Abstract
The FCGR3A gene encodes for the receptor important for antibody-dependent natural killer cell-mediated cytotoxicity. FCGR3A gene polymorphisms could affect the success of monoclonal antibody therapy. Although polymorphisms, such as the FcγRIIIA-V158F and -48L/R/H, have been studied extensively, an overview of other polymorphisms within this gene is lacking. To provide an overview of FCGR3A polymorphisms, we analysed the 1000 Genomes project database and found a total of 234 polymorphisms within the FCGR3A gene, of which 69%, 16%, and 15% occur in the intron, UTR, and exon regions respectively. Additionally, only 16% of all polymorphisms had a minor allele frequency (MAF) > 0.01. To facilitate (full-length) analysis of FCGR3A gene polymorphism, we developed a FCGR3A gene-specific amplification and sequencing protocol for Sanger sequencing and MinION (Nanopore Technologies). First, we used the Sanger sequencing protocol to study the presence of the V158F polymorphism in 76 individuals resulting in frequencies of 38% homozygous T/T, 7% homozygous G/G and 55% heterozygous. Next, we performed a pilot with both Sanger sequencing and MinION based sequencing of 14 DNA samples which showed a good concordance between Sanger- and MinION sequencing. Additionally, we detected 13 SNPs listed in the 1000 Genome Project, from which 11 had MAF > 0.01, and 10 SNPs were not listed in 1000 Genome Project. In summary, we demonstrated that FCGR3A gene is more polymorphic than previously described. As most novel polymorphisms are located in non-coding regions, their functional relevance needs to be studied in future functional studies.
Introduction
Natural killer (NK) cells are innate lymphocytes and pivotal players in the defence against malignant- or virally-infected cells1. NK cells can produce cytokines and kill target cells2. Moreover, NK cells mediate antibody-dependent cell-mediated cytotoxicity (ADCC) via the ligation of their low affinity Fc receptor, FcγRIIIa, also known as CD16a, with an antibody bound to a potential target cell1,3.
As reviewed recently, the strength of the ADCC response could be determined by several factors, amongst them the isotype-, fucosylation- and glycosylation- characteristics of the antibody as well as genotypic variation of the FcγRIIIa receptor itself4. A clear example of the latter is the single nucleotide substitution (SNP) from G to T at cDNA nucleotide position 559 of the FCGR3A gene generating two different FcγRIIIa allotypes: one with a valine (V) and one with a phenylalanine (F) at amino acid position 158, known as FcγRIIIA-V158F polymorphism (rs396991)5–7. The presence of a valine (V/V or V/F) has been shown to enhance the NK cell’s binding affinity to an IgG1 or IgG3 antibody as compared to the presence of a homozygous phenylalanine genotype (F/F), resulting in a higher level of NK cell-mediated ADCC6–8.
In antibody-based immunotherapy, NK cell-mediated ADCC is one of the mechanisms underlying the anti-cancer effects of frequently used antibodies like rituximab, trastuzumab, and cetuximab. Several clinical studies provided evidence for the functional relevance of the V158F polymorphism in this setting: in non-Hodgkin lymphoma, HER-2/neu-positive metastatic breast cancer, metastatic colorectal cancer or head and neck cancer, patients with V/V polymorphism appeared to have an improved progression-free survival as compared to patients with F/F phenotype9–13. Moreover, a study examining rituximab and ADCC in healthy donors suggested that the expression of at least one valine at FcγRIIIa-158 could explain the improved clinical outcome14. Nonetheless, two other studies15,16 did not find any correlation between the V158F polymorphism and the clinical outcome, possibly due to sample size limitation.
The characterization of the FCGR3A gene polymorphism may also be relevant in the solid organ transplantation setting where, in the presence of antibodies against a renal graft, NK cells have been shown to mediate ADCC contributing to graft rejection17,18. A recent study on cardiac allograft showed that patients with V/V genotype had an enhanced CD16a expression and were associated with a higher risk of developing vasculopathy and eventually allograft rejection19.
Interestingly, a study on bone marrow transplantation for myeloid malignancies suggested that the V158F polymorphism in recipients could predict transplant outcomes and the presence of V/V genotype in recipients was associated with a significantly reduced risk of acute and chronic graft-versus-host disease as well as better overall survival20. Furthermore, patients with F/F or V/F genotype have been shown to have a higher predisposition to an increased incidence of infection after liver transplantation21.
In addition to the V158F polymorphism, several additional polymorphisms in the FCGR3A gene have been identified: (1) the FcγRIIIA-48L/R/H polymorphism (rs10127939), where a single nucleotide substitution from T to G is responsible for a leucine (L) to an arginine (R) substitution and T to A is responsible for a leucine (L) to a histidine (H) at amino acid position 48. Both these substitutions have been reported to have an enhanced binding to the IgG1, IgG3, and IgG422. This polymorphism has also been demonstrated to be linked to the FcγRIIIA-V158F polymorphism6 where the FcγRIIIA-48L/R/H polymorphism influenced ligand binding capacity in the presence of the FcγRIIIA-V158F polymorphism23. The presence of R or H allele and at least one copy of V allele provided a higher binding capacity. (2) A homozygous missense mutation in the FCGR3A gene encoding a L48H substitution causing a defect in NK cell cytotoxicity due to a reduced surface expression of CD2, a co-activation receptor, while preserving an intact ADCC24. (3) two SNPs (rs4656317 and rs12071048) located within the enhancer region of the FCGR3A gene that are in strong linkage disequilibrium with the FcγRIIIA-V158F polymorphism and strongly affected NK cell ADCC activity where the major alleles had a higher ADCC activity than those with minor alleles25, (4) a 3-SNP/1-indel FCGR3A intragenic haplotype which was associated with increased FcγRIIIa expression26. (5) Several other polymorphisms in the FCGR3A gene, i.e. rs209968427; rs1091954328; and rs44550929, that have been found to be associated with arteritis27,28 and chronic periodontitis29.
The above mentioned studies highlighted the potential relevance of FCGR3A polymorphisms for NK cell effector function and their potential clinical relevance. However, the analysis is frequently complicated by the presence of FCGR3B gene, encoding the inhibitory FcɣRIIIb receptor, as the FCGR3B gene is highly homologous to the FCGR3A gene except that it has a T at nucleotide 531 of the cDNA instead of a C7,30. Another issue is that previous methods were focused on sequencing particular exons of the gene7,31 while extended polymorphism in for example 5′ or 3′ UTR or in introns could also influence CD16a expression e.g. by influencing micro-RNA binding or alternative splicing32. To facilitate future studies to unravel the functional consequences of CD16a polymorphism, we established a standardized way to determine V158F gene polymorphism using Sanger sequencing and we tested a new full-length gene single molecule sequencing method for the identification of polymorphism in the FCGR3A gene using MinION (a Nanopore technology). We subsequently used these methods, combined with the data present for the FCGR3A gene in the database of phase 3 of the 1000 Genomes project (1KGP), to generate a more comprehensive overview of full-length CD16a polymorphism.
