Significance
Evolutionary and comparative genomics, combined with reverse genetics, have the power to identify and characterize new biology. Here, we use these approaches in several nontraditional model species of budding yeasts to characterize a budding yeast secondary metabolite gene cluster, a set of genes responsible for production and reutilization of the siderophore pulcherrimin. We also use this information to assign roles in pulcherrimin utilization for two previously uncharacterized Saccharomyces cerevisiae genes. The evolution of this gene cluster in budding yeasts suggests an ecological role for pulcherrimin akin to other microbial public goods systems.
Keywords: secondary metabolism, gene cluster, siderophore, budding yeasts, cyclodipeptide synthase
Abstract
Secondary metabolites are key in how organisms from all domains of life interact with their environment and each other. The iron-binding molecule pulcherrimin was described a century ago, but the genes responsible for its production in budding yeasts have remained uncharacterized. Here, we used phylogenomic footprinting on 90 genomes across the budding yeast subphylum Saccharomycotina to identify the gene cluster associated with pulcherrimin production. Using targeted gene replacements in Kluyveromyces lactis, we characterized the four genes that make up the cluster, which likely encode two pulcherriminic acid biosynthesis enzymes, a pulcherrimin transporter, and a transcription factor involved in both biosynthesis and transport. The requirement of a functional putative transporter to utilize extracellular pulcherrimin-complexed iron demonstrates that pulcherriminic acid is a siderophore, a chelator that binds iron outside the cell for subsequent uptake. Surprisingly, we identified homologs of the putative transporter and transcription factor genes in multiple yeast genera that lacked the biosynthesis genes and could not make pulcherrimin, including the model yeast Saccharomyces cerevisiae. We deleted these previously uncharacterized genes and showed they are also required for pulcherrimin utilization in S. cerevisiae, raising the possibility that other genes of unknown function are linked to secondary metabolism. Phylogenetic analyses of this gene cluster suggest that pulcherrimin biosynthesis and utilization were ancestral to budding yeasts, but the biosynthesis genes and, subsequently, the utilization genes, were lost in many lineages, mirroring other microbial public goods systems that lead to the rise of cheater organisms.
The production of secondary metabolites is found in organisms from all domains of life (1–3). Fungi are particularly well known for their production of secondary metabolites, whose functions can include antibiotics, virulence factors, pigments, toxins, quorum-sensing molecules, and siderophores (chelators produced by organisms to capture extracellular iron) (3, 4). These molecules are generally synthesized by proteins encoded in gene clusters, typically containing a synthase protein belonging to one of several families: a nonribosomal peptide synthetase (NRPS), polyketide synthase (PKS), terpene cyclase, dimethylallyl tryptophan synthetase, or cyclodipeptide synthase (CDPS) (2, 5–7). The absence of known genes encoding any of these functions in sequenced budding yeast genomes has precluded an understanding of secondary metabolism in the budding yeast subphylum Saccharomycotina (8).
The pigment pulcherrimin has long been known to be synthesized by a small number of budding yeast species, including various Metschnikowia spp. and some Kluyveromyces spp. (9–12). Pulcherrimin is a red iron-containing pigment composed of two cyclized and modified leucine molecules (13). The ecological role of pulcherrimin is not well understood, but several studies have shown it to mediate interspecific antagonistic interactions, possibly due to its ability to sequester ferric iron from the growth medium (14–16). The ability to sequester free iron from the environment from competitors could be counterproductive if the producer species were also unable to utilize the pulcherrimin-bound iron, raising the possibility that pulcherrimin producers can also reutilize the compound as a siderophore. Pulcherrimin may not be the only secondary metabolite produced by budding yeasts; there is evidence for ferrichrome production by some Lipomyces and Dipodascopsis spp. (17), but the genes responsible for production of both secondary metabolites in yeasts are unknown. Biochemical work in the pulcherrimin-producing bacterium Bacillus subtilis has characterized a two-step biosynthetic pathway: Two leucine molecules are cyclized via a CDPS, and the resulting diketopiperazine is oxidized by a cytochrome P450 oxidase to generate pulcherriminic acid (PA), which can then spontaneously bind iron to form pulcherrimin (18–20). The formation of diketopiperazines, such as the cyclo-Leu-Leu precursor to PA, is often catalyzed by biochemical pathways containing NRPS or CDPS proteins (21). Thus, the absence of known homologs of these genes in budding yeasts raises the possibilities of highly divergent homologs or novel biochemical pathways for pulcherrimin production.
Here, we use a comparative genomic approach to identify a putative gene cluster involved in pulcherrimin production. Using targeted gene replacements in Kluyveromyces lactis, we characterize this four-gene cluster, finding genes responsible for the biosynthesis and reutilization of pulcherrimin. We also find that several species, including Saccharomyces cerevisiae, contain a partial gene cluster comprised of only the two utilization genes. Using targeted gene replacements in S. cerevisiae, we assign functions and standard names to these previously uncharacterized genes. Finally, we infer that the gene cluster was ancestral to all budding yeasts, but repeated gene loss led to its patchy species distribution, as well as to the rise of cheater organisms that exploit the public good of pulcherrimin without the cost of production.
Results
A Secondary Metabolite Gene Cluster Is Responsible for Production of the Siderophore Pulcherrimin.
