Abstract
Pathological cardiac hypertrophy is associated with the accumulation of lipid peroxidation-derived aldehydes such as 4-hydroxy-trans-2-nonenal (HNE) and acrolein in the heart. These aldehydes are metabolized via several pathways, of which aldose reductase (AR) represents a broad-specificity route for their elimination. We tested the hypothesis that by preventing aldehyde removal, AR deficiency accentuates the pathological effects of transverse aortic constriction (TAC). We found that the levels of AR in the heart were increased in mice subjected to TAC for 2 weeks. In comparison with wild-type (WT), AR-null mice showed lower ejection fraction, which was exacerbated 2 weeks after TAC. Levels of atrial natriuretic peptide and myosin heavy chain were higher in AR-null than in WT TAC hearts. Deficiency of AR decreased urinary levels of the acrolein metabolite, 3-hydroxypropylmercapturic acid. Deletion of AR did not affect the levels of the other aldehyde-metabolizing enzyme - aldehyde dehydrogenase 2 in the heart, or its urinary product - (N-Acetyl-S-(2-carboxyethyl)-L-cystiene). AR-null hearts subjected to TAC showed increased accumulation of HNE- and acrolein-modified proteins, as well as increased AMPK phosphorylation and autophagy. Superfusion with HNE led to a greater increase in p62, LC3II formation, and GFP-LC3-II punctae formation in AR-null than WT cardiac myocytes. Pharmacological inactivation of JNK decreased HNE-induced autophagy in AR-null cardiac myocytes. Collectively, these results suggest that during hypertrophy the accumulation of lipid peroxidation derived aldehydes promotes pathological remodeling via excessive autophagy, and that metabolic detoxification of these aldehydes by AR may be essential for maintaining cardiac function during early stages of pressure overload.
Keywords: acrolein, autophagy, carnosine, hydroxynonenal, hypertrophy
1. Introduction
Heart failure is one of the leading causes of death worldwide [1]. Although there have been significant medical and surgical advances in the treatment of heart failure, mortality associated with this condition remains high [2]. Development of overt heart failure could result from hypertension and pathologic hypertrophy, but the mechanisms underlying the transition from hypertrophy to heart failure are unclear, therefore, it is difficult to directly target the processes or diminish its progression. Previous work has shown that one of the major determinants of pathological myocardial remodeling due to hemodynamic stress is an increase in generation of reactive oxygen species (ROS) [3, 4]; however, clinical trials with antioxidant therapies failed to prevent cardiovascular diseases [5, 6]. Hence, to develop effective therapeutic interventions, there is a need to develop a deeper understanding of processes that generate ROS and those that mediate the adaptive or the pathologic effects of oxidative stress in the failing heart.
It is generally recognized that unquenched ROS participates in rather indiscriminate reactions with nucleophilic centers in lipids, proteins, and nucleic acids [7–11]. Because phospholipids maintain 6- to 8-fold higher concentrations of O2 [12] and contain multiple unsaturated conjugated centers that can stabilize free radicals, they are the most vulnerable targets of ROS. Oxidation of polyunsaturated fatty acids in phospholipids results in the development of several highly reactive secondary products that amplify and propagate damage initiated by ROS. Due to their ability to spread oxidative damage, products of lipid peroxidation act as “second messengers of ROS toxicity” [12–14]. Numerous studies have shown that the products of lipid peroxidation - such as 4-hydroxy trans-2-nonenal (HNE), malondialdehyde (MDA), and acrolein-accumulate in experimental models of myocardial hypertrophy. Additionally, HNE bound to proteins has been detected in hearts of cardiomyopathic hamsters [15], spontaneously hypertensive rats [16], rats infused with isoproterenol [17], and mice subjected to transverse aortic constriction [18, 19]. Similarly, proteins adducted to acrolein accumulate in failing hearts [20], and proteins modified by lipid peroxidation-derived aldehydes have been detected in myocardial tissue from humans with end stage congestive heart failure [21] and hypertrophic cardiomyopathy [22]
Despite the strong association between the accumulation of lipid peroxidation-derived aldehydes and heart failure, the pathologic significance of such products remains unclear. Because of their high chemical reactivity, lipid-derived aldehydes can react with, and modify multiple cellular targets and interfere with their normal function. In addition, proteins extensively modified by aldehydes, which are poor proteasomal substrates [23] could trigger the unfolded protein response or autophagy [24, 25], which could contribute to widespread changes in myocardial structure and function; however, there is no direct evidence linking lipid peroxidation products to pathologic remodeling or autophagy in hypertrophic hearts. Hence, we examined whether changes in the metabolism or detoxification of lipid peroxidation products would affect cardiac function and autophagy during pressure-induced myocardial hypertrophy.
In the heart, aldehydes generated by lipid peroxidation are removed or metabolized by several enzymatic processes, which are catalyzed by glutathione S-transferases, aldehyde dehydrogenases, and aldo-keto reductases (AKRs) using peptidyl antioxidants such as glutathione and carnosine (β-alanine-histidine) [26–28]. Of these, aldose reductase (AR), a member of the AKR1B family plays a critical role in the metabolism of lipid peroxidation-derived aldehydes [29–31]. The enzyme catalyzes reduction of several lipid-derived aldehydes such as HNE and acrolein, and their glutathione and carnosine conjugates with high efficiency [32]. Inhibition of AR increases the accumulation and toxicity of HNE [33] and acrolein in inflamed arteries [34] and ischemic hearts [35]. Inhibition of the enzyme has also been shown to abolish ischemic preconditioning [36]. The increase in AR levels in the failing human hearts [37] might be an adaptive response to minimize aldehyde toxicity. Nevertheless, the contribution of AR or its substrates to pathological cardiac remodeling has not been assessed. We hypothesized that increased production of lipid peroxidation in a failing heart upregulates AR, and that removal of HNE and related aldehydes by AR protects the heart from the deleterious consequences of such aldehydes. To test this hypothesis, we examined the role of AR in mouse hearts subjected to transverse aortic constriction (TAC). Our results show that AR deficiency accelerates contractile dysfunction, increases the accumulation of aldehyde modified proteins, and promotes maladaptive autophagy in mice subjected to TAC. These findings reveal a new link between lipid peroxidation-derived aldehydes, autophagy and pathological remodeling, and support the concept that detoxification of reactive aldehydes by AR is a pivotal pathway in maintaining cardiac function during pressure overload.
2. Materials and methods
2.1. Animals
Adult male C57BL/6 mice were obtained from The Jackson Laboratory (Bar Harbor, ME). AR-null mice were obtained from Dr. Stephen S. Chung, University of Hong Kong. All treatments and protocols were approved by the University of Louisville Institutional Animal Care and Use Committee.
2.2. Transverse aortic constriction and assessment of hypertrophy
TAC was performed as described previously [38] and in accordance with the University of Louisville Animal Care and Use Committee. Briefly, 12–16-week-old mice were anesthetized with ketamine (50 mg/kg intraperitoneal) and pentobarbital (50 mg/kg, intraperitoneal); orally intubated with a polyethylene-60 tubing; and ventilated (Harvard Apparatus Rodent Ventilator, model 845) with oxygen supplementation. Tidal volumes and breathing rates were set based on standard allometeric equations. The aorta was visualized through an intercostal incision. A 7–0 nylon suture was placed around the transverse aorta between the brachiocephalic and left common carotid arteries. The suture was tied around a blunt needle (27 gauge; to constrict the aorta to a reproducible diameter), which was subsequently removed, leaving a discrete region of stenosis; the chest was closed. Mice were extubated upon recovery in warm, clean cages supplemented with oxygen. Analgesia (ketoprofen, 5 mg/kg, subcutaneous) was given before mice recovered from anesthesia. Sham surgeries were done with the same procedure except the suture was only passed underneath the aorta and not tied. At the end of study, both sham and TAC operated mice were euthanized and the hearts were rapidly removed. Heart weight, body weight, left and right ventricular weight, and tibia length were measured. Left and right ventricles were frozen in liquid nitrogen.
2.3. Echocardiographic analysis of cardiac function
Transthoracic echocardiography of the left ventricle was performed using a Vevo 770 echocardiography system as described previously [39, 40]. Body temperature was maintained (36.5 –37.5°C) using a rectal thermometer interfaced with a servo-controlled heat lamp. Mice were anesthetized with 2% isoflurane, maintained under anesthesia with 1.5% isoflurane. The chest was shaved and the mouse was placed chest up on an examination board interfaced with the Vevo 770. The 707-B (30 MHZ) scan head was used to obtain 2D images of the parasternal long axis. M-modes were taken from the same anatomical position. The probe was then rotated to acquire a short axis view of the heart. Stroke volume (SV) was calculated as: Diastolic Volume-Systolic Volume. Ejection fraction was calculated as (SV/Diastolic Volume) *100%. Cardiac output was determined by: SV*HR. All images were acquired with the Vevo 770’s rail system to maintain probe placement and allow for precise adjustments of position. Left ventricular diameters during diastole (LVIDd) and left ventricular diameter during systole (LVIDs) and heart rate (HR) were determined from long axis M-modes. Left ventricular fractional shortening (%FS) was calculated as ((LVIDd-LVIDs)/LVIDd) *100%.
2.4. Quantitative RT-PCR
Total RNA was isolated from frozen LV by using the RNeasy Fibrous Tissue Mini Kit (Qiagen). Expression of the genes encoding atrial natriuretic peptide (ANP) and myosin heavy chain (β-MHC) was determined by quantitative real time PCR. Results were normalized to 18s rRNA and expressed according to the comparative Ct method. The Ct values of gene of interest from TAC were compared with the sham operated hearts. The Ct values of both the calibrator and the gene of interest are normalized to 18 srRNA or GAPDH.
