Significance
Cell adhesion molecules (CAMs) provide highly specific cell-surface recognition signals by which developing neurons interact with specific partners. However, these CAMs are common between neurons of the same type, and without a mechanism of self-avoidance or of homotypic avoidance, developing neurons will excessively adhere with themselves. This self-avoidance can be promoted by extreme molecular diversity, such as that of Dscam1 (Down syndrome cell adhesion molecule 1) in flies, which gives neurons distinct barcodes. Mouse Dscam, on the other hand, promotes self-avoidance without molecular diversity. Here, we provide evidence that DSCAM can functionally interact with other CAMs, called cadherins and protocadherins, to act like a general “nonstick” signal. Through this “adhesive masking” mechanism, DSCAM allows neurons to develop their appropriate shapes, positions, and connections.
Keywords: DSCAMs, dendrite fasciculation, autism, cell identity, cadherins
Abstract
During neural development, self-avoidance ensures that a neuron’s processes arborize to evenly fill a particular spatial domain. At the individual cell level, self-avoidance is promoted by genes encoding cell-surface molecules capable of generating thousands of diverse isoforms, such as Dscam1 (Down syndrome cell adhesion molecule 1) in Drosophila. Isoform choice differs between neighboring cells, allowing neurons to distinguish “self” from “nonself”. In the mouse retina, Dscam promotes self-avoidance at the level of cell types, but without extreme isoform diversity. Therefore, we hypothesize that DSCAM is a general self-avoidance cue that “masks” other cell type-specific adhesion systems to prevent overly exuberant adhesion. Here, we provide in vivo and in vitro evidence that DSCAM masks the functions of members of the cadherin superfamily, supporting this hypothesis. Thus, unlike the isoform-rich molecules tasked with self-avoidance at the individual cell level, here the diversity resides on the adhesive side, positioning DSCAM as a generalized modulator of cell adhesion during neural development.
The specification of cell body position, dendritic arbor morphology, axonal targeting, and synaptic connectivity requires a complex system of recognition steps. To mediate these recognition events, a given neuronal cell type expresses multiple cell adhesion molecules (CAMs), many of which are distinct from those expressed by neighboring cell types (1). Each CAM displays a clear ligand preference, be it homophilic or heterophilic, for molecules in the extracellular matrix or at the surface of other cells, providing each cell type with a unique repertoire of interactions with the extracellular environment. This array of adhesive interactions is balanced by self-avoidance, which prevents close association as developing neurites extend to sample available interactions.
Self-avoidance occurs on at least two levels: Sister neurites from the same cell (i.e., “self”) recognize and avoid each other to promote appropriate arbor formation; and cells of the same subtype (i.e., “homotypic”) space themselves nonrandomly relative to each other (2). This relative spacing can be completely nonoverlapping (called “tiling”) or can involve extensive overlap of neighboring neurites with “mosaic” spacing of cell bodies, as is the case in the vertebrate retina (3–5).
One strategy to allow self-avoidance at the individual cell level with extensive overlap between neighboring neurons is to use diverse molecular signals to distinguish self from nonself. This is typified by Dscam1 (Down syndrome cell adhesion molecule 1) in Drosophila (6). Dscam1 encodes a member of the Ig superfamily of CAMs capable of generating 19,008 distinct, homophilic recognition molecules through alternative exon usage (7). Each neuron expresses a handful of isoforms, allowing neurites to recognize and repel other self neurites while still contacting and interacting with nonself neurites (8–11). In some mammalian cell types, such as starburst amacrine cells (SACs) or cerebellar Purkinje cells, γ-protocadherins (γ-Pcdhs; from the Pcdhg gene) serve analogous functions by generating diverse protein multimers with homophilic recognition specificity (12–18). For both Drosophila Dscam1 and mammalian Pcdhg, the molecular diversity is essential for normal self/nonself discrimination needed for the proper self-avoidance of an individual neuron (8, 14).
However, not all self-avoidance requires extreme recognition diversity: Semaphorin6A and PlexinA4 regulate self-avoidance in horizontal cells in the mammalian retina (19), and mammalian Dscams (Dscam and Dscaml1) promote self-avoidance at the individual cell level and between homotypic neurons without generating multiple isoforms (20, 21). Dscam and Dscaml1 are expressed in nonoverlapping cell types in the retina. In mice mutant for either Dscam, neurons lose their normal mosaic spacing and uniform dendritic coverage and, instead, cluster, with their processes forming fascicles with neighboring homotypic neurons (20, 21). Drosophila Dscam1 functions through direct repulsion (9–11); however, this is likely not the case for mammalian Dscams. Cell types do not tile into discrete territories, and while nearly all retinal ganglion cells (RGCs) express the single Dscam isoform, the position of one RGC type has no relationship to the position of other RGC types (21). Thus, Dscam-expressing cells are “indifferent” to one another, rather than actively repellent.
Furthermore, we have shown that different cell types have differing dependence on the Dscams’ PDZ-interacting C termini, indicating that individual cell types require distinct intracellular interactions for Dscam-mediated self-avoidance (22). This, together with the cell type-specific nature of clustering and fasciculation in mutant retinas (20, 21), leads us to hypothesize that Dscams serve as general “nonstick” signals that “mask” multiple cell type-specific adhesion mechanisms to promote self-avoidance by active indifference rather than repulsion. Here, we focus just on Dscam (not Dscaml1) to test this hypothesis. Through a series of double mutants of Dscam with members of the cadherin superfamily, we show that reducing adhesion is able to rescue neurite fasciculation in Dscam−/−. Conversely, driving ectopic expression of an individual cadherin in the absence of DSCAM causes a random collection of cells to fasciculate with each other as if they were homotypic. Lastly, we show that trans DSCAM interactions acutely attenuate adhesive responses.
Results
Classical Cadherins Are Candidates to Be Masked by DSCAM.
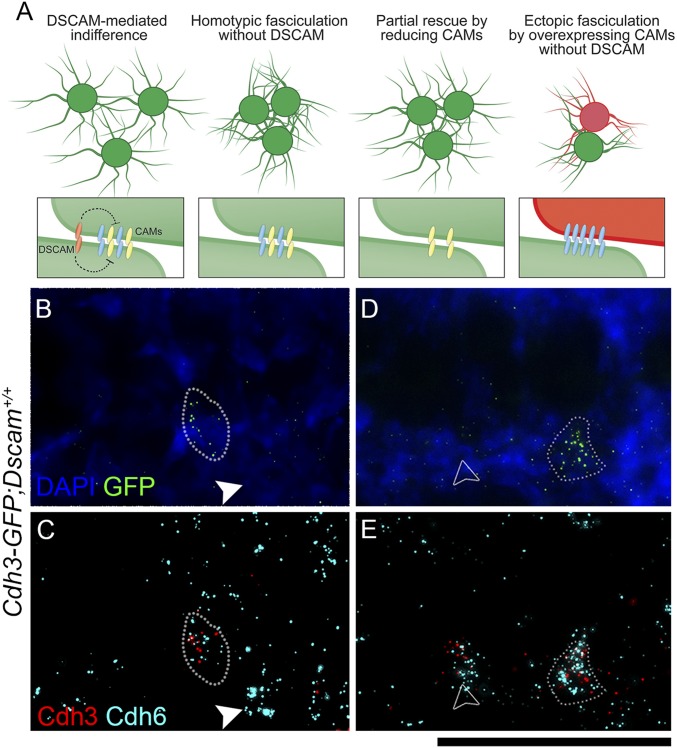
We hypothesize that DSCAM masks cell type-specific adhesion systems to balance adhesive forces during development in the mouse retina. This offers an explanation for the homotypic nature of clustering and fasciculation in Dscam−/− mutants. Our hypothesis predicts that if unopposed adhesion drives this fasciculation, then reducing the complement of CAMs in a cell type would partially rescue the fasciculation and clustering phenotype. Furthermore, this also predicts that overexpressing an ectopic CAM in a random subset of neurons would cause these cells to behave as if they were homotypic and fasciculate with each other in the absence of DSCAM (Fig. 1A).
Fig. 1.
Cadherin expression in Cdh3-GFP-RGCs. (A) We propose that DSCAM masks inappropriate adhesion, allowing indifference between homotypic neurites. Without DSCAM, unmasked adhesion drives clustering and fasciculation, predicting that reducing adhesion will partially rescue fasciculation, and that ectopic overexpressing of a CAM will make random cells fasciculate together as if they were homotypic. (B–E) In situ hybridization (RNAscope), with probes against GFP (green), Cdh3 (red), and Cdh6 (cyan), performed on P0 Cdh3-GFP retinas. Forty-eight of 50 GFP-positive cells were positive for both Cdh3 and Cdh6 (dotted lines). Many GFP-negative cells were Cdh6 positive (filled arrowheads in B and C), while occasional cells that were GFP-negative but Cdh3/Cdh6 double positive were also observed (open arrowheads in D and E, n = 6 retinas). (Scale bar: 50 μm.)