Pulcherrimin biosynthesis in B. subtilis consists of the cyclization of two leucine molecules via a CDPS encoded by yvmC, followed by oxidation via a cytochrome P450 oxidase encoded by cypX (18–20). When we searched a custom BLAST database composed of all publicly available Saccharomycotina genome assemblies (y1000plus.org/blast) (22), we found no homologs for yvmC and only one significant hit (e < 0.001) for cypX: the widely conserved sterol biosynthesis gene ERG11 (23). Thus, the B. subtilis genes did not suggest candidates for the budding yeast pathway. We reasoned that a more divergent cytochrome P450 oxidase could be involved in pulcherrimin biosynthesis, so we searched the genome of the pulcherrimin producer K. lactis for proteins with a P450 conserved domain using NCBI’s CDD tool and BLASTp. This approach yielded five P450 superfamily genes, one of which had no homologs in S. cerevisiae and was annotated in GenBank as a “gliC homolog.” gliC encodes a cytochrome P450 oxidase involved in secondary metabolism in some species of filamentous fungi, including the biosynthesis of gliotoxin by Aspergillus spp. (24, 25). We searched the Saccharomycotina BLAST database for homologs of this protein and found significant hits (e < 0.001) in the genomes of several Kluyveromyces spp., Candida auris, and Metschnikowia fructicola.
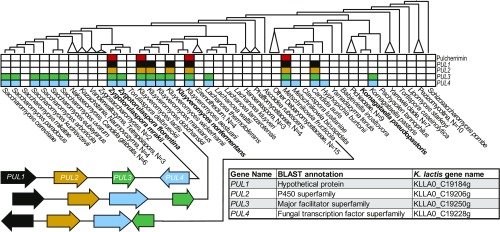
Among these species, we also found conservation of three genes surrounding the cytochrome P450 oxidase homolog, hereafter termed the PUL (PULcherrimin) gene cluster, with genes labeled PUL1-4 (Fig. 1 and SI Appendix, Fig. S1). In addition to K. lactis (10), Kluyveromyces aestuarii and M. fructicola had been described as producing pulcherrimin (11, 12). We sequenced the genomes of two more budding yeast species: one previously described as producing an unknown red pigment, Zygotorulaspora mrakii (26), from which we identified a PUL gene cluster; one from the other recognized species of this genus, Zygotorulaspora florentina, a nonpigmented strain that did not contain a complete PUL cluster. Thus, in each of the three genera, Kluyveromyces, Metschnikowia, and Zygotorulaspora, we found that red-pigmented species always have complete PUL clusters, whereas species lacking the complete PUL cluster were never pigmented (Fig. 1). A handful of species have complete PUL clusters but did not readily produce pulcherrimin during laboratory growth, nor did their species descriptions include a positive pulcherrimin trait. These species may be cryptic producers that make pulcherrimin under specific, yet untested, conditions (e.g., C. auris, and some Kluyveromyces spp.).
Fig. 1.
The PUL gene cluster and its phylogenetic distribution. The genome-scale cladogram was constructed using alignments of 1,037 conserved BUSCO genes (22) with newly sequenced genomes in bold. Collapsed clades with multiple (N) species are shown as triangles, and the complete tree, with bootstrap values, is shown in SI Appendix, Fig. S1. Red filled boxes indicate pulcherrimin production has been observed in that species, and filled boxes for PUL1-4 indicate presence or absence of the corresponding PUL gene. Gene arrow diagrams show the arrangement of the four PUL genes in representative species that produce pulcherrimin. The gene table contains putative annotations for the four genes before this study.
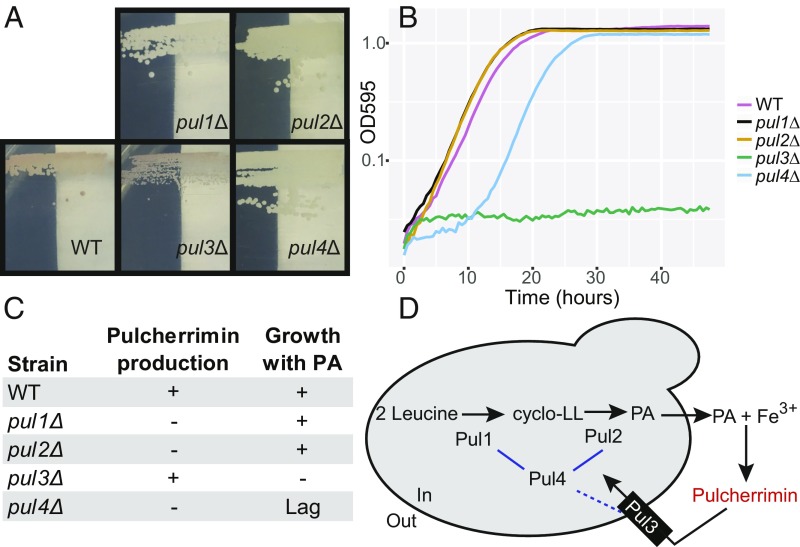
To directly test whether this gene cluster was responsible for pulcherrimin production, we generated targeted gene replacements in the genetically tractable, pulcherrimin-producing yeast K. lactis (27, 28). We replaced each of the four PUL genes individually with an antibiotic-resistance cassette and tested the mutant strains for pulcherrimin production on a defined, synthetic complete (SC) agar medium. Under these conditions, the wild-type strain produced a red pigment, a trait that was abolished in pul1Δ, pul2Δ, and pul4Δ mutants (Fig. 2A). Complementation by reinsertion of each deleted gene restored pigmentation to all mutants (SI Appendix, Fig. S2). To further characterize these genes, we performed cross-feeding experiments between mutants. A pul2Δ strain constitutively expressing PUL1 conferred pigmentation to a pul1Δ strain constitutively expressing PUL2, but not vice versa, strongly suggesting that PUL1 is upstream of PUL2 in the biochemical pathway (SI Appendix, Fig. S3). Further, while pul4Δ strains constitutively expressing PUL1 or PUL2 were not pigmented on their own, we observed the same cross-feeding relationship in this deletion strain, indicating that PUL4 is not required for pulcherrimin biosynthesis when PUL1 or PUL2 are expressed heterologously (SI Appendix, Fig. S3). We also attempted to express the B. subtilis yvmC and cypX genes in pul1Δ and pul2Δ strains, respectively, but they did not successfully complement the K. lactis mutants (SI Appendix, Fig. S4). Given these data and the putative annotations (Fig. 1), we hypothesize that PUL4 encodes a transcription factor that regulates PUL1 and PUL2, which we hypothesize encode the enzymes responsible for the two-step pulcherrimin biosynthesis pathway (Fig. 2D).