Results
FCGR3A gene variability beyond the V158F polymorphism
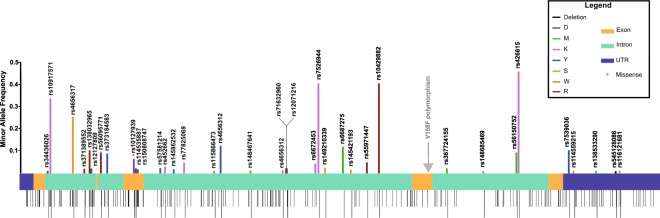
To study the magnitude of FCGR3A gene polymorphism, we analysed the nucleotide variability data available in the 1KGP for this gene and mapped all the polymorphisms identified in 1KGP based on the location (gene region) and the minor allele frequency (MAF) (Fig. 1). The polymorphic index (PI), the number of polymorphic positions divided by the length of the whole gene and of the individual introns/exons/UTR, was calculated (Table 1). This illustrated that exon 3 is the most polymorphic region in the gene, with a PI of 0.066, while exon 2 has the lowest PI. The gene sequence of exon 2–5 encodes for the FcγRIIIa receptor, which consists of an extracellular domain with two Ig-like domains (exon 3 and 4) and five potential N-glycosylation sites (three in exon 3 and two in exon 4), a transmembrane domain (exon 5) and a cytoplasmic domain (exon 5).
Figure 1.
Schematic illustration of FCGR3A gene and its 234 polymorphisms according to the 1000 Genome database. The stripes present underneath represent different polymorphisms. All polymorphisms with MAF > 0.01 are shown on the upper part of the scheme and the rs number is shown. Different colors denote the amino acid changes as shown in the legend. The grey arrow points at the location of the V/F polymorphism.
Table 1.
Number of polymorphisms present in the FCGR3A gene described in the 1KGP database.
| Location | Bases | Polymorphisms | PI |
|---|---|---|---|
| 5′UTR | 183 | 5 | 0.027 |
| Exon 1 | 147 | 8 | 0.054 |
| Intron 1 | 664 | 30 | 0.045 |
| Exon 2 | 20 | 0 | 0.000 |
| Intron 2 | 331 | 13 | 0.039 |
| Exon 3 | 257 | 17 | 0.066 |
| Intron 3 | 3461 | 87 | 0.025 |
| Exon 4 | 257 | 4 | 0.016 |
| Intron 4 | 1501 | 32 | 0.021 |
| Exon 5 | 186 | 5 | 0.027 |
| 3′UTR | 1252 | 33 | 0.026 |
| Coding region | 867 | 34 | 0.039 |
| Noncoding region | 7392 | 200 | 0.027 |
| Whole gene | 8259 | 234 | 0.028 |
The table reports the number of polymorphisms per location and the polymorphic index. PI = Polymorphic index.
A total of 234 polymorphisms (3% of the entire gene, SNP density: 2.83 SNP/100 bp) were identified, of which 34 (15%) are present in the exons, 162 (69%) in the introns, and 38 (16%) in the untranslated regions (UTRs) (Fig. 2). Of note, only 36 of these 234 polymorphisms have a MAF higher than 1% (Table 2). A relatively high number of these polymorphisms with a MAF higher than 1% are located in intron 3 as compared to the other regions (16 out of 36).
Figure 2.
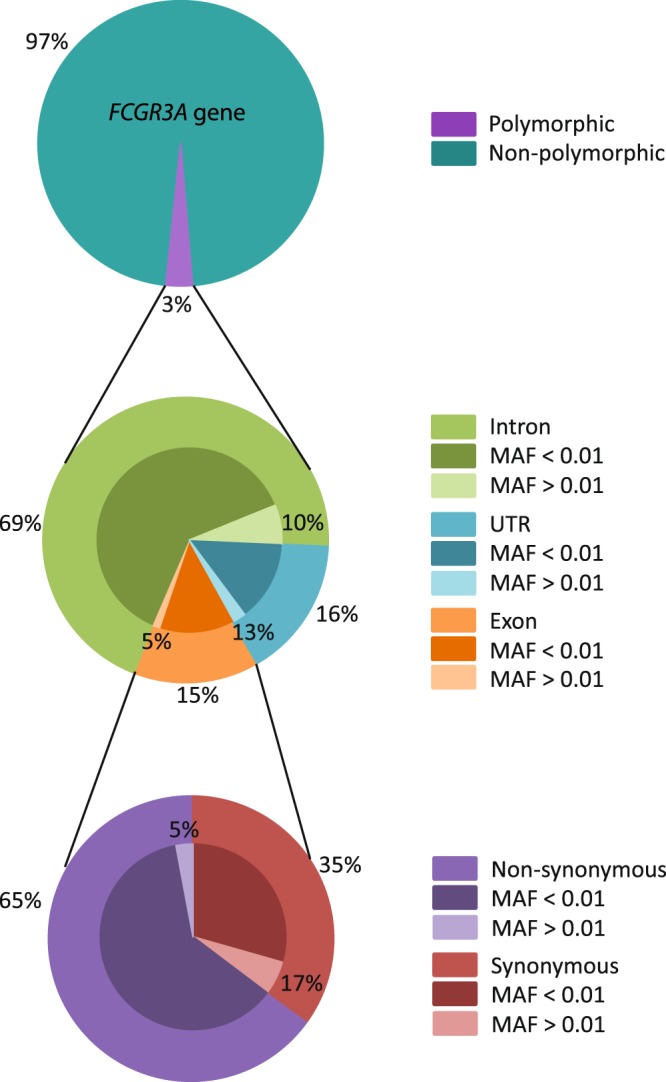
Schematic overview of overall polymorphisms in the FCGR3A gene in the 1KGP database. The upper circle depicts the variability percentages of the whole FCGR3A gene. The middle circle represents the variability in the specific gene regions and the lower circle shows the exonic variability. The percentages of polymorphisms found with a Minor Allele Frequency (MAF) lower or higher than 1% are specifically illustrated.
Table 2.