To begin testing this hypothesis, we focused on the RGCs labeled in Cdh3-GFP BAC transgenic mice (23, 24). We chose these cells because they require Dscam for self-avoidance (22) and are reported to express classical cadherins, including Cdh3 and Cdh6 (24). Cdh3-GFP-RGCs comprise more than one cell type (25). To ask how many of these cells express Cdh3 and Cdh6, and to verify that they are expressed during the time frame when dendrite fasciculation occurs in Dscam mutants, we performed RNAscope in situ hybridization on P0 Cdh3-GFP retinas (Fig. 1 B–E). Of 50 GFP-positive cells in the retinal ganglion layer, 48 were positive for both Cdh3 and Cdh6 and two were positive only for Cdh6. Cdh6 was also expressed in GFP-negative cells within the retinal ganglion layer and the inner nuclear layer, consistent with previous reports (Fig. 1 B and C, filled arrowheads) (26). There was also a smaller population of Cdh3/Cdh6 double-positive cells that were not clearly GFP positive (Fig. 1 D and E, open arrowheads). Thus, while multiple RGC subtypes are labeled in Cdh3-GFP retinas (25), the majority of GFP-positive cells express Cdh6 and Cdh3.
Classical Cadherin-Mediated Adhesion Contributes to Dendrite Fasciculation in Cdh3-GFP-RGCs in Dscam−/− Mutants.
Genetic analysis of double mutants has proved useful for parsing opposing adhesive and repulsive signals in neural development (27). Therefore, we tested whether reducing adhesion in Cdh3-GFP-RGCs by eliminating cadherin-3 or cadherin-6 would partially rescue the fasciculation observed in Dscam−/− null mice, directly testing our masking hypothesis (Fig. 1A). We generated double mutants for Dscam with Cdh3 or Cdh6 in the presence of the Cdh3-GFP transgene. To more broadly disrupt cadherin-mediated adhesion, we also used cdf mice (cerebellar deficient folia), a mouse line in which a large deletion removed Ctnna2, the gene encoding αN-catenin (28), which participates in the cytoplasmic complex important for classical cadherin function in neurons. We did not recover Dscam−/−;Ctnna2CDF/CDF double homozygous mutants, so we analyzed Dscam−/−;Ctnna2CDF/+ heterozygous mice. Dscam;Cadherin double mutants had reduced viability by 1 wk of age, and we have previously shown that RGC fasciculation begins embryonically (29). Therefore, we analyzed mutants at postnatal day 4. Retinas from these three groups of double-mutant mice, along with wild-type and single-mutant controls, were immunostained in whole mount for GFP and imaged en face by confocal microscopy. We analyzed cell number and cell body spacing and did not find any effect of the individual cadherin mutations at this age (SI Appendix, Fig. S1).
To analyze dendrite fasciculation independent of cell body spacing, we made confocal projections through the inner plexiform layer of the retina containing the dendritic arbors of these cells, but excluding cell bodies and axons (Fig. 2 A–H). We compared fasciculation in these images using two independent techniques. First, we generated a fasciculation score (FS) using the metric space technique (MST). The MST is an image analysis methodology developed and used in astrophysics (30–36). In the MST, information is extracted from the images in the form of output functions, in which a one-dimensional function represents a profile of some physically meaningful quantity. To quantify the differences between images, a metric is defined and used to give information on “how far” the images are from each other by calculating the metric distance between the images’ output functions. We explored many possible uses and combinations of output functions to best differentiate between Cdh3-GFP dendrite fasciculation in Dscam+/+ and Dscam−/− retinas. The best discrimination was found when the distribution of density and the distribution of filament indices were combined (30–35). The distribution of density is reduced in images from Dscam−/− mutants, because as dendrites fasciculate, they leave more unoccupied space (SI Appendix, Fig. S2A). Conversely, images from Dscam−/− mutants have an increased distribution of filament indices. This is a measure of continuous, elongated image features, which become more prominent as dendrites fasciculate (SI Appendix, Fig. S2B). These measures were calculated over multiple thresholds and then combined by division to yield the FS for each image. A higher FS indicates a higher degree of fasciculation (SI Appendix, Fig. S2C). A detailed description is in SI Appendix, Supplementary Methods.
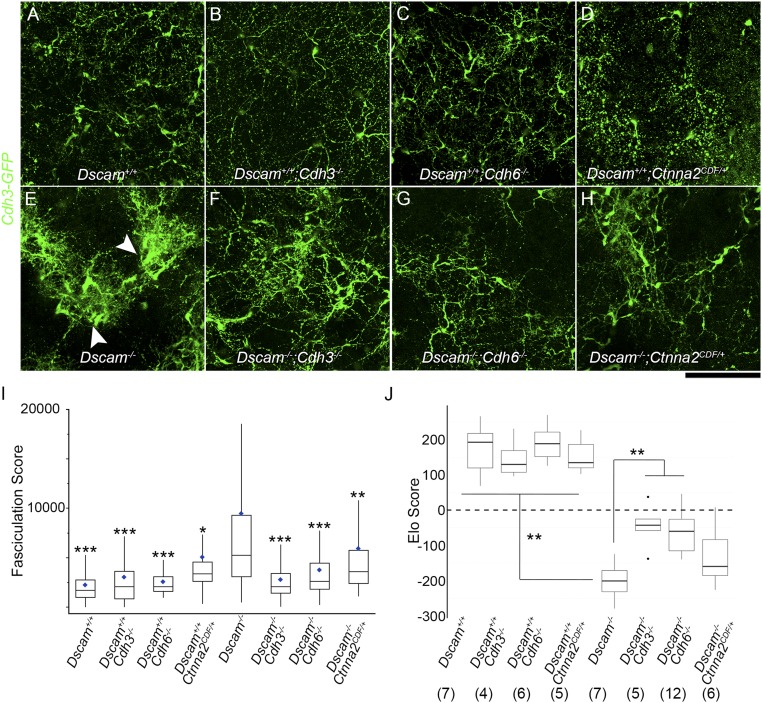
Fig. 2.
Cadherin-mediated adhesion contributes to Cdh3-GFP-RGC fasciculation. Confocal image projections through Cdh3-GFP-RGC dendrites in whole-mount retinas from wild-type (A), Cdh3−/− (B), Cdh6−/− (C), and Ctnna2CDF/+ (D) mice and from all genotypes in combination with Dscam−/− (E–H) demonstrate that dendrite fascicles are less severe in double mutants than in Dscam−/− alone (arrowheads). Images were quantified using the FS (I) and the Elo score (J). n = 4 to 12 retinas per genotype (actual n values are noted in J) over one to four microscope fields of view (median, 3 fields per retina). Box plots represent the median, first and third quartiles, range, and outliers. Blue diamonds in I represent the means. *P < 0.05, **P < 0.01, and ***P < 0.001 by pairwise Wilcoxon rank sum test compared with Dscam−/−. (Scale bar: 100 μm.)
While the single cadherin mutants did not differ from the control, FSs for Dscam−/− images were significantly higher than any other genotype, including Dscam−/−;Cdh3−/− (P = 10−11), Dscam−/−;Cdh6−/− (P = 10−9), and Dscam−/−;Ctnna2CDF/+ (P = 0.006), indicating that the cadherin mutations were able to partially rescue the fasciculation in Dscam−/− retinas (Fig. 2I). Furthermore, when we defined a threshold FS at two SDs above the mean in the distribution of Dscam+/+ images, a significantly higher proportion of Dscam−/− images exceeded this threshold compared with images from any other genotype (SI Appendix, Fig. S1K).
As an independent verification of this method, we also used a qualitative scoring system dubbed Image Echelon. This system is based on iterative, head-to-head, forced-choice comparisons and an Elo algorithm, which uses these win–loss matchups to efficiently sort the images into groups (Elo score; SI Appendix, Supplementary Methods and Fig. S2 D and E). Importantly, the rankings of images of different genotypes from the Elo analysis largely agreed with the FS (SI Appendix, Fig. S2F). Retinas lacking Cdh3 alone or Cdh6 alone or that were heterozygous for Ctnna2 were indistinguishable from wild-type, and all were significantly better than Dscam−/−. In double-mutant combinations, Dscam−/−;Cdh3−/− images and Dscam−/−;Cdh6−/− images showed significantly less fasciculation than Dscam−/− images (P = 0.018 and P = 0.008 respectively, Fig. 2J). Dscam−/−;Ctnna2CDF/+ images trended toward rescue, but the difference from Dscam−/− did not reach statistical significance. These results show that cadherins contribute to the excessive adhesion and fasciculation between Cdh3-GFP RGCs in Dscam−/− mice, consistent with our hypothesis that Dscam masks such cell type-specific adhesion mechanisms.