2.5. Measurement of urinary metabolites
Urinary metabolites of acrolein-3-hydroxypropyl mercapturic acid (3-HPMA) and N-Acetyl-S-(2-carboxyethyl)-L-cystiene (CEMA) were analyzed using ACQUITY UPLC Quattro Premier XE triple quadrupole mass spectrometer operating in multiple reaction-monitoring (MRM) mode. Briefly, mouse urine samples were spiked with internal standards d3-3HPMA, d3-CEMA and diluted with solvent A (15 mM ammonium acetate, pH 6.8). Separations were performed by using a Acquity UPLC HSS T3 column (150 mm × 2.1 mm, 1.8 µm) (Waters Inc, MA). The elution of metabolites was achieved by using a binary gradient (Solvent A) and acetonitrile at a flow rate of 0.45 mL/min. The data for 3HPMA, CEMA and internal standards were acquired by monitoring the following transitions: 220→91, 220→89 (3HPMA); 223→91 (d3-3HPMA); 234→162, 234→105 (CEMA) and 237→165 (d3-CEMA). Analytes in urine samples were quantified using peak area ratio based on an 8 point-standard curve. TargetLynx quantification application manager software (Waters Inc., MA) was used for peak integration, calibration, and quantification. 3HPMA and CEMA levels were normalized to creatinine as previously reported [41].
2.7. Western blot analysis
Left ventricles (LVs) of the sham and TAC mice were homogenized and separated by SDS-PAGE. Acrolein and HNE protein adducts were analyzed using polyclonal anti-acrolein (0.25 µg/mL, Novus Biological) and anti-HNE (0.5 µg/mL, Abcam) antibodies. Immuno-blots were developed using p62 (0.027 µg/mL, Novus Biological), LC3-II (0.026 µg/mL, Cell Signaling), ALDH2 (1 µg/mL, Novus Biological), monoclonal anti-AR (0.1 µg/mL, Santa Cruz), AMPK (0.026 µg/mL, Cell Signaling), pAMPK (0.027 µg/mL, Cell Signaling) antibodies. Western blots were developed using ECL Plus reagent and detected with a Typhoon 9400 variable mode imager. Band intensity was quantified by using Image Quant TL software and bands were normalized to amido-black. Western blots shown are representative of at least n=3–4 samples per group.
2.8. Proteomic analysis of 4-hydroxynonenal (HNE) immunoprecipitation (IP) samples
To identify HNE-modified proteins in AR-null TAC hearts protiens were immunoprecipated (IP) using a protein-HNE adduct antibody (Abcam). LC/MS based proteomic studies were conducted as described previously [28] [42]. HNE IP eluates were thawed and diluted with an equal volume of 18 MΩ water to achieve a final concentration of 50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol. To stabilize HNE protein adducts by reductive amination the eluates were adjusted to a final 50 mM sodium cyanoborohydride (Sigma-Aldrich, St. Louis, MO) concentration using a 0.5 M sodium cyanoborohydride stock solution freshly prepared in 0.1 M NaOH. After a 4 h room temperature incubation, the reaction was quenched by addition of 10-molar equivalents of acetone and incubated for 30 min and subsequent 30 min incubation with 10-molar equivalents of acetone (Sigma-Aldrich, St. Louis, MO). The reaction was transferred to a Microcon-10 (EMD Millipore, Billerica, MA) 10 kDa centrifugal filter device for proteolytic digestion using trypsin (Promega, Madison, WI) by the FASP method modified with inclusion of a Bicine buffer (Sigma-Aldrich, St. Louis, MO) buffer [43].
Tryptic digests were concentrated and desalted in two steps with a first reversed phase (RP) procedure with C18 PROTOTM Ultra MicroSpin columns (Nest Group, Inc, Southborough, MA) as previously described [43]. Next, the samples were cleaned of polyethylene glycol-based polymers using a Strata X-C plate (Phenomenex, Torrance, CA) according to the manufacturer’s guidelines. Samples were lyophilized, and re-dissolved into 2% acetonitrile/0.1% formic acid prior to LC/MS analysis. Digests were separated using an EASY n-LC (Thermo) UHPLC system with a 2-cm Dionex Acclaim PepMap 100 (C18, 3 µm, 100Å) trap column (Dionex, Sunnyvale, CA) and a 50 cm Dionex Acclaim PepMap RSLC (C18, 2 µm, 100 Å) separating column (Dionex) thermostated to 50°C. Tandem mass spectrometry data were generated by nanospray ionization into an LTQ Orbitrap ELITE mass spectrometer (Thermo-Fisher Scientific) after separation was accomplished with a 90-min gradient from 100 % Buffer A (2% acetonitrile/0.1% formic acid) to 50%, Buffer B (80% acetonitrile/0.1% formic acid).
Data were acquired using the LTQ-Orbitrap ELITE mass spectrometer as an Nth Order Double Play (Xcalibur v2.2) with scan event one to obtain an FTMS MS1 scan (normal mass range; 240,000 resolutions, full scan type, positive polarity, profile data type) for the range 300–2000 m/z. Scan event two obtained ITMS MS2 scans (normal mass range, rapid scan rate, centroid data type) on up to twenty peaks that had a minimum signal threshold of 5,000 counts from scan event one. The lock mass option was enabled (0% lock mass abundance) using the 371.101236 m/z polysiloxane peak as an internal calibrant.
The acquired Thermo RAW files containing the FTMS and ITMS data were analyzed in Peaks 7.5 Studio considering the following: Denovo parent mass error 50.0 ppm, fragment mass error 1.0 Da, trypsin protease, variable modifications of Oxidation (M, +15.99 Da), 4-hydroxynonenal or “HNE” (C/H/K, +156.12 Da), dehydrated HNE (C/H/K, +138.1 Da), reduced HNE (C/H/K, +158.13 Da), up to 3 variable PTM per peptide, and 2 maximum missed cleavages including one non-canonical tryptic cleavage. The data were searched against the Mouse Reviewed (170321) database (24,981 entries). False discovery rate (FDR) estimation was enabled and results filtered at the 1% FDR level. Peaks denovo scoring (ALC%) threshold minimum was ≥80, peptide hit threshold (−10logP) was set ≥25.6, and protein hit threshold (−10logP) was set ≥20.
2.9. Isolation of cardiac myocytes and exposure to HNE
Mouse cardiac myocytes were isolated by using modified Langendorff perfusion and collagenase digestion, as described previously [44]. Briefly, the mouse hearts were excised rinsed with physiological saline and perfused with oxygenated (95% O2–5% CO2), Ca2+-free modified Tyrode bicarbonate buffer (buffer A in mM: NaCl 126, KCl 4.4, MgCl2 1.0, NaHCO3 18, glucose 11, HEPES 4, 2,3-butanedione monoxime 10, taurine 30, pH 7.35) at 37°C for 5 min. The extracellular matrix was digested by buffer containing buffer A with Liberase Blendzyme type 1 0.25 mg/mL, 0.14 mg/mL trypsin in the 50 mL of recirculating digestion buffer (buffer A with 12.5 µM CaCl2) for 12–15 min. Hearts were separated in mincing buffer (10 mL digestion buffer with 9 mg/mL albumin, and Liberase) and cells were allowed to settle. CaCl2 was reintroduced in a graded fashion at 5-min intervals (five total steps) to sequentially increase the Ca2+ concentration to 500 µM. After the final step, isolated myocytes were superfused with 1, 5, 10 and 15 µM of HNE and cardiac myocytes were constantly monitored under the microscope at ×20 magnification. For inhibiting jun N-terminal kinase (JNK), isolated WT and AR-null cardiac myocytes were pretreated with or without JNK inhibitor (SP6000125; 25 µM for 30 min) prior to superfusion with HNE (15 µM for 40 min). After superfusion the cells were scraped in RIPA buffer and homogenates were immunoblotted and developed with LC3II, phospho-c-JNK (P-JNK) and JNK antibodies (Cell Signaling).
2.10. Adenoviral gene transfection and autophagosome formation in isolated cardiac myocytes
The adenovirus GFP-LC3 was generated as described previously [45]. Briefly, the GFP-LC3 plasmid (a gift from Prof. Roberta Gottlieb) was amplified in E. coli and excised from the backbone. The GFP-LC3 virus was developed at Vector Biolabs, (PA, USA) by cloning the GFP-LC3 into pAd5-dE1/E3 vector for viral packaging and subsequent amplification in HEK-293 cells. Adenoviruses were used at a multiplicity of infection (MOI) of 100 and GFP expression was used to ascertain the efficiency. Transduced cells were perfused with varying concentrations of HNE (10–20 µM) and LC3 punctae were visualized using an EVOS fluorescence microscope.
2.11. Statistical analysis
Data are mean±SEM. Sham and TAC groups were analyzed with one-way analysis of variance followed by Bonferroni test or Student’s t test. Statistical significance was accepted at p<0.05.