Fig. 2.
Characterization of the PUL genes. (A) Cells grown on defined medium produced a red pigment when the complete PULcherrimin pathway was present, but they lacked pigmentation when key genes were deleted. Complementation tests can be found in SI Appendix, Fig. S2. (B) Representative growth curves of individual pul mutants in PA-treated medium, where cells must uptake pulcherrimin to grow. Complementation tests can be found in SI Appendix, Fig. S5B. (C) Summary table of pulcherrimin production and growth in PA-treated medium. (D) Model of the putative roles of the PUL genes in siderophore production and reuptake based on our genetic data and previous biochemistry data (18–20); however, the yeast proteins currently lack direct biochemical assays. Blue lines represent regulation of PUL gene expression by Pul4.
The PUL Cluster also Contains Genes Responsible for Pulcherrimin-Complexed Iron Utilization.
While the pul3Δ mutant still produced pulcherrimin, it grew poorly on SC (Fig. 2). Several observations led us to hypothesize that the pul3Δ mutant was unable to reutilize the pulcherrimin it produced: growth on SC often gave rise to Pul− suppressors that grew normally but did not produce pulcherrimin, and a pul1Δ pul3Δ double mutant grew normally but did not produce pulcherrimin (SI Appendix, Fig. S5). Since the pul3Δ mutant strains grew poorly when they produced pulcherrimin, we hypothesized that the PUL3 gene was required to bring pulcherrimin-complexed iron back into the cell, perhaps by a mechanism similar to the siderophore-iron transporters characterized in S. cerevisiae, which are encoded by genes that include ENB1, SIT1, ARN1, and ARN2 (29).
To test the hypothesis that PUL3 is required for the uptake of iron using pulcherrimin, we devised a growth assay where strains were grown in liquid SC, with and without the addition of exogenous PA. PA is pulcherrimin that has been extracted away from iron using strong base and readily binds iron in growth media. Thus, addition of PA to growth medium sequesters available iron in the form of pulcherrimin. Growth rates for wild-type K. lactis in PA-treated medium were not significantly different from growth rates in untreated control medium (SI Appendix, Fig. S6). In PA-treated medium, wild-type, pul1Δ, and pul2Δ mutants grew normally, implying that PUL1 and PUL2 play a role specific to pulcherrimin biosynthesis and not reutilization (Fig. 2). Although the pul3Δ mutant grew normally in YPD liquid, it had a lower maximum cell density in SC than the wild-type strain, in line with the observations on solid medium (SI Appendix, Fig. S6). The pul3Δ strain grew negligibly compared with wild type in PA-treated medium (Student’s t test, P = 0.001; Fig. 2B), demonstrating that PUL3 is required for utilization of pulcherrimin-complexed iron.
Along with the role of PUL4 in pulcherrimin biosynthesis (Fig. 2D), the pul4Δ mutant also showed a significant lag during growth in PA-treated medium compared with wild-type cells (Student’s t test, P = 0.04). After the lag, the pul4Δ strain did not show a significant growth decrease compared with wild-type cells (Student’s t test, P = 0.15; Fig. 2B). These results suggest that PUL4 also positively regulates PUL3, but there are likely other ways for the cells to activate PUL3 expression in K. lactis (Fig. 2B).
