecular dynamics to probe the structural basis for enhanced stability in thermal stable cytochromes P450. Biochemistry. 2010;49:6680–6686. doi: 10.1021/bi100929x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishida CR, Ortiz de Montellano PR. Thermophilic cytochrome P450 enzymes. Biochem Biophys Res Commun. 2005;338:437–445. doi: 10.1016/j.bbrc.2005.08.093. [DOI] [PubMed] [Google Scholar]
- 29.Brandman R, Lampe JN, Brandman Y, de Montellano PRO. Active-site residues move independently from the rest of the protein in a 200 ns molecular dynamics simulation of cytochrome P450 CYP119. Arch Biochem Biophys. 2011;509:127–132. doi: 10.1016/j.abb.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colthart AM, et al. Detection of substrate-dependent conformational changes in the P450 fold by nuclear magnetic resonance. Sci Rep. 2016;6:22035. doi: 10.1038/srep22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Navrátilová V, et al. Molecular insights into the role of a distal F240A mutation that alters CYP1A1 activity towards persistent organic pollutants. Biochim Biophys Acta. 2017;1861:2852–2860. doi: 10.1016/j.bbagen.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Tietz DR, Podust LM, Sherman DH, Pochapsky TC. Solution conformations and dynamics of substrate-bound cytochrome P450 MycG. Biochemistry. 2017;56:2701–2714. doi: 10.1021/acs.biochem.7b00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasutake Y, Kameda T, Tamura T. Structural insights into the mechanism of the drastic changes in enzymatic activity of the cytochrome P450 vitamin D3 hydroxylase (CYP107BR1) caused by a mutation distant from the active site. Acta Crystallogr F Struct Biol Commun. 2017;73:266–275. doi: 10.1107/S2053230X17004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asciutto EK, Pochapsky TC. Some surprising implications of NMR-directed simulations of substrate recognition and binding by cytochrome P450cam (CYP101A1) J Mol Biol. 2018;430:1295–1310. doi: 10.1016/j.jmb.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu S, et al. Altering the regioselectivity of cytochrome P450 BM-3 by saturation mutagenesis for the biosynthesis of indirubin. J Mol Catal B Enzym. 2010;67:29–35. [Google Scholar]
- 36.Hong L, et al. Determination of functional collective motions in a protein at atomic resolution using coherent neutron scattering. Sci Adv. 2016;2:e1600886. doi: 10.1126/sciadv.1600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hernández G, Jenney FE, Jr, Adams MWW, LeMaster DM. Millisecond time scale conformational flexibility in a hyperthermophile protein at ambient temperature. Proc Natl Acad Sci USA. 2000;97:3166–3170. doi: 10.1073/pnas.040569697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaenicke R. Do ultrastable proteins from hyperthermophiles have high or low conformational rigidity? Proc Natl Acad Sci USA. 2000;97:2962–2964. doi: 10.1073/pnas.97.7.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Danciulescu C, Ladenstein R, Nilsson L. Dynamic arrangement of ion pairs and individual contributions to the thermal stability of the cofactor-binding domain of glutamate dehydrogenase from Thermotoga maritima. Biochemistry. 2007;46:8537–8549. doi: 10.1021/bi7004398. [DOI] [PubMed] [Google Scholar]
- 40.Lee Y-T, Wilson RF, Rupniewski I, Goodin DB. P450cam visits an open conformation in the absence of substrate. Biochemistry. 2010;49:3412–3419. doi: 10.1021/bi100183g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yano JK, et al. Preliminary characterization and crystal structure of a thermostable cytochrome P450 from Thermus thermophilus. J Biol Chem. 2003;278:608–616. doi: 10.1074/jbc.M206568200. [DOI] [PubMed] [Google Scholar]
- 42.Krishna SN, et al. A fluorescence-based thermal shift assay identifies inhibitors of mitogen activated protein kinase kinase 4. PLoS One. 2013;8:e81504. doi: 10.1371/journal.pone.0081504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 44.Rasmussen BF, Stock AM, Ringe D, Petsko GA. Crystalline ribonuclease A loses function below the dynamical transition at 220 K. Nature. 1992;357:423–424. doi: 10.1038/357423a0. [DOI] [PubMed] [Google Scholar]