3. Results
3.1. TAC induces cardiac AR expression
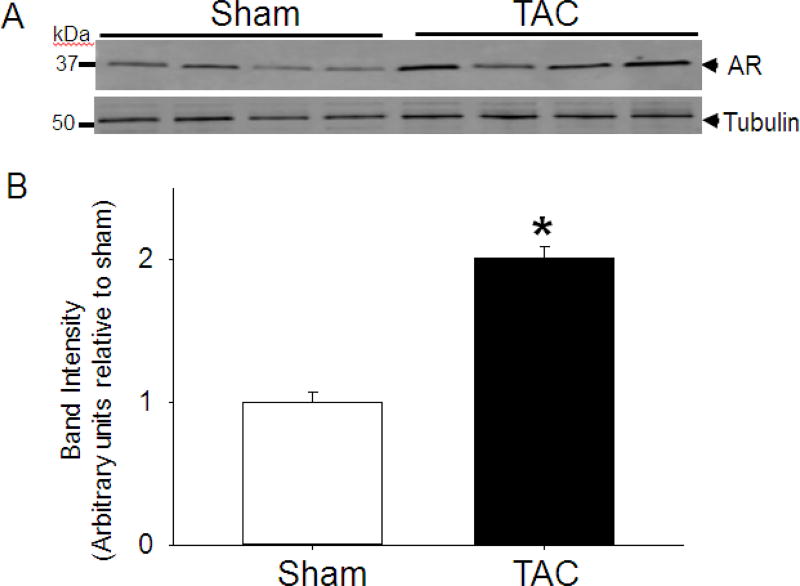
Previous work has shown an increase in mRNA levels of AR in myocardial tissue samples from patients with ischemic and dilated cardiomyopathy [37] and hence to determine whether the AR expression is also increased in murine model of heart failure, we subjected WT C57 mice to TAC and quantified the levels of LV AR protein expression. Our results showed that in comparison with sham-operated mice, there was a nearly 2-fold increase in LV AR protein abundance in mice subjected to TAC for 2 weeks (Fig.1). This increase in AR protein suggests that this may be a compensatory mechanism to attenuate aldehyde toxicity in pathological hypertrophy. To examine this possibility, we subjected WT and AR-null mice to TAC.
Fig. 1. Transverse aortic constriction (TAC) increase the abundance of aldose reductase in the heart.
(A) Representative Western blots of tissue homogenates prepared from the left ventricles of WT mice hearts that were subjected to TAC for 2 weeks. AR was detected using the anti-AR antibody. (B) Bar graph shows the intensity of immunopositive bands normalized to tubulin. Data are mean ± SEM *p<0.05 vs sham; n=4 mice.
3.2. AR deletion exacerbates TAC induced pathological cardiac hypertrophy
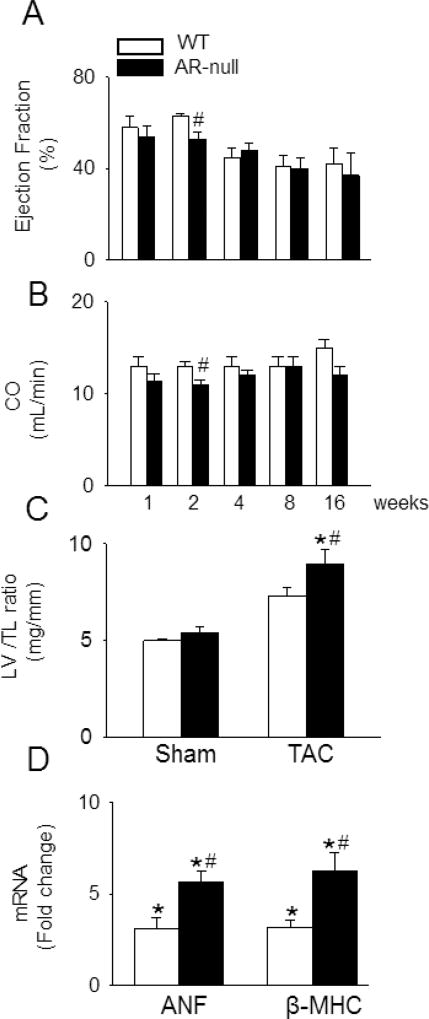
AR-null mice bred normally, maintained normal body weight, showed normal growth behavior and have normal heart weight and heart weight/tibia length ratio; however, in comparison with WT mice, they have slightly lower ejection fraction (age 14–16 weeks; 63±1% vs 69±1%; Supplemental Table I). To examine whether deletion of AR exacerbates TAC induced cardiac dysfunction, we performed serial echocardiography at 1, 2, 4, 8 and 16 weeks after TAC surgery. Echocardiography of WT mice 1 week after TAC surgery showed no difference in cardiac function in comparison with the sham-operated mice, but the ejection fraction in the AR-null TAC mice was reduced compared with the sham operated WT and AR-null mice (Table 1). No differences in cardiac function was observed between the WT and AR-null TAC mice hearts at this time point (Fig. 2A; Table 1). Echocardiography of WT mice at 2 weeks after TAC surgery showed no differences in LVIDd and ejection fraction, when compared with sham-operated mice (Table 1); however, as shown in Fig. 2A, in AR-null mice, TAC decreased ejection fraction to 53±3%, which was significantly lower than that measured in WT hearts subjected to TAC (64±2%) or sham-operated AR-null hearts (67±3%; Fig. 2A, Table 1). In the AR-null hearts, this LV dysfunction was also associated with decreased cardiac output (11.88±0.52 mL/min), which was lower than the cardiac output of WT mice subjected to TAC (13.45±0.47 mL/min; Fig. 2B). Associated with these changes, we also observed that the LV mass to tibia length ratio was significantly higher in WT mice subjected to TAC in comparison with sham-operated hearts, indicating that TAC induces cardiac hypertrophy in WT mice; however, this increase in LV mass to tibia length was more prominent in AR-null mice compared with WT TAC hearts (Fig. 2C). No change was observed in the right ventricular mass between the TAC and sham operated hearts. The expression of hypertrophic markers, ANP and β-MHC, was significantly higher in the AR-null hearts in comparison with WT hearts after 2 weeks of TAC (Fig. 2D). No difference in cardiac function between WT and AR-null mice was observed after 4, 8, or 16 weeks of TAC (Fig. 2 A, B; Supplemental Table I). Collectively, these data indicate that lack of AR exacerbates early cardiac dysfunction and hypertrophy in response to pressure overload.
Table 1. Echocardiography of WT and AR-null mice after 1 and 2 weeks of sham and TAC surgery.
Data are presented as mean ± SEM, n=12-15 mice per group.
| Sham | 1 week | TAC | 1 week | Sham | 2 weeks | TAC | 2 weeks | |
|---|---|---|---|---|---|---|---|---|
| WT | AR-null | WT | AR-null | WT | AR-null | WT | AR-null | |
| LVI Dd (mm) | 3.87±0.01 | 3.75±0.09 | 3.56±0.05 | 3.60±0.06 | 3.69±0.06 | 3.81±0.12 | 3.80±0.05 | 3.87±0.08 |
| LVI Ds (mm) | 2.29±0.53 | 2.18±0.13 | 2.35± 0.15 | 2.68±0.89 | 2.22±0.07 | 2.27±0.14 | 2.42±0.09 | 2.70±0.11 |
| End o EDV | 50±6 | 42±5 | 40±2 | 41±2 | 42±1 | 45±3 | 45±1 | 47±2 |
| End o ESV | 15±3 | 11±3 | 17±2 | 19±3 | 13±1 | 15±2 | 18±2 | 23±2 |
| EF (%) | 70±3 | 73±2 | 58±5 | 54±5*$ | 68±1 | 67±3 | 64±2 | 53±3*$# |
| Heart rate | 566±16 | 511±20 | 518±14 | 518±15 | 474±6 | 480±6 | 495±6 | 491±6 |
| SV | 35±3 | 31±4 | 23±3 | 19±2 | 29±1 | 29±2 | 27±1 | 24±1 |
| CO | 19.48±1.55 | 15.48±0.43 | 12.95±1.07 | 11.37±0.79 | 13.81±0.51 | 14.22±1.14 | 13.45±0.47 | 11.88±0.52# |
p<0.05 vs WT sham
p<0.05 vs WT TAC
p<0.05 vs AR-null sham.
Fig. 2. Deletion of AR exacerbates early contractile dysfunction in mice subjected to transverse aortic constriction.
WT and AR-null mice were subjected to TAC or sham surgery and long axis m-mode echocardiographic data were acquired at the indicated times after TAC. (A) Ejection fraction and (B) cardiac output (CO), at 1, 2, 4, 8 and 16 weeks after TAC. (C) Ratio of left ventricle (LV) to tibia length (TL), (D) fold change in ANF and β-MHC mRNA expression in LV of TAC hearts compared with WT sham after 2 weeks of surgery. Data are mean ± SEM, n=12–15 mice per group. *p<0.05 vs WT sham and #p<0.05 vs WT TAC.
3.3. Loss of AR impairs aldehyde removal
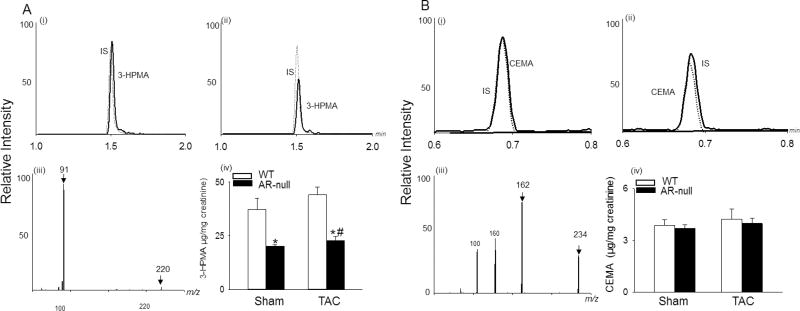
Because several studies have shown that AR detoxifies a wide range of reactive aldehydes [17, 31, 46–50], we next sought to determine whether the removal of aldehydes is impaired in AR-null mice. For this, we measured urinary metabolites of the lipid peroxidation product acrolein from WT and AR null mice that were subjected to 2 weeks of TAC. We found that urinary levels of the acrolein metabolite 3-HPMA, were significantly lower in AR-null mice than in WT mice, indicating that deletion of AR decreases glutathione-linked acrolein metabolism; however, TAC did not affect urinary levels of 3-HPMA. Levels of the ALDH2 - derived metabolite, CEMA [51] remained unchanged in WT and AR-null mice under basal and TAC conditons (Fig. 3 A, B). Similarly, there was no difference in the protein abundance of ALDH2 between WT and AR-null sham and TAC hearts (Supplemental Fig. I A, B).