Polymorphisms with a minor allele frequency (MAF) higher than 1% in the 1KGP.
| SNP | Position in gene | Location | Polymorphism | MAF | Amino Acid Change |
|---|---|---|---|---|---|
| rs34436026 | 195 | Intron 1 | R | A: 0.011 | |
| rs10917571 | 224 | Intron 1 | K | T: 0.340 | |
| rs4656317 | 516 | Intron 1 | S | G: 0.257 | |
| rs371389552 | 664 | Intron 1 | R | A: 0.021 | |
| rs138032965 | 727 | Intron 1 | R | A: 0.106 | |
| rs12127809 | 756 | Intron 1 | Y | C: 0.022 | |
| rs56095771 | 878 | Intron 2 | R | G: 0.095 | |
| rs373184583 | 959 | Intron 2 | Y | T: 0.090 | |
| rs10127939 | 1302 | Exon 3 | D | G: 0.039, A: 0.027 |
L/R/H |
| rs114535887 | 1321 | Exon 3 | R | A: 0.019 | |
| rs150808747 | 1336 | Exon 3 | Y | T: 0.012 | |
| rs57581214 | 1641 | Intron 3 | Y | T: 0.024 | |
| rs452662 | 1696 | Intron 3 | K | T: 0.030 | |
| rs145862532 | 1816 | Intron 3 | Y | T: 0.019 | |
| rs77825069 | 1950 | Intron 3 | K | T: 0.048 | |
| rs115866473 | 2336 | Intron 3 | W | T: 0.012 | |
| rs4656312 | 2418 | Intron 3 | Y | T: 0.126 | |
| rs148467641 | 2802 | Intron 3 | W | T: 0.014 | |
| rs145392761 | 3213 | Intron 3 | K | T: 0.014 | |
| rs71632960 | 3275 | Intron 3 | Y | T: 0.023 | |
| rs12071216 | 3278 | Intron 3 | R | G: 0.018 | |
| rs7526944 | 3678 | Intron 3 | K | T: 0.408 | |
| rs149210339 | 3763 | Intron 3 | S | C: 0.025 | |
| rs6687275 | 3997 | Intron 3 | M | C: 0.121 | |
| rs145421193 | 4098 | Intron 3 | S | G: 0.017 | |
| rs55971447 | 4308 | Intron 3 | R | A: 0.050 | |
| rs10429882 | 4459 | Intron 3 | R | G: 0.408 | |
| rs367724155 | 5333 | Intron 4 | M | C: 0.023 | |
| rs148685469 | 5803 | Intron 4 | K | G: 0.011 | |
| rs56150752 | 6229 | Intron 4 | M | C: 0.094 | |
| rs426615 | 6258 | Intron 4 | K | G: 0.462 | |
| rs7539036 | 6904 | 3′UTR | Y | T: 0.107 | |
| rs114559215 | 6975 | 3′UTR | S | C: 0.013 | |
| rs138533290 | 7256 | 3′UTR | M | C: 0.012 | |
| rs545128086 | 7507 | 3′UTR | Deletion (T/−) | (-): 0.010 | |
| rs116121681 | 7558 | 3′UTR | S | C: 0.012 |
Of the 34 polymorphisms identified in the exons, 22 (65%) are non-synonymous and 12 (35%) are synonymous. Only one non-synonymous (rs10127939 C/T) and two synonymous polymorphisms (rs114535887 and rs150808747) have a MAF greater than 1%. The non-synonymous polymorphism is located at nucleotide position 1302 in exon 3 with three different nucleotides possible (T, G, and A), resulting in three different amino acids: leucine (L, MAF 0.09), arginine (R, MAF 0.039), and histidine (H, MAF 0.027) and three different alleles. The synonymous polymorphisms are located at nucleotide position 1321 and 1336 and have a MAF of 0.019, and 0.012 respectively.
The V158F polymorphism at nucleotide position 5093 (rs396991) is not documented in the 1KGP because it did not reach the quality control threshold, most probably because of its location in a homopolymer-rich region, and thus no frequency information of this polymorphism was attainable. For this reason, Fig. 1 shows an arrow demonstrating the location of the V/F polymorphism, but provides no further information.
Sanger sequencing for detection of V158F polymorphism in the FCGR3A gene
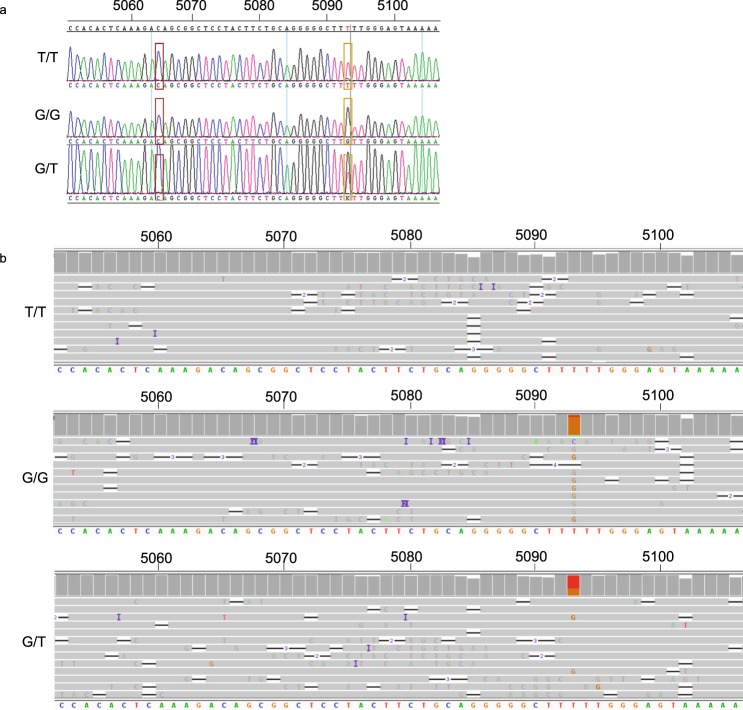
To establish a convenient method to analyse FcγRIIIa-158 polymorphism, while excluding the highly homologous FCGR3B gene, we explored a Sanger sequencing based approach that would also enable analysis of extended polymorphism. The FCGR3A gene sequence of the 1KGP database was used as a reference for the sequencing analysis. First, we focused the analysis on the FcγRIIIa-158 polymorphism and, in our sequence data, we identified the FCGR3A gene by the presence of a C nucleotide at the position 5065 while a T would be identified in case of an FCGR3B gene (Fig. 3a). The FcγRIIIa-158 polymorphism variants at nucleotide position 5093 (T/T, T/G, and G/G) could be distinguished by analysis of the chromatograms. The T/T genotype would result in an F/F phenotype (low affinity), T/G in a V/F phenotype, whereas G/G would result in a V/V phenotype (high affinity). We subsequently analysed a total of 76 samples for the V/F polymorphism and a total of 29 samples were found to be homozygous for T (F/F), 42 samples were heterozygous (V/F), and 5 samples were homozygous for G (V/V). Hence, the T allele was overall the most prevalent; 66% T compared to 34% G. To validate the results obtained from the sequencing, we used sequence-specific primers (SSPs) to specifically amplify the variants of the FCGR3A gene separately. For this validation, we selected a total of 11 samples. The gel electrophoresis results confirmed the sequencing results (Supplementary Fig. S1).
Figure 3.