Cadherin-3 Can Drive Neurite Fasciculation in the Absence of DSCAM.
Our masking hypothesis also predicts that ectopic overexpression of a CAM in Dscam mutants will drive neurons to fasciculate with each other, even if they are not homotypic (Fig. 1A). To test this, we needed a method to overexpress Cdh3 in multiple cell types that require DSCAM for self-avoidance. We chose the in vivo electroporation of Cre-dependent Cdh3 expression constructs in Vgat-Cre mice (37). In the retinas of these mice, Cre is expressed in GABAergic horizontal cells and amacrine cells. There are at least 12 types of GABAergic amacrine cells (38), many of which express Dscam (39). Horizontal cells do not express Dscam and are not affected in mutant retinas, but are easy to exclude from our analyses because of their laminar position and because they are inefficiently transduced by electroporation postnatally (39, 40). We chose to focus on Cdh3 because, in our RNAscope results, we observed more limited expression of Cdh3 than of Cdh6 in the inner nuclear layer (Fig. 1 B–E). To verify that Cre is expressed early enough developmentally to recombine a Dscam conditional allele (DscamF) (29) and induce clustering and fasciculation, we analyzed two amacrine cell types in Vgat-Cre:DscamF/F retinas (hereafter DscamcKO/cKO). Both tyrosine hydroxylase-positive (TH+) and brain NOS+ amacrine cells showed homotypic clustering and fasciculation in DscamcKO/cKO mutants (SI Appendix, Fig. S3 A and B), as had been previously described for these cell types (20).
We electroporated Vgat-Cre mice at P0 with Cre-dependent DsRed (pCALNL-DsRed) (41) and Cdh3 and, after 2 wk, observed coexpression of DsRed and cadherin-3 (identified with a Myc epitope tag) in neurons morphologically consistent with amacrine cells (SI Appendix, Fig. S3 C and D). When DsRed alone was electroporated into mice wild-type for Dscam (Vgat-Cre positive), labeling was consistent with a stochastic subset of amacrine cells (Fig. 3A). Likewise, when DsRed was introduced into DscamcKO/cKO retinas, labeled cells were distributed across the electroporated area (Fig. 3B). Importantly, neurons were rarely seen fasciculated with each other, as we saw for homotypic cells (SI Appendix, Fig. S3), indicating that multiple types of GABAergic cells were labeled. When we introduced DsRed with Cdh3 into retinas wild-type for Dscam, there was no appreciable increase in fasciculation (Fig. 3C). However, when we electroporated the same DsRed and Cdh3 constructs into DscamcKO/cKO mutants, neurites formed tight fascicles with each other, behaving as if they were homotypic (Fig. 3D). These images were compared using Image Echelon to score fasciculation between neurons blind to genotype or condition. DscamcKO/cKO retinas electroporated with DsRed and Cdh3 were significantly more fasciculated than the other three conditions (Fig. 3E).
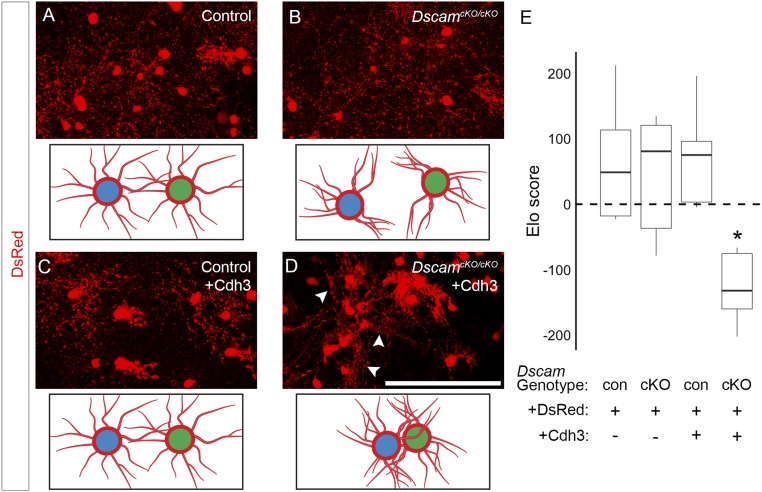
Fig. 3.
Cdh3 overexpression can drive fasciculation between nonhomotypic cells. Retinas of Vgat-Cre mice were electroporated in vivo at P0 with the Cre-inducible expression construct pCALNL-DsRed, alone or along with pCALNL-Cdh3. After 2 wk, these were fixed, stained for DsRed, and imaged en face. (A and B) Mice wild-type for Dscam (A) and DscamcKO/cKO mutants with conditional mutation in GABAergic amacrine cells (B) were electroporated with DsRed alone (Upper). There was no appreciable fasciculation (depicted in Lower) between electroporated cells. (C) Likewise, when retinas wild-type for Dscam were coelectroporated with DsRed and Cdh3 (Upper), no fasciculation (depicted in Lower) was observed. (D) In contrast, when DsRed and Cdh3 were together introduced into DscamcKO/cKO mutant retinas (Upper), significant fasciculation (depicted in Lower) was observed (arrowheads). (E) Images were compared for fasciculation using the Elo score. P = 0.025 by pairwise Wilcoxon rank sum test compared with each other condition. n = 6 mice per condition. (Scale bar: 100 μm.)
DSCAM Masks a Classical Cadherin Adhesive Response.
Our in vivo loss-of-function and overexpression experiments demonstrate the balance between cadherin-mediated adhesion and DSCAM-mediated self-avoidance. To test whether DSCAM can indeed prevent the accumulation of cadherins, we developed an in vitro assay using cultured neurons in which these interactions could be manipulated. The ectodomain of a homophilic CAM, presented on a bead to a neuron, will induce clustering of that CAM in the neuron at the point of contact (42). Using cadherin-3 as an example, we found that trans DSCAM interactions could prevent this clustering. Neurons from the cerebral cortex of wild-type or Dscam−/− mice were transfected with constructs expressing FLAG-tagged cadherin-3, and after 8 d in vitro, were incubated for 1 h with beads cocoated with cadherin-3 and DSCAM ectodomains (Fig. 4 and SI Appendix, Fig. S4). Beads cocoated with DSCAM and cadherin-3 did not induce accumulation of cadherin-3 (FLAG) at the point of contact with wild-type neurons, consistent with DSCAM masking cadherin-3 homophilic recognition (Fig. 4 A and C). In contrast, when the same beads were presented to Dscam-deficient neurons, cadherin-3 aggregated at the point of contact (Fig. 4 B and C). Similarly, cadherin-3 accumulated at the point of contact when DSCAM was not on the bead or when DSCAM was neither on the bead nor in the neuron (Fig. 4 D–F). No cadherin-3 clustering was observed when beads were coated with DSCAM only (SI Appendix, Fig. S4B). Thus, cadherin-3–mediated clustering was masked only when DSCAM was present on the bead as well as in the neuron, indicating that trans DSCAM interactions are needed to prevent cadherin-3 accumulation in the neuron at the point of contact with the bead.
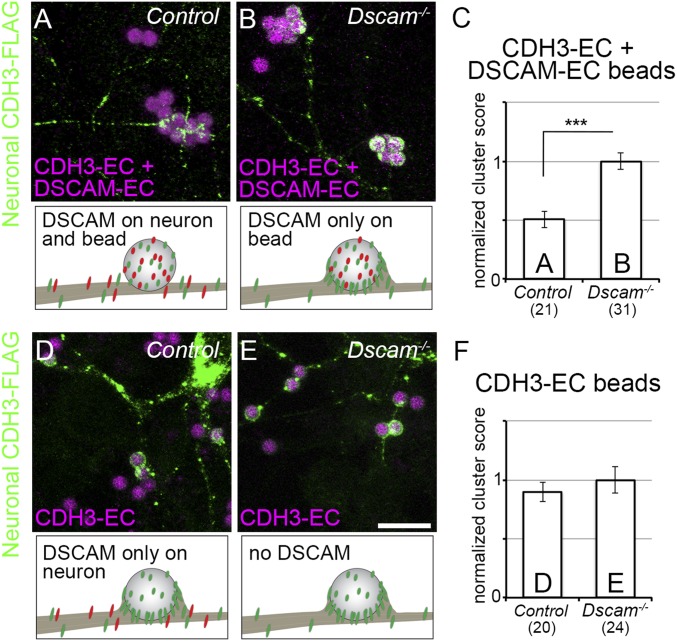
Fig. 4.