Fig. 3. Deletion of AR prevents acrolein metabolism.
(A) Representative LC spectra of the acrolein metabolite, 3-hydroxypropyl mercapturic acid (3-HPMA; solid line), internal standard (IS, dotted line) detected in urine of (i) WT (ii) AR-null mice, and (iii) ESI+/ MS spectra of 3-HPMA. (B) Representative LC spectra of N-Acetyl-S-(2-carboxyethyl)-L-cystiene (CEMA; solid line), IS (dotted line) in (i) WT and (ii) AR-null mice urine, and (iii) ESI+/MS spectra of CEMA. (A, iv; B, iv) Bar graphs shows 3-HPMA and CEMA levels detected in the WT and AR-null urine. Data are mean ± SEM. *p<0.05 vs WT sham and #p<0.05 vs WT TAC, n=5–6 mice.
3.4. Increased accumulation of aldehyde-protein adducts in hypertrophic hearts
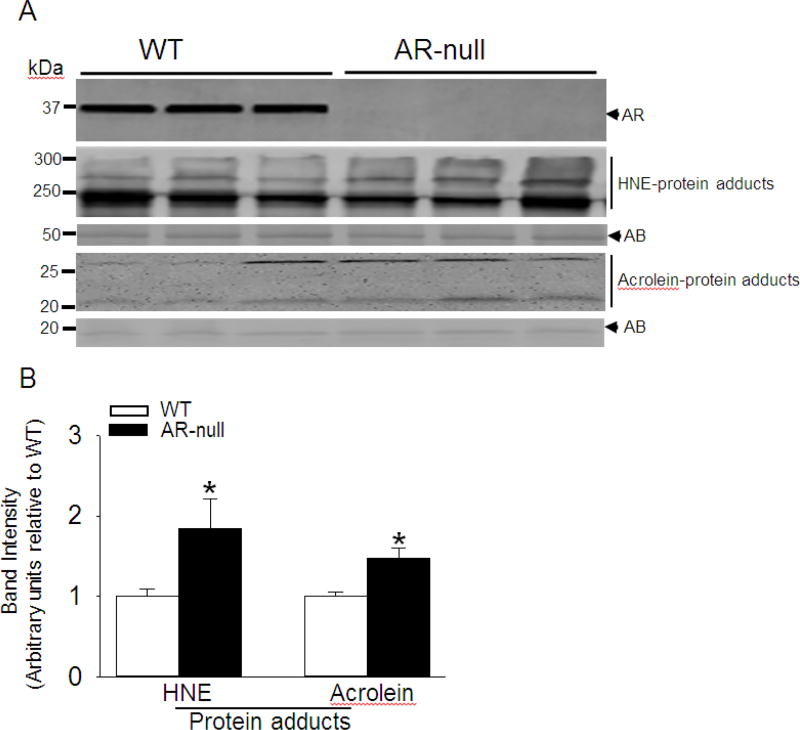
Because the removal of reactive aldehydes is impaired in AR-null mice and that increased levels of HNE or HNE-modified proteins have been detected in most models of cardiac hypertrophy or failing hearts [15–17], we next examined whether the lack of AR increases the levels of aldehyde-modified proteins, after 1 and 2 weeks of TAC. To assess the accumulation of aldehyde modified protein adducts, we performed Western blots of LV homogenates using antibodies that recognize proteins adducted to HNE or acrolein. As shown in Fig. 4, the accumulation of protein-HNE and protein-acrolein adducts were significantly higher at 2 weeks after TAC in the AR-null hearts compared with WT hearts, whereas no change in these proteins adducts was observed between WT and AR-null hearts at 1 week after TAC (Supplemental Fig. I, C, D, E, F). Taken together, these results indicate that loss of AR increases the accumulation of aldehyde-modified proteins in hearts at 2 weeks after TAC.
Fig. 4. Increased accumulation of protein adducts in aldose reductase (AR)-null hearts subjected to transverse aortic constriction (TAC).
(A) Representative Western blots developed from homogenates of the left ventricle of WT and AR-null mice subjected to TAC for 2 weeks. The blots were developed using anti-AR antibody, anti-HNE and anti-acrolein protein adduct antibodies as indicated. (B) Bar graphs showing the intensity of bands normalized to amido-black (AB). Data are mean ± SEM, *p<0.05 vs WT TAC, n=3–5 mice hearts.
3.5. Analysis of HNE-modified protein by LC-MS/MS
To identify which proteins were modified by HNE, proteins from AR-null TAC hearts were immunoprecipitated using anti-HNE protein adduct antibody. Immunoprecipitated proteins were subjected to trypsin digestion, and MS analysis was performed using LTQ Orbitrap ELITE mass spectrometer. MS analysis identified 182 proteins, which included functionally important proteins such as ATP synthase subunit α, glucose regulated protein 78 (GRP78), histones, electron transfer flavoprotein subunit alpha, and myosin-6/7 (Supplemental Table 2). Further analysis of the MS data identified 6 specific proteins that had a high confidence of lysine-HNE modified peptides, indicating direct binding of HNE to these proteins (Table 2). As shown in Supplemental Fig. 2 the fragmentation pattern of peptide in myosin-6/7 (MYH6/MYH7) had an unreduced HNE (m/z 156 Da) bound to the lysine residue of peptide. Similarly, reduced HNE was also found to be adducted with keratin type II cytoskeletal 1 (K2C1) proteins. Analysis of the collected data showed that 3 proteins; circadian protein homolog (PER2), gamma-2A chain C region (GCAB), and keratin type II cytoskeletal 4 (K2C4) had an unreduced HNE adduct (m/z 158 Da) bound to the lysine residues of peptide. In addition, we also identified a single dehydrated HNE (m/z 136 Da) adducted to the lysine on the C-terminal end of calcium/calmodulin-dependent 3′,5′-cyclic nucleotide phosphodiesterase 1C (PDE1C; Table 2). Together, these data support the notion that loss of AR is accompanied by increased accumulation of several aldehyde-modified cardiac proteins, such as those involved in energy regulation and protein folding.
Table 2. Proteins with high confidence of lysine-HNE in AR-null TAC hearts.
Site specific modification of lysine (k) residues by HNE are noted with bold letters and lower case.
| Sequence number |
Accession number |
Proteins | m/z | Peptide sequence |
|---|---|---|---|---|
| 1 | O54943 | Period circadian protein homolog 2 | 158 | GAGSSDTSHTSk |
| 2 | P01864 | Ig gamma-2A chain C region | 158 | YVLPPPAEEMTk |
| 3 | P07744 | Keratin,type II, cytoskeletal 4 | 158 | TAAENDFVVLk |
| 4 | Q02566/Q91Z83 | Myosin-6/Myosin-7 | 156 | SNAAAAALDk |
| 5 | P04104 | Keratin,type II cytoskeletal 1 | 156 | TNAENEFVTIk |
| 6 | Q64338 | Calcium/calmodulin-dependent 3’5’ nucleotide phosphodiesterase 1C | 138 | SQAEQGTTSKGEk |
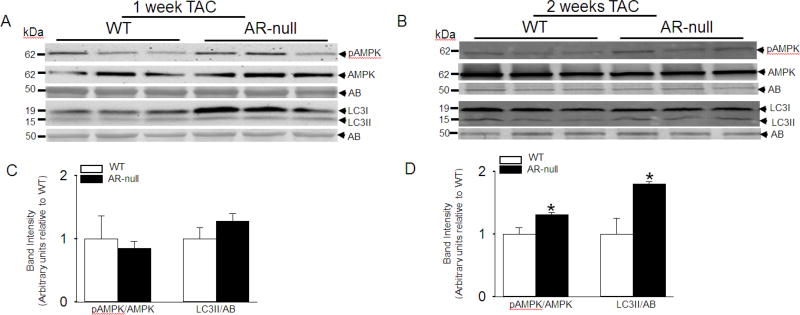
3.6. AR deletion enhances autophagy in TAC hearts
Because accumulation of aldehyde-modified proteins was increased in the hearts of AR-null mice subjected to TAC, we next investigated whether the accumulation of such proteins in TAC hearts could trigger autophagy. We measured the relative abundance of autophagy markers LC3II and pAMPK, in WT and AR-null hearts after 1 and 2 weeks of TAC. Our results show that there is no difference in LC3II and pAMPK levels in the hearts that were analyzed after 1 week of TAC (Fig. 5 A, C); however, we found that formation of LC3II was higher in AR-null than WT hearts after 2 weeks of TAC. Similarly, the AR-null hearts showed greater abundance of pAMPK than WT hearts after 2 weeks of TAC (Fig. 5 B, D). These results provide evidence that TAC induces a greater increase in autophagy in AR-null than WT hearts.
Fig. 5. Increased cardiac autophagy in AR-null hearts after pressure overload.
Representative immuno-blots of left ventricular homogenates from WT and AR-null hearts that were subjected to (A) 1 week (B) 2 weeks of transverse aortic constriction. The blots were developed by using, pAMPK, AMPK, LC3 antibodies and normalized to amido-black (AB) staining. Relative Band intensities of pAMPK normalized to AMPK and levels of LC3II normalized to AB after (C) 1 and (D) 2 weeks of TAC. Data are mean ± S.E.M. *p<0.05 vs WT TAC, n=3–4 mice.