Detection of V158F polymorphism by showing 3 different genotypes, homozygous T, homozygous G, and heterozygous. (a) Electropherograms show the Sanger-based sequencing result around the V158F polymorphism. The red square indicates nucleotide position 5064, which is used to check whether the FCGR3B gene is co-amplified. The yellow square indicates nucleotide position 5093, used for determining the V158F polymorphism. Nucleotide code K indicates that both T and G are present. (b) MinION sequencing result around the V158F polymorphism. Dark grey bars on the top show the sequence coverage identical to the consensus sequence. If the sequence is not identical to the consensus the bars will have the color of the corresponding nucleotide. The light grey lines show a small part of the reads obtained with the MinION run and the sequence at the bottom shows the consensus sequence. The first result represents a sample homozygous for T at position nucleotide 5093 (coverage: A: 0%: C: 2% G: 2% T: 96%), which was also used as consensus, the second sample is homozygous for G (coverage: A: 3% C: 2% G: 85% T: 9%), and the third is heterozygous at nucleotide position 5093 (coverage: A: 1% C: 3% G: 32% T: 64%).
Altogether, these data showed that the developed Sanger sequencing approach is reliable to identify polymorphisms in the FCGR3A gene.
Detection of extended, full-length polymorphisms is feasible using Sanger- and MinION Nanopore-based sequencing
The results from the 1KGP database analysis on FCGR3A gene polymorphisms revealed that there were more polymorphisms within the FCGR3A gene than previously described. Additionally, the 1KGP polymorphism frequency database showed that some of these polymorphisms occurred in the worldwide population with a frequency higher than 1.0%. We therefore envisioned that detection of extended full-length polymorphisms, including the non-coding regions, in this gene could facilitate future studies on the functional relevance of the FcγRIIIa receptor polymorphism. To investigate the feasibility of detecting polymorphisms in the FCGR3A gene, we set up a pilot study and amplified the whole FCGR3A gene region for 14 DNA samples and subsequently sequenced using two approaches: Sanger- and MinION sequencing (Oxford Nanopore Technologies). Despite full-length amplification, we did not perform full length sequencing for Sanger sequencing for this pilot and used primers covering a part of intron 3, exon 4, intron 4, and 3′UTR region. MinION is a novel portable real-time single molecule sequencing device developed to sequence long regions with ultra-long reads. With this technique, we were therefore able to sequence the complete full-length gene, also including all non-coding gene regions. MinION amplification primers were also tagged enabling us to barcode and sequence multiple samples simultaneously. After sequencing, we analysed the sequencing results and compared the results obtained by Sanger sequencing with those obtained by MinION and with the data from the 1KGP.
In this pilot study, we detected 23 SNPs in the FCGR3A gene of the 14 individuals (Table 3). Of these 23 SNPs, 13 were also identified by the 1KGP and two of these SNPs (G3121A and T3155C) were found with a MAF < 0.01. The only exonic SNP T5093G (V158F polymorphism) is listed in 1KGP database as “failed variant” and the allelic frequency data is not available in this database. We therefore used the MAF data from other databases (GO-ESP and ExAC) in Table 3. The 10 SNPs not identified by the 1KGP were located in the non-coding intron 3, intron 4 or 3′UTR region.
Table 3.
SNPs found within the FCGR3A gene detected by Sanger sequencing and MinION, compared to 1KGP.
| SNP | Chromosomal position | Gene position | Gene location | Detected by | SNP name | MAF | ||
|---|---|---|---|---|---|---|---|---|
| Sanger | MinION | 1KGP | ||||||
| G224T | 1:161519411 | 224 | Intron 1 | No* | Yes | Yes | rs10917571 | 0,34 |
| C516G | 1:161519119 | 516 | Intron 1 | No* | Yes | Yes | rs4656317 | 0,26 |
| G727A | 1:161518908 | 727 | Intron 1 | No* | Yes | Yes | rs138032965 | 0,11 |
| G1463A | 1:161518172 | 1463 | Intron 3 | No* | Yes | No | n/a | n/a |
| G1793C | 1:161517842 | 1793 | Intron 3 | No* | Yes | No | n/a | n/a |
| C2418T | 1:161517217 | 2418 | Intron 3 | No* | Yes | Yes | rs4656312 | 0,13 |
| A2967G | 1:161516668 | 2967 | Intron 3 | No* | Yes | No | n/a | n/a |
| G3121A | 1:161516514 | 3121 | Intron 3 | No* | Yes | Yes | rs545876704 | <0,01 |
| T3155C | 1:161516480 | 3155 | Intron 3 | No* | Yes | Yes | rs180923798 | <0,01 |
| A3187G | 1:161516448 | 3187 | Intron 3 | No* | Yes | No | n/a | n/a |
| G3624T | 1:161516011 | 3624 | Intron 3 | Yes | Yes | Yes | rs6672453 | 0,11 |
| G3683T | 1:161515952 | 3683 | Intron 3 | Yes | Yes | Yes | rs7526944 | 0,41 |
| G3763C | 1:161515872 | 3763 | Intron 3 | Yes | Yes | Yes | rs149210339 | 0,03 |
| A4083G | 1:161515552 | 4083 | Intron 3 | Yes | Yes | No | n/a | n/a |
| A4327C | 1:161515308 | 4327 | Intron 3 | Yes | Yes | No | n/a | n/a |
| A4459G | 1:161515176 | 4459 | Intron 3 | Yes | Yes | Yes | rs10429882 | 0,41 |
| T5093G | 1:161514542 | 5093 | Exon 4 | Yes | Yes | Yes | rs396991 | 0.27**–0.33*** |
| T5728C | 1:161513907 | 5728 | Intron 4 | Yes | Yes | No | n/a | n/a |
| C5876G | 1:161513759 | 5876 | Intron 4 | No* | Yes | No | n/a | n/a |
| T6187C | 1:161513448 | 6187 | Intron 4 | No* | Yes | No | n/a | n/a |
| T6258G | 1:161513377 | 6258 | Intron 4 | No* | Yes | Yes | rs426615 | 0.46 |
| C6904T | 1:161512731 | 6904 | 3′UTR | Yes | Yes | Yes | rs7539036 | 0.11 |
| G8054C | 1:161511581 | 8054 | 3′UTR | No* | Yes | No | n/a | n/a |
Using the same amplification primers, 14 DNA samples were sequenced using Sanger and MinION. MAF represents the Minor Allele Frequency data from 2504 individuals obtained from the 1KGP database except for rs396991 (F158V) were the MAF was obtained from the GO-ESP** and ExAC*** database. No* = this position was not included within the Sanger sequence region.
Of the 23 detected SNPs, nine were identified by both the Sanger sequencing and MinION technique. Since the other 14 SNPs were located in regions outside the Sanger sequence area, these were only identified by MinION. This result demonstrates that MinION sequencing can be used to determine full-length FCGR3A polymorphism. Although the results of MinION sequencing were similar to Sanger sequencing, some cautions should be taken when reading the sequencing results in the region where V158F polymorphism is located (Fig. 3b). We observed that MinION could misreport the presence of heterozygous G/T where it would be reported as a homozygous T/T genotype (depending on the analysis settings/percentage of nucleotides present), most likely due to the presence of a homopolymer sequence within the region. This might actually be the reason why it is reported as a “failed variant” in the 1KGP, since all NGS methods encounter difficulties in analysing homopolymer regions.