Trans DSCAM interactions mask the CDH3 adhesive response. Cortical neurons from wild-type (A and D) and Dscam−/− (B and E) mice were transfected with constructs encoding CDH3 with a C-terminal FLAG tag (green) and then incubated for 1 h with beads coated with CDH3 and DSCAM ectodomains (A and B, magenta) or CDH3-EC alone (D and E, magenta). The accumulation of CDH3-FLAG at sites of contact between beads and neurons was quantified (C and F). In all four conditions, CDH3 was present both in the neuron and on the bead. While neuronal CDH3-FLAG was largely indifferent to the beads when DSCAM was both in the neuron and on the bead (A), CDH3 accumulated at these sites when DSCAM was present only on the bead (B), only in the neuron (D), or completely absent (E), demonstrating that trans DSCAM interactions masked this accumulation. Means ± SEM are presented in C and F. n = 20 to 31 neurons per condition, from cultures separately prepared from three different mice per genotype (actual n values are noted in C and F). ***P < 0.001 by two-tailed Student’s t test. (Scale bar: 10 μm.)
Fasciculation Is Not Rescued by Reducing NRCAM-Mediated Adhesion.
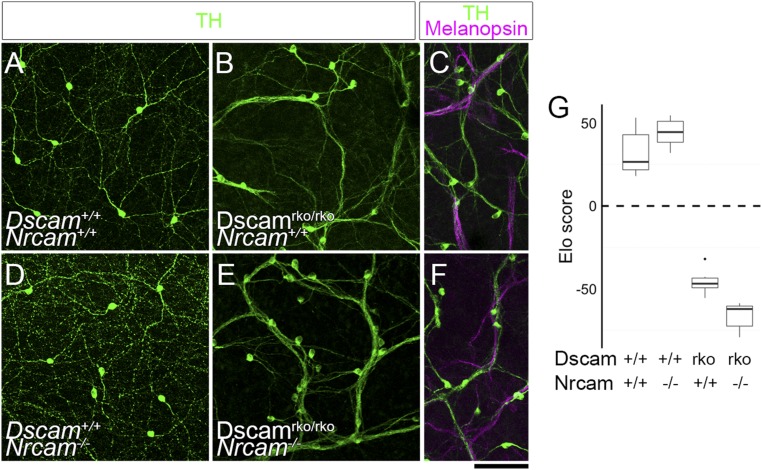
One possible interpretation of these data is that any reduction in cell adhesion will rescue fasciculation. To address this, we next focused on NRCAM (neuronal cell adhesion molecule), an Ig superfamily CAM with a canonical PDZ-interacting domain that mediates heterophilic adhesion with a variety of ligands (e.g., contactins, neurofascin, and L1) in addition to homophilic adhesion (43). Nrcam expression is enriched in dopaminergic amacrine (DA) cells and intrinsically photosensitive RGCs (ipRGCs) (SI Appendix, Fig. S5). Interestingly, DA cells require DSCAM–PDZ interactions for self-avoidance and to prevent cofasciculation with ipRGCs (22), making NRCAM a plausible target for DSCAM-mediated masking in DA cells, although the loss of Nrcam alone does not alter cell number, spacing, or dendrite arborization in these cells (SI Appendix, Fig. S5). We crossed Nrcam null mutant mice (44) to conditional floxed Dscam mutants, with recombination restricted to the retina by Pax6α-Cre (Dscamrko/rko; rko for retinal knockout), and analyzed DA cell spacing and neurite fasciculation at P14. We did not find any reduction in fasciculation in Dscamrko/rko;Nrcam−/− mutants compared with Dscamrko/rko mutants alone (Fig. 5 A, B, D, E, and G), nor did we find reduced cofasciculation between DA cells and ipRGCs, which also express Nrcam (Fig. 5 C and F). NRCAM may not significantly contribute to the fasciculation and clustering of DA cells in Dscam mutants, or it may be redundant with other CAMs, but this result shows that not every reduction in the cellular complement of CAMs is sufficient to mitigate clustering and fasciculation in the absence of DSCAM.
Fig. 5.
Removing NRCAM-mediated adhesion is not sufficient to reduce DA cell fasciculation. DA cells (TH+, green) were imaged in whole-mount retinas from wild-type (A), Dscamrko/rko (B), Nrcam−/− (D), and Dscamrko/rko;Nrcam−/− (E) mice. (G) Loss of NRCAM-mediated adhesion did not affect fasciculation between homotypic DA cell neurites (C and F), nor did it reduce cofasciculation between DA cells and ipRGC dendrites. n = 6 retinas per genotype over two to four microscope fields of view (median, 3 fields per retina). Box plots represent the median, first and third quartiles, range, and outliers. (Scale bar: 100 μm.)
DSCAM Masks γ-Pcdh–Mediated Adhesion.
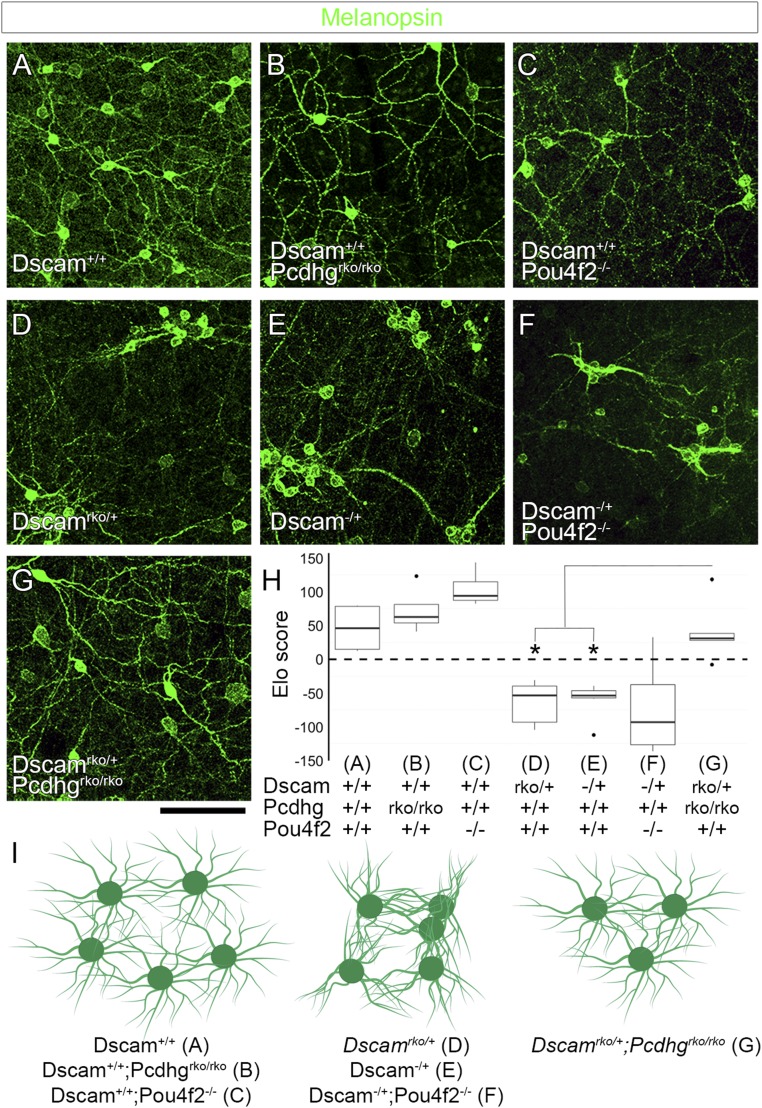
The functions of Dscams and γ-Pcdhs vary with their cell-type context. γ-Pcdhs promote self/nonself discrimination and self-avoidance in SACs (13, 14), but there are no obvious self-avoidance defects in other retinal cell types in Pcdhg mutants, including ipRGCs (16, 45), and other neurons in the central nervous system (CNS) have phenotypes more consistent with adhesive functions (46–48). Therefore, we asked whether DSCAM is masking γ-Pcdh–mediated adhesion in ipRGCs. In the wild-type retina, ipRGCs normally have extensive dendritic overlap with their neighbors (Fig. 6 and SI Appendix, Fig. S6) but undergo significant cell death in the absence of Pcdhg (Pcdhgrko/rko, Fig. 6B) (45). This reduction in cell number was similar to that observed in Pou4f2−/− mutants, a gene essential for RGC differentiation, allowing its use as a control for changes in cell density (Fig. 6C). In Dscam null retinas, ipRGCs form very tight clusters and fascicles (21, 22), and even heterozygotes have a significant, although less severe, phenotype (Fig. 6 D and E). We focused on this heterozygous phenotype, as any partial rescue was predicted to be more clearly discerned than in the background of extreme fasciculation and clumping seen in homozygous mutants (21, 22, 49). Importantly, in Dscam−/+;Pou4f2−/− double mutants, clustering and fasciculation was not reduced compared with Dscam−/+ mutants alone (Fig. 6F), indicating that reduced cell number is not sufficient to rescue these self-avoidance defects. To see whether reducing γ-Pcdh–mediated adhesion could rescue, we crossed Pcdhg floxed conditional mutants with Dscam conditional mutants under the control of Pax6α-Cre to generate Dscamrko/+;Pcdhgrko/rko mutants (Fig. 6G). We found that there was, indeed, less clustering and fasciculation at P14 in these double mutants than in Dscamrko/+ alone (Fig. 6H). Thus, the excessive adhesion of ipRGCs seen in Dscam mutants is partially alleviated by deletion of Pcdhg, independent of changes in cell number.