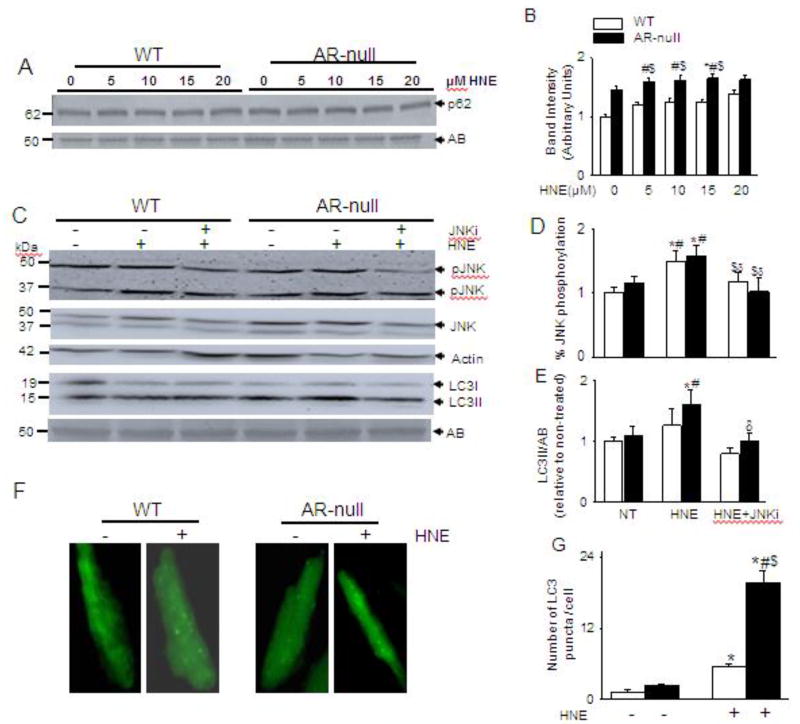
3.7. AR regulates autophagic flux
Because HNE-modified proteins accumulated in AR-null TAC hearts, and protein aggregates in cardiac myocytes stimulate autophagy by activating p62 [52], we next examined whether in the absence of TAC, HNE-protein adducts were sufficient to trigger p62 mediated autophagy. For this, we superfused isolated cardiac myocytes from WT and AR-null hearts with varying concentrations (5–20 µM) of HNE. As shown in Fig. 6 A and B, superfusion of AR-null cardiac myocytes with HNE for 40 min enhanced p62 levels compared with non-treated WT and AR-null cardiac myocytes. Previously, we had reported that HNE could also directly trigger autophagy by activating JNK [53]. To further validate this mechanism, cardiac myocytes from WT and AR-null mice were pretreated with JNK inhibitor (JNKi) SP600125 (25 µM) for 30 min and then superfused with 15 µM HNE for 40 min. The HNE superfusion significantly increased phosphorylation of JNK that was abrogated in the presence of JNKi in both the WT and AR-null cardiac myocytes (Fig. 6 C, D). To confirm that the absence of AR enhances autophagy, we also determined the LC3II formation. Our results show that superfusion of WT cardiac myocytes with HNE led to an increase in LC3II levels which did not reach to statistical significance; whereas AR-null cardiac myocytes superfused with the same dose of HNE showed a greater increase in the LC3II levels indicating that absence of AR results in higher levels of autophagy. Formation of LC3II in AR-null cardiac myocytes was decreased in the presence of JNKi, suggesting that HNE induced autophagy in AR-null cardiac myocytes, is partially, mediated via JNK activation (Fig. 6 C, E). Taken together, these observations suggest that the lack of AR potentiates HNE-induced autophagy, which is mediated by p62 and JNK activation.
Fig. 6. HNE-induced increase in autophagy is dependent on P62 and JNK.
Isolated cardiac myocytes from WT and AR-null mice were superfused with varying concentrations of HNE (5–20 µM) for 40 min and analyzed for (A) p62 levels. (C) Cardiac myocytes isolated from WT and AR-null cardiac myocytes were pretreated with or or without JNK inhibitor SP 600125 (25 µM for 30 min) and then superfused with HNE (15 µM) for 40 min. Cell lysate were immunoblotted with (C) pJNK and LC3 antibodies. (F) Representative images of isolated adult cardiac myocytes from WT and AR-null hearts transduced with Ad-GFP-LC3 and then perfused with 15 µM HNE for 60 min. (B, D, E,G) Data are mean ± SEM. *p < 0.05 vs non-treated WT, #p < 0.05 vs non-treated AR-null, $ p < 0.01 vs HNE-treated WT cardiac myocytes, δp<0.05 vs AR-null HNE treated cardiac myocytes, n=3–4 mice in each group.
Furthermore, to corroborate the role of AR in HNE-induced autophagosome formation, we transduced WT and AR-null isolated cardiac myocytes with AdGFP-LC3 and then treated them with HNE (15 µM) for 60 min. As shown in Fig.6 F and G, treatment with HNE led to a nearly 3-fold increase in the formation of discrete green punctae in WT cardiac myocytes. Notably, similar treatment of AR-null myocytes with HNE led to a nearly 20-fold increase in punctae formation.
4. Discussion
The major finding of this study is that metabolism of lipid peroxidation products, such as HNE, by AR is essential for maintenance of normal cardiac function and deficiency of AR exacerbates the early response to pressure overload induced cardiac dysfunction. Involvement of AR following TAC is supported by our results showing that AR is upregulated in response to TAC and that lack of AR accentuates TAC-induced LV hypertrophy. We found that the lack of AR was associated with decreased formation of 3-HPMA in AR-null mice, generation of aldehyde protein adducts, and excessive autophagy in TAC hearts. Our observations that deletion of AR increases p62 in HNE superfused cardiac myocytes suggests that HNE protein adducts could activate p62 mediated autophagy. Moreover, that inhibition of JNK decreased LC3II formation in AR-null cardiac myocytes superfused with HNE supports the notion that JNK may be essential for triggering HNE mediated autophagy. Taken together, these finding suggest that impaired removal of aldehydes in AR-null hearts could accentuate the effects of TAC-induced pathological remodeling. Collectively, these studies reveal a new axis that links aldehydes, autophagy, and pathological remodeling, and identifies AR as an important regulator of these processes.
AR plays a pivotal role in the metabolism and detoxification of reactive aldehydes [32]. The kinetic and structural properties of AR are well suited for the removal of a wide range of saturated and unsaturated aldehydes, phospholipid aldehydes [32], peptide-aldehyde conjugates [28], and glutathione-aldehyde conjugates [49, 54]. Most of these reactive aldehydes are downstream signaling molecules of oxidative stress, and, like their free radical precursors, are highly active. Therefore, reduction by AR could be considered a protective mechanism that prevents tissue injury, dysfunction, and damage induced by secondary products of lipid peroxidation. The expression of AR has been shown to be increased by oxidative stress induced in myocardial ischemia-reperfusion [36], atherogenesis [31], vasculitis [34], and restenosis [55]. In agreement with these observations, we found that AR expression was increased in TAC hearts, indicating that AR may be an important modulator of the toxic effects of aldehydes in hypertrophic hearts.
Our results show that the urinary levels of 3-HPMA were decreased in AR-null mice. The urinary metabolite, 3-HPMA, is derived from the conjugate of acrolein with glutathione, which is further reduced by AR and then metabolized to the mercapturic acids [56]. Recent work from our laboratory has shown that 3-HPMA levels in humans are positively associated with cardiovascular disease risk [57]. Therefore, reduction of acrolein-glutathione conjugates by AR may be an essential metabolic transformation to maintain cardiac function in the postnatal heart. Previous reports had shown that aldehyde dehydrogenase (ALDH2) activation or overexpression improves cardiac function during heart failure and ischemia/reperfusion injury by removal of reactive aldehydes [58] [59] [60]. Our observation that the levels of ALDH2 in the heart and it metabolite CEMA [51] in the urine were not different between WT and AR-null mice under either basal conditions or TAC, indicate that there were no compensatory changes in the ALDH2 pathway due to AR deletion.
Consistent with the detoxification capacity of AR, numerous studies have shown that the inhibition of AR augments the modification of proteins in inflamed arteries [34], ischemic hearts [36], atherosclerotic lesions [31], aged hearts [28], diabetic hearts [61], and adipocytes from obese mice [61, 62]. Extensive evidence suggests that the accumulation of oxidized products of lipids e.g., HNE [15–19, 63] and acrolein [20], is a significant feature of pathological hypertrophy and heart failure; however, a causal relationship between reactive aldehydes and cardiac dysfunction has not been established. Therefore, our observations showing that deletion of AR accentuates TAC-induced pathological remodeling and increases the accumulation of a wide range of aldehyde modified proteins indicates that the reactive aldehydes generated in hypertrophic hearts may contribute to its pathological remodeling; however, our observations that deficiency of AR exacerbates early stages of the hypertrophic response (Fig. 2), but does not affect the progression of TAC-induced cardiac dysfunction, indicates that the removal of these aldehydes may be essential during the early stages of pathological remodeling. Yet, at later stages, other compensatory mechanisms, such as removal of aldehydes via carnosine or other metabolic pathways, may be able to compensate for the loss of AR.