S. cerevisiae and Some Other Yeast Species Have Pulcherrimin-Complexed Iron Utilization Genes.
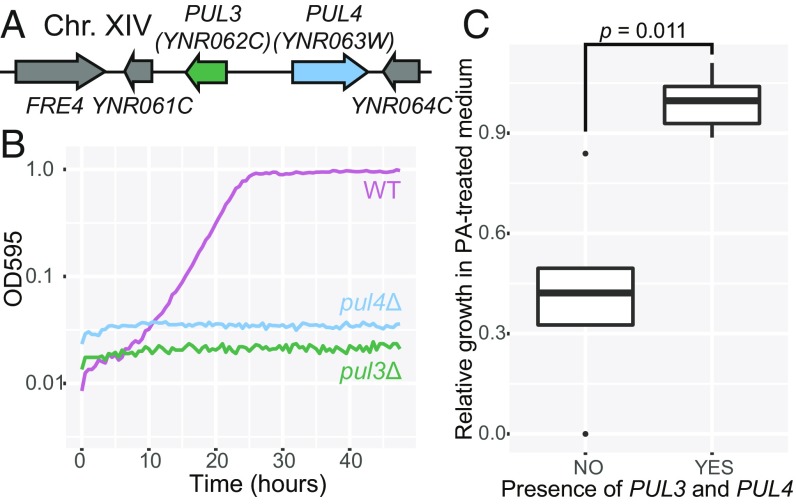
In addition to the complete PUL clusters present in all pulcherrimin-producing yeast species, we also found that 12 species contain partial PUL gene clusters composed of PUL3 and PUL4, or only PUL4 (Fig. 1). We hypothesized that organisms containing PUL3, all of which also contain PUL4, might be able to utilize exogeneous pulcherrimin, but not make their own. One such species is the model yeast S. cerevisiae, and its PUL3 and PUL4 homologs are the adjacent and previously uncharacterized genes YNR062C and YNR063W, respectively (Fig. 3A). To test whether these genes also play a role in pulcherrimin-complexed iron utilization in S. cerevisiae, we individually deleted YNR062C and YNR063W and grew both mutant strains in PA-treated media. Wild-type S. cerevisiae grew similarly in PA-treated and untreated growth media (SI Appendix, Fig. S7), but both the pul3Δ and pul4Δ mutant strains failed to grow in PA-treated medium (Fig. 3B). PUL4 has been shown to bind DNA in S. cerevisiae via protein-binding microarrays (30), and we therefore hypothesize, as in K. lactis, that PUL4 encodes a transcription factor that regulates expression of the putative transporter PUL3.
Fig. 3.
(A) Gene arrow diagram depicting the location and arrangement of the partial PUL gene cluster in S. cerevisiae. (B) Representative growth curves of pul mutants of S. cerevisiae in PA-treated medium, where cells must uptake pulcherrimin to grow. (C) PA-treated:untreated growth rate ratios for pairs of congeneric species containing or lacking PUL3 and PUL4 homologs. Boxplots contain the mean growth rate ratio from three biological replicates for each of the five species in each group. The “YES” group is composed of species containing PUL3 and PUL4, and the “NO” group is comprised of closely related species lacking one or both genes. P is the result of a paired Student’s t test.
To determine whether the presence or absence of PUL3 and PUL4 homologs was correlated with growth in PA-treated medium for other, less genetically tractable yeast species, we examined several monophyletic genera where species varied in the presence of PUL3 and PUL4. Five genera contain examples of species that contain PUL3 and PUL4 and species that lack both genes, or at least PUL3. For each genus, we grew one species containing PUL3 and PUL4 and one species lacking these genes in media with and without PA treatment. Species lacking PUL3 and PUL4 showed significantly slower growth in PA-treated medium, relative to growth in untreated medium, compared with closely related species containing the genes (Student’s t test, P = 0.011; Fig. 3C), suggesting that these genes enable pulcherrimin-mediated iron uptake across a broad range of budding yeast taxa.
The Rarity of Pulcherrimin Production Is the Result of Pervasive Gene Loss.
The sparse distribution of the PUL gene cluster within budding yeasts implied either that the cluster was ancestral to the subphylum Saccharomycotina and lost in most extant lineages or that it was gained via several horizontal gene transfer events. Horizontal gene transfer, including transfer of entire metabolic gene clusters, has been observed in both budding yeasts and fungi in general, but it is considered to be quite rare (31–34). We identified significant (e < 0.001) BLAST hits in the GenBank nonredundant (nr) protein sequence database for all four PUL genes in the Pezizomycotina and Basidiomycota, ranging between 22 and 30% identity at the amino acid level. In other fungi, the gene neighborhoods of PUL1 homologs were often predicted to encode cytochrome P450 oxidases, MFS transporters, and transcription factors, indicating that the PUL cluster may exist in other fungi (SI Appendix, Fig. S8). When we constructed maximum-likelihood phylogenies for each of the fungal PUL genes, all four gene trees supported a single Saccharomycotina clade with 100% bootstrap support, showing a shared origin for all Saccharomycotina PUL gene copies (SI Appendix, Figs. S9–S12). Thus, there is no evidence that individual species within Saccharomycotina obtained the PUL genes from a known source outside of the subphylum.
We also tested for HGT within the subphylum by using one-sided Kishino–Hasegawa (35) and approximately unbiased tests (36) to test for significant differences between likelihood values for the gene trees and corresponding trees constrained on the genome-wide species topology (Fig. 1). None of these tests identified significant differences between the gene trees and species tree (SI Appendix, Table S3), meaning the phylogenies are fully compatible with accepted yeast relationships and suggest no HGT within the subphylum. In the absence of evidence for HGT, and given the rarity of HGT in fungi, we infer that the PUL gene cluster was present in the common ancestor of Saccharomycotina. Its patchy distribution is best explained by parallel loss events during evolution, a feature commonly observed in secondary metabole gene clusters (33, 34), rather than rare acquisitions via HGT.
Discussion
The Power of Nontraditional Model Systems.
We have discovered and characterized a secondary metabolite gene cluster in budding yeasts. This gene cluster is responsible for the production of the siderophore pulcherrimin to capture iron from the environment. The rarity of pulcherrimin production among yeast species made the identification and characterization of the underlying genes dependent on knowledge of the genome sequences of diverse yeast species for comparative genomics and, fortunately, a genetically tractable system in K. lactis. Our study underscores the importance of efforts to sequence the genomes of diverse species (37–39), as well as to explore the biology of both established and nontraditional model systems. In fact, understanding the pulcherrimin system in K. lactis was essential to characterizing PUL3 and PUL4 (YNR062C and YNR063W) in S. cerevisiae, two genes that had previously evaded characterization by several high-throughput screens of gene function (40, 41).