Altogether, we demonstrated that using full-length Sanger-based and MinION-based sequencing methods we could detect both known as well as new polymorphisms within the FCGR3A gene.
Discussion
NK cells are the principal mediator of ADCC due to the high expression of the activating FcγRIIIa and the absence of the inhibitory FcγRIIIb on their surface3. The large availability of clinical grade antibodies triggering ADCC against cancer cells has put increased focus on NK cell-mediated ADCC and emphasizes the relevance of the FcγRIIIa for cancer immunotherapy4. In addition, a few studies underlined the functional relevance of FcγRIIIa in the transplantation setting by showing that NK cell mediated ADCC could play a role in allograft rejection33. Albeit several FCGR3A gene polymorphisms have been shown to impact NK cell mediated ADCC, full-length gene polymorphism has not been determined. Hence, we provide here an overview of FCGR3A gene polymorphisms, as well as two improved sequencing methods for further gene exploration.
In this study, with 234 polymorphisms identified, we demonstrated that FCGR3A gene is more polymorphic than currently known; 34 SNPs were located in the exons and only 3 of them had a MAF > 0.01. We identified two non-synonymous SNPs either by Sanger sequencing/MinION sequencing or in 1KGP. The first one was rs10127939, representing the FcγRIIIA-48L/R/H previously shown to influence ADCC22,23. We did not detect this polymorphism in our full-length sequencing samples presumably because of our limited sample size and the fact that the frequency of this polymorphism is relatively low in the population (MAF = 0.039 and 0.027). The second non-synonymous SNP in the coding region was rs396991, representing the V158F polymorphism which we detected both by Sanger- and by MinION sequencing. In our test panel the V/F phenotype (G/T genotype) is the most common (55%) followed by F/F (T/T genotype, 38%) and V/V (G/G genotype, 7%). The presence of V158F polymorphism has been previously investigated in individuals from different populations, including ethnic groups from Singapore34, the Netherlands, Great Britain, Norway35 and Japan36. Overall these studies reported the V/F or F/F phenotype as the most frequent, whereas the V/V was the least frequent phenotype in all populations, which is comparable to our results and could suggest some kind of selective pressure on FCGR3A. Our study set up did not allow us to reliably compare V158F gene- and allele frequencies between the 34 samples from the Guadaloupe population vs the 42 samples from our institute or with the results from the 1KGP. The major reason for this was the lack of information on the exact ethnic background of the individuals and the low sample size. Given the known highly heterogeneous background of the Guadaloupe population, it would, however, be highly interesting to compare this population with other populations in a future study.
In this study, we demonstrated that the majority of FCGR3A gene polymorphism is located in the non-coding regions and at least 33 of the 200 identified non-coding SNPs have a MAF > 0.01 in the 2504 individuals of the 1KGP. As introns have been demonstrated to be involved in gene regulation37 and many intronic polymorphisms could exhibit functional significance38, it might be worthwhile to perform additional functional studies. SNPs located in the intron regions could potentially affect RNA splicing by altering the sequences of the 5′/3′ splice site, branch point, polypyrimidine tract or intronic splicing enhancer/silencer motifs. A study on FCGR2C gene interestingly showed that a mutation in an intronic splice site introduced novel stop codons resulting in a loss of FcγRIIc expression39. In this study we have investigated the two consensus splice site sequences on the 5′ and 3′ end of the intron (GT on 5′ and AG on 3′) and we already observed one SNP (rs544630563) at the 3′ of intron 4, turning AG into GG, although it was found with low frequency in the 1KGP database (MAF < 0.01). Additionally, as in a recent paper reviewing different studies on different disease genes, several mutations deep within the introns (for example 100 base pairs upstream exon-intron boundary) were identified as being associated to multiple diseases40. In line with our data, where we observed intronic polymorphisms located upstream the exon-intron boundary, it would be interesting to look at the association of these polymorphisms with the functionality of the FcγRIIIa receptor.
In the present study, we successfully set up a Sanger- and a MinION-based protocol to sequence the FCGR3A gene. We subsequently demonstrated that both Sanger sequencing and MinION were able to identify FCGR3A polymorphisms present in the 1KGP database. While Sanger sequencing is based on the capillary electrophoresis, MinION technology consists of nanopores embedded in an electrically resistant membrane through which a current is applied, causing a potential which flows through the aperture of the nanopores. The changes observed in the current correspond with 5 to 6 nucleotides passing through the nanopores. This electrical signal is translated into reads that can be analysed and by this technology, MinION can sequence reads up to hundreds of kilo base pairs. For both techniques, we performed identical full-length amplification of the gene. However, MinION has the advantage of directly generating full-length gene reads and phasing of the two variants is possible without group-specific amplification. Although MinION allows full-gene sequencing of various samples in a relatively short time, this technology is not yet widely implemented. Compared to conventional sequencing approaches MinION has a lower accuracy and sensitivity and therefore more reads must be generated. We demonstrated a good concordance between Sanger sequencing and MinION, and were able to identify the V158F polymorphism in all samples using both MinION and Sanger. Nonetheless, a homopolymer region, such as the sequence around V158F, is known to be problematic in all next generation sequencing approaches, apparently including MinION and presumably this also explains the lack of data for this region in the 1KGP. The challenge of analyzing such homopolymer regions with MinION sequencing has been described in several other studies as well41–43. Hence despite its usefulness for full-length gene analysis, Sanger sequencing for now seems the preferred method when only analysis of V158F polymorphism is required.
In summary, we showed that FCGR3A gene is highly polymorphic especially in the non-coding regions of the gene requiring functional studies to investigate the functional consequences. Additionally, we demonstrated that our Sanger- and MinION-based sequencing approaches can be used to identify the extended polymorphisms of the gene. Although further optimization and validation is warranted, we also identified MinION as a powerful method to perform direct full-length FCGR3A gene sequencing.
Material and Methods
Subjects
FCGR3A sequences were studied in a test panel consisting of 76 distinct samples with unknown DNA sequence, 42 of them were volunteers from the institute and 36 were individuals from the Guadeloupe islands44. Samples were left over from diagnostic procedures which does not require ethical approval in the Netherlands under the Dutch Code for Proper Secondary Use of Human.
DNA isolation
Genomic DNA was extracted from ethylenediamine tetraacetic acid (EDTA) blood samples using the QIAamp DNA blood mini kit (Qiagen, Hilden, Germany). DNA concentrations were measured using the NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, Delaware).
Amplification of the FCGR3A gene for Sanger- and MinION sequencing
Amplification primers
Primers specific for the FCGR3A gene were designed by comparing the sequences of the FCGR3A and FCGR3B genes, including their polymorphisms, and finding the discrepancies among them. Due to the extreme homology of the genes, some generic primers (not specific for the FCGR3A gene) were also designed as a control and always used in combination with a specific primer.