To further understand the potential contribution of HNE-modified proteins in pathological remodeling, we used an unbiased global analysis to identify which proteins are targeted by HNE in AR-null mice during pathological remodeling. We found that an extensive set of myocardial proteins were modified in these hearts. In particular, the modification of glucose regulated protein (GRP78) and elongation factor 1-alpha 2 (elf2-α) may be particularly significant. These proteins play a pivotal role to maintain proper protein folding during unfolded protein response [64]. Previously, we showed that these proteins are highly vulnerable to HNE modification [53] and our current results further extend these findings. Hence, modification of these proteins may be harmful not only by disrupting their functions, but also by affecting protein folding pathways. In the current study, we also identified mitochondrial proteins, such as isocitrate dehydrogenase (ICDH) are adducted with HNE. This enzyme is predominantly confined to cardiac myocytes [65–67] and catalyzes reversible interconversion between isocitrate and α-ketoglutarate [68]. It has been reported that HNE adduction of ICDH is associated with pathophysiological events that could lead to cardiac hypertrophy [16]. Similarly, we found that ATP synthase is modified by HNE in AR-null TAC hearts. Previously, Zhao et al [69] reported that the activity of ATP synthase is reduced on adduction with HNE in doxorubicin treated mice hearts. Thus, we could speculate that HNE modification of ATP synthase in AR-null TAC hearts could deplete myocardial ATP during cardiac remodeling and exacerbate contractile dysfunction in AR-null mice after TAC. Based on these observations it appears that proteins modified by lipid peroxidation products could trigger pathways contributing to pathological cardiac remodeling.
Increased generation of lipid peroxidation-derived aldehydes in the hypertrophic hearts could exacerbate myocardial injury or dysfunction by a variety of mechanisms such as glutathione and carnosine depletion; however, a key feature of their toxicity may be widespread modification of myocardial proteins. Yet, the mechanisms by which aldehyde modified protein adducts could trigger pathological cardiac remodeling remains unclear. It has been reported that misfolded proteins in cardiac myocytes upregulates transcript and proteins levels of p62, which acts as a downstream effector of autophagy [52]. Similarly, we had shown that autophagy is the significant pathway for the removal of HNE-modified proteins in vascular smooth muscle cells [24]. Autophagy is a highly regulated and a well-studied feature of cardiac hypertrophy. In the heart, autophagy is essential for maintaining normal cardiac function under basal conditions, but under stress, excessive or defective autophagy could lead to cardiac abnormalities [70]. The reasons for such divergent outcomes remain unclear, in part because triggers and mediators of autophagy under basal and stressed conditions have not been identified. Nevertheless, whether aldehyde-modified proteins trigger autophagy during pathological remodeling in cardiac tissues is not clearly defined. Our observations showing that the lack of AR increases autophagy as well as the accumulation of aldehyde-modified proteins in the heart are consistent with the view that extensive modification of proteins and lipids by secondary products of lipid peroxidation may be a significant trigger of autophagy in stressed hearts. Furthermore, our results showing that p62 and LC3II levels were increased in HNE- superfused AR-null cardiac myocytes suggests that p62 is the downstream signaling mechanism by which HNE protein adducts could induce autophagy.
Previously, we had reported that HNE treatment activated autophagy even in unstressed cells and pharmacological inhibition of JNK prevented LC3II formation [24]. Consistent with these observations, our results demonstrate that HNE superfusion activates JNK in WT and AR-null cardiac myocytes and increases LC3II formation in AR-null cardiac myocytes. Pharmacological inhibition of JNK decreased HNE-induced autophagy in AR-null cardiac myocytes. Taken together, these observations suggest that both HNE-modified proteins and HNE could trigger autophagy, which in part is mediated by p62 and JNK, respectively. Although additional work is required to fully understand the role of lipid peroxidation in myocardial autophagy, our current observation that HNE triggers autophagy and deletion of AR increases protein-HNE accumulation and autophagy in hypertrophic hearts, supports the view that accumulation of these aldehydes may be an important mechanism for the stimulation of autophagy in stressed hearts. Our results also suggest that excessive autophagy could have deleterious consequences in the stressed heart. We found that increased autophagy in AR-null hearts was associated with diminished cardiac function and increased pathological remodeling, suggesting that excessive autophagy may be maladaptive in hypertrophic hearts. Further work is required to identify if other antioxidant pathways that prevent ROS generation or promote the removal of lipid peroxidation products have similar effects on myocardial remodeling and dysfunction in heart failure.
5. Conclusion
In conclusion, deficiency of AR promotes pathological remodeling in hearts subjected to pressure overload. The deleterious effects of AR deletion could be attributed, at least in part, to an increase in the accumulation of lipid peroxidation products such as HNE, aldehyde modified protein adducts, and an increase in autophagy. Overall, our findings support the view that reactive aldehydes generated by lipid peroxidation could contribute to the development of LV hypertrophy and maladaptive autophagy, and AR is a critical component of maintaining normal cardiac function.
Supplementary Material
Supplemental Fig. I Expression of aldehyde dehydrogenase (ALDH2) and protein adducts in WT and aldose reductase (AR)-null mice hearts. (A) Representative Western blots of ALDH2 expression in left ventricles of WT and AR-null mice after 2 weeks of sham or TAC surgery. Representative Western blots of (C) HNE and (E) acrolein protein adducts in left ventricles of WT and AR-null hearts after 1 week of TAC. (B, D, F) Bar graphs showing the intensity of bands normalized to amido-black (AB). Data are mean ± SEM, n=3–5 mice hearts.
Supplemental Fig. II Mass spectrometry-based identification of HNE modified peptide derived common to myosin-6 (MYH6) and myosin-7 (MYH7). Representative fragmentation spectrum for the +2-charged ion with a monoisotopic m/z of 544.2903 millimass unit (mmu) was observed using collision induced dissociation in the linear ion trap of an Orbtrap ELITE with the Orbitrap operating at maximum resolution (240,000 (FWHM) at m/z 400). Analysis of the data by Peaks Studio (v7.5) identified a peptide (−10logP score = 34.29) with the sequence SNAAAAALDk (+156.12) and a MH+ of 1087.5992 at a precursor error of <25 ppm. Fragmentation of the peptide yielded a combined continuous b- and y- ion cover of the peptide amino acid sequence including the HNE modified lysine denoted ask (+156.12).
Supplemental Table I: Serial echocardiography data of WT and AR-null mice. Echocardiographic parameters under baseline and after 4, 8 and 16 weeks of TAC surgery. Data are mean ± SEM, n=12–15 mice per group. #p<0.05 vs WT and *p<0.05 vs WT sham.
Supplemental Table II: Proteins immuno-precipitated from AR-null TAC hearts by the anti-HNE protein adduct antibody. Immunoprecipitated proteins were identified by LC-LTQ-Orbitrap-ELITE mass spectrometer.
Highlights.
Aldose reductase expression, aldehydes and autophagy are enhanced in heart during early pressure overload.
Acknowledgments
We would like to thank Linda Harrison for expert technical assistance.
Funding Sources
This work was supported by grants from the NIH (R01 HL083320, R01 HL094419, R01 HL131647, P01 HL078825, R01 HL122581, R01 HL55477, P20 GM103492) and the American Heart Association (BGIA7720034).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None.
References
- 1.Writing Group M. Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, et al. American Heart Association Statistics C., Stroke Statistics S., Heart Disease and Stroke Statistics-2016 Update: A Report From the American Heart Association. Circulation. 2016;133(4):e38–360. doi: 10.1161/CIR.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 2.Roger VL, Go AS, Lloyd-Jones DM, Adams RJ, Berry JD, Brown TM, et al. American Heart Association Statistics C., Stroke Statistics S., Heart disease and stroke statistics--2011 update: a report from the American Heart Association. Circulation. 2011;123(4):e18–e209. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giordano FJ. Oxygen, oxidative stress, hypoxia, and heart failure. J Clin Invest. 2005;115(3):500–8. doi: 10.1172/JCI200524408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawyer DB, Siwik DA, Xiao L, Pimentel DR, Singh K, Colucci WS. Role of oxidative stress in myocardial hypertrophy and failure. J Mol Cell Cardiol. 2002;34(4):379–88. doi: 10.1006/jmcc.2002.1526. [DOI] [PubMed] [Google Scholar]
- 5.Hennekens CH, Buring JE, Manson JE, Stampfer M, Rosner B, Cook NR, et al. Lack of effect of long-term supplementation with beta carotene on the incidence of malignant neoplasms and cardiovascular disease. N Engl J Med. 1996;334(18):1145–9. doi: 10.1056/NEJM199605023341801. [DOI] [PubMed] [Google Scholar]