Unlike the other three genes required for pulcherrimin biosynthesis, the predicted coding sequence of PUL1 lacks obvious sequence similarity to known families. The biochemical function of Pul1 remains unverified, as direct complementation of pul1Δ by expressing bacterial yvmC was unsuccessful and attempts at recombinant protein expression for biochemical characterization so far have been unsuccessful. The lack of complementation observed from the bacterial genes could result from lack of recognition of yeast tRNA-Leu, different cofactor requirements, or trivial differences that prevent protein expression, stability, or function. The possibility also remains that the biochemistry of the yeast pulcherrimin biosynthesis pathway differs from that of bacteria, and further work will be required to elucidate this.
We have also identified homologs of PUL1 in filamentous fungi, where their gene neighborhoods frequently resemble the PUL clusters characterized in this study and many other fungal secondary metabolite gene clusters. It is unknown whether these PUL1-containing clusters in diverse fungi produce pulcherrimin, but it seems likely that they are involved in secondary metabolite production. Searching for secondary metabolite gene clusters is often centered around identifying genes encoding NRPS or PKS using computational programs, such as antiSMASH (42) and SMURF (43), and genes encoding CDPS have not been comprehensively incorporated into these tools. Incorporating known CDPS and the PUL1 gene into future scans could help identify new secondary metabolite gene clusters (42, 44).
The Ecological Role of Pulcherrimin.
The rarity of pulcherrimin production among budding yeast species has also made understanding its ecological role difficult. However, by characterizing the genes in the PUL locus, we have identified several cryptic producers that have full PUL clusters but do not produce pulcherrimin in laboratory conditions, including some Kluyveromyces spp. and the emerging pathogen C. auris. Also, several species contain partial clusters that confer the ability to utilize pulcherrimin without producing it. Ultimately, we infer that the ancestor of all budding yeasts contained a complete PUL cluster, but the genes were lost in parallel in most lineages during yeast evolution.
Pulcherrimin has been previously described as mediating antagonistic interactions toward nonproducing yeast species, a property proposed to be linked to the sequestration of free iron from the environment (14, 15). However, without the ability to reutilize this pulcherrimin-bound iron, producers would inhibit their own growth, as observed in the K. lactis pul3Δ mutant. Therefore, in this study, we have expanded the ecological and functional role of pulcherrimin from solely an antagonistic compound to also include a role as an iron-binding siderophore. The pulcherrimin iron-chelation system is peculiar in budding yeasts; most characterized bacterial siderophores are used to scavenge for scarce iron in the environment (45), but many yeasts produce pulcherrimin even under high-iron conditions. Thus, pulcherrimin’s role in budding yeasts may not be in iron scavenging, but rather iron monopolizing.
Gene Loss and the Public Goods Problem.
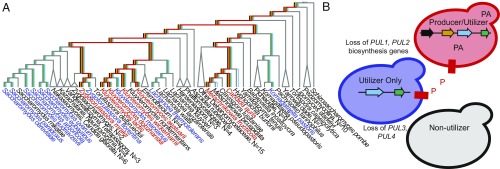
We have inferred that the PUL gene cluster was present in the common ancestor of Saccharomycotina, and the resulting sparse distribution of species containing the PUL genes, including the presence of partial PUL gene clusters, was the result of many gene loss events. We identified four different genotypes: (i) the presence of all four PUL genes, (ii) the presence of PUL3 and PUL4, (iii) the presence of PUL4 only, and (iv) the absence of PUL genes. We hypothesize that gene loss has occurred, and continues to occur, in order from 1 to 4 (Fig. 4). This ordered loss would be consistent with the observation of a growth defect in the K. lactis pul3Δ mutant. While many of these loss events likely occurred in the deep branches of the phylogeny, comparisons with close relatives suggest several recent loss events between species, which may or may not reflect entire species given the use of a single strain for these analyses. The PUL genes are localized to subtelomeres, which are often associated with gene content variation, as well as sources of evolutionary novelty (46, 47).
Fig. 4.
(A) Repeated loss of the PUL cluster in Saccharomycotina evolution. Branch colors indicate the hypothesized presence or absence of each PUL gene for the lineage, reconstructed using Dollo parsimony (i.e., only allowing gene loss because there is no evidence of gene gain by HGT). Black names indicate the absence of PUL genes; blue names indicate the presence of PUL3 and PUL4 but not PUL1 and PUL2; and red names indicate all four PUL genes are present. (B) Model for the order of PUL gene loss and how resulting phenotypes interact relevant to the public goods problem.
In characterizing the phenotypes resulting from gene loss events, we propose a simple model. The minimal cost of pulcherrimin production is the production of two leucine molecules and the likely expenditure of ATP to cyclize them. There is no apparent cost to utilizing pulcherrimin, except for production of the Pul3 putative transporter and Pul4 transcription factor. Therefore, we predict that cells that lose the biosynthesis genes avoid the costs of pulcherrimin production, but still reap the potential benefits of pulcherrimin production from neighboring cells. Similar public goods dynamics are common in microbes, in which cells producing a good are exploited by nonproducing cells that consume the good without the costs of production. Similar scenarios of gene loss in siderophore biosynthesis genes, with retention of the utilization genes, have been observed and described in this context in Vibrio and Pseudomonas spp. (48, 49). By combining the public goods problem with the patterns of gene loss without regain, we propose an evolutionary model where the initial loss of the biosynthesis genes allows a lineage to exploit closely related producers and rise in frequency. However, if they outcompete the producers too strongly and replace them, there is no more pulcherrimin to exploit, and the remaining utilization genes will eventually be lost. As more lineages lose pulcherrimin utilization, the rare lineages that have retained production may regain the upper hand. These dynamics may explain the many independent PUL gene cluster losses during budding yeast evolution.