Polymerase Chain Reaction
The entire FCGR3A gene, including the 5′UTR and 3′UTR, was amplified using an FCGR3A gene-specific forward primer and a generic reverse primer, producing a 9654 bases long polymerase chain reaction (PCR) product. The PCR reaction contained 300 ng of genomic DNA, 67 mM Tris-HCl (pH 8.8) (Merck, Darmstadt, Germany), 16.6 mM ammonium sulfate (Merck), 0.01% Tween 20 (Merck), 1.5 mM MgCl2 (Life Technologies, Austin, Texas), 0.2 mM of each dNTP (GE Healthcare, Diegem, Belgium), 0.1 µg/µl cresol red (Sigma-Aldrich, St. Louis, Missouri), 5% glycerol (Alfa Aesar, Karlsruhe, Germany), 15 pmol of each primer (Sigma-Aldrich) and 2.5 U of Expand Long Template PCR System (Roche, Basel, Switzerland) with a final volume of 30 µl. The PCR program consisted of an initial denaturation step of 2 minutes at 94 °C; followed by 10 cycles of 15 seconds at 94 °C, 30 seconds at 63 °C and 4 minutes at 68 °C; then 10 cycles of 15 seconds at 94 °C, 30 seconds at 60 °C and 6 minutes at 68 °C; afterwards 10 cycles of 15 seconds at 94 °C, 30 seconds at 60 °C and 10 minutes at 68 °C; and a final elongation step of 7 minutes at 68 °C. The PCR products were checked by electrophoresis using a 1.5% agarose gel containing 0.5 µg/µl ethidium bromide (Sigma-Aldrich).
MinION amplification
The same amplification primers used for Sanger sequencing were used for the MinION sequencing mixture, with a tag-sequence (indicated as italic and red) added to the ends to enable barcoding amplification for identification of different samples after all samples were pooled.
Sanger sequencing of V158F region
Amplicons obtained from all 76 DNA samples were purified by ExoSAP-IT (Affymetrix, Santa Clara, California) following the manufacturer’s protocol.
Purified amplicons were sequenced using ABI BigDye Terminator Chemistry (Life Technologies) and an ABI 3730 sequencer (Life Technologies) with a forward and a reverse sequencing primer. For FCGR3A gene sequencing using Sanger, several sequencing primers were used to cover different locations in the gene. The sequencing mixture consisted of 1 µl purified PCR product, 0.5 µl sequencing primer (5 pmol, Sigma-Aldrich), 1 µl of BigDye Terminator v1.1 mix, 1.5 µl 5x Big Dye Terminator sequencing buffer and 6 µl distilled water. The PCR program consisted of: 1 minute at 95 °C, followed by 25 cycles of 10 seconds at 95 °C, 5 seconds at 50 °C, and 4 minutes at 60 °C. Successively, the mixtures were purified by Sephadex G-50 Fine (GE Healthcare Life Sciences, Little Chalfont, UK) and placed in the ABI 3730 sequencer for capillary electrophoresis sequencing. The chromatograms were aligned with a reference sequence obtained from the 1KGP and analysed using DNASTAR Lasergene SeqMan Pro (DNASTAR Lasergene, Madison, Wisconsin).
MinION Nanopore-based sequencing
Amplicons obtained from 14 DNA samples from the cohort of individuals present in the institute, were barcoded and sequenced following Oxford Nanopore’s instructions (NSK-LSK208). In short, we purified the amplicons using CleanPCR beads (GC Biotech, Alphen aan den Rijn, the Netherlands) followed by determining DNA concentration using a DS-11 spectrophotometer (DeNovix, Delaware, USA). Next, 48 ng of amplicon was barcoded using the PCR barcoding Kit 1 (Oxford Nanopore Technologies, Oxford, UK) and LongAmp Taq2x (New England Biolabs, Massachusetts, USA) followed by purification of the barcoded PCR product using CleanPCR beads and determination of DNA concentration. The barcoded DNA samples were pooled to an end volume of 1 µg in 45 µl and an endrepair/dA-tailing was performed (NEBNext Ultra II End-Repair/dA-tailing module, New England Biolabs) followed by a purification step using AMPure XP beads (Beckman Coulter, California, USA). After that, DNA adapter ligation was performed using NEB Blunt/TA ligase master mix (New England Biolabs) and samples were purified using MyOne C1 Dynabeads (Thermo Fisher Scientific, Massachusetts, USA). Of this adapter library, 75 µl was loaded into a FLO-MIN106 flow cell. Sequencing run was performed and base calling was done using Albacore software (V1.2.4, Oxford Nanopore Technologies). Sequencing data was analysed using in-house software and Integrative Genomics Viewer (IGV)45.
PCR amplification using sequence specific primers for Sanger sequencing validation
The sequence specific primers (SSPs) consisted of one primer specific for the T allele and one for the G allele. An FCGR3A gene-specific primer was used in combination with the SSPs to assure specific amplification of the FCGR3A gene. The PCR program was almost identical to the Sanger sequencing amplification protocol described in this article, except that the annealing temperature used for SSP PCR was 63 °C during the first 10 cycles.
1000 Genome Project Data Analysis
Based on the publicly available data present in the third phase of the 1KGP (http://phase3browser.1000genomes.org/index.html), including 2504 individuals originating from 26 different populations, the FCGR3A gene comprised 8259 bp located on chromosome 1: 161511549-161519818 (reverse direction). This sequence corresponds to the FCGR3A-001 protein coding transcript and the start of exon 1 (position 1:161519634) was used as nucleotide position 1 in this paper. We recorded the polymorphism and its location on the chromosome as well as the gene location (position of nucleotide and region (i.e. UTR, exon, or intron)). The population genetic tool was used to acquire an overview of the overall allele frequencies and the frequencies within different population.