- 6.Yusuf S, Dagenais G, Pogue J, Bosch J, Sleight P. Vitamin E supplementation and cardiovascular events in high-risk patients. The Heart Outcomes Prevention Evaluation Study Investigators. N Engl J Med. 2000;342(3):154–60. doi: 10.1056/NEJM200001203420302. [DOI] [PubMed] [Google Scholar]
- 7.Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact. 2006;160(1):1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Marnett LJ. Lipid peroxidation-DNA damage by malondialdehyde. Mutat Res. 1999;424(1–2):83–95. doi: 10.1016/s0027-5107(99)00010-x. [DOI] [PubMed] [Google Scholar]
- 9.Siems WG, Grune T, Esterbauer H. 4-Hydroxynonenal formation during ischemia and reperfusion of rat small intestine. Life Sci. 1995;57(8):785–9. doi: 10.1016/0024-3205(95)02006-5. [DOI] [PubMed] [Google Scholar]
- 10.Stadtman ER. Role of oxidant species in aging. Curr Med Chem. 2004;11(9):1105–12. doi: 10.2174/0929867043365341. [DOI] [PubMed] [Google Scholar]
- 11.Halliwell BGJ. Oxford University Press. New York: 1999. [Google Scholar]
- 12.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11(1):81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 13.Awashti YC. Toxicology of Glutathione Transferases. Taylor & Francis; Galveston, TX: 2007. [Google Scholar]
- 14.Singh M, Kapoor A, Bhatnagar A. Oxidative and reductive metabolism of lipid-peroxidation derived carbonyls. Chemico-biological interactions. 2015;234:261–73. doi: 10.1016/j.cbi.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato Y, Iwase M, Ichihara S, Kanazawa H, Hashimoto K, Noda A, et al. Beneficial effects of growth hormone-releasing peptide on myocardial oxidative stress and left ventricular dysfunction in dilated cardiomyopathic hamsters. Circulation journal : official journal of the Japanese Circulation Society. 2010;74(1):163–70. doi: 10.1253/circj.cj-09-0378. [DOI] [PubMed] [Google Scholar]
- 16.Benderdour M, Charron G, DeBlois D, Comte B, Des Rosiers C. Cardiac mitochondrial NADP+-isocitrate dehydrogenase is inactivated through 4-hydroxynonenal adduct formation: an event that precedes hypertrophy development. J Biol Chem. 2003;278(46):45154–9. doi: 10.1074/jbc.M306285200. [DOI] [PubMed] [Google Scholar]
- 17.Srivastava S, Chandrasekar B, Guo Y, Luo J, Hamid T, Hill BG, et al. Downregulation of CuZn-superoxide dismutase contributes to beta-adrenergic receptor-mediated oxidative stress in the heart. Cardiovascular research. 2007;74(3):445–55. doi: 10.1016/j.cardiores.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 18.Zhang P, Xu X, Hu X, van Deel ED, Zhu G, Chen Y. Inducible nitric oxide synthase deficiency protects the heart from systolic overload-induced ventricular hypertrophy and congestive heart failure. Circ Res. 2007;100(7):1089–98. doi: 10.1161/01.RES.0000264081.78659.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu YH, Carretero OA, Cingolani OH, Liao TD, Sun Y, Xu J, et al. Role of inducible nitric oxide synthase in cardiac function and remodeling in mice with heart failure due to myocardial infarction. American journal of physiology. Heart and circulatory physiology. 2005;289(6):H2616–23. doi: 10.1152/ajpheart.00546.2005. [DOI] [PubMed] [Google Scholar]
- 20.Vasilyev N, Williams T, Brennan ML, Unzek S, Zhou X, Heinecke JW. Myeloperoxidase-generated oxidants modulate left ventricular remodeling but not infarct size after myocardial infarction. Circulation. 2005;112(18):2812–20. doi: 10.1161/CIRCULATIONAHA.105.542340. [DOI] [PubMed] [Google Scholar]
- 21.Lu Z, Xu X, Hu X, Lee S, Traverse JH, Zhu G, et al. Oxidative stress regulates left ventricular PDE5 expression in the failing heart. Circulation. 2010;121(13):1474–83. doi: 10.1161/CIRCULATIONAHA.109.906818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura K, Kusano K, Nakamura Y, Kakishita M, Ohta K, Nagase S, et al. Carvedilol decreases elevated oxidative stress in human failing myocardium. Circulation. 2002;105(24):2867–71. doi: 10.1161/01.cir.0000018605.14470.dd. [DOI] [PubMed] [Google Scholar]
- 23.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill BG, Haberzettl P, Ahmed Y, Srivastava S, Bhatnagar A. Unsaturated lipid peroxidation-derived aldehydes activate autophagy in vascular smooth-muscle cells. Biochem J. 2008;410(3):525–34. doi: 10.1042/BJ20071063. [DOI] [PubMed] [Google Scholar]
- 25.Sun A, Cheng Y, Zhang Y, Zhang Q, Wang S, Tian S, et al. Aldehyde dehydrogenase 2 ameliorates doxorubicin-induced myocardial dysfunction through detoxification of 4-HNE and suppression of autophagy. J Mol Cell Cardiol. 2014;71:92–104. doi: 10.1016/j.yjmcc.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava S, Chandra A, Bhatnagar A, Srivastava SK, Ansari NH. Lipid peroxidation product, 4-hydroxynonenal and its conjugate with GSH are excellent substrates of bovine lens aldose reductase. Biochem Biophys Res Commun. 1995;217(3):741–6. doi: 10.1006/bbrc.1995.2835. [DOI] [PubMed] [Google Scholar]
- 27.Barski OA, Tipparaju SM, Bhatnagar A. The aldo-keto reductase superfamily and its role in drug metabolism and detoxification. Drug Metab Rev. 2008;40(4):553–624. doi: 10.1080/03602530802431439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baba SP, Hoetker JD, Merchant M, Klein JB, Cai J, Barski OA, et al. Role of aldose reductase in the metabolism and detoxification of carnosine-acrolein conjugates. J Biol Chem. 2013;288(39):28163–79. doi: 10.1074/jbc.M113.504753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivastava S, Chandra A, Ansari NH, Srivastava SK, Bhatnagar A. Identification of cardiac oxidoreductase(s) involved in the metabolism of the lipid peroxidation-derived aldehyde-4-hydroxynonenal. Biochem J. 1998;329(Pt 3):469–75. doi: 10.1042/bj3290469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srivastava S, Chandrasekar B, Bhatnagar A, Prabhu SD. Lipid peroxidation-derived aldehydes and oxidative stress in the failing heart: role of aldose reductase. Am J Physiol Heart Circ Physiol. 2002;283(6):H2612–9. doi: 10.1152/ajpheart.00592.2002. [DOI] [PubMed] [Google Scholar]
- 31.Srivastava S, Vladykovskaya E, Barski OA, Spite M, Kaiserova K, Petrash JM, et al. Aldose reductase protects against early atherosclerotic lesion formation in apolipoprotein E-null mice. Circ Res. 2009;105(8):793–802. doi: 10.1161/CIRCRESAHA.109.200568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivastava SK, Ramana KV, Bhatnagar A. Role of aldose reductase and oxidative damage in diabetes and the consequent potential for therapeutic options. Endocr Rev. 2005;26(3):380–92. doi: 10.1210/er.2004-0028. [DOI] [PubMed] [Google Scholar]
- 33.Yadav UC, Ramana KV, Awasthi YC, Srivastava SK. Glutathione level regulates HNE-induced genotoxicity in human erythroleukemia cells. Toxicol Appl Pharmacol. 2008;227(2):257–64. doi: 10.1016/j.taap.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rittner HL, Hafner V, Klimiuk PA, Szweda LI, Goronzy JJ, Weyand CM. Aldose reductase functions as a detoxification system for lipid peroxidation products in vasculitis. J Clin Invest. 1999;103(7):1007–13. doi: 10.1172/JCI4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hill BG, Awe SO, Vladykovskaya E, Ahmed Y, Liu SQ, Bhatnagar A, et al. Myocardial ischaemia inhibits mitochondrial metabolism of 4-hydroxy-trans-2-nonenal. Biochem J. 2009;417(2):513–24. doi: 10.1042/BJ20081615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shinmura K, Bolli R, Liu SQ, Tang XL, Kodani E, Xuan YT, et al. Aldose reductase is an obligatory mediator of the late phase of ischemic preconditioning. Circ Res. 2002;91(3):240–6. doi: 10.1161/01.res.0000029970.97247.57. [DOI] [PubMed] [Google Scholar]
- 37.Yang J, Moravec CS, Sussman MA, DiPaola NR, Fu D, Hawthorn L, et al. Decreased SLIM1 expression and increased gelsolin expression in failing human hearts measured by high-density oligonucleotide arrays. Circulation. 2000;102(25):3046–52. doi: 10.1161/01.cir.102.25.3046. [DOI] [PubMed] [Google Scholar]