A similar public goods scenario has been studied in laboratory experiments using S. cerevisiae, where suc2 mutants are able to exploit the invertase production of cells with functional SUC2 genes by utilizing the byproducts of sucrose digestion in mixed culture (50). However, a study of wild isolates did not identify any naturally occurring “cheaters” that lacked invertase production (51). Here, we showed that genetic variation in the PUL locus exists between species, but we have only investigated a single strain for each species. Further investigations of the biology of distinct strains within species will be required to test evolutionary hypotheses at more recent timescales. For example, a single strain of Kluyveromyces marxianus, UFS-Y2791 (52), contains homologs of PUL1 and PUL2, differing from the type strain, which lacks them.
The Power of Exploring Biodiversity.
The clustered arrangement of genes encoding secondary metabolite functions is common in other fungi, and the PULcherrimin gene cluster in budding yeasts is exceptional mainly because it is found in a group of fungi previously thought to lack such clusters. As the abundance of genome-scale data continues to increase in the subphylum Saccharomycotina (37), we predict that the genetic pathways for additional metabolites will be discovered and characterized in these diverse yeasts as it becomes clear that budding yeasts harbor many traits that the model yeast S. cerevisiae lacks. Exploring novel biology in nontraditional model organisms will continue to shed light on uncharacterized aspects of even well-studied model organisms, enhance our understanding of yeast ecology, and reveal both novel and general biology.
Materials and Methods
Detailed materials and methods can be found in the SI Appendix.
Strains, Media, and Oligonucleotides Used in This Study.
All genetic work was performed in a MATa ku80-Δ nonhomologous end joining-deficient background derived from K. lactis CBS2359 (27, 28) and in a MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 background of S. cerevisiae (BY4741) (53). Most yeast species used were obtained from the ARS Culture Collection [Northern Regional Research Laboratory (NRRL)]. Media formulations can be found in SI Appendix. A complete list of strains and oligonucleotides used can be found in SI Appendix, Tables S1 and S2. Genetic manipulations in K. lactis and S. cerevisiae were performed according to standard protocols.
Pulcherriminic Acid Extraction and Treatment.
Extraction of PA was performed similarly to previously described protocols (9) using a strain of Metschnikowia sp., yHQL305. This strain is an avid pulcherrimin producer that we isolated from Garry oak (Quercus garryana) leaves from the Standish–Hickey State Recreation Area in California using established protocols (54). Extraction details can be found in SI Appendix.
Growth Experiments and Analysis.
Growth experiments were performed in 96-well plates on a FLUOstar Omega plate reader (BMG Labtech). Experiments using only S. cerevisiae and K. lactis were performed at 30 °C, while experiments using several yeast species were performed at 22 °C to permit the growth of species that prefer cooler temperatures. Three biological replicates were performed of all growth experiments. Growth data were analyzed using the grofit package in R under the logistic model of growth (55); plots were generated with ggplot2 (56). Spline-fitted values were used when growth was negligible, causing the model to fail. Representative growth curves shown in Results are the biological replicate whose growth rate is closest to the mean growth rate for the set of replicates. Statistical comparisons of grofit parameters were performed using paired Student’s t tests with all pairs grown concurrently on the same plate using the same reagent batch.
Sequence Alignment, Phylogeny Construction, and Topology Testing.
Phylogenetic analyses were performed according to standard methods. Details can be found in SI Appendix, SI Methods.
Data Availability.
Genome assemblies for Z. mrakii, Z. florentina, Kluyveromyces nonfermentans, and Ko. pseudopastoris can be found under NCBI accession numbers PPHZ00000000, PPJY00000000, PPKX00000000, and QYLQ00000000.
Supplementary Material
Acknowledgments
We thank the University of Wisconsin Biotechnology Center DNA Sequencing Facility for providing Illumina and Sanger sequencing facilities and services, Russell Wrobel for helpful suggestions on oligonucleotide design, Gilles Fischer for providing Lachancea thermotolerans Y-8284, Kevin Myers and Audrey Gasch for providing S. cerevisiae strains, Jue Wang for providing Bacillus subtilis genomic DNA, Maitreya Dunham for providing the pIL68 plasmid, and William Vagt for collecting the sample from which yHQL305 was isolated. This material is partly based on work supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research under Award DE-SC0018409; work funded by the DOE Great Lakes Bioenergy Research Center under DOE BER Office of Science Grant DE-FC02-07ER64494; and National Science Foundation Grant DEB-1442148 (to C.T.H. and C.P.K.), DEB-1442113 (to A.R.), and DEB-1253634 (to C.T.H.). C.T.H. is a Pew Scholar in the Biomedical Sciences and Vilas Faculty Early Career Investigator, supported by the Pew Charitable Trusts and the Vilas Trust Estate, respectively. Q.K.L. was supported by a National Science Foundation Graduate Research Fellowship DGE-126259 and the National Institutes of Health Predoctoral Training Program for the University of Wisconsin–Madison Laboratory of Genetics from Grant 5T32GM007133. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture (USDA). USDA is an equal opportunity provider and employer.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Genome assemblies for Zygotorulaspora mrakii, Zygotorulaspora florentina, Kluyveromyces nonfermentans, and Komagataella pseudopastoris can be found under National Center for Biotechnology Information (NCBI) accession nos. PPHZ00000000, PPJY00000000, PPKX00000000, and QYLQ00000000.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806268115/-/DCSupplemental.