List of primers
| Sanger sequencing Amplification Primers | ||
|---|---|---|
| Direction | Sequence (5′ to 3′) | Location 1000 Genomes |
| FW | GCTGCCTGGGTTCATTTCCA | 1:161520918-161520938 |
| RV | CCTCTGCCCAGGCCTCTA | 1:161511283-161511301 |
| MinION Amplification Primers | ||
|---|---|---|
| Direction | Sequence (5′ to 3′) | Location 1000 Genomes |
| FW | TTTCTGTTGGTGCTGATATTGCGCTGCCTGGGTTCATTTCCA | 1:161520918-161520938 |
| RV | ACTTGCCTGTCGCTCTATCTTCCCTCTGCCCAGGCCTCTA | 1:161511283-161511301 |
| Sanger sequencing V158F region Sequencing Primers | ||
|---|---|---|
| Direction | Sequence (5′ to 3′) | Location 1000 Genomes |
| FW | GTGTTCAAGGAGGAAGACC | 1:161514701-1:161514719 |
| RV | ACTCAACTTCCCAGTGTGATT | 1:161511283-161511301 |
| SSP Amplification Primers | |||
|---|---|---|---|
| Direction | Sequence (5′ to 3′) | Location 1000 Genomes | Specificity |
| RV | AAGACACATTTTTACTCCCAAA | 1:161514521-1:161514542 | T allele |
| RV | AAGACACATTTTTACTCCCAAC | 1:161514521-1:161514542 | G allele |
| FW | GCTGCCTGGGTTCATTTCCA | 1:161520918-161520938 | FCGR3A |
| Sanger sequencing FCGR3A gene Sequencing Primers | |||
|---|---|---|---|
| Direction | Sequence (5′ to 3′) | Location 1000 Genomes | Specificity |
| FW | CTAATAATGATTCATCTCTYTGC | 1:161525783 - 1:161525805 | Intron 3 |
| FW | TGCTKAAAAAGTAAGTGGWTAG | 1:161525803 - 1:161525824 | Intron 3 |
| RV | GGTAAGTATTATAATGGCAYAAG | 1:161526243 - 1:161526260 | Intron 3 |
| RV | TTATAGGTAAGTATTATAATGGC | 1:161526248 - 1:161526265 | Intron 3 |
| FW | KTTTGGCAGTGYCAACCWTC | 1:161528867 - 1:161528886 | Exon 5/3′ UTR |
| FW | TCCACCTGGGTACCAAGTC | 1:161528898 - 1:161528916 | Exon 5/3′ UTR |
| RV | TTCTATGTTTCCTGCTGCTTG | 1:161529146 - 1:161529166 | Exon 5/3′ UTR |
| RV | RGGATCTGGCTCTGAGTTC | 1:161529163 - 1:161529182 | Exon 5/3′ UTR |
| FW | GTGTTCAAGGAGGAAGACC | 1:161514701-1:161514719 | V158F region |
| RV | ACTCAACTTCCCAGTGTGATT | 1:161514701-1:161514719 | V158F region |
Electronic supplementary material
Author Contributions
N.M.M., T.I.O., co-wrote the main manuscript text. L.W. and C.E.M.V. supervised the writing of the manuscript. I.O.R., S.J.J.M., were responsible for the practical part and data acquisition. N.M.M. generated the figures. M.G. was responsible for the bio-informatics part of the project. N.M.M. and T.I.O. were responsible for data analysis and interpretation of the data. C.E.M.V. and L.W. supervised the interpretation of the data. Critical reviews were given by G.M.J.B., M.G.J.T., C.E.M.V. and L.W. Final approval was given by all authors.
Data Availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information file).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-34258-1.
References
- 1.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat. Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 2.Campbell KS, Hasegawa J. Natural killer cell biology: An update and future directions. J. Allergy Clin. Immunol. 2013;132:536–544. doi: 10.1016/j.jaci.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seidel UJE, Schlegel P, Lang P. Natural Killer Cell Mediated Antibody-Dependent Cellular Cytotoxicity in Tumor Immunotherapy with Therapeutic Antibodies. Front. Immunol. 2013;4:76. doi: 10.3389/fimmu.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang W, Erbe AK, Hank JA, Morris ZS, Sondel PM. NK Cell-Mediated Antibody-Dependent Cellular Cytotoxicity in Cancer Immunotherapy. Front. Immunol. 2015;6:368. doi: 10.3389/fimmu.2015.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowles JA, Weiner GJ. CD16 polymorphisms and NK activation induced by monoclonal antibody-coated target cells. J. Immunol. Methods. 2005;304:88–99. doi: 10.1016/j.jim.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 6.Koene HR, et al. Fc gammaRIIIa-158V/F polymorphism influences the binding of IgG by natural killer cell Fc gammaRIIIa, independently of the Fc gammaRIIIa-48L/R/H phenotype. Blood. 1997;90:1109–14. [PubMed] [Google Scholar]
- 7.Wu J, et al. A novel polymorphism of FcγRIIIa (CD16) alters receptor function and predisposes to autoimmune disease. J. Clin. Invest. 1997;100:1059–1070. doi: 10.1172/JCI119616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Congy-Jolivet N, et al. Fc gamma RIIIa expression is not increased on natural killer cells expressing the Fc gamma RIIIa-158V allotype. Cancer Res. 2008;68:976–80. doi: 10.1158/0008-5472.CAN-07-6523. [DOI] [PubMed] [Google Scholar]
- 9.Weng W-K, Levy R. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J. Clin. Oncol. 2003;21:3940–7. doi: 10.1200/JCO.2003.05.013. [DOI] [PubMed] [Google Scholar]
- 10.Cartron G, et al. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood. 2002;99:754–758. doi: 10.1182/blood.V99.3.754. [DOI] [PubMed] [Google Scholar]
- 11.Musolino A, et al. Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of trastuzumab-based therapy in patients with HER-2/neu-positive metastatic breast cancer. J. Clin. Oncol. 2008;26:1789–96. doi: 10.1200/JCO.2007.14.8957. [DOI] [PubMed] [Google Scholar]
- 12.Bibeau F, et al. Impact of FcγRIIa-FcγRIIIa Polymorphisms and KRAS Mutations on the Clinical Outcome of Patients With Metastatic Colorectal Cancer Treated With Cetuximab Plus Irinotecan. J. Clin. Oncol. 2009;27:1122–1129. doi: 10.1200/JCO.2008.18.0463. [DOI] [PubMed] [Google Scholar]
- 13.Taylor RJ, et al. FcγRIIIa polymorphisms and cetuximab induced cytotoxicity in squamous cell carcinoma of the head and neck. Cancer Immunol. Immunother. 2009;58:997–1006. doi: 10.1007/s00262-008-0613-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatjiharissi E, et al. Increased natural killer cell expression of CD16, augmented binding and ADCC activity to rituximab among individuals expressing the Fc γ Brief report Increased natural killer cell expression of CD16, augmented binding and ADCC activity to rituximab amon. Blood J. 2012;110:2561–2564. doi: 10.1182/blood-2007-01-070656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K, et al. Fc R2A and 3A polymorphisms predict clinical outcome of trastuzumab in both neoadjuvant and metastatic settings in patients with HER2-positive breast cancer. Ann. Oncol. 2011;22:1302–1307. doi: 10.1093/annonc/mdq585. [DOI] [PubMed] [Google Scholar]
- 16.Botticelli A, et al. FCGRs Polymorphisms and Response to Trastuzumab in Patients With HER2-Positive Breast Cancer: Far From Predictive Value. World J. Oncol. 2015;6:437–440. doi: 10.14740/wjon934w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hidalgo LG, et al. NK Cell Transcripts and NK Cells in Kidney Biopsies from Patients with Donor-Specific Antibodies: Evidence for NK Cell Involvement in Antibody-Mediated Rejection. Am. J. Transplant. 2010;10:1812–1822. doi: 10.1111/j.1600-6143.2010.03201.x. [DOI] [PubMed] [Google Scholar]