- 45.Basudhar D, et al. Analysis of cytochrome P450 CYP119 ligand-dependent conformational dynamics by two-dimensional NMR and X-ray crystallography. J Biol Chem. 2015;290:10000–10017. doi: 10.1074/jbc.M114.627935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lampe JN, et al. Ligand-induced conformational heterogeneity of cytochrome P450 CYP119 identified by 2D NMR spectroscopy with the unnatural amino acid (13)C-p-methoxyphenylalanine. J Am Chem Soc. 2008;130:16168–16169. doi: 10.1021/ja8071463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pochapsky TC, Kazanis S, Dang M. Conformational plasticity and structure/function relationships in cytochromes P450. Antioxid Redox Signal. 2010;13:1273–1296. doi: 10.1089/ars.2010.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc Natl Acad Sci USA. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puchkaev AV, Koo LS, Ortiz de Montellano PR. Aromatic stacking as a determinant of the thermal stability of CYP119 from Sulfolobus solfataricus. Arch Biochem Biophys. 2003;409:52–58. doi: 10.1016/s0003-9861(02)00402-2. [DOI] [PubMed] [Google Scholar]
- 50.Maves SA, Sligar SG. Understanding thermostability in cytochrome P450 by combinatorial mutagenesis. Protein Sci. 2001;10:161–168. doi: 10.1110/ps.17601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lazaridis T, Archontis G, Karplus M. Enthalpic contribution to protein stability: Insights from atom-based calculations and statistical mechanics. In: Anfinsen CB, Richards FM, Edsall JT, Eisenberg DS, editors. Advances in Protein Chemistry. Vol 47. Academic; New York: 1995. pp. 231–306. [DOI] [PubMed] [Google Scholar]
- 52.Sawle L, Ghosh K. How do thermophilic proteins and proteomes withstand high temperature? Biophys J. 2011;101:217–227. doi: 10.1016/j.bpj.2011.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pica A, Graziano G. Shedding light on the extra thermal stability of thermophilic proteins. Biopolymers. 2016;105:856–863. doi: 10.1002/bip.22923. [DOI] [PubMed] [Google Scholar]
- 54.Matthews BW, Nicholson H, Becktel WJ. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc Natl Acad Sci USA. 1987;84:6663–6667. doi: 10.1073/pnas.84.19.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu C-C, LiCata VJ. The stability of Taq DNA polymerase results from a reduced entropic folding penalty; identification of other thermophilic proteins with similar folding thermodynamics. Proteins. 2014;82:785–793. doi: 10.1002/prot.24458. [DOI] [PubMed] [Google Scholar]
- 56.Butterwick JA, et al. Multiple time scale backbone dynamics of homologous thermophilic and mesophilic ribonuclease HI enzymes. J Mol Biol. 2004;339:855–871. doi: 10.1016/j.jmb.2004.03.055. [DOI] [PubMed] [Google Scholar]
- 57.Tehei M, Madern D, Franzetti B, Zaccai G. Neutron scattering reveals the dynamic basis of protein adaptation to extreme temperature. J Biol Chem. 2005;280:40974–40979. doi: 10.1074/jbc.M508417200. [DOI] [PubMed] [Google Scholar]
- 58.Meinhold L, et al. Protein dynamics and stability: The distribution of atomic fluctuations in thermophilic and mesophilic dihydrofolate reductase derived using elastic incoherent neutron scattering. Biophys J. 2008;94:4812–4818. doi: 10.1529/biophysj.107.121418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Missimer JH, et al. Configurational entropy elucidates the role of salt-bridge networks in protein thermostability. Protein Sci. 2007;16:1349–1359. doi: 10.1110/ps.062542907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seewald MJ, et al. The role of backbone conformational heat capacity in protein stability: Temperature dependent dynamics of the B1 domain of streptococcal protein G. Protein Sci. 2000;9:1177–1193. doi: 10.1110/ps.9.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rader AJ. 2002. Protein rigidity and flexibility: Application to folding and thermostability. Doctor of Philosophy Doctoral thesis (Michigan State Univ, Ann Arbor, MI)
- 62.Rader AJ. Thermostability in rubredoxin and its relationship to mechanical rigidity. Phys Biol. 2009;7:16002. doi: 10.1088/1478-3975/7/1/016002. [DOI] [PubMed] [Google Scholar]
- 63.Santoro MM, Bolen DW. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 64.Khodadadi S, et al. Dynamics of biological macromolecules: Not a simple slaving by hydration water. Biophys J. 2010;98:1321–1326. doi: 10.1016/j.bpj.2009.12.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gabel F, et al. Protein dynamics studied by neutron scattering. Q Rev Biophys. 2002;35:327–367. doi: 10.1017/s0033583502003840. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.