Fig. 6.
γ-Pcdh–mediated adhesion contributes to ipRGC fasciculation. (A) Confocal images from whole-mount retinas stained for melanopsin to label ipRGCs show normal spacing and dendritic coverage in wild-type animals. (B and C) Retina-specific deletion of Pcdhg or deletion of Pou4f2 reduces ipRGC cell number. (D and E) Retinas heterozygous for Dscam [Dscamrko/+ (D) or Dscam−/+ (E)] show significant ipRGC clustering and fasciculation. (F) This is not rescued by reducing cell number in Dscam−/+;Pou4f2−/− double mutants. (G) However, this loss of self-avoidance is rescued by reducing γ-Pcdh–mediated adhesion in Dscamrko/+;Pcdhgrko/rko double mutants. Image quantification is presented in H. * in H denotes P < 0.05 by pairwise Wilcoxon rank sum test compared to Dscamrko/+;Pcdhgrko/rko. These results are represented in I: Fasciculation is not observed in controls, Pcdhg mutants, or Pou4f2 mutants, although the latter two reduce cell number. Fasciculation is observed in Dscamrko/+, Dscam−/+, and Dscam−/+;Pou4f2−/−, despite changes in cell number, but these are rescued in Dscamrko/+;Pcdhgrko/rko double mutants. n = 6 retinas per genotype over two microscope fields of view per retina. Images of whole-retina quadrants were used for quantification (SI Appendix, Fig. S6). Box plots represent the median, first and third quartiles, range, and outliers. (Scale bar: 100 μm.)
Discussion
Here, we provide evidence that DSCAM’s self-avoidance function in the mouse retina is to counteract cell type-specific adhesion mechanisms. We term this function masking. In this hypothesis, homotypic neurons share a repertoire of cell adhesion molecules through which they interact with their environment to assume their proper position, find the targets for their axons and dendrites, and form synaptic connections with appropriate partners (26, 50). Many of these CAMs are homophilic and will cause homotypic eurons to adhere to each other, resulting in fasciculated dendrites and clustered cell bodies without a mechanism to counteract, or mask, this adhesion. DSCAM provides this balancing force by locally preventing adhesion where it is not desirable. We have presented genetic and morphological data indicating that Dscam masks adhesion mediated by classical cadherins and protocadherins. Homophilic DSCAM interactions in trans prevent their adhesion, but loss of Dscam allows these adhesion systems to function unopposed, resulting in cell type-specific clustering and fasciculation. This study provides direct evidence in support of our adhesive masking hypothesis.
Self-avoidance in the mouse retina occurs on at least two levels: between sister neurites of a single cell, and between neurites of homotypic cells. DSCAM is required for self-avoidance at both levels; in DA cells (a more sparse population), individual self-crossings can be observed before clustering and fasciculation between neighboring neurons (20). It may be formally possible for a neuron to have deficient individual self-avoidance with normal homotypic avoidance (or vice versa). However, because of the cell density of neuronal subtypes examined here, we have not separated these two but have focused at the level of homotypic avoidance. We presume that DSCAM allows individual self-avoidance through the same adhesive masking mechanism, although this is presently untested. It also remains unclear whether this is truly different from the mechanisms of tiling, in which neurons actively repel each other to occupy distinct domains. However, as future studies define the molecular mechanisms of adhesive masking, we will be able to better distinguish between these processes. For example, the cellular indifference observed in overlapping mosaic neurons may reflect a lower gain on signaling that could also lead to repulsion.
These findings, together with our previous work (22), offer an explanation for self-avoidance without requiring isoform diversity. At the individual cell level, the well-described repulsive mediators of self-avoidance function by generating thousands of distinctly homophilic recognition units (51). Dscam1 in Drosophila uses three banks of alternatively spliced exons to produce 19,008 isoforms with distinct extracellular domains (6), and the vertebrate Pcdhg cluster generates thousands of distinctly homophilic recognition multimers (12, 18). Differential isoform expression gives each neuron a distinct fingerprint, allowing it to recognize and avoid self through repulsion while still interacting with its neighbors, a process called self/nonself discrimination (9–11). As might be expected with such a mechanism in which each cell is uniquely identified, vast isoform diversity is required for this function (8, 13, 14, 52). Without extensive isoform diversity, vertebrate Dscam is not competent to provide this individualized level of self-recognition. Rather, in the adhesive masking hypothesis, cell identity or cellular subtypes are conferred by the repertoire of CAMs expressed, whereas self-avoidance between all of the cells of that subtype is provided by the single DSCAM isoform (4, 5, 53). This form of self-avoidance does not allow the cell to differentiate self from self-type, but permits homotypic neurites to be indifferent to each other.
In many retinal neuron types, this regulated homotypic indifference is sufficient for normal field coverage. Conversely, self-avoidance in SACs requires self/nonself discrimination. SACs are exceptional among retinal neurons for how they use γ-Pcdh recognition molecules: To date, they are the only retinal cell type described to have self-avoidance defects in Pcdhg mutants. Other neuronal cell types in the retina undergo excessive cell death in the absence of Pcdhg but do not exhibit the self-crossings found in individual SACs (16, 45). Interestingly, in addition to SACs, horizontal cells are spared this excess of cell death in Pcdhg mutants. Horizontal cells depend on neither Dscams nor γ-Pcdhs for self-avoidance but use plexin/semaphorin cues (19). Consistent with previous studies (16), we found no deficiencies in self-avoidance in ipRGCs in Pcdhg mutants (Fig. 6). Indeed, our findings indicate that γ-Pcdhs may contribute to adhesion in this cell type. Outside of Purkinje cells, other cell types in the CNS do not obviously depend on Pcdhg for self-avoidance and, in fact, mediate interactions between different cells (47, 48, 54–58). Conversely, both Drosophila and vertebrate Dscams can promote neurite recognition through adhesive mechanisms in some neuron types (50, 59), but generally promote self-avoidance in the retina, illustrating that the roles for these molecules can vary considerably with cellular context. SACs express neither Dscam nor Dscaml1, raising the interesting possibility that coexpression (or lack thereof) between the Dscams and the clustered protocadherins could determine their functions in different cell types.
Interestingly, we found that Nrcam mutation did not rescue fasciculation in the cell types that most strongly express it. This could be because (i) NRCAM does not contribute to the adhesion that drives fasciculation in these cells, (ii) DSCAM is not competent to mask all CAMs, or (iii) there is more adhesive redundancy in these cells. It will be important to expand our analyses beyond the cadherin superfamily to test DSCAM masking of other types of CAMs in future studies.
Members of the cadherin superfamily have emerged as key contributors to neurodevelopmental disorders, including autism, schizophrenia, bipolar disease, and intellectual disability (60–62). We have shown that Dscam can regulate the function of cadherins and protocadherins, making it a candidate target for these disabilities. Indeed, de novo mutations in Dscam have been linked to autism spectrum disorder in three families (63). Further investigation of the cell types in which DSCAM functions and the molecular mechanisms by which DSCAM masks adhesion will be instructive for understanding both normal development and the potential mechanisms underlying neurodevelopmental disorders.
Materials and Methods
Mouse Strains.
All animals were housed in the research animal facility at The Jackson Laboratory under standard housing conditions with a 12 h/12 h light/dark cycle and food and water ad libitum. All procedures using animals were performed in accordance with The Guide for the Care and Use of Laboratory Animals (64) and were reviewed and approved by the The Jackson Laboratory Institutional Animal Care and Use Committee. All experiments included a mix of male and female animals. Previously described strains were as follows: Dscam−/− is Dscamdel17/Rwb, RRID:IMSR_JAX:008000 (20); DscamF is Dscamtm1Pfu, MGI:5305022 (29); Cdh3-GFP, RRID:MMRRC_000236-UNC, courtesy of Andrew Huberman, Stanford University, Stanford, CA (24); Cdh3−/− is Cdh3tm1Hyn/J, RRID:IMSR_JAX:003180 (65); Cdh6−/− is Cdh6tm1Sma/J, RRID:IMSR_JAX:003742 (66); Ctnna2CDF is Ctnna2cdf/J, RRID:IMSR_JAX:002235 (28); Vgat-Cre is Slc32a1tm2(cre)Lowl/MwarJ, RRID:IMSR_JAX:028862 (37); PcdhgF is Pcdhgtm2Xzw/J, RRID:IMSR_JAX:012644 (56); Pax6a-Cre is Tg(Pax6 Cre,GFP)/2Pgr, RRID:MGI:3845671, courtesy of Peter Gruss, Max Planck Institute for Biophysical Chemistry, Gottingen, Germany (67); Pou4f2−/− is Pou4f2tm1Nat, RRID:MGI:3641269, courtesy of Lin Gan, University of Rochester Medical Center, Rochester, NY (68); Nrcam−/− is NrcammiJ/GrsrJRwb, MGI:5882344 (44); and Opn4-Tau-Lacz is Opn4tm1.1Yau, RRID:IMSR_JAX:021153 (69).