- 38.Facundo HT, Brainard RE, Watson LJ, Ngoh GA, Hamid T, Prabhu SD, et al. O-GlcNAc signaling is essential for NFAT-mediated transcriptional reprogramming during cardiomyocyte hypertrophy, American journal of physiology. Heart and circulatory physiology. 2012;302(10):H2122–30. doi: 10.1152/ajpheart.00775.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watson LJ, Facundo HT, Ngoh GA, Ameen M, Brainard RE, Lemma KM. O-linked beta-N-acetylglucosamine transferase is indispensable in the failing heart. Proc Natl Acad Sci U S A. 2010;107(41):17797–802. doi: 10.1073/pnas.1001907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sansbury BE, DeMartino AM, Xie Z, Brooks AC, Brainard RE, Watson LJ, et al. Metabolomic analysis of pressure-overloaded and infarcted mouse hearts, Circulation. Heart failure. 2014;7(4):634–42. doi: 10.1161/CIRCHEARTFAILURE.114.001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conklin DJ, Guo Y, Jagatheesan G, Kilfoil PJ, Haberzettl P, Hill BG, et al. Velayutham M, Zweier JL, Hoetker JD, Riggs DW, Srivastava S, Bolli R, Bhatnagar A. Genetic Deficiency of Glutathione S-Transferase P Increases Myocardial Sensitivity to Ischemia-Reperfusion Injury. Circ Res. 2015;117(5):437–49. doi: 10.1161/CIRCRESAHA.114.305518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conklin DJ, Haberzettl P, Jagatheesan G, Baba S, Merchant ML, Prough RA, et al. Glutathione S-transferase P protects against cyclophosphamide-induced cardiotoxicity in mice. Toxicol Appl Pharmacol. 2015;285(2):136–48. doi: 10.1016/j.taap.2015.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hobeika L, Barati MT, Caster DJ, McLeish KR, Merchant ML. Characterization of glomerular extracellular matrix by proteomic analysis of laser-captured microdissected glomeruli. Kidney Int. 2017;91(2):501–511. doi: 10.1016/j.kint.2016.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keith RJ, Haberzettl P, Vladykovskaya E, Hill BG, Kaiserova K, Srivastava S, et al. Aldose reductase decreases endoplasmic reticulum stress in ischemic hearts. Chem Biol Interact. 2009;178(1–3):242–9. doi: 10.1016/j.cbi.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salabei JK, Cummins TD, Singh M, Jones SP, Bhatnagar A, Hill BG. PDGF-mediated autophagy regulates vascular smooth muscle cell phenotype and resistance to oxidative stress. Biochem J. 2013;451(3):375–88. doi: 10.1042/BJ20121344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Srivastava S, Chandra A, Wang LF, Seifert WE, Jr, DaGue BB, Ansari NH, et al. Metabolism of the lipid peroxidation product, 4-hydroxy-trans-2-nonenal, in isolated perfused rat heart. J Biol Chem. 1998;273(18):10893–900. doi: 10.1074/jbc.273.18.10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Srivastava S, Dixit BL, Cai J, Sharma S, Hurst HE, Bhatnagar A, Srivastava SK. Metabolism of lipid peroxidation product, 4-hydroxynonenal (HNE) in rat erythrocytes: role of aldose reductase. Free Radic Biol Med. 2000;29(7):642–51. doi: 10.1016/s0891-5849(00)00351-8. [DOI] [PubMed] [Google Scholar]
- 48.Srivastava S, Harter TM, Chandra A, Bhatnagar A, Srivastava SK, Petrash JM. Kinetic studies of FR-1, a growth factor-inducible aldo-keto reductase. Biochemistry. 1998;37(37):12909–17. doi: 10.1021/bi9804333. [DOI] [PubMed] [Google Scholar]
- 49.Srivastava S, Watowich SJ, Petrash JM, Srivastava SK, Bhatnagar A. Structural and kinetic determinants of aldehyde reduction by aldose reductase. Biochemistry. 1999;38(1):42–54. doi: 10.1021/bi981794l. [DOI] [PubMed] [Google Scholar]
- 50.Srivastava SK, Ramana KV, Bhatnagar A. Role of aldose reductase and oxidative damage in diabetes and the consequent potential for therapeutic options. Endocr Rev. 2005;26(3):380–92. doi: 10.1210/er.2004-0028. [DOI] [PubMed] [Google Scholar]
- 51.Abraham K, Andres S, Palavinskas R, Berg K, Appel KE, Lampen A. Toxicology and risk assessment of acrolein in food. Mol Nutr Food Res. 2011;55(9):1277–90. doi: 10.1002/mnfr.201100481. [DOI] [PubMed] [Google Scholar]
- 52.Zheng Q, Su H, Ranek MJ, Wang X. Autophagy and p62 in cardiac proteinopathy. Circ Res. 2011;109(3):296–308. doi: 10.1161/CIRCRESAHA.111.244707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haberzettl P, Hill BG. Oxidized lipids activate autophagy in a JNK-dependent manner by stimulating the endoplasmic reticulum stress response. Redox Biol. 2013;1:56–64. doi: 10.1016/j.redox.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramana KV, Dixit BL, Srivastava S, Balendiran GK, Srivastava SK, Bhatnagar A. Selective recognition of glutathiolated aldehydes by aldose reductase. Biochemistry. 2000;39(40):12172–80. doi: 10.1021/bi000796e. [DOI] [PubMed] [Google Scholar]
- 55.Ruef J, Liu SQ, Bode C, Tocchi M, Srivastava S, Runge MS, Bhatnagar A. Involvement of aldose reductase in vascular smooth muscle cell growth and lesion formation after arterial injury. Arterioscler Thromb Vasc Biol. 2000;20(7):1745–52. doi: 10.1161/01.atv.20.7.1745. [DOI] [PubMed] [Google Scholar]
- 56.Parent RA, Paust DE, Schrimpf MK, Talaat RE, Doane RA, Caravello HE, et al. Metabolism and distribution of [2,3-14C]acrolein in Sprague-Dawley rats. II. Identification of urinary and fecal metabolites. Toxicol Sci. 1998;43(2):110–20. doi: 10.1006/toxs.1998.2462. [DOI] [PubMed] [Google Scholar]
- 57.DeJarnett N, Yeager R, Conklin DJ, Lee J, O'Toole TE, McCracken J, et al. Residential Proximity to Major Roadways Is Associated With Increased Levels of AC133+ Circulating Angiogenic Cells. Arterioscler Thromb Vasc Biol. 2015;35(11):2468–77. doi: 10.1161/ATVBAHA.115.305724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gomes KM, Campos JC, Bechara LR, Queliconi B, Lima VM, Disatnik MH, et al. Aldehyde dehydrogenase 2 activation in heart failure restores mitochondrial function and improves ventricular function and remodelling. Cardiovascular research. 2014;103(4):498–508. doi: 10.1093/cvr/cvu125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen CH, Budas GR, Churchill EN, Disatnik MH, Hurley TD, Mochly-Rosen D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321(5895):1493–5. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Endo J, Sano M, Katayama T, Hishiki T, Shinmura K, Morizane S, et al. Metabolic remodeling induced by mitochondrial aldehyde stress stimulates tolerance to oxidative stress in the heart. Circ Res. 2009;105(11):1118–27. doi: 10.1161/CIRCRESAHA.109.206607. [DOI] [PubMed] [Google Scholar]
- 61.Baba SP, Barski OA, Ahmed Y, O'Toole TE, Conklin DJ, Bhatnagar A, et al. Reductive metabolism of AGE precursors: a metabolic route for preventing AGE accumulation in cardiovascular tissue. Diabetes. 2009;58(11):2486–97. doi: 10.2337/db09-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baba SP, Hellmann J, Srivastava S, Bhatnagar A. Aldose reductase (AKR1B3) regulates the accumulation of advanced glycosylation end products (AGEs) and the expression of AGE receptor (RAGE) Chem Biol Interact. 2011;191(1–3):357–63. doi: 10.1016/j.cbi.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jungsuwadee P, Cole MP, Sultana R, Joshi G, Tangpong J, Butterfield DA, et al. Increase in Mrp1 expression and 4-hydroxy-2-nonenal adduction in heart tissue of Adriamycin-treated C57BL/6 mice. Molecular cancer therapeutics. 2006;5(11):2851–60. doi: 10.1158/1535-7163.MCT-06-0297. [DOI] [PubMed] [Google Scholar]
- 64.Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115(10):2656–64. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang L, Luo H, Vinay P, Wu J. Molecular cloning of the cDNA of mouse mitochondrial NADP-dependent isocitrate dehydrogenase and the expression of the gene during lymphocyte activation. J Cell Biochem. 1996;60(3):400–10. doi: 10.1002/(SICI)1097-4644(19960301)60:3%3C400::AID-JCB11%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 66.Jo SH, Son MK, Koh HJ, Lee SM, Song IH, Kim YO, et al. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J Biol Chem. 2001;276(19):16168–76. doi: 10.1074/jbc.M010120200. [DOI] [PubMed] [Google Scholar]
- 67.Haraguchi CM, Mabuchi T, Yokota S. Localization of a mitochondrial type of NADP-dependent isocitrate dehydrogenase in kidney and heart of rat: an immunocytochemical and biochemical study. J Histochem Cytochem. 2003;51(2):215–26. doi: 10.1177/002215540305100210. [DOI] [PubMed] [Google Scholar]
- 68.Thorsness PE, Koshland DE., Jr Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J Biol Chem. 1987;262(22):10422–5. [PubMed] [Google Scholar]
- 69.Zhao Y, Miriyala S, Miao L, Mitov M, Schnell D, Dhar SK, et al. Redox proteomic identification of HNE-bound mitochondrial proteins in cardiac tissues reveals a systemic effect on energy metabolism after doxorubicin treatment. Free Radic Biol Med. 2014;72:55–65. doi: 10.1016/j.freeradbiomed.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rothermel BA, Hill JA. Myocyte autophagy in heart disease: friend or foe? Autophagy. 2007;3(6):632–4. doi: 10.4161/auto.4913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. I Expression of aldehyde dehydrogenase (ALDH2) and protein adducts in WT and aldose reductase (AR)-null mice hearts. (A) Representative Western blots of ALDH2 expression in left ventricles of WT and AR-null mice after 2 weeks of sham or TAC surgery. Representative Western blots of (C) HNE and (E) acrolein protein adducts in left ventricles of WT and AR-null hearts after 1 week of TAC. (B, D, F) Bar graphs showing the intensity of bands normalized to amido-black (AB). Data are mean ± SEM, n=3–5 mice hearts.
Supplemental Fig. II Mass spectrometry-based identification of HNE modified peptide derived common to myosin-6 (MYH6) and myosin-7 (MYH7). Representative fragmentation spectrum for the +2-charged ion with a monoisotopic m/z of 544.2903 millimass unit (mmu) was observed using collision induced dissociation in the linear ion trap of an Orbtrap ELITE with the Orbitrap operating at maximum resolution (240,000 (FWHM) at m/z 400). Analysis of the data by Peaks Studio (v7.5) identified a peptide (−10logP score = 34.29) with the sequence SNAAAAALDk (+156.12) and a MH+ of 1087.5992 at a precursor error of <25 ppm. Fragmentation of the peptide yielded a combined continuous b- and y- ion cover of the peptide amino acid sequence including the HNE modified lysine denoted ask (+156.12).
Supplemental Table I: Serial echocardiography data of WT and AR-null mice. Echocardiographic parameters under baseline and after 4, 8 and 16 weeks of TAC surgery. Data are mean ± SEM, n=12–15 mice per group. #p<0.05 vs WT and *p<0.05 vs WT sham.
Supplemental Table II: Proteins immuno-precipitated from AR-null TAC hearts by the anti-HNE protein adduct antibody. Immunoprecipitated proteins were identified by LC-LTQ-Orbitrap-ELITE mass spectrometer.