References
- 1.Charlesworth JC, Burns BP. Untapped resources: Biotechnological potential of peptides and secondary metabolites in Archaea. Archaea. 2015;2015:282035. doi: 10.1155/2015/282035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donadio S, Monciardini P, Sosio M. Polyketide synthases and nonribosomal peptide synthetases: The emerging view from bacterial genomics. Nat Prod Rep. 2007;24:1073–1109. doi: 10.1039/b514050c. [DOI] [PubMed] [Google Scholar]
- 3.Macheleidt J, et al. Regulation and role of fungal secondary metabolites. Annu Rev Genet. 2016;50:371–392. doi: 10.1146/annurev-genet-120215-035203. [DOI] [PubMed] [Google Scholar]
- 4.Keller NP, Turner G, Bennett JW. Fungal secondary metabolism–From biochemistry to genomics. Nat Rev Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 5.Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides. Annu Rev Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 6.Weissman KJ, Leadlay PF. Combinatorial biosynthesis of reduced polyketides. Nat Rev Microbiol. 2005;3:925–936. doi: 10.1038/nrmicro1287. [DOI] [PubMed] [Google Scholar]
- 7.Gondry M, et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat Chem Biol. 2009;5:414–420. doi: 10.1038/nchembio.175. [DOI] [PubMed] [Google Scholar]
- 8.Bushley KE, Turgeon BG. Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships. BMC Evol Biol. 2010;10:26. doi: 10.1186/1471-2148-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kluyver AJ, van der Walt JP, van Triet AJ. Pulcherrimin, the pigment of Candida pulcherrima. Proc Natl Acad Sci USA. 1953;39:583–593. doi: 10.1073/pnas.39.7.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurtzman C, Fell JW, Boekhout T. The Yeasts: A Taxonomic Study. Elsevier; London: 2011. [Google Scholar]
- 11.Lachance MA. Metschnikowia: Half tetrads, a regicide and the fountain of youth. Yeast. 2016;33:563–574. doi: 10.1002/yea.3208. [DOI] [PubMed] [Google Scholar]
- 12.Araujo FV, Hagler AN. Kluyveromyces aestuarii, a potential environmental quality indicator yeast for mangroves in the State of Rio de Janeiro, Brazil. Braz J Microbiol. 2011;42:954–958. doi: 10.1590/S1517-838220110003000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacDonald JC. Biosynthesis of pulcherriminic acid. Biochem J. 1965;96:533–538. doi: 10.1042/bj0960533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Türkel S, Ener B. Isolation and characterization of new Metschnikowia pulcherrima strains as producers of the antimicrobial pigment pulcherrimin. Z Naturforsch C. 2009;64:405–410. doi: 10.1515/znc-2009-5-618. [DOI] [PubMed] [Google Scholar]
- 15.Sipiczki M. Metschnikowia strains isolated from botrytized grapes antagonize fungal and bacterial growth by iron depletion. Appl Environ Microbiol. 2006;72:6716–6724. doi: 10.1128/AEM.01275-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oro L, Ciani M, Comitini F. Antimicrobial activity of Metschnikowia pulcherrima on wine yeasts. J Appl Microbiol. 2014;116:1209–1217. doi: 10.1111/jam.12446. [DOI] [PubMed] [Google Scholar]
- 17.Van Der Walt JP, Botha A, Eicker A. Ferrichrome production by Lipomycetaceae. Syst Appl Microbiol. 1990;13:131–135. [Google Scholar]
- 18.Bonnefond L, et al. Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog. Proc Natl Acad Sci USA. 2011;108:3912–3917. doi: 10.1073/pnas.1019480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Randazzo P, Aubert-Frambourg A, Guillot A, Auger S. The MarR-like protein PchR (YvmB) regulates expression of genes involved in pulcherriminic acid biosynthesis and in the initiation of sporulation in Bacillus subtilis. BMC Microbiol. 2016;16:190. doi: 10.1186/s12866-016-0807-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cryle MJ, Bell SG, Schlichting I. Structural and biochemical characterization of the cytochrome P450 CypX (CYP134A1) from Bacillus subtilis: A cyclo-L-leucyl-L-leucyl dipeptide oxidase. Biochemistry. 2010;49:7282–7296. doi: 10.1021/bi100910y. [DOI] [PubMed] [Google Scholar]
- 21.Gu B, He S, Yan X, Zhang L. Tentative biosynthetic pathways of some microbial diketopiperazines. Appl Microbiol Biotechnol. 2013;97:8439–8453. doi: 10.1007/s00253-013-5175-4. [DOI] [PubMed] [Google Scholar]
- 22.Shen XX, et al. Reconstructing the backbone of the Saccharomycotina yeast phylogeny using genome-scale data. G3 (Bethesda) 2016;6:3927–3939. doi: 10.1534/g3.116.034744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karst F, Lacroute F. Ertosterol biosynthesis in Saccharomyces cerevisiae: Mutants deficient in the early steps of the pathway. Mol Gen Genet. 1977;154:269–277. doi: 10.1007/BF00571282. [DOI] [PubMed] [Google Scholar]
- 24.Gardiner DM, Howlett BJ. Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbiol Lett. 2005;248:241–248. doi: 10.1016/j.femsle.2005.05.046. [DOI] [PubMed] [Google Scholar]
- 25.Gardiner DM, Cozijnsen AJ, Wilson LM, Pedras MSC, Howlett BJ. The sirodesmin biosynthetic gene cluster of the plant pathogenic fungus Leptosphaeria maculans. Mol Microbiol. 2004;53:1307–1318. doi: 10.1111/j.1365-2958.2004.04215.x. [DOI] [PubMed] [Google Scholar]
- 26.Capriotti A. Zygosaccharomyces mrakii nova species. Arch Mikrobiol. 1958;30:387–392. doi: 10.1007/BF00411232. [DOI] [PubMed] [Google Scholar]