Because the BAC used to make the Cdh3-GFP line included the entire Cdh3 transcription unit, it was not possible to genotype Cdh3GFP;Cdh3−/− mice using the published PCR primers, because primers against the wild-type Cdh3 sequence also detected the transgene. To genotype for Cdh3−/− homozygosity, we used PCR simple-length polymorphism markers to detect strain-specific differences on chromosome 8 flanking the endogenous Cdh3 gene but beyond the BAC used for making the transgene. These polymorphisms differ between 129S and C57BL/6, the strain background originally targeted to make the Cdh3 mutation and the strain in which we were working, respectively. The pairs D8MIT12 and D8MIT15 were particularly informative. Dscam;Cdh double mutants and the associated controls were analyzed at P4, because of the low viability of older animals. All other experiments were performed at P14 unless otherwise noted.
Primary Neuron Cultures.
As described previously (55, 70), cerebral cortices were isolated from P0 mice, and the meninges were carefully removed. Isolated cortices were chopped into 1 mm pieces and then digested in papain for 40 min at 37 °C. The tissue was quenched in trypsin inhibitor and BSA (1% each in HBSS), rinsed in plating media (Basal Medium Eagle, 5% FBS, N2 Supplement, glutaMAX, and Pen/Strep), and lightly triturated. Cells were plated at 250,000 cells per well onto 9-mm glass coverslips coated with matrigel (1:50 in Neurobasal) in a 24-well dish. After 4 h and every subsequent 2 d, media was changed to serum-free media (Neurobasal, B27 Supplement, glutaMAX, and Pen/Strep). All tissue culture media and reagents were obtained from Gibco, unless otherwise noted.
Cell Lines.
HEK293T cells were obtained from American Type Culture Collection (lot 62312975) where they were tested free of mycoplasma and their identity was verified by short tandem repeat analysis. These cells were maintained in DMEM, 10% FBS, glutaMAX, and Pen/Strep.
In Situ Hybridization.
Eyes were collected from P0 Cdh3-GFP mice and immediately frozen in Tissue-Tek OCT (Sakura) in 2-methylbutane cooled in a dry ice/ethanol bath. Sections were cut at 12 μm and processed for in situ hybridization using the RNAscope Fluorescent Multiplex Reagent Kit (Advanced Cellular Diagnostics) according to the manufacturer’s instructions for fresh-frozen tissue. The following probes were used: EGFP-C3 (400281-C3); Mm-Cdh3 (514591); and Mm-Cdh6-C2 (519541-C2). The final amplification was performed using the Amp 4 Alt C option.
Immunofluorescence.
Whole retinas were isolated and fixed in 4% paraformaldehyde for 4 to 8 h. Retinas were stained free-floating in 2.5% BSA with 0.5% Triton X-100 in the indicated antibodies for 48 to 72 h at 4 °C. After washing off unbound primary antibodies, secondary antibodies were applied in the same solution overnight at 4 °C. For sectioning, lenses were removed from enucleated eyes. Eyecups were fixed, cryopreserved in 30% sucrose, and frozen in Tissue-Tek OCT (Sakura). Cryosections were cut at 12 μm and immunostained on the slide. Primary antibodies were applied overnight in blocking solution at 4 °C, and secondary antibodies for 1 h at room temperature. Sections and whole retinas were imaged on a Leica SP5 confocal microscope.
Antibodies.
The following antibodies were used: Rabbit anti-GFP [1:500, RRID:AB_91337 (AB3080; Millipore)]; Rabbit anti-mCherry [1:500, RRID:AB_2552323 (PA5-34974; Thermo Fisher Scientific)]; Sheep anti-tyrosine hydroxylase [1:500, RRID:AB_11213126 (AB1542; Millipore)]; Rabbit anti-melanopsin [1:10,000, RRID:AB_1266795 (IT-44-100; Advanced Targeting Systems)]; Rabbit anti-NRCAM [1:250, RRID:AB_448024 (ab24344; ABCAM)]; Mouse anti-Cadherin6 [1:150, RRID:AB_907139 (MAB2715; R&D Systems)]; Mouse anti-FLAG, M2 [1:500, RRID:AB_262044 (F1804; Sigma-Aldrich)]; Chicken anti-beta-Galactosidase [1:10,000, RRID:AB_2313507 (BGL-1040; Aves Labs)]; Goat anti-Human IgG Fc Secondary Antibody, HRP [RRID:AB_2535606 (A18829; Thermo Fisher Scientific)]; and Alexa Fluor-conjugated secondary antibodies [1:500 (Thermo Fisher Scientific)].
DNA Constructs.
An expression construct containing Myc-DDK–tagged cadherin-3 (pLenti-Cdh3) was obtained from Origene (MR227582L1). This construct was also used as a PCR template to isolate the sequence encoding cadherin-3 ectodomain (CDH3-EC) using the following primers: ATATGCTAGCACCACCCTTCCAGGGTCTGGGGCAGTC and ATATAGATCTATGGAGCTTCTTAGTGGGCCTCAC. The PCR product was cloned in-frame with human IgG1-FC (the fragment crystallizable region of an antibody; binds to protein G) in a pShuttle-CMV vector into BglII and NheI sites. The sequence encoding DSCAM ectodomain (DSCAM-EC) was similarly PCR-cloned from pCAG-Dscam (71) in-frame with human IgG1-FC into pShuttle-CMV using the following primers: TTTGGGGCTAGCCTTGAGCCCTTCGTTGGTTGTCAGCC and GAATTCATGTGGATACTGGCTCTCTCC. To generate Cre-inducible expression of Cdh3, full-length Cdh3 was isolated by PCR using the following primers: TCGGTACGATTTAAATTGAATTCATGGAGCTTCTTAGTGGGCCTCACGC and GGCAGCCTGCACCTGAGGAGTGCGGCCGCTAAACCTTATCGTCGTCATCC. pCALNL-DsRed (41) was digested with EcoRI and NotI to remove DsRed, and the Cdh3 PCR product was inserted by Gibson assembly.
In Vitro Bead Assay.
Beads were coated with protein ectodomains in a protocol adapted from ref. 72. HEK293T cells were transfected with constructs encoding DSCAM-EC or CDH3-EC (both with C-terminal FC domains added) with Lipofectamine 3000 according to the manufacturer’s instructions. Twenty-four hours after transfection, medium was replaced with unsupplemented DMEM. After 1 h, this was replaced with fresh DMEM. Forty-eight hours later, the medium was removed, filtered, and concentrated with an Amicon Ultra 30 KDa spin filter. Ectodomains were verified by Western blot (SI Appendix, Fig. S4). Concentrated medium from CDH3-EC cells alone, or mixed 1:1 with medium from DSCAM-EC cells, was incubated with protein G Dynabeads for 4 h at 4 °C, with rotation (0.3 μL of beads for each well of neurons). Beads were then rinsed, resuspended in Neurobasal media, and used immediately in the assay.
Cortical neurons cultured for 6 d in vitro were transfected using NeuroMag (Oz Biosciences) according to the manufacturer’s protocol. For each well, 0.75 μg of pLenti-Cdh3 DNA was combined with 1.5 μL of NeuroMag reagent in 100 μL of DMEM at room temperature for 20 min, and then applied to the cells. The 24-well plate was immediately placed on a magnetic plate for 20 min at 37°.
The bead assay of masking was performed 48 h after the neuron transfection. Beads coated with CDH3-EC alone, or CDH3-EC with DSCAM-EC, were applied to each well for 1 h at 37 °C. Cultures were immediately fixed in 4% paraformaldehyde in PBS for 10 min at room temperature, and stained for FLAG. Transfected neurons contacting beads were imaged on a Leica SP5 microscope. Beads fluoresced when excited by laser light at 633 nm.
In Vivo Electroporation.