- 18.Venner JM, Hidalgo LG, Famulski KS, Chang J, Halloran PF. The molecular landscape of antibody-mediated kidney transplant rejection: Evidence for NK involvement through CD16a Fc receptors. Am. J. Transplant. 2015;15:1336–1348. doi: 10.1111/ajt.13115. [DOI] [PubMed] [Google Scholar]
- 19.Paul P, et al. Genetic and Functional Profiling of CD16-Dependent Natural Killer Activation Identifies Patients at Higher Risk of Cardiac Allograft Vasculopathy. Circulation. 2018;137:1049–1059. doi: 10.1161/CIRCULATIONAHA.117.030435. [DOI] [PubMed] [Google Scholar]
- 20.Takami A, et al. A single-nucleotide polymorphism of the Fcγ receptor type IIIA gene in the recipient predicts transplant outcomes after HLA fully matched unrelated BMT for myeloid malignancies. Bone Marrow Transplant. 2011;46:238–43. doi: 10.1038/bmt.2010.88. [DOI] [PubMed] [Google Scholar]
- 21.Shimizu S, et al. Fc-Gamma Receptor Polymorphisms Predispose Patients to Infectious Complications After Liver Transplantation. Am. J. Transplant. 2016;16:625–633. doi: 10.1111/ajt.13492. [DOI] [PubMed] [Google Scholar]
- 22.de Haas M, et al. A triallelic Fc gamma receptor type IIIA polymorphism influences the binding of human IgG by NK cell Fc gamma RIIIa. J. Immunol. 1996;156:2948–55. [PubMed] [Google Scholar]
- 23.Dong C, et al. Fcγ Receptor IIIa Single-Nucleotide Polymorphisms and Haplotypes Affect Human IgG Binding and Are Associated With Lupus Nephritis in African Americans. Arthritis Rheumatol. 2014;66:1291–1299. doi: 10.1002/art.38337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grier JT, et al. Human immunodeficiency-causing mutation defines CD16 in spontaneous NK cell cytotoxicity. J. Clin. Invest. 2012;122:3769–3780. doi: 10.1172/JCI64837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oboshi W, et al. SNPs rs4656317 and rs12071048 located within an enhancer in FCGR3A are in strong linkage disequilibrium with rs396991 and influence NK cell-mediated ADCC by transcriptional regulation. Hum. Immunol. 2016;77:997–1003. doi: 10.1016/j.humimm.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 26.Lassauniere R, Shalekoff S, Tiemessen CT. A novel FCGR3A intragenic haplotype is associated with increased FcgammaRIIIa/CD16a cell surface density and population differences. Hum Immunol. 2013;74:627–634. doi: 10.1016/j.humimm.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 27.Chen S, et al. Association of FCGR2A/FCGR3A variant rs2099684 with Takayasu arteritis in the Han Chinese population. Oncotarget. 2017;8:17239–17245. doi: 10.18632/oncotarget.12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin F, et al. Single nucleotide polymorphism rs10919543 in FCGR2A/FCGR3A region confers susceptibility to takayasu arteritis in chinese population. Chin. Med. J. (Engl). 2016;129:854–859. doi: 10.4103/0366-6999.178965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chai L, Song Y-Q, Zee K-Y, Leung WK. SNPs of Fc-gamma receptor genes and chronic periodontitis. J. Dent. Res. 2010;89:705–10. doi: 10.1177/0022034510365444. [DOI] [PubMed] [Google Scholar]
- 30.Ravetch JV, Perussia B. Alternative membrane forms of Fc gamma RIII(CD16) on human natural killer cells and neutrophils. Cell type-specific expression of two genes that differ in single nucleotide substitutions. J. Exp. Med. 1989;170:481–97. doi: 10.1084/jem.170.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dall’Ozzo Sé, Andres C, Bardos P, Watier H, Thibault G. Rapid single-step FCGR3A genotyping based on SYBR Green I fluorescence in real-time multiplex allele-specific PCR. J. Immunol. Methods. 2003;277:185–192. doi: 10.1016/S0022-1759(03)00123-6. [DOI] [PubMed] [Google Scholar]
- 32.Rosales C. Fcγ receptor heterogeneity in leukocyte functional responses. Front. Immunol. 2017;8:1–13. doi: 10.3389/fimmu.2017.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valenzuela, N. M., Hickey, M. J. & Reed, E. F. Antibody subclass repertoire and graft outcome following solid organ transplantation. Front. Immunol. 7 (2016). [DOI] [PMC free article] [PubMed]
- 34.Chong KT, Ho WF, Koo SH, Thompson P, Lee EJD. Distribution of the FcgammaRIIIa 176 F/V polymorphism amongst healthy Chinese, Malays and Asian Indians in Singapore. Br. J. Clin. Pharmacol. 2007;63:328–32. doi: 10.1111/j.1365-2125.2006.02771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Sorge NM, et al. Severity of Guillain-Barré syndrome is associated with Fcγ Receptor III polymorphisms. J. Neuroimmunol. 2005;162:157–164. doi: 10.1016/j.jneuroim.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 36.Van Der Pol WL, et al. Evidence for non-random distribution of Fcγ receptor genotype combinations. Immunogenetics. 2003;55:240–246. doi: 10.1007/s00251-003-0574-9. [DOI] [PubMed] [Google Scholar]
- 37.Chorev M, Carmel L. The Function of Introns. Front. Genet. 2012;3:55. doi: 10.3389/fgene.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooper DN. Functional intronic polymorphisms: Buried treasure awaiting discovery within our genes. Hum. Genomics. 2010;4:284. doi: 10.1186/1479-7364-4-5-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagelkerke, S. Q. & Kuijpers, T. W. Immunomodulation by IVIg and the role of Fc-gamma receptors: Classic mechanisms of action after all? Front. Immunol. 6 (2015). [DOI] [PMC free article] [PubMed]
- 40.Vaz-Drago R, Custódio N, Carmo-Fonseca M. Deep intronic mutations and human disease. Hum. Genet. 2017;136:1093–1111. doi: 10.1007/s00439-017-1809-4. [DOI] [PubMed] [Google Scholar]
- 41.Ip, C. L. C. et al. MinION Analysis and Reference Consortium: Phase 1 data release and analysis. F1000Research, 10.12688/f1000research.7201.1 (2015). [DOI] [PMC free article] [PubMed]
- 42.Lu H, Giordano F, Ning Z. Oxford Nanopore MinION Sequencing and Genome Assembly. Genomics, Proteomics Bioinforma. 2016;14:265–279. doi: 10.1016/j.gpb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cornelis S, Gansemans Y, Deleye L, Deforce D, Van Nieuwerburgh F. Forensic SNP Genotyping using Nanopore MinION Sequencing. Sci. Rep. 2017;7:41759. doi: 10.1038/srep41759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voorter CEM, et al. Allele and haplotype frequencies of HLA-DPA1 and -DPB1 in the population of Guadeloupe. Tissue Antigens. 2014;83:147–153. doi: 10.1111/tan.12271. [DOI] [PubMed] [Google Scholar]
- 45.Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information file).