- 27.Hittinger CT, Carroll SB. Gene duplication and the adaptive evolution of a classic genetic switch. Nature. 2007;449:677–681. doi: 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- 28.Kooistra R, Hooykaas PJJ, Steensma HY. Efficient gene targeting in Kluyveromyces lactis. Yeast. 2004;21:781–792. doi: 10.1002/yea.1131. [DOI] [PubMed] [Google Scholar]
- 29.Philpott CC. Iron uptake in fungi: A system for every source. Biochim Biophys Acta. 2006;1763:636–645. doi: 10.1016/j.bbamcr.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Zhu C, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fitzpatrick DA. Horizontal gene transfer in fungi. FEMS Microbiol Lett. 2012;329:1–8. doi: 10.1111/j.1574-6968.2011.02465.x. [DOI] [PubMed] [Google Scholar]
- 32.Novo M, et al. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc Natl Acad Sci USA. 2009;106:16333–16338. doi: 10.1073/pnas.0904673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wisecaver JH, Slot JC, Rokas A. The evolution of fungal metabolic pathways. PLoS Genet. 2014;10:e1004816. doi: 10.1371/journal.pgen.1004816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wisecaver JH, Rokas A. Fungal metabolic gene clusters-caravans traveling across genomes and environments. Front Microbiol. 2015;6:161. doi: 10.3389/fmicb.2015.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldman N, Anderson JP, Rodrigo AG. Likelihood-based tests of topologies in phylogenetics. Syst Biol. 2000;49:652–670. doi: 10.1080/106351500750049752. [DOI] [PubMed] [Google Scholar]
- 36.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 37.Hittinger CT, et al. Genomics and the making of yeast biodiversity. Curr Opin Genet Dev. 2015;35:100–109. doi: 10.1016/j.gde.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu D, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grigoriev IV, et al. Fueling the future with fungal genomics. Mycology. 2011;2:192–209. [Google Scholar]
- 40.Costanzo M, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353:aaf1420. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Usaj M, et al. TheCellMap.org: A web-accessible database for visualizing and mining the global yeast genetic interaction network. G3 (Bethesda) 2017;7:1539–1549. doi: 10.1534/g3.117.040220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blin K, et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017;45:W36–W41. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khaldi N, et al. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dejong CA, et al. Polyketide and nonribosomal peptide retro-biosynthesis and global gene cluster matching. Nat Chem Biol. 2016;12:1007–1014. doi: 10.1038/nchembio.2188. [DOI] [PubMed] [Google Scholar]
- 45.Neilands JB. Siderophores: Structure and function of microbial iron transport compounds. J Biol Chem. 1995;270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 46.Brown CA, Murray AW, Verstrepen KJ. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr Biol. 2010;20:895–903. doi: 10.1016/j.cub.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snoek T, Voordeckers K, Verstrepen KJ. Subtelomeres. Springer; Berlin: 2014. Subtelomeric regions promote evolutionary innovation of gene families in yeast. [Google Scholar]
- 48.Cordero OX, Ventouras LA, DeLong EF, Polz MF. Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc Natl Acad Sci USA. 2012;109:20059–20064. doi: 10.1073/pnas.1213344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Butaitė E, Baumgartner M, Wyder S, Kümmerli R. Siderophore cheating and cheating resistance shape competition for iron in soil and freshwater Pseudomonas communities. Nat Commun. 2017;8:414. doi: 10.1038/s41467-017-00509-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greig D, Travisano M. The Prisoner’s Dilemma and polymorphism in yeast SUC genes. Proc Biol Sci. 2004;271:S25–S26. doi: 10.1098/rsbl.2003.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bozdag GO, Greig D. The genetics of a putative social trait in natural populations of yeast. Mol Ecol. 2014;23:5061–5071. doi: 10.1111/mec.12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schabort TW, Letebele PK, Steyn L, Kilian SG, du Preez JC. Differential RNA-seq, multi-network analysis and metabolic regulation analysis of Kluyveromyces marxianus reveals a compartmentalised response to xylose. PLoS One. 2016;11:e0156242. doi: 10.1371/journal.pone.0156242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 54.Sylvester K, et al. Temperature and host preferences drive the diversification of Saccharomyces and other yeasts: A survey and the discovery of eight new yeast species. FEMS Yeast Res. 2015;15:fov002. doi: 10.1093/femsyr/fov002. [DOI] [PubMed] [Google Scholar]
- 55.Kahm M, et al. grofit: Fitting biological growth curves with R. J Stat Softw. 2010;33:1–21. [Google Scholar]
- 56.Wickham H. Ggplot2: Elegant Graphics for Data Analysis. Springer; New York: 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome assemblies for Z. mrakii, Z. florentina, Kluyveromyces nonfermentans, and Ko. pseudopastoris can be found under NCBI accession numbers PPHZ00000000, PPJY00000000, PPKX00000000, and QYLQ00000000.