P0 pups were electroporated as described previously (73). Mice were anesthetized by hypothermia. Only the right eye was electroporated. Eyelids were opened using a 30-gauge needle to make an incision along the fused junctional epithelium, and a small puncture was made in the sclera below the junction with the cornea. A blunt 33-gauge needle in a Hamilton syringe was inserted into the opening and through the retina at the back of the eye, allowing subretinal injection of 0.3 μL of plasmid DNA (4 μg/μL). Mice were immediately electroporated with tweezer electrodes placed across the head, with the positive pole over the injected eye. Five pulses were delivered for 50 ms at 80 V, with 950-ms intervals. Retinas were assessed by immunofluorescence 14 d after electroporation.
Image Analysis.
Adhesion molecule clustering.
To quantify clustering at contact points in the bead assay, an annulus-shaped region of interest (ROI) was defined with an inner diameter of 1.9 μm and an outer diameter of 3.9 μm (∼9 μm2 ROI area). When centered on a bead, this ROI corresponded to the region within 1 μm of the bead. The average fluorescence intensity within this ROI was measured (i) centered on each bead that contacted the transfected neuron (contact), (ii) on the region of the neuron directly adjacent to the contact ROI (adjacent), and (iii) on 10 beads within the image that did not contact the transfected neuron. The values from these 10 beads were averaged to calculate background. The cluster score for each site of contact was then calculated according to the following formula:
The cluster scores from all contact points on a single neuron were averaged. Within each bead preparation, these per-neuron scores were normalized to the average score on Dscam−/− neurons. Normalized scores from multiple bead preparations were combined, and scores on control neurons were compared with those on Dscam−/− neurons by two-sided Student’s t test. Similarity of variances was tested using an F-test.
FS.
The MST is an image analysis methodology that has been developed and used in astrophysics (30). Here, we combined two output functions: the distribution of density and the distribution of filament indices (30–35) to create a new output function called the distribution of fasciculation. This was calculated across multiple thresholds by dividing the square of the average filament index of sufficiently large (>50 pixels) isolated components by the square of the density (i.e., the ratio of the summation of all pixel values greater than the threshold value to the total sum of pixel values for the image). The summation of the distribution of fasciculation yields a unique number, coined the FS. A higher FS is associated with a higher degree of fasciculation. The algorithmic implementations were done via in-house software for which the mathematical calculations are coded in C routines wrapped in a Tcl/Tk interpreter. The development of the FS is described in detail in SI Appendix, Supplementary Methods. FSs from each genotype were compared using a pairwise Wilcoxon ranked sum test.
Elo score.
Images were compared using a custom web-based program (Image Echelon, code available at https://github.com/TheJacksonLaboratory/ImageEchelon) that uses an Elo ranking algorithm, as described previously (22, 74). Each image set was scored by >10 users blind to genotype. Scores from each individual retina were averaged (1 to 4 images per retina), and these retinal mean scores were used to compare across genotypes using a pairwise Wilcoxon ranked sum test. This method is described in detail and compared with FSs in SI Appendix, Supplementary Methods. For comparisons involving Dscam heterozygous mutants (Fig. 6), composite images of whole-retina quadrants were used for scoring due to the more heterogeneous nature of clustering and fasciculation in these animals. Representative composites of quadrants are in SI Appendix, Fig. S6. The data are presented in box plots prepared in R using ggplot2 and represent the median, first and third quartiles, range [encompassing values within 1.5 times the interquartile range (IQR) from the first or third quartile], and outliers (any values beyond 1.5 IQR from the first or third quartile).
Cell Spacing.
The density recovery profile (DRP) and overall cell density were calculated in WinDRP as described previously (20, 22). For each image, the DRPs were normalized so that the overall density was set at 1. This allowed for direct comparison of relative spacing independent of overall cell number. Values from multiple images per retina were averaged to find a retinal mean. These means were compared across genotype by ANOVA with pairwise Tukey post hoc tests.
Supplementary Material
Acknowledgments
We thank Keith Sheppard for assistance with Image Echelon, and our colleagues at The Jackson Laboratory for performing image comparisons. We also thank Dr. Joshua Sanes and Dr. Joshua Weiner and members of the R.W.B. laboratory for helpful discussion of the manuscript. This work was supported by NIH Grant R01 NS054154 (to R.W.B.). A.M.G. was supported by NIH Grants F32EY021942 and T3200HD7065. The scientific services at The Jackson Laboratory are supported in part by NIH Grant CA34196.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809430115/-/DCSupplemental.
References
- 1.Honjo M, et al. Differential expression of cadherin adhesion receptors in neural retina of the postnatal mouse. Invest Ophthalmol Vis Sci. 2000;41:546–551. [PubMed] [Google Scholar]
- 2.Zipursky SL, Sanes JR. Chemoaffinity revisited: Dscams, protocadherins, and neural circuit assembly. Cell. 2010;143:343–353. doi: 10.1016/j.cell.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Cook JE, Chalupa LM. Retinal mosaics: New insights into an old concept. Trends Neurosci. 2000;23:26–34. doi: 10.1016/s0166-2236(99)01487-3. [DOI] [PubMed] [Google Scholar]
- 4.Fuerst PG, Burgess RW. Adhesion molecules in establishing retinal circuitry. Curr Opin Neurobiol. 2009;19:389–394. doi: 10.1016/j.conb.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garrett AM, Burgess RW. Candidate molecular mechanisms for establishing cell identity in the developing retina. Dev Neurobiol. 2011;71:1258–1272. doi: 10.1002/dneu.20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmucker D, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 7.Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hattori D, et al. Robust discrimination between self and non-self neurites requires thousands of Dscam1 isoforms. Nature. 2009;461:644–648. doi: 10.1038/nature08431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes ME, et al. Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron. 2007;54:417–427. doi: 10.1016/j.neuron.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthews BJ, et al. Dendrite self-avoidance is controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 11.Soba P, et al. Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron. 2007;54:403–416. doi: 10.1016/j.neuron.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schreiner D, Weiner JA. Combinatorial homophilic interaction between gamma-protocadherin multimers greatly expands the molecular diversity of cell adhesion. Proc Natl Acad Sci USA. 2010;107:14893–14898. doi: 10.1073/pnas.1004526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature. 2012;488:517–521. doi: 10.1038/nature11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kostadinov D, Sanes JR. Protocadherin-dependent dendritic self-avoidance regulates neural connectivity and circuit function. eLife. 2015;4:e08964. doi: 10.7554/eLife.08964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodman KM, et al. Protocadherin cis-dimer architecture and recognition unit diversity. Proc Natl Acad Sci USA. 2017;114:E9829–E9837. doi: 10.1073/pnas.1713449114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ing-Esteves S, et al. Combinatorial effects of alpha- and gamma-protocadherins on neuronal survival and dendritic self-avoidance. J Neurosci. 2018;38:2713–2729. doi: 10.1523/JNEUROSCI.3035-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubinstein R, et al. Molecular logic of neuronal self-recognition through protocadherin domain interactions. Cell. 2015;163:629–642. doi: 10.1016/j.cell.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thu CA, et al. Single-cell identity generated by combinatorial homophilic interactions between α, β, and γ protocadherins. Cell. 2014;158:1045–1059. doi: 10.1016/j.cell.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuoka RL, et al. Guidance-cue control of horizontal cell morphology, lamination, and synapse formation in the mammalian outer retina. J Neurosci. 2012;32:6859–6868. doi: 10.1523/JNEUROSCI.0267-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuerst PG, Koizumi A, Masland RH, Burgess RW. Neurite arborization and mosaic spacing in the mouse retina require DSCAM. Nature. 2008;451:470–474. doi: 10.1038/nature06514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuerst PG, et al. DSCAM and DSCAML1 function in self-avoidance in multiple cell types in the developing mouse retina. Neuron. 2009;64:484–497. doi: 10.1016/j.neuron.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrett AM, Tadenev ALD, Hammond YT, Fuerst PG, Burgess RW. Replacing the PDZ-interacting C-termini of DSCAM and DSCAML1 with epitope tags causes different phenotypic severity in different cell populations. eLife. 2016;5:e16144. doi: 10.7554/eLife.16144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 24.Osterhout JA, et al. Cadherin-6 mediates axon-target matching in a non-image-forming visual circuit. Neuron. 2011;71:632–639. doi: 10.1016/j.neuron.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sümbül U, et al. A genetic and computational approach to structurally classify neuronal types. Nat Commun. 2014;5:3512. doi: 10.1038/ncomms4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duan X, Krishnaswamy A, De la Huerta I, Sanes JR. Type II cadherins guide assembly of a direction-selective retinal circuit. Cell. 2014;158:793–807. doi: 10.1016/j.cell.2014.06.047. [DOI] [PubMed] [Google Scholar]
- 27.Yu HH, Huang AS, Kolodkin AL. Semaphorin-1a acts in concert with the cell adhesion molecules fasciclin II and connectin to regulate axon fasciculation in Drosophila. Genetics. 2000;156:723–731. doi: 10.1093/genetics/156.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park C, Falls W, Finger JH, Longo-Guess CM, Ackerman SL. Deletion in Catna2, encoding alpha N-catenin, causes cerebellar and hippocampal lamination defects and impaired startle modulation. Nat Genet. 2002;31:279–284. doi: 10.1038/ng908. [DOI] [PubMed] [Google Scholar]
- 29.Fuerst PG, Bruce F, Rounds RP, Erskine L, Burgess RW. Cell autonomy of DSCAM function in retinal development. Dev Biol. 2012;361:326–337. doi: 10.1016/j.ydbio.2011.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams FC. A topological/geometrical approach to the study of astrophysical maps. Astrophys J. 1992;387:572–590. [Google Scholar]
- 31.Adams FC, Wiseman J. Formal results regarding metric space techniques for the study of astrophysical maps. Astrophys J. 1994;435:693–707. [Google Scholar]
- 32.Wiseman J, Adams FC. A quantitative analysis of IRAS maps of molecular clouds. Astrophys J. 1994;435:708–721. [Google Scholar]
- 33.Khalil A, Joncas G, Nekka F. Morphological analysis of HI features. I. Metric space technique. Astrophys J. 2004;601:352–364. [Google Scholar]
- 34.Robitaille JF, Joncas G, Khalil A. Morphological analysis of HI features–III. Metric space technique revisited. Mon Not R Astron Soc. 2010;405:638–656. [Google Scholar]
- 35.Wu Y, Batuski DJ, Khalil A. Multi-scale morphological analysis of SDSS DR5 survey using the metric space technique. Astrophys J. 2009;707:1160–1167. [Google Scholar]
- 36.Wu Y, Batuski DJ, Khalil A. Three-dimensional filamentation analysis of SDSS DR5 survey. IRSN Astron Astrophys. 2012;2012:171829. [Google Scholar]
- 37.Vong L, et al. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron. 2011;71:142–154. doi: 10.1016/j.neuron.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Andrade GB, Long SS, Fleming H, Li W, Fuerst PG. DSCAM localization and function at the mouse cone synapse. J Comp Neurol. 2014;522:2609–2633. doi: 10.1002/cne.23552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li S, et al. DSCAM promotes refinement in the mouse retina through cell death and restriction of exploring dendrites. J Neurosci. 2015;35:5640–5654. doi: 10.1523/JNEUROSCI.2202-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci USA. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lambert M, Padilla F, Mège RM. Immobilized dimers of N-cadherin-Fc chimera mimic cadherin-mediated cell contact formation: Contribution of both outside-in and inside-out signals. J Cell Sci. 2000;113:2207–2219. doi: 10.1242/jcs.113.12.2207. [DOI] [PubMed] [Google Scholar]
- 43.Hortsch M. Structural and functional evolution of the L1 family: Are four adhesion molecules better than one? Mol Cell Neurosci. 2000;15:1–10. doi: 10.1006/mcne.1999.0809. [DOI] [PubMed] [Google Scholar]
- 44.Morelli KH, et al. Severity of demyelinating and axonal neuropathy mouse models is modified by genes affecting structure and function of peripheral nodes. Cell Rep. 2017;18:3178–3191. doi: 10.1016/j.celrep.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lefebvre JL, Zhang Y, Meister M, Wang X, Sanes JR. Gamma-protocadherins regulate neuronal survival but are dispensable for circuit formation in retina. Development. 2008;135:4141–4151. doi: 10.1242/dev.027912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tarusawa E, et al. Establishment of high reciprocal connectivity between clonal cortical neurons is regulated by the Dnmt3b DNA methyltransferase and clustered protocadherins. BMC Biol. 2016;14:103. doi: 10.1186/s12915-016-0326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molumby MJ, Keeler AB, Weiner JA. Homophilic protocadherin cell-cell interactions promote dendrite complexity. Cell Rep. 2016;15:1037–1050. doi: 10.1016/j.celrep.2016.03.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garrett AM, Weiner JA. Control of CNS synapse development by gamma-protocadherin-mediated astrocyte-neuron contact. J Neurosci. 2009;29:11723–11731. doi: 10.1523/JNEUROSCI.2818-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keeley PW, et al. Neuronal clustering and fasciculation phenotype in Dscam- and Bax-deficient mouse retinas. J Comp Neurol. 2012;520:1349–1364. doi: 10.1002/cne.23033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamagata M, Sanes JR. Dscam and sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature. 2008;451:465–469. doi: 10.1038/nature06469. [DOI] [PubMed] [Google Scholar]
- 51.Zipursky SL, Grueber WB. The molecular basis of self-avoidance. Annu Rev Neurosci. 2013;36:547–568. doi: 10.1146/annurev-neuro-062111-150414. [DOI] [PubMed] [Google Scholar]
- 52.Hattori D, et al. Dscam diversity is essential for neuronal wiring and self-recognition. Nature. 2007;449:223–227. doi: 10.1038/nature06099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garrett AM, Tadenev AL, Burgess RW. DSCAMs: Restoring balance to developmental forces. Front Mol Neurosci. 2012;5:86. doi: 10.3389/fnmol.2012.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Molumby MJ, et al. γ-protocadherins interact with neuroligin-1 and negatively regulate dendritic spine morphogenesis. Cell Rep. 2017;18:2702–2714. doi: 10.1016/j.celrep.2017.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garrett AM, Schreiner D, Lobas MA, Weiner JA. γ-protocadherins control cortical dendrite arborization by regulating the activity of a FAK/PKC/MARCKS signaling pathway. Neuron. 2012;74:269–276. doi: 10.1016/j.neuron.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prasad T, Wang X, Gray PA, Weiner JA. A differential developmental pattern of spinal interneuron apoptosis during synaptogenesis: Insights from genetic analyses of the protocadherin-gamma gene cluster. Development. 2008;135:4153–4164. doi: 10.1242/dev.026807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weiner JA, Wang X, Tapia JC, Sanes JR. Gamma protocadherins are required for synaptic development in the spinal cord. Proc Natl Acad Sci USA. 2005;102:8–14. doi: 10.1073/pnas.0407931101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X, et al. Gamma protocadherins are required for survival of spinal interneurons. Neuron. 2002;36:843–854. doi: 10.1016/s0896-6273(02)01090-5. [DOI] [PubMed] [Google Scholar]
- 59.Tadros W, et al. Dscam proteins direct dendritic targeting through adhesion. Neuron. 2016;89:480–493. doi: 10.1016/j.neuron.2015.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Redies C, Hertel N, Hübner CA. Cadherins and neuropsychiatric disorders. Brain Res. 2012;1470:130–144. doi: 10.1016/j.brainres.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 61.Corvin AP. Neuronal cell adhesion genes: Key players in risk for schizophrenia, bipolar disorder and other neurodevelopmental brain disorders? Cell Adhes Migr. 2010;4:511–514. doi: 10.4161/cam.4.4.12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naskar T, et al. Ancestral variations of the PCDHG gene cluster predispose to dyslexia in a multiplex family. EBioMedicine. 2018;28:168–179. doi: 10.1016/j.ebiom.2017.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
- 65.Radice GL, et al. Precocious mammary gland development in P-cadherin-deficient mice. J Cell Biol. 1997;139:1025–1032. doi: 10.1083/jcb.139.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mah SP, Saueressig H, Goulding M, Kintner C, Dressler GR. Kidney development in cadherin-6 mutants: Delayed mesenchyme-to-epithelial conversion and loss of nephrons. Dev Biol. 2000;223:38–53. doi: 10.1006/dbio.2000.9738. [DOI] [PubMed] [Google Scholar]
- 67.Marquardt T, et al. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 68.Gan L, et al. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci USA. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hattar S, Liao HW, Takao M, Berson DM, Yau KW. Melanopsin-containing retinal ganglion cells: Architecture, projections, and intrinsic photosensitivity. Science. 2002;295:1065–1070. doi: 10.1126/science.1069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ghosh A, Greenberg ME. Distinct roles for bFGF and NT-3 in the regulation of cortical neurogenesis. Neuron. 1995;15:89–103. doi: 10.1016/0896-6273(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 71.Schramm RD, et al. A novel mouse Dscam mutation inhibits localization and shedding of DSCAM. PLoS One. 2012;7:e52652. doi: 10.1371/journal.pone.0052652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Emond MR, Jontes JD. Bead aggregation assays for the characterization of putative cell adhesion molecules. J Vis Exp. 2014:e51762. doi: 10.3791/51762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Melo J, Blackshaw S. In vivo electroporation of developing mouse retina. J Vis Exp. 2011:2847. doi: 10.3791/2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Elo AE. 1978. The Rating of Chessplayers: Past and Present (Arco Publishing, New York), 208 